Abstract
Circulating hemocytes of the snail Biomphalaria glabrata, a major intermediate host for the blood fluke Schistosoma mansoni, represent the primary immune effector cells comprising the host's internal defense system. Within hours of miracidial entry into resistant B. glabrata strains, hemocytes infiltrate around developing sporocysts forming multi-layered cellular capsules that results in larval death, typically within 24–48 h post-infection. Using an in vitro model of hemocyte-sporocyst encapsulation that recapitulates in vivo events, we conducted a comparative proteomic analysis on the responses of hemocytes from inbred B. glabrata strains during the encapsulation of S. mansoni primary sporocysts. This was accomplished by a combination of Laser-capture microdissection (LCM) to isolate sections of hemocyte capsules both in the presence and absence of sporocysts, in conjunction with mass spectrometric analyses to establish protein expression profiles. Comparison of susceptible NMRI snail hemocytes in the presence and absence of sporocysts revealed a dramatic downregulation of proteins in during larval encapsulation, especially those involved in protein/CHO metabolism, immune-related, redox and signaling pathways. One of 4 upregulated proteins was arginase, competitor of nitric oxide synthetase and inhibitor of larval-killing NO production. By contrast, when compared to control capsules, sporocyst-encapsulating hemocytes of resistant BS-90 B. glabrata exhibited a more balanced profile with enhanced expression of shared proteins involved in protein synthesis/processing, immunity, and redox, and unique expression of anti-microbial/anti-parasite proteins. A final comparison of NMRI and BS-90 host hemocyte responses to co-cultured sporocysts demonstrated a decrease or downregulation of 77% of shared proteins by NMRI cells during encapsulation compared to those of the BS-90 strain, including lipopolysaccharide-binding protein, thioredoxin reductase 1 and hemoglobins 1 and 2. Overall, using this in vitro model, results of our proteomic analyses demonstrate striking differences in proteins expressed by susceptible NMRI and resistant BS-90 snail hemocytes to S. mansoni sporocysts during active encapsulation, with NMRI hemocytes exhibiting extensive downregulation of protein expression and a lower level of constitutively expressed immune-relevant proteins (e.g., FREP2) compared to BS-90. Our data suggest that snail strain differences in hemocyte protein expression during the encapsulation process account for observed differences in their cytotoxic capacity to interact with and kill sporocysts.
Keywords: innate immunity, mollusk, Biomphalaria glabrata, Schistosoma mansoni, hemocytic encapsulation, sporocyst, proteomic response, in vitro cytotoxicity assay
Introduction
Circulating hemocytes of Biomphalaria snail species represent the primary immune effector cells of the host, and, when encountering primary sporocysts of incompatible strains of the human blood fluke Schistosoma mansoni, respond by rapid encapsulation and destruction of early developing larvae (1–5). Cellular encapsulation responses typically are characterized by infiltration of circulating hemocytes around the newly transforming primary sporocysts, commencing usually within 2–3 h post-infection, followed by adherence and spreading of hemocytes in multiple layers around the parasite that result in the killing of encapsulated larvae within 24–48 h post-infection (6, 7). An in vitro model of larval encapsulation has been developed for the B. glabrata-S. mansoni system that closely parallels the events observed under natural in vivo infection conditions (8). In this cell-mediated cytotoxicity (CMC) assay, live in vitro transformed primary sporocysts (9) were co-incubated in 1-mL tubes with freshly extracted hemolymph isolated from inbred susceptible and resistant strains of B. glabrata, resulting in the formation hemocytic capsules around sporocysts. Although hemocytes of both susceptible and resistant snail strains formed cellular encapsulations, significantly enhanced killing/damage of sporocysts was observed in reactions with resistant snail cells when compared to those involving hemocytes from susceptible snails. Follow-up experiments showed that components from plasma (cell-free hemolymph) alone also may be involved in triggering the CMC response, although the humoral factor(s) and their functional role in hemocyte reactivity have yet to be identified and described (7, 10). Additional support that hemocytes are the primary mediators of anti-schistosome resistance recently has been demonstrated in vivo (11) using a resistant Guadeloupean stain of B. glabrata exhibiting allelic variation in a gene complex designated the Guadeloupe resistance complex (GRC) (12, 13). Notably, RNAi knockdown of a novel transmembrane protein encoded within the GRC appears to influence the resistance phenotype (no development of S. mansoni) (13) but the mechanism(s) by which this is accomplished remains unknown.
Although comparative transcriptomic analyses have been performed on whole snails or circulating hemocytes from different B. glabrata strains (14–17) and/or on snails in response to schistosome infection (18–22), evaluation of parasite-induced changes in hemocyte protein expression, specifically in hemocytes actively participating in sporocyst encapsulation reactions, have not been examined in detail (23). Targeted investigations of specific hemolymph proteins, such as Cu/Zn superoxide dismutase SOD1 (24), HSP 70 and 90 (25, 26), fibrinogen-related proteins (27, 28), thioester-containing proteins (29), MIF (30), biomphalysin protease (31), Toll-like receptor (TLR) (32), hemopoeitic factor granularin (33), and an unknown protein encoded by gene Grctm6 (13) have demonstrated functional associations with snail immunity to schistosome infection. The diversity of protein mediators identified in these studies suggest that the molecular events transpiring prior to and during the hemocyte encapsulation reactions likely are complex, involving multiple protein components that differ depending on the specific snail-schistosome strain combination under investigation (3, 34, 35).
In the present study we performed a comparative proteomic analysis on hemocytes from inbred schistosome-susceptible (NMRI) and—resistant (BS-90) strains of B. glabrata during their active participation in in vitro encapsulation reactions with primary sporocysts of S. mansoni (NMRI strain). Specifically, we addressed the questions: What proteins are expressed by hemocytes directly participating in encapsulation reactions? Do susceptible and resistant snail hemocytes exhibit differential protein expression responses in the presence of encapsulated sporocysts? In order to address these questions we combined two powerful analytical methods, laser-capture microdissection (LCM) microscopy and nano-LC tandem mass spectrometry, to provide the first comparative proteomic analysis of schistosome-susceptible and—resistant hemocytes during active in vitro encapsulation of S. mansoni sporocysts.
Materials and methods
Ethics statement
Mice were maintained in an AAALAC-approved animal housing facility (Charmany Instructional Facility, University of Wisconsin-Madison) using standard-of-care cage housing and feeding. All animal handling and experimental protocols were approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee (IACUC) under Animal Welfare Assurance No. A3368-01.
Maintenance of B. glabrata snails
Two inbred laboratory strains of B. glabrata snails, the S. mansoni-susceptible NMRI and -resistant BS-90 strains, were used in the present study. Snails were maintained in 20-gal aquaria at 26°C under a 12 h light:12 h dark photoperiod and fed leaf-lettuce ad libitum supplemented with Tetramin® fish food and chalk.
Isolation of miracidia and sporocyst preparation
Mice, infected with the NMRI strain of S. mansoni, were provided by the Biomedical Research Institute (Rockville, MD) under a NIAID contract to BEI Resources (Manassas, VA). At 7 weeks post-infection, mice were euthanized and necropsied for removal of livers and axenic egg isolation (36).
Miracidia were isolated under axenic conditions as previously described (36, 37) and placed into in vitro culture in 24-well tissue culture plates (Corning Costar, NY) containing Chernin's balanced salt solution (CBSS) (38) under normoxic conditions at 26°C. The CBSS was supplemented with penicillin (100 U/mL), streptomycin sulfate (0.1 mg/mL) and 1 g/L each of glucose and trehalose. Following miracidial transformation (24 h), primary sporocysts were maintained in culture for an additional 24 h at 26°C, after which they were washed 5 times in CBSS by gravity sedimentation to remove the shed epidermal plates and other cellular debris.
Hemocyte preparation
Hemolymph (HL) from 120 B. glabrata snails (12–15 mm shell diameter) of each strain was removed by the headfoot retraction method (39). Briefly, prior to bleeding, snail shells were wiped with cotton swabs soaked in 70% ethanol and air dried, followed by carefully peeling back the shell to expose the headfoot region. A rapid poking of the tissue by forceps forced the headfoot to retract while extruding HL from the hemal pore. The HL was then collected by micropipettor using siliconized, sterilized tips, and dispensed onto a cold sterile glass plate for 15–20 s to allow for remove any shell debris or mucus before transfer to a sterile 15-mL conical centrifuge tube containing cold CBSS. The tube was kept on ice during HL collection, with the final HL: CBSS ratio after collection equaling ~1:1. All procedures outlined above for HL extraction were performed in a biosafety cabinet to minimize microbial contamination. Hemocyte concentrations of pooled samples were determined by Neubauer hemocytometer (318 ± 131 and 304 ± 56 cells/μL in BS-90 and NMRI, respectively).
Hemocyte-sporocyst cell-mediated cytotoxicity (CMC) assay
The CMC assay was modified from that reported by Bayne et al. (8) and Hahn et al. (40). After adjusting cell concentrations for NMRI and BS-90 hemocyte preparations, HL was transferred to 1.5 mL siliconized, sterile microcentrifuge tubes containing approximately 100 in vitro transformed S. mansoni primary sporocysts. Each tube contained a cushion of 0.5% agarose dissolved in CBSS at its bottom to provide a substrate for hemocytes and sporocysts to interact. Tubes were centrifuged at 1,000 rpm (Eppendorf 5415D; Brinkmann Instruments, Westbury, NY) for 10 min to bring hemocytes into contact with the sporocysts at the bottom of tubes. The control groups consisted of introducing pooled HL to identical tubes containing agarose plugs, but in the absence of sporocysts. In this case, hemocyte aggregates, resembling larval encapsulations, spontaneously formed at the agarose surface. These were processed identically to larval-containing cell preparations.
Sample collection and cryosectioning
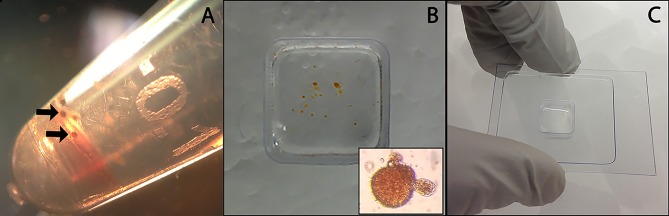
Hemocyte/sporocyst and hemocyte only cultures were incubated undisturbed for 18 h at 26°C under normoxic conditions. Following incubation, cell clumps (Figure 1A) were washed 5x with CBSS, transferred to cryomolds, and allowed to settled (Figure 1B). CBSS was then removed and replaced by the cryosectioning embedding medium, OCT (Lab-Tek Products, Naperville, IL) (Figure 1C), followed immediately by freezing on dry ice and storage at −80°C until sectioning. Frozen sections (10 μm thickness) of OTC-embedded NMRI and BS-90 hemocyte aggregates were cut using a Leica CM 1900 cryostat (Leica) at −28°C, placed on membrane-coated glass slides (Molecular Machines & Industries-MMI microslides, Haslett, MI) and maintained at −80°C before LCM. Slide-mounted cryosections were fixed in 70% EtOH for 30 sec, stained with Mayer's hematoxylin solution (30 s) (Sigma-Aldrich), wash 2x in Milli-Q water (15 s), and dehydrated in a graded EtOH series consisting of 70% (10 s), 2 × 95% (10 s), and 2 × 100% (10 and 30 s) absolute EtOH. The stained tissue sections were air-dried in a laminar flow hood for 15 min and then subjected to LCM.
Figure 1.
Preparation of control B. glabrata hemocyte aggregates and hemocyte-encapsulated S. mansoni sporocysts for cryohistology. Hemocyte-sporocyst encapsulations (arrows) following larval-hemolymph coculture for 18 h and washing in CBSS (A); Isolated hemocyte samples placed in a cryo-mold (insert hemocyte-sporocyst aggregate) (B); Quick freezing of cellular encapsulations in OTC prior to cryosectioning (C).
