Abstract
Mutations in PINK1 and Parkin result in autosomal recessive Parkinson's disease (PD). Cell culture and in vitro studies have elaborated the PINK1-dependent regulation of Parkin and defined how this dyad orchestrates the elimination of damaged mitochondria via mitophagy. PINK1 phosphorylates ubiquitin at serine 65 (Ser65) and Parkin at an equivalent Ser65 residue located within its N-terminal ubiquitin-like domain, resulting in activation; however, the physiological significance of Parkin Ser65 phosphorylation in vivo in mammals remains unknown. To address this, we generated a Parkin Ser65Ala (S65A) knock-in mouse model. We observe endogenous Parkin Ser65 phosphorylation and activation in mature primary neurons following mitochondrial depolarization and reveal this is disrupted in ParkinS65A/S65A neurons. Phenotypically, ParkinS65A/S65A mice exhibit selective motor dysfunction in the absence of any overt neurodegeneration or alterations in nigrostriatal mitophagy. The clinical relevance of our findings is substantiated by the discovery of homozygous PARKIN (PARK2) p.S65N mutations in two unrelated patients with PD. Moreover, biochemical and structural analysis demonstrates that the ParkinS65N/S65N mutant is pathogenic and cannot be activated by PINK1. Our findings highlight the central role of Parkin Ser65 phosphorylation in health and disease.
Keywords: mitochondria; mitophagy; neurodegeneration; Parkin, Parkinson's disease; PINK1
1. Introduction
Mutations in genes encoding PTEN-induced kinase 1 (PINK1) (human PARK6) and the ubiquitin E3 ligase Parkin (human PARK2) are causal for early onset Parkinson's disease (PD) with clinically indistinguishable phenotypes [1,2]. Patients typically exhibit a motor syndrome associated with nigrostriatal pathology that can occur in the absence or the presence of Lewy body inclusions [3]. Under specific conditions of mitotoxic stress in cultured cells, PINK1 and Parkin converge in a common signal transduction pathway to eliminate damaged mitochondria via autophagy, known as mitophagy [4–6]. Over the last decade, this pathway has been intensively studied to understand the mechanisms by which these enzymes regulate mitochondrial homeostasis [4–6]. In cell culture, PINK1 is activated upon mitochondrial depolarization that can be induced using mitochondrial uncoupling agents, e.g. antimycin A/oligomycin (A/O). This results in the recruitment and activation of Parkin at the outer mitochondrial membrane (OMM) [4–6]. PINK1 directly phosphorylates Parkin at a highly conserved serine 65 (Ser65) residue within its N-terminal ubiquitin-like domain (Ubl), as well as the equivalent Ser65 residue of ubiquitin (Ub). Phosphorylation of both Parkin and ubiquitin is required for maximal activation of Parkin E3 ligase activity [7–11]. Upon activation, Parkin ubiquitylates multiple substrates at the OMM leading to both de novo assembly and elongation of existing ubiquitin chains that are, in turn, phosphorylated by PINK1. Together, this generates a feed-forward enhancement of Parkin activation and mitochondrial ubiquitylation that heralds the recruitment of selective autophagy adaptors necessary for the completion of mitophagy [12–15].
Our present understanding of PINK1-dependent Parkin activation is largely predicated on in vitro observations and cell culture studies, which often exploit the over-expression of exogenous Parkin and/or PINK1 at supra-physiological levels [4–6]. These include the previous demonstration that Parkin Ser65 phosphorylation may not be essential for its complete E3 ligase activity [8,12] or its depolarization-induced mitochondrial translocation [7,10]. Despite a concerted body of work in this area from many laboratories, the physiological significance of Parkin Ser65 phosphorylation by PINK1 in vivo remains largely enigmatic. Furthermore, additional substrates for PINK1 have been widely reported [16] but, to date, it is unknown whether inactivation of PINK1-dependent substrate phosphorylation is sufficient to recapitulate neurodegeneration in vivo. Models of germ-line Parkin knockout mice only exhibit mild phenotypes, with chronic mitotoxicity or extreme stress required to elicit PD-relevant neurological phenotypes [17–21]. To test the hypothesis that Parkin Ser65 phosphorylation is critical for its activation in vivo, we generated a Park2 Ser65Ala knock-in mouse (ParkinS65A/S65A) and performed an extensive multi-parametric characterization of this model. We demonstrate that phosphorylation of Ser65 is critical for endogenous Parkin activation and Ser65-phosphorylated ubiquitin (hereafter referred to as phospho-ubiquitin) accumulation in primary cells, including neurons. Loss of endogenous Parkin Ser65 phosphorylation and activity results in selective locomotor impairments, accompanied by a mild striatal-specific mitochondrial defect in vivo. Furthermore, crossing our ParkinS65A/S65A mice with the recently described mito-QC mitophagy reporter model [22] revealed that basal mitophagy in nigrostriatal dopamine neurons proceeds normally in ParkinS65A/S65A mice, suggesting that the regulation of basal mitophagy is unaffected by the loss of Parkin E3 ligase activity in dopaminergic neurons. The pathophysiological significance of Parkin Ser65 phosphorylation in mammals is ultimately substantiated by the clinical discovery of two unrelated patients with relatively early onset PD harbouring a homozygous PARK2 Ser65Asn (ParkinS65N) mutation. Characterization of patient-derived primary cells demonstrates that the Parkin S65N mutation is inactive, suggesting that the loss of PINK1-dependent Parkin Ser65 phosphorylation and subsequent inactivation in humans is sufficient to cause PD. Taken together, our data demonstrate that the phosphorylation state of Parkin at Ser65 is crucial for mitochondrial integrity and human nigrostriatal function.
2. Results
2.1. Endogenous Parkin activation is abolished in ParkinS65A/S65A knock-in mice
To test the hypothesis that Parkin Ser65 phosphorylation is central to Parkin activation and neuronal homeostasis in vivo, we generated a knock-in mouse model in which the codon encoding Parkin Ser65 was altered by homologous recombination to alanine to prevent Parkin protein phosphorylation by PINK1 (figure 1a). Homozygous ParkinS65A/S65A mice were born at expected Mendelian frequencies and wild-type and mutant mice were indistinguishable in terms of gross measures of growth, weight and development (data not shown).
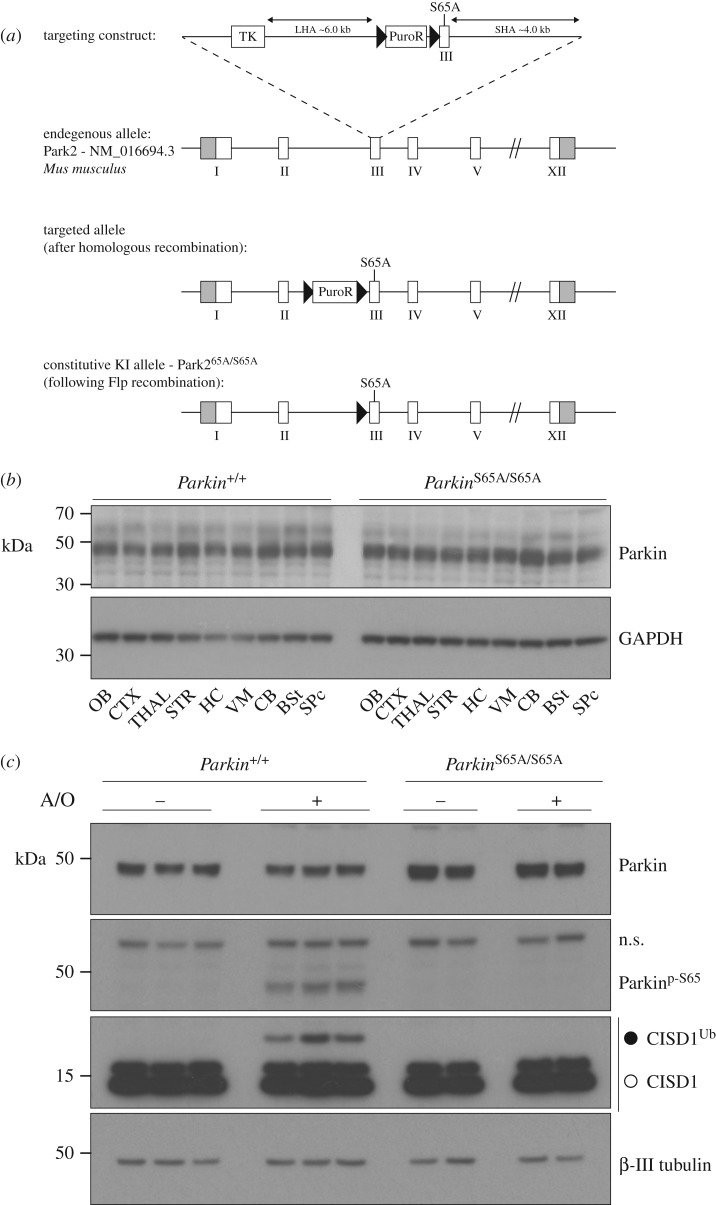
Figure 1.
Generation and validation of the ParkinS65A/S65A mouse. (a) Depiction of targeting strategy to generate a constitutive knock-in of Ser65Ala (S65A) point mutation in the Mus musculus Park2 (Parkin) gene (targeting strategy based on NCBI transcript NM_016694.3). Exon 1 contains the translation initiation codon. The S65A mutation was introduced into exon 3. The Park2S65A/S65A knock-in (KI) allele was generated following Flp-mediated recombination. (b) Rostro-caudal expression analysis of Parkin protein expression in the mouse central nervous system (CNS) under basal conditions. Immunoblot analysis of sub-dissected CNS regions from adult wild-type and ParkinS65A/S65A mice. OB, olfactory bulb; CTX, neocortex; THAL, thalamus; STR, striatum; HC, hippocampus; VM, ventral midbrain; CB, cerebellum; BSt, brainstem; SPc, spinal cord. (c) Parkin Ser65 phosphorylation is essential for Parkin activation in primary neurons. Mature (21 DIV) primary cortical neuron cultures were established from wild-type and ParkinS65A/S65A mice, and stimulated for 3 h with a combination of antimycin A (10 µM)/oligomycin (1 µM). Whole-cell extracts were subjected to SDS-PAGE and immunoblot analysis with anti-Parkin, anti-phospho-Ser65 Parkin, anti-CISD1 and anti-β-III tubulin antibodies.
