Abstract
The transcription factor Gata3 is crucial for the development of several tissues and cell lineages both during development as well as postnatally. This importance is apparent from the early embryonic lethality following germline Gata3 deletion, with embryos displaying a number of phenotypes, and from the fact that Gata3 has been implicated in several cancer types. It often acts at the level of stem and progenitor cells in which it controls the expression of key lineage-determining factors as well as cell cycle genes, thus being one of the main drivers of cell fate choice and tissue morphogenesis. Gata3 is involved at various stages of haematopoiesis both in the adult as well as during development. This review summarizes the various contributions of Gata3 to haematopoiesis with a particular focus on the emergence of the first haematopoietic stem cells in the embryo—a process that appears to be influenced by Gata3 at various levels, thus highlighting the complex nature of Gata3 action.
Keywords: Gata3, haematopoiesis, development
1. Introduction
Haematopoiesis refers to the formation of blood cellular components. All these components originate from multipotent haematopoietic stem cells (HSCs), which form the foundation of this process. The system is traditionally seen as hierarchical, with the multipotent HSC being the mother cell that gives rise to and differentiates into multipotent and unipotent intermediate progenitor cells, resulting in the production of functional mature blood cells, although in recent years exceptions have been discovered that have an HSC-independent origin (reviewed in [1]). As such, HSCs have unique characteristics that in combination distinguish them from other more mature cells: (i) self-renewal ability; (ii) high proliferation ability; (iii) long-term activity; and (iv) potential to differentiate into all the different haematopoietic lineages. All these characteristics make those cells the most clinically relevant cells for transplants.
Haematopoiesis is a complex and intricate process that is governed by a large number of signalling pathways and transcription factors. The transcription factors themselves are often organized in multi-gene families and play essential roles in activating target genes of specific cell fates and in repressing target genes of alternative cell fates. The GATA family of transcription factors are such master regulators. This family has six members in vertebrates, and the disruption of each of the Gata genes, with exception of Gata5, causes embryonic lethality in mice. They are grouped into haematopoietic (Gata1, Gata2, Gata3), and endodermal (Gata4, Gata5, Gata6) subgroups (recently reviewed in [2]). Each GATA transcription factor is highly conserved across vertebrates. For example, Gata3 homologues are found in human, mouse, rat, chimpanzee, dog, chicken, frog and zebrafish. Gata3 shares 97% of its amino acid identity between mouse and human. Within the GATA family, members share varying degrees of homology. For example, while GATA2 and GATA3 are about 55% homologous at the amino acid level, GATA3 and GATA4 are only 20% homologous.
The largest degree of conservation is found in the zinc finger domains, which are about 80% homologous among all six members. The two zinc fingers bind to different sequences and each has a unique function. The C-finger binds to the GATA consensus site, WGATAR, although in a genome-wide ChIP-seq experiment, Fujiwara et al. [3] reported that GATA proteins preferred binding to WGATAA sequences in vivo. However, the abundance of these motifs is such that the binding of GATA factors to specific DNA loci cannot be inferred from the presence of the sequence alone. Instead, it has become clear that occupancy of specific DNA sites by GATA factors is dependent on the presence of other co-regulatory proteins which can facilitate recruitment of GATA factors to specific protein complexes or regulate GATA activity through post-translational modifications (reviewed in [4]).
The N-finger facilitates the interaction with such co-regulatory proteins, but can also bind specific, yet distinct, DNA sequences [3,5–7]. This combination of DNA sequence-specific binding and recruitment to chromatin via a dynamic range of protein complexes resulting in both transcriptional activation and repression allows GATA factors to participate in a large repertoire of different processes that are highly cell context-dependent. In the case of Gata3, this has been extensively studied in lymphoid development, regulated by Gata3 at various stages, which has recently been comprehensively reviewed [8]. In addition, in a crystallographic structural study by Bates et al. [9] for Gata3, it was observed that GATA family members could bind to DNA by either homo- or heterodimerizing, by forming a dimer with other GATA factors through their two C-fingers.
All GATA factors have both distinct and common biological roles and biochemical characteristics, and all have a restricted expression pattern, which is controlled by tissue-specific enhancer elements. The regulatory elements that control Gata3 expression have been well characterized through the identification of enhancers that drive its expression in the urogenital system, the central nervous system, the endocardium and in natural killer (NK)/T-cells, with the NK/T-cell-specific element being located 280 kb downstream of the Gata3 gene [10,11]. However, in some cases, the functions of GATA factors are interchangeable [12]. For example, Gata1, Gata2, Gata3 and Gata4 can activate interleukin-4 (Il4) and Il5 expression in T-cells, which are classically target genes for Gata3, and repress the activation of interferon-γ [13]. Moreover, a Gata3 knock-in can partially rescue erythrocyte defects in Gata1 null mice; however, Gata3 cannot fully rescue the phenotype of Gata1 null mice, indicating that each GATA factor maintains its unique functions [14,15].
2. The three haematopoietic GATAs
While Gata4, Gata5 and Gata6 drive differentiation of mesoderm- and endoderm-derived tissues and are therefore critical for the development of heart and lung, the first three members of the GATA family are involved in the differentiation of mesoderm- and ectoderm-derived tissues and play essential roles in the development and maintenance of the haematopoietic system. Very broadly speaking, the main function of Gata1 is cell fate determination at an early branch point in the haematopoietic tree, where it induces megakaryocyte and erythrocyte development, while preventing granulocyte-monocyte and lymphoid commitment. However, it is also expressed further downstream in common lymphoid and myeloid progenitors, mast cells and eosinophils [16,17]. The most critical role of Gata2 is the formation and maintenance of HSCs [18,19], although it has additional functions in specific blood lineages as discussed below. Gata3 is crucial for the development of several lymphoid lineages (reviewed in [8]) and early definite haematopoietic stem and progenitor cells [20,21], which will be discussed further below.
The haematopoietic group within the GATA factors control each other's expression during development in different cells, and are capable of functioning consecutively during cell specification and lineage commitment in a process called a GATA switch. GATA switch refers to instances where one GATA factor is replaced by another GATA at the chromatin site. GATA switches occur at many functionally critical loci during development, including those that control the expression of regulators of haematopoiesis, such as Gata2 itself [22]. Gata2 is a direct target of Gata1; however, in the absence of Gata1, it can bind to a target region upstream of its own promoter, which activates transcription and induces histone acetylation. However, when Gata1 is expressed, Gata2 is displaced by Gata1 at its chromatin site, which activates erythropoiesis [23,24] (and reviewed in [4,22]).
2.1. Gata1
The essential role of Gata1 in erythropoiesis was demonstrated in Gata1-deficient mice which suffer from early embryonic death (E10.5–11.5) and an inablility to complete primitive and definitive erythroid differentiation [25,26]. Gata1 is expressed in HSCs and common myeloid and/or lymphoid progenitors. It is also crucial for the development of the megakaryocyte lineage [27] and for the survival of erythrocyte precursors [28,29]. Gata1 downregulates cofactors that are necessary for granulocyte–monocyte and lymphoid development, including Spi1 (PU.1), Il7 and Pax5 [30,31], while promoting megakaryocyte and erythrocyte commitment. Gata1 is also expressed in eosinophils and mast cells, where it plays a role in their terminal differentiation [16,17]. Functionally, Gata1 is involved in cell cycle regulation. In the context of erythroid maturation, it was shown to induce G1 arrest by targeting a number of key cell cycle regulators, which allows the cells to undergo maturation, driven by a Gata1-dependent erythroid gene expression programme [32].
2.2. Gata2
Gata2 is a master regulator of haematopoiesis. It is expressed in HSCs, multipotent haematopoietic progenitors, megakaryocytes, erythroid precursors, eosinophils and mast cells. Its deletion leads to embryonic death at E10.5 and a complete disruption of definitive haematopoiesis [33]. This is at the level of HSCs, as Gata2 is required for their emergence (as discussed further below) and their subsequent survival in a dose-dependent fashion [18,19,34]. However, while Gata2 is required for the proliferation and survival of multipotent haematopoietic progenitors and mast cell formation, it is dispensable for the terminal differentiation of erythroid cells and macrophages [35].
