Abstract
Natural history museums and the specimen collections they curate are vital scientific infrastructure, a fact as true today as it was when biologists began collecting and preserving specimens over 200 years ago. The importance of museum specimens in studies of taxonomy, systematics, ecology and evolutionary biology is evidenced by a rich and abundant literature, yet creative and novel uses of specimens are constantly broadening the impact of natural history collections on biodiversity science and global sustainability. Excellent examples of the critical importance of specimens come from their use in documenting the consequences of environmental change, which is particularly relevant considering the alarming rate at which we now modify our planet in the Anthropocene. In this review, we highlight the important role of bird, mammal and amphibian specimens in documenting the Anthropocene and provide examples that underscore the need for continued collection of museum specimens.
This article is part of the theme issue ‘Biological collections for understanding biodiversity in the Anthropocene’.
Keywords: museum specimens, Anthropocene, climate change, contamination, emergent disease
1. Introduction
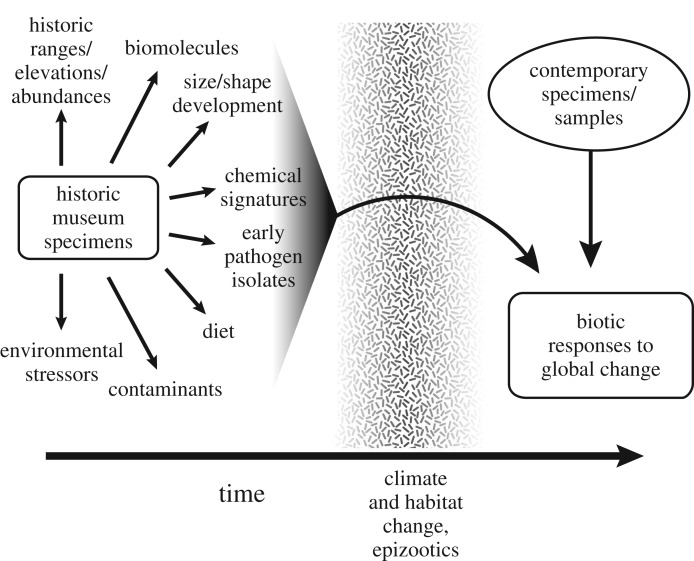
Abundant evidence of human-driven environmental change supports the designation the Anthropocene as a new geological epoch [1]. Notable signatures of the Anthropocene include precipitous increases in global temperature and temperature anomalies (i.e. climate change) [2,3], contamination [4–6], emergence of infectious diseases [7], species' declines [8] and many others [1]. Prominent among indicators of environmental change are specimens curated in natural history museums [9–11]. Museum specimens are a particularly powerful resource for documenting change in the environment because they offer scientists snapshots of the Earth across spatial, temporal and taxonomic scales [12–16]. In this paper, we highlight the use of museum specimens of birds, mammals and amphibians as sensitive indicators of environmental change in the Anthropocene, reviewing exemplary studies that have employed museum specimens to document changes in emergent diseases, contamination, isotope/hormone ecology and climate. Central to our argument is the view that a well-curated museum specimen represents a multidimensional snapshot of the environment at a specific time and place in the past (figure 1, [17]). Ample evidence now exists to show that natural history museum specimens can be used for diverse applications of value to society, yet continued support of museums by funding agencies and dedication to specimen collection by museums are needed to build and maintain this critical scientific resource moving forward.
Figure 1.
Vertebrate museum specimens as historical snapshots of the total environment. Diverse data types stemming from museum specimens (left of the figure) provide historical data that can be compared to data sampled from contemporary specimens or samples (right of the figure). Together these temporal datasets can shed light on how anthropogenic change (stippled bar at the centre of figure) drives diverse physiological, morphological, genetic and behavioural changes in vertebrate populations.