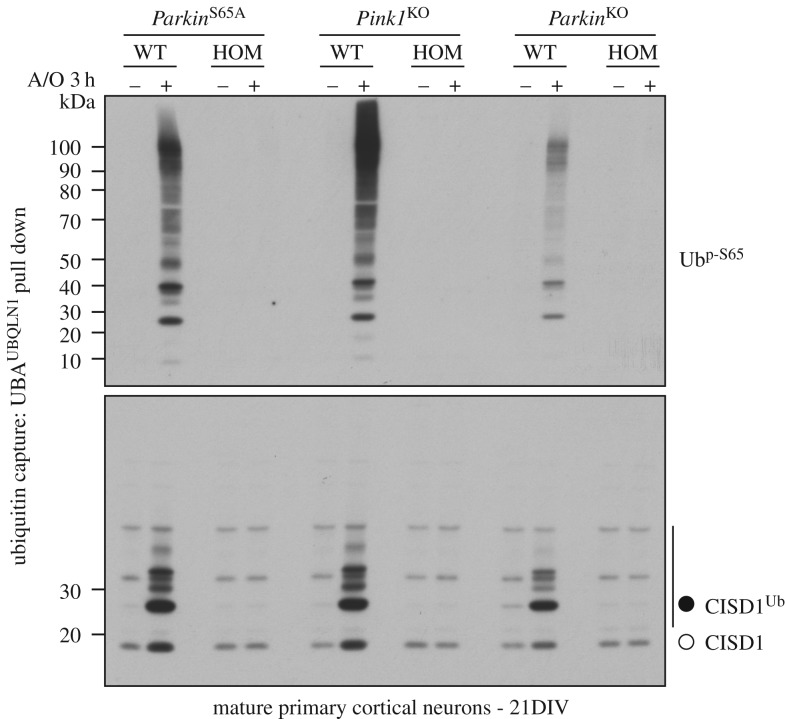
The expression and stability of the endogenous mutant ParkinS65A protein in ParkinS65A/S65A mice was comparable to wild-type mice across all brain regions (olfactory bulb, cortex, thalamus, striatum, hippocampus, ventral midbrain, cerebellum, brainstem and spinal cord) and extra-neural tissues (heart, spleen, lung and pancreas) (figure 1b and electronic supplementary material, figure S1). To assess how the S65A mutation affects Parkin ubiquitin E3 ligase activity, we undertook biochemical analysis of endogenous substrate ubiquitylation in cultured primary neurons. Given the availability of reliable reagents and building upon our previous work in this area [11,23], we focused our attention on the mitochondrial Fe/S domain-containing protein CISD1, a well-described Parkin substrate, in addition to monitoring endogenous PINK1-mediated Parkin Ser65 phosphorylation. We have previously observed that endogenous Parkin is expressed at low levels in mouse embryonic fibroblasts (MEFs) which limits the robust detection of endogenous Parkin signalling [23]. However, in primary cortical neuron cultures established from E16.5 mouse embryos cultured to maturity for 21 days in vitro (DIV), endogenous Parkin phosphorylation and substrate ubiquitylation are reliably detectable after combined antimycin A and oligomycin stimulation (to trigger mitochondrial depolarization; hereafter referred to as A/O) [24]. Upon treatment of mature (21 DIV) cortical neurons with A/O for three hours (3 h), we observed complete loss of CISD1 ubiquitylation in ParkinS65A/S65A neurons compared to wild-type, as measured by anti-CISD1 immunoblotting (figure 1c). Immunoblot analysis using anti-phospho-Ser65 Parkin antibodies confirmed the loss of Parkin phosphorylation at Ser65 in mutant neurons (figure 1c). We also assessed endogenous ubiquitylation status of additional Parkin substrates including Mitofusin2, Miro2 and VDAC1; however, we were unable to detect robust signal in neurons in whole-cell lysates or following HALO-Ubiquilin1 UBA-domain tetramer (UBAUBQLN1) TUBE-pulldown experiments (electronic supplementary material, figure S2). This may be due to their low stoichiometry of ubiquitylation, demonstrated recently in human neurons by quantitative mass spectrometry [25]. We next asked whether loss of Parkin activity in ParkinS65A/S65A neurons influenced the PINK1-dependent phosphorylation of ubiquitin at Ser65 and its subsequent accumulation. We undertook comparative biochemical analyses of mature primary neuron cultures established from homozygous ParkinS65A/S65A, Pink1 knockout and Parkin knockout mice with their respective corresponding wild-type littermate controls. Consistent with previous studies in proliferating cell lines, we observed complete loss of Ser65-phosphorylated ubiquitin (phospho-ubiquitin) and CISD1 ubiquitylation in Pink1 knockout neurons, as judged by immunoblotting of HALO-UBAUBQLN1 pulldowns with anti-phospho-Ser65 ubiquitin and anti-CISD1 antibodies (figure 2). Strikingly, we did not observe phospho-ubiquitin accumulation in either ParkinS65A/S65A or Parkin knockout neurons that were associated with loss of CISD1 substrate ubiquitylation (figure 2). These data support the Parkin-dependent feed-forward model of phospho-ubiquitin accumulation [12,15]. Similar findings were recently reported in CRISPR/Cas9-mediated S65A mutant human embryonic stem (ES) cell-derived neurons stimulated with A/O in which phospho-ubiquitin was not detectable by immunoblotting, although the authors were able to detect phospho-ubiquitin by mass spectrometry [25]. To confirm this finding in other primary mouse cell types, we exploited a tractable culture paradigm of mouse primary adult lung fibroblast cultures [26]. We stimulated adult primary fibroblast cultures from ParkinS65A/S65A and Pink1 knockout mice with the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) for 18 h, which dissipates mitochondrial membrane potential similar to A/O. As Parkin protein levels were lower in primary lung fibroblasts compared with neurons, we performed an additional mitochondrial enrichment step prior to ubiquitin capture by HALO-UBAUBQLN1 pulldown. Consistent with our results in neurons, we did not observe the accumulation of phospho-ubiquitin or CISD1 ubiquitylation in either ParkinS65A/S65A or Pink1 knockout fibroblasts compared with wild-type cultures (electronic supplementary material, figure S3). Overall, our data indicate that phosphorylation of Parkin Ser65 by PINK1 is essential for both activation of its E3 ligase activity and the robust generation of phospho-ubiquitin in mature mammalian neurons.
Figure 2.
Phospho-ubiquitin levels are diminished in ParkinS65A/S65A mature primary neurons. Immunoblots showing comparative analysis of phospho-Ser65 ubiquitin levels in mature (21 DIV) primary cortical neuron cultures from the following groups: ParkinS65A/S65A mice and matched wild-type controls; Pink1 knockout (KO) and wild-type mice and Parkin knockout and wild-type mice. Cultures were stimulated with A/O for 3 h before lysis and membrane enrichment. Protein extracts were enriched for ubiquitylated substrates by incubating with ubiquitin-binding resin derived from His-Halo-Ubiquilin UBA-domain tetramer (HALO-UBAUBQLN1). Enriched lysates were subjected to immunoblotting with anti-phospho-Ser65 ubiquitin and anti-CISD1 antibodies.
2.2. ParkinS65A/S65A knock-in mice exhibit selective motor impairments
Since our ParkinS65A/S65A mice have been engineered with an inactivating point mutation in the endogenous Park2 locus and express comparable levels of Parkin protein, we speculated that our model may bypass epistatic compensation induced by the loss of total protein in traditional knockout models. We therefore undertook a comprehensive behavioural characterization of our ParkinS65A/S65A mice to investigate if the loss of endogenous Parkin phosphorylation could influence striatal function in vivo. Overall, ParkinS65A/S65A mice exhibited normal gross development and ageing in terms of neurological and behavioural function. We employed the raised balance beam as a more sensitive and powerful test of voluntary locomotor function, balance and coordinated limb use in our mice. Strikingly, we observed that ParkinS65A/S65A mice at 12 and 18 months of age exhibited consistent impairments in all measures of balance beam performance compared with wild-type control mice (figure 3a). Both the latency to turn towards the goal box when placed on the beam end (AGE × GENO: F1,32 = 11.41, p < 0.01), and the subsequent latency to cross the beam (AGE × GENO × SEX: F1,32 = 12.82, p < 0.01) were slower for ParkinS65A/S65A mice compared with wild-type controls, with mutant males being slower to traverse the beam than their female counterparts (figure 3a). Furthermore, quantitation of foot-slips while traversing the balance beam demonstrated that the number of forelimb (GENO: F1,47 = 29.08, p < 0.01) or hindlimb (GENO: F1,47 = 38.52, p < 0.01) slips made by ParkinS65A/S65A mice was greater than controls (figure 3a), but this phenotype was not exacerbated at 18 months. In addition to deficits in motor function, the impairments observed in ParkinS65A/S65A mice to complete the task, particularly during the ‘beam turn’ aspect, could also indicate an underlying, yet subtle defect in cognitive planning (figure 3a). Rotarod testing at 12 and 18 months of age did not reveal any substantial differences between ParkinS65A/S65A and wild-type mice (AGE × GENO: p = 0.058) (electronic supplementary material, figure S4). Additionally, Parkin mutant mice did not exhibit any impairments in gait in comparison to wild-type mice in terms of stride length and stride width (electronic supplementary material, figure S4). Although the rotarod is routinely used to assess locomotor function in rodents, its lack of sensitivity and confounding variables are well recognized [27]. Overall, these data demonstrate that endogenous Parkin inactivation in vivo results in selective deficits in a sensitive test of voluntary motor function. Consistent with our results, deficits in fine motor coordination were reported in one previous characterization of Parkin knockout mice generated by targeted deletion of exon 3 [17]; however, in another independently generated exon 3 deleted knockout model, no motor deficits were reported although these mice exhibited learning and memory deficits [28]. More recently, similar fine motor deficits were found in Pink1 knockout mice in the absence of nigrostriatal degenerative pathology [29], further underscoring the importance of performing more sensitive behavioural testing in mouse models of PD. In our study, we were unable to determine the age-of-onset of the motor deficits, although it was reported that Pink1 knockout mice take approximately six months to exhibit detectable locomotor dysfunction [29].
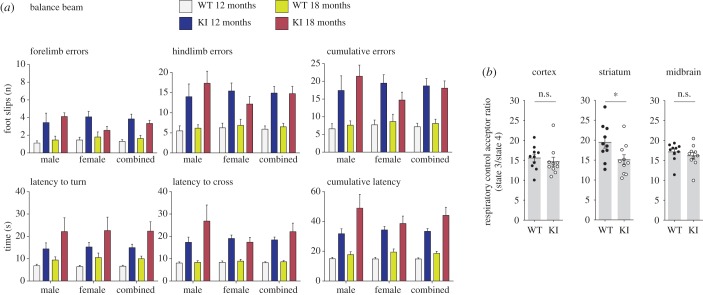
Figure 3.
ParkinS65A/S65A mice exhibit selective impairments on a sensitive task of voluntary motor function. (a) Balance beam performance in ParkinS65A/S65A (KI) and wild-type mice at 12 months and 18 months of age. Animals were assessed by their ability to reach a platform by competently traversing a raised and tapered beam. The performance was recorded by the number of errors scored as slips (forelimb, hindlimb and combined). ParkinS65A/S65A mice made significantly more forelimb errors and hindlimb errors at 12 and 18 months (p < 0.01) than their WT littermates. The ParkinS65A/S65A mice also demonstrated an age-related decline in their ability to orientate themselves on the beam end and cross the beam, resulting in longer latencies on both measures compared to WT littermates (p < 0.01). Error bars represent the standard error of the mean. (b) Striatal RCR is affected in ParkinS65A/S65A mice. Mitochondrial respiratory acceptor control ratios in the striatum, midbrain, and cortex of 1-year-old aged wild-type and ParkinS65A/S65A mice (n = 10 per group).
Given the mitochondrial-centric functions ascribed to the PINK1–Parkin dyad, we next investigated if the selective motor phenotype in our ParkinS65A/S65A mice could be attributed to mitochondrial dysfunction. We assayed mitochondrial respiration in discrete adult brain regions from 3-month old and 12-month-old wild-type and mutant mice using high-resolution (Oroboros Oxygraph-2k) respirometry [20]. In 3-month-old mice, we observed no difference in the respiratory control ratio (RCR) defined as State3ADP/State4 that reflects mitochondrial oxidative phosphorylation coupling efficiency independently of mitochondrial content [30,31] (electronic supplementary material, figure S5a). By contrast, we detected a highly selective and consistent difference in mitochondrial RCR between the striatum of 12-month-old ParkinS65A/S65A and wild-type mice but not in other brain regions including cortex (figure 3b and electronic supplementary material, figure S5b). This is consistent with previous reports of impaired mitochondrial respiration in select brain regions of aged Parkin knockout mice [20], and suggests that Parkin Ser65 phosphorylation contributes to mitochondrial integrity in this brain region.
2.3. Nigrostriatal integrity and basal mitophagy levels are normal in ParkinS65A/S65A knock-in mice
The observed locomotor and striatal mitochondrial dysfunction in ParkinS65A/S65A mice suggested that endogenous Parkin inactivation could also negatively impact nigrostriatal integrity in vivo. We next performed histological analysis of the substantia nigra of aged (approx. 18 months) ParkinS65A/S65A and wild-type mice to investigate if the impaired viability of nigrostriatal dopaminergic (DA) neurons might account for the selective motor defects in ParkinS65A/S65A mice. Inactivation of endogenous Parkin did not influence striatal anatomy or volume in aged mice (figure 4a–c). Nigrostriatal projections are highly complex, with a single DA neuron capable of innervating up to 2% of its terminal target field [5,32]. Although total DA neuron numbers were comparable between ParkinS65A/S65A mice and controls (data not shown), it is conceivable that differences in target-field arborization could account for the observed motor defect in our mutant animals. At a gross neuroanatomical level, conventional chromogenic tyrosine hydroxlase (TH)-immunohistochemistry revealed indistinguishable striatal innervation between genotypes (figure 4a); however, this approach does not permit a high-resolution analysis of the nigrostriatal projection. To determine whether Parkin inactivation could impact the ontogeny and arborization of DA neurons in vivo, we employed whole volume imaging of the entire nigrostriatal pathway within intact, optically cleared adult brains using iDISCO+ [22,33,34]. This approach enabled utilization of confocal microscopy to assess the degree of innervation and overall connectivity between aged ParkinS65A/S65A mice and littermate control mice at high resolution. Using this approach, we successfully resolved DA projections in wild-type and ParkinS65A/S65A mice; however, we observed no differences between groups (figure 4b). Dysfunction in non-neuronal cells is also a reported feature of PD mouse models [35]. However, immunohistochemical labelling of microglia and astroglia by Iba1 and GFAP, respectively, revealed no overt differences between wild-type and ParkinS65A/S65A mice (data not shown). To further investigate nigrostriatal function, we next profiled catecholamine neurotransmission in our mutant mice. We rapidly dissected striatal tissue from age-matched wild-type and ParkinS65A/S65A littermates and subjected these to HPLC analysis. We observed comparable levels of DA and DA turnover in wild-type and ParkinS65A/S65A striata (figure 4c), indicating that in our mouse model, genetic ablation of endogenous Parkin phosphorylation and activation is not sufficient to impact DA neurotransmission in vivo. Collectively, these data demonstrate that endogenous Parkin inactivation results in a selective locomotor defect in the absence of structural nigrostriatal dysfunction in vivo. Importantly, our findings highlight the importance of Parkin activation under steady-state conditions in vivo.
