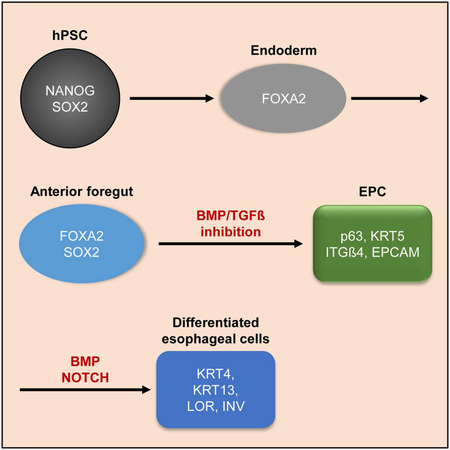
SUMMARY
Pluripotent stem cells (PSCs) could provide a powerful system to model development of the human esophagus, whose distinct tissue organization compared to rodent esophagus suggests that developmental mechanisms may not be conserved between species. We therefore established an efficient protocol for generating esophageal epithelial progenitors (EPCs) from human PSCs. We found that inhibition of TGFß and BMP signaling is required for sequential specification of EPCs, which can be further purified using cell surface markers. These EPCs resemble their human fetal counterparts and can recapitulate normal development of esophageal stratified squamous epithelium during in vitro 3D cultures and in vivo. Importantly, combining hPSC differentiation strategies with mouse genetics elucidated a critical role for Notch signaling in the formation of this epithelium. These studies therefore not only provide an efficient approach to generate EPCs, but also offer a model system to study the regulatory mechanisms underlying development of the human esophagus.
Keywords: human embryonic stem cells, human induced pluripotent stem cell, basal cells, esophagus, organoids, BMP, TGFß, WNT, NOTCH
Graphical Abstract
In Brief (eTOC statement):
Que, Zhang and colleagues established an efficient approach to differentiate human pluripotent stem cells (hPSCs) into esophageal progenitor cells (EPCs). Combined use of hPSC-derived EPCs and mouse genetic models demonstrates the important role of BMP and NOTCH signaling in promoting squamous differentiation of EPCs.
INTRODUCTION
Differentiation of human pluripotent stem cells (hPSCs) including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) has offered avenues to directly study human organ development and disease mechanisms (Huang et al., 2014; Longmire et al., 2012; McCracken et al., 2014; Mou et al., 2012; Pagliuca et al., 2014; Shi et al., 2017; Spence et al., 2011; Wong et al., 2012). In addition, these hPSC-derived cells hold the potential to provide unlimited sources for clinical uses and pharmacological applications (Trounson and DeWitt, 2016). Generation of lineage-specific progenitor cells from hPSCs leverages the knowledge of signaling mechanisms obtained from studying other species mostly mice. Multiple signaling pathways such as WNT and BMP have been shown to play essential roles in the development of different organs including the lung and esophagus (Domyan et al., 2011; Goss et al., 2009; Harris-Johnson et al., 2009; Que et al., 2006). Much of this knowledge has now been utilized to successfully promote the differentiation of hPSCs into various cell lineages in tissues like the lung and thyroid where the same epithelial types are shared among rodents and humans (Huang et al., 2014; Longmire et al., 2012). By contrast, for the esophagus where the epithelial structure is distinct between rodents (keratinized) and humans (non-keratinized), it is unknown whether the developmental mechanisms are conserved. Consequently, thus far derivation of esophageal epithelium from hPSCs has not been successful.
The esophagus is established from the dorsal side of the anterior foregut endoderm (AFE) at around 4 weeks of development in humans and embryonic (E) 9.5 in mice. By contrast, the ventral foregut endoderm gives rise to the thyroid, lung and trachea (Jacobs et al., 2012). Studies of mouse genetic models have shown that establishment of these foregut organs involves a dorsal-ventral patterning of transcription factors and signaling pathways (Que, 2015). For example, the transcription factors NKX2.1 and SOX2 are preferentially expressed in the ventral and dorsal side of the AFE, respectively. Disruption of the Nkx2.1 or Sox2 gene leads to abnormal formation of the lung and esophagus (Minoo et al., 1999; Que et al., 2007). Furthermore, BMP and WNT signaling are preferentially activated in the ventral foregut, and disruption of the signaling pathways also leads to abnormal lung specification and agenesis (Domyan et al., 2011; Goss et al., 2009; Harris-Johnson et al., 2009; Que et al., 2006). Accordingly, activation of the WNT pathway using the GSK3ß inhibitor CHIR99021 is instrumental for coaxing the differentiation of hPSCs towards lung epithelium (Huang et al., 2014; McCauley et al., 2017). We previously showed that the BMP inhibitor Noggin is enriched in the dorsal side of the early foregut. Deletion of the Nog gene leads to failed separation of the esophagus from the foregut, resulting in birth defects including esophageal atresia with tracheoesophageal fistula (EA/TEF) (Que et al., 2006). Our further studies showed that Noggin-mediated inhibition of BMP signaling continues to play important roles for epithelial morphogenesis in the developing esophagus. Nog deletion results in failed conversion of simple columnar cell into stratified squamous epithelium and the esophagus becomes lined by a mucin-producing glandular epithelium (Rodriguez et al., 2010). Moreover, our recent studies suggested that BMP inhibition is required for the maintenance of basal cells, progenitor cells of the stratified squamous epithelium in the esophagus (Jiang et al., 2015). Of note is that there are several distinct characteristics between the mouse and human esophagus. For example, similar to the skin, the mouse esophageal epithelium is keratinized in contrast to the non-keratinized human esophagus (Jacobs et al., 2012). Therefore, it remains unknown whether the activities of the relevant signaling pathways (e.g. BMP) are similarly involved in the specification of human esophageal epithelium. It is also unknown whether other signaling pathway(s) are required for epithelial morphogenesis.
Here, we report an efficient method to induce differentiation of hPSCs towards esophageal progenitor cells (EPCs) which can be further purified with the cell surface markers EPCAM and ITGß4. The hPSC-derived EPCs express genes that are enriched in the human fetal esophagus, and they are able to recapitulate human esophageal developmental processes, forming the stratified squamous epithelium in three-dimensional (3D) organoids and kidney capsule xenografts. Notably, using a combination of hPSC differentiation and mouse genetics we further identified a conserved role for NOTCH signaling in esophageal development in human and mouse.
RESULTS
Sequential differentiation of hPSCs towards esophageal progenitor cells through the inhibition of BMP and TGFβ signaling.
We previously demonstrated that Noggin expression is localized in the dorsal foregut endoderm where progenitor cells for the esophageal epithelium arise (Que et al., 2006). The unique expression of Noggin in the dorsal foregut is maintained at E10.5 and E11.5, but it is absent at E12.5 (Figure 1A). We also used a BRE-lacZ transgenic reporter mouse line in which ß-gal expression is regulated by BMP response elements (BREs) from the ID1 gene to determine BMP activity (Blank et al., 2008). Consistently, BMP activation is limited to the ventral side of the anterior foregut where the lung and trachea arise (Figure 1A). These findings suggest that inhibition of BMP signaling is required for the specification of EPCs from the AFE. In addition, a previous study showed that BMP/TGFß dual inhibition promotes the expansion of mouse esophageal basal cells in vitro (Mou et al., 2016). These findings prompted us to test whether inhibition of BMP and TGFß signaling promotes the specification of AFE towards EPCs.
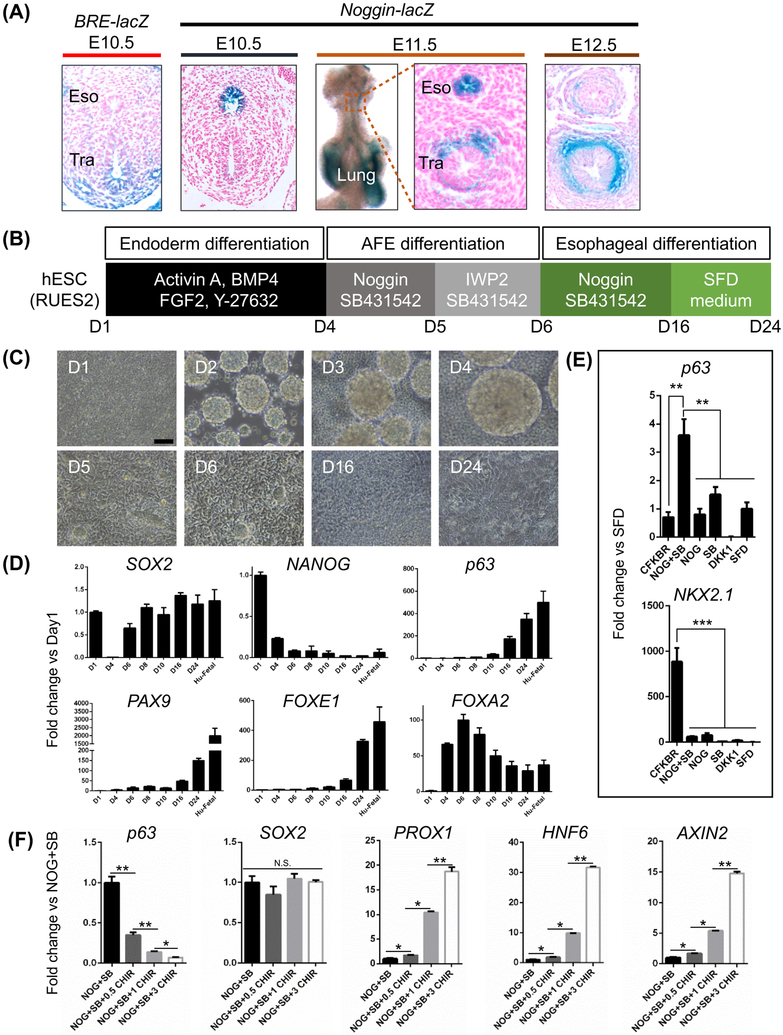
Figure 1. Derivation of esophageal progenitor cells (EPCs) from human embryonic stem cells (hESCs) by inhibiting TGFβ and BMP signaling.