Laser-capture microdissection (LCM)
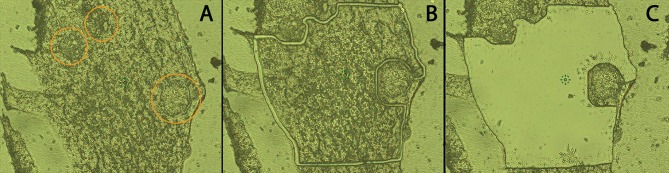
LCM was used to separate and isolate hemocytes (Hc) from encapsulated sporocysts in order to enrich for Hc-associated proteins in Hc-sporocyst co-cultivation samples. The MMI CellCut® Nikon microscope (Molecular Machines & Industries, Haslett, MI) was used to image and microdissect encapsulating tissue. Using the MMI CellCut® software, regions of interest were selected in the tissue images (Figure 2A) and microdissected with a laser (Figures 2B,C). The dissected tissue sections were then collected in MMI IsolationCap® tubes for subsequent proteomic analysis. Approximately 3 × 106 μm2 of sectioned tissue per sample were collected with one experiment consisting of four sample collections: BS-90 Hc only (BS90), BS-90 Hc + sporocysts (BS90-SP), NMRI Hc only (NMRI), NMRI Hc + sporocysts (NMRI-SP). Two independent experiments were conducted, each consisting of fresh pooled hemolymph and a new batch of transformed sporocysts.
Figure 2.
Cryosection of in vitro hemocyte encapsulation containing S. mansoni sporocysts. Following location of encapsulated sporocysts (A; circled), hemocyte and sporocyst tissue sections were dissected by laser beam using LCM (B), and hemocyte tissue subsequently transferred to ISOLATION-CAP™ tubes (C) for protease digestion and MS processing (20x).
Proteomic analysis
Sectioned microdissected tissue in microcap tubes were solubilized and protein denatured in 12 μL of a chaotropic detergent containing solution [6M Urea/0.5% ProteaseMAX™ surfactant (Promega Corp., Madison, WI) in 50 mM NH4HCO3 (pH 8.5)] for 1 min at 24°C then switched to 42°C for 10 min and subsequently diluted to 60 μL in a reduction step by addition of 2.5 μL of 25 mM dithiothreitol (DTT), 5 μL MeOH and 40.5 μL of 25 mM NH4HCO3 (pH 8.5). Samples were then incubated at 52°C for 15 min, cooled on ice to 22°C, followed by addition of 3 μL of 55 mM iodoacetamide for alkylation in the dark for 15 min at 22°C. Reactions were quenched by adding 6 μL of 25 mM DTT. Protease digestion was initiated by adding 4 μL of Trypsin/LysC solution [10 ng/μL Trypsin/LysC mix (Promega) in 25 mM NH4HCO3] and 27 μL of 25 mM NH4HCO3 (pH 8.5) to each sample tube for 2 h at 42°C, followed by the addition of 3 μL more of Trypsin/LysC solution and continued overnight digestion at 37°C. Reactions were terminated by acidification with 2.5% trifluoroacetic acid (TFA, Sigma-Aldrich) to 0.3% final concentration.
Digests were cleaned up using OMIX C18 SPE cartridges (Agilent Technologies, Palo Alto, CA) per the manufacturer's protocol and eluted in 10 μL of 70/30/0.1% acetonitrile (ACN)/H2O/TFA, dried to completion in a speed-vac centrifuge and finally reconstituted to 20 μL in 0.1% formic acid. Peptides were analyzed by nanoLC-MS/MS using an Agilent 1100 nanoflow system connected to a hybrid linear ion trap-orbitrap mass spectrometer (LTQ-Orbitrap XL, Thermo Fisher Scientific) equipped with a nanoelectrospray ion source. Chromatography of peptides prior to mass spectral analysis was accomplished using a C18 reverse-phase HPLC trap column (Zorbax 300SB-C18, 5 μM, 5 × 0.3 mm, Agilent Technologies) and capillary emitter column (in-house packed with MAGIC C18, 3-μM, 150 × 0.075 mm; Michrom Bioresources, Inc., Sacramento, CA) onto which 8 μL of digest was automatically loaded. A nanoHPLC system delivered solvents A (0.1% (v/v) formic acid) and B (95% (v/v) ACN, 0.1% (v/v) formic acid) at a rate of 10 μL/min to load the peptides (over 45 min) and 0.2 μL/min to elute peptides directly into the nano-electrospray with a continuous gradient from 0% (v/v) B to 40% (v/v) B over the next 145 min. This was followed by a fast gradient elution from 40% (v/v) B to 60% (v/v) B for 20 min and finally a 5 min flash-out from 50 to 95% (v/v) B. As peptides eluted from the HPLC-column/electrospray source, survey MS scans were acquired in the Orbitrap with a resolution of 100,000, and up to 5 most intense peptides per scan were fragmented and detected in the ion trap over the 300–2,000 m/z, with redundancy being limited by dynamic exclusion.
Bioinformatic analyses
Raw MS/MS data were converted to mgf file format using the Trans-Proteomic Pipeline (http://tools.proteomecenter.org/software.php; Seattle Proteome Center, Seattle, WA). Resulting mgf files were used to search against concatenated B. glabrata and S. mansoni protein databases consisting of RefSeq non-redundant entries plus common lab contaminants and decoy entries for false discover rate (FDR) calculations (96,790 total entries) with cysteine carbamidomethylation as fixed modification plus methionine oxidation and asparagine/glutamine deamidation as variable modifications. Peptide mass tolerance was set at 15 ppm and fragment mass at 0.6 Da. Thermo Proteome Discoverer version 1.4.1.14 was used to integrate results from dual search engines (Sequest and Mascot) and Scaffold (version Scaffold_4.3.2, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >84.0% probability to achieve an FDR < 1.0% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they achieved an FDR < 1.0% and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (41). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The complete raw dataset used in bioinformatics analyses of the hemocyte proteomic data are available as project #1492 available at chorusproject.org SMART (Simple Modular Architecture Research Tool) was employed to identify protein domains using normal mode, including outlier homologs and PFAM domains (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1). Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) (42) was used for multiple-sequence alignments. Unknown/uncharacterized/hypothetical proteins were subjected to BLASTp searches of the nrNCBI db (as of May 2018) to acquire related protein identifications. The entire experiment was performed twice on different dates using different pools of hemolymph and in vitro cultured sporocysts. The results generated in both experiments were combined into a single dataset to provide an overall assessment of the proteomic response of encapsulating hemocyte in the presence and absence of larval S. mansoni. The combined dataset from each snail strains was then subjected to pair-wise comparison (NMRI vs. NMRI-SP, BS-90 vs. BS-90-SP, and BS-90-SP vs. NMRI-SP) of unique and commonly-occurring proteins identified in each combination. All proteins identified as S. mansoni were removed from the analyses. Unique proteins (identified in only one of the pair) were further trimmed by removing all proteins with a total normalized spectral counts of <2. The remaining unique proteins were then identified and gene ontology analyses were performed using Uniprot. Similarly, proteins shared in common in each paired comparison were further processed by removing proteins with total normalized spectral counts of <4. In anticipation of analyzing these common proteins for potential differential expression, we verified that each experiment had comparable protein sample sizes using normalized spectral counts of actin (XP_013070997.1, XP_013083901.1, XP_013083930.1, XP_013083929.1) as a housekeeping protein. By comparing the average actin spectral counts identified within each proteomic database, we observed count differences ranging from 1.2 to 18% between paired samples (BS-90 vs. BS-90-SP: 18%; NMRI vs. NMRI-SP: 1.2%; and BS-90-SP vs. NMRI-SP: 7%). Given this range of sampling, for proteins shared in common between groups, a fourfold difference in total normalized spectral counts was imposed as a conservative cut-off for determining differential protein expression in pairwise treatment comparisons.
Results and discussion
In an effort to better understand the molecular events associated with the differences observed at the host-parasite interface of B. glabrata and S. mansoni, a proteomic analysis of hemocytes actively engaged in the encapsulation of S. mansoni primary sporocyst was conducted. A MALDI-MS/MS approach was undertaken to analyze the expressed protein profile of hemocytes of inbred resistant BS-90 and susceptible NMRI B. glabrata snail strains involved in the encapsulation of S. mansoni (NMRI strain) primary sporocyst. The main goal of the study was to show the differential expression pattern of proteins expressed in hemocytes during the encapsulation of S. mansoni sporocyst, and to compare the proteins expressed in hemocytes of these snail strains in order to provide a better understanding of the molecular events associated with the innate immune response of B. glabrata snails to early developing sporocysts. Using this comparative proteomics approach, a total of 1061 B. glabrata hemocyte proteins were identified from cellular capsules in the presence and/or absence (control) of in vitro co-cultured S. mansoni sporocysts. Although an additional 155 S. mansoni proteins were identified, the current study will focus on hemocyte responses during parasitic encapsulation. All identified B. glabrata and S. mansoni proteins are listed in Supplementary Table 1 and include normalized spectral counts per protein for each treatment. Raw MS proteomics data can be downloaded at the website address:
https://chorusproject.org/anonymous/download/experiment/defa50bf549a41dda7a156928ca0a03f under project reference number 1492.
Proteomic response of hemocytes to encapsulated sporocysts
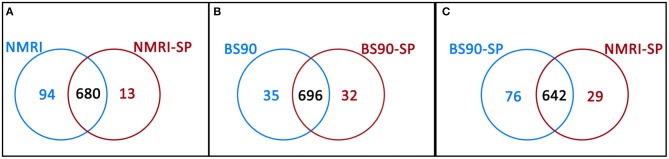
Hemocytes of both the NMRI and BS-90 B. glabrata strains readily encapsulated S. mansoni sporocysts (SP) in vitro forming cellular aggregation of various sizes (Figures 1A,B). In the absence of sporocysts (controls), hemocytes of both snail strains also spontaneously formed cellular capsules of similar size and number as corresponding co-cultures. This design permitted simultaneous comparisons of expressed hemocyte proteins between snail strains in the presence and absence of sporocysts. As shown in Venn diagrams (Figure 3), following application of filtering criteria, the numbers of unique and common hemocyte proteins identified in control/sporocyst (NMRI/NMRI-SP, BS90/BS90-SP) and sporocyst/sporocyst (NMRI-SP/BS90-SP) combinations for each snail strain are presented. Table 1 highlights a list of selected identified unique proteins of particular interest in this study and are designated as presence (+), absence (−) or presence, but below the defined threshold cutoff (+/−) for each host/parasite comparison listed above. Similarly, proteins found to be shared in common among all compared treatments and strains, and presenting with a 4-fold difference in total normalized spectral counts, are listed in Tables 2–4.
Figure 3.
Venn diagrams illustrating the number of unique proteins and proteins shared in common between (A) NMRI control (NMRI) and NMRI-sporocyst (NMRI-SP) encapsulations; (B) BS-90 control (BS90) and BS-90-sporocyst (BS90-SP) encapsulations and (C) sporocyst encapsulations by NMRI and BS-90 B. glabrata hemocytes (NMRI-SP and BS90-SP).
Table 1.