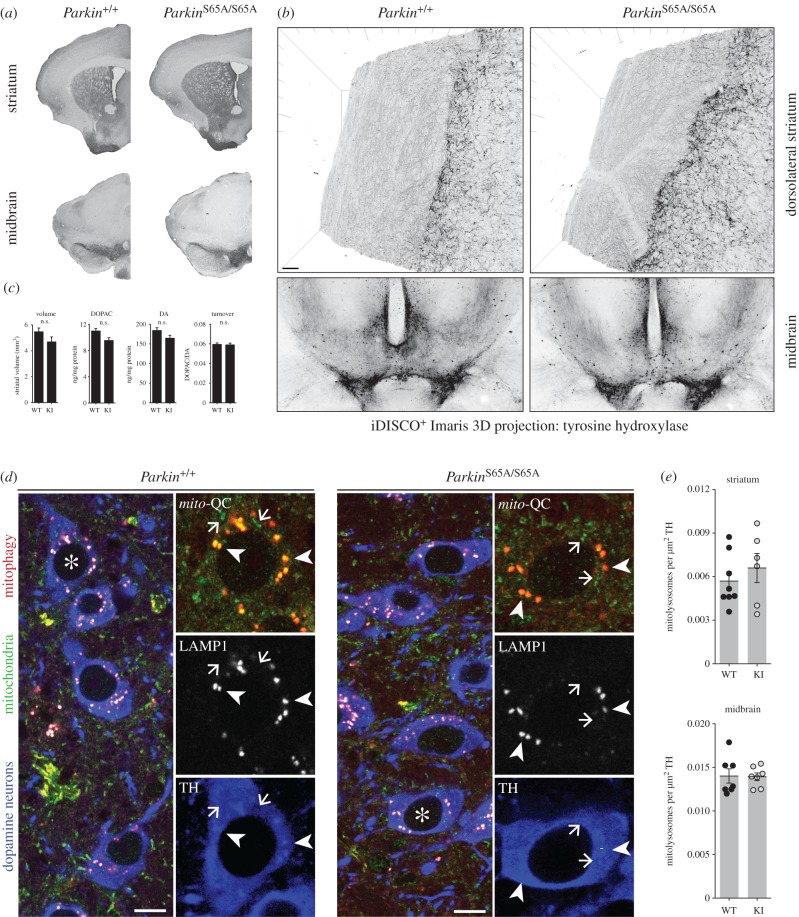
Figure 4.
ParkinS65A/S65A mice do not exhibit nigrostriatal degeneration or defective mitophagy. (a–c) Striatal innervation is indistinguishable between wild-type and ParkinS65A/S65A (KI) mice. (a) No differences in neuron number or striatal innervation were observed by anti-tyrosine hydroxylase (TH)-based immunohistochemistry analysis of midbrain and striatum between genotypes. (b) Striatal innervation analysis of TH-positive DA neurons within intact brains processed by iDISCO+. Arborization of DA neurons is indistinguishable between wild-type and KI mice. (c) No differences were observed in striatal volume between genotypes. Error bars represent standard errors of the mean. Striatal dopamine levels are unaltered in ParkinS65A/S65A mice. HPLC analysis revealed no differences in dopamine (DA) and 3,4-dihydroxyphenylacetic acid (3,4-DOPAC) levels between wild-type and KI mice (n = 10 per genotype). Data represent mean values ±s.e.m. n.s., not significant. (d) Basal mitophagy is unaltered by endogenous Parkin activation in vivo. Maximum z-projections of midbrain DA neurons from mito-QC wild-type and KI mice immunolabelled with antibodies to TH (blue) and LAMP1 (greyscale). Arrows point to mitochondria and arrowheads point to mitochondria associated with (autophago)lysosomes/mitolysosomes as defined by co-localization of LAMP1 and mito-QC staining. Mitochondrial delivery to lysosomes (mitophagy) is associated with quenching of GFP signal of the mito-QC reporter and residual mCherry only signal intensity. Scale bar 5 µm. Asterisks indicates cells within insets. (e) Quantitation revealed no differences between mitophagy in DA cell bodies or axons. Results are expressed as mean values ± s.e.m.
Parkin has received most attention for its role in modulating mitochondrial quality control [5]. Maximal activation of Parkin E3 ligase activity leads to ubiquitylation of myriad substrates on the mitochondrial outer membrane, that engage autophagy receptors and drive the elimination of damaged mitochondria via mitophagy [4–6]. This widely accepted and reproducible model is largely based on in vitro findings, however recent advances in mouse genetics using the mito-QC and mt-Keima reporter models have revealed the physiological nature of basal mitophagy in vivo [22,36,37]. Furthermore, DA neurons have recently been shown to undergo high levels of mitophagy in vivo which occurs independently of PINK1 [38,39]. To investigate the regulation of basal mitophagy in the context of endogenous Parkin inactivation in mammals, we crossed ParkinS65A/S65A mice with the mito-QC mitophagy reporter model to generate ParkinS65A/S65A-mito-QC mice. An assessment of wild-type and S65A mito-QC animals revealed that basal mitophagy in nigrostriatal DA cell bodies and projections was indistinguishable between wild-type and mutant mice (figure 4d,e). These data reveal that nigrostriatal mitophagy is unaffected in a mouse model of endogenous Parkin inactivation exhibiting selective defects in motor and mitochondrial function.
2.4. PARK2S65N mutations cause human Parkinson's disease via direct disruption of the PINK1–Parkin axis
Human patients with PARK2 mutations typically present with early onset PD displaying slow progression and sustained response to L-DOPA. Mutations in several key regulatory residues of Parkin have previously been identified, including Cys431Phe which disrupts the catalytic acceptor cysteine within the RING2 domain [40]. However, to date, no mutations affecting the Ser65 phosphorylation site have been reported. Here we report the first clinical and genetic evidence of PD associated with homozygous mutations at p.S65N, the critical node of the PINK1–Parkin pathway.
2.5. Case series
2.5.1. Case 1
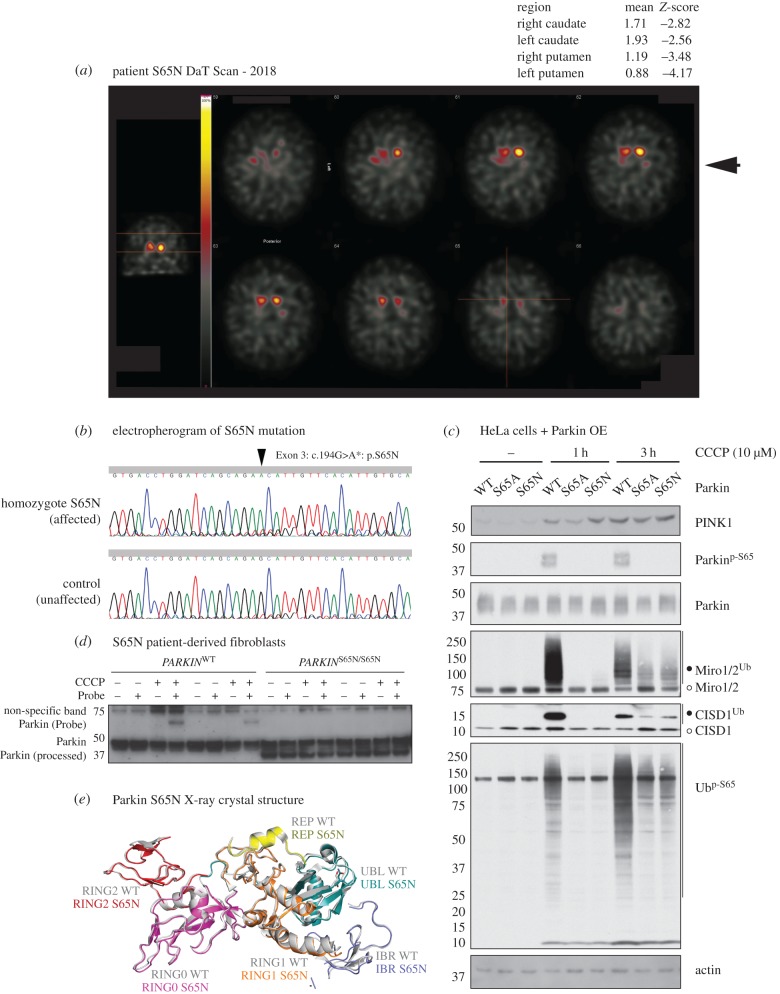
We report a 71-year-old Finnish male, diagnosed with early onset PD at the age of 40 years with no reported family history of Parkinson's. His symptomatic presentation included rigidity of the lower limbs that was initially more marked on the left-hand side, and he exhibited shortness of steps. He has continued to experience rigidity throughout his disease course, without the development of tremor. Recently, the patient has suffered from a mild gait disturbance with occasional freezing and postural instability. Overall, the progression of his illness has been exceptionally slow with a sustained response to medication that currently comprises: levodopa (300–400 mg d−1), pramipexole (2.1 mg d−1) and selegiline (10 mg d−1). On examination during the years 2016–2017, he was categorized at stage 2.5 of the modified Hoehn and Yahr Scale, with a United Parkinson's Disease Rating Scale (UPDRS) score of 41/199 (electronic supplementary material, table S1) Dopamine transporter (DaT) single-photon emission computerized tomography (SPECT) of the brain demonstrated reduced density of DA synaptic terminals in the caudate and putamen consistent with degenerative PD (figure 5a).
Figure 5.
Discovery and characterization of case 1: a human PD-causing Parkin mutation at Ser65. (a) Dopamine transporter imaging (DaT scan) results with region-based semi-quantitative analysis. 123I-FP-CIT brain single-photon emission computed tomography (SPECT) or DaT scan imaging measures DAT density in presynaptic terminals of the DA neurons projecting from the substantia nigra to dorsal striatum (caudate nucleus and putamen) thereby enabling assessment of the structural integrity of the nigrostriatal pathway in humans. DaT SPECT imaging was performed three hours after an intravenous injection of 186 MBq 123I-FP-CIT using a double head gamma camera with a fan beam collimator. Thyroidal uptake was blocked by 300 mg of oral potassium perchlorate before the tracer injection. Occipital lobe was used as a reference region of non-specific binding. The Z-scores in the top right of the figure indicate significant deviation from the population mean values for both the left and right caudate and putamen. In the putamen, the DAT density was slightly more reduced, which is typical of the rostro-caudal pattern of uptake loss in PD. (b) Electropherogram detailing S65N mutation data (Exon 3 G > A). (c) Parkin S65A and S65N mutants display reduced Parkin activation in response to PINK1 activation. Flp-In T-Rex-HeLa cells stably expressing wild-type (WT), S65A and S65N Parkin were induced with 0.1 µg ml–1 doxycycline for 24 h prior to stimulation with DMSO or 10 µM CCCP at indicated timepoints (1 h or 3 h). Whole-cell lysates were subjected to immunoblot analysis using the indicated antibodies. For detection of ubiquitylation (marked by open circles), lysates were subjected to ubiquitylated-protein capture by His-Halo-Ubiquilin1 UBA-domain tetramer (HALO-UBAUBQLN1), prior to immunoblot with anti-CISD1 (Proteintech Europe) and anti-Miro1/2 (DSTT, S531D, 5th bleed). (d) Activity-based profiling of Parkinson's disease associated Parkin S65N patient-derived fibroblasts. Primary fibroblasts derived from skin biopsies from a healthy subject (WT) and a PD patient harbouring a Parkin S65N homozygous mutation were profiled with E2-ubiquitin-based probe to monitor Parkin E3 transthiolation activity as described previously [41]. Mitochondrial depolarization does not activate the PINK1–Parkin pathway in Parkin S65N patient cells (CCCP treatment (3 h, 10 µM)). (e) Composite schematic illustrating the X-ray crystal structure of the Parkin S65N disease mutant, superimposed over the wild-type structure (PDB ID: 5C1Z, Grey). ParkinS65N structure is depicted in colour: UBL (teal), RING0 (magenta), RING1 (orange), IBR (blue), REP (yellow) and RING2/Rcat (red).
Targeted next-generation sequencing (NGS) revealed that the patient carried a novel homozygous c.194G > A variant in exon 3 of the PARK2 gene causing an amino acid change p.Ser65Asn (S65N) in the PARKIN protein (figure 5b). The variant is very rare: only two heterozygous carriers among 122 271 subjects were found in the Genome Aggregation Database (gnomAD) with an allelic frequency of 8.2 × 10−6. In the Exome Aggregation Consortium (ExAC) database, two heterozygotes were found among 60 691 subjects (allele frequency 1.6 × 10−5). Furthermore, the variant site is highly conserved in vertebrates (electronic supplementary material, figure S6a), and in silico analysis of the variant by the method of Combined Annotation Dependent Depletion (CADD) [42] predicted that the mutation is deleterious with a CADD C-score of 25.5. We analysed coding variants in 82 selected PD-associated loci (electronic supplementary material, table S2) with a CADD C-score of greater than 20 and population carrier frequency less than 1%, but did not find any other likely pathogenic variant in S65N-M70.
2.5.2. Case 2
A 60 year-old Caucasian female diagnosed with PD at the age of 54 was identified from the Parkinson's Progression Markers Initiative (PPMI). Her initial clinical features were bradykinesia and gait difficulty on the right side. She exhibited characteristic but mild motor symptoms for PD and no atypical features have been observed. Twelve months following her diagnosis, she was commenced on pramipexole resulting in a positive and sustained response (current dosing 2.25 mg of pramipexole/day). At her latest examination, the patient was categorized at Stage 2 of the Hoehn and Yahr Scale, and she has also been examined using the Unified Parkinson's Disease Rating Scale (MDS-UPDRS) for 5 years (scores from latest examination are provided in electronic supplementary material, table S1).
The patient has also been followed up by DaT imaging for four years, which has demonstrated reduced density of DA synaptic terminals in the caudate and putamen consistent with DA denervation (striatal binding ratio (SBR) calculations from the latest session are provided in electronic supplementary material, table S1).