(A) Noggin-mediated inhibition of BMP signaling in mouse esophageal progenitor cells. BMP signaling is active in the ventral but not dorsal foregut where Nog-lacZ is expressed at E10.5. Note Nog-lacZ expression in the tracheal mesenchyme at E11.5 and E12.5. (B-C) Sequential differentiation of the human ES cell line RUES2 into EPCs. Scale bar: 100 μm. (D) Increased expression of EPC proteins during RUES2 differentiation. Note SOX2 levels were reduced at day4 but increased at day6. The fold change (Y axis) was generated by normalizing the transcript levels to those of day 1 (D1) hESC. Human fetal esophagus (Hu-Fetal) was included as control. Data represent mean ± SEM (n = 3). (E) Expression of p63 and NKX2.1 in cultures treated in parallel with either Noggin/SB-431542 or other factors from day6 to day16. The transcript levels of p63 and NKX2.1 were represented by the fold change compared to SFD condition. Data represent mean ± SEM (n = 3). **p < 0.01, ***p < 0.001 by unpaired, two-tailed Student’s t test. (F) Ectopic WNT activation inhibits AFE commitment to EPCs. The GSK3 inhibitor CHIR99021 (0.5, 1 or 3 μM) was added to the culture from day6 to day16 along with Noggin (NOG) and SB431542 (SB). The expression of p63, AXIN2, PROX1, HNF6 and SOX2 was quantified by qPCR and reported as the fold change compared to NOG+SB. Data represent mean ± SEM (n = 3). *p < 0.05, **p < 0.01 by unpaired, two-tailed Student’s t test. Abbreviation: Eso, esophagus; Tra, trachea; NOG, Noggin; SB, SB431542; CFKBR=CHIR99021, FGF10, KGF, BMP4 and Retinoic acid; SFD, serum free medium. N.S., not significant. See also Figures S1-S4.
We first followed a previously published protocol to differentiate the ES cell line RUES2 into the endoderm (CXCR4+ c-KIT+) with a combination of Activin A, BMP4, FGF2 and the ROCK inhibitor Y-27632 (day1-4) (Figures 1B-C and S1A) (Huang et al., 2014). Noggin and SB431542 were then used to block BMP and TGFß signaling, respectively (day4-5), followed by a one-day treatment of SB431542 and the WNT/ß-catenin inhibitor IWP2 to promote the formation of AFE (Huang et al., 2014). From day6 to day16 Noggin and SB431542 were reapplied to the culture to block BMP and TGFß signaling. Cells were then maintained in the serum-free differentiation medium (SFD) to allow full commitment until day24 (Figures 1B-C). Noggin and SB431542 treatments led to significant reduction in the levels of the BMP signaling targets ID2 and JUNB and TGFß targets p15, p21, COL1A1 and TGFBI (Figure S1B). During the initial differentiation the transcript levels of the pluripotent markers NANOG and SOX2 decreased concomitantly with increased expression of the endodermal marker FOXA2 post endodermal induction by Activin A (day4) (Figure 1D). However, the levels of SOX2 increased at day6 and maintained during EPC differentiation, indicating that BMP/TGFß dual inhibition promotes the specification of AFE towards the dorsal foregut endoderm which serves as progenitor cells for the esophagus (Figure 1D). Consistently, the transcript levels of the esophageal basal cell marker p63, PAX9 and FOXE1 increased upon differentiation (Dathan et al., 2002; Peters et al., 1998) (Figure 1D). Of note is that the transcripts of these genes are all similarly enriched in the epithelium of human fetal esophagus (Figure 1D). p63+ cells were first detected by immunostaining at day10 of differentiation, co-expressing FOXA2, PAX9 and FOXE1 (Figures S1D-E). The transcript levels of KLF4, KLF5 and WNT5A, critical factors in regulating esophageal epithelial proliferation and differentiation (Goldstein et al., 2007; Okano et al., 2000; Tetreault et al., 2016), also increased upon differentiation (Figure S1C). Notably, treatment with Noggin or SB431542 alone was insufficient to promote AFE differentiation into p63+ NKX2.1− EPCs (Figure 1E). Taken together, these results emphasize the importance of BMP and TGFß dual inhibition in promoting AFE towards EPCs.
WNT has been shown to play an important role in the foregut patterning into different organs including thyroid, lung, stomach, liver and pancreas in mouse development and hPSC differentiation (Goss et al., 2009; Harris-Johnson et al., 2009; Jacob et al., 2017; Longmire et al., 2012; McCracken et al., 2017; Ober et al., 2006; Wells et al., 2007). Previous studies revealed that WNT signaling is inactive during dorsal foregut commitment to the esophagus at E9.5 (Jacobs et al., 2012). Consistently, the transcript levels of the WNT downstream targets AXIN2, LEF1 and NKD1 were low (Figure S2A), suggesting low WNT signaling activities during the specification of EPCs. In addition, treatment with the WNT inhibitor DKK1 alone was insufficient to promote p63 expression in hPSCs (Figure 1E), and the presence of WNT inhibitor IWP2 in culture treated with Noggin and SB431542 at day6-16 did not further promote p63 expression (Figure S2B). These data are consistent with the finding that WNT loss of function does not affect esophageal development in mice (Goss et al., 2009; Harris-Johnson et al., 2009). By contrast, activation of WNT signaling reduced p63 expression in a dose-dependent manner (Figures 1F and S2C), which is consistent with the finding that ectopic WNT activation inhibits squamous cell specification in the mouse esophagus and forestomach which is also lined by the stratified squamous epithelium (Goss et al., 2009; Harris-Johnson et al., 2009). Notably, while the levels of the general foregut marker SOX2 remained unchanged, the levels of the posterior foregut markers PROX1 and HNF6 increased upon WNT activation (Figure 1F) (Burke and Oliver, 2002; Rausa et al., 1997), indicating that ectopic WNT activation in the presence of Noggin and SB431542 posteriorizes the foregut reminiscent of the role of WNT signaling in promoting the development of posterior foregut organs such as liver and pancreas (Figure S2D) (Ober et al., 2006; Wells et al., 2007). Together these data suggest that maintaining WNT at low activities facilitates the specification of AFE towards the EPC lineage.
hPSC-derived EPCs express SOX9 and KRT7, characteristics of the developing human and mouse esophagus
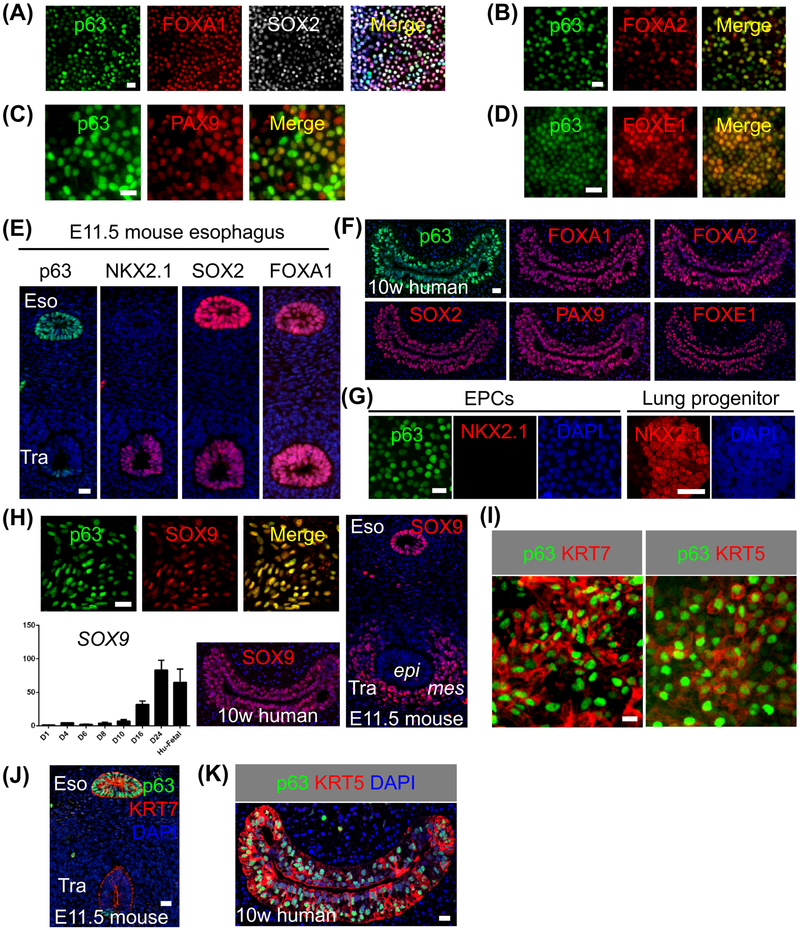
Our transcript analysis showed that hESC-derived EPCs expressed endodermal and esophageal progenitor markers such as p63, SOX2 and FOXA2 (Figure 1D). Immunostaining confirmed strong expression of these transcription factors at day24 (Figures 2A-B and S3A). In addition, the transcription factors PAX9 and FOXE1 that are present in the mouse embryonic esophageal epithelium (Dathan et al., 2002; Peters et al., 1998), were also expressed by hESC-derived EPCs at the protein levels (Figures 2C-D). These transcription factors were also present in the epithelium of the developing mouse and human fetal esophagus (Figure 2E-F and data not shown). Of note, NKX2.1 was not detected in EPCs (Figure 2G), excluding the lung and thyroid lineages. A previous study showed that NANOG is expressed in the basal cells of the adult mouse esophagus (Piazzolla et al., 2014). However, we did not see the expression in EPCs (Figure S3B). Intriguingly, immunostaining analysis indicated that the hESC-derived EPCs also expressed SOX9 (Figure 2H), a transcription factor considered as a specific marker for the distal lung epithelium and tracheal cartilage cells (Chang et al., 2013; Rockich et al., 2013). We found SOX9 proteins are also present in the developing mouse and human fetal esophagus (Figure 2H), but not in their adult counterpart (Figure S3C). hESC-derived EPCs also express high levels of the intermediate filament protein KRT7 and KRT5 (Figure 2I). While KRT7 is specifically expressed in the embryonic but not adult esophagus (Figures 2J and S3D), KRT5 is expressed in both the fetal and adult esophagus (Figures 2K and S3E). These findings suggest that the hESC-derived EPCs mimic human esophageal progenitor cells at the embryonic stage. Furthermore, EPCs can also be reproducibly generated from the hESC cell line H9 using the same protocol (Figure S3F).
Figure 2. Human ESC-derived EPCs express embryonic esophageal markers.
(A-D) hESC RUES2-derived EPCs express multiple markers when examined at day24 of differentiation (n = 5). (E-F) The epithelia in E11.5 mouse esophagus and 10-week human fetal esophagus express p63 etc. (G) RUES2-derived EPCs do not express NKX2.1. (H) SOX9 is expressed in RUES2-derived EPCs and epithelial progenitors in E11.5 mouse and 10-week human esophagus. Note the increased expression of SOX9 transcripts during hESC differentiation and SOX9 expression in the tracheal mesenchyme. Human fetal esophagus (Hu-Fetal) was included as control. (I-K) RUES2-derived EPCs express the intermediate filament protein KRT7 that is transiently present in the embryonic esophagus. Note EPCs also express the basal cell marker KRT5 (I, K). Abbreviation: Eso, esophagus; Tra, trachea; epi, epithelium; mes, mesenchyme. Scale bars: 20 μm. See also Figures S1-S3.
