Abstract
Purpose
In genomics, the return of negative screening results for rare, medically actionable conditions in large unselected populations with low prior risk of disease is novel and may involve important and nuanced concerns for communicating their meaning. Recruitment may result in self-selection because of participants’ personal or family history, changing the characteristics of the screened population and interpretation of both positive and negative findings; prior motivations may also affect responses to results.
Methods
Using data from GeneScreen, an exploratory adult screening project that targets 17 genes related to 11 medically actionable conditions, we address four questions: 1) Do participants self-select based on actual or perceived risk for one of the conditions? 2) Do participants understand negative results? 3) What are their psychosocial responses? 4) Are negative results related to changes in reported health-related behaviors?
Results
We found disproportionate enrollment of individuals at elevated prior risk for conditions being screened, and a need to improve communication about the nature of screening and meaning of negative screening results. Participants expressed no decision regret and did not report intention to change health-related behaviors.
Conclusion
This study illuminates critical challenges to overcome if genomic screening is to benefit the general population.
Keywords: genomic sequencing, returning results, population screening, ethical, legal, social issues
INTRODUCTION
Medical screening, which aims to identify individuals at risk for developing disease before the onset of symptoms in order to prevent or treat it early,1 is a cornerstone of clinical practice and public health. Given declining costs of DNA sequencing, there is growing enthusiasm among academic, health system, and commercial entities for pursuing primary genomic screening in the general population.2–4 Examples include calls to sequence BRCA1/2 in women over age 305 and efforts such as those at Geisinger, which screens biobank participants’ DNA for variants related to 27 medically-actionable genetic conditions.6
The rationale for targeted genomic screening is the presumed health benefit of identifying pathogenic variants for which prevention or treatment changes the course of a serious condition. As in many screening programs, the intention is to screen an unselected population—a population not pre-determined to be at heightened risk for a specific disease. However, recruitment may result in self-selection by those who participate because of personal or family histories, changing the characteristics of the sample and the interpretation of positive and negative findings. Not only are these individuals potentially different from the general population, but their responses to screening results may also be affected by their motivations.
Research is needed on returning negative screening results in genomics, the risks for harm, and procedures that mitigate those risks. Though the standard of care in medical screening is to return negative results, the return of negative genomic screening results for rare, medically actionable conditions in a large population with low prior risk of disease is novel and may involve important and nuanced concerns for communicating their meaning.
One particular concern is misunderstanding the risk for false-negative results, a risk in any screening test. In genomic screening, false-negative results can arise from: not testing for all genes associated with a given phenotype; the inherent inability to test currently unknown genes that cause a particular phenotype; not assaying for specific types of pathogenic variants (e.g., insertions/deletions); and the fact that variants of uncertain significance are functionally interpreted as negative, but some will likely eventually be shown to be pathogenic.
It is also possible that individuals motivated to participate in a screening program have an elevated a priori risk for a condition because of personal or family histories and are therefore at greater risk for a false-negative result. These participants may receive false reassurance, which may lead to a reduced adherence to necessary evaluations or surveillance7. Furthermore, a false sense of security may lead them to ignore other risk factors that contribute to disease, or engage in potentially negative health behaviors.8–10 Research has shown that unrealistically optimistic risk perceptions are common and can lead to lower motivation to engage in health-protective behavior.11
We investigated these issues in GeneScreen, an exploratory screening project for healthy adults that targeted 17 genes that confer risk for 11 rare, medically actionable conditions, most of which are related to a high risk for cancer or cardiovascular disease.12 These genes were selected with a semi-quantitative metric developed in the context of the Clinical Sequencing Exploratory Research program (See Table 1).13,14 Participants provided a saliva sample or permitted the use of their previously banked DNA for screening and completed a baseline survey, and a second survey after return of results. Most (94.7%) received negative results, returned via an online report. As a pilot project intended to guide development of a much larger, longitudinal study, we anticipated formulating hypotheses to test with appropriately powered samples and well-defined measures. In this stage, we report examination of four key questions: 1) Do a substantial number of GeneScreen participants self-select based on actual or perceived risk for one of the 11 conditions? 2) After receiving negative results online, do participants understand them? 3) What are participants’ psychosocial responses to negative results? 4) Is receipt of negative results related to changes in intention to engage in appropriate medical screening or other health-related behaviors? Our findings have important implications for the future of targeted genomic population screening and set an agenda for new standards in communication of negative genomic screening results.
Table 1.