2.3. Gata3
Gata3 has been extensively studied in the context of innate and adaptive lymphoid development, where it regulates differentiation and cell fate determination at various levels (for an extensive recent review see [8]). Specifically, it was found to be essential for the development, maintenance, survival and proliferation of early T-cell progenitors, as ES cells with a deleted Gata3 gene were able to contribute to the B-cell, myelomonocytic and erythroid lineages but not thymocytic or T-cell lineages in chimera studies [36]. It was subsequently demonstrated that Gata3 is required to induce and seal the T-cell fate in early lymphoid progenitors, while repressing the B lymphoid programme [37,38]. Within the T-cell lineage, Gata3 is a master regulator of T helper type 2 cells (Th2). It regulates the differentiation of Th2 cells by controlling genes that encode Th2 cytokines Il4, Il5 and Il13 [39]. It appears, however, that Gata3 levels need to be carefully controlled throughout thymocyte development as levels that are too high are cytotoxic, and levels that are too low cause developmental failure [40]. Indeed, enforced expression of Gata3 in the T-cell lineage caused a maturation arrest in the cytotoxic T-cell lineage and promoted the formation of thymic lymphoblastoid tumours [41], and overexpression of Gata3 at early fetal thymocyte stages redirected their differentiation towards the mast cell lineage [42].
Within the branch of innate immunity, Gata3 is central to the development of the recently discovered innate lymphoid cells (ILCs), especially the ILC2 lineage [43,44], a subset of ILC3 cells [45] and a subset of ILC1-derived, tissue-resident NK cells [46,47]. Thus, Gata3 is not necessary for the development of classic (NK) cells, but is crucial for a specialized subset of them. It is important for the terminal differentiation of NK cells and their exit from the bone marrow, and is crucial for the maintenance of liver-resident NK cells [48].
The importance of GATA3 in lymphoid development and function is further highlighted by the fact that GATA3 has also been implicated in T-cell acute lymphoblastic leukaemia (T-ALL). Together, T-cell acute lymphocytic leukaemia 1 (TAL1), RUNX1 and GATA3 form a positive interconnected auto-regulatory loop that directly activates the MYB oncogene, thus reinforcing and stabilizing the oncogenic programme that contributes to malignant transformation [49]. In addition, whole genome sequencing of patients with early T-cell precursor acute lymphoblastic leukaemia (ETP-ALL), an aggressive subtype of T-ALL, has revealed GATA3 inactivating lesions disrupting haematopoietic development [50]. GATA3 has also been linked to other types of lymphoid malignancies. In a genomic profiling study, a GATA3 single-nucleotide polymorphism genotype has been identified in a subtype of childhood acute lymphoblastic leukaemia (ALL), Philadelphia chromosome-positive ALL (Ph-like ALL), that has been associated with early treatment response, higher risk of relapse and overall poor prognosis [51]. And in anaplastic large cell lymphoma, the absence of the GATA3 protein in addition to the presence of suppressive histone (H3K27) trimethylation at the GATA3 promoter suggests epigenetic regulation of GATA3 as a mechanism involved in disease pathogenesis [52].
Gata3 is also highly expressed in the long-term repopulating HSC (LT-HSC) population [53–55]. Using Gata3-null mice (deleted postnatally with Mx1-Cre), Ku et al. [56] have shown that Gata3 deletion results in the production of lower numbers of adult LT-HSCs, and that a lower number of these Gata3-deficient LT-HSCs are in cycle. This suggests that Gata3 is necessary for maintaining normal numbers of LT-HSCs, and that it regulates their entry into the cell cycle. However, by using a conditional Gata3 knockout mouse line crossed to a Vav-Cre line, Buza-Vidas et al. [57] have shown that deletion of Gata3 from HSCs after their emergence in the embryo does not affect the ability of HSCs to expand normally and that their numbers remain unaffected in the bone marrow after birth. Moreover, they reported that Gata3 deletion does not affect the ability of HSCs to self-renew [57].
More recently, Frelin et al. [58] published data suggesting that Gata3 controls the balance of LT-HSC self-renewal and differentiation by regulating their reprogramming from LT-HSC to intermediate term HSCs (IT-HSC). IT-HSCs are important for maintaining blood counts at steady state. They differ from LT-HSCs in that they are able to generate myeloid and erythroid progeny for 12 weeks [53,54,59], are more abundant than LT-HSCs (three times higher) [53,60], are more proliferative, and exit the quiescent state of the cell cycle more frequently than LT-HSCs: every 10–20 days compared to 50–100 days in LT-HSCs [61]. However, the precise function of Gata3 in adult HSCs requires further investigation.
3. Gata3 in development
Gata3 plays a major role in cell lineage specification and development in a variety of cells, tissues and organs during embryogenesis (table 1), including adipocytes [67], kidney [65], mammary gland [69], skin [62,73] and sympathetic nervous system (SNS) [63,71,74]. Not only does the study of Gata3 activity in these different systems provide us with important clues about the molecular details of Gata3 function, it has also demonstrated that due to the close proximity of developing systems in the embryo, Gata3 action in one tissue can influence the development of a neighbouring tissue [20].
Table 1.
Tissue-specific functions of Gata3.
| system | function | reference |
|---|---|---|
| skin and hair | generation of skin selective barrier promotion of progenitor differentiation in the hair follicle and proper hair structure |
[62–64] |
| kidney | nephric duct development | [65,66] |
| fat | inhibition of adipocyte differentiation | [67] |
| inner ear | cochlea morphology | [68] |
| mammary gland | mammary gland morphology | [69,70] |
| haematopoietic system | T-cell development Th2 commitment ILCs NK subsets Cell cycle regulation in adult HSCs non-cell autonomous role in embryonic HSC production |
[36] [39] [43–45] [46–48] [56,58] [20] |
| SNS | essential for the production of catecholamines promotes survival of sympathetic neurons in both adults and embryos |
[71,72] |
3.1. Skin and hair
Gata3 is essential for stem cell lineage determination in the skin. It is expressed at the onset of inner root sheath (IRS) cell specification in hair follicles. When Gata3 was deleted using a LacZ knock-in, IRS progenitors failed to differentiate to form IRS, leading to the production of a defective hair structure [73]. In addition, Gata3 is the most highly expressed member of the GATA family in the interfollicular epidermis. The specific deletion of Gata3 from the epidermal layer, using a keratin-14-Cre (K14-Cre) mouse line, proved to be prenatally lethal due to impairment of the skin selective barrier. Those mice showed defects in skin differentiation, abnormal hair follicle organization and delayed hair growth and maintenance. Genomic analysis of the mice revealed defective lipid biosynthesis. This could be attributed to the loss of lipid acetyltransferase gene (Agpat5), a gene that is a direct target of Gata3 [62,64]. Interestingly, Gata3 has previously been linked to adipogenesis where it was shown to inhibit adipocyte differentiation [67].
3.2. Kidney
Gata3 is the only GATA factor that is expressed in the urogenital system prior to E12.5. It is necessary for the normal development of the nephric duct. Using a HoxB7-Cre transgenic line, Grote et al. [65,66] specifically deleted Gata3 from the nephric duct, which resulted in severe abnormalities in the urogenital system and revealed that Gata3 is required to prevent ectopic metanephric kidney duct formation and premature cell differentiation. Additionally, it was reported that Gata3 haploinsufficiency resulted in renal dysplasia.
Gata3 has also been implicated in clear cell renal cell carcinoma (cc-RCC), the most common subtype of RCC. Cooper et al. [75] uncovered that when Gata3 expression was downregulated by promoter hypermethylation, it resulted in decreased expression of type III TGF-β receptor (TβRIII), which is a betaglycan protein with tumour suppressor features [75].
3.3. Inner ear
Gata3 is widely expressed in several cell types during ear development, including inner hair cells, outer hair cells as well as supportive cells [76–78]. As a consequence, the entire cochlea of the inner ear shows significant degeneration in mice heterozygous for Gata3, which leads to hearing loss [68]. This is mirrored in patients with HDR syndrome, who have only one functional copy of GATA3 and who suffer from hypoparathyroidism, deafness and renal defects [79,80].