2. Museum specimens document the origin and spread of emergent infectious diseases
One characteristic of the Anthropocene is an acceleration in the emergence of infectious diseases [7]. Emergent diseases in natural populations are now increasing consistently [18] and will pose challenges for wildlife populations, humans and our domesticated or crop species in the near future [19–21]. Museum collections, including biorepositories of frozen vertebrate tissue, have played a central role in the discovery of previously unknown pathogens in the past 25 years [22–24]. That role is sure to increase in the future if we build the temporal, spatial and taxonomic breadth of these collections through continued fieldwork [25]. Through retrospective screening of museum samples, new insights in pathobiology include the rapid identification of key attributes of newly emergent pathogens, such as taxonomic identity, temporal dynamics and critical aspects related to spatial distribution [26,27]. In aggregate, museum collections often provide the detailed sampling of vertebrate hosts necessary to refine the potential spatial distribution of a pathogen and to confirm whether a pathogen is narrowly confined to a single host or instead ranges widely across multiple hosts [28,29]. These critical diagnostic features allow investigators to predict conditions associated with emergence [30], and also to design effective public health response efforts. More detailed assessments, using phylogeographic perspectives, for example, allow us to probe the dynamics of the historical association of both the pathogen and host. Importantly, because up to 75% of the emerging pathogens responsible for the most serious human disease outbreaks are zoonotic in origin [31], museum collections can play a critical role in public health [32], but only if this critical resource continues to be developed so it can serve as a temporal and spatial database of potential human diseases in animal hosts.
(a). Documenting fungal pathogens in declining amphibian populations
One of the best examples of the use of museum collections in documenting the spread of epizootics is the tracking the origin and spread of the chytrid fungus Batrachochytrium dendrobatidis (Bd), the cause of devastating declines in amphibian populations globally. Chytrid was first brought to the attention of the international community as a possible cause of global amphibian declines owing to chytridiomycosis in 1998 [33], over a decade after the first reports of amphibian declines in the mid-1980s [34]. Typically, amphibians and reptile specimens are stored in formalin and transferred to ethanol for long-term curation. These specimens are ideal for histology or dissection-based diagnostic tests; however, given the detrimental effects of formalin on the long-term quality of DNA [35], retrieving DNA sequences from formalin-fixed specimens has been a challenge. Recent advances in DNA extraction methods and next-generation sequencing have overcome significant obstacles to molecular analysis of amphibian specimens in museum collections [35–38]. Thus, museum collections proved critical in establishing Bd as the causative agent in amphibian declines and in tracing the routes by which this pathogen spread across the globe, both through morphological and genetic analyses of historical amphibian samples across diverse localities and habitats [26–28].
Soon after the discovery of the emergence of Bd, researchers turned to archived museum collections to better understand the origins, transmission and spread of the pathogen. These specimens yielded diverse types of useful data, such as multilocus genotypes of chytrid from skin swabs of diverse frog species and populations [39,40]; quantitative PCR measurement of pathogen levels [41]; and whole genome sequences of Bd isolates [42,43]. Bd has been retrieved from museum specimens collected in the late nineteenth [26] and early twentieth centuries [44] and has been documented on multiple continents with the aid of museum collections [45,46]. The most recent surveys using whole genome sequencing, aided by museum collections, suggest substantial diversity of Bd in the Korean peninsula, suggesting that East Asia may have been the source of the ongoing epizootic and that international trade facilitated its spread [43].
A newly described chytrid fungus, Batrachochytrium salamandrivorans (Bsal), infecting primarily newts (salamanders in the family Salamandridae) has also emerged from Asia and is now threatening salamanders in western Europe and the New World [47,48]. The global scale of transport that characterizes the Anthropocene makes it likely that this pathogen will continue to spread to areas where native salamanders exist and have never been exposed to this pathogen before. To date, this newly emergent chytrid has been identified in museum specimens and natural populations from Asia [48,49], but has not been detected elsewhere, confirming that we are faced with an invasive emergent pathogen. As with frogs in the Bd invasion, salamander specimens in natural history collections will be a baseline for diagnoses of pathogen presence and absence for this newly emerging pathogen.
(b). Characterizing the Sin Nombre hantavirus
Museum collections and museum-based fieldwork played a central role in identifying, tracking and mitigating the deadly Sin Nombre hantavirus (Hantaviridae: Order Bunyavirales). The Sin Nombre virus [50] emerged in the Four Corners region of the American Southwest in the spring of 1993, killing 27 people that year. Because the virus was previously unknown, this emergence challenged our public health system, with local clinics and healthcare providers unable to diagnose, treat or in some cases even admit suspected cases [22]. Speculation about the origin of this pulmonary illness ranged wildly [51] but the availability of frozen archives of tissues from wild rodents with known collection dates and localities at the Museum of Southwestern Biology (MSB) allowed investigators from the Centers for Disease Control and University of New Mexico Hospital to quickly identify the pathogen as a hantavirus with a wild reservoir traced to the common deer mouse (Peromyscus maniculatus). In addition to identifying Sin Nombre as a hantavirus and demonstrating its zoonotic source, historic specimens also showed that this virus had been circulating in deer mouse populations since at least the earliest collections archived a decade earlier [22].