GeneScreen Conditions and Genes
| Condition | Gene |
|---|---|
| Cancer risk syndromes: | |
|
| |
| Familial adenomatous polyposis (FAP) | APC |
| Hereditary breast and ovarian cancer syndrome (HBOC) | BRCA1, BRCA2 |
| Lynch syndrome | MLH1, MSH2, MSH6, PMS2 |
| Multiple endocrine neoplasia, type 2 (MEN2) | RET |
| MUTYH-associated polyposis (MAP) | MUTYH |
|
| |
| Connective tissue and heart disease: | |
|
| |
| Marfan syndrome | FBN1 |
|
| |
| Heart rhythm problems: | |
|
| |
| Long QT syndrome | KCNQ1, KCNH2, SCN5A |
|
| |
| Heart disease: | |
|
| |
| Familial hypercholesterolemia (FH) | LDLR |
|
| |
| Lung disease: | |
|
| |
| Alpha-1-antitrypsin deficiency | SERPINA1 |
|
| |
| Liver disease: | |
|
| |
| Hereditary hemochromatosis | HFE |
|
| |
| Harmful reaction to anesthesia during surgery: | |
|
| |
| Malignant hyperthermia | RYR1 |
METHODS
Participants and procedures
Recruitment letters were mailed to potential participants at two sites: 1) a hospital-based general medicine clinic at the University of North Carolina-Chapel Hill (UNC) (n = 436) and 2) a research biobank associated with Kaiser Permanente Northwest (KPNW) (n = 650). Details of recruitment and enrollment are described elsewhere12 and in Figure 1. For each site, we sent letters to equal numbers of men and women and, where possible, equal numbers from selected racial/ethnic groups (Black, White, Other) and adult age groups (18–40, 41–60, 61 and over). Given the demographics of its region, Kaiser’s recruitment of Blacks was quite low. Further, because recruitment was based on medical record reports of race, which in some cases did not match self-reported race, this complicated calculation of response rates (see data in supplemental materials). The study website’s education and consent materials provided information about how genes affect health, the study goals, the screening test and the GeneScreen conditions; it provided a decision aid and summarized study features required for informed consent. We chose to use the term “mutation” rather than “pathogenic variant” since the former seemed more understandable to the public, and the study began prior to recent recommendations.15 Twenty-four percent of those recruited subsequently enrolled, and then completed the 30-minute baseline (BL) survey assessing decision making, personal and family health history, health status, demographic characteristics, perceived risk of having a GeneScreen mutation, medical screening behavior, genetic knowledge, and knowledge of the study. GeneScreen was approved by the IRBs of UNC and KPNW.
Figure 1.
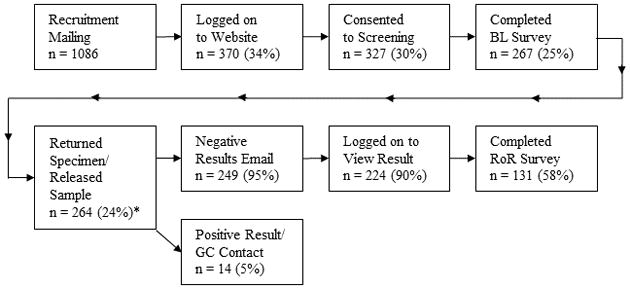
Sequencing and return of results
Enrollees’ DNA samples were sequenced at the UNC High Throughput Genomic Sequencing Facility. Fourteen screened positive16. Results were confirmed in a CLIA-approved lab and communicated by phone to participants by geneticists and/or genetic counselors who developed a care plan with participants and their providers. Among these individuals, five already knew of their genetic condition. The other 249 had a normal/negative result, in line with population prevalence of deleterious variants in the genes analyzed. These individuals were invited to log on to a website to see their results. The website text read: “Your results are normal/negative,” followed by a section titled “Questions You May Have” (see Table 2, and online supplement Table 2), including the possibility of false negatives.
Table 2.