3.4. Mammary gland
Gata3 is crucial for mammary gland development. It is the transcription factor with the highest expression in the mammary epithelium as shown by genome-wide transcript analysis [70]. When Gata3 was specifically deleted from the mammary epithelium at the onset of puberty, using the murine mammary tumour virus (MMTV) promoter-Cre recombinase (MMTV-Cre), the mammary glands failed to develop terminal end buds (TEBs), resulting in abnormal ductal structures [69,70].
Its crucial role in the mammary gland is supported by the detection of GATA3 mutations in around 10% of human breast cancers. While the range of somatic mutations is varied, they cluster mainly in the highly conserved C-terminal second zinc finger [81]. Data from in vitro and in vivo studies suggest that GATA3 acts as a tumour suppressor gene. In a murine luminal breast cancer model, the loss of Gata3 resulted in tumour progression and tumour dissemination [82]. More specifically, GATA3 expression was shown to inhibit breast cancer growth and pulmonary metastasis by repressing metastasis-associated genes such as ID1, ID3, KRTHB1, LY6E and RARRES3 [83], and restoration of Gata3 expression in a breast cancer mouse model induced breast cancer differentiation and supressed its dissemination [82]. Moreover, GATA3 was found to promote the expression of microRNA-29b (miR-29b), which in turn induces differentiation, suppresses metastasis and changes the tumour microenvironment [84]. In addition, low expression of GATA3 was associated with a poor survival rate and more aggressive disease, whereas GATA3-expressing breast cancer patients had a better prognosis, were less likely to relapse, and had a better overall survival rate [85]. The involvement of GATA3 in breast cancer, however, is complex as it was also shown to promote the growth of oestrogen-responsive tumours through direct binding to and activation of the oestrogen receptor α (ERα) gene [86]. ERα-positive tumours display a more differentiated phenotype and are generally less aggressive, which may be another reason why GATA3 expression in breast cancer is associated with a more favourable prognosis [87–89].
3.5. Sympathetic nervous system
Gata3 is essential for the development of the SNS [63,71,72,74,90]. In fact, this is the reason why Gata3 deletion is embryonically lethal at around E11.5. This lethality was attributed to noradrenaline deficiency in the SNS and could be pharmacologically rescued by feeding the mothers DOPS, a synthetic catecholamine intermediate [63]. It was subsequently confirmed that Gata3 is essential for the production of catecholamines, the SNS mediators, through controlling the expression of tyrosine hydroxylase, the enzyme required for catecholamine synthesis. It also plays a major role in the survival of sympathetic neurons in both adults and embryos [71,72].
4. Gata3 in haematopoietic stem cell emergence
The process of HSC emergence is highly conserved in vertebrates and is closely linked to vascular development (reviewed in [91,92]). It has been most intensely studied in the intra-embryonic aorta–gonad–mesonephros (AGM) region, where the first directly transplantable HSCs emerge at E10.5 in the mouse [93,94]. Haematopoietic stem and progenitor cell emergence involves activation of a haematopoietic transcriptional programme in a subset of endothelial cells, termed haemogenic endothelial cells, within the major arterial vessels of the embryo, such as the dorsal aorta within the AGM [95–97]. These haemogenic endothelial cells then undergo major structural and morphological changes that allow them to round up and detach from the endothelium as haematopoietic stem and progenitor cells. Localized production of de novo blood cells can be detected in tissue sections as clusters of cells co-expressing endothelial and haematopoietic markers that are attached to the endothelium, and has recently been captured by live imaging [98–100]. This endothelial-to-haematopoietic transition (EHT) is acutely dependent on the transcription factor Runx1 [101]. In its absence, haemogenic endothelial cells undergo apoptosis [100], no intra-aortic clusters are formed [102–104] and the generation of definitive haematopoietic stem and progenitor cells is disrupted [105,106].
The recent development of an elegant ex vivo co-aggregation explant culture system has revealed additional maturation stages that haemogenic endothelial cells have to undergo before they become fully functional HSCs [107–109]. There are at least three intermediate states as haemogenic endothelial cells mature first into pro-HSCs, then type I pre-HSCs and eventually via type II pre-HSCs into adult-type HSCs that can directly repopulate adult recipients. These stages can be distinguished temporally and through the sequential upregulation of the haematopoiesis-associated cell surface markers CD41, CD43 and CD45 [107].
A day after their emergence in the dorsal aorta and associated vitelline and umbilical arteries, HSCs are also detected in the yolk sac and the placenta [93,110–112]. Their presence in these tissues is only temporary as they eventually go on to colonize the fetal liver which becomes the major haematopoietic organ in the embryo from E12.5 until birth when HSCs relocate to the bone marrow.
Gata3 is expressed in the sites of intraembryonic haematopoietic cell generation in the mouse (AGM and its precursor, the para-aortic splanchnopleura) [20,113] (figure 1), zebrafish [115], chicken [116], human [117] and in Xenopus [118,119]. In the mouse embryo, Gata3 is expressed at low level at E8.5 in the splanchnic mesoderm [113]. However, by E11.5 Gata3 is expressed at high levels throughout the embryo. At this stage, Gata3 deletion was embryonically lethal, death occurring concomitantly with the onset of definitive haematopoiesis in the fetal liver [21]. Gata3 knockout embryos were shown to have growth retardation, along with severe deformities in spinal cord and brain, massive internal haemorrhage, anaemia and defective liver haematopoiesis, i.e. definitive haematopoiesis, suggesting that Gata3 is essential for the development of various systems [21]. Yolk sac (YS) haematopoiesis was normal, which corresponds with the fact the Gata3 is not expressed in the YS [113]. Specifically, in an in vitro culture system, the colonies that resulted from the YS of Gata3 knockout embryos, compared to their wild-type and heterozygous littermates, were normal, indicating the maintenance of primitive erythropoiesis. However, the colony numbers from the fetal liver of the knockout embryos were low compared to the wild-type and heterozygous littermates, indicating that Gata3 disruption severely affects definitive haematopoiesis [21].
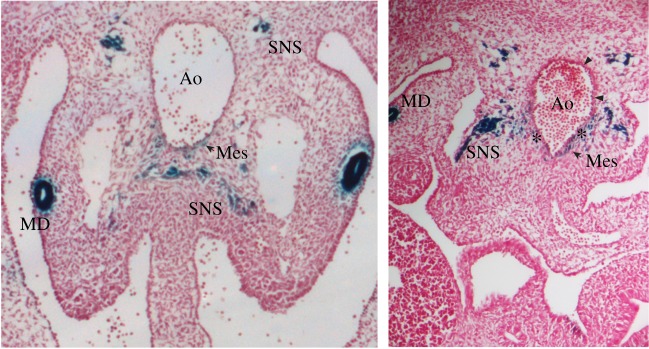
Figure 1.
Gata3 expression in the AGM region. Images show cryosections of Gata3+/Lz [114] E11.5 embryos stained with X-Gal for β-galactosidase activity (blue) and counterstained with Neutral Red. Ao, aorta; MD, mesonephric duct; Mes, mesenchyme; SNS, sympathetic nervous system; asterisks indicate Gata3 staining underneath haematopoietic clusters; arrowheads point to individual Gata3-expressing endothelial cells.
4.1. The sympathetic nervous system as part of the haematopoietic stem cell niche
The observation that definitive haematopoiesis is affected in Gata3−/− embryos [21] and that Gata3 is expressed in the AGM [113] (figure 1) suggested that Gata3 may also be involved in HSC emergence. Furthermore, expression analyses and sorting of specific cell populations followed by transplantation into immunocompromised mice led to the proposal that defined structures located ventrally to the dorsal aorta, termed sub-aortic patches, contain HSCs and/or their precursors and that these expressed Gata3 [113,120]. The cells in these patches were also found to co-express Gata2.