Since the identification of Sin Nombre virus in the early 1990s, our knowledge of this group of pathogens has been radically reshaped thanks to progress stimulated by access to extensive frozen (now more than 138 000 rodents, each with multiple tissues at MSB) and ethanol-preserved samples in museums [52]. More than 25 additional hantaviruses have been described in the Western Hemisphere in the past 25 years [52–55], some of which lead to high mortality in humans [23,24]. Further research based on intensive field surveys has also detected hantaviruses in mammals other than rodents [52,56,57], including moles (Talpidae), shrews (Soricidae) and bats (Nycteridae, Vespertilionidae, Pteropodidae). The discovery of diverse new hosts radically changes the way we think about these potentially zoonotic viruses [58], especially because some host species are commensal with humans and pose a critical public health issue [59,60]. Additionally, comprehensive phylogenetic analyses have clarified the evolutionary relationships of hantaviruses [61,62], and a growing number of cases of co-circulation of multiple hantaviruses opens the possibility of rapid virus evolution owing to reassortment among their tri-partite genomes.
Importantly, in response to the emergence of chytrid and hantavirus, several museum collections facilitated productive collaborations among public health agencies, virologists, evolutionary biologists and conservation biologists that have reshaped interdisciplinary approaches to pathogen discovery [32,43,52,63]. Temporally deep and taxonomically diverse biorepositories of vertebrates and their tissues allow scientists to search for and discover novel pathogens, essentially providing basic infrastructure for rapid and efficient assessment, prevention and mitigation of emerging diseases. Moving forward, we need to recognize that biorepositories and associated databases are critical infrastructure in pathobiology research [63]. Biorepositories have the potential to reveal key aspects of the biology of newly emergent diseases and their hosts, including some that could reduce the impact of catastrophic events [64].
3. Tracking the spread of contaminants across time and space
Environmental contamination is one of the most pervasive anthropogenic impacts and has obvious implications for ecosystem and human health. Museum specimens are a powerful and increasingly used resource for documenting anthropogenic contamination in the environment [65–72]. Vertebrates accumulate contaminants in their integument, feathers and organs, through physical contact with the environment and also through bioaccumulation of pollutants in their diet [73]. Many of these contaminants remain as traces in host tissues, sometimes for decades or centuries [74]. Accordingly, the amount of contamination on museum specimens or in associated tissue samples provides a sensitive index of ongoing contamination and a timeline of historical trends in the presence and concentration of particular contaminants in the environment. Importantly, collection data of museum specimens are spatially and temporally explicit, allowing researchers to investigate contamination in a geographical and temporal context. Two recent studies [75,76] demonstrate how temporally well-sampled museum specimens of birds are sensitive indicators of anthropogenic emissions of black carbon and mercury.
(a). Trends in black carbon pollution measured from bird specimens
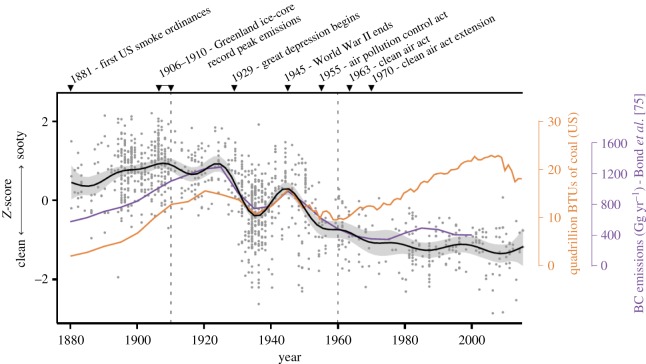
Black carbon, or soot, is an atmospheric pollutant that arises from the inefficient combustion of fossil fuels and has negative impacts on human health and climate change [77–81]. Birds accumulate black carbon on their feathers, rendering otherwise white feathers a sooty grey colour. For example, a bird's plumage can become soiled by black carbon through physical contact with substrates where black carbon has precipitated from the air. The yearly replacement of a bird's plumage, or moulting, adds to the sensitivity of bird specimens as indicators of change because pollutant deposition on or within a feather provides an environmental snapshot from the specific year in which the specimen was collected [75]. Although ornithologists have long been aware of the existence of sooty bird specimens in natural history museums, only recently have these specimens been examined in detail. DuBay & Fuldner [75] used scanning electron microscopy to confirm that the feathers of sooty specimens from the US Manufacturing Belt (Wisconsin, Illinois, Michigan, Indiana, Ohio and Pennsylvania) are covered in black carbon particulates, while melanin pigments in feathers are similar. This demonstrated that the variation in plumage of birds was a result of black carbon soilage rather than adaptive increases in melanin pigmentation such as in cases of industrial melanism. Using a time series of 1347 bird specimens collected between 1880 and 2015, the authors quantified the reflectance of breast and belly feathers as a relative measure of the amount of black carbon deposition on each specimen. They discovered that the amount of black carbon deposited on feathers decreased through time, closely tracking the implementation of environmental policies over that time period (figure 2). In addition, DuBay & Fuldner [75] showed that models of black carbon from the late 1880s [78] likely underestimate black carbon levels, underscoring the utility of museum specimens as inventories of environmental contaminants.