Text of GeneScreen Negative-Results Report
| Question | Response |
|---|---|
| What do normal/negative results mean? | This means that in the 17 genes that were tested, we found no medically important mutations. |
| How do these results impact my healthcare? | Your usual medical care should not change based on these normal/negative GeneScreen results. You should continue to follow your doctors’ recommendations for health screening based on general population guidelines and your personal and family medical history. If your personal or family history suggests that you may have a risk for any genetic condition, it is recommended that you see a clinical genetic specialist. |
| What type of test was done? | We conducted targeted sequencing and analysis of the 17 genes on the GeneScreen Research Study panel. |
| How accurate is this test? | The technology used in GeneScreen is expected to detect more than 90% of the people who have a mutation associated with one of the GeneScreen conditions. Thus, in rare cases, a person may have normal/negative GeneScreen results, but still have a GeneScreen condition. This is due to the limitations of the test used in this research study. If the GeneScreen research test had identified a mutation, a second test would have been used to confirm the presence of the abnormal/positive result. This would help to ensure the highest accuracy of an abnormal/positive GeneScreen result. |
| What are the limitations of this test? | This test only looks at certain preventable genetic health conditions. There are other genetic health conditions that this test does not look for. This test looks for mutations in 17 genes known to be associated with the GeneScreen conditions. It does not find all the possible causes of these conditions. Only mutations that are currently known to be harmful and medically important are reported. |
Most logged on to the results website. One week later they were invited to participate in an on-line survey (return of results survey: RoR) for which they were offered a $30 incentive. Just over half completed the survey. Additional reminders were sent to 25 participants who had not logged in to the results website; two were confirmed deceased and the remaining 23 were mailed their results with a cover letter.
The 221 participants who logged in to see their results were younger than those who did not log in (M = 58.50, SD = 15.28 versus M = 65.83, SD = 16.52; t (1, 244) = 2.22, p = 0.028). There were no other demographic differences. Likewise, there were no statistically significant demographic differences between the 131 who completed the RoR survey and the 112 who did not.
Measures
Unless otherwise referenced, measures were created for this study.
1) Do a substantial number of GeneScreen participants self-select based on actual or perceived risk for one of the 11 conditions?
To assess actual risk, we created questions that asked whether participants had family or personal histories of health conditions (e.g., colon cancer) associated with GeneScreen genetic conditions (e.g., Lynch Syndrome). For each health condition, we asked if the participant had ever been diagnosed. They could answer: “No, I’ve never had this” = 0 or “Yes, I have this now or in the past” =1. They were also asked if anyone in their family had ever been diagnosed with the health condition, to which they could answer, “No one in my family has ever had this” = 0, “At least one person in my family has had this” = 1, or “I don’t know” = missing. We excluded responses regarding heart disease, high cholesterol, and chronic obstructive pulmonary disease from our analyses because these health conditions are very commonly environmental or multifactorial and thus rarely caused by the genetic conditions screened for by GeneScreen. We included responses concerning aortic aneurysm, breast cancer, uterine cancer, ovarian cancer, colon cancer, thyroid cancer, serious reaction to general anesthesia, and hemochromatosis. If participants indicated that they had personal or family histories of any of these health conditions, they were given a score of 1; otherwise they received a score of 0.
We measured perceived risk17 for having a GeneScreen mutation with one item that asked, “How likely do you think it is that you have one of the GeneScreen mutations?” Responses were given on a scale of 1 = “Extremely unlikely” to 6 = “Extremely likely.”
2) After receiving negative results online, do participants understand them?
We used two variables to assess understanding of results, and one to assess perceived understanding. First, we asked, “Which statement best describes your understanding of your GeneScreen test results?” Options ranged from “I definitely do not have a GeneScreen mutation” = 1 to “I definitely do have a GeneScreen mutation” = 6. Second, to assess nuance in understanding, we analyzed responses to three open-ended questions about worries, positive reactions, and how they described their results to family. Two team members independently analyzed and coded these responses such that 0 = indicated problematic understanding; 1= stated that their results were negative/normal with no further detail; 2 = showed a nuanced understanding of their screening results, such as “low likelihood of a mutation.” Third, we measured perceived understanding of test results with a three-item scale (e.g., “I understand what my test results mean for my health.”). Responses ranged from “Strongly disagree” = 1 to “Strongly agree” = 6. Items were averaged so that higher scores indicated greater perceived understanding. The scale had adequate internal reliability (Cronbach’s α= 0.833).
We measured knowledge of genetics and knowledge of GeneScreen features to determine whether these variables predicted understanding a negative-results report. Knowledge of genetics was assessed with 15 items from the UNC Genomic Knowledge Scale, which measures knowledge in three domains thought critical for informed decision-making in genomic sequencing.18 Knowledge of GeneScreen features was assessed with five statements that described important study components. Participants could answer “true,” “false,” or “don’t know.” Correct responses were scored as 1 and incorrect and “don’t know” were scored as 0.12
3) What are participants’ psychosocial responses to receiving negative results?