Gata3 was also identified as a potential AGM haematopoiesis regulator in expression profiling studies [121]. This study comprised three comparisons: (i) HSC containing region (middle part of the dorsal aorta) versus a region without HSCs (caudal and rostral part of the aorta); (ii) the microenvironment of HSCs before and after their emergence, i.e. the aorta with the immediate mesenchyme of E9–E10 versus E11; and (iii) using Ly-6A GFP transgenic embryos, which express GFP in all embryonic HSCs and their precursors, populations enriched for HSCs (E11 Ly-6A GFP+ cells) or their precursors (E9 Ly-6A GFP+ cells) were compared. Gata3 was found to be upregulated in two of the three comparisons (ii and iii), i.e. in tissues surrounding the dorsal aorta specifically at the time of HSC emergence in the AGM and in HSC-enriched populations: E11 Ly-6A GFP+ cells.
A role for Gata3 in HSC production in the AGM was subsequently confirmed [20]. Germline-deleted Gata3−/− AGMs contained fewer Ly-6A-GFP+ aortic endothelial cells and showed reduced intra-aortic cluster formation. Most importantly, HSC activity in transplantation assays was severely reduced in Gata3−/− and Gata3+/− AGMs. Interestingly, transplantation of Gata3-LacZ+ and Gata3-Lacz− AGM cells clearly assigned repopulation activity to the Gata3-LacZ− population, strongly suggesting that Gata3 is not expressed in newly formed HSCs, but performs a non-cell autonomous role via the AGM haematopoietic microenvironment [20].
One of the components of the AGM haematopoietic microenvironment turned out to be the co-developing SNS. It had previously been reported that the SNS plays a major role in the mobilization [122,123] and proliferation [124] of adult HSCs. The fact that Gata3 deletion affected both the SNS [63] as well as HSC production [20] in the AGM suggested the intriguing possibility that a functional interplay between the haematopoietic system and SNS already occurred at the time when these first develop during embryogenesis. Indeed, it was then demonstrated that external provision of catecholamines to Gata3-deficient embryos rescued the HSC defect, confirming that Gata3 regulates HSC numbers through catecholamine production [20]. This indicates that HSC emergence in the AGM should be investigated as a part of a whole developmental process that is influenced by neighbouring tissues [20,121]. It also suggests that the previously described sub-aortic patches [113,120] may, in fact, have been cells of the SNS as they are known to co-express Gata3 and Gata2 [20,71] (figures 1 and 2). This does not rule out, however, that there may be individual Gata2 and Gata3 expressing mesenchymal HSC precursors [125].
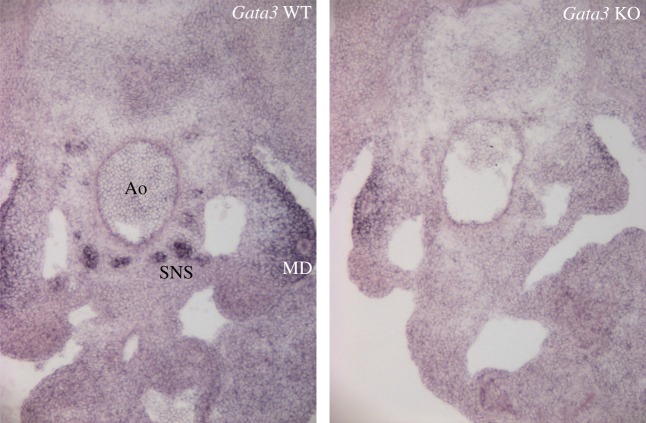
Figure 2.
Gata2 expression in Gata3 wild-type (WT) and knockout (KO) AGMs. Images of in situ hybridization with a Gata2 riboprobe on cryosections from E11.5 Gata3+/+ (left) and Gata3−/− [21] (right) embryos. Ao, aorta; MD, mesonephric duct; SNS, sympathetic nervous system.
4.2. A role for Gata3 in haematopoietic stem cell precursors
In addition to its role in the AGM HSC microenvironment (via the SNS) described above, there is also evidence that Gata3 may contribute to the specification of the definitive haematopoietic lineage. Manaia et al. [113] analysed the expression of Gata3 and Lmo2 in mouse embryonic development to understand the mechanisms involved in the generation of definitive HSCs. Interestingly, they found that Gata3 and Lmo2 are expressed concomitantly in the caudal embryonic mesoderm where haematopoietic cluster-bearing vessels develop, suggesting an involvement in cell fate determination. Another observation of their study was that Gata3 expression is restricted to sites involved in definitive haematopoiesis. Gata3 is expressed in the environment from which intraembryonic precursors emerge, and in the developing haematopoietic sites before their colonization. Gata3 was expressed both in the thymic rudiment until the first migrants arrived, and in the septum transversum before it gives rise to the fetal liver. However, no CD45+ haematopoietic cells were present within those sites at that developmental stage [113].
More recently, Gata3 was also found to be upregulated in a subset of E10.5 endothelial cells that express GFP under the control of the Runx1 + 23 enhancer element (23GFP+) that was described to mark haemogenic endothelial cells [97], indicating a possible involvement in HSC and progenitor generation. In addition, Gata3 expression increased upon Notch1 signalling induction, which expands the haemogenic endothelial population, and enhanced their haematopoietic potential [126]. Indeed, we found Gata3 expression in individual endothelial cells, often in the vicinity of haematopoietic clusters (figure 1); however, further studies are required to determine if Gata3 plays a functional role in haemogenic endothelium and the EHT.
4.3. Gata3 expression in other compartments of the embryonic haematopoietic stem cell microenvironment
Gata3 expression is also found in individual cells in the subaortic mesenchyme of the AGM region (figure 1). Expression in this stromal compartment is restricted to the ventral side of the dorsal aorta, where HSCs are preferentially located [127]. In fact, Gata3-expressing cells were often detected just underneath intra-aortic haematopoietic cell clusters [20]. This mesenchymal expression disappears after E12.5, when intraembryonic clusters cease to be generated [113]. A number of known regulators of HSC emergence are expressed in cells of the sub-aortic ventral mesenchyme including Bmp4, Scf, Runx1, Bmper, Thpo and Dlk1, demonstrating that this cell compartment forms an important embryonic HSC niche [128–133]. Furthermore, cells with mesenchymal stem/stromal cell potential have been detected specifically in the AGM at the time of HSC emergence [134]. The sub-aortic mesenchymal cell compartment, however, is a very heterogeneous population, and it is currently unknown which cells participate in the niche for emerging HSCs and whether this compartment contains and may even be maintained by mesenchymal stem cells. It is also currently not clear whether Gata3 plays an important role in the mesenchymal stroma of the AGM.
Gata3 is expressed in one further cell compartment in the AGM region, the mesonephric ducts within the urogenital ridges (UGRs) [20] (figure 1). It was recently demonstrated that UGRs do not contain HSCs or their precursors, but their presence promotes HSC formation in the neighbouring aorta in co-aggregation studies, indicating that UGRs also form part of the HSC supportive microenvironment [133]. Yet, while it has been established that Gata3 expression in the UGRs is required for kidney development [65], it is currently unknown whether it promotes the production of HSC-supportive factors in these structures. However, the fact that Gata3 is expressed in several different cell types in the AGM relevant to haematopoiesis highlights its complex involvement in the production of the first HSCs.
5. Concluding remarks
Gata3 is essential for the development of several types of cells, organs and tissues (table 1), and its disruption during development results in severe defects and impairment in those systems, leading to embryonic death at midgestation following germline deletion [21]. Analysis of the phenotypes of Gata3 deletion in these different systems has revealed that Gata3 is often expressed in the stem and progenitor compartment where it regulates cell fate determination and differentiation. In the developing kidney [65,66] and preadipocytes [67], it seems to prevent premature differentiation, whereas in skin [73] and mammary gland [69] it promotes progenitor differentiation. Overall, however, its function is to ensure correct tissue morphogenesis. Considering this crucial function, it is therefore not surprising that Gata3 has been implicated in a number of different cancer types.