Genetic analysis of the individual revealed a homozygous Parkin p.S65N mutation. Analysis of 82 PD-associated loci revealed three other gene variants (in POLG, MC1R and Glucocerebrosidase (GBA) (electronic supplementary material, table S3) that passed our filtering criteria (CADD C-score greater than 20, carrier frequency less than 1%). A heterozygous recessive variant (p.G268A) in POLG has been linked to autosomal recessive/sporadic progressive external ophthalmoplegia (PEO) in compound heterozygous or homozygous form [43,44], but also described as single-heterozygous in a child with a syndrome including Parkinsonism born from consanguineous parents (probable autosomal recessive mode of inheritance) [45]. She also carried another POLG variant, a rare inframe deletion (exon2:c.153_158del:p.Gln54_Gln55del) in a tandem repeat region of POLG; however, this is likely a benign variant, given a CADD C-score of 6.2. Overall, we did not consider the heterozygous p.G268A POLG mutation as disease-causing in the patient. A rare heterozygous variant (p.R142H) was detected in the MC1R gene. This gene was initially found to be associated with PD, but subsequent studies with large patient cohorts have not supported this finding [46–49]. Furthermore, the p.R142H variant has to date only been associated with red hair and not PD [46]. Thus, we did not consider p.R142H being a disease-causing variant in this patient. By contrast, a heterozygous p.N409S variant in GBA was detected, which is a recognized susceptibility factor for PD [50] although the variant does not always lead to PD [51]. Therefore, we conclude that the PARKIN p.S65N mutation is likely to be the major disease-causing variant in the patient, although we cannot exclude that the GBA p.N409S variant may contribute to pre-disposition and/or the clinical phenotype of the patient.
2.5.3. Functional analysis of the PARKINS65N/S65N mutation
We initially assessed the pathogenicity of the ParkinS65N/S65N mutation by assaying PINK1–Parkin signalling in Flp-In TRex HeLa cells, engineered with stable expression of either wild-type Parkin, a non-phosphorylatable S65A mutant and the S65N mutant of Parkin. Following the induction of Parkin expression, cells were treated with 10 µM CCCP or DMSO for 1 or 3 h. Immunoblotting of protein extracts demonstrated that the S65N mutation was functionally equivalent to the S65A mutation, leading to both a loss of both Parkin activation (as judged by reduced CISD1 and Miro1/2 substrate ubiquitylation) and reduced phospho-ubiquitin accumulation (figure 5c). We next interrogated the influence of the S65N mutation on endogenous Parkin function, by assaying its activity and downstream signalling in patient-derived cells upon mitochondrial depolarization. Primary fibroblast cultures were established from a skin biopsy of the Finnish patient with homozygous Parkin S65N mutation, and an unaffected human control. Cultures were treated with 10 µM CCCP or DMSO for 3 h to induce PINK1 stabilization and activation (electronic supplementary material, figure S6b). Immunoblotting with anti-phospho-Ser65 Parkin antibodies demonstrated that Parkin phosphorylation was abolished in cells expressing S65N (electronic supplementary material, figure S6b). This was associated with substantial reduction of Parkin E3 ligase activity as judged directly by ubiquitylation of CISD1 in PARKINS65N/S65N fibroblasts (electronic supplementary material, figure S6b). Furthermore, we observed reduced accumulation of phospho-ubiquitin in patient fibroblasts compared with control cells, indicating reduced Parkin E3 ligase activity (electronic supplementary material, figure S6b). We further confirmed loss of Parkin E3 activity in patient-derived primary cultures by deploying a chemical probe that measures transthiolation activity of Parkin via covalent labelling of its active site cysteine residue Cys431 [41]. The probe revealed robust labelling of activated wild-type human Parkin in control fibroblasts, whereas this was completely disrupted in PARKINS65N/S65N mutant fibroblasts (figure 5d). Interestingly upon mitochondrial depolarization, we observed a partial reduction in mitochondrial PINK1 stabilization in the PARKINS65N/S65N mutant fibroblasts (electronic supplementary material, figure S7) and this is consistent with similar findings observed in CRISPR/Cas9 mediated S65A mutant human ES cell-derived neurons stimulated with A/O [25].
We next investigated the effect of the endogenous PARKINS65N/S65N mutation on basal and depolarization-induced mitophagy. We first generated a stable mitophagy reporter cell line via retroviral transduction of mito-QC [39,52] in both control and PARKINS65N/S65N patient fibroblast lines. To induce substantial levels of mitophagy in culture, patient and control cells expressing mito-QC were stimulated with A/O for 24 h. Upon A/O stimulation, we observed no difference in mitophagy between control and PARKINS65N/S65N mito-QC cells (electronic supplementary material, figure S8). These data are consistent with a previous report demonstrating that endogenous Parkin is dispensable for basal and depolarization-induced mitophagy in patient-derived primary fibroblasts [52].
2.5.4. Structural analysis of the ParkinS65N mutant protein
To understand precisely how the pathogenic S65N mutation affects Parkin activity, we crystallized and solved the structure of the novel human Parkin PD mutant (UBLR0RBR, Δ84–143) at 2.9 Å. Comparative analyses of wild-type and ParkinS65N x-ray crystal structures did not reveal any conformational differences between the wild-type and mutant protein (figure 5e and electronic supplementary material, figure S6c), indicating that the S65N mutation does not significantly perturb Parkin structure. However, as the S65N mutation prevents incorporation of a phosphate group at this position, the absence of a negative charge at the UBL-IBR interface inhibits the ability of ParkinS65N to adopt the open conformation required for full E3 activity. This contrasts to wild-type Parkin, where phosphorylation at Ser65 induces key conformational changes and new interactions [53–56].
3. Discussion
Combining a novel mouse knock-in model and unique human clinical case studies, we present the most compelling evidence to date demonstrating the physiological and clinical significance of PINK1-dependent Parkin Ser65 phosphorylation. Furthermore, despite over 20 putative substrates reported in the literature [57,58], our research indicates that inactivation of this PINK1 phosphorylation site on Parkin alone is sufficient for humans to develop early onset Parkinson's disease (PD).
Understanding the mechanistic basis of DA neuronal loss in mammalian models of PINK1 and Parkin has proved challenging, because it has been difficult to recapitulate a PD-like neurodegenerative phenotype in rodents, with the notable exceptions of mutant Parkin over-expression in mice and rats [59,60]; the Pink1-null rat model [61]; and exposure of Parkin-null mice to chronic mitotoxic stress [21]. While ParkinS65A/S65A mice have an anatomically intact nigrostriatal pathway with no detectable loss of viability, denervation or dopamine depletion even after ageing for 18 months (figure 4a–c), we observed selective locomotor dysfunction particularly on balance beam performance (figure 3a). Our results are consistent with previous reports of motor deficits reported in Parkin knockout mice [17] and Pink1 knockout mice [29] in the absence of nigrostriatal pathology.
The neurobiological basis of the fine motor defects emerging in Parkin and Pink1 genetic models is poorly understood. We did not observe any substantial changes in the absolute levels of dopamine in Parkin knock-in mice. Interestingly, multiple lines of evidence suggest that Parkin knockout mice exhibit compartment defects with reduced striatal synaptic release of dopamine, associated with reduced expression of both the dopamine transporter (DAT) and vesicular monoamine transporter (VMAT2) and higher levels of extracellular dopamine in the striatum [17,18]. Mild mitochondrial defects have also been reported in 24-month old Parkin knockout mice that are not observed in younger mice (9 months of age) [20]. These effects were restricted to the striatum, and not detected in other brain regions including the midbrain or cortex [20]. Using a similar approach, our analysis revealed a mild, age-dependent defect in mitochondrial respiration in the striatum of Parkin knock-in mice (figure 3b). Interestingly, this was not associated with any change in basal nigrostriatal mitophagy in Parkin knock-in mice (figure 4d,e). Our current findings are also consistent with our recent demonstration of PINK1-independent mitophagy in vivo [39]. Recent analysis of Drosophila melanogaster models expressing fluorescent mitophagy reporters has also revealed that basal mitophagy proceeds independently of both PINK1 and Parkin in vivo [38]. Nevertheless, because cell culture studies in vitro provide a compelling case for Parkin activation in stress-evoked mitochondrial quality control pathways [4–6], it will be vital to investigate if nigrostriatal mitophagy is specifically affected in models of constitutive mitotoxicity reporting PD-relevant pathology, e.g. Polγ mutator mouse crossed with Parkin knockout mice [21,62]. Ultimately, our findings highlight the importance of uncovering the mitophagy-independent pathways by which endogenous Parkin sustains striatal function and mitostasis during ageing. Our data also adds to the growing body of evidence that suggests multiple mitophagy pathways operate to sustain mitochondrial quality control during specific contexts i.e. under basal conditions compared to extreme or chronic stress.
At the molecular level, the feed-forward mechanism of Parkin activation stipulates that (i) PINK1-dependent generation of phospho-ubiquitin stimulates Parkin recruitment to damaged mitochondria, (ii) this primes Parkin for phosphorylation by PINK1 and thus maximal activation, (iii) leading to further phospho-ubiquitin accumulation [4–6]. Evidence for this model includes the demonstration that the Parkin His302Ala (H302A) mutant, that cannot bind phospho-ubiquitin but is catalytically competent, exhibits reduced mitochondrial recruitment, Parkin Ser65 phosphorylation and ubiquitylation of substrates in HeLa cells [11,41]. However, HeLa studies also suggest that the initial amount of phospho-ubiquitin generated by PINK1 (independent of Parkin activity) is likely to be low because over-expression of the catalytically inactive Parkin Cys431Ser mutant does not result in significantly detectable phospho-ubiquitin accumulation [12,15]. Consistent with this latter observation, we did not observe any phospho-ubiquitin accumulation in mature primary cortical neurons (figure 2) or adult primary lung fibroblasts (electronic supplementary material, figure S3) subjected to mitochondrial depolarization from ParkinS65A/S65A and Parkin knockout mice. This is consistent with recent analysis of CRISPR/Cas9-mediated S65A mutant human ES cell-derived neurons stimulated with A/O [25]. By contrast, we did detect phospho-ubiquitin accumulation in ParkinS65N mutant patient-derived fibroblasts and HeLa cells (figure 5c and electronic supplementary material, figure S6b), albeit at lower levels than wild-type cells, suggesting potential redundancy of Parkin in these cell types with both Parkin-dependent and Parkin-independent formation of phospho-ubiquitin. The MUL1 E3 ligase has recently been reported to operate in a parallel pathway to Parkin in non-neuronal cells, and could account for the phospho-ubiquitin we observe in our human cell systems [63], although further work is required to substantiate this.
We and others have previously demonstrated that phospho-ubiquitin can partially activate recombinant non-phosphorylatable Parkin S65A protein in vitro [8,12]. Despite detectable accumulation of phospho-ubiquitin in fibroblasts, we observed complete loss of E3 activity in PARKINS65N/S65N mutant cells. The interaction affinity between the non-phosphorylated Parkin-phospho-ubiquitin complex with its charged E2 ∼ Ub is approximately 20-fold lower than that for the Phospho-Ser65-Parkin and phospho-ubiquitin complex [64], which may partly explain the lack of activation of endogenous ParkinS65N (and similarly ParkinS65A) protein. Our results therefore highlight the importance of studying signalling pathways at endogenous levels in physiologically relevant systems. Moreover, our data may also be explained by recent structural analysis of Parkin activation mechanisms. Initial binding of phospho-ubiquitin triggers a conformational shift that exposes a donor ubiquitin-binding site and partial activation of Parkin catalytic activity [54]. The subsequent PINK1-dependent phosphorylation of Parkin Ser65 ultimately drives Parkin activation by a series of structural alterations. In particular, displacement of the phospho-Ubl domain maintains interaction of the ubiquitin-charged E2 to the RING1 domain, and the phospho-Ubl subsequently binds to a basic phosphate-binding site on RING0 (flanked by residues Lys161 and Lys211 as well as Arg163) [53,55,56]. The binding of phospho-Ubl to RING0 leads to the release of the catalytic RING2 domain thereby exposing C431 to promote full Parkin activation and enhancing interactions with E2 [53,55,56]. Collectively, our endogenous neuronal results combined with recent structural insights highlight the pre-eminent status of Parkin Ser65 phosphorylation in modulating its catalytic activation.
The clinical importance of endogenous PINK1-mediated Parkin phosphorylation is substantiated by our discovery of two unrelated PD patients harbouring novel mutations at Parkin Ser65. Numerically, mutations in Parkin are associated with early onset PD, accounting for approximately 40% of cases with age of onset below 45 years; however, patients with pathogenic Parkin inactivating mutations have been reported with mid-age of onset that would be consistent with the second case [65]. In line with our mouse data, we found that basal and depolarization-induced mitophagy in S65N patient-derived primary fibroblasts was not diminished, despite a loss of Parkin E3 ligase activity (electronic supplementary material, figure S8). In the future, it will be exciting to address whether this is the case in a more neural context, by generating dopamine neurons derived from human pluripotent stem cells (hPSCs).