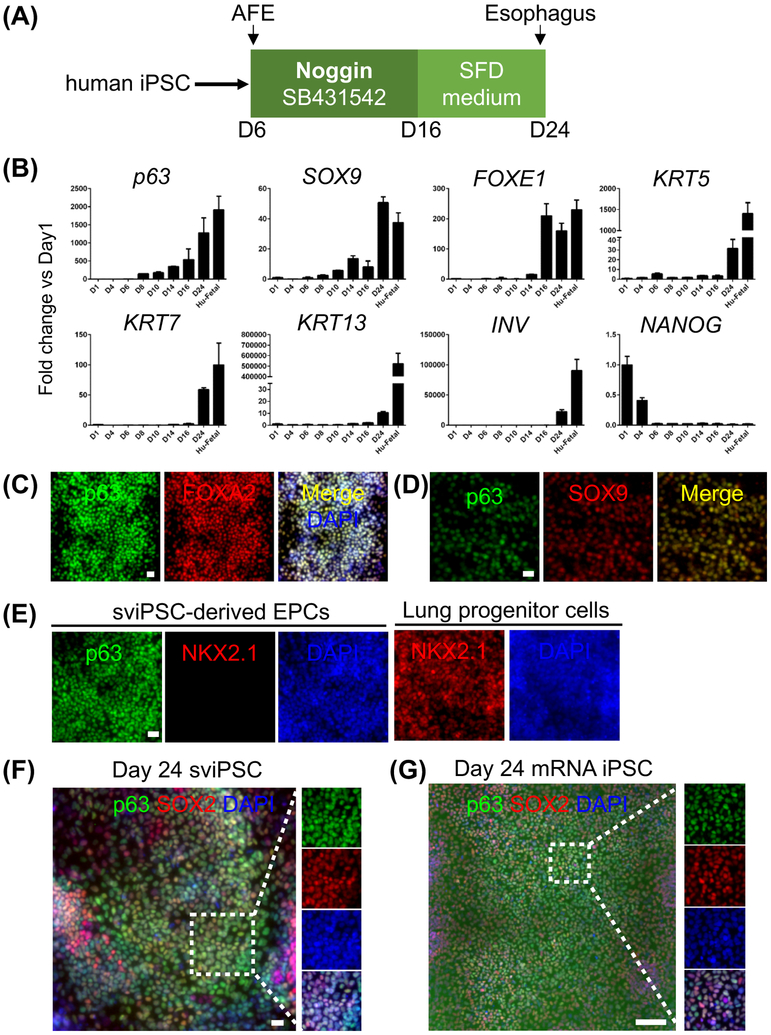
We also asked whether EPCs can be derived from iPSCs and exhibit similar characteristics. We tested two iPSC cell lines, sviPSC and mRNA iPSC using the same protocol above (Figure 3A). Both cell lines gave rise to EPCs which express increased levels of p63, SOX2, FOXA2, SOX9, KRT5, KRT7, KRT13, and INVOLUCRIN (INV) but reduced levels of NANOG during differentiation (Figures 3B-D, F-G and data not shown) as observed in hESC-derived EPCs. Consistently, NKX2.1 expression was not detected in the iPSC-derived EPCs (Figure 3E), confirming that these EPCs are not lung and thyroid cells.
Figure 3. Derivation of esophageal epithelial progenitor cells from two induced human pluripotent stem cell lines, sviPS and mRNA iPS.
(A) Schematic diagram depicts the differentiation of EPCs from human iPS cell lines. (B) Increased expression of p63, SOX9, FOXE1, KRT5, KRT7, KRT13 and INV but reduced NANOG expression during the commitment of sviPSC towards EPCs. Human fetal esophagus (Hu-Fetal) was included as control. The transcript levels were represented by the fold change compared to day 1 (D1) sviPSC. Data represent mean ± SEM (n = 3). (C-E) Similar to hESC-derived EPCs, sviPSC-derived EPCs express FOXA2 and SOX9 but not NKX2.1. (F-G) Representative tile scan images of sviPSC- and mRNA iPSC-derived EPCs co-expressing p63 and SOX2. Scale bars: (C-F) 20 μm, (G) 100 μm. See also Figures S3-S4.
Purification of hPSC-derived EPCs with the cell surface markers EPCAM and ITGβ4
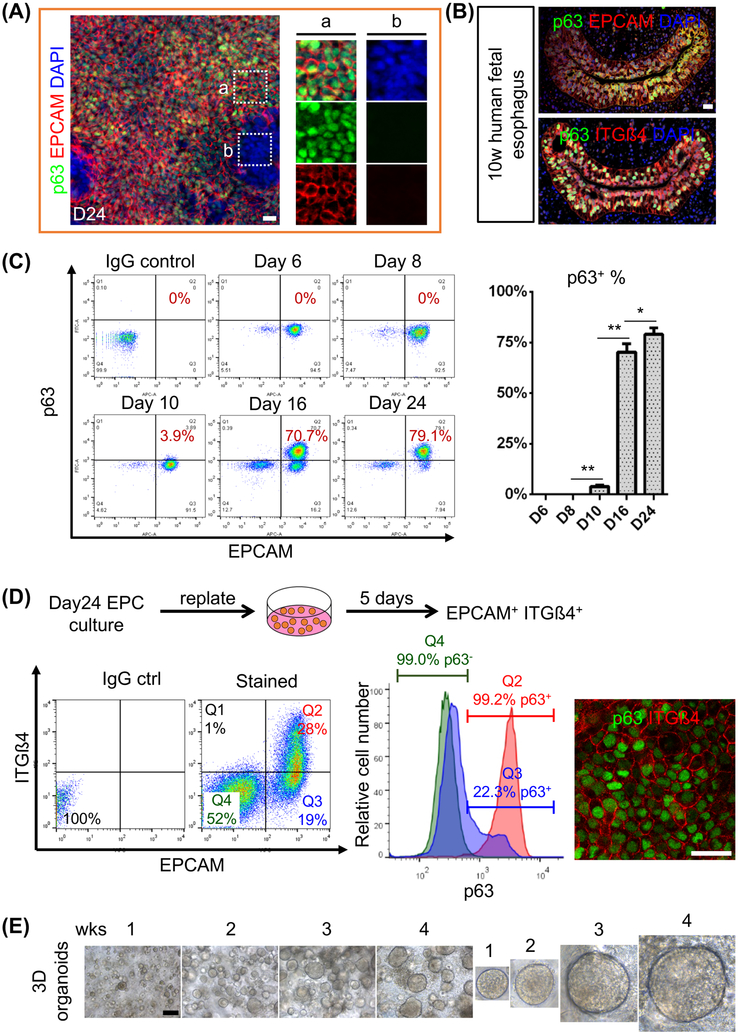
We noticed a mixture of epithelial (EPCAM+) and non-epithelial (EPCAM−) cells in the culture differentiated for 24 days (Figure 4A). If the epithelial cells were not isolated, the highly proliferative non-epithelial cells which were mainly neuroectodermal cells (PAX6+ Vimentin+) quickly overgrew (Figures 4D and S4A-B). EPCAM is co-expressed with p63 in both mouse and human fetal esophageal progenitor cells and the hPSC-derived EPCs (Figures 4A-B and S4C). We therefore initially used FACs to determine the percentage of p63+ EPCAM+ cells in both ESC and iPSC culture at day24 differentiation. In RUES2 culture, the percentage of p63+ EPCAM+ cells gradually increased to 79±3.2% from day10 to day24 (Figure 4C), which is consistent with the increased expression of p63 transcripts along differentiation (Figure 1D). Similarly, the yield of EPCAM+ p63+ cells in H9 hESC culture was 80.6±4.2% (Figure S4D). The EPC derivation efficiency for iPSC lines was also high although the efficiency for mRNA iPSC was relatively lower (Figures S4E-F). At day24 of differentiation the yield of p63+ EPCAM+ cells for sviPSC and mRNA iPSC was 80.2±6.5% and 50.1±3.5%, respectively (Figure S4E-F). Previously the cell surface markers CD47 and CD26 were used for the prospective isolation of hPSC-derived lung progenitor cells during differentiation (Hawkins et al., 2017). We asked whether hESC-derived EPCs could also be isolated with cell surface proteins. Multiple markers including p75, integrin α6, Bß1 and ß4, CD34 and CD73 have been used to enrich stem/progenitor cells in the adult mouse and human esophagus (Barbera et al., 2015; DeWard et al., 2014; Kalabis et al., 2008). Some of these markers including p75 are also expressed in other cell lineages (e.g. neural) which introduced contamination in FACS analysis (data not shown). Interestingly, the epithelial cells (EPCAM+) in the human fetal esophagus express integrin ß4 (ITGß4) (Figure 4B). We therefore combined EPCAM and ITGß4 to further purify EPCs from the mixed culture (Figure 4D). Significantly, approximately 100% epithelial cells purified with EPCAM and ITGß4 expressed p63 while nearly all EPCAM− ITGß4− cells did not express p63 (Figure 4D). However, we also observed a small percentage of EPCAM+ ITGß4− expressed p63 (Figure 4D), which is consistent with previous finding that a small subpopulation of EPCs expressed low levels of ITß4 in the esophagus (DeWard et al., 2014). The FACS-purified EPCs (EPCAM+ ITGß4+) were able to grow and form 3D organoids (Figures 4E and S5C). Overall these findings indicate that use of the cell surface markers EPCAM and ITGß4 allows the purification of hESC-derived EPCs for further application.
Figure 4. Purification of RUES2-derived EPCs with the cell surface markers EPCAM and ITG෴.