Unique hemocyte proteins of interest identified in paired comparisons of susceptible (NMRI) and resistant (BS90) strains of B. glabrata in the presence and absence of S. mansoni sporocysts (SP) (NMRI vs. NMRI-SP; BS90 vs. BS90-SP; NMRI-SP vs. BS90-SP).
| UNIQUE proteins | NMRI | NMRI-SP | BS90 | BS90-SP | NMRI-SP | BS90-SP | |
|---|---|---|---|---|---|---|---|
| NCBI acc. no. | Protein identification | ||||||
| IMMUNE RELATED | |||||||
| XP_013062219.1 | Integrin alpha-PS1-like [Bg] | + | − | − | − | − | − |
| XP_013064668.1 | */Fibrinogen-related protein F precursor [Bg] AQX34583.1 (96%) | + | − | + | +/− | − | +/− |
| XP_013070670.1 | lipopolysaccharide-binding protein-like isoform X1 [Bg] | − | − | +/− | + | − | + |
| XP_013064836.1 | */ Interleukin-1 receptor-associated kinase -a [Mytilus galloprovincialis] AHI17298.1 (31%) | − | + | − | − | + | − |
| XP_013089248.1 | Piwi-like protein 1 [Bg] | + | − | + | − | − | − |
| XP_013068459.1 | Protein argonaute-2-like isoform X1 [Bg] | − | − | + | − | − | − |
| XP_013079233.1 | Protein argonaute 18-like [Bg] | − | + | − | +/− | + | +/− |
| XP_013060966.1 | Histone H2A.V [Bg] | − | + | + | + | + | + |
| XP_013088376.1 | Bactericidal permeability-increasing protein-like [Bg] | +/− | + | − | + | + | + |
| XP_013095937.1 | Bactericidal permeability-increasing protein-like [Bg] | +/− | − | +/− | + | − | + |
| XP_013064309.1 | Fibrinogen-like protein A [Bg] | − | − | + | + | − | + |
| XP_013065165.1 | */ CD209 antigen-like protein 2 [Bg] XP_013095847.1 (56%) | + | + | +/− | − | + | − |
| XP_013064709.1 | */Pannexin10 [Aplysia californica] NP_001191462.1 (24%) | +/− | + | − | − | + | − |
| XP_013075978.1 | Perilipin-2-like [Bg] | + | − | +/− | + | − | + |
| XP_013069899.1 | GRB2-associated-binding protein 1-like isoform X1 [Bg] | + | − | − | − | − | − |
| XP_013077735.1 | Alpha-L-fucosidase-like [Bg] | + | − | − | − | − | − |
| XP_013066054.1 | Sequestosome-1-like [Bg] | + | − | + | +/− | − | +/− |
| XP_013067929.1 | Leucine-rich repeat-containing protein 47-like [Bg] | + | − | − | − | − | − |
| REDOX | |||||||
| XP_013077425.1 | Ferric-chelate reductase 1 [Bg] | + | − | + | +/− | − | +/− |
| XP_013084576.1 | Glutaredoxin-3-like [Bg] | + | − | − | +/− | − | +/− |
| XP_013091598.1 | Peroxidasin homolog [Bg] | + | − | − | − | − | − |
| XP_013077074.1 | SH3 and PX domain-containing protein 2A-like isoform X1 [Bg] | + | + | − | − | + | − |
| XP_013076408.1 | Glutarate-semialdehyde dehydrogenase DavD-like, partial [Bg] | + | + | + | − | + | − |
| XP_013068651.1 | Microsomal glutathione S-transferase 3-like [Bg] | + | +/− | + | − | +/− | − |
| XP_013091634.1 | D-arabinono-1,4-lactone oxidase-like [Bg] | + | + | − | + | + | + |
| XP_013070344.1 | Superoxide dismutase [Cu-Zn]-like [Bg] | +/− | − | − | + | − | + |
| XP_013084558.1 | Dual oxidase-like [Bg] | − | − | +/− | + | − | + |
| XP_013060671.1 | Glutathione S-transferase 4-like [Bg] | +/− | − | + | + | − | + |
| XP_013090296.1 | Nicotinamide phosphoribosyltransferase-lie, partial, [Bg] | − | − | − | + | − | + |
| XP_013085832.1 | Laccase-3-like isoform X1 [Bg] | +/− | − | − | + | − | + |
| XP_013073837.1 | Isocitrate dehydrogenase [NADP] cytoplasmic-like isoform X1 [Bg] | + | + | + | − | + | − |
| XP_013083111.1 | NADPH−cytochrome P450 reductase-like [Bg] | + | − | +/− | + | − | + |
| NP_976169.1 | Cytochrome c oxidase subunit II (mitochondrion) [Bg] | + | + | + | − | + | − |
| XP_013067188.1 | Cytochrome P450 2J3-like [Bg] | − | − | + | + | − | + |
| APOPTOSIS | |||||||
| XP_013071242.1 | */ E3 ubiquitin-protein ligase MIB1-like [Bg]; XP_013087470.1 (51%) | − | +/− | + | − | +/− | − |
| XP_013086341.1 | Death-associated protein 1-like [Bg] | − | − | − | + | − | + |
| XP_013065969.1 | Bcl-2-related protein A1-like [Bg] | + | − | + | + | − | + |
| XP_013094855.1 | Ubiquitin-like modifier-activating enzyme 6 isoform X1 [Bg] | + | +/− | − | + | +/− | + |
| XP_013088836.1 | TAR DNA-binding protein 43-like [Bg] | − | + | + | − | + | − |
| STRESS RESPONSE | |||||||
| XP_013064401.1 | T-complex protein 1 subunit gamma-like, partial [Bg] | + | − | − | + | − | + |
| XP_013064658.1 | Hsc70-interacting protein-like [Bg] | − | − | +/− | + | − | + |
| XP_013061307.1 | Prefoldin subunit 1-like [Bg] | + | + | − | − | + | − |
| XP_013068854.1 | Parkin coregulated gene protein homolog isoform X1 [Bg] | − | + | − | − | + | − |
| XP_013077814.1 | CDGSH iron-sulfur domain-containing protein 2-like [Bg] | − | − | − | + | − | + |
| XP_013094352.1 | Serine/threonine-protein kinase fray2-like [Bg] | − | + | − | − | + | − |
| SIGNALING | |||||||
| XP_013077238.1 | Coronin-1A-like [Bg] | + | + | + | − | + | − |
| XP_013070025.1 | Rap1 GTPase-activating protein 1-like isoform X4 [Bg] | + | − | +/− | − | − | − |
| XP_013074990.1 | COP9 signalosome complex subunit 6-like [Bg] | + | − | +/− | − | − | − |
| XP_013086090.1 | COP9 signalosome complex subunit 4-like isoform X1 [Bg] | + | − | + | + | − | + |
| XP_013078600.1 | SH3 domain-containing kinase-binding protein 1-like isoform X1 [Bg] | + | − | +/− | +/− | − | +/− |
| XP_013087123.1 | SHC-transforming protein 1-like [Bg] | − | − | + | − | − | − |
| XP_013088835.1 | */SH3 Domain-containing guanine exchange factor [Crassostrea gigas]; EKC39783.1 (48%) | + | − | +/− | + | − | + |
| XP_013087149.1 | */Tensin-1-like, partial [Aplysia californica] (49%) | + | − | +/− | +/− | − | +/− |
| XP_013078554.1 | */Receptor-type tyrosine-protein phosphatase T-like isoform X1 [Bg]; XP_013072072.1 (40%) | + | − | + | + | − | + |
| CALCIUM BINDING | |||||||
| XP_013077729.1 | Calmodulin-like, partial [Bg] | − | − | + | + | − | + |
| XP_013079182.1 | */Microtubule-actin cross-linking factor 1-like isoform X13 [Bg]; XP_013079259.1 (99%) | +/− | − | + | − | − | − |
| XP_013083369.1 | Clathrin interactor 1-like isoform X3 [Bg] | + | + | − | − | + | − |
| XP_013091491.1 | 16 kDa calcium-binding protein-like [Bg] | − | − | +/− | + | − | + |
| XP_013072348.1 | Calcineurin subunit B type 1 [Bg] | − | − | − | + | − | + |
| XP_013060775.1 | Calumenin-like [Bg] | − | − | +/− | + | − | + |
| XP_013066201.1 | Gelsolin-like protein 1, partial [Bg] | − | − | +/− | + | − | + |
| XP_013068775.1 | Gelsolin-like protein 1 [Bg] | + | − | − | + | − | + |
| XP_013088705.1 | Peflin-like [Bg] | + | + | + | − | + | − |
| XP_013086544.1 | Nucleobindin-2-like [Bg] | +/− | +/− | − | + | +/− | + |
| TRANSCRIPTION/TRANSLATION PROCESS | |||||||
| XP_013063581.1 | 60S Ribosomal protein L14-like [Bg] | +/− | − | + | + | − | + |
| XP_013075095.1 | 60S Ribosomal protein L31-like [Bg] | + | − | + | + | − | + |
| XP_013089238.1 | 60S Ribosomal protein L36-like [Bg] | + | − | + | +/− | − | +/− |
| XP_013096652.1 | 60S Ribosomal protein L37a-like [Bg] | + | − | + | + | − | + |
| XP_013091387.1 | Host cell factor-like isoform X1 [Bg] | + | − | + | − | − | − |
| XP_013062003.1 | LIM and senescent cell antigen-like-containing domain protein1 isoform X1 [Bg] | − | +/− | − | + | +/− | + |
| XP_013062476.1 | Signal recognition particle subunit SRP72-like [Bg] | + | − | + | + | − | + |
| XP_013093252.1 | */Histone-lysine N-methyltransferase MLL4 [Crassostrea gigas]; EKC19620.1 (61%) | + | +/− | + | − | +/− | − |
| XP_013072871.1 | Krueppel homolog 1-like isoform X1 [Bg] | + | − | − | − | − | − |
| METABOLISM | |||||||
| XP_013083732.1 | Inter-alpha-trypsin inhibitor heavy chain H3-like isoform X1 [Bg] | + | + | − | − | + | − |
| XP_013079110.1 | Alpha-N-acetylglucosaminidase-like isoform X1 [Bg] | + | − | + | +/− | − | +/− |
| XP_013080405.1 | Gamma-glutamyl hydrolase A-like [Bg] | − | − | − | + | − | + |
| TRAFFICKING | |||||||
| XP_013083454.1 | Intersectin-1-like [Bg] | + | − | +/− | − | − | − |
| XP_013094430.1 | Nuclear pore membrane glycoprotein 210-like [Bg] | +/− | − | + | − | − | − |
| XP_013083696.1 | AP-3 complex subunit delta-1-like isoform X1 [Bg] | + | − | +/− | +/− | − | +/− |
| XP_013060735.1 | Sorting nexin-33-like, partial [Bg] | + | − | + | +/− | − | +/− |
| MISC. | |||||||
| XP_013092372.1 | Endothelin-converting enzyme 2-like [Bg] | − | − | + | − | − | − |
| XP_013063554.1 | Basement membrane-specific heparan sulfate proteoglycan core protein-like [Bg] | + | + | + | − | + | − |
Unique proteins are organized by functional catagory and NCBI accession numbers and protein identification are listed. For each paired comparison, (+) indicates the presence of the protein; (−) indicates the absence of the protein and (+/−) indicates the protein presence, but the threshold of less than 2 peptides was not met. Uncharacterized protein are indicated (*/), followed by their closest NCBI Blastp IDs and ID percentages.
Table 2.
Proteins shared in common between control NMRI and sporocyst-encapsulating (NMRI-SP) hemocytes that present at least a 4-fold difference in total normalized spectral counts.