We assessed decision regret19 using a 5-item scale (e.g., “The choice did me a lot of harm,” and “It was the right decision.”). Response options ranged from “Strongly disagree” = 1 to “Strongly agree” = 6. All five items were averaged so that higher scores indicated greater decision regret (α = .815).
We also assessed variables related to family support and family conflict regarding the GeneScreen test using two items from the MICRA scale.20 Specifically, family support was measured with one item that asked participants how much they have been “feeling that your family has been supportive during the genetic screening process.” Family conflict was measured with one item that asked how much participants had been “feeling worried that the genetic screening process has caused conflict in your family.” Both items had response options of 0 = “Not at all” to 3 = “A lot.” We measured family interactions with an item asking, “How likely is it that you will tell at least one family member your GeneScreen test results?” Participants could respond, “I have already told someone” or could rate likelihood on a scale of 1 = “not at all likely” to 5 = “extremely likely.”
4) Is receipt of negative results related to changes in intention to engage in regular medical screening or other health-related behaviors?
In the BL survey, we asked, “How often do you think you need regular medical screening tests compared to other people your age and gender?” In the RoR survey, we asked, “What do you think your GeneScreen test results mean for how often you need regular medical screening compared to other people your age and gender?” For both questions, 1 = “much less often”; 3 = “about as often”; and 5 = “much more often.” We used the change score (RoR survey – BL survey) to examine change in perceived need for medical screening.
We examined motivation to change health-related behaviors using the stem, “Do your GeneScreen test results make you feel less or more motivated or cause no change in motivation….” Participants reported changes in motivation regarding several health behaviors (e.g., diet, exercise). More motivation was coded as 1, no change as 0, and less motivation as −1.
Analysis plan
First, we computed descriptive statistics to evaluate the distributional and psychometric properties of each variable. Income was the only variable with significant missing data (n =8); this was handled with an expectation-maximization imputation.21 For other variables with fewer than 3% of cases missing, we used mean replacement. To address specific research questions, we used descriptive statistics and correlations.
RESULTS
Descriptive statistics
As shown in Table 3, most participants were women (68.7% at BL and 66.4% at RoR), White (78.7% and 80.2%), older, relatively well-educated, and affluent.
Table 3.
Participant Characteristics
| Variable | Baseline (n=262)* | RoR (n= 131) |
|---|---|---|
| Age | ||
| 18–40 | 43 (16.4%) | 24 (18.3%) |
| 41–60 | 89 (34.0%) | 44 (33.6%) |
| 61+ | 130 (49.6%) | 63 (48.1%) |
| Male | 82 (31.3%) | 44 (33.6%) |
| Female | 180 (68.7%) | 87 (66.4%) |
| Education | ||
| Less than high school | 1 (.4%) | 0 (0%) |
| High school or GED | 24 (9.2%) | 9 (6.9%) |
| Some college | 75 (28.6%) | 33 (25.2%) |
| Four-year college degree | 63 (24.0%) | 36 (27.5%) |
| Graduate or professional degree | 99 (37.8%) | 53 (40.5%) |
| Household Income | ||
| Less than $50,000 | 55 (22.0%) | 26 (21.1%) |
| $50,000 to $99,999 | 90 (36.0%) | 43 (35%) |
| $100,000 to $149,999 | 53 (21.2%) | 30 (24.4%) |
| $150,000 or more | 52 (20.8%) | 24 (19.5%) |
| Missing | 12 | 8 |
| Ethnicity | ||
| Not Hispanic | 236 (90.4%) | 121 (92.4%) |
| Hispanic | 25 (9.6%) | 10 (7.6%) |
| Missing | 1 | 0 |
| Race | ||
| White | 207 (78.7%) | 105 (80.2%) |
| African-American | 13 (4.9%) | 7 (5.3%) |
| American Indian | 2 (0.8%) | 1 (0.8%) |
| Asian-American | 17 (6.5%) | 7 (5.3%) |
| Mixed | 17 (6.5%) | 10 (7.6%) |
| Other | 5 (1.9%) | 1 (0.8%) |
| Missing | 1 | 0 |
| Institution | ||
| KPNW | 195 (74.4%) | 92 (70.2%) |
| UNC | 67 (25.6%) | 39 (29.8%) |
Two hundred sixty-four participants submitted the joiner survey and sent in saliva (UNC) or permitted the use of banked DNA (KPNW) for sequencing. Two participants completed less than 50% of the survey, so their data were not included in these analyses.