(A) Day24 culture contains a mixture of epithelium (EPCAM+) and non-epithelium (EPCAM−) cells. Note that most of EPCAM+ cells are also p63+ (n = 5). (B) ITGß4 is co-expressed with EPCAM and p63 in the epithelium of the human fetal esophagus. (C) Gradual increase of p63+ EPCAM+ cells during RUES2 differentiation. Data represent mean ± SEM (n = 3). *p < 0.05, **p< 0.01 by unpaired, two-tailed Student’s t test. Note that a minor population (3.9%) of p63+ cells began to appear at day 10. (D) RUES2-derived EPCs can be further purified with EPCAM and ITGß4 as confirmed by immunostaining. Day24 cultures were detached, replated and maintained in the SFD medium supplemented with 5% FBS, 20 ng/ml EGF, 20 ng/ml FGF2 and 10 μM ROCK inhibitor Y-27632 for another five days prior to FACs sorting. Note that non-epithelium (EPCAM−) cells outgrow EPCAM+ cells during the five-day culture as shown in FACS analysis. In one presentative of five experiments, 99.2% EPCAM+ ITGß4+ (Q2) cells expressed p63 while 99.0% EPCAM− ITGß4− (Q4) cells were p63 negative. We also noticed 22.3% of EPCAM+ITGß4− (Q3) expressed p63 which is consistent with previous finding that a small subpopulation EPCs expressed low levels of ITGß4. (E) Gradual expansion of organoids formed from single RUES2-derived EPCs that were sorted with EPCAM and ITGß4. Scale bars: (A-B and D) 20 μm, (E) 100 μm. See also Figures S4-S5
hPSC-derived EPCs are capable of recapitulating the stratified squamous epithelium both in vitro and in vivo
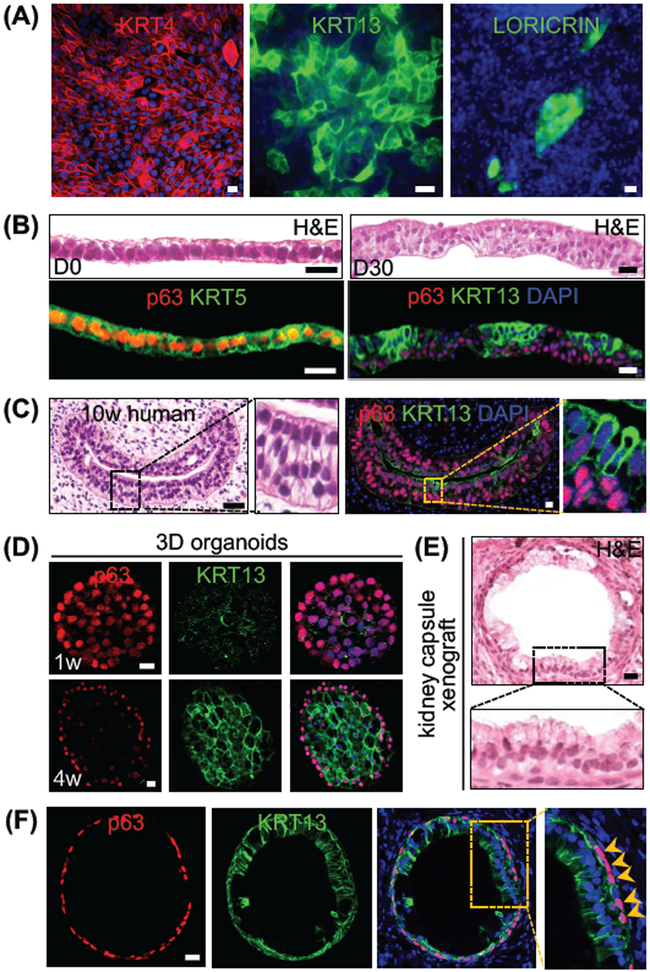
Next, we asked whether the hPSC-derived EPCs can undergo normal squamous differentiation. Interestingly, hESC-derived EPCs expressed the squamous differentiation proteins KRT4 and KRT13 when they were further cultured for 10 days in the SFD medium supplemented with 5% FBS, 20 ng/ml EGF, 20 ng/ml FGF2 and 10 μM ROCK inhibitors (Figure 5A). We also observed that a very minor population of epithelial cells expressed the terminal differentiation marker LORICRIN (Figure 5A). Consistently, while KRT4 and KRT13 were readily detected in the differentiating suprabasal cells (Figures 5C and S5A), LORICRIN is barely detected in the embryonic mouse and human esophagus (Figure S5B and data not shown). The nascent esophagus is initially lined by a simple layer of cells that are replaced by stratified squamous epithelia during development (Zhang et al., 2017). To test whether hPSC-derived EPCs are able to recapitulate the morphogenetic process we used air-liquid interface (ALI) culture (Kalabis et al., 2012). The seeded EPCs initially formed a simple columnar epithelium (p63+ KRT5+) which proliferated and differentiated to form a stratified squamous epithelium composed of basal cells (p63+) and differentiating suprabasal cells (KRT13+) (Figure 5B). The epithelium resembled the cells lining the 10-week human fetal esophagus where p63 expression was enriched in the basal layers and diminished in the suprabasal cells (KRT13+) cells (Figure 5C).
Figure 5. hESC RUES2-derived EPCs reconstitute the stratified squamous epithelium, mimicking human esophageal development.
(A) Differentiating EPCs express KRT4 and KRT13. A minor population also expresses LORICRIN. (B) Reconstitution of the stratified squamous epithelium in air-liquid interface (ALI) culture of EPCAM+ ITGß4+ EPCs. Note conversion of simple layer of epithelium to stratified squamous epithelium with basal cells (p63+) and suprabasal cells (KRT13+). (C) 10-week human fetal esophagus expressing KRT13. (D) Differentiation of EPCAM+ ITGß4+ EPCs in 3D organoid culture. Note epithelial cells in the center of the sphere lose p63 expression while gaining KRT13 at 4 weeks. (E-F) EPCs form tubular structure lined by a mixture of simple columnar and stratified epithelium (KRT13+) upon transplantation into the mouse kidney capsule for 30 days (n=3). Note that basal cells (p63+, arrowheads) surround the tube. Scale bars: 20 μm. See also Figure S5.
We and others previously showed that mouse esophageal progenitor cells formed organoid (esophageospheres) when cultured in Matrigel (DeWard et al., 2014; Giroux et al., 2017; Liu et al., 2013). hPSC-derived EPCs also formed 3D organoids when cultured in Matrigel (Figures 4E, 5D and S5C-D). The sphere at week 1 consisted of undifferentiated cells expressing high levels of p63 and KRT5 and low levels of KRT7 (Figures 5D and S5D). At week 4, p63+ cells were limited to the peripheral regions of the spheres, and cells in the center expressed high levels of KRT13 (Figure 5D). Thus hPSC-derived EPCs undergo squamous differentiation to recapitulate the stratified epithelium.
Next we tested whether these EPCs are capable of differentiation into the stratified squamous epithelium in an in vivo setting, and we delivered Matrigel implants containing EPCs into the kidney capsule. An esophagus-like tubular structure developed one month after implantation, and the lumen was lined by a stratified epithelium with underlying p63+ cells (Figures 5E-F). Of note is that the differentiation process did not seem synchronized and unified throughout the tube which was also noticed in the developing mouse esophagus (Wang et al., 2011). While the epithelium was stratified with 2-3 layers of cells in certain parts of the tube, a large portion of the epithelium remained columnar-like (Figures 5E-F) mimicking a 10-week human fetal esophagus (Figure 5C). Similar to ALI and organoid culture, the differentiating cells located at the top layers expressed KRT13 (Figures 5F).
Combined use of hPSC differentiation and mouse genetic models identified a conserved role for NOTCH signaling in esophageal development
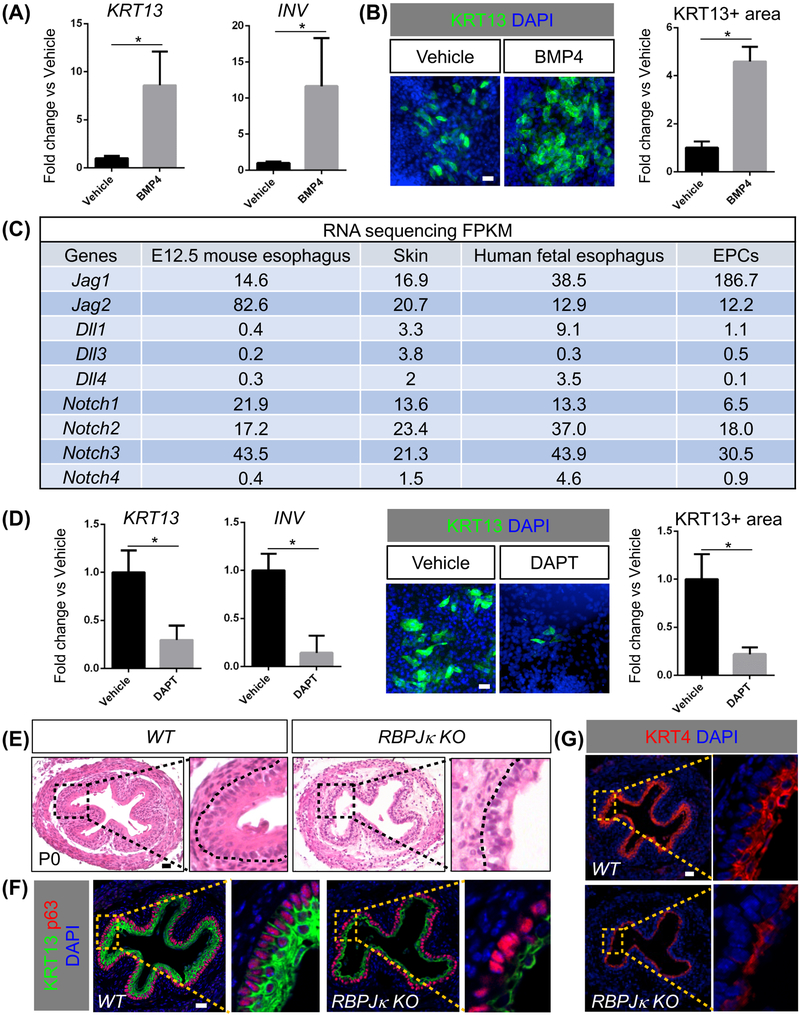
We previously showed that BMP signaling activation promotes terminal differentiation of esophageal progenitors in the developing and adult esophagus (Jiang et al., 2015; Rodriguez et al., 2010). We asked whether purified hPSCs-derived EPCs respond to BMP activation in a similar manner, and found that BMP4 treatment significantly increased the levels of KRT13 and INV (Figures 6A-B). We reasoned that our hPSC differentiation system will allow quick and efficient functional tests of candidate pathway(s) by adding chemical stimulators/inhibitors. Our RNA-seq analysis revealed that the major components (e.g. Jag 1, 2, Notch1, 2, 3) of the NOTCH signaling pathway were enriched in hPSC-derived EPCs and human fetal esophageal epithelia (Figure 6C). Interestingly, these components including Jag1 and Jag2 were also enriched in the epithelium of E12.5 mouse esophagus as compared to the skin (Figure 6C). RNA in situ hybridization confirmed Jag1 and 2 expression in the E12.5 mouse esophagus and skin (Figure S6A) and E18.5 esophagus (Figure S6B). By contrast, Dll1, 3 and 4 were expressed at very low levels in hPSC-derived EPCs, mouse and human fetal esophageal epithelium (Figure 6C). The expression of Notch 1 Intracellular Domain (NICD1) seemed correlated with the differentiation of esophageal progenitors. While NICD1 was too low to be detected at E12.5, at E18.5 the expression was limited to the differentiated suprabasal cells (Figure S6C). We then tested whether NOTCH signaling is involved in the differentiation of hPSC-derived EPCs. We treated the hPSC-derived EPCs with DAPT to block NOTCH signaling. As expected, treatment with DAPT led to downregulation of HES5, HEY1 and HEY2, the downstream targets of NOTCH signaling (Kopan and Ilagan, 2009) (Figure S6D). More importantly, inhibition of NOTCH signaling led to the reduced expression of KRT13 and INV at the transcription and protein levels (Figure 6D), supporting that NOTCH is required for the differentiation of the hPSC-derived EPCs. We also tested whether inhibition of NOTCH signaling affected the specification of EPCs during PSC differentiation, and we found that application of DAPT did not affect initial EPC commitment from the AFE (Figure S6E). This finding is consistent with the normal specification of esophageal progenitor cells (p63+ Sox2+ NKX2.1−) in the E11.5 Shh-Cre; RBPjκloxp/loxp mouse mutants where the transcriptional regulator RBPjκ of canonical Notch signaling was ablated in the early AFE (Figure S6F), confirming that NOTCH signaling is not required for EPC specification from AFE in both human and mouse.