| NMRI vs. NMRI-SP: 4 fold difference | ||||
|---|---|---|---|---|
| NCBI acc. no. | Protein identification | NMRI | NMRI-SP | Function |
| IMMUNE RELATED | ||||
| XP_013070669.1 | Lipopolysaccharide-binding protein-like [Bg] | 9.57 | 1.01 | Cellular defense |
| XP_013070436.1 | Syntenin-1-like isoform X1 [Bg] | 11.98 | 0.96 | Immunomodulation |
| XP_013082155.1 | Apolipophorins-like isoform X1 [Bg] | 9.57 | 0.96 | Detox LPS |
| XP_013067749.1 | Lysosomal alpha-mannosidase-like, partial [Bg] | 5.30 | 0.96 | Killing of foreign material |
| XP_013066053.1 | */ Sequestosome-1-like [Limulus polyphemus] XP_013792674.1 (36%) | 18.78 | 0.96 | Autophagy |
| XP_013093397.1 | Lysosomal alpha-glucosidase-like isoform X1 [Bg] | 7.44 | 1.01 | Degradation of Phagocytosed materials |
| XP_013088376.1 | Bactericidal permeability-increasing protein-like [Bg] | 1.11 | 4.89 | Anti-microbial |
| XP_013074089.1 | Arginase-1-like isoform X3 [Bg] | 15.05 | 65.36 | Immune response |
| REDOX | ||||
| XP_013077724.1 | */ Carbonyl reductase [NADPH] 1-like [Bg] XP_013077725.1 (74%) | 4.27 | 0.96 | Redox |
| XP_013076465.1 | 3-Oxo-5-alpha-steroid 4-dehydrogenase 1-like [Bg] | 4.37 | 1.01 | NADPH binding/Redox |
| XP_013093038.1 | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial-like [Bg] | 1.02 | 6.86 | Mitochondrial electron transport |
| XP_013073195.1 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial-like [Bg] | 1.11 | 4.84 | Mitochondrial electron transport |
| STRESS RESPONSE | ||||
| XP_013091012.1 | Stromal cell-derived factor 2-like isoform X1 [Bg] | 4.27 | 1.01 | Regulation apoptosis/ chaperonin |
| XP_013065387.1 | Protein FAM107B-like, partial [Bg] | 4.27 | 0.96 | Similarity with heat-shock proteins |
| SIGNALING | ||||
| XP_013088940.1 | G-protein coupled receptor 64-like isoform X1 [Bg] | 4.18 | 0.96 | Signaling proteins |
| XP_013077039.1 | */ Receptor-type tyrosine-protein phosphatase α-like [Bg] XP_013063115.1 (42%) | 4.27 | 0.96 | MAPK cascade |
| XP_013077041.1 | Receptor-type tyrosine-protein phosphatase alpha-like, partial [Bg] | 5.30 | 0.96 | MAPK cascade |
| XP_013081619.1 | Serine/threonine-protein kinase PAK 2-like [Bg] | 8.55 | 1.97 | Signaling |
| CELL ADHESION | ||||
| XP_013081823.1 | Collagen alpha-1(XVIII) chain-like [Bg] | 8.55 | 1.91 | Cell adhesion |
| XP_013086335.1 | */ Neurexin-1-like [Aplysia californica] XP_012941838.1 (48%) | 38.20 | 1.91 | Cell adhesion |
| TRANSCRIPTION/TRANSLATION PROCESS | ||||
| XP_013061577.1 | 40S Ribosomal protein S25-like [Bg] | 4.27 | 0.96 | Ribosomal |
| XP_013084756.1 | 60S Ribosomal protein L19-like, partial [Bg] | 4.18 | 1.01 | Ribosomal |
| XP_013071682.1 | 60S Ribosomal protein L30-like [Bg] | 4.37 | 0.96 | Ribosomal |
| XP_013073467.1 | 60S Ribosomal protein L34-like [Bg] | 5.39 | 1.01 | Ribosomal |
| XP_013063478.1 | 60S Ribosomal protein L8 [Bg] | 21.55 | 4.95 | Ribosomal |
| XP_013084934.1 | Heterogeneous nuclear ribonucleoprotein U-like protein 1 isoform X1 [Bg] | 8.28 | 1.97 | Transcription regulation |
| XP_013070062.1 | Protein LZIC-like [Bg] | 5.39 | 1.01 | B-catenin binding |
| XP_013081352.1 | Protein TFG-like isoform X1 [Bg] | 4.37 | 0.96 | Regulation of NFKB signaling |
| XP_013063363.1 | Ribonuclease UK114-like [Bg] | 5.30 | 1.01 | Translation inhibition |
| XP_013062948.1 | RNA-binding protein lark-like [Bg] | 5.39 | 0.96 | Translation regulation |
| XP_013075272.1 | Serine/arginine repetitive matrix protein 2-like [Bg] | 4.37 | 0.96 | Pre-mRNA splicing |
| XP_013089331.1 | Serine/arginine-rich splicing factor 7-like [Bg] | 4.18 | 1.01 | Pre-mRNA splicing |
| METABOLISM PROCESS | ||||
| XP_013069940.1 | Alpha-galactosidase A-like isoform X1 [Bg] | 5.48 | 1.01 | Glycosidase |
| XP_013076662.1 | Succinyl-CoA:3-ketoacid coenz. A transferase 1, mitochondrial-like [Bg] | 8.37 | 1.01 | Ketone body catabolism |
| XP_013062675.1 | UTP–glucose-1-phosphate uridylyltransferase-like [Bg] | 9.75 | 0.96 | Cellular metabolic pathways |
| XP_013061266.1 | 1,4-alpha-glucan-branching enzyme-like [Bg] | 4.27 | 0.96 | Glycogen biosynthesis |
| XP_013092915.1 | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial-like [Bg] | 8.64 | 1.91 | Cellular amino acid metabolism |
| XP_013094977.1 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG-like [Bg] | 5.48 | 1.01 | Fatty acid biosynthesis |
| PROTEIN SORTING AND TRAFFICKING | ||||
| XP_013089878.1 | AP-1 complex subunit beta-1-like [Bg] | 9.85 | 1.91 | Clathrin binding/ transporter activity |
| XP_013095772.1 | Charged multivesicular body protein 4b-like [Bg] | 12.73 | 2.92 | Multivesicular body pathway |
| XP_013061300.1 | FK506-binding protein 15-like [Bg] | 5.39 | 1.01 | Transport of early endosomes |
| XP_013067947.1 | Huntingtin-interacting protein 1-like isoform X1 [Bg] | 4.18 | 0.96 | Trafficking |
| XP_013082360.1 | Vacuolar protein sorting-associated protein 27 isoform X1 [Bg] | 5.30 | 0.96 | Trafficking |
| XP_013080513.1 | Vacuolar protein sorting-associated protein 35-like [Bg] | 6.50 | 0.96 | Trafficking |
| MISC | ||||
| XP_013080947.1 | */Coiled-coil protein 90B, mitochondrial [Crassostrea gigas] EKC25784.1 (50%) | 6.41 | 0.96 | Transmembrane protein |
| XP_013080174.1 | */voltage-dependent calcium channel γ2 subunit-like [Bg] XP_013080170.1 (40%) | 5.39 | 1.01 | Voltage-dependent calcium channel |
| XP_013069538.1 | V-type proton ATPase 116 kDa subunit a isoform 1 [Bg] | 4.27 | 0.96 | Proton transporter |
NCBI accession numbers and protein identification are listed. Color backgrounds indicate an increased spectral count for proteins in the corresponding treatment. Uncharacterized proteins (*/) are followed by their closest NCBI Blastp IDs and ID percentages.
Table 4.
Proteins shared in common between sporocyst (SP)-encapsulating hemocytes of susceptible NMRI and resistant BS90 B. glabrata that present at least a 4-fold difference in total normalized spectral counts.
| BS90-SP vs. NMRI-SP: 4 fold difference | ||||
|---|---|---|---|---|
| NCBI acc. no. | Protein identification | BS90-SP | NMRI-SP | Function |
| IMMUNE RELATED | ||||
| XP_013070436.1 | Syntenin-1-like isoform X1 [Bg] | 3.96603 | 0.95609 | Trafficking/ immunomodulation |
| XP_013067749.1 | Lysosomal alpha-mannosidase-like, partial [Bg] | 3.8991 | 0.95609 | Oligosaccharide catabolism |
| XP_013070669.1 | Lipopolysaccharide-binding protein-like [Bg] | 14.923 | 1.0127 | Innate immune response |
| XP_013078814.1 | */ Proteoglycan 4-like [Aplysia californica] XP_005092461.1 (45%) | 8.6727 | 1.96879 | Immune response |
| XP_013079231.1 | N-acetylgalactosamine kinase-like [Bg] | 3.8991 | 0.95609 | Galactose binding |
| XP_013064208.1 | Reelin-like [Bg] | 2.8909 | 16.3097 | Lipoprotein receptor binding |
| REDOX | ||||
| XP_013077724.1 | */Carbonyl reductase [NADPH] 1-like [Bg] XP_013077725.1 (74%) | 19.6296 | 0.95609 | Redox |
| XP_013080583.1 | Thioredoxin reductase 1, cytoplasmic-like isoform X1 [Bg] | 7.99883 | 0.95609 | Redox |
| XP_013069507.1 | */Phytanoyl-CoA dioxygenase domain-containing protein 1-like [Pomacea canaliculata] XP_025105244.1 (70%) | 7.7982 | 1.0127 | Redox |
| XP_013060614.1 | */ Hemoglobin type 1 [Bg] CAJ44466.1 (58%) | 13.179 | 1.9122 | O2 binding |
| XP_013062383.1 | */Hemoglobin type 1 [Bg] CAJ44466.1 (82%) | 44.5771 | 8.718 | O2 binding |
| XP_013062584.1 | */ Hemoglobin type 2 CAJ44467.1 (99%) | 103.608 | 13.5552 | O2 binding |
| XP_013075721.1 | D-amino-acid oxidase-like [Bg] | 1.8827 | 8.8879 | Redox |
| XP_013062841.1 | Prolyl 4-hydroxylase subunit alpha-1-like isoform X2 [Bg] | 0.94133 | 5.7931 | Redox |
| XP_013080374.1 | Electron transfer flavoprotein subunit alpha, mitochondrial-like [Bg] | 2.0164 | 8.8879 | Redox |
| APOPTOSIS | ||||
| XP_013066053.1 | */sequestosome-1-like [Limulus polyphemus] XP_013792674.1 (36%) | 4.0329 | 0.95609 | Immune/Autophagy |
| XP_013091656.1 | Apoptosis-inducing factor 3-like isoform X1 [Bg] | 1.0082 | 4.8937 | Apoptosis |
| STRESS RESPONSE | ||||
| XP_013075913.1 | Endoplasmin-like [Bg] | 3.8322 | 0.95609 | Chaperone |
| XP_013079911.1 | Heterochromatin protein 1-binding protein 3-like isoform X1 [Bg] | 4.8404 | 0.95609 | Cellular response to hypoxia |
| XP_013065461.1 | Protein deglycase DJ-1zDJ-1-like [Bg] | 7.932 | 1.96879 | Chaperone/Stress response |
| SIGNALING | ||||
| XP_013077041.1 | Receptor-type tyrosine-protein phosphatase alpha-like, partial [Bg] | 4.7066 | 0.95609 | MAPK cascade |
| XP_013077039.1 | */ Receptor-type tyrosine-protein phosphatase alpha-like [Bg] XP_013063115.1 (42%) | 5.7817 | 0.95609 | MAPK cascade |
| XP_013081619.1 | Serine/threonine-protein kinase PAK 2-like [Bg] | 13.6468 | 1.96879 | Signaling |
| XP_013087690.1 | Formin-like protein CG32138 [Bg] | 6.79 | 0.95609 | Rac GTPase binding |
| XP_013088191.1 | Guanine nucleotide-binding protein G(o) subunit alpha [Bg] | 4.7735 | 0.95609 | GTP binding |
| TRANSCRIPTION/TRANSLATION PROCESS | ||||
| XP_013061577.1 | 40S Ribosomal protein S25-like [Bg] | 4.8404 | 0.95609 | Translation initiation |
| XP_013070062.1 | Protein LZIC-like [Bg] | 4.9074 | 1.0127 | Transcription factor |
| XP_013062948.1 | RNA-binding protein lark-like [Bg] | 5.7148 | 0.95609 | Translation regulation |
| XP_013063676.1 | */ Nucleolin-like [Aplysia californica] XP_005109234.1 (31%) | 5.0411 | 0.95609 | Translation regulation |
| XP_013063201.1 | Histone H1-delta-like [Bg] | 5.8487 | 1.0127 | Transcription regulation |
| XP_013084934.1 | Heterogeneous nuclear ribonucleoprotein U-like protein 1 isoform X1 [Bg] | 14.5881 | 1.96879 | Transcription regulation |
| METABOLISM/CATABOLISM | ||||
| XP_013068834.1 | 3-Hydroxyacyl-CoA dehydrogenase type-2-like [Bg] | 4.7735 | 0.95609 | Lipid metabolism |
| XP_013072118.1 | Glycogen debranching enzyme-like [Bg] | 4.7735 | 0.95609 | Catabolic glycogen process |
| XP_013062675.1 | UTP–glucose-1-phosphate uridylyltransferase-like [Bg] | 7.7313 | 0.95609 | Glucose metabolism |
| XP_013081380.1 | Fumarate hydratase class I, aerobic-like [Bg] | 0.94133 | 3.881 | Carbohydrate metabolism |
| XP_013069189.1 | 2-Aminoethylphosphonate–pyruvate transaminase-like [Bg] | 1.94953 | 8.8313 | Phosphonate Catabolism |
| XP_013076947.1 | Fatty acid synthase-like [Bg] | 0.94133 | 4.837 | Lipid metabolism |
| MISC | ||||
| XP_013088283.1 | */Cartilage matrix protein-like [Aplysia californica] XP_005102985.2 (50%) | 5.98243 | 0.95609 | Collagen binding |
| XP_013075555.1 | ATP synthase subunit f, mitochondrial [Bg] | 1.0082 | 6.9192 | Proton transport |
NCBI accession numbers and protein identification are listed. Color backgrounds indicate an increased spectral count for proteins in the corresponding treatment. Uncharacterized proteins (*/) are followed by their closest NCBI Blastp IDs and ID percentages.