We used data from the BL survey to address our first research question, and the RoR survey for the other questions.
1) Do a substantial number of GeneScreen participants self-select based on actual or perceived risk for one of the 11 conditions?
Nearly three-quarters (72.5%) of GeneScreen participants reported personal or family histories of eight health conditions on the GeneScreen panel (See Table 1 in supplemental materials). In the BL survey, almost 13% of participants (n=33) believed it was at least somewhat likely they had a GeneScreen mutation, despite online information that these conditions affect only 1–2% of the population.
2) After receiving their negative results online, do participants understand them?
Most participants who completed the RoR survey believed they understood their results. More than 50% scored themselves at least 5 out of 6 and 35% scored themselves 6, meaning they answered “strongly agree” to all three questions assessing perceived understanding. However, participants may not have understood the nuance of their results. As shown in Table 4, 44.3%, indicated they definitely did not have a GeneScreen mutation, whereas 51.9% indicated it was extremely unlikely they had a GeneScreen mutation—a more accurate response. The coded open-ended responses revealed approximately 80% of participants simply wrote that they had negative results or they did not have a mutation; the remaining responses mentioned complex causes of disease or the small possibility of false negatives (e.g., “Tests came back normal, doesn’t rule out everything”); four responses were problematic (e.g., “I was recruited because my doctor thought I had a mutation”).
Table 4.
Participants’ Understanding of the Negative GeneScreen Test Result.
| Response | Frequency | Percent |
|---|---|---|
| 1) I definitely do not have a GeneScreen mutation. | 58 | 44.3 |
| 2) It is extremely unlikely that I have a GeneScreen mutation. | 68 | 51.9 |
| 3) It is somewhat unlikely that I have a GeneScreen mutation. | 3 | 2.3 |
| 4) It is somewhat likely that I have a GeneScreen mutation. | 1 | 0.8 |
| 5) It is extremely likely that I have a GeneScreen mutation. | 1 | 0.8 |
| 6) I definitely do have a GeneScreen mutation. | 0 | 0.0 |
We examined potential correlates of these ways of measuring participants’ understanding, including demographic variables, knowledge of genetics, knowledge of the GeneScreen study, personal or family histories of a GeneScreen condition, and perceived risk. Because of the slight possibility of a false negative, the knowledge variable was recoded such that 1, 4, 5, and 6 were coded as incorrect and 2 and 3 were coded as correct. Since we examined 24 correlations, Bonferroni correction resulted in a significance cut-off of < 0.002. There was only one significant correlation: Knowledge of genetics was positively correlated with nuance (r = 0.27, p. = 0.002).
3) What are participants’ psychosocial responses to receiving negative results?
Participants’ psychosocial reactions were largely positive. They did not experience decision regret. On the 1–6 scale (with 1 indicating no regret), the mean response was 1.40 (SD = 0.58, range 1–4). Most participants appeared to feel comfortable talking with family about GeneScreen. More than half (64.1%) had already told someone in their family about their GeneScreen results and another 14.5% reported that it was either extremely or very likely that they would tell a family member. Likewise, participants reported high levels of family support during the genetic screening process (M = 2.14, SD = 1.08) and low levels of family conflict (M = 0.06, SD = 0.35).
4: Is receipt of negative results related to changes in intention to engage in regular medical screening or other health-related behaviors?
Most participants reported they would not change their health-related behaviors in response to their results. Change in perceived need for medical screening was very close to 0 (M = −0.21, SD = 1.09). Most (82.4%) reported they needed screening about as often as people their age and gender and that their GeneScreen results did not affect their motivation to change other health-related behaviors.
DISCUSSION
GeneScreen participants are typical of “early adopters.”22 Despite efforts to recruit individuals with no increased risk of the genetic conditions being screened, we found that at least some saw themselves at higher risk for genetic conditions and may have been motivated to join by personal or family histories of health conditions related to the genetic conditions in the GeneScreen panel. We also found that although most participants who accessed their results and completed the RoR survey were very confident they understood the meaning of their “negative/normal” result, fewer than 20% offered statements demonstrating nuanced understanding of the standard limitations of a negative test, outlined in the return of results report. We found little or no decisional regret for joining the study. Rather, participants expressed that they made the right decision in joining. Our measures of self-reported intentions regarding health-related behaviors, including medical screening behaviors, did not change following receipt of negative results. Thus, at least in the short term, reassurance did not lead to intentions to change.