Figure 6. Conserved roles for BMP and NOTCH signaling in the differentiation of hPSC-derived EPCs.
(A-B) BMP4 treatment promotes the squamous differentiation of RUES2-derived EPCs. Data represent mean ± SEM (n = 4). *p < 0.05 by unpaired, two-tailed Student’s t test. (C) Enrichment of Jag1, Jag2 and receptors in the epithelium of E12.5 mouse esophagus (n=3) and skin, human fetal esophageal epithelia (n=4) and RUES2-derived EPCs (n=3). FPKM, Fragments Per Kilobase of transcript per Million mapped reads. (D) DAPT treatment leads to the reduced expression of KRT13 and INVOLUCRIN in EPCs. Data represent mean ± SEM (n = 3). *p < 0.05 by unpaired, two-tailed Student’s t test. (E-G) Deletion of RBPjκblocks the formation of the stratified squamous epithelium in the esophagus of Shh-Cre; RBPjκloxp/loxp mutants (n = 4). Note the reduced thickness of the suprabasal layers (KRT13+ KRT4+) in mutants. Scale bars: 20 μm. See also Figure S6.
Next we asked whether the NOTCH pathway played a conserved role in the differentiation of the mouse esophagus. We first examined the esophagus of Shh-Cre; RBPjκloxp/loxp mutants at P0 (Figure 6E). Strikingly, the epithelial morphogenesis was severely disrupted, and the number of epithelial layers was decreased following RBPjκ deletion (Figures 6E and S6G). RBPjκ deletion also resulted in decreased thickness of the differentiating suprabasal cells (KRT13+) (Figures 6F and S6H). In keeping with this finding, the numbers of differentiating cells (KRT4+) were also decreased (Figure 6G). We next performed combined deletion of Jag1 and Jag2 in Shh-Cre; Jag1loxp/loxp; Jag2loxp/loxp mutants. Loss of Jag1 and Jag2 also blocked the squamous differentiation of progenitor cells in the developing esophagus (Figure 7A), and thickness of the differentiating suprabasal layer (KRT13+ KRT4+) was significantly reduced (Figures 7B-C and S6I). Notably, while the impact on epithelial differentiation was apparent in the mutant esophagus lacking Jag2, epithelial differentiation is minimally affected in mutants lacking Jag1 only (Figures 7A-C). Taken together, these studies suggest that hPSC differentiation and mouse genetics studies complement each other, providing an efficient platform to identify the important role of the NOTCH pathway in the morphogenesis of the esophageal epithelium.
Figure 7. Conditional deletion of the NOTCH ligands Jag1 and Jag2 impairs the squamous differentiation of esophageal progenitor cells.
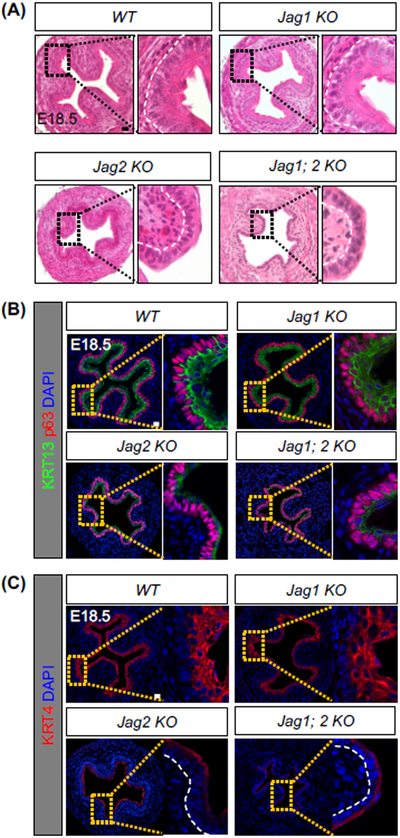
(A) Shh-Cre mediated deletion of Jag2 but not Jag1 blocks the formation of the stratified squamous epithelium. Note the comparable phenotypes in mutants lacking Jag2 only and Jag1; Jag2. (B-C) Impaired squamous differentiation in the esophagus of Shh-Cre; Jag2loxp/loxp (Jag 2 KO) and Shh-Cre; Jag1loxp/loxp ;Jag2loxp/loxp (Jag1; 2 KO) compound mutants. Note the significant loss of KRT4 and KRT13 expression in the mutant esophagus (n = 4). Scale bars: 20 μm. See also Figure S6.
DISCUSSION
Here, we provide a robust protocol to generate human EPCs through sequential differentiation of hPSCs including hESCs and iPSCs. We further identified EPCAM and ITGß4 as the cell surface markers to allow purification of hPSC-derived EPCs. Functionally, these purified human EPCs are capable of undergoing normal differentiation to establish the stratified squamous epithelium both in vitro and in vivo. This differentiation system combined with genetic mouse models allowed us to identify a critical role of the NOTCH pathway in esophageal development. Our approach therefore provides a unique and efficient platform to study the developmental mechanisms of the human esophagus. Complimentary findings reported by Dr. Trisno et al. also suggest that hPSC-derived EPCs are useful for studying the molecular mechanisms promoting esophageal versus tracheal lineage (Trisno et al., 2018).
We built on our previous findings and used the BMP inhibitor Noggin to promote the commitment of AFE towards EPCs (p63+ SOX2+ NKX2.1−). This is in contrast to the effect of WNTs which promote the differentiation of AFE into lung epithelial progenitors (McCauley et al., 2017). Consistently, we and others showed that WNT signaling promotes lung specification while BMP signaling needs to be inhibited by Noggin to allow generation of the esophagus in mice. Deletion of β-catenin or Nog leads to abnormal formation of the lung and esophagus, respectively (Domyan et al., 2011; Goss et al., 2009; Harris-Johnson et al., 2009; Que et al., 2006). Furthermore, ectopic WNT activation in genetically engineered mice suppresses the formation of the squamous epithelium in both forestomach and esophagus (Goss et al., 2009; Harris-Johnson et al., 2009). Here, we found low WNT signaling activities during the specification of EPCs. Ectopic WNT activation in combination with BMP and TGFß dual inhibition represses EPC specification accompanied by increased expression of PROX1 and HNF6, which are expressed in the posterior foregut-derived organs e.g. liver and pancreas (Burke and Oliver, 2002; Rausa et al., 1997), suggesting ectopic WNT signaling posteriorizes the foregut (Ober et al., 2006; Wells et al., 2007). BMP signaling is required for the terminal differentiation of epithelial progenitor cells after the esophagus is established from the early foregut (Jiang et al., 2015; Rodriguez et al., 2010). Consistently, we found that BMP4 treatment promotes the squamous differentiation of hPSC-derived EPCs. Therefore, function of BMP signaling in esophageal morphogenesis seems conserved in mouse and human.
The hPSC differentiation system has offered an avenue to study the mechanisms regulating the development of the esophagus. Here, we combined hPSCs and moue genetic models to identify the role of NOTCH signaling in the differentiation of epithelial progenitor cells in the developing esophagus. We identified that the Notch ligands (Jag1 and Jag2) are enriched in the esophageal epithelium and hPSC-derived EPCs. NOTCH1 and 3 previously have been shown to be expressed in the adult human esophageal epithelium (Ohashi et al., 2011; Ohashi et al., 2010). Here, we found that blocking NOTCH signaling in both mouse genetic models and hPSC-derived EPCs leads to reduced squamous differentiation of esophageal progenitor cells. These studies prove that a combination of mouse genetics and hPSCs differentiation is powerful for elucidating the developmental mechanisms conserved between species.
Notably, through the differentiation of hPSCs we identified for the first time that SOX9 is expressed in the early esophageal progenitor cells. We further showed that SOX9 is also expressed in the epithelium of the developing mouse esophagus but the expression is lost in adults. SOX9 is similarly expressed in the lung epithelium in the early stage of development, but the expression falls to undetectable levels in the adult lung (Chang et al., 2013; Rockich et al., 2013). Conditional deletion of SOX9 disrupts lung branching morphogenesis and epithelial differentiation (Chang et al., 2013; Rockich et al., 2013). Although the role of SOX9 in the developing esophagus remains unknown, re-expression of SOX9 accompanied by high levels of KRT7 has been found in Barrett’s esophagus (also known as intestinal metaplasia) and esophageal adenocarcinoma (Jiang et al., 2017; Song et al., 2014; Wang et al., 2014). How SOX9 is involved in the pathogenesis of Barrett’s esophagus and tumorigenesis remains unknown. Study of the hPSC-derived EPCs (SOX9+) may provide insights into this issue.
In summary, building on our previous understanding of mouse esophageal development we established a robust protocol to derive EPCs from both hESCs and iPSCs. The hPSC-derived EPCs are capable of undergoing normal differentiation and generating the stratified squamous epithelium from simple columnar cells both in vitro and in vivo. More importantly, we employed the differentiation system in combination with mouse genetic models to identify the conserved roles of the BMP and NOTCH pathways in the morphogenesis of the esophagus. Our findings therefore provide a useful platform for studying regulatory mechanisms underlying human esophageal development and diseases.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by, the lead contact, Jianwen Que (jq2240@cumc.columbia.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Shh-Cre (Harfe et al., 2004), RBPjκloxp/loxp (Han et al., 2002), Jag1loxp/loxp (Brooker et al., 2006), Jag2 loxp/loxp (Xu et al., 2010), BRE-lacZ (Blank et al., 2008), Noggin-lacZ (McMahon et al., 1998), NOD.Cg-Prkdcscid.Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory) mouse strains were maintained on a C57BL/6 and 129SvEv mixed background, 8 to 24 weeks of age, and of both sexes. All animals were genotyped by PCR of tail DNA. Mice were housed in a specific pathogen-free mouse facility with 12-hour light/dark cycle and provided with food and water ad libitum according to Columbia University IACUC. Mice used had no known health/immune concerns, were not involved in previous procedures, and were drug or test naive. Experimental procedures and animal care were performed in accordance with the protocols approved by The Columbia University Institutional Animal Care and Use Committee.