Proteomic response of susceptible (NMRI) snail hemocytes to encapsulated sporocysts
In the susceptible NMRI strain of B. glabrata, 680 hemocyte proteins were shared between the control and sporocyst-encapsulated groups, while 94 proteins were unique to the NMRI-control group and 13 to the NMRI-SP group (Figure 3A). Of the hemocyte proteins shared in common, 47 showed at least a 4-fold difference between control and sporocyst encapsulations. Of these 47 common proteins, the vast majority (43/47; 91%) exhibited a striking reduction in specific proteins in the presence of sporocysts, suggesting a downregulation of expression in susceptible snail cells in response to parasite exposure (Table 2). This parasite-associated decrease in NMRI hemocyte protein expression is also mirrored in the number of unique proteins identified in control (94 proteins) compared to sporocyst (13 proteins) encapsulations.
The majority of proteins (4-fold and unique) observed to be down-regulated in presence of parasites appears to involve protein synthesis/trafficking, metabolism, cell signaling, or oxidation-reduction (redox) activities (Table 2), all of which may have implications for an impaired immune capability. Among the down-regulated NMRI hemocyte proteins identified as potentially involved in immune-related functions in the presence of encapsulated sporocysts include the following: LPS binding protein (LBP), an antimicrobial protein capable of interacting with the larval tegument and secretions (43); apolipophorin, an immune-stimulating molecule in insects (44), found to be downregulated in snails exposed to microbial antigens (LPS, FCN, PGN) (45); and sequestosome 1/p62, a protein involved in various mechanisms, including apoptosis, innate immunity and autophagy (46). In addition to binding ubiquinated proteins for basal cellular autophagy, p62 may also target intracellular pathogens for degradation through a p62-dependent selective autophagy mechanisms and participate in host defense. For example, in murine macrophages, activation of this selective autophagic mechanism by LPS-triggered TLR4 requires upregulation of p62 expression, suggesting a role of p62 in innate immune responses to pathogens (47). Of relevance here, TLRs have been implicated in B. glabrata-S. mansoni interactions as demonstrated by a differential increase in BgTLR protein levels associated with snail strains exhibiting resistance to S. mansoni (32). Our finding that p62/sequestosome1 is dramatically decreased in NMRI-SP capsules, suggests an impairment of hemocyte p62-mediated selective autophagy by a parasite-induced targeting of p62. A similar disruption of p62 has been postulated as a survival mechanism by some pathogens (46).
By contrast, 4 immune-related NMRI hemocyte proteins exhibited an increase in expression in the presence of parasites: An uncharacterized protein, containing a N-terminal death domain of interleukin-1 receptor-associated kinases (IRAK) followed by a catalytic domain of the serine/threonine kinases, argonaute 18, bactericidal permeability increasing protein (BPI) and arginase 1. Although uncharacterized, the presence of the death-domain of IRAK and serine/threonine kinase suggests this protein belongs to the IRAK superfamily, with a potential involvement in the Toll pathway (48). The presence of argonaute 18 suggests the involvement of small RNAs, possibly in gene expression regulation via RISC (RNA-induced silencing complex). B. glabrata snails (Belo Horizonte strain) exposed to S. mansoni exhibited an upregulation of argonaute gene expression at 24 h post-infection, consistent with our current proteomic data (49). Our observed increase in BPI, a pattern recognition protein known to bind to lipopolysaccharides of Gram-negative bacteria (50), in hemocytes of both NMRI and BS-90 snails participating in larval encapsulations is in contrast to a reported decrease in BPI transcript response to the echinostome Echinostoma caproni in resistant B. glabrata (51), suggesting a potential pathogen specificity for BPI expression between S. mansoni-resistant B. glabrata strains. Finally, the finding that arginase1 expression is highly increased in the presence of sporocysts in susceptible host capsules may be of functional significance. Arginase is known to inhibit the production of NO by directly competing with nitric oxide synthase (NOS) for L-arginine (substrate), resulting in reduced levels of larval-killing NO production (52). Therefore, a parasite-induced triggering of hemocyte arginase production, as shown in the present study, may be serving as an anti-immune survival mechanism by creating a detoxified cellular microenvironment for encapsulated sporocysts.
Although proteins involved in redox reactions were found to be mostly down-regulated in presence of parasites (3-oxo-5-alpha-steroid 4-dehydrogenase 1; NADPH-cytochrome P450 reductase; peroxidasin; glutaredoxin-3; ferric-chelate reductase 1), two others, cytochrome c oxidase (Cyt c) and succinate dehydrogenase (ubiquinone-containing complex) exhibited increased protein expression. Cyt c and ubiquinone are part of the mitochondrial electron transport chain that, in addition to generating ATP, also are involved in the production of ROS. However, it is not known whether metabolically-generated ROS from this source may be serving a host immune effector role. More likely, the increased expression of these proteins probably reflects an enhancement of metabolic stress of host cells in response to sporocysts.
Overall, hemocytes of susceptible NMRI snails in the presence of S. mansoni sporocysts appear to have 2 distinct responses: A general down-regulation of proteomic activity across various protein groups, and a discrete increase of selected proteins, suggesting that different mechanisms potentially may be initiated either by the host or parasite. Although ROS, BPI and the death domain-IRAK containing protein increases are likely to be initiated by the host in response to a pathogen invasion, the increase of arginase, and with it, a resulting decrease in toxic NO production, or the general protein knockdown observed in NMRI hemocytes, could be a direct result of host modulation by the parasite to favor its own survival.
Proteomic response of resistant (BS-90) snail hemocytes to encapsulated sporocysts
As a result of proteomic analysis of BS-90 hemocyte capsules, 696 proteins were identified as shared in common between unexposed (BS90) and S. mansoni sporocyst-encapsulated (BS90-SP) hemocyte groups, while 35 and 32 unique proteins were identified in BS90 and BS90-SP hemocyte samples, respectively (Figure 3B). Similar to the NMRI treatment groups, 44 of the identified common proteins met the 4-fold minimum threshold difference (Table 3). However, in contrast to susceptible snail hemocytes, approximatively the same number of sporocyst-encapsulating BS-90 cell proteins were observed to be up and down-regulated in categories such as protein processing, metabolism, signaling, redox or immune-related proteins, with 59% of the proteins exhibiting a 4-fold decrease in presence of parasite.
Table 3.
Proteins shared in common between control BS90 and sporocyst-encapsulating (BS90-SP) hemocytes that present at least a 4-fold difference in total normalized spectral counts.
| Common BS90 vs. BS90-SP: 4 fold difference | ||||
|---|---|---|---|---|
| NCBI acc. no. | Protein identification | BS90 | BS90-SP | Function |
| IMMUNE-RELATED | ||||
| XP_013095968.1 | */Proteasome subunit alpha type-5-like [Aplysia californica] XP_005110071.1 (91%) | 3.938 | 0.94133 | Proteasome |
| XP_013066054.1 | Sequestosome-1-like [Bg] | 7.4474 | 1.0082 | Autophagy |
| XP_013066053.1 | */Sequestosome-1-like [Limulus polyphemus] XP_013792674.1(36%) | 29.823 | 4.0329 | Autophagy |
| XP_013064208.1 | Reelin-like [Bg] | 20.299 | 2.8909 | Lipoprotein receptor binding |
| XP_013079231.1 | N-acetylgalactosamine kinase-like [Bg] | 0.9309 | 3.8991 | Galactose binding |
| XP_013062216.1 | 26S proteasome non-ATPase regulatory subunit 12-like [Bg] | 1.0381 | 4.7735 | Proteasome |
| REDOX | ||||
| XP_013075526.1 | Glutathione S-transferase 7 [Bg] | 8.6998 | 2.0164 | Detoxification of ROS |
| XP_013089814.1 | Alcohol dehydrogenase class-3-like [Bg] | 3.938 | 0.94133 | Redox |
| XP_013077425.1 | Ferric-chelate reductase 1 [Bg] | 4.8689 | 1.0082 | Redox |
| XP_013073942.1 | Aldehyde dehydrogenase family 16 member A1-like [Bg] | 10.883 | 1.94953 | Redox |
| XP_013086335.1 | */Neurexin-1 [Aplysia californica] XP_012941838.1 (48%) | 20.621 | 4.8404 | Redox |
| XP_013069507.1 | */Phytanoyl-CoA dioxygenase 2-like [Saccoglossus kowalevskii] XP_006814546.1 (56%) | 1.8619 | 7.7982 | Fatty acid alpha-oxidation |
| XP_013084558.1 | Dual oxidase-like [Bg] | 0.9309 | 4.0329 | Generates ROS |
| APOPTOSIS AND REGULATION | ||||
| XP_013091656.1 | Apoptosis-inducing factor 3-like isoform X1 [Bg] | 7.6617 | 1.0082 | Pro-apoptosis |
| XP_013062216.1 | 26S proteasome non-ATPase regulatory subunit 12-like [Bg] | 1.0381 | 4.7735 | Proteasome |
| XP_013084356.1 | Acidic leucine-rich nuclear phosphoprotein 32 family member B-like [Bg] | 1.0381 | 8.8064 | Anti-apoptotic |
| STRESS RESPONSE | ||||
| XP_013094041.1 | Universal stress protein Slr1101-like [Bg] | 3.8309 | 0.94133 | Response to stress |
| SIGNALING | ||||
| XP_013069708.1 | Protein Mo25-like [Bg] | 0.9309 | 3.7653 | Calcium binding protein |
| CELL ADHESION/MOVEMENT | ||||
| XP_013073410.1 | Sushi, von Willebrand factor A, EGF /pentraxin domain-containing protein 1-like[Bg] | 4.8689 | 0.94133 | Cell attachment process |
| XP_013092731.1 | Triple functional domain protein-like, partial [Bg] | 4.1522 | 1.0082 | Actin remodeling |
| XP_013068207.1 | Myosin heavy chain, striated muscle-like isoform X1 [Bg] | 5.1903 | 1.0082 | Muscle contraction |
| TRANSCRIPTION/TRANSLATION PROCESS | ||||
| XP_013089238.1 | 60S Ribosomal protein L36-like [Bg] | 3.938 | 0.94133 | Pibosomal |
| XP_013082582.1 | Eukaryotic translation initiation factor 3 subunit K-like [Bg] | 4.9761 | 0.94133 | Translation regulation |
| XP_013096529.1 | Eukaryotic peptide chain release factor subunit 1 [Bg] | 3.8309 | 0.94133 | Translation regulation |
| XP_013089019.1 | Protein disulfide-isomerase A4-like [Bg] | 4.0451 | 1.0082 | Protein folding |
| XP_013093243.1 | */DNA ligase 1-like isoform X3 [Bg] XP_013093256.1 (100%) | 16.683 | 3.8322 | DNA binding/ repair |
| XP_013078837.1 | 40S Ribosomal protein S12-like [Bg] | 1.0381 | 5.7817 | Ribosomal |
| XP_013061577.1 | 40S Ribosomal protein S25-like [Bg] | 1.0381 | 4.8404 | Ribosomal |
| XP_013073661.1 | Cleavage and polyadenylation specificity factor subunit 5-like [Bg] | 1.969 | 9.6809 | Polyadenylation |
| XP_013080627.1 | Bifunctional glutamate/proline–tRNA ligase-like [Bg] | 1.0381 | 4.8404 | Translation regulation |
| XP_013060775.1 | Calumenin-like [Bg] | 0.9309 | 6.5893 | Calcium binding |
| METABOLISM | ||||
| XP_013093293.1 | Trifunctional nucleotide phosphoesterase protein YfkN-like, partial [Bg] | 4.7618 | 0.94133 | Nucleotide catabolic process |
| XP_013083683.1 | Sulfotransferase family cytosolic 1B member 1-like, partial [Bg] | 4.9761 | 1.0082 | Lipid metabolism |
| XP_013095124.1 | Stomatin-like protein 2, mitochondrial [Bg] | 4.9761 | 1.0082 | Lipid binding |
| XP_013069854.1 | Phosphonoacetaldehyde hydrolase-like [Bg] | 0.9309 | 8.8064 | Phosphonate catabolism |
| XP_013070092.1 | Ethanolamine-phosphate cytidylyltransferase-like isoform X1 [Bg] | 0.9309 | 3.8991 | Lipid metabolism |
| XP_013094998.1 | Enoyl-CoA hydratase, mitochondrial-like isoform X1 [Bg] | 1.0381 | 4.7735 | Lipid metabolism |
| XP_013075978.1 | Perilipin-2-like [Bg] | 1.0381 | 4.8404 | Lipid metabolism |
| PROTEIN SORTING AND TRAFFICKING | ||||
| XP_013077091.1 | ATP synthase subunit g, mitochondrial-like, partial [Bg] | 4.9761 | 0.94133 | Proton transporter |
| XP_013085659.1 | Coatomer subunit beta'-like [Bg] | 5.7998 | 1.0082 | Protein transport |
| XP_013086081.1 | */Tripartite motif-containing protein 3-like isoform X1 [Pomacea canaliculata] (25%) | 4.0451 | 0.94133 | Vesicular trafficking |
| XP_013073443.1 | dynamin-1-like [Bg] | 8.0902 | 1.0082 | Endocytosis. |
| MISC | ||||
| XP_013062313.1 | Unch. prot. LOC106051655 [Bg] | 4.0451 | 28.369 | Unknown |
| XP_013089315.1 | */Caffeoyl-CoA O-methyltransferase 1 [Bg] XP_013087214.1 (59%) | 0.9309 | 3.96603 | Methyl transferase |
NCBI accession numbers and protein identification are listed. Color backgrounds indicate an increased spectral count for proteins in the corresponding treatment. Uncharacterized proteins (*/) are followed by their closest NCBI Blastp IDs and ID percentages.