How Gata3 functions at the molecular level is not well understood in many of these systems. One common underlying theme may be regulation of the cell cycle as has been suggested for the role of Gata3 in adult HSCs [56,58]. Gata3 may allow tissue-specific progenitors to differentiate by blocking their cell cycle, and this may also be how haemogenic endothelial cells can then undergo the morphological changes as they become blood cells. However, it is also clear that Gata3 activates tissue-specific genes such as tyrosine hydroxylase in the SNS and lipid acetyltransferase in the skin. Some of the genes that Gata3 activates during metastasis may also be relevant for the EHT.
GATA factors have often been observed in common complexes and have been seen to cross-regulate each other, with the GATA switch being such an example. Within the AGM, Gata2 and Gata3 expression overlaps in several cell compartments (figures 1 and 2) and they are both involved in HSC production in the AGM, but the nature of their interaction appears to be tissue-specific. Both are expressed in the urogenital ridges, but while Gata3 is expressed specifically in the mesonephric ducts, Gata2 expression is found in the tissue directly surrounding the ducts. Interestingly, however, the expression of Gata2 around the ducts disappears in Gata3−/− embryos [20] (figure 2). Both have been detected in the haemogenic endothelium, but the deletion of Gata2 affects the development of definitive haematopoietic stem and progenitor cells much more severely, suggesting that it may act upstream of Gata3 here or performs a much more crucial function in the EHT. In the SNS, on the other hand, Gata3 is clearly upstream of Gata2, as Gata2 expression in sympathoadrenal cells disappears in Gata3-null embryos, while its expression in the aortic endothelium remains [20] (figure 2).
Very little is currently known about the upstream regulators of Gata3 in the different AGM cell compartments. Phox2b is required for Gata3 expression in the SNS [71]. Considering the crucial roles of Gata2 and Runx1 in the EHT, these two transcription factors may well be upstream of Gata3 in haemogenic endothelial cells. In addition, Notch1 was shown to induce Gata3 in an embryonic stem cell model of the EHT [126]. As Notch1 is also crucial for HSC emergence in the AGM [135], it is likely that Gata3 dependence on Notch signalling is conserved in vivo.
Current data have shown that Gata3 plays several roles in the embryonic and adult haematopoietic system. However, the similarities and differences in these roles require further dissection. For example, in development, Gata3 was shown to regulate HSCs by means of controlling SNS development and the secretion of HSC-supportive catecholamines [20]; however, its expression patterns in the HSC microenvironment suggest a more complex role in haematopoietic stem and progenitor cell regulation (figure 3). In the sub-aortic mesenchymal compartment, Gata3 expression is restricted to a few scattered cells on the ventral side. The fact that it has been associated with the stem and progenitor compartment in several tissues makes it tempting to speculate that its expression marks mesenchymal stem cells. Dissecting its function in each cell type and identifying its role in HSC precursors and the HSC regulatory microenvironment is an intricate process and will require tissue-specific deletion.
Figure 3.
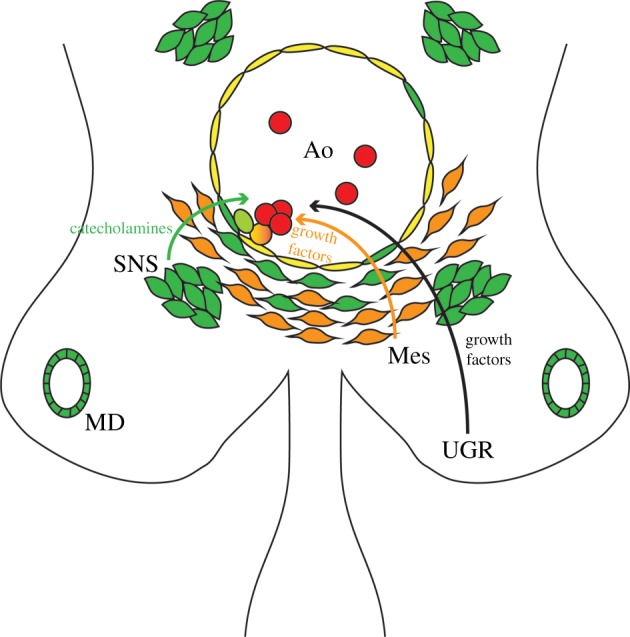
Gata3 involvement in AGM haematopoiesis. Schematic diagram of a transverse section through an E11.5 AGM region, highlighting the cell compartments that express Gata3. Gata3-positive cells (green) are found within the endothelial layer (yellow) of the dorsal aorta (Ao), in the mesonephric duct (MD), within the subaortic mesenchyme (orange; Mes) and in the sympathetic nervous system (SNS). Blood cells are shown in red. The light green cell depicts the putative involvement of Gata3 in the endothelial-to-haematopoietic transition. Curly arrows illustrate contributions made by the different components of the microenvironment to EHT/HSC support, of which only catecholamines are currently known to be Gata3-dependent. UGR, urogenital ridges.
In addition, the scarcity of these cells and the dynamic nature of the developing embryo add more challenges to identifying the role of Gata3 in the emerging definitive haematopoietic system. However, the availability of powerful tools such as RNA-Seq, which can now be performed on a single-cell level, will help to identify the genetic programme of these individual cell types and contribute to a better understanding of the nature and function of these cells and how Gata3 may influence their function. It is, however, very likely that the haematopoietic phenotype described in germline-deleted Gata3-null embryos [20,21] is a compound phenotype resulting from the effects of Gata3 deletion in the various AGM cell types.
Supplementary Material
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
Our Gata3-related work was supported by an Intermediate Fellowship from the Kay Kendall Leukaemia Fund (K.O.) and by a fellowship from the King Abdullah International Medical Research Centre (KAIMRC), Ministry of National Guard (N.Z.).