Human Fetal Esophagus
This study was approved by the Medical Ethical Committee of the Leiden Medical University Center (P08.087) Informed consent was obtained and the study was conducted in accordance with the Declaration of Helsinki by the World Medical Association. 10-week-old human fetal esophagi were obtained from abortion material (vacuum aspiration) without medical indication (Roost et al., 2015). De-identified human fetal esophagi between 14-18 weeks of gestation were obtained under IRB approvals at CHLA and USC (USC-HS-13-0399 and CHLA-14-2211) after signed informed consent was granted. Tissues were collected in cold HBSS and processed in the lab within an hour of collection. Tissues from samples with known structural or chromosomal anomalies were excluded from this study.
Maintenance of hPSCs
RUES2 (Rockefeller University Embryonic Stem Cell Line 2, NIH approval number NIHhESC-09-0013), and Sendai virus and modified mRNA generated human dermal fibroblasts iPSC lines (sviPSC) and mRNA iPSC were kindly provided by the Mount Sinai Stem Cell Core facility and cultured as previously described (Huang et al., 2015; Huang et al., 2014). H9 was purchased from WiCell. hPSCs lines were maintained on mouse embryonic fibroblasts (MEFs) feeder cells. Briefly, CF-1 MEF (MTI-GlobalStem) mitotically arrested by irradiation were plated at a density of ~25,000 cells/cm2. hPSCs were plated on the fibroblasts and cultured in the maintenance medium: 400 ml of DMEM/F12 (Thermo Fisher Scientific), 100 ml of KnockOut Serum Replacement (Thermo Fisher Scientific), 5 ml of GlutaMAX (Thermo Fisher Scientific), 5 ml of MEM non-essential amino acids solution (Thermo Fisher Scientific), 3.5 μl of 2-mercaptoethanol (Sigma-Aldrich), 1 ml of primocin (Thermo Fisher Scientific), and FGF2 (R&D Systems) with a final concentration of 20 ng/ml to make a total of ~500 ml of medium. For passaging cells were detached with Accutase/EDTA (Innovative Cell Technologies) and replated at a ratio of 1:24. Cells were maintained in an incubator with 95% humidity, 95% air and 5% CO2 at 37°C. Human ES/iPS cell research was conducted under the approval of the Columbia University Human Embryonic and Human Embryonic Stem Cell Research Committee.
METHOD DETAILS
Endoderm and Anterior Foregut Endoderm Differentiation
We followed a previously described protocol to differentiate hPSCs into endodermal and anterior foregut (Huang et al., 2015; Huang et al., 2014). Serum-Free Differentiation (SFD) medium was prepared as following: 750 ml of reconstituted IMDM (Thermo Fisher Scientific), 250 ml of F- 12 (Corning), 7.5 ml of 7.5% Bovine Albumin Fraction V Solution (Thermo Fisher Scientific), 10 ml of GlutaMAX (Thermo Fisher Scientific), 5 ml of N2 (Thermo Fisher Scientific), 10 ml of B27 (Thermo Fisher Scientific) and 10 ml of Penicillin/Streptomycin (Thermo Fisher Scientific), and adding L-Ascorbic acid (Sigma-Aldrich) and MTG (Sigma-Aldrich) on the day of use to obtain a final concentration of 50 μg/ml and 0.04 μl/ml, respectively. To generate endoderm, hPSCs were detached by Accutase/EDTA and cultured in the SFD medium plus 10 μM Rock inhibitor Y-27632 (Tocris), 100 ng/ml Activin A, 2.5 ng FGF2 and 0.5 ng/ml BMP4 (R&D Systems) in 6-well Ultra-Low-Attachment plates (Corning) for 72 hours (day1-4). At day4 anterior foregut progenitor cells were further induced by culturing endoderm in the SFD medium plus 10 μM SB431542 (Tocris) and 100 ng/ml Noggin (R&D Systems) for 24 hours (day4-5) and the SFD medium plus 10 μM SB431542 and 1 μM IWP2 (Tocris) for another 24 hours (day5-6). Cells were maintained at 5% O2/95% N2/5% CO2 from day1-6.
Esophageal and Lung Progenitor Cell Differentiation
To induce esophageal progenitor cell differentiation, anterior foregut progenitor cells were cultured from day6 to day16 in the SFD medium plus 10 μM SB431542 and 100 ng/ml Noggin. From day16 to day24, cells were maintained in the SFD medium. We followed a previously described protocol to generate lung progenitor cells from AFE (Huang et al., 2015; Huang et al., 2014) in which AFE cells were cultured in the SFD medium supplemented with 3 μM CHIR99021 (Tocris), 10 ng/ml human FGF10 (R&D Systems), 10 ng/ml human KGF/FGF7 (R&D Systems), 10 ng/ml human BMP4 and 50 nM retinoid acid (Sigma) (day6-16). Cells were cultured at 5% O2/95% N2/5% CO2 at day6-7 and maintained at 95% air/5% CO2 from day7 onwards.
3D Organoid Culture and Air-liquid interface (ALI) culture
20,000 sorted hPSC-derived EPCs (ITGß4+ EPCAM+) were suspended in 75 μl medium and mixed with 75 μl Matrigel (Corning). The mixture was plated in 24-well cell culture inserts (Falcon), and the medium was added to the bottom and top chambers after Matrigel solidified. The organoid culture medium including the SFD medium supplemented with 10 μM Y27632, 100 ng/ml Noggin, 10 μM SB431542, 3 μM CHIR99021, 20 ng/ml FGF2, 200 ng/ml EGF was modified from previous studies (DeWard et al., 2014; Giroux et al., 2017; Liu et al., 2013). For ALI culture, 20,000 sorted EPCs (ITGß4+ EPCAM+) were cultured in Matrigel-coated 24-well inserts (Falcon) in the SFD medium supplemented with 5% FBS, 20 ng/ml EGF, 20 ng/ml FGF2 and 10 μM Y27632. When cells were confluent, medium was removed from the upper chamber to create air-liquid interface, and the culture was further maintained for one month.
Kidney Capsule Implantation
For the kidney transplantation assay, one million RUES2-derived esophageal progenitor cells (day24 differentiation) were mixed with Matrigel (Coring) at 1:1 ratio and implanted under the kidney capsule as previously described (Chen et al., 2017). Grafts harvested from the kidney capsules one month after transplantation were embedded in paraffin and subjected to histological analysis.
Immunofluorescence, X-gal Staining and Microscopy Imaging
For immunofluorescence staining, cells were fixed in 4% paraformaldehyde (PFA) for 15 minutes at room temperature and washed with 1× PBS for three times. Cells were permeabilized with 0.3% Triton X-100 in 1× PBS for 15 minutes. Then cells were incubated in blocking solution (0.3% Triton X-100 plus 2% donkey serum in 1× PBS) for 1 hour. Primary antibodies were added into blocking solution and incubated at 4°C overnight. The next day, cells were washed with 1× PBS for three times. Secondary antibodies conjugated to Alexa Fluor (Thermo Fisher Scientific) were incubated for 1 hour. Images were taken using Leica DMI6000 B (Leica Microsystems) or DMi8 (Leica Microsystems) and a Zeiss LSM700 confocal laser scanning microscope. Bright field images were acquired using a Nikon Labophot 2 microscope equipped with a Nikon Digital Sight DS-Ri1 charge-coupled device camera. The thickness and areas composed of KRT13+ cells were calculated by ImageJ (National Institutes of Health). Primary antibodies are listed in the Key Resources Table. For X-gal staining, tissues were fixed in 4% paraformaldehyde for 30 minutes and incubated in X-gal solution overnight at 37°C as previously described (Que et al., 2006).
RNA in situ Hybridization
RNA in situ hybridization was performed as previously described (Que et al., 2006). Briefly, embryos were fixed in 4% PFA overnight and embedded in OCT. Cryo-sections were hybridized with specific digoxigenin-labeled riboprobe at 65°C in a moist chamber, overnight. Sections were then washed in high-stringency conditions and incubated with alkaline phosphatase-conjugated anti-digoxigenin antibody overnight at 4°C. Following a chromogenic reaction with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate, in situ gene expression was indicated by the blue color of specific tissue regions. Jag1 and Jag2 probes were kindly provided by Dr. Doris K. Wu (National Institute on Deafness and Other Communicative Disorders) and Dr. Thomas Gridley (Maine Medical Center Research Institute), respectively (Chang et al., 2008; Kiernan et al., 2005).
Flow Cytometric Analysis and Cell Sorting
To perform cell surface marker staining, cells were disassociated with 0.05% Trypsin-EDTA (Thermo Fisher Scientific) and stained with fluorophore conjugated antibodies in FACS buffer (1× PBS, 2% FBS, 0.2mM EDTA) for 30 min with live/dead staining dye (LIVE/DEAD™ Fixable Violet Dead Cell Stain Kit, Thermo Fisher Scientific) to exclude dead cells. Cell surface marker antibodies are listed in Key Resources Table. Intracellular staining cells were performed according to the manufacturer’s instructions in eBioscience™ Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific). Fixation and permeabilization was performed at room temperature for 1 hour followed by incubation of primary antibody for 1 hour. Cells were washed with 1× PBS and fluorophore-conjugated secondary antibodies were incubated for 1 hour. Stained cells were analyzed BD FACSCanto (BD Biosciences) data were analyzed with FlowJo software (Ashland, Oregon). Sorted EPCAM+ ITGß4+ cells were maintained in the SFD medium supplemented with 5% FBS, 20 ng/ml EGF, 20 ng/ml FGF2 and 10 μM Rock inhibitor Y27632.
Mouse and human fetal esophageal epithelium isolation
Muscle layers were stripped off the esophagi using forceps and the remaining tissue (epithelium and mesenchyme) was incubated in 50 U/ml Dispase (Corning) in 1xPBS for 10 minutes at room temperature for mouse esophagi and 16 U/ml Dispase for 8 minutes at room temperature for human fetal esophagi. Epithelium was peeled off from mesenchyme with forceps and subjected to RNA purification.