Although PINK1–Parkin signalling constitutes a clear pathway that modulates depolarization-induced mitophagy in vitro, basal mitophagy is unaffected in both Pink1 knockout and Parkin S65A knock-in mice. Though the clinical significance of PINK1–Parkin signalling is incontrovertible, the contribution of this pathway to mitophagy in vivo remains unclear, as does the contribution of dysregulated mitophagy to PD [66]. We predict that distinct mitophagy pathways orchestrate mitochondrial homeostasis under steady-state and stress-response scenarios.
In summary, this study is the first to establish both the physiological and clinical significance of PINK1-mediated phosphorylation of Parkin at serine 65. Our findings shed much-needed light on endogenous Parkin function and should provoke a reassessment of how it orchestrates cell-specific mitostasis in vivo.
4. Material and methods
4.1. Generation and validation of ParkinS65A/S65A knock-in mice
ParkinS65A/S65A mice were generated by TaconicArtemis GmbH, using a targeting strategy to introduce a constitutive knock-in of a point mutation (KI-PM) in the Park2 gene by homologous recombination in mouse ES cells. The targeting strategy was based on NCBI transcript NM_016694.3. Exon 1 contains the translation initiation codon, and the S65A mutation was introduced into exon 3. A positive selection marker (puromycin resistance, PuroR) flanked by FRT sites was inserted into intron 2. The targeting vector was generated using BAC clones from the C57BL/6 J RPCIB-731 BAC library and transfected into the TaconicArtemis C57BL/6N Tac ES cell line. Homologous recombinant clones were isolated using positive and negative (thymidine kinase, TK) selection, and the constitutive KI-PM allele was obtained after Flp-mediated removal of the selection marker. This KI-PM allele was predicted to express the mutated Park2 S65A protein product. The remaining recombination site will be located in a non-conserved region of the genome. The knock-in mice were generated and maintained on an inbred C57BL/6J background. Genotyping was performed by endpoint PCR using genomic DNA isolated from tails using the following primer sets at an annealing temperature of 60°C: Primer 1 (5′GAACAAGATAGGAGCACAGAAGG 3′) and Primer 2 (5′GACCAATTTACCTCTCGAGTGC 3′) that enabled detection of the wild-type (290 bp) and S65A knock-in (365 bp) alleles. The mito-QC mouse model used in this study was generated as previously described on a C57BL/6N background strain [22] and crossed with Parkin S65A knock-in heterozygote animals to produce homozygous knock-in animals and wild-type littermate controls with endogenous mito-QC reporter.
4.2. Materials and reagents
HaloLink resin was purchased from Promega. All mutagenesis was carried out using the QuikChange site-directed mutagenesis method (Stratagene) with KOD polymerase (Novagen). All DNA constructs were verified by the MRC Protein Phosphorylation and Ubiquitylation Unit (PPU) DNA Sequencing Service, School of Life Sciences, University of Dundee, using DYEnamic ET terminator chemistry (Amersham Biosciences) on Applied Biosystems automated DNA sequencers. DNA for bacterial protein expression was transformed into Escherichia coli BL21 DE3 RIL (codon plus) cells (Stratagene). Stock solutions of carbonyl cyanide m-chlorophenyl hydrazone (CCCP; Sigma), antimycin A (Sigma) and oligomycin (Sigma) were used for experiments in cells. Unless otherwise specified, general reagents and chemicals were from Sigma-Aldrich (Merck) and cell culture reagents were from Gibco/Invitrogen (Thermo Fisher Scientific). All cDNA plasmids, antibodies and recombinant proteins generated in house for this study are available on request through our dedicated reagents website: https://mrcppureagents.dundee.ac.uk/
4.3. Mouse motor and behavioural studies
In total, 51 PARK2 S65A mice were used for behavioural experiments in groups which consisted of 25 wild-type (14 males and 11 females) and 26 Park2S65A/S65A mice (11 males and 15 females). At 18 months 35 mice remained, 16 wild-type (10 males and 6 females) and 19 Park2S65A/S65A mice (9 males and 10 females). Mice were housed in sex-matched littermate groups of between two and five animals per cage with lights adjusted to a 12 L : 12 D cycle (lights on at 06.30) and with a constant ambient room temperature of 21° ± 1°C. Animals had ad libitum access to food and water throughout. The mice underwent behavioural testing at 12 months and 18 months of age and all experiments were conducted in accordance with the 2013 European Union Directive 2010/63/EU and the UK Animals (Scientific Procedures) Act of 1986 and local ethical review at the University of Cardiff.
The motor test battery has been described previously elsewhere [67], but briefly mice were tested for simple neurological deficits using: clasping and reaching assays; accelerating rotarod and balance beam test to measure motor coordination and balance; 30 min and 24 h open field activity; footprint analysis to measure changes in gait; and inverted grid task to measure grip strength. For the rotarod test (Ugo Basile, Varese, Italy), mice are placed on a rotating beam which accelerates. Initial training consisted of training the mice at different speeds for 2 min (4, 8 and 12 r.p.m.). Data were collected from five sessions at the 12-month time point and two sessions at the 18-month time point whereby the mice were placed on the rod that accelerated from 4 to 40 r.p.m. over 5 min. The latency of the mice to fall from the beam was recorded and used as a measure of motor coordination. For the balance beam test (see above reference for dimensions), each mouse was trained to run the beam prior to the test session. This consisted of starting the mice in the goal box at the top of the inclined beam and over repeated exposures moving the starting point on the beam to the lower end (start point) such that the mice ran further as the training progressed. Training was complete when the mouse was placed on the lower end of the beam (facing away from the beam) and the mouse turned and ran the length of the beam unprompted. Foot-slips were recorded when a foot slipped from the beam onto the narrow ledge that ran either side of the length of the beam. Each mouse provided data (fore and hind limb foot-slips, latency to turn and cross the beam) from two runs for analysis. Gait analysis was run using a footprint test whereby the feet of the mice were painted with red (front feet) and blue (rear feet) non-toxic water-based paint, and the mice were permitted to run a 1-m corridor that was fitted with paper on the floor such that they left coloured paw prints. The paw print patterns permit the measurement of variation in gait: stride length; base width (distance between left and right front/hind limbs); front/hind paw overlap (in normal locomotion the hind paw placement overlaps closely with the location where the fore paw had previously landed; gait impairment is manifest by an increase in the magnitude and variability of the separation. For the motor activity testing using an automated system (Med Associates Inc, Vermont, USA), mice were placed in an open field arena for 32 h with free access to food and water throughout. Data were collected automatically with activity being measured as the number of infrared beam breaks. Activity was measured as the initial 30 min period (transfer activity) and a 24 h period (15.00 to 15.00) to capture a circadian cycle. In addition, several simple behavioural assessments were run: clasping and reaching reflexes where the mice is suspended by the tail and lowered to the benchtop with clasping and reaching measured through observation and recorded as present or absent; negative geotaxis, measuring the natural inclination of the mouse to face upwards when placed head down on a vertical grid, with the mouse being allowed 30 s to correct its orientation to the upright position; and horizontal grid grip strength, whereby the mouse is placed on a flat grid surface which is then rotated 180° along the horizontal axis such that the mouse is clinging to the underside of the grid, the maximum time for the mouse to remain upturned being 60 s without falling.
4.3.1. Statistical analyses of mice behavioural studies
Parametric split-plot analyses of variance using age as the within groups factor and genotype and sex as between-group factors were used where applicable as three-way ANOVAs. All statistical analyses were conducted on Genstat 17 (VSN International, Hemel Hempstead, UK). Missing values were corrected with an inbuilt linear interpolation routine. For non-parametric data, χ2 was used to determine significance. Significance level was taken at 95% confidence.
4.4. Immunohistochemistry, iDISCO+ and microscopy
Transcardial perfusion was performed under terminal general anaesthesia to obtain tissues for histological analyses. Histology, immunohistochemistry, immunocytochemistry and confocal microscopy were performed as previously described [22,39,68,69], with minor modifications. Following terminal anaesthesia via intraperitoneal administration of Euthatal, adult animals were trans-cardially perfused with phosphate-buffered saline (PBS) to remove excess blood. Tissues were rapidly harvested and processed by immersion fixation in freshly prepared 3.7% PFA at pH 7.0 in 0.2 M HEPES. For mito-QC animals, transverse brain sections (200 mm) were acquired using a vibratome (Leica). For chromogenic immunohistochemistry and stereological analyses, coronal brain sections (40 mm) were cut on a sledge microtome equipped with a freezing stage (Leitz) and processed for free-floating immunohistochemistry. The following primary antibodies were used: rat monoclonal anti-lysosomal associated membrane protein 1 (LAMP1) (1D4B) (Cat#: sc-19992 RRID: AB_2134495; Santa Cruz Biotechnology, Inc.); rabbit, sheep and mouse anti-Tyrosine Hydroxylase (rabbit: Cat#: AB152; RRID: AB_390204; sheep: Cat#: AB1542; RRID: AB_90755; mouse: Cat#: MAB318; RRID: AB_2201528; Millipore); rabbit polyclonal anti-Ionized calcium binding adaptor molecule 1 (Iba1) (Cat#: 019-19741; RRID: AB_839504 - Wako), chicken anti-GFP (Cat# GFP-1020, RRID:AB_10000240; Aves Labs; Cat# ab13970, RRID:AB_300798; Abcam). Alexa-Fluor conjugated secondary antibodies were obtained from Life Technologies (Molecular Probes/Thermo Fisher Scientific) as described previously [39]. VECTASHIELD Antifade Mounting Medium H-1000 was used to mount tissue sections on slides (Leica Surgipath). For cell counts and tinctorial histochemistry, Cresyl fast violet (Nissl) stain was obtained from Sigma-Aldrich (C5042-10G). For whole-mount imaging of the nigrostriatal pathway in intact mouse brains, specimens were processed, stained using rabbit-TH (AB152) and optically cleared using the detailed protocol specified for iDISCO+ [33] (https://idisco.info). Following the acquisition of images from intact whole brains (dorsal and ventral surfaces), cleared brain specimens were hemisected along the midline and subsequently bisected to facilitate deeper imaging in the sagittal and coronal planes, respectively. Images were acquired using a Zeiss LSM 710 laser scanning microscope (Plan-Neofuar 340 objective, NA 1.30; Plan Apochromat 363 objective NA 1.4; Plan Apochromat 320 objective, NA 0.8), or a Zeiss LSM880 Airyscan confocal scanning microscope (ZEISS; Plan Apochromat 363 objective, NA 1.4) and processed using ZEISS Zen software/Adobe Photoshop or Imaris (Bitplane) for 3D isosurface rendering. Images were digitally altered within linear parameters, with minimal adjustments to levels and linear contrast applied to all images.
4.5. Quantification and statistical analysis
Semi-automated quantitation images were processed with Volocity 6.3 image analysis software (PerkinElmer) using algorithms developed to analyse object overlap and count individual structures. For all analyses, we obtained images using uniform random sampling by an experimenter blind to all conditions. All images in each experimental group were processed as a batch using identical protocols. All images were prefiltered to suppress noise (3 × 3 median filter). The same strategy was used to quantify all non-immunolabelled mito-QC-labelled images using auto-thresholding to identify objects (mean intensity + 3 standard deviations). Objects were filtered using a minimum size cutoff (0.16 mm2) and touching objects were separated using a guide size (0.4 mm2). Mitolysosomes were typically identified on the basis of thresholded red signal not overlapping with green (this condition was relaxed in the case of samples where dequenching of the mito-QC probe was observed). Where dequenching or spectral-overlap was observed in some instances, we employed LAMP1 immunohistochemistry to verify the lysosomal nature of mitolysosomes and green channel/combined with LAMP1 immunostaining was used as a proxy for mitolysosomes. In the case of immunolabels used to identify cells of interest in brain sections (TH, Iba1), we employed an autothresholding criterion of mean intensity + 1.5 standard deviations. A minimum size of 0.16 mm2 was required, and a ‘Fill Holes in Objects' processing step was applied.
Cell counts were conducted using two-dimensional stereology with an automated stage and stereology module ‘NEWCAST TM’ from Visiopharm integrator system software (Visiopharm) on an Olympus Bx50 microscope (Olympus Optical Co. Ltd, Tokyo, Japan). For NeuN immunohistochemistry, cell counts were conducted on a 1 : 12 series throughout the entire striatum (left and right sides), with the counting objective set at 100× and the randomized sampling counting frame area at 285 µm2 and corrected using the Abercrombie formula [70]. For the TH and Calbindin double labelling, cell counts were carried out on a 1 : 12 series through the substantia nigra with the counting objective set at 40×, and all visible cells were counted within this area with double-stained cells denoting A10 dopamine cells and cells only stained for TH denoted as A9 dopamine cells.