Contrary to the near complete shutdown of protein processing/metabolism in larval-encapsulating NMRI hemocytes, we observed an increase in 40S ribosomal proteins and others involved in mRNA/tRNA processing in BS-90 capsules indicating a marked enhancement of cellular translation processes in response to contact with encapsulated larvae (Table 3). By contrast, members of the argonaute family proteins (piwi and argonaute2), potentially involved in gene expression regulation via mi/piRNA were decreased in presence of sporocysts, suggesting a decrease in transcription mechanisms. Apoptosis also appears to be downregulated with a decrease in apoptosis-inducing factor 3, and a concurrent increase in an anti-apoptotic protein (acidic leucine-rich nuclear phosphoprotein 32), suggesting an overall reduction of the apoptosis mechanism in presence of parasites. Host modulation of hemocyte apoptosis in response to direct contact with the parasite, would likely prevent premature cell death in effector hemocytes thereby sustaining a more effective immune response. Interestingly, like NMRI hemocytes, sequestosome 1/p62 also was sharply down-regulated in sporocyst-encapsulated BS-90 cells (Table 3) suggesting that, although clearly a response to larval contact, hemocytes apparently do not require this protein for their cytotoxic activity. Upregulation of other immune-related genes such as T-complex protein 1, BPI (Table 1) and N-acetylgalactosamine kinase, involved in GalNAc binding interactions (Table 3) would be consistent with a functioning immune system.
The snail's redox system plays a central role, not only in maintaining the host's own redox balance, but as a primary effector response against larval helminths through production of ROS and reactive nitrogen species (RNS) (53). In response to sporocysts, encapsulating BS-90 hemocytes exhibited both up- and down-regulation of redox proteins. Those showing significant decreases include glutathione S-transferase (GST) 7, ferric-chelate reductase 1, aldehyde dehydrogenase family (Table 3), as well as glutarate-semialdehyde, isocitrate dehydrogenase and microsomal GST 3 found expressed as unique proteins (Table 1). However, down-regulation of these redox-related proteins is countered by increases in hydrogen peroxide-generating dual oxidase and phytanoyl-CoA dioxygenase 2 (Table 3), and uniquely expressed Cu-Zn superoxide dismutase (SOD1), D-arabinono-1,4-lactone oxidase and laccase-3 in the presence of parasites (Table 1). Dual oxidase, a transmembrane protein and member of the NADPH oxidase family, produces ROS, specifically hydrogen peroxide (54). Proteins similarly involved in H2O2 production include the peroxisomal enzyme, phytanoyl-CoA dioxygenase, SOD 1, and D-arabinono-1,4-lactone oxidase. Laccase3 is a phenoloxidase-related enzyme known to be active in B. glabrata hemolymph (55) and involved in molluscan immune defenses (56). The involvement of ROS in mediating killing of S. mansoni sporocysts is well-documented (53), as is the association of SOD1 gene expression with snail resistance to larval schistosome infection (57). Our finding that ROS-associated enzymes are upregulated in resistant snail hemocytes during encapsulation reactions is consistent with these reported observations.
Overall, the proteomic response of BS-90 snail hemocytes to the presence of sporocysts appears to be more balanced than in the susceptible snails in terms of maintaining metabolic functions, while at the same time mounting a potentially robust immune response against encapsulated larvae through upregulated expression of both shared and unique proteins associated with cellular metabolism and various immune functions.
Host strain comparison of NMRI and BS-90 B. glabrata hemocyte responses to sporocysts
Paired comparison of the proteomic responses of B. glabrata strains to the presence of S. mansoni sporocysts (NMRI-SP vs. BS-90-SP) revealed 76 unique proteins in the BS90-SP group, 29 unique proteins in NMRI-SP group and 642 commonly expressed proteins (Figure 3C). A total of 39 proteins shared between the two strains, presented a minimum of 4-fold differences. A large majority of these proteins (9/39, 77%) were found to be decreased/downregulated in NMRI-SP compared to BS90-SP suggesting either a suppressive parasite effect on the susceptible snail host and/or an enhanced response by the resistant host strain to encapsulated parasite (Table 4). As for unique proteins of interest, all of those found earlier in BS90-SP capsules when compared to BS-90 controls were also uniquely present in BS90-SP when compared to NMRI-SP, suggesting either a lack/delay in the recognition of, or response to, sporocysts by NMRI hemocytes at this time point and/or a parasite-mediated suppression exclusively in hemocyte of the NMRI snail strain. In total, we observed 30 proteins uniquely expressed in BS90-SP compared to NMRI-SP that appear to be constitutively present in hemocytes of the BS-90 B. glabrata strain, which could be an important contributing factor to earlier parasite recognition/response in resistant snails.
Among those specific proteins with reduced expression in NMRI-SP compared to BS-90-SP, a high proportion were associated with immune, redox and cell signaling functions. For immune-related proteins, those exhibiting decreased expression include LPS binding protein (LBP), syntenin1, and lysosomal alpha-mannosidase (Table 4), as well as uniquely identified perilipin 2, BPI protein, and fibrinogen-related protein A precursor (FREP A precursor) (Table 1). As previously mentioned, LBP from snail plasma has been found to interact with the tegument and secretions of S. mansoni sporocysts (43) and therefore, differences may reflect lower NMRI plasma LBP concentrations and/or decreased hemocyte LBP synthesis. Syntenin 1 is an adapter protein, containing PDZ (postsynaptic density, discs large, zona-occludens) domains, allowing for multiprotein complex assembly (58) involved in various mechanisms including apoptosis regulation (eukaryotic translation initiation factor 5A) and immunity (alpha-2-macroglobulin) (59, 60). Alpha-mannosidase-like is a lysosomal hydrolase that, similar to other known hydrolases in B. glabrata hemolymph (61), can be released from hemocytes to degrade pathogen-associated glycans (62), while peripilin 2, a component of lipid droplets (LD), may potentially be involved in immune reactivity to microbial pathogens as shown in Aedes mosquitoes (63).
FREP A precursor protein, which presents a 96% identity with FREP 2.1, was found constitutively expressed solely in hemocytes of the BS-90 resistant strain. FREPs represent a diverse group of lectin PRRs (27, 64) that are closely linked to the snail's immune response against larval S. mansoni (65). Consistent with our current data, FREP2 transcripts were found to be dramatically increased following S. mansoni exposure of BS-90, but not susceptible M-line, snails (66). Although FREP2 is present in the plasma of both susceptible and resistant B. glabrata strains, and is reactive with glycoproteins [e.g., polymorphic mucins (67)] of larval S. mansoni, our finding of these immune proteins in BS90/BS-90-SP hemocyte capsules, but not those of NMRI/NMRI-SP, suggests a specific interaction of a FREP2 subfamily variant with BS-90 hemocytes during cell-cell or cell-parasite aggregation.
Similarly, we observed numerous redox proteins that were reduced or downregulated in NMRI-SP capsules when compared to those of BS90-SP, including SOD1, thioredoxin reductase 1, GST4, dual oxidase, laccase 3 and hemoglobins 1 and 2. H2O2 and NO production have previously been shown to be critical to in vitro killing of S. mansoni sporocysts in the snail (53). Hemocyte SOD1, especially important as it catalyzes the conversion of to H2O2 during encapsulation, has been shown to have differential activity and transcriptomic levels according to the resistance/susceptibility of selected snail strains (57). Similarly, dual oxidase and laccase 3, as described in the previous section, also are involved in H2O2 production and phenoxidase activity, respectively, thereby contributing to the innate immune defenses in the host (56). Thioredoxin1 (TRX1) and GST4 are antioxidants likely contributing to cellular redox homeostasis that would be required by the hemocytes to regulate their own production of ROS in response of the parasite, thereby preventing intracellular damage while also engaging in cytotoxic activities. In NMRI-SP encapsulations, a similar homeostatic mechanism may not be required since NMRI hemocytes were not found to release as much ROS in presence of parasites. GST was previously reported to be upregulated in snails exposed to sublethal doses of molluscicides, suggesting a stress-related response more than a specific immune response to a pathogen (68).
One explanation for the comparatively weak response of encapsulating NMRI hemocytes to sporocysts is that susceptible snail strains are, in general, immunologically inferior to the resistant strains. However, previous studies have shown that a single strain of B. glabrata can be both susceptible to one strain of S. mansoni while at the same time resistant to a different S. mansoni strain (34, 35), indicating that parasite infectivity factors are equally important in determining host-parasite compatibility as the genetic makeup of the snail host (2–4). This also holds true for snails confronted with other non-schistosome pathogens. For example, Echinostoma paraensei has been shown to infect both R and S strains of B. glabrata (66, 69), while bacterial pathogens Paenibacillus (70) and Vibrio parahaemolyticus (71) infect all exposed B. glabrata strains regardless of their schistosome-susceptibility status. This strongly suggests that the S and R phenotypes exhibited by these model B. glabrata/S. mansoni systems are host/parasite strain-specific, and generalizations as to their immune status or susceptibility to other pathogens cannot be made.
Finally, we demonstrated that NMRI and BS-90 snail hemocytes, irrespective of the presence or absence of sporocysts, consistently differed in their levels of hemoglobins (Hb)1 and 2, with BS-90 capsules containing more protein (based on fourfold higher total normalized spectral counts) than NMRI (Supplementary Table 2). This difference is most likely due to different levels of plasma Hb binding to hemocytes (72) and/or sporocysts (73, 74), and not due to the synthesis and differential expression by encapsulating cells. The immune-relevance of hemocyte-bound Hb is presently unclear. However, it has previously been demonstrated that snail Hb was toxic to schistosome sporocysts in vitro (75) and that reactions of Hb-conjugated Fe2+ with NO can lead to the generation of peroxynitrites, a powerful RNS with potential pathogen killing capacity (76). Recently, a role for Hb2 as a parasite immune “sensing” protein has been postulated (74), thereby opening up the possibility that higher constitutive levels of cell-bound Hb (as shown in the present study) may contribute to an early response/killing by resistant BS-90 snail hemocytes in this model system. In addition to Hb, other B. glabrata plasma proteins with sporocyst membrane-binding activity (43, 67) were found to be associated with capsular hemocytes including several with known or proposed immune function: LBP, fibrinogen-like protein A (FREP 2.1), FREP F precursor, alpha-amylase and an actin2 protein (4, 50, 64, 67, 74). Although hemocytes themselves may be the source of these proteins, it is more likely that they are of plasma origin and possess hemocyte and/or sporocyst binding activity.