RNA Sequencing
RNA was extracted from RUES2-derived esophageal progenitor cells, human fetal (14-18 weeks) and E12.5 mouse esophageal epithelium and skin using the PicoPure™ RNA Isolation Kit (Thermo Fisher Scientific). RNA concentration was measured by 2100 Bioanalyzer (Agilent Technologies). Libraries were prepared using Illumina TruSeq RNA prep kit (Illumina) and sequenced by the Illumina HiSeq4000 (Illumina) at the Columbia Genome Center. We multiplexed samples in each lane, which yields targeted number of single-end/paired-end 100 bp reads for each sample, as a fraction of 180 million reads for the whole lane. We used RTA (Illumina) for base calling and bcl2fastq (version 1.8.4) for converting BCL to fastq format, coupled with adaptor trimming. We mapped the reads to a reference genome (Mouse: UCSC/mm9) and (Human: NCBI/build37.2) using Tophat (version 2.1.0) with 4 mismatches and 10 maximum multiple hits. To tackle the mapping issue of reads that are from exon-exon junctions, Tophat inferred novel exon-exon junctions ab initio, and combined them with junctions from known mRNA sequences as the reference annotation. We estimated the relative abundance/expression level of genes and splice isoforms using Cufflinks (version 2.0.2) with default settings. Estimated normalized expression level Fragments Per Kilobase of transcript per Million (FPKM) of known genes and transcripts were presented.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Cells were lysed with TRIzol (Invitrogen) and RNA was purified using the RNeasy Mini Kit (QIAGEN). RNA was reverse transcribed to cDNA by the SuperScript III First-Strand SuperMix (Invitrogen). cDNA was quantified by real-time PCR using the iQ SYBR Green Supermix (Bio-Rad) and StepOnePlus™ Real-Time PCR System (Applied Biosystems). The transcript level of each gene was normalized to the ß-actin control using 2(−ΔΔCT) method. Relative gene expression was calculated and reported as fold change compared to the indicated samples using ß-actin-normalized transcript level. All qRT-PCR experiments were performed at least triplicate. PCR primers were designed and produced by Integrated DNA Technologies and primer sequences were summarized in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as the mean ± SEM using GraphPad Software Prism 6. Statistical significance was determined by Student's t tests. When more than two groups are compared, multiple comparisons were performed using one-way ANOVA followed by Bonferroni correction. For each analysis, at least 3 biological replicates were included. Representative pictures shown are indicated in the legends. P-values of 0.05 or less were considered to be statistically significant.
DATA AND SOFTWARE AVAILABILITY
The accession number for the published RNA-sequencing data reported in this paper is GEO: GSE116272.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal APC anti-human CD184 (CXCR4) | BioLegend | Cat#306510; RRID:AB_314616 |
| Mouse monoclonal PE anti-human CD184 (CXCR4) | BioLegend | Cat#306505; RRID:AB_314611 |
| Mouse monoclonal APC anti-human CD326 (EPCAM) | BioLegend | Cat#324208; RRID:AB_756082 |
| Mouse monoclonal PE anti-human CD117 (c-KIT) | BioLegend | Cat#313204; RRID:AB_314983 |
| Mouse monoclonal PE anti-human CD104 (ITGß4) | BioLegend | Cat#327807; RRID:AB_2129147 |
| Rabbit polyclonal anti-EPCAM | abcam | Cat#ab71916; RRID: AB_1603782 |
| Rabbit monoclonal anti-P63 | Santa Cruz | Cat#sc-8343; RRID:AB_653763 |
| Mouse monoclonal anti-P63 | BioLegend | Cat#687202; RRID:AB_2616941 |
| Rabbit monoclonal anti-P63-α | Cell Signaling | Cat#13109; RRID:AB_2637091 |
| Mouse monoclonal anti-NKX2.1 | Thermo Fisher Scientific | Cat#MA5-13961; RRID: AB_10984070 |
| Rabbit monoclonal anti-NKX2.1 | abcam | Cat#ab76013; RRID: AB_1310784 |
| Rat monoclonal anti-SOX2 | Thermo Fisher Scientific | Cat#14-9811-82;RRID: AB_11219471 |
| Goat polyclonal anti-FOXA2 | Santa Cruz | Cat#sc-6554; RRID:AB_2262810 |
| Mouse monoclonal anti-FOXA1 | Santa Cruz | Cat#sc-101058;RRID: AB_1124659 |
| Goat polyclonal anti-SOX9 | R&D Systems | Cat#AF3075; RRID: AB_2194160 |
| Mouse monoclonal anti-PAX9 | BioLegend | Cat#658202; RRID: AB_2562877 |
| Goat polyclonal anti-FOXE1 | abcam | Cat#ab5080; RRID: AB_304738 |
| Mouse monoclonal anti-KRT4 | abcam | Cat#ab9004; RRID: AB_306932 |
| Chicken polyclonal anti-KRT5 | BioLegend | Cat#905901; RRID: AB_2565054 |
| Mouse monoclonal anti-KRT7 | abcam | Cat#ab9021; RRID: AB_306947 |
| Rabbit monoclonal anti-KRT13 | abcam | Cat#ab92551; RRID: AB_2134681 |
| Mouse monoclonal anti-KRT14 | abcam | Cat#ab7800; RRID: AB_306091 |
| Rabbit polyclonal anti-LORICRIN | BioLegend | Cat#9051011; RRID: AB_2565046 |
| Mouse monoclonal anti-ITGα6 | abcam | Cat#Ab20142; RRID: AB_445361 |
| Rabbit monoclonal anti-NOTCH3 | Cell Signaling | Cat#5276S; RRID: AB_10560515 |
| Rabbit monoclonal anti-NICD1/Cleaved NOTCH1 (Val1744) | Cell signaling | Cat#4147S; RRID: AB_2153348 |
| Rabbit monoclonal anti-RBPjκ | Cell Signaling | Cat#5313S; RRID: AB_2665555 |
| Goat polyclonal anti-NANOG | R&D Systems | Cat#AF1997; RRID: AB_355097 |
| Rabbit polyclonal anti-PAX6 | BioLegend | Cat#901301; RRID: AB_2565003 |
| Mouse monoclonal anti-Vimentin | Santa Cruz | Cat#sc-6260; RRID: AB_628437 |
| Biological Samples | ||
| Primary human fetal esophagus | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ham's F12 | Corning | Cat#10-080-CV |
| Iscove's Modified Dulbecco's Medium (IMDM) | Corning | Cat#10-016-CV |
| DMEM/F12 | Corning | Cat#10-092-CV |
| Growth factor reduced matrigel | Corning | Cat#354231 |
| 6-well Ultra-Low-Attachment plates | Corning | Cat#3471 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| 1xPBS | Thermo Fisher Scientific | Cat#10010049 |
| KnockOut Serum Replacement | Thermo Fisher Scientific | Cat#10828010 |
| N2 | Thermo Fisher Scientific | Cat#17502048 |
| B27 | Thermo Fisher Scientific | Cat#17504044 |
| 7.5% Bovine Albumin Fraction V Solution | Thermo Fisher Scientific | Cat#15260037 |
| GlutaMAX | Thermo Fisher Scientific | Cat#35050061 |
| MEM non-essential amino acids solution | Thermo Fisher Scientific | Cat#11140050 |
| Accutase/EDTA | Innovative Cell Technologies | Cat#AT104 |
| 0.05% Trypsin/EDTA | Thermo Fisher Scientific | Cat#25300120 |
| 0.5M EDTA | Thermo Fisher Scientific | Cat#15575020 |
| Primocin | InvivoGen | Cat#ant-pm-1 |
| Mouse embryonic fibroblasts | MTI-GlobalStem | Cat#GSC-6201G |
| Fibronectin | Sigma | Cat#F0895 |
| L-Ascorbic acid | Sigma | Cat#A4544 |
| 1-Thioglycerol (MTG) | Sigma | Cat#M6145 |
| 2-mercaptoethanol | Sigma | Cat#M3148 |
| BMP4 | R&D System | Cat#314-BP |
| FGF2 | R&D System | Cat#233-FB |
| Activin A | R&D System | Cat#338-AC |
| FGF10 | R&D System | Cat#345-FG |
| KGF/FGF7 | R&D System | Cat#251-KG |
| All-trans Retinoic acid | R&D System | Cat#0695 |
| Noggin | R&D System | Cat#6057-NG |
| SB431542 | Tocris | Cat#1614 |
| IWP2 | Tocris | Cat#3533 |
| Y-27632 | Tocris | Cat#1254 |
| CHIR99021 | Tocris | Cat#4423 |
| Deposited data | ||
| RNA sequencing of E12.5 mouse esophagus and skin, human fetal esophagus and RUES2-derived EPCs | This paper | GEO: GSE116272 |
| Experimental Models: Cell Lines | ||
| Human: RUES2 embryonic stem cell line | Rockefeller University | National Institutes of Health (NIH) approval number: NIHhESC-09-0013 |
| Human: H9 embryonic stem cell line | WiCell | Cat#WA09 |
| Human: Sendai virus-generated iPSC (sviPSC) line | Sunita D'Souza in Icahn School of Medicine at Mount Sinai (Huang et al., 2014) | N/A |
| Human: modified mRNA transfection-generated iPSC line | Sunita D'Souza in Icahn School of Medicine at Mount Sinai (Huang et al., 2014) | N/A |
| Experimental Models: Organisms/Strains | ||
| NOD.Cg-Prkdcscid.Il2rgtm1Wjl/SzJ (NSG) | The Jackson Laboratory | Stock#005557 |
| Shh-Cre | (Harfe et al., 2004) | N/A |
| RBPjkloxp/loxp | (Han et al., 2002) | N/A |
| Jag1loxp/loxp | (Brooker et al., 2006) | N/A |
| Jag2loxp/loxp | (Xu et al., 2010) | N/A |
| BRE-lacZ | (Blank et al., 2008) | N/A |
| Noggin-lacZ | (Harfe et al., 2004) | N/A |
| Recombinant DNA | ||
| Plasmid for Jag1 in situ probe | Dr. Doris K. Wu (National Institute on Deafness and Other Communication Disorders)(Chang et al., 2008) | |
| Plasmid for Jag2 in situ probe | Dr. Thomas Gridley (Maine Medical Center Research Institute)(Kiernan et al., 2005) | |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Prism 6 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
Highlights:
BMP/TGFß dual inhibition promotes hPSC differentiation into EPCs
hPSC-derived EPCs can be purified with EPCAM and ITGß4
hPSC-derived EPCs recapitulate human esophageal epithelial development
BMP and NOTCH pathways are required for EPCs differentiation
Acknowledgments
We thank members in the Que and Wellington laboratory for critical reading of the manuscript and helpful discussion. This work in the Que lab is partly supported by R01DK113144, R01DK100342, R01HL132996, National Natural Science Foundation of China 81728001 (J.Q) and the Price Family Foundation. S.D. acknowledges a fellowship support from the Hastings center for pulmonary research (USC). Flow cytometry was performed in the Columbia Center for Translational Immunology (CCTI) Flow Cytometry Core at Columbia University Medical Center, supported in part by the Office of the Director, National Institutes of Health under awards S10OD020056. We would also like to thank Matthew Thornton, Drs. Brendan Grubbs, Melissa L. Wilson (Department of Preventive Medicine, University of Southern California) and Rachel Steward (Family Planning Associates) for coordinating fetal tissue collection.