References
- 1.Dzierzak E, Bigas A. 2018. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell 22, 639–651. ( 10.1016/j.stem.2018.04.015) [DOI] [PubMed] [Google Scholar]
- 2.Lentjes MH, Niessen HE, Akiyama Y, de Bruine AP, Melotte V, van Engeland M. 2016. The emerging role of GATA transcription factors in development and disease. Expert Rev. Mol. Med. 18, e3 ( 10.1017/erm.2016.2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681. ( 10.1016/j.molcel.2009.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katsumura KR, Bresnick EH, Group GFM. 2017. The GATA factor revolution in hematology. Blood 129, 2092–2102. ( 10.1182/blood-2016-09-687871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin DI, Orkin SH. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4, 1886–1898. ( 10.1101/gad.4.11.1886) [DOI] [PubMed] [Google Scholar]
- 6.Newton A, Mackay J, Crossley M. 2001. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem. 276, 35 794–35 801. ( 10.1074/jbc.M106256200) [DOI] [PubMed] [Google Scholar]
- 7.Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. 1996. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol. Cell Biol. 16, 2238–2247. ( 10.1128/MCB.16.5.2238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tindemans I, Serafini N, Di Santo JP, Hendriks RW. 2014. GATA-3 function in innate and adaptive immunity. Immunity 41, 191–206. ( 10.1016/j.immuni.2014.06.006) [DOI] [PubMed] [Google Scholar]
- 9.Bates DL, Chen Y, Kim G, Guo L, Chen L. 2008. Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J. Mol. Biol. 381, 1292–1306. ( 10.1016/j.jmb.2008.06.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosoya-Ohmura S, Lin YH, Herrmann M, Kuroha T, Rao A, Moriguchi T, Lim KC, Hosoya T, Engel JD. 2011. An NK and T cell enhancer lies 280 kilobase pairs 3′ to the gata3 structural gene. Mol. Cell Biol. 31, 1894–1904. ( 10.1128/MCB.05065-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakshmanan G, Lieuw KH, Lim KC, Gu Y, Grosveld F, Engel JD, Karis A. 1999. Localization of distant urogenital system-, central nervous system-, and endocardium-specific transcriptional regulatory elements in the GATA-3 locus. Mol. Cell Biol. 19, 1558–1568. ( 10.1128/MCB.19.2.1558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burch JB. 2005. Regulation of GATA gene expression during vertebrate development. Semin. Cell Dev. Biol. 16, 71–81. ( 10.1016/j.semcdb.2004.10.002) [DOI] [PubMed] [Google Scholar]
- 13.Ranganath S, Murphy KM. 2001. Structure and specificity of GATA proteins in Th2 development. Mol. Cell Biol. 21, 2716–2725. ( 10.1128/MCB.21.8.2716-2725.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi S, et al. 2000. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood 96, 910–916. [PubMed] [Google Scholar]
- 15.Tsai FY, Browne CP, Orkin SH. 1998. Knock-in mutation of transcription factor GATA-3 into the GATA-1 locus: partial rescue of GATA-1 loss of function in erythroid cells. Dev. Biol. 196, 218–227. ( 10.1006/dbio.1997.8842) [DOI] [PubMed] [Google Scholar]
- 16.Harigae H, et al. 1998. Differential roles of GATA-1 and GATA-2 in growth and differentiation of mast cells. Genes Cells 3, 39–50. ( 10.1046/j.1365-2443.1998.00166.x) [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki H, Mizuno S, Wells RA, Cantor AB, Watanabe S, Akashi K. 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity 19, 451–462. ( 10.1016/S1074-7613(03)00242-5) [DOI] [PubMed] [Google Scholar]
- 18.de Pater E, et al. 2013. Gata2 is required for HSC generation and survival. J. Exp. Med. 210, 2843–2850. ( 10.1084/jem.20130751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X, Johnson KD, Chang YI, Boyer ME, Dewey CN, Zhang J, Bresnick EH. 2013. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J. Exp. Med. 210, 2833–2842. ( 10.1084/jem.20130733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitch SR, Kimber GM, Wilson NK, Parker A, Mirshekar-Syahkal B, Gottgens B, Medvinsky A, Dzierzak E, Ottersbach K. 2012. Signaling from the sympathetic nervous system regulates hematopoietic stem cell emergence during embryogenesis. Cell Stem Cell 11, 554–566. ( 10.1016/j.stem.2012.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Engel JD, Lindenbaum MH. 1995. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat. Genet. 11, 40–44. ( 10.1038/ng0995-40) [DOI] [PubMed] [Google Scholar]
- 22.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. 2010. GATA switches as developmental drivers. J. Biol. Chem. 285, 31 087–31 093. ( 10.1074/jbc.R110.159079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl Acad. Sci. USA 100, 8811–8816. ( 10.1073/pnas.1432147100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki M, et al. 2013. GATA factor switching from GATA2 to GATA1 contributes to erythroid differentiation. Genes Cells 18, 921–933. ( 10.1111/gtc.12086) [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl Acad. Sci. USA 93, 12 355–12 358. ( 10.1073/pnas.93.22.12355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349, 257–260. ( 10.1038/349257a0) [DOI] [PubMed] [Google Scholar]
- 27.Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH. 2002. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl Acad. Sci. USA 99, 9237–9242. ( 10.1073/pnas.142302099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiba T, Nagata Y, Kishi A, Sakamaki K, Miyajima A, Yamamoto M, Engel JD, Todokoro K. 1993. Induction of erythroid-specific gene expression in lymphoid cells. Proc. Natl Acad. Sci. USA 90, 11 593–11 597. ( 10.1073/pnas.90.24.11593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. 1999. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood 94, 87–96. [PubMed] [Google Scholar]
- 30.Heavey B, Charalambous C, Cobaleda C, Busslinger M. 2003. Myeloid lineage switch of Pax5 mutant but not wild-type B cell progenitors by C/EBPα and GATA factors. EMBO J. 22, 3887–3897. ( 10.1093/emboj/cdg380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nerlov C, Querfurth E, Kulessa H, Graf T. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95, 2543–2551. [PubMed] [Google Scholar]
- 32.Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ. 2003. GATA-1-mediated proliferation arrest during erythroid maturation. Mol. Cell Biol. 23, 5031–5042. ( 10.1128/MCB.23.14.5031-5042.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226. ( 10.1038/371221a0) [DOI] [PubMed] [Google Scholar]
- 34.Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. 2004. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200, 871–882. ( 10.1084/jem.20031556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai FY, Orkin SH. 1997. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 89, 3636–3643. [PubMed] [Google Scholar]
- 36.Ting CN, Olson MC, Barton KP, Leiden JM. 1996. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384, 474–478. ( 10.1038/384474a0) [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Ojeda ME, Klein Wolterink RG, Lemaitre F, Richard-Le Goff O, Hasan M, Hendriks RW, Cumano A, Di Santo JP. 2013. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood 121, 1749–1759. ( 10.1182/blood-2012-06-440065) [DOI] [PubMed] [Google Scholar]
- 38.Rothenberg EV. 2013. GATA-3 locks the door to the B-cell option. Blood 121, 1673–1674. ( 10.1182/blood-2013-01-477737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee GR, Fields PE, Flavell RA. 2001. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity 14, 447–459. ( 10.1016/S1074-7613(01)00125-X) [DOI] [PubMed] [Google Scholar]
- 40.Ho IC, Tai TS, Pai SY. 2009. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 9, 125–135. ( 10.1038/nri2476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nawijn MC, Ferreira R, Dingjan GM, Kahre O, Drabek D, Karis A, Grosveld F, Hendriks RW. 2001. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J. Immunol. 167, 715–723. ( 10.4049/jimmunol.167.2.715) [DOI] [PubMed] [Google Scholar]
- 42.Taghon T, Yui MA, Rothenberg EV. 2007. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 8, 845–855. ( 10.1038/ni1486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37, 634–648. ( 10.1016/j.immuni.2012.06.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein WRG, Serafini N, van Nimwegen M, Vosshenrich CA, de Bruijn MJ, Fonseca Pereira D, Veiga Fernandes H, Hendriks RW, Di Santo JP. 2013. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc. Natl Acad. Sci. USA 110, 10 240–10 245. ( 10.1073/pnas.1217158110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, Di Santo JP. 2014. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J. Exp. Med. 211, 199–208. ( 10.1084/jem.20131038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samson SI, et al. 2003. GATA-3 promotes maturation, IFN-γ production, and liver-specific homing of NK cells. Immunity 19, 701–711. ( 10.1016/S1074-7613(03)00294-2) [DOI] [PubMed] [Google Scholar]
- 47.Vosshenrich CA, et al. 2006. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 7, 1217–1224. ( 10.1038/ni1395) [DOI] [PubMed] [Google Scholar]