Potential hazards from misunderstanding the implications of negative results most clearly and seriously manifest in the direct-to-consumer (DTC) setting where limited (or no) professional engagement exists. Indeed, the acceleration of DTC genomic analyses increases the relevance and importance of our findings. Furthermore, in a clinical setting, negative results are usually delivered via a very brief, templated summary, and when coupled with limited genetic expertise among medical providers, may result in abundant opportunity for misunderstanding.
Despite these concerns, skillful risk communication and improved language use may reduce potential misunderstandings,23–26 and increase the proportion of individuals able to recognize the limits of a negative-results report. Haga and colleagues (2014) cite a CDC recommendation for reports to state “no mutation detected” rather than “normal,” arguing that patients may interpret “normal” as a true negative result when in fact it may largely but not completely rule out mutations in one of many possible genes for a genetically heterogeneous disease.26,27 Indeed, in a screening setting, most “negative” results are best thought of as “not positive.” Although communication may be improved to a standard where a short phone call or online report can adequately convey these messages,28 it is critical to test understandability of communication, particularly in populations more diverse than our participants. Haga and colleagues emphasize that individuals’ understanding of test results is influenced by the context of their “motivational and emotional state, underlying expectations, and family members’ experience with an illness.”26 Thus, future research should include evaluation of factors not fully assessed in GeneScreen.
People who enrolled in GeneScreen may have been motivated to join due to prior health history, given that nearly three-quarters had personal or family histories of health conditions associated with eight conditions in the GeneScreen panel. These data do not prove that participants were at increased risk for GeneScreen conditions, but some participants may have joined GeneScreen for diagnostic purposes because they were at increased risk or perceived themselves to be. This highlights a potential threat to genomic screening efforts that must be addressed: due to the inherent lack of complete sensitivity in any screening test, individuals at higher prior probability of disease (e.g., due to positive family history) will be at elevated risk of a false-negative result in a screening context. This may lead to false reassurance and the potential for deferring necessary diagnostic evaluation or enhanced screening. Post hoc analyses found that responses to the question about the need for screening were not associated with reporting personal or family histories of GeneScreen conditions, which may indicate that people are not well-informed about how risk impacts the need for screening. On the other hand, the limitations of our measures must be considered as a possible explanation. Identifying participants who may be vulnerable to misunderstanding negative results due to medical or family history is challenging but can be addressed. Although gathering such data is too time consuming for large-scale public health screening programs, efforts to leverage technology may make these tasks more feasible in the future.29 Collecting these data will identify individuals at increased risk for a false negative result and allow appropriate framing of negative results to guide further evaluation, if necessary. Building such information into the enrollment process of any targeted genomic screening program will also prove valuable in the interpretation of both positive and negative results.
LIMITATIONS
There are limitations to our study. First, despite our recruitment efforts, GeneScreen participants were not diverse. Answers to our four research questions must be considered in light of selection biases, particularly when considering the potential impact of lower socioeconomic status on understanding, psychosocial responses, subsequent behaviors, and possible disinhibition due to false reassurance.8,30,31
Other limitations arose as we addressed each research question. For instance, each measure used to explore whether participants were at higher risk for testing positive had significant limitations. Personal or family history queries focused on health conditions related to genetic conditions in the GeneScreen panel (e.g., thyroid cancer and multiple endocrine neoplasia, type 2 [RET gene]). For others, such as familial hypercholesterolemia [LDLR gene], high cholesterol is so prevalent in the general population that a more detailed personal or family history would be needed to indicate increased prior risk. Similarly, the question about perceived risk of having a GeneScreen condition is not an objective indicator of actual increased prior risk. Our central recommendation moving forward with targeted genomic screening programs is to use existing, validated risk assessment tools to collect information about relevant personal and family histories.
Our second research question revealed significant limitations in measuring understanding, but more importantly, revealed poor communication about the meaning of negative results. We recommend that negative-results reports clearly communicate the small likelihood of a false negative result and why that might occur. The GeneScreen negative-results report heading should not have simply stated “Your results are normal/negative,” but rather a message such as: “No mutation was found, but in rare cases this result may not be accurate.” Developing and evaluating more sensitive measures for optimal communication about negative results would advance future recommendations.
Our investigation of psychosocial responses to receiving negative results used well-validated measures; however, qualitative research with a diverse population of people receiving negative results could refine our understanding of psychosocial responses, thereby guiding development of measures specific to the genomic screening context.