4.6. Mitochondrial respiration assays
Wild-type and Parkin S65A knock-in mice (10 per genotype) were killed by decapitation without prior anaesthesia. A fragment of the tail was retrieved for confirmation of the genotype, determined by PCR amplification, as previously described [71]. The brains were rapidly removed and the regions of interest (striatum, midbrain and cortex) dissected on ice and weighed. Post-nuclear supernatants were obtained by manual homogenization of brain samples in nine volumes of isolation buffer (300 mM sucrose, 1 mM EGTA, 5 mM Tris–HCl pH 7.4), using a glass Dounce homogenizer with a glass pestle, followed by centrifugation at 1000g, 4°C, for 10 min. The post-nuclear supernatants were directly used for respiration and protein assays, as well as for the analysis of citrate synthase (CS) activity. The post-nuclear supernatants were analysed in a thermostatically controlled oxygraphic chamber at 25°C with continuous stirring (Oxygraph-2k, Oroboros Instruments, Innsbruck, Austria) as previously described [20]. In brief, the samples were resuspended in 2 ml of mitochondrial respiration medium (100 mM KCl, 40 mM sucrose, 10 mM TES, 5 mM MgCl2, 1 mM EGTA, 0.4% w/v fatty acid free-BSA (bovine serum albumin), pH 7.2) and the protein concentration was determined (Bradford protein assay, Bio-Rad). Each respiration assay was run with a constant amount of protein from one wild-type and one knock-in mouse (0.3 mg for striatum, 0.6 mg from midbrain and cortex). The results were expressed in nmol of O2/min mg−1 of total protein. The respiration rate was recorded sequentially: (i) basal respiration in the presence of complex I substrates (10 mM glutamate, 5 mM malate); (ii) state 3 respiration with addition of 1 mM ADP; (iii) state 4 respiration with 1 µg ml−1 oligomycin; and (iv) maximal respiration rate was determined by progressive additions of 1.25 µM carbonyl cyanide m-chlorophenyl hydrazone (CCCP). The respiratory reserve capacity was calculated as maximal respiration − basal respiration. Respiratory control ratio or RCR was calculated as state 3/state 4.
4.7. Determination of citrate synthase activity
CS activity was determined using a colorimetric assay based on the reaction between 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) and CoA-SH to form 5-thio-2-nitrobenzoic acid (TNB), which exhibits maximum absorbance at 412 nm, according to the manufacturer's instructions (Sigma, CS0720). In brief, 20 µl of cold CelLytic M supplemented with protease inhibitors (protease inhibitor cocktail, Thermo Scientific 1861279) was added to 10 µl of post-nuclear supernatant. The sample was homogenized and centrifuged at 12 000g for 10 min. The supernatant was collected, and protein concentration was determined by the Bradford method (BioRad). Fifteen to 20 µg of protein was added to a 200 µl reaction mixture containing 1× assay buffer, 300 µM acetyl CoA and 100 µM DTNB. The reaction was initiated by addition of oxaloacetate (OAA) to a final concentration of 0.5 mM. CS activity was evaluated by following the absorbance of the reaction mixture at 412 nm (SpectraMax M4) before and after addition of OAA every 10 s, for 2 min. Each sample was run in duplicate and CS activity was expressed as units/min mg−1 of total protein.
4.8. Dopamine HPLC analysis
Dopamine and its metabolite 3,4-dihydroxyphenylacetic acid (DOPAC) concentrations were measured in the dorsal striatum by reverse phase high-performance liquid chromatography coupled with electrochemical detection (Coulochem III, Thermo Scientific) as described earlier [72]. Briefly, 18-month-old wild-type and ParkinS65A/S65A mice were killed by cervical dislocation. Brains were quickly removed, pre-commissural dorsal striatum was dissected on an ice-cold Petri dish and freshly homogenized in 0.4 M percloric acid. Following centrifugation and filtration of the supernatant, dopamine and DOPAC were separated using a C18 reverse phase column (Thermo Scientific). The mobile phase consisted of 10% acetonitrile, 75 mM NaH2PO4·H2O, 0.17 mM octanesulfonic acid, 2.5 mM triethylamine and 25 mM EDTA, adjusted to pH 3.0 with orthophosphoric acid, and was delivered at a flow rate of 0.6 ml min−1. Protein levels were determined by Lowry assay from pellets of the homogenates and catecholamine concentrations are expressed per milligram protein.
4.9. Antibodies for biochemical studies
The following primary antibodies were used: total Parkin immunoblotting (sc32282, Santa Cruz), mouse Parkin immunoprecipitation (in-house, S328D, 5th bleed), βI-tubulin (Sigma), β-actin (Sigma), Calbindin (c9848Sigma), GAPDH (Santa Cruz), CISD1 (Proteintech), NeuN (MAB377, Millipore), PINK1 immunoblotting (BC100-494, Novus), PINK1 immunoprecipitation (in-house S085D, 3rd bleed), Mitofusin2 (MFN2) (in-house, S781D, 4th bleed), OPA-1 (#612607, BD Biosciences), RHOT2 (Miro2) (H00089941, Abnova), COX IV (#4850, Cell signalling technology), PDH (#2784, Cell signalling technology), Tyrosine Hydroxylase (ab112, Abcam), VDAC1 (ab15895, Abcam), Vinculin (#4650, Cell signalling technology). Horseradish-peroxidase (HRP)-conjugated secondary antibodies (Sigma) were used. Anti-Parkin phospho-Ser65 rabbit monoclonal antibody was raised by Abcam in collaboration with the Michael J. Fox Foundation for Research. Anti-phospho-Ser65-ubiquitin rabbit polyclonal antibodies were generated by 21st Century Biochemicals Inc. (Marlborough, MA, USA) using two phosphorylated peptides, C-Ahx-YNIQKE[pS]TLHLVL-amide and Ac-YNIQKE[pS]TLHLVL-Ahx-C-amide, that correspond to amino acid residues 59–71 of ubiquitin. Antibodies were affinity purified using phosphorylated and non-phosphorylated versions of the peptide immunogen (electronic supplementary material, figure S9a). A fragment of human Miro1 fusion protein encompassing residues 1–592 with N-terminal His-SUMO tag was used as an immunogen to raise a sheep polyclonal anti-Miro1 antibody (S531D). Antibodies were affinity purified from antisera and characterized indicating cross-reactivity with Miro1/2 as described in the electronic supplementary material, figure S9b.
4.10. Tissue culture: proliferating and primary cells
Flp-In T-Rex HeLa stable cell lines were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FBS (fetal bovine serum), 2 mM l-glutamine, 1× penicillin/streptomycin 1× non-essential amino acids (Life Technologies), 15 µg ml−1 of blasticidin and 100 µg ml−1 of hygromycin. Cell transfections of untagged wild-type or mutant Parkin were performed using polyethylene method [73]. Cultures were induced to express protein by addition of 0.1 µg ml−1 of doxycycline to the medium for 24 h. To depolarize or uncouple mitochondrial membrane potential, proliferating cells were treated with 10 µM antimycin (Sigma) and 1 µM oligomycin dissolved in DMSO. Primary mouse cortical neurons were isolated from the brains of wild-type or mutant embryos of either sex at E16.5. Embryonic cortices were collected in HBSS, and cells were dissociated by incubation with trypsin-EDTA (#25300-054, Gibco) at 37°C. Cells were then diluted in Neurobasal medium containing B27 supplement, Glutamax, penicillin/streptomycin and plated at a density of 6.0 × 106 cells/well on 6-well plates coated with 0.1 mg ml−1 poly-l-lysine (PLL; Sigma). Neurons were cultured at 37°C in a humidified incubator with 5% CO2. Every 7 days, the medium was replaced with fresh medium containing B27. To depolarize or uncouple mitochondrial membrane potential in neurons, cultures were treated with 10 µM antimycin (Sigma) and 1 µM oligomycin dissolved in DMSO at 37°C.
Primary lung fibroblasts were derived from adult mice as previously described and maintained in DMEM/20%FBS/penicillin–streptomycin at 37°C/5% CO2 [26]. Primary human skin fibroblast cultures at a low passage from the patient and one matched control were established from a minimally invasive skin biopsy sample according to routine culture procedures. The cells were cultured in 5% CO2 and 90% humidity in high-glucose DMEM media (Gibco) supplemented with 10% FBS (LSP #S-001-BR), 1× non-essential amino acids, 1 mM sodium pyruvate, 100 Units/ml penicillin, 100 µg ml−1 streptomycin and 2 mM l-glutamine. The retrovirus for analysis of mito-QC in human fibroblasts was generated as previously described [52]. The cDNA for the mito-QC reporter (mCherry, GFP and residues 101–152 of human FIS1) was cloned into a pBABE.hygro vector or pBABE.puro vector. The construct was co-transfected into 293FT cells with GAG/POL and VSV-G expression plasmids (Clontech, Saint-Germain-en-Laye, France) for retrovirus production using Lipofectamine 2000 (Life Technologies) in accordance with the manufacturer's instructions. Virus was harvested 48 h after transfection and applied to cells in the presence of 10 mg ml−1 polybrene. Cells were selected with either 500 µg ml−1 hygromycin B or 2 µg ml−1 puromycin. Selected cells were seeded and incubated for 24 h into glass bottom dishes (Ibidi μ-Dish 35 mm) with media free of sodium pyruvate and non-essential amino acids. Mitophagy was stimulated for 24 h with 10 µM antimycin A (Sigma #A8674) and 5 µM oligomycin A (Sigma #75251), and controls were treated at equal volumes of DMSO. Cells were fixed for 20 min with 3.7% PFA (Sigma #P6148) pH 7.0, and subsequently washed twice with 2 ml of DMEM + 10 mM HEPES pH 7.0 and incubated for at least 10 min. Images were acquired using a ZEISS LSM 710 confocal microscope and quantified using Volocity 6.3 image analysis software as previous described [39].
4.11. Tissue and cell lysis
For analysis of endogenous Parkin protein expression in vivo, fresh brains were rapidly excised, placed in cold PBS and microdissected with ultrafine microknives under stereomicroscopy. Upon isolation, each brain region was collected in a single 1.5 ml microcentrifuge tube and immediately plunged into dry ice. Tissue samples were stored at −80°C until ready for processing. To make protein extracts, all tissues (neural subregions and extra-neural tissues) were weighed and defrosted on wet ice in fivefold mass excess of freshly prepared, ice-cold lysis buffer containing: 50 mM Tris/HCl pH 7.5, 1 mM EDTA pH 8.0, 1 mM EGTA pH 8.0, 1% Triton X-100, 0.25 M sucrose, 150 mM NaCl, 2 mM sodium orthovanadate, 1 mM NaF, 10 mM sodium glycerolphosphate, 1.15 mM sodium molybdate, 4 mM sodium tartrate dehydrate, 100 mM 2-chloroacetamide, 1 mM DTT and complete protease inhibitor cocktail (Roche). All inhibitors and DTT were added immediately prior to use. Tissue homogenization was performed using a probe sonicator at 4°C (Branson Instruments). Crude lysates were incubated at 4°C on wet ice for 30–45 min, before clarification by centrifugation at 14 000 r.p.m. for 30 min at 4°C. Supernatants used for subsequent steps were carefully removed and either used for downstream biochemical analyses or snap-frozen and stored at −80°C.
Primary neurons or Hela cells were sonicated in lysis buffer containing 50 mM Tris–HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% (w/v) Triton, 1 mM sodium orthovanadate, 10 mM sodium glycerophosphate, 50 mM sodium fluoride, 10 mM sodium pyrophosphate, 0.25 M sucrose, 1 mM benzamidine, 0.1 mM PMSF and protease inhibitor cocktail (Roche). Following the sonication, lysates were incubated for 30 min on ice. Samples were spun at 20 800g in an Eppendorf 5417R centrifuge for 30 min. Supernatants were collected and protein concentration was determined using the Bradford kit (Pierce). Primary human skin fibroblasts were lysed with ice-cold lysis buffer containing 50 mM Tris–HCl pH 7.5, 1% (v/v) Triton X-100, 1 mM EGTA, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 10 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 0.1 µg ml−1 microcystin-LR (Enzo Life Sciences), 270 mM sucrose, complete EDTA-free protease inhibitor cocktail (Sigma Aldrich Cat # 11836170001). Lysates were centrifuged at 20 800g for 10 min at 4°C. Supernatants were collected, quantified by Bradford assay (Thermo Scientific) and subjected to immunoblot analysis.
4.12. Membrane fraction enrichment
Cells were collected in ice-cold PBS containing 200 mM chloroacetamide. They were then lysed in buffer containing 250 mM sucrose, 20 mM HEPES, 3 mM EDTA, 1% (w/v) 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM NaF, 5 mM sodium pyrophosphate, pH 7.5 and protease inhibitor cocktail (Roche) supplemented with 100 mM chloroacetamide at 4°C. Cells were disrupted using a glass hand-held homogenizer (40 passes) and the lysates were clarified by centrifugation (800g at 4°C for 10 min). The supernatant was harvested and subjected to an additional centrifugation step at 16 600g for 10 min at 4°C. The resulting pellet containing the mitochondrial-enriched fraction was resuspended in the above buffer containing 1% Triton X-100 and centrifuged at 13 000 r.p.m. for 10 min at 4°C. The supernatant arising from this step contained solubilized mitochondrial proteins.