Figure 2.
Black carbon soiling of museum specimens (black line) closely tracks previous estimates of black carbon emissions (purple line) [78] between 1880 and 2015. Estimates of black carbon decouple from coal consumption in the United States following progressive clean air legislation. Figure used with permission from DuBay & Fuldner [75]. BC, black carbon; BTUs, British thermal units. (Online version in colour.)
(b). Museum specimens track organic mercury contamination
Mercury is a significant environmental pollutant and an anthropogenic increase of mercury in the environment is a well-established trend that results directly from human discharge of mercury into the Earth's ecosystems [5,6]. Although IHg (inorganic mercury) is the predominant form of mercury pollution in the environment, it is converted into organic or methyl mercury (MeHg) by sulfate-reducing bacteria [82]. MeHg is an environmental toxin that bioaccumulates through food webs and builds up in animal tissues, including bird feathers [83,84]. Few studies have investigated whether anthropogenic increases in IHg have resulted in increased bioaccumulation of MeHg, much less the potential effects of increased MeHg on avian communities or physiology [85]. Novel studies have used museum specimens to demonstrate increases in the amount of MeHg deposited in tissues of fish [65], birds [67–71,86] and bats [87]. One recent study used museum specimens to document MeHg contamination in black-footed albatross (Phoebastria nigripes), a seabird formerly designated as vulnerable and now near-threatened [88]. Vo et al. [76] measured both IHg and MeHg in the feathers of P. nigripes specimens collected in the Pacific Ocean across a 120-year period and documented a statistically significant increase in MeHg. Crucially, unlike most previous studies, the authors were able to distinguish between curator-mediated IHg and environmentally accumulated MeHg through creative use of control populations and by using a gas chromatography inductively coupled plasma mass spectrometry (GC-ICP-MS) approach, which can chemically distinguish between IHg and MeHg. Furthermore, Vo et al. [76] showed that a significant number of specimens collected after 1990 had MeHg concentrations above thresholds considered by the EPA to be adverse, highlighting the severity of mercury contamination in the Pacific Ocean. Museum specimens of birds and other vertebrates will undoubtedly have a prominent role in further documenting mercury and other contaminants across the planet's ecosystems, a critical first step in mitigating our impact on the environment.
4. Museum specimens record historical changes in diet, migration routes, environmental stress and morphology
In addition to providing detailed records of environmental pollutants, museum specimens can be used as sensitive indicators of changes in organisms' diet, changing migratory routes and stress as a result of changing environments [72]. These insights stem from the fact that stable isotopes [89,90] and hormones [91,92] remain relatively inert in museum specimens and can be detected reliably over periods of decades or centuries. Stable isotopes have been used to trace evolving diets of fish [93], mammals [94–96] and birds [89,97–99], with ratios of isotopic nitrogen and carbon, in particular, providing insight into shifts in dietary regime over the past several decades. Isotopes can also be used to create ‘isoscapes’, maps of isotope isoclines across the landscape, allowing researchers to connect breeding and wintering ranges of migratory species [100], among other inferences such as elevational migration [101]. For example, Hobson et al. [100] used museum specimens to identify a migratory divide between western and eastern populations of rusty blackbird (Euphagus carolinus). Levels of corticosterone, an indicator of stress in natural populations, are increasingly being used to assess the activity of the hypothalamic–pituitary–adrenal axis, a measure of organisms' response to environmental pollutants and other stressors [98]. In addition, multiple studies have documented consistent trends in morphological traits such as body size and wing-pointedness, often in response to anthropogenic change [102] and increasing habitat patchiness [103], although the precise drivers are still debated [104]. Although detection of such trends is not without biases in terms of location or temporal period sampled, museum specimens are providing unprecedented windows into organisms' responses to environmental change, without which we would know far less about the impact of anthropogenic change on natural populations.