Investigating possible harms of screening for health behaviors depended on measures that centered on false reassurance, which we theorized would produce less attention to appropriate medical screening, and reassurance that could be risky in the case of false negative results. Although we did not find evidence of such harms, returning negative results in a large and more diverse population screening program needs further study over a longer time. Furthermore, for individuals with perceived personal or family histories of GeneScreen conditions, it is not clear what the best answer would be. As recommended above, such research should not rely on self-reports, but use data from medical records or other valid personal and family history data.
CONCLUSION
Targeted genomic screening of populations to detect individuals at high risk of rare but preventable medical problems holds great promise. This study illuminates some challenges, including enrollment of individuals at elevated prior risk for the diseases being screened and a need to help individuals develop a better understanding of the nature of screening and the meaning of negative screening results. These must be overcome if targeted genomic screening is to benefit the general population.
Supplementary Material
Acknowledgments
The authors would like to thank those who kindly participated in the GeneScreen study. Thank you also to Katherine J. Souris, MPH, for her contributions to the manuscript. Research for this study was funded by the National Institutes of Health (NIH) (grant No. 2P50HG004488 to G.E.H. for the Center for Genomics and Society (CGS). The views expressed are those of the authors alone and do not necessarily reflect views of the NIH or all CGS investigators. Access to the NW Biobank resource was made possible by support from the Oregon Clinical and Translational Research Institute, grant No. UL1 RR024140 from the National Center for Research Resources, a component of the NIH, the NIH Roadmap for Medical Research, the MJ Murdock Charitable Trust, and institutional support from the Center for Health Research and Kaiser Permanente Northwest (KPNW).
Footnotes
Conflict of Interest Notification
None of the authors have any conflicts of interest to disclose.
References
- 1.Wilson JMG, Jungner G. Principles and Practice of Screening for Disease. France: Geneva: World Health Organization; 1968. [Accessed March 14, 2018]. Available at: http://www.who.int/ionizing_radiation/medical_radiation_exposure/munich-WHO-1968-Screening-Disease.pdf. [Google Scholar]
- 2.Lewis MA, Stine A, Paquin RS, et al. Parental preferences toward genomic sequencing for non-medically actionable conditions in children: a discrete-choice experiment. Genet Med. 2017 doi: 10.1038/gim.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldenberg AJ, Dodson DS, Davis MM, Tarini BA. Parents’ interest in whole-genome sequencing of newborns. Genet Med. 2014;16(1):78–84. doi: 10.1038/gim.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster MW, Mulvihill JJ, Sharp RR. Evaluating the utility of personal genomic information. Genet Med. 2009;11(8):570–574. doi: 10.1097/GIM.0b013e3181a2743e. [DOI] [PubMed] [Google Scholar]
- 5.King M-C, Levy-Lahad E, Lahad A. Population-Based Screening for BRCA1 and BRCA2. JAMA. 2014;312(11):1091. doi: 10.1001/jama.2014.12483. [DOI] [PubMed] [Google Scholar]
- 6.Carey DJ, Fetterolf SN, Davis FD, et al. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18(9):906–913. doi: 10.1038/gim.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siu AL U S. Preventive Services Task Force. Screening for breast cancer: U.S. preventive services task force recommendation statement. Ann Intern Med. 2016;164(4):279–296. doi: 10.7326/M15-2886. [DOI] [PubMed] [Google Scholar]
- 8.Yurgelun MB, Hiller E, Garber JE. Population-Wide Screening for Germline BRCA1 and BRCA2 Mutations: Too Much of a Good Thing? J Clin Oncol. 2015;33(28):3092–3095. doi: 10.1200/JCO.2015.60.8596. [DOI] [PubMed] [Google Scholar]