4.13. Immunoblotting and immunoprecipitation
For immunoprecipitation of endogenous PINK1 from human skin fibroblasts, 150 µg of whole-cell lysate was incubated overnight at 4°C with 5 µg of protein G pre-bound to PINK1 antibody. The immunoprecipitations were washed four times with lysis buffer containing 150 mM NaCl, then twice with buffer containing 50 mM Tris–HCl pH 7.5, 0.1 mM EGTA, and eluted by resuspending in 20 µl of 1× SDS sample buffer. Denatured samples were subjected to SDS-PAGE (4–12% Bis-Tris gels) and were transferred on to Protran 0.2 NC nitrocellulose membranes (Amersham). Membranes were blocked for 1 h at room temperature with 5% non-fat milk (Marvel) or BSA in TBST (Tris-buffered saline (50 mM Tris/HCl and 150 mM NaCl, pH 7.5) containing 0.1% Tween-20) in PBS pH 7.4 and incubated with the indicated antibodies overnight at 4°C with agitation. Detection was performed using HRP-conjugated secondary antibodies (Goat anti-Rabbit IgG (H + L) Secondary Antibody, HRP conjugate, #31460, Thermo Scientific; Rabbit anti-Mouse IgG (H + L) Secondary Antibody, HRP conjugate, #31450, Thermo Scientific; Rabbit anti-Sheep IgG (H + L) Secondary Antibody, HRP conjugate, #31480, Thermo Scientific). Membranes were developed using standard chemiluminescence with ECL (Amersham) and exposure to hyperfilm (GE Healthcare).
4.14. Activity-based profiling of Parkin
Transthiolation activity of Parkin activity in primary human skin fibroblasts was measured as described previously [41]. Briefly, Parkin probe (produced by Dr. Satpal Virdee, MRC PPU) was added to 50 µg of whole-cell lysate at a final concentration of 5 µM. Reactions were incubated at 30°C for 4 h and quenched by the addition of LDS sample buffer with β-mercaptoethanol, before analysis via SDS-PAGE (4–12% Bis-Tris Gels, Novex) and immunoblotting as previously described.
4.15. Ubiquitin enrichment
His-Halo-Ubiquilin UBA-domain tetramer (HALO-UBAUBQLN1) was expressed in E. coli BL21 cells, affinity purified on Ni-NTA-agarose and dialysed into 50 mM HEPES pH 7.5, 10% glycerol, 150 mM NaCl, 1 mM DTT [11]. For ubiquitin capture, 200–400 µg of whole-cell extract or 1 mg of membrane-enriched fraction was used for pulldown with HALO-UBAUBQLN1 [11]. HALO-UBAUBQLN1 was incubated with 200 µl of HaloLink resin (Promega) in binding buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% NP-40) overnight at 4°C under agitation [11]. HALO-TUBE beads (20 µl) were added to lysates and incubated at 4°C for 4 h under agitation. Beads were washed three times with lysis buffer containing 0.25 M NaCl, and eluted by resuspending in 20 µl of 1× LDS sample buffer containing 1 mM DTT.
4.16. Protein purification, crystallization and structure determination
Parkin S65N (ParkinS65N) (Δ84–143) was cloned into pET156P vector (DU56345) and expressed as His-SUMO fusion in E. coli BL21 cells, using conditions similar to those described in Chaugule et al. [74]. His-tagged ParkinS65N was purified using Ni-NTA resin. Protein eluted from Ni resin was incubated overnight at 4°C with SENP1 protease to remove the His-SUMO tag, and dialysed in 20 mM Tris pH 7.5, 75 mM NaCl and 250 µM TCEP buffer. Dialysed protein was further incubated with Ni-NTA resin and tag-free ParkinS65N was collected in the flow through, purified on Mono-Q and Superdex-75 columns (as described in [64]) and concentrated to 8 mg ml−1 for crystallization. ParkinS65N was crystallized at 4°C in 100 mM BIS-TRIS pH 5.5, 200 mM LiSO4 and 20% PEG3350 by using a vapour diffusion method under sitting drop. Crystals were flash frozen in 20% glycerol, the native dataset was collected at ID30A-1 beamline, European Synchrotron Radiation Facility (ESRF, Grenoble). Data were processed using Mosflm and Aimless in CCP4 (CCP4, 1994). ParkinS65N structure was determined by molecular replacement using Phaser in CCP4, WT-Parkin (pdb id: 5C1Z) was used as the search model. The initial model obtained from Phaser was further constructed and refined using Coot [75] and REFMAC5 in CCP4 [76], respectively. The final model of ParkinS65N shows excellent geometry (electronic supplementary material, table S4).
4.17. Human genetic analysis
4.17.1. Case 1 (S65N-M70)
Patient DNA samples were screened for variants using HaloPlex targeted sequencing of 82 selected PD-associated loci (Agilent Technologies, Santa Clara, CA, USA). DNA was extracted from peripheral blood by standard procedures. Sequencing was performed with a MiSeq sequencer (Illumina, San Diego, CA, USA), variant calling with Genome Analysis Toolkit [77] and the annotation with ANNOVAR [78]. The variants were confirmed by independent DNA sequencing method, Sanger sequencing. For the c.194G > A mutation in PARK2 we used the following PCR and sequencing primers for PCR and sequencing: 5′-CTCGCATTTCATGTTTGACATTTCC; 5′-ATGCTCCATGCAGACTGCACTAAAG; sequencing 5′-CCTGCTGTCAGTGTGCAGAATG. The frequency of variants was screened from ExAC (http://exac.broadinstitute.org/) and gnomAD browsers (http://gnomad.broadinstitute.org) [79], the potential pathogenic effects were evaluated using CADD C-score [42] and conservation using UniProt [80].
4.17.2. Case 2 (S65N-F60)
We downloaded the clinical data of case 2 (ID 3185) from the Parkinson's Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data) on 13 August 2018. PPMI is a multi-centre, on-going longitudinal and observational study, which contains clinical, imaging and biological data from 454 PD patients and 215 control patients currently stored at the Laboratory of Neuro Imaging (LONI) database at University of California, Los Angles (UCLA). The cohort consists of 620 females and 828 males. The open-access repository is available online after registration at http://www.ppmi-info.org/. Informed consent was signed by the patients enrolled in the PPMI study. The patient's DNA whole-genome sequence data were downloaded from PPMI repository on 8–11 September 2018 and we filtered all coding variants in 82 PD-associated loci (used in-targeted sequencing panel of patient S65N-M70 (ID3185) excluding mitochondrial DNA) with a CADD C-score of over 20 and a carrier frequency less than 1%. Polyphen-2 [81] HVAR-based predictions are provided for each variant.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We wish to thank the Finnish Institute for Molecular Medicine (FIMM) for sequencing services. We are grateful to Tuula Manninen (University of Helsinki, Finland) for technical assistance with human fibroblasts cultures and Sanger sequencing. PPMI is a public–private partnership funded by the Michael J. Fox Foundation for Parkinson's Research and funding partners including: Abbvie, Avid Radiopharmaceuticals, Biogen Idec, Bristol-Myers Squibb, Covance, Eli Lilly & Co., F. Hoffman-La Roche, Ltd., GE Healthcare, Genentech, GlaxoSmithKline, Lundbeck, Merck, MesoScale, Piramal, Pfizer and UCB. Informed consent was signed by the patients enrolled in the PPMI study. We thank the PPMI Data & Publications (DPC) for reviewing the manuscript prior to submission and to Ms Alyssa Reimer and Dr Shalini Padmanabhan of the Michael J. Fox Foundation for advice on obtaining PPMI data. The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about. The authors would like to thank the Exome Aggregation Consortium. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. We are grateful to Dr Xiao Tian and Prof. Vera Gorbunova (University of Rochester, USA) for guidance on establishing adult primary fibroblast cultures. We are grateful to Dr Wolfdieter Springer (Mayo Clinic, USA) for providing an initial aliquot of anti-phospho-ubiquitin antibody for optimization studies. We thank Dr Shane Grealish (Lund University, Sweden) and Dr Fumi Suomi (University of Helsinki, Finland) for critical reading of the manuscript.
Ethics
All animal studies and breeding in Dundee were approved by the University of Dundee ethical review committee, and further subjected to approved study plans by the Named Veterinary Surgeon and Compliance Officer (Dr. Ngaire Dennison) and performed under a UK Home Office project licence in accordance with the Animal Scientific Procedures Act (ASPA, 1986). Associated collaborative institutes which contributed to the present study (mouse behaviour and histology—Cardiff University, UK; mitochondrial respiration analyses—CNRS, Paris, FR) also received institutional ethical approval. Informed consent was obtained from Case 1. Ethical approval for the study (Dnro 432/E9/07) was given by the Ethics Review Board of Helsinki University Central Hospital. We used UPDRS and Modified Hoehn and Yahr scales to evaluate current progression of the disease. Informed consent was signed by the patients enrolled in the PPMI study. The authors have agreed to PPMI Data Use Agreement. The manuscript was reviewed by PPMI Data & Publications Committee (DPC) prior to submission.
Data accessibility
Coordinates and structure factors have been deposited with the PDB database (PDB ID 6HUE).
Authors' contributions
Conceptualization, M.M.K.M. and T.G.M.; investigation, T.G.M., E.B., R.P-P., S.P.B., F.S., S.B., K.B., A.K., L.M.-G., A.R.P., S.M., F.M-L., G.B., R.H., A.K., A.U., J.T., O.A., K.F., L.S., R.B., E.P., M.V., J.E-R.; writing: original draft, T.G.M. and M.M.K.M.; writing: reviewing and editing, T.G.M., R.P-P., S.P.B., F.S., S.B., K.B., A.K., L.M-G., A.R.P., A.U., M.V., J.E-R., P.T., O.C., S.P.B., S.B.D., I.G., A.S. and M.M.K.M.; funding acquisition, I.G.G., A.S., M.M.K.M.; supervision, P.T., O.C., S.P.B., S.B.D., I.G., A.S. and M.M.K.M.
Competing interests
We declare we have no competing interests.
Funding
We thank our colleagues from the MRC PPU support teams for assistance with tissue culture and genotyping and our colleagues in Biological Services at both the University of Dundee and Cardiff University for excellent animal husbandry and care. M.M.K.M. is funded by a Wellcome Trust Senior Research Fellowship in Clinical Science (101022/Z/13/Z). This work was supported by the Wellcome Trust; Medical Research Council; Parkinson's UK; the Michael J. Fox Foundation for Parkinson's disease research; and a EMBO Young Investigator Programme (YIP) award. A. J. Macdonald Menzies Charitable Trust Prize Studentship funds K.B. O.A. is funded by a Rosetrees Trust PhD Studentship. G.B. is supported by an MRC Next Generation Optical Microscopy Award (MR/K015869/1) and Dundee Imaging Facility is supported by the ‘Wellcome Trust Technology Platform’ award (097945/B/11/Z). We are grateful to the sequencing service (School of Life Sciences, University of Dundee); and the MRC PPU Reagents and Services antibody purification teams (coordinated by James Hastie and Hilary McLauchlan) for excellent technical support. O.C. and colleagues (France) are supported by Fondation Institut du Cerveau et de la Moelle Epinière and Agence Nationale pour la Recherche (‘Investissements d'avenir’, grant ANR-10-IAIHU-06) and Innovative Medicines Initiative Joint Undertaking under grant agreement no.115568, resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies' in kind contribution. AS and colleagues (Finland) would like to thank the following funding sources: Sigrid Juselius Foundation; Academy of Finland, Helsinki University Central Hospital, and University of Helsinki (to A.S.), Biomedicum Helsinki Foundation, The Finnish Cultural Foundation and The Finnish Medical Foundation (to R.P-P.). Work by S.P.B. and colleagues (Cardiff) was supported by a Ser Cymru PhD studentship, Life Sciences Research Network Wales (K.F.) and a Wellcome Trust Studentship (L.S.).