5. Measuring the effects of climate change on ecological communities using museum specimens
Understanding the current and future impact of climate change on Earth's ecosystems is a fundamental challenge for humanity [105–107]. One critical goal is predicting how organisms and ecosystems will shift across landscapes in response to climate change. In general, climate change is linked to significant range shifts [108–110], extinctions [111] and potentially community reorganization [112]; however, the predictive power of models is often hampered by uncertainties in climate change trajectories [113] as well as limited knowledge of the distributions of species prior to the Anthropocene. Museum collections span the onset of rapid climate change, and thus have the potential to increase the resolution of climate change studies with precise records of where and when organisms occurred in the past as well as demographic information such as age distributions [114]. Furthermore, museum specimens and associated tissue samples provide opportunities to study both morphological and genetic responses to climate and habitat change [115].
(a). Museum specimens of small mammals reveal climate-associated range shifts
One of the best examples of the potential of museum collections to document climate change is the Grinnell Resurvey Project. From 1904 to 1940, Joseph Grinnell and colleagues from the Museum of Vertebrate Zoology (MVZ) at the University of California, Berkeley, collected and thoroughly surveyed the terrestrial ecosystems of California and the western United States to document vertebrate diversity in time and space. Their legacy of specimens, meticulous field notes and photographs are a veritable ‘gold mine for investigations of species' responses to climate change, changes in human land use and other stressors’ [116]. Grinnell and colleagues knew that their efforts would establish a baseline for future studies, although it would be nearly a century before the power of their work would be fully realized. In the early 2000s, the Grinnell Resurvey Project (GRP) was started by MVZ researchers and collaborators who set out to follow in Grinnell's footsteps by collecting and resurveying sites that had been visited in the early twentieth century. As of today, the GRP has produced many publications and resources that illustrate the importance of scientific specimens in documenting climate change [109,117–120].
One influential study resulting from the GRP investigated distributional changes of small-mammals across a 3000 m elevational gradient in California's Sierra Nevada Mountains. Moritz et al. [109] minimized the confounding effects of land-use change on small-mammal distributions over time by repeating Grinnell's surveys and collecting efforts across Yosemite National Park, a protected and pristine landscape established in 1890. Furthermore, standardized field protocols and detailed field notes from historical and contemporary surveys allowed the researchers to robustly estimate species' absences through occupancy models. On average, the ranges of 14 species shifted 500 m upwards across the Yosemite transect over nearly a century. The shift in species' ranges was consistent with a 3°C increase in temperature recorded between historical and contemporary surveys, with high-elevation species exhibiting range contractions and low-elevation species exhibiting range expansions. Overall, the observed pattern of upslope range increases and associated range contractions does not bode well for high-elevation species whose distributions may become highly fragmented or potentially ‘pushed’ off of mountain tops [121,122].
The genetic consequences of such historic range contractions have been revealed in spectacular detail owing to methodological and technological innovations over the past 20 years in extracting and sequencing highly degraded DNA from historical specimens [123]. A salient example of the use of specimens and novel next-generation sequencing in documenting the impacts of climate change is a study of alpine chipmunks (Tamias alpinus) [117], a species that has experienced significant range contractions since the surveys of Grinnell and colleagues in the early 1900s [109]. Using a novel exon capture protocol and robust bioinformatic pipelines, Bi et al. [117] investigated the impact of range contractions on the genetic diversity and population structure of T. alpinus. Following extensive data filtering and DNA damage corrections of single nucleotide polymorphisms from 10 583 exons, the authors [117] found no change in genetic diversity between historic samples collected by Grinnell and colleagues and modern samples collected by the GRP; however, they uncovered newly arisen population subdivision as a result of range retractions, a pattern also found in other alpine species [124]. Such studies illustrate how museum specimens and their associated tissue samples are invaluable resources for documenting climate change and its impacts on biodiversity.