- 48.Ali AK, Oh JS, Vivier E, Busslinger M, Lee SH. 2016. NK cell-specific gata3 ablation identifies the maturation program required for bone marrow exit and control of proliferation. J. Immunol. 196, 1753–1767. ( 10.4049/jimmunol.1501593) [DOI] [PubMed] [Google Scholar]
- 49.Sanda T, et al. 2012. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell 22, 209–221. ( 10.1016/j.ccr.2012.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163. ( 10.1038/nature10725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez-Andreu V, et al. 2013. Inherited GATA3 variants are associated with pH-like childhood acute lymphoblastic leukemia and risk of relapse. Nat. Genet. 45, 1494–1498. ( 10.1038/ng.2803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joosten M, Seitz V, Zimmermann K, Sommerfeld A, Berg E, Lenze D, Leser U, Stein H, Hummel M. 2013. Histone acetylation and DNA demethylation of T cells result in an anaplastic large cell lymphoma-like phenotype. Haematologica 98, 247–254. ( 10.3324/haematol.2011.054619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyam D, Iscove NN. 2010. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell 6, 48–58. ( 10.1016/j.stem.2009.11.014) [DOI] [PubMed] [Google Scholar]
- 54.Kent DG, et al. 2009. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood 113, 6342–6350. ( 10.1182/blood-2008-12-192054) [DOI] [PubMed] [Google Scholar]
- 55.Zhong JF, et al. 2005. Gene expression profile of murine long-term reconstituting vs. short-term reconstituting hematopoietic stem cells. Proc. Natl Acad. Sci. USA 102, 2448–2453. ( 10.1073/pnas.0409459102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ku CJ, Hosoya T, Maillard I, Engel JD. 2012. GATA-3 regulates hematopoietic stem cell maintenance and cell-cycle entry. Blood 119, 2242–2251. ( 10.1182/blood-2011-07-366070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buza-Vidas N, Duarte S, Luc S, Bouriez-Jones T, Woll PS, Jacobsen SE. 2011. GATA3 is redundant for maintenance and self-renewal of hematopoietic stem cells. Blood 118, 1291–1293. ( 10.1182/blood-2011-02-338046) [DOI] [PubMed] [Google Scholar]
- 58.Frelin C, et al. 2013. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat. Immunol. 14, 1037–1044. ( 10.1038/ni.2692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee SJ, Brinkman R, Eaves C. 2007. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 1, 218–229. ( 10.1016/j.stem.2007.05.015) [DOI] [PubMed] [Google Scholar]
- 60.Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, Hock H. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27, 84–90. ( 10.1038/nbt.1517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson A, et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129. ( 10.1016/j.cell.2008.10.048) [DOI] [PubMed] [Google Scholar]
- 62.Kurek D, Garinis GA, van Doorninck JH, van der Wees J, Grosveld FG. 2007. Transcriptome and phenotypic analysis reveals Gata3-dependent signalling pathways in murine hair follicles. Development 134, 261–272. ( 10.1242/dev.02721) [DOI] [PubMed] [Google Scholar]
- 63.Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. 2000. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat. Genet. 25, 209–212. ( 10.1038/76080) [DOI] [PubMed] [Google Scholar]
- 64.de Guzman Strong C, et al. 2006. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J. Cell Biol. 175, 661–670. ( 10.1083/jcb.200605057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grote D, Souabni A, Busslinger M, Bouchard M. 2006. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development 133, 53–61. ( 10.1242/dev.02184) [DOI] [PubMed] [Google Scholar]
- 66.Grote D, Boualia SK, Souabni A, Merkel C, Chi X, Costantini F, Carroll T, Bouchard M. 2008. Gata3 acts downstream of β-catenin signaling to prevent ectopic metanephric kidney induction. PLoS Genet. 4, e1000316 ( 10.1371/journal.pgen.1000316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290, 134–138. ( 10.1126/science.290.5489.134) [DOI] [PubMed] [Google Scholar]
- 68.van der Wees J, et al. 2004. Hearing loss following Gata3 haploinsufficiency is caused by cochlear disorder. Neurobiol Dis. 16, 169–178. ( 10.1016/j.nbd.2004.02.004) [DOI] [PubMed] [Google Scholar]
- 69.Asselin-Labat ML, et al. 2007. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209. ( 10.1038/ncb1530) [DOI] [PubMed] [Google Scholar]
- 70.Kouros-Mehr H, Werb Z. 2006. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev. Dyn. 235, 3404–3412. ( 10.1002/dvdy.20978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsarovina K, Pattyn A, Stubbusch J, Muller F, van der Wees J, Schneider C, Brunet JF, Rohrer H. 2004. Essential role of Gata transcription factors in sympathetic neuron development. Development 131, 4775–4786. ( 10.1242/dev.01370) [DOI] [PubMed] [Google Scholar]
- 72.Tsarovina K, Reiff T, Stubbusch J, Kurek D, Grosveld FG, Parlato R, Schutz G, Rohrer H. 2010. The Gata3 transcription factor is required for the survival of embryonic and adult sympathetic neurons. J. Neurosci. 30, 10 833–10 843. ( 10.1523/JNEUROSCI.0175-10.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, Lim KC, Dai X, Alegre ML, Fuchs E. 2003. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 17, 2108–2122. ( 10.1101/gad.1115203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moriguchi T, et al. 2006. Gata3 participates in a complex transcriptional feedback network to regulate sympathoadrenal differentiation. Development 133, 3871–3881. ( 10.1242/dev.02553) [DOI] [PubMed] [Google Scholar]
- 75.Cooper SJ, et al. 2010. Loss of type III transforming growth factor-β receptor expression is due to methylation silencing of the transcription factor GATA3 in renal cell carcinoma. Oncogene 29, 2905–2915. ( 10.1038/onc.2010.64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, de Caprona D, Fritzsch B. 2001. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J. Comp. Neurol. 429, 615–630. ( 10.1002/1096-9861(20010122)429:4%3C615::AID-CNE8%3E3.0.CO;2-F) [DOI] [PubMed] [Google Scholar]
- 77.Lawoko-Kerali G, Rivolta MN, Holley M. 2002. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J. Comp. Neurol. 442, 378–391. ( 10.1002/cne.10088) [DOI] [PubMed] [Google Scholar]
- 78.Rivolta MN, Holley MC. 1998. GATA3 is downregulated during hair cell differentiation in the mouse cochlea. J. Neurocytol. 27, 637–647. ( 10.1023/A:1006951813063) [DOI] [PubMed] [Google Scholar]
- 79.Muroya K, et al. 2001. GATA3 abnormalities and the phenotypic spectrum of HDR syndrome. J. Med. Genet. 38, 374–380. ( 10.1136/jmg.38.6.374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Esch H, et al. 2000. GATA3 haplo-insufficiency causes human HDR syndrome. Nature 406, 419–422. ( 10.1038/35019088) [DOI] [PubMed] [Google Scholar]
- 81.Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. ( 10.1038/nature11412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kouros-Mehr H, Bechis SK, Slorach EM, Littlepage LE, Egeblad M, Ewald AJ, Pai SY, Ho IC, Werb Z. 2008. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell 13, 141–152. ( 10.1016/j.ccr.2008.01.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dydensborg AB, Rose AA, Wilson BJ, Grote D, Paquet M, Giguere V, Siegel PM, Bouchard M. 2009. GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 28, 2634–2642. ( 10.1038/onc.2009.126) [DOI] [PubMed] [Google Scholar]
- 84.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. 2013. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 15, 201–213. ( 10.1038/ncb2672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oh DS, Troester MA, Usary J, Hu Z, He X, Fan C, Wu J, Carey LA, Perou CM. 2006. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J. Clin. Oncol. 24, 1656–1664. ( 10.1200/JCO.2005.03.2755) [DOI] [PubMed] [Google Scholar]
- 86.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res. 67, 6477–6483. ( 10.1158/0008-5472.CAN-07-0746) [DOI] [PubMed] [Google Scholar]
- 87.Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D, Chinnaiyan AM, Kleer CG. 2005. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 65, 11 259–11 264. ( 10.1158/0008-5472.CAN-05-2495) [DOI] [PubMed] [Google Scholar]
- 88.Parikh P, Palazzo JP, Rose LJ, Daskalakis C, Weigel RJ. 2005. GATA-3 expression as a predictor of hormone response in breast cancer. J. Am. Coll. Surg. 200, 705–710. ( 10.1016/j.jamcollsurg.2004.12.025) [DOI] [PubMed] [Google Scholar]
- 89.Sorlie T, et al. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10 869–10 874. ( 10.1073/pnas.191367098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hong SJ, Choi HJ, Hong S, Huh Y, Chae H, Kim KS. 2008. Transcription factor GATA-3 regulates the transcriptional activity of dopamine β-hydroxylase by interacting with Sp1 and AP4. Neurochem. Res. 33, 1821–1831. ( 10.1007/s11064-008-9639-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ciau-Uitz A, Monteiro R, Kirmizitas A, Patient R. 2014. Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp. Hematol. 42, 669–683. ( 10.1016/j.exphem.2014.06.001) [DOI] [PubMed] [Google Scholar]