References
- 1.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. 1998. Mutations in the Parkin gene cause autosomal recessive juvenile Parkinsonism. Nature 392, 605–608. ( 10.1038/33416) [DOI] [PubMed] [Google Scholar]
- 2.Valente EM, et al. 2004. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160. ( 10.1126/science.1096284) [DOI] [PubMed] [Google Scholar]
- 3.Walden H, Muqit MM. 2017. Ubiquitin and Parkinson's disease through the looking glass of genetics. Biochem. J. 474, 1439–1451. ( 10.1042/BCJ20160498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickrell AM, Youle RJ. 2015. The roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson's disease. Neuron 85, 257–273. ( 10.1016/j.neuron.2014.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McWilliams TG, Muqit MM. 2017. PINK1 and Parkin: emerging themes in mitochondrial homeostasis. Curr. Opin. Cell Biol. 45, 83–91. ( 10.1016/j.ceb.2017.03.013) [DOI] [PubMed] [Google Scholar]
- 6.Harper JW, Ordureau A, Heo JM. 2018. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93–108. ( 10.1038/nrm.2017.129) [DOI] [PubMed] [Google Scholar]
- 7.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. 2014. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153. ( 10.1083/jcb.201402104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazlauskaite A, et al. 2014. Phosphorylation of Parkin at Serine65 is essential for activation: elaboration of a Miro1 substrate-based assay of Parkin E3 ligase activity. Open Biol. 4, 130213 ( 10.1098/rsob.130213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazlauskaite A, et al. 2014. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139. ( 10.1042/BJ20140334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koyano F, et al. 2014. Ubiquitin is phosphorylated by PINK1 to activate Parkin. Nature 510, 162–166. ( 10.1038/nature13392) [DOI] [PubMed] [Google Scholar]
- 11.Kazlauskaite A, et al. 2015. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 16, 939–954. ( 10.15252/embr.201540352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ordureau A, et al. 2014. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375. ( 10.1016/j.molcel.2014.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. 2015. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 60, 7–20. ( 10.1016/j.molcel.2015.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. ( 10.1038/nature14893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ordureau A, Heo JM, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, Rinehart J, Schulman BA, Harper JW. 2015. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl Acad. Sci. USA 112, 6637–6642. ( 10.1073/pnas.1506593112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truban D, Hou X, Caulfield TR, Fiesel FC, Springer W. 2017. PINK1, Parkin, and mitochondrial quality control: what can we learn about Parkinson's disease pathobiology? J. Parkinsons Dis. 7, 13–29. ( 10.3233/JPD-160989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg MS, et al. 2003. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 278, 43 628–43 635. ( 10.1074/jbc.M308947200) [DOI] [PubMed] [Google Scholar]
- 18.Itier J, et al. 2003. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum. Mol. Genet. 12, 2277–2291. ( 10.1093/hmg/ddg239) [DOI] [PubMed] [Google Scholar]
- 19.Kitada T, et al. 2009. Impaired dopamine release and synaptic plasticity in the striatum of Parkin-/- mice. J. Neurochem. 110, 613–621. ( 10.1111/j.1471-4159.2009.06152.x) [DOI] [PubMed] [Google Scholar]
- 20.Damiano M, et al. 2014. Tissue- and cell-specific mitochondrial defect in Parkin-deficient mice. PLoS ONE 9, e99898 ( 10.1371/journal.pone.0099898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, Youle RJ. 2015. Endogenous Parkin preserves dopaminergic substantia nigral neurons following mitochondrial DNA mutagenic stress. Neuron 87, 371–381. ( 10.1016/j.neuron.2015.06.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, Muqit MM, Ganley IG. 2016. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333–345. ( 10.1083/jcb.201603039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai YC, et al. 2015. Phosphoproteomic screening identifies Rab GTPases as novel downstream targets of PINK1. EMBO J. 34, 2840–2861. ( 10.15252/embj.201591593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barini E, et al. 2018. The anthelmintic drug niclosamide and its analogues activate the Parkinson's disease associated protein kinase PINK1. ChemBioChem 19, 425–429. ( 10.1002/cbic.201700500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ordureau A, et al. 2018. Dynamics of PARKIN-dependent mitochondrial ubiquitylation in induced neurons and model systems revealed by digital snapshot proteomics. Mol. Cell 70, 211–227 e218. ( 10.1016/j.molcel.2018.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian X, et al. 2013. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349. ( 10.1038/nature12234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks SP, Dunnett SB. 2009. Tests to assess motor phenotype in mice: a user's guide. Nat. Rev. Neurosci. 10, 519–529. ( 10.1038/nrn2652) [DOI] [PubMed] [Google Scholar]
- 28.Rial D, et al. 2014. Behavioral phenotyping of Parkin-deficient mice: looking for early preclinical features of Parkinson's disease. PLoS ONE 9, e114216 ( 10.1371/journal.pone.0114216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelm-Nelson CA, et al. 2018. Characterization of early-onset motor deficits in the Pink1-/- mouse model of Parkinson disease. Brain Res. 1680, 1–12. ( 10.1016/j.brainres.2017.12.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chance B, Williams GR. 1955. Respiratory enzymes in oxidative phosphorylation. I. Kinetics of oxygen utilization. J. Biol. Chem. 217, 383–393. [PubMed] [Google Scholar]
- 31.Brand MD, Nicholls DG. 2011. Assessing mitochondrial dysfunction in cells. Biochem. J. 435, 297–312. ( 10.1042/BJ20110162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuda W, Furuta T, Nakamura KC, Hioki H, Fujiyama F, Arai R, Kaneko T. 2009. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J. Neurosci. 29, 444–453. ( 10.1523/JNEUROSCI.4029-08.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. 2014. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. ( 10.1016/j.cell.2014.10.010) [DOI] [PubMed] [Google Scholar]
- 34.McWilliams TG, Howard L, Wyatt S, Davies AM. 2017. TNF superfamily member APRIL enhances midbrain dopaminergic axon growth and contributes to the nigrostriatal projection in vivo. Exp. Neurol. 298, 97–103. ( 10.1016/j.expneurol.2017.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vingill S, et al. 2016. Loss of FBXO7 (PARK15) results in reduced proteasome activity and models a Parkinsonism-like phenotype in mice. EMBO J. 35, 2008–2025. ( 10.15252/embj.201593585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun N, et al. 2015. Measuring in vivo mitophagy. Mol. Cell 60, 685–696. ( 10.1016/j.molcel.2015.10.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuma A, Komatsu M, Mizushima N. 2017. Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619–1628. ( 10.1080/15548627.2017.1343770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JJ, Sanchez-Martinez A, Zarate AM, Beninca C, Mayor U, Clague MJ, Whitworth AJ. 2018. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or Parkin. J. Cell Biol. 217, 1613–1622. ( 10.1083/jcb.201801044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, Ganley IG. 2018. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439– 449 e435 ( 10.1016/j.cmet.2017.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruyama M, et al. 2000. Novel mutations, pseudo-dominant inheritance, and possible familial affects in patients with autosomal recessive juvenile Parkinsonism. Ann. Neurol. 48, 245–250. ( 10.1002/1531-8249(200008)48:2%3C245::AID-ANA15%3E3.0.CO;2-2) [DOI] [PubMed] [Google Scholar]
- 41.Pao KC, et al. 2016. Probes of ubiquitin E3 ligases enable systematic dissection of Parkin activation. Nat. Chem. Biol. 12, 324–331. ( 10.1038/nchembio.2045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315. ( 10.1038/ng.2892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Fonzo A, Bordoni A, Crimi M, Sara G, Del Bo R, Bresolin N, Comi GP. 2003. POLG mutations in sporadic mitochondrial disorders with multiple mtDNA deletions. Hum. Mutat. 22, 498–499. ( 10.1002/humu.9203) [DOI] [PubMed] [Google Scholar]
- 44.Graziewicz MA, Longley MJ, Copeland WC. 2006. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 106, 383–405. ( 10.1021/cr040463d) [DOI] [PubMed] [Google Scholar]
- 45.Blok MJ, et al. 2009. The unfolding clinical spectrum of POLG mutations. J. Med. Genet. 46, 776–785. ( 10.1136/jmg.2009.067686) [DOI] [PubMed] [Google Scholar]
- 46.Flanagan N, Healy E, Ray A, Philips S, Todd C, Jackson IJ, Birch-Machin MA, Rees JL. 2000. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum. Mol. Genet. 9, 2531–2537. ( 10.1093/hmg/9.17.2531) [DOI] [PubMed] [Google Scholar]
- 47.Gao X, Simon KC, Han J, Schwarzschild MA, Ascherio A. 2009. Genetic determinants of hair color and Parkinson's disease risk. Ann. Neurol. 65, 76–82. ( 10.1002/ana.21535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong J, Gao J, Nalls M, Gao X, Huang X, Han J, Singleton AB, Chen H. 2014. Susceptibility loci for pigmentation and melanoma in relation to Parkinson's disease. Neurobiol. Aging 35, 1512.e5–1512.e10. ( 10.1016/j.neurobiolaging.2013.12.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lubbe SJ, et al. 2016. Is the MC1R variant p.R160 W associated with Parkinson's? Ann. Neurol. 79, 159–161. ( 10.1002/ana.24527) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sidransky E, et al. 2009. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 361, 1651–1661. ( 10.1056/NEJMoa0901281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gegg ME, Schapira AHV. 2018. The role of glucocerebrosidase in Parkinson disease pathogenesis. FEBS J. 285, 3591–3603. ( 10.1111/febs.14393) [DOI] [PubMed] [Google Scholar]
- 52.Allen GF, Toth R, James J, Ganley IG. 2013. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 14, 1127–1135. ( 10.1038/embor.2013.168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wauer T, Simicek M, Schubert A, Komander D. 2015. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524, 370–374. ( 10.1038/nature14879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar A, et al. 2017. Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity. Nat. Struct. Mol. Biol. 24, 475–483. ( 10.1038/nsmb.3400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gladkova C, Maslen SL, Skehel JM, Komander D. 2018. Mechanism of Parkin activation by PINK1. Nature 559, 410–414. ( 10.1038/s41586-018-0224-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauve V, Sung G, Soya N, Kozlov G, Blaimschein N, Miotto LS, Trempe JF, Lukacs GL, Gehring K. 2018. Mechanism of Parkin activation by phosphorylation. Nat. Struct. Mol. Biol. 25, 623–630. ( 10.1038/s41594-018-0088-7) [DOI] [PubMed] [Google Scholar]
- 57.Deas E, Plun-Favreau H, Wood NW. 2009. PINK1 function in health and disease. EMBO Mol. Med. 1, 152–165. ( 10.1002/emmm.200900024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuda S, Kitagishi Y, Kobayashi M. 2013. Function and characteristics of PINK1 in mitochondria. Oxid. Med. Cell Longev. 2013, 601587 ( 10.1155/2013/601587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu XH, et al. 2009. Bacterial artificial chromosome transgenic mice expressing a truncated mutant Parkin exhibit age-dependent hypokinetic motor deficits, dopaminergic neuron degeneration, and accumulation of proteinase K-resistant alpha-synuclein. J. Neurosci. 29, 1962–1976. ( 10.1523/JNEUROSCI.5351-08.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Rompuy AS, Lobbestael E, Van der Perren A, Van den Haute C, Baekelandt V. 2014. Long-term overexpression of human wild-type and T240R mutant Parkin in rat substantia nigra induces progressive dopaminergic neurodegeneration. J. Neuropathol. Exp. Neurol. 73, 159–174. ( 10.1097/NEN.0000000000000039) [DOI] [PubMed] [Google Scholar]
- 61.Dave KD, et al. 2014. Phenotypic characterization of recessive gene knockout rat models of Parkinson's disease. Neurobiol. Dis. 70, 190–203. ( 10.1016/j.nbd.2014.06.009) [DOI] [PubMed] [Google Scholar]
- 62.Sliter DA, et al. 2018. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. ( 10.1038/s41586-018-0448-9) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Rojansky R, Cha MY, Chan DC. 2016. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife 5, e17896 ( 10.7554/eLife.17896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar A, et al. 2015. Disruption of the autoinhibited state primes the E3 ligase Parkin for activation and catalysis. EMBO J. 34, 2506–2521. ( 10.15252/embj.201592337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hattori N, Mizuno Y. 2017. Twenty years since the discovery of the Parkin gene. J. Neural Transm. (Vienna) 124, 1037–1054. ( 10.1007/s00702-017-1742-7) [DOI] [PubMed] [Google Scholar]
- 66.Jang JY, Blum A, Liu J, Finkel T. 2018. The role of mitochondria in aging. J. Clin. Invest. 128, 3662–3670. ( 10.1172/JCI120842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brooks SP, Trueman RC, Dunnett SB. 2012. Assessment of motor coordination and balance in mice using the rotarod, elevated bridge, and footprint tests. Curr. Protoc. Mouse Biol. 2, 37–53. ( 10.1002/9780470942390.mo110165) [DOI] [PubMed] [Google Scholar]
- 68.Bayram-Weston Z, Jones L, Dunnett SB, Brooks SP. 2012. Light and electron microscopic characterization of the evolution of cellular pathology in the R6/1 Huntington's disease transgenic mice. Brain Res. Bull. 88, 104–112. ( 10.1016/j.brainresbull.2011.07.009) [DOI] [PubMed] [Google Scholar]
- 69.Bayram-Weston Z, Jones L, Dunnett SB, Brooks SP. 2016. Comparison of mHTT antibodies in Huntington's disease mouse models reveal specific binding profiles and steady-state ubiquitin levels with disease development. PLoS ONE 11, e0155834 ( 10.1371/journal.pone.0155834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abercrombie M. 1946. Estimation of nuclear population from microtome sections. Anat. Rec. 94, 239–247. ( 10.1002/ar.1090940210) [DOI] [PubMed] [Google Scholar]
- 71.Fournier M, et al. 2013. Parkin depletion delays motor decline dose-dependently without overtly affecting neuropathology in alpha-synuclein transgenic mice. BMC Neurosci. 14, 135 ( 10.1186/1471-2202-14-135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kilpatrick IC, Jones MW, Phillipson OT. 1986. A semiautomated analysis method for catecholamines, indoleamines, and some prominent metabolites in microdissected regions of the nervous system: an isocratic HPLC technique employing coulometric detection and minimal sample preparation. J. Neurochem. 46, 1865–1876. ( 10.1111/j.1471-4159.1986.tb08506.x) [DOI] [PubMed] [Google Scholar]
- 73.Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods 138, 85–98. ( 10.1016/j.jviromet.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 74.Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. 2011. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867. ( 10.1038/emboj.2011.204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60(Pt 12 Pt 1), 2126–2132. ( 10.1107/S0907444904019158) [DOI] [PubMed] [Google Scholar]
- 76.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367. ( 10.1107/S0907444911001314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 ( 10.1093/nar/gkq603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lek M, et al. 2016. Analysis of protein-coding genetic variation in 60 706 humans. Nature 536, 285–291. ( 10.1038/nature19057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Apweiler R, et al. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32(Database issue), D115–D119. ( 10.1093/nar/gkh131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249. ( 10.1038/nmeth0410-248) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited with the PDB database (PDB ID 6HUE).