In summary, we have investigated the proteomic response of B. glabrata hemocytes during in vitro encapsulation of S. mansoni sporocysts by comparing the proteomic profiles of reactive hemocytes both within and between susceptible and resistant snail strains. In the susceptible NMRI B. glabrata strain, we observed a dramatic downregulation of proteins during parasite encapsulation, many of which are related to immune, redox and autophagic function. Of the few upregulated proteins, one (arginase 1) appeared to be induced by the parasite to favor its survival during encapsulation. A more balance proteomic response was exhibited by resistance BS-90 snails, supporting a more robust immune capacity. These included several mechanisms involved in parasite killing such as an upregulation of proteins associated with H2O2 production (SOD1, dual oxidase) and antioxidants to counter intracellular oxidative damage to hemocytes. Interestingly, the NMRI/BS-90 strain comparison highlighted the potential importance of hemocyte proteins constitutively expressed in the resistant snails, but absent in the susceptible strain. These included FREP2, mannosidase and a potential NO-dependent role of hemoglobin in the resistant snail. The constitutive presence of these, and other larval-responsive, immune-related proteins provide important insights into effector mechanisms of encapsulating hemocytes that characterize the resistance phenotype in this model host-parasite system.
Author contributions
MC and TY developed study concept and, with ND, created the experimental design. MC performed experiments. MC, ND, X-JW, and UB-W data analysis and interpretation. GS generated proteomic data. GS, ND, and UB-W bioinformatic analyses. ND, MC, GS, and TY manuscript writing and editing. ND and TY critical review and final editing. All authors approved the final submitted draft.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer CA declared a past co-authorship with one of the authors TY, to the handling Editor.
Acknowledgments
The authors thank T. Pier and J. Kenecht (UW-Madison LCM facility) and M. Gehring for technical support.
Footnotes
Funding. Snails and mice used in this study were provided by BEI Resources under NIH/NIAID contract HHSN2722010000051 to the Biomedical Research Institute (Rockville, MD). This project was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) - Programa Ciências sem Fronteiras (Brazil) to MC and the NIAID AI015503 to TY.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02773/full#supplementary-material
: Biomphalaria glabrata hemocyte proteins identified in two replicate experiments. Each entry lists the identified protein, NCBI accession number, molecular weight and normalized spectral counts for each treatment: control hemocyte capsules (BS90, NMRI), hemocyte capsules with sporocysts (BS90-SP, NMRI-SP). Numbers 1 and 2 (e.g., BS90 1) represent the first and second independent replicate, respectively.
: List of proteins showing high identities to Biomphalaria glabrata hemoglobins (Hb) 1 or 2. NCBI accession numbers and molecular weights are listed for each entry (NMRI, NMRI-SP, BS90, BS90-SP) with their corresponding total normalized spectral counts. Initial protein identifications were returned as “uncharacterized protein.” However, subsequent BlastP searches of these sequences revealed highest identities to B. glabrata Hb 1 and 2.
References
- 1.Adema CM, Loker ES. Digenean-gastropod host associations inform on aspects of specific immunity in snails. Dev Comp. Immunol. (2015) 48:275–283. 10.1016/j.dci.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coustau C, Gourbal B, Duval D, Yoshino TP, Adema CM, Mitta G. Advances in gastropod immunity from the study of the interaction between the snail Biomphalaria glabrata and its parasites: a review of research progress over the last decade. Fish Shellfish Immunol. (2015) 46:5–16. 10.1016/j.fsi.2015.01.036 [DOI] [PubMed] [Google Scholar]
- 3.Mitta G, Gourbal B, Grunau C, Knight M, Bridger JM, Théron A. The compatibility between Biomphalaria glabrata snails and Schistosoma mansoni: An increasingly complex puzzle. Adv Parasitol. (2017) 97:112–44. 10.1016/bs.apar.2016.08.006 [DOI] [PubMed] [Google Scholar]
- 4.Pila EA, Li H, Hambrook JR, Wu X, Hanington PC. Schistosomiasis from a snail's perspective: advances in snail immunity. Trends Parasitol. (2017) 33:845–57. 10.1016/j.pt.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 5.Yoshino TP, Gourbal B, Théron A. Schistosoma sporocysts. In: Jamieson BGM, editor. Schistosoma: Biology, Pathology and Control. Boca Raton, FL: CRC Press; (2017). p. 118–48. [Google Scholar]
- 6.Kassim OO, Richards CS. Host reactions in Biomphalaria glabrata to Schistosoma mansoni miracidia, involving variations in parasite strains, numbers and sequence of exposures. Int J Parasitol. (1979) 9:565–70. [DOI] [PubMed] [Google Scholar]
- 7.Loker ES, Bayne CJ. In vitro encounters between Schistosoma mansoni primary sporocysts and hemolymph components of susceptible and resistant strains of Biomphalaria glabrata. Am J Trop Med Hyg. (1982) 31:999–1005. [DOI] [PubMed] [Google Scholar]
- 8.Bayne CJ, Buckley PM, DeWan PC. Macrophage-like hemocytes of resistant Biomphalaria glabrata are cytotoxic for sporocysts of Schistosoma mansoni in vitro. J Parasitol. (1980) 66:413–19. [PubMed] [Google Scholar]
- 9.Basch PF, DiConza JJ. The miracidium-sporocyst transition in Schistosoma mansoni: surface changes in vitro with ultrastructural correlation. J Parasitol. (1974) 60:935–41. 10.2307/3278518 [DOI] [PubMed] [Google Scholar]
- 10.Bayne CJ, Buckley PM, DeWan PC. Schistosoma mansoni: cytotoxicity of hemocytes from susceptible snail hosts for sporocysts in plasma from resistant Biomphalaria glabrata. Exp Parasitol. (1980) 50:409–16. [DOI] [PubMed] [Google Scholar]
- 11.Allan ERO, Gourbal B, Dores CB, Portet A, Bayne CJ, Blouin MS. Clearance of schistosome parasites by resistant genotypes at a single genomic region in Biomphalaria glabrata snails involves cellular components of the hemolymph. Internatl J Parasitol. (2018) 48:387–93. 10.1016/j.ijpara.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tennessen JA, Théron A, Marine M, Yeh JY, Rognon A, Blouin MS. Hyperdiverse gene cluster in snail host conveys resistance to human schistosome parasites. PLoS Genet (2015) 11:e1005067. 10.1371/journal.pgen.1005067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allan ERO, Tennessen JA, Bollmann SR, Hanington PC, Bayne CJ, Blouin MS. Schistosome infectivity in the snail, Biomphalaria glabrata, is partially dependent on the expression of Grctm6, a Guadeloupe Resistance Complex protein. PLoS Negl Trop Dis (2017) 11:e0005362. 10.1371/journal.pntd.0005362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitta G, Galinier R, Tisseyre P, Allienne J-F, Girerd-Chambaz Y, Guillou F, et al. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. (2005) 29:393–407. 10.1016/j.dci.2004.10.002 [DOI] [PubMed] [Google Scholar]
- 15.Lockyer AE, Spinks J, Noble LR, Rollinson D, Jones CS. Identification of genes involved in interactions between Biomphalaria glabrata and Schistosoma mansoni by suppression subtractive hybridization. Mol Biochem Parasitol. (2007) 151:18–27. 10.1016/j.molbiopara.2006.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lockyer AE, Spinks J, Kane RA, Hoffman KF, Fitzpatrick JM, Rollinson D, et al. Biomphalaria glabrata transcriptome: cDNA microarray profiling identifies resistant- and susceptible-specific gene expression in haemocytes from snail strains exposed to Schistosoma mansoni. BMC Genomics (2008) 9:634. 10.1186/1471-2164-9-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny NJ, Truchado-García M, Grande C. Deep, multi-stage transcriptome of the schistosomiasis vector Biomphalaria glabrata provides platform for understanding molluscan disease-related pathways. BMC Infect Dis. (2016) 16:618. 10.1186/s12879-016-1944-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raghavan N, Miller AN, Gardner M, FitzGerald PC, Kerlavage AR, Johnston DA, et al. Comparative gene analysis of Biomphalaria glabrata hemocytes pre- and post-exposure to miracidia of Schistosoma mansoni. Mol Biochem Parasitol. (2003) 126:181–91. 10.1016/S0166-6851(02)00272-4 [DOI] [PubMed] [Google Scholar]
- 19.Guillou Mitta Galinier Coustau C. Identification and expression of gene transcripts generated during an anti-parasitic response in Biomphalaria glabrata. Dev Comp Immunol. (2007) 31:657–71. 10.1016/j.dci.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 20.Adema CM, Hanington PC, Lun CM, Rosenberg GH, Aragon AD, Stout BA, et al. Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes). Mol Immunol. (2010) 47:849–60. 10.1016/j.molimm.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lockyer AE, Emery AM, Kane RA, Walker AJ, Mayer CD, Mitta G, et al. Early differential gene expression in haemocytes from resistant and susceptible Biomphalaria glabrata strains in response to Schistosoma mansoni. PLoS ONE (2012) 7:e51102. 10.1371/journal.pone.0051102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahoor Z, Lockyer AE, Davies AJ, Kirk RS, Emery AM, Rollinson D, et al. Differences in the gene expression profiles of haemocytes from schistosome-susceptible and -resistant Biomphalaria glabrata exposed to Schistosoma mansoni excretory-secretory products. PloS ONE (2014) 9:e93215. 10.1371/journal.pone.0093215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouchut A, Sautiere PE, Coustau C, Mitta G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: Potential involvement of proteins from hemocytes revealed by a proteomic approach. Acta Trop (2006) 98:234–46. 10.1016/j.actatropica.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 24.Bender RC, Goodall CP, Blouin MS, Bayne CJ. Variation in expression of Biomphalaria glabrata SOD1: a potential controlling factor in susceptibility/resistance to Schistosoma mansoni. Dev Comp Immunol. (2007) 31:874e878. 10.1016/j.dci.2006.12.005 [DOI] [PubMed] [Google Scholar]
- 25.Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ. Larval excretory-secretory products from the parasite Schistosoma mansoni modulate HSP 70 protein expression in defence cells of its snail host, Biomphalaria glabrata. Cell Stress Chaperones (2010) 15:639–50. 10.1007/s12192-010-0176-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ittiprasert W, Knight M. Reversing the resistance phenotype of the Biomphalaria glabrata snail host Schistosoma mansoni infection by temperature modulation. PLoS Pathog. (2012) 8:e1002677. 10.1371/journal.ppat.1002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanington PC, Forys MA, Dragoo JW, Zhang SM, Adema CM, Loker ES. Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc Natl Acad Sci USA. (2010) 107:21087–92. 10.1073/pnas.1011242107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanington PC, Forys MA, Loker ES. A somatically diversified defense factor, FREP3, is a determinant of snail resistance to schistosome infection. PLoS Negl Trop Dis. (2012) 6:e1591. 10.1371/journal.pntd.0001591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Portet A, Galinier R, Pinaud S, Portela J, Nowacki F, Gourbal B, et al. BgTEP: An antiprotease involved in innate immune sensing in Biomphalaria glabrata. Front Immunol. (2018) 9:1206. 10.3389/fimmu.2018.01206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baeza-Garcia A, Pierce RJ, Gourbal B, Werkmeister E, Colinet D, Reichhart JM, et al. Involvement of the cytokine MIF in the snail host immune response to the parasite Schistosoma mansoni. PLoS Pathog. (2010) 6:e1001115. 10.1371/journal.ppat.1001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galinier R, Portela J, Moné Y, Allienne JF, Henri H, Delbecq S, et al. Biomphalysin, a new pore-forming toxin involved in Biomphalaria glabrata immune defense against Schistosoma mansoni. PLoS Pathog. (2013) 9:e1003216. 10.1371/journal.ppat.1003216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pila EA, Tarrabain M, Kabore AL, Hanington PC. A Novel Toll-Like receptor (TLR) influences compatibility between the gastropod Biomphalaria glabrata, and the digenean trematode Schistosoma mansoni. PLoS Pathog. (2016) 12:e1005513. 10.1371/journal.ppat.1005513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pila EA, Gordy MA, Phillips VK, Kabore AL, Rudko SP, Hanington PC. Endogenous growth factor stimulation of hemocyte proliferation induces resistance to Schistosoma mansoni challenge in the snail host. Proc Natl Acad Sci USA. (2016) 113:5305–10. 10.1073/pnas.1521239113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Théron A, Rognon A, Gourbal B, Mitta G. Multi-parasite host susceptibility and multi-host parasite infectivity: a new approach of the Biomphalaria glabrata/Schistosoma mansoni compatibility polymorphism. Infect Genet Evol. (2014) 26:80–8. 10.1016/j.meegid.2014.04.025 [DOI] [PubMed] [Google Scholar]