- 92.Medvinsky A, Rybtsov S, Taoudi S. 2011. Embryonic origin of the adult hematopoietic system: advances and questions. Development 138, 1017–1031. ( 10.1242/dev.040998) [DOI] [PubMed] [Google Scholar]
- 93.Medvinsky A, Dzierzak E. 1996. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906. ( 10.1016/S0092-8674(00)80165-8) [DOI] [PubMed] [Google Scholar]
- 94.Muller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. 1994. Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1, 291–301. ( 10.1016/1074-7613(94)90081-7) [DOI] [PubMed] [Google Scholar]
- 95.Li Y, et al. 2014. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28, 2597–2612. ( 10.1101/gad.253302.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Solaimani KP, et al. 2015. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J. Exp. Med. 212, 93–106. ( 10.1084/jem.20140767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Swiers G, et al. 2013. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 4, 2924 ( 10.1038/ncomms3924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. 2010. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111. ( 10.1038/nature08738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120. ( 10.1038/nature08764) [DOI] [PubMed] [Google Scholar]
- 100.Kissa K, Herbomel P. 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115. ( 10.1038/nature08761) [DOI] [PubMed] [Google Scholar]
- 101.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891. ( 10.1038/nature07619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing RJ, Dzierzak E. 2000. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13, 423–431. ( 10.1016/S1074-7613(00)00042-X) [DOI] [PubMed] [Google Scholar]
- 103.North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126, 2563–2575. [DOI] [PubMed] [Google Scholar]
- 104.Yokomizo T, Dzierzak E. 2010. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development 137, 3651–3661. ( 10.1242/dev.051094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330. ( 10.1016/S0092-8674(00)80986-1) [DOI] [PubMed] [Google Scholar]
- 106.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA 93, 3444–3449. ( 10.1073/pnas.93.8.3444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rybtsov S, Batsivari A, Bilotkach K, Paruzina D, Senserrich J, Nerushev O, Medvinsky A. 2014. Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43− embryonic precursor. Stem Cell Reports 3, 489–501. ( 10.1016/j.stemcr.2014.07.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rybtsov S, et al. 2011. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med. 208, 1305–1315. ( 10.1084/jem.20102419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taoudi S, Gonneau C, Moore K, Sheridan JM, Blackburn CC, Taylor E, Medvinsky A. 2008. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-Cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell 3, 99–108. ( 10.1016/j.stem.2008.06.004) [DOI] [PubMed] [Google Scholar]
- 110.de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. 2000. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19, 2465–2474. ( 10.1093/emboj/19.11.2465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. 2005. The placenta is a niche for hematopoietic stem cells. Dev. Cell 8, 365–375. ( 10.1016/j.devcel.2004.12.016) [DOI] [PubMed] [Google Scholar]
- 112.Ottersbach K, Dzierzak E. 2005. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev. Cell 8, 377–387. ( 10.1016/j.devcel.2005.02.001) [DOI] [PubMed] [Google Scholar]
- 113.Manaia A, Lemarchandel V, Klaine M, Max-Audit I, Romeo P, Dieterlen-Lievre F, Godin I. 2000. Lmo2 and GATA-3 associated expression in intraembryonic hemogenic sites. Development 127, 643–653. [DOI] [PubMed] [Google Scholar]
- 114.van Doorninck JH, van Der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M, Grosveld F, De Zeeuw CI. 1999. GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei. J. Neurosci. 19, RC12 ( 10.1523/JNEUROSCI.19-12-j0002.1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Neave B, Rodaway A, Wilson SW, Patient R, Holder N. 1995. Expression of zebrafish GATA 3 (gta3) during gastrulation and neurulation suggests a role in the specification of cell fate. Mech. Dev. 51, 169–182. ( 10.1016/0925-4773(95)00351-7) [DOI] [PubMed] [Google Scholar]
- 116.Leonard MW, Lim KC, Engel JD. 1993. Expression of the chicken GATA factor family during early erythroid development and differentiation. Development 119, 519–531. [DOI] [PubMed] [Google Scholar]
- 117.Labastie MC, Cortes F, Romeo PH, Dulac C, Peault B. 1998. Molecular identity of hematopoietic precursor cells emerging in the human embryo. Blood 92, 3624–3635. [PubMed] [Google Scholar]
- 118.Bertwistle D, Walmsley ME, Read EM, Pizzey JA, Patient RK. 1996. GATA factors and the origins of adult and embryonic blood in Xenopus: responses to retinoic acid. Mech. Dev. 57, 199–214. ( 10.1016/0925-4773(96)00547-3) [DOI] [PubMed] [Google Scholar]
- 119.Turpen JB, Kelley CM, Mead PE, Zon LI. 1997. Bipotential primitive-definitive hematopoietic progenitors in the vertebrate embryo. Immunity 7, 325–334. ( 10.1016/S1074-7613(00)80354-4) [DOI] [PubMed] [Google Scholar]
- 120.Bertrand JY, Giroux S, Golub R, Klaine M, Jalil A, Boucontet L, Godin I, Cumano A. 2005. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc. Natl Acad. Sci. USA 102, 134–139. ( 10.1073/pnas.0402270102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mascarenhas MI, Parker A, Dzierzak E, Ottersbach K. 2009. Identification of novel regulators of hematopoietic stem cell development through refinement of stem cell localization and expression profiling. Blood 114, 4645–4653. ( 10.1182/blood-2009-06-230037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Katayama Y, Battista M, Kao WM, Hidalgo A, Peired AJ, Thomas SA, Frenette PS. 2006. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell 124, 407–421. ( 10.1016/j.cell.2005.10.041) [DOI] [PubMed] [Google Scholar]
- 123.Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. 2008. Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447. ( 10.1038/nature06685) [DOI] [PubMed] [Google Scholar]
- 124.Spiegel A, et al. 2007. Catecholaminergic neurotransmitters regulate migration and repopulation of immature human CD34+ cells through Wnt signaling. Nat. Immunol. 8, 1123–1131. ( 10.1038/ni1509) [DOI] [PubMed] [Google Scholar]
- 125.Zovein AC, et al. 2008. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3, 625–636. ( 10.1016/j.stem.2008.09.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jang IH, et al. 2015. Notch1 acts via Foxc2 to promote definitive hematopoiesis via effects on hemogenic endothelium. Blood 125, 1418–1426. ( 10.1182/blood-2014-04-568170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Taoudi S, Medvinsky A. 2007. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc. Natl Acad. Sci. USA 104, 9399–9403. ( 10.1073/pnas.0700984104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Durand C, Robin C, Bollerot K, Baron MH, Ottersbach K, Dzierzak E. 2007. Embryonic stromal clones reveal developmental regulators of definitive hematopoietic stem cells. Proc. Natl Acad. Sci. USA 104, 20 838–20 843. ( 10.1073/pnas.0706923105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mascarenhas MI, Bacon WA, Kapeni C, Fitch SR, Kimber G, Cheng SW, Li J, Green AR, Ottersbach K. 2016. Analysis of Jak2 signaling reveals resistance of mouse embryonic hematopoietic stem cells to myeloproliferative disease mutation. Blood 127, 2298–2309. ( 10.1182/blood-2015-08-664631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McGarvey AC, et al. 2017. A molecular roadmap of the AGM region reveals BMPER as a novel regulator of HSC maturation. J. Exp. Med. 214, 3731–3751. ( 10.1084/jem.20162012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mirshekar-Syahkal B, et al. 2013. Dlk1 is a negative regulator of emerging hematopoietic stem and progenitor cells. Haematologica 98, 163–171. ( 10.3324/haematol.2012.070789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Peeters M, Ottersbach K, Bollerot K, Orelio C, de Bruijn M, Wijgerde M, Dzierzak E. 2009. Ventral embryonic tissues and Hedgehog proteins induce early AGM hematopoietic stem cell development. Development 136, 2613–2621. ( 10.1242/dev.034728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Souilhol C, et al. 2016. Inductive interactions mediated by interplay of asymmetric signalling underlie development of adult haematopoietic stem cells. Nat. Commun. 7, 10784 ( 10.1038/ncomms10784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mendes SC, Robin C, Dzierzak E. 2005. Mesenchymal progenitor cells localize within hematopoietic sites throughout ontogeny. Development 132, 1127–1136. ( 10.1242/dev.01615) [DOI] [PubMed] [Google Scholar]
- 135.Kumano K, et al. 2003. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity 18, 699–711. ( 10.1016/S1074-7613(03)00117-1) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.