Footnotes
Declaration of Interests
A related patent application titled Method to generate basal cells from human pluripotent stem cells has been filed by Columbia Technology Ventures at Columbia University.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Barbera M, di Pietro M, Walker E, Brierley C, MacRae S, Simons BD, Jones PH, Stingl J, and Fitzgerald RC (2015). The human squamous oesophagus has widespread capacity for clonal expansion from cells at diverse stages of differentiation. Gut 64, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U, Seto ML, Adams DC, Wojchowski DM, Karolak MJ, and Oxburgh L (2008). An in vivo reporter of BMP signaling in organogenesis reveals targets in the developing kidney. BMC Dev Biol 8, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, and Lewis J (2006). Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development 133, 1277–1286. [DOI] [PubMed] [Google Scholar]
- Burke Z, and Oliver G (2002). Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Develop 118, 147–155. [DOI] [PubMed] [Google Scholar]
- Chang DR, Martinez Alanis D, Miller RK, Ji H, Akiyama H, McCrea PD, and Chen J (2013). Lung epithelial branching program antagonizes alveolar differentiation. Proc Natl Acad Sci U S A 110, 18042–18051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WI, Lin ZS, Kulessa H, Hebert J, Hogan BLM, and Wu DK (2008). Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. Plos Genetics 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, Huang SX, de Carvalho A, Ho SH, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova JL, Bhattacharya J, et al. (2017). A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19, 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathan N, Parlato R, Rosica A, De Felice M, and Di Lauro R (2002). Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev Dyn 224, 450–456. [DOI] [PubMed] [Google Scholar]
- DeWard AD, Cramer J, and Lagasse E (2014). Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep 9, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, and Sun X (2011). Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development 138, 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux V, Lento AA, Islam M, Pitarresi JR, Kharbanda A, Hamilton KE, Whelan KA, Long A, Rhoades B, Tang Q, et al. (2017). Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J Clin Invest 127, 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BG, Chao HH, Yang Y, Yermolina YA, Tobias JW, and Katz JP (2007). Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol 292, G1784–1792. [DOI] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, and Morrisey EE (2009). Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell 17, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, and Honjo T (2002). Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol 14, 637–645. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, and Tabin CJ (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528. [DOI] [PubMed] [Google Scholar]
- Harris-Johnson KS, Domyan ET, Vezina CM, and Sun X (2009). beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci U S A 106, 16287–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins F, Kramer P, Jacob A, Driver I, Thomas DC, McCauley KB, Skvir N, Crane AM, Kurmann AA, Hollenberg AN, et al. (2017). Prospective isolation of NKX2-1-expressing human lung progenitors derived from pluripotent stem cells. J Clin Invest 127, 2277–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Green MD, de Carvalho AT, Mumau M, Chen YW, D'Souza SL, and Snoeck HW (2015). The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat Protoc 10, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Islam MN, O'Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, et al. (2014). Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol 32, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A, Morley M, Hawkins F, McCauley KB, Jean JC, Heins H, Na CL, Weaver TE, Vedaie M, Hurley K, et al. (2017). Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell 21, 472–488.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs IJ, Ku WY, and Que J (2012). Genetic and cellular mechanisms regulating anterior foregut and esophageal development. Dev Biol 369, 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Ku WY, Zhou Z, Dellon ES, Falk GW, Nakagawa H, Wang ML, Liu K, Wang J, Katzka DA, et al. (2015). BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J Clin Invest 125, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Li H, Zhang Y, Yang Y, Lu R, Liu K, Lin S, Lan X, Wang H, Wu H, et al. (2017). Transitional basal cells at the squamous-columnar junction generate Barrett’s oesophagus. Nature Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabis J, Oyama K, Okawa T, Nakagawa H, Michaylira CZ, Stairs DB, Figueiredo JL, Mahmood U, Diehl JA, Herlyn M, et al. (2008). A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J Clin Invest 118, 3860–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabis J, Wong GS, Vega ME, Natsuizaka M, Robertson ES, Herlyn M, Nakagawa H, and Rustgi AK (2012). Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc 7, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Cordes R, Kopan R, Gossler A, and Gridley T (2005). The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development 132, 4353–4362. [DOI] [PubMed] [Google Scholar]
- Kopan R, and Ilagan MX (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S, et al. (2013). Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell 12, 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire TA, Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, Kwok LW, Mou H, Rajagopal J, Shen SS, et al. (2012). Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, and Kotton DN (2017). Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 20, 844–857 e846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Aihara E, Martin B, Crawford CM, Broda T, Treguier J, Zhang X, Shannon JM, Montrose MH, and Wells JM (2017). Wnt/beta-catenin promotes gastric fundus specification in mice and humans. Nature 541, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, and McMahon AP (1998). Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev 12, 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, and Kimura S (1999). Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol 209, 60–71. [DOI] [PubMed] [Google Scholar]
- Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, et al. (2016). Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell 19, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Zhao R, Sherwood R, Ahfeldt T, Lapey A, Wain J, Sicilian L, Izvolsky K, Musunuru K, Cowan C, et al. (2012). Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell 10, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, and Stainier DY (2006). Mesodermal Wnt2b signalling positively regulates liver specification. Nature 442, 688–691. [DOI] [PubMed] [Google Scholar]
- Ohashi S, Natsuizaka M, Naganuma S, Kagawa S, Kimura S, Itoh H, Kalman RA, Nakagawa M, Darling DS, Basu D, et al. (2011). A NOTCH3-mediated squamous cell differentiation program limits expansion of EMT-competent cells that express the ZEB transcription factors. Cancer Res 71, 6836–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi S, Natsuizaka M, Yashiro-Ohtani Y, Kalman RA, Nakagawa M, Wu L, Klein-Szanto AJ, Herlyn M, Diehl JA, Katz JP, et al. (2010). NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology 139, 2113–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano J, Opitz OG, Nakagawa H, Jenkins TD, Friedman SL, and Rustgi AK (2000). The Kruppel-like transcriptional factors Zf9 and GKLF coactivate the human keratin 4 promoter and physically interact. FEBS Lett 473, 95–100. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, and Melton DA (2014). Generation of functional human pancreatic beta cells in vitro. Cell 159, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, and Balling R (1998). Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev 12, 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazzolla D, Palla AR, Pantoja C, Canamero M, de Castro IP, Ortega S, Gomez-Lopez G, Dominguez O, Megias D, Roncador G, et al. (2014). Lineage-restricted function of the pluripotency factor NANOG in stratified epithelia. Nat Commun 5, 4226. [DOI] [PubMed] [Google Scholar]
- Que J (2015). The initial establishment and epithelial morphogenesis of the esophagus: a new model of tracheal-esophageal separation and transition of simple columnar into stratified squamous epithelium in the developing esophagus. Wiley Interdiscip Rev Dev Biol 4, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Choi M, Ziel JW, Klingensmith J, and Hogan BL (2006). Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation 74, 422–437. [DOI] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, and Hogan BL (2007). Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausa F, Samadani U, Ye HG, Lim L, Fletcher CF, Jenkins NA, Copeland NG, and Costa RH (1997). The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3 beta in the developing murine liver and pancreas. Developmental Biology 192, 228–246. [DOI] [PubMed] [Google Scholar]
- Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MA, Kopp JL, Sander M, Wellik DM, and Spence JR (2013). Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci U S A 110, E4456–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Da Silva S, Oxburgh L, Wang F, Hogan BL, and Que J (2010). BMP signaling in the development of the mouse esophagus and forestomach. Development 137, 4171–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost MS, van Iperen L, Ariyurek Y, Buermans HP, Arindrarto W, Devalla HD, Passier R, Mummery CL, Carlotti F, de Koning EJP, et al. (2015). KeyGenes, a Tool to Probe Tissue Differentiation Using a Human Fetal Transcriptional Atlas. Stem Cell Rep 4, 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ZD, Lee K, Yang D, Amin S, Verma N, Li QV, Zhu Z, Soh CL, Kumar R, Evans T, et al. (2017). Genome Editing in hPSCs Reveals GATA6 Haploinsufficiency and a Genetic Interaction with GATA4 in Human Pancreatic Development. Cell Stem Cell 20, 675–688 e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Ajani JA, Honjo S, Maru DM, Chen Q, Scott AW, Heallen TR, Xiao L, Hofstetter WL, Weston B, et al. (2014). Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res 74, 4170–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–U120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault MP, Weinblatt D, Shaverdashvili K, Yang Y, and Katz JP (2016). KLF4 transcriptionally activates non-canonical WNT5A to control epithelial stratification. Sci Rep 6, 26130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A, and DeWitt ND (2016). Pluripotent stem cells progressing to the clinic. Nat Rev Mol Cell Biol 17, 194–200. [DOI] [PubMed] [Google Scholar]
- Wang DH, Tiwari A, Kim ME, Clemons NJ, Regmi NL, Hodges WA, Berman DM, Montgomery EA, Watkins DN, Zhang X, et al. (2014). Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett's metaplasia. J Clin Invest 124, 3767–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ouyang H, Yamamoto Y, Kumar PA, Wei TS, Dagher R, Vincent M, Lu X, Bellizzi AM, Ho KY, et al. (2011). Residual embryonic cells as precursors of a Barrett's-like metaplasia. Cell 145, 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM, Esni F, Boivin GP, Aronow BJ, Stuart W, Combs C, Sklenka A, Leach SD, and Lowy AM (2007). Wnt/beta-catenin signaling is required for development of the exocrine pancreas. Bmc Developmental Biology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, Ratjen F, Ellis J, and Rossant J (2012). Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nature Biotechnology 30, 876–U108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Krebs LT, and Gridley T (2010). Generation of mice with a conditional null allele of the Jagged2 gene. Genesis 48, 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jiang M, Kim E, Lin S, Liu K, Lan X, and Que J (2017). Development and stem cells of the esophagus. Semin Cell Dev Biol 66, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.