- 9.Linderman MD, Nielsen DE, Green RC. Personal genome sequencing in ostensibly healthy individuals and the peopleseq consortium. J Pers Med. 2016;6(2) doi: 10.3390/jpm6020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker F, van El CG, Ibarreta D, et al. Genetic testing and common disorders in a public health framework: how to assess relevance and possibilities. Background Document to the ESHG recommendations on genetic testing and common disorders. Eur J Hum Genet. 2011;19(Suppl 1):S6–44. doi: 10.1038/ejhg.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrer R, Klein WM. Risk perceptions and health behavior. Curr Opin Psychol. 2015;5:85–89. doi: 10.1016/j.copsyc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadigan RJ, Butterfield R, Rini C, et al. Online Education and e-Consent for GeneScreen, a Preventive Genomic Screening Study. Public Health Genomics. 2017;20:235–46. doi: 10.1159/000481359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg JS, Foreman AKM, O’Daniel JM, et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med. 2016;18(5):467–475. doi: 10.1038/gim.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lázaro-Muñoz G, Conley JM, Davis AM, Prince AER, Cadigan RJ. Which results to return: subjective judgments in selecting medically actionable genes. Genet Test Mol Biomarkers. 2017;21(3):184–194. doi: 10.1089/gtmb.2016.0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvik GP, Evans JP. Mastering genomic terminology. Genet Med. 2017;19(5):491–492. doi: 10.1038/gim.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muessig KR, Kucznski K, Himes P, et al. Targeted genomic screening in unselected adults. Poster presented at the American Society of Human Genetics (ASHG) Annual Meeting; October, 2017; Orlando, FL. [Accessed March 15, 2018]. https://ep70.eventpilotadmin.com/web/page.php?page=IntHtml&project=ASHG17&id=170120967. [Google Scholar]
- 17.Janz NK, Becker MH. The Health Belief Model: a decade later. Health Educ Q. 1984;11(1):1–47. doi: 10.1177/109019818401100101. [DOI] [PubMed] [Google Scholar]
- 18.Langer MM, Roche MI, Brewer NT, et al. Development and Validation of a Genomic Knowledge Scale to Advance Informed Decision-Making Research in Genomic Sequencing. MDM Policy & Practice. 2017;2(1):238146831769258. doi: 10.1177/2381468317692582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brehaut JC, O’Connor AM, Wood TJ, et al. Validation of a decision regret scale. Med Decis Making. 2003;23(4):281–292. doi: 10.1177/0272989X03256005. [DOI] [PubMed] [Google Scholar]
- 20.Cella D, Hughes C, Peterman A, et al. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21(6):564–572. [PubMed] [Google Scholar]
- 21.Newman DA. Missing Data. Organizational Research Methods. 2014;17(4):372–411. doi: 10.1177/1094428114548590. [DOI] [Google Scholar]
- 22.Lewis KL, Han PKJ, Hooker GW, Klein WMP, Biesecker LG, Biesecker BB. Characterizing participants in the clinseq genome sequencing cohort as early adopters of a new health technology. PLoS ONE. 2015;10(7):e0132690. doi: 10.1371/journal.pone.0132690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dicke JM, Van Duyne L, Bradshaw R. The utilization and choices of aneuploidy screening in a midwestern population. J Genet Couns. 2014;23(5):874–880. doi: 10.1007/s10897-014-9711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipkus IM. Numeric, verbal, and visual formats of conveying health risks: suggested best practices and future recommendations. Med Decis Making. 2007;27(5):696–713. doi: 10.1177/0272989X07307271. [DOI] [PubMed] [Google Scholar]
- 25.O’Doherty K, Suthers GK. Risky communication: pitfalls in counseling about risk, and how to avoid them. J Genet Couns. 2007;16(4):409–417. doi: 10.1007/s10897-006-9077-9. [DOI] [PubMed] [Google Scholar]
- 26.Haga SB, Mills R, Pollak KI, et al. Developing patient-friendly genetic and genomic test reports: formats to promote patient engagement and understanding. Genome Med. 2014;6(7):58. doi: 10.1186/s13073-014-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B, Gagnon M, Shahangian S, et al. Good laboratory practices for molecular genetic testing for heritable diseases and conditions. MMWR Recomm Rep. 2009;58(RR-6):1–37. quiz CE. [PubMed] [Google Scholar]
- 28.Horowitz CR, Abul-Husn NS, Ellis S, et al. Determining the effects and challenges of incorporating genetic testing into primary care management of hypertensive patients with African ancestry. Contemp Clin Trials. 2016;47:101–108. doi: 10.1016/j.cct.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Facio FM, Feero WG, Linn A, Oden N, Manickam K, Biesecker LG. Validation of My Family Health Portrait for six common heritable conditions. Genet Med. 2010;12(6):370–375. doi: 10.1097/GIM.0b013e3181e15bd5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantwill S, Monestel-Umaña S, Schulz PJ. The Relationship between Health Literacy and Health Disparities: A Systematic Review. PLoS ONE. 2015;10(12):e0145455. doi: 10.1371/journal.pone.0145455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith SK, Dixon A, Trevena L, Nutbeam D, McCaffery KJ. Exploring patient involvement in healthcare decision making across different education and functional health literacy groups. Soc Sci Med. 2009;69(12):1805–1812. doi: 10.1016/j.socscimed.2009.09.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