- 35.Galinier R, Roger E, Moné Y, Duval D, Portet A, Pinaud S, et al. A multistrain approach to studying the mechanisms underlying compatibility in the interaction between Biomphalaria glabrata and Schistosoma mansoni. PLoS Negl Trop Dis. (2017) 11:e0005398. 10.1371/journal.pntd.0005398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshino TP, Laursen JR. Production of Schistosoma mansoni daughter sporocysts from mother sporocysts maintained in synxenic culture with Biomphalaria glabrata embryonic Bge cells. J Parasitol. (1995) 81:714–22. 10.2307/3283960 [DOI] [PubMed] [Google Scholar]
- 37.Dinguirard N, Heinemann C, Yoshino TP. Mass isolation and in vitro cultivation of intramolluscan stages of the human blood fluke Schistosoma mansoni. J Vis Exp. (2018) 131:e56345 10.3791/56345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chernin E. Observations on hearts explanted in vitro from the snail Australorbis glabratus. J Parasitol. (1963) 49:353–64. 10.2307/3275797 [DOI] [PubMed] [Google Scholar]
- 39.Sminia T, Barendsen LA. Comparative morphological and enzyme biochemical study of blood cells of the freshwater snails Lymnaea stagnalis, Biomphalaria glabrata, and Bulinus truncates. J Morphol. (1980) 165:31–9. 10.1002/jmor.1051650104 [DOI] [PubMed] [Google Scholar]
- 40.Hahn UK, Bender RC, Bayne CJ. Killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata: role of reactive oxygen species. J Parasitol. (2001) 87:292–9. 10.1645/0022-3395(2001)087[0292:KOSMSB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 41.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. (2003) 75:4646–58. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 42.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. (2011) 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu XJ, Dinguirard N, Sabat G, Lui HD, Gonzalez L, Gehring M, et al. Proteomic analysis of Biomphalaria glabrata plasma proteins with binding affinity to those expressed by early developing larval Schistosoma mansoni. PloS Pathog. (2017) 13:e1006081. 10.1371/journal.ppat.1006081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staczek S, Zdybicka-Barabas A, Mak P, Sowa-Jasiłek A, Kedracka-Krok S, Jankowska U, et al. Studies on localization and protein ligands of Galleria mellonella apolipophorin III during immune response against different pathogens. J Insect Physiol. (2018) 105:18–27. 10.1016/j.jinsphys.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 45.Zhang SM, Loker ES, Sullivan JT. Pathogen-associated molecular patterns activate expression of genes involved in cell proliferation, immunity and detoxification in the amebocyte-producing organ of the snail Biomphalaria glabrata. Dev Comp Immunol. (2016) 56:25–36. 10.1016/j.dci.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lippai M, Low P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int. (2014) 2014:832704. 10.1155/2014/832704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujita K, Srinivasula SM. TLR4-mediated autophagy in macrophages is a p62-dependent type of selective autophagy of aggresome-like induced structures (ALIS). Autophagy (2011) 7:552–4. 10.4161/auto.7.5.15101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gan L, Li L. Regulations and roles of the interleukin-1 receptor associated kinases (IRAKs) in innate and adaptive immunity. Immunol Res. (2006) 35:295. 10.1385/IR:35:3:295 [DOI] [PubMed] [Google Scholar]
- 49.Queiroz FR, Silva LM, Jeremias WJ, Babá ÉH, Caldeira RL, Coelho PMZ, et al. Differential expression of small RNA pathway genes associated with the Biomphalaria glabrata/Schistosoma mansoni interaction. PLoS ONE (2017) 12:e0181483. 10.1371/journal.pone.0181483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baron OL, Deleury E, Reichhart JM, Coustau C. The LBP/BPI multigenic family in invertebrates: Evolutionary history and evidences of specialization in mollusks. Dev Comp Immunol. (2016) 57:20–30. 10.1016/j.dci.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 51.Guillou F, Roger E, Moné Y, Rognon A, Grunau C, Théron A, et al. Excretory-secretory proteome of larval Schistosoma mansoni and Echinostoma caproni, two parasites of Biomphalaria glabrata. Mol Biochem Parasitol. (2007) 155:45–56. 10.1016/j.molbiopara.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 52.Hahn UK, Bender RC, Bayne CJ. Role of nitric oxide in killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata. J Parasitol. (2001) 87:778–85. 10.1645/0022-3395(2001)087[0778:IONOIK]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 53.Bayne CJ, Hahn UK, Bender RC. Mechanisms of molluscan host resistance and of parasite strategies for survival. Parasitology (2001) 123:S159–67. 10.1017/S0031182001008137 [DOI] [PubMed] [Google Scholar]
- 54.Yang X, Smith AA, Williams MS, Pal U. A dityrosine network mediated by dual oxidase and peroxidase influences the persistence of Lyme disease pathogens within the vector. J Biol Chem. (2014) 289:12813–22. 10.1074/jbc.M113.538272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Clec'h W, Anderson TJC, Chevalier FD. Characterization of hemolymph phenoloxidase activity in two Biomphalaria snail species and impact of Schistosoma mansoni infection. Parasit Vectors (2016) 9:32. 10.1186/s13071-016-1319-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luna-Acosta A, Breitwieser M, Renault T, Thomas-Guyon H. Recent findings on phenoloxidases in bivalves. Mar Pollut Bull. (2017) 122:5–16. 10.1016/j.marpolbul.2017.06.031 [DOI] [PubMed] [Google Scholar]
- 57.Goodall CP, Bender RC, Brooks JK, Bayne CJ. Biomphalaria glabrata cytosolic copper/zinc superoxide dismutase (SOD1) gene: association of SOD1 alleles with resistance/susceptibility to Schistosoma mansoni. Mol Biochem Parasitol. (2006) 147:207–10. 10.1016/j.molbiopara.2006.02.009 [DOI] [PubMed] [Google Scholar]
- 58.Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Invest. (1999) 103:767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li AL, Li HY, Jin BF, Ye QN, Zhou T, Yu XD, et al. A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J Biol Chem. (2004) 279:49251–8. 10.1074/jbc.M407165200 [DOI] [PubMed] [Google Scholar]
- 60.Tonganunt M, Phongdara A, Chotigeat W, Fujise K. Identification and characterization of syntenin binding protein in the black tiger shrimp Penaeus monodon. J Biotechnol. (2005) 120:135–45. 10.1016/j.jbiotec.2005.06.006 [DOI] [PubMed] [Google Scholar]
- 61.Granath WO, Jr, Yoshino TP. Lysosomal enzyme activities in susceptible and refractory strains of Biomphalaria glabrata during the course of infection with Schistosoma mansoni. J Parasitol. (1983) 69:1018–26. [PubMed] [Google Scholar]
- 62.Mohandas A, Cheng TC, Cheng JB. Mechanism of lysosomal enzyme release from Mercenaria mercenaria granulocytes: a scanning electron microscope study. J Invertebr Pathol. (1985) 46:189–97. [DOI] [PubMed] [Google Scholar]
- 63.Barletta AB, Alves LR, Silva MC, Sim S, Dimopoulos G, Liechocki S, et al. Emerging role of lipid droplets in Aedes aegypti immune response against bacteria and Dengue virus. Sci Rep. (2016) 6:19928. 10.1038/srep19928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science (2004) 305:251–4. 10.1126/science.1088069 [DOI] [PubMed] [Google Scholar]
- 65.Gordy MA, Pila EA, Hanington PC. The role of FREPs fibrinogen-related proteins in the gastropod immune response. Fish Shellfish Immunol. (2015) 46:39–49. 10.1016/j.fsi.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 66.Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei. Dev Comp Immunol. (2005) 29:295–303. 10.1016/j.dci.2004.08.003 [DOI] [PubMed] [Google Scholar]
- 67.Moné Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S, Mitta G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl Trop Dis. (2010) 4:e813. 10.1371/journal.pntd.0000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang S-M, Buddenborg SK, Adema CM, Sullivan JT, Loker ES. Altered gene expression in the schistosome-transmitting snail Biomphalaria glabrata following exposure to niclosamide, the active ingredient in the widely used molluscicide Bayluscide. PLoS Negl Trop Dis. (2015) 9:e0004131. 10.1371/journal.pntd.0004131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia JS, Maldonado Junior A, Bidau CJ, Corrêa Ldos R, Lanfredi RM, Coelho PM. The effect of early infection with Echinostoma paraensei on the interaction of Schistosoma mansoni with Biomphalaria glabrata and Biomphalaria tenagophila. Mem Inst Oswaldo Cruz (2010) 105:499–503. 10.1590/S0074-02762010000400026 [DOI] [PubMed] [Google Scholar]
- 70.Duval D, Galinier R, Mouahid G, Toulza E, Allienne JF, Portela J, et al. A novel bacterial pathogen of Biomphalaria glabrata: a potential weapon for schistosomiasis control? PLoS Negl Trop Dis. (2015) 9:e0003489 10.1371/journal.pntd.0003489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ducklow HW, Tarraza HM, Jr, Mitchell R. Experimental pathogenicity of Vibrio parahaemolyticus for the schistosome-bearing snail Biomphalaria glabrata. Can J Microbiol. (1980) 26:503–6. [DOI] [PubMed] [Google Scholar]
- 72.Yoshino TP. Surface antigens of Biomphalaria glabrata (Gastropoda) hemocytes: occurrence of membrane-associated hemolymph-like factors antigenically related to snail hemoglobin. J Invertebr Pathol. (1983) 41:310–20. [DOI] [PubMed] [Google Scholar]
- 73.Bayne CJ, Hull CJ. The host-parasite interface in molluscan schistosomiasis: biotin as a probe for sporocyst and hemocyte surface peptides. Vet Parasitol. (1988) 29:131–42. 10.1016/0304-4017(88)90121-5 [DOI] [PubMed] [Google Scholar]
- 74.Tetreau G, Pinaud S, Portet A, Galinier R, Gourbal B, Duval D. Specific pathogen recognition by multiple innate immune sensors in an invertebrate. Front Immunol. (2017) 8:1249. 10.3389/fimmu.2017.01249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bender RC, Bixler LM, Lerner JP, Bayne CJ. Schistosoma mansoni sporocysts in culture: host plasma hemoglobin contributes to in vitro oxidative stress. J Parasitol. (2002) 88:14–18. 10.1645/0022-3395(2002)088[0014:SMSICH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 76.Coates CJ, Decker H. Immunological properties of oxygen-transport proteins: hemoglobin, hemocyanin and hemerythrin. Cell Mol Life Sci. (2017) 74:293–317. 10.1007/s00018-016-2326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
: Biomphalaria glabrata hemocyte proteins identified in two replicate experiments. Each entry lists the identified protein, NCBI accession number, molecular weight and normalized spectral counts for each treatment: control hemocyte capsules (BS90, NMRI), hemocyte capsules with sporocysts (BS90-SP, NMRI-SP). Numbers 1 and 2 (e.g., BS90 1) represent the first and second independent replicate, respectively.
: List of proteins showing high identities to Biomphalaria glabrata hemoglobins (Hb) 1 or 2. NCBI accession numbers and molecular weights are listed for each entry (NMRI, NMRI-SP, BS90, BS90-SP) with their corresponding total normalized spectral counts. Initial protein identifications were returned as “uncharacterized protein.” However, subsequent BlastP searches of these sequences revealed highest identities to B. glabrata Hb 1 and 2.