6. Discussion
(a). Maximizing the utility of vertebrate specimens for the Anthropocene
We have reviewed exemplary studies that use museum specimens to document anthropogenic change and conclude by discussing the importance of continued collecting of specimens, as well as best practices for museum scientists moving forward. Although it is widely accepted that existing specimens curated in natural history museums document the past, continued dedication to collecting new specimens is often criticized and overlooked as an invaluable investment in the future [125]. We argue that continued collection of new specimens is a necessity without which future scientists will be severely limited in their abilities to document and predict the impact of climate change and other rapidly intensifying anthropogenic pressures on biodiversity and the environment [15,126]. It is imperative that biodiversity scientists continue to direct their efforts to collect specimens across time, space and taxonomic diversity with sufficient sample sizes, metadata and breadth so as to ensure maximum impact across multiple disciplines [13,14].
A concerted effort to maximize the potential of museum collections to document the changing planet will depend critically on greater collaboration between field ecologists, government permitting agencies, citizen and museum scientists. Different questions and goals for documenting the Anthropocene will require different breadths and intensities of sampling and different modes of preservation. For example, the National Ecological Observatory Network (NEON) gives unprecedented opportunities to provide snapshots of continent-wide ecosystems via the collection of animal populations [127,128], and there is ongoing discussion about what to collect and how best to preserve it [25,129]. Collection of vertebrate populations has always been challenging in terms of the labour, cost and museum space required to document diversity adequately. These challenges are exacerbated because we do not know what types of specimens and samples will allow us to monitor future change in multiple dimensions, and because infrastructural constraints place limits on the breadth of samples that can be collected from any given specimen. To the extent that it can be accommodated by the museum community, the ‘total specimen’ [130]—one that maximizes the preservation and availability of the phenotypic and genotypic variation in lineages across space and time—is our best option for providing an ongoing record of biodiversity on a rapidly changing planet.
It is already clear that in order to maximize specimen value for tracking environmental change, extensive re-collecting of high quality, data-rich specimens will be necessary, such as envisioned by the Global Genome Initiative [131]. Best practices for museums moving forward include preserving high-quality tissue samples, associated parasites and multi-part specimens [130]. First, we encourage museums to preserve tissue samples capable of yielding high-molecular-weight DNA for whole-genome sequencing. Similarly, we recommend that RNA-quality tissues of multiple organs be preserved for transcriptomic and epigenomics studies [132]. Parasites associated with host specimens should also be preserved to foster integration across diverse sets of questions, such as whether hosts and parasites respond concordantly to anthropogenic change [25]. We also advocate preserving diverse voucher specimens per individual, such as skeletons and skins from the same individual, spread-wings and other non-traditional vouchers [133] or fluid-preserved specimens commonplace in herpetological collections; such specimens have tremendous potential to be analysed with modern scanning technologies [134,135]. Ongoing salvage will continue to be a mainstay for vertebrate collections, but ultimately active, research-grade collections will prove the most valuable. Finally, we stress that online access to original specimen data linked to research data subsequently derived from each specimen (e.g. gene sequences, CT scans, photographs, sound recordings, etc.) provides a powerful nexus for integrating diverse investigations [74,136–138]. The responsibility for reporting the detailed provenance of and metadata associated with specimens used in scientific research falls equally on researchers, museum curators, authors, reviewers and most critically on the editors of journals that publish the research. The reporting of many types of data archiving, such as DNA sequences and phylogenetic trees, is now standard across many journals in the biological sciences, but the reporting of original specimen information from such data, or links to them, is haphazard and inconsistent at best.
(b). The role of the public and the evolving perception of museums
Paramount in the effort to document changing biodiversity through time is the involvement of the general public, which often grossly misunderstands the intent behind vertebrate collections and their value to society [139]. It is often challenging for the public to separate the negative consequences for individual animals incurred by collecting from the often imperceptible impact at the population level. Not only the general public but also often scientists and wildlife managers themselves oppose continued collecting, often owing to misconceptions about the perceived negative impact of museum collecting on vertebrate populations [140,141]. Involvement of the public through citizen science in ongoing specimen-based documentation of biodiversity, for example, through ‘BioBlitzes’, and in decision making as to what to collect, will be essential [142]. Specimens and associated data can readily be incorporated into educational enterprises and provide powerful opportunities for student-led, inquiry-driven lessons about diverse aspects of our natural world, including climate change and the Anthropocene [136,143,144].
The public and non-museum research communities often have the mistaken perception that museum specimens are collected for a narrow range of uses in the fields of taxonomy, systematics and biogeography, or perhaps that specimens collected for such these fields will have little use outside them. By highlighting the utility of both historical and modern museum specimens in documenting change in the environment, we showcase uses of museum collections in research and teaching that may be less well known outside the museum community. The examples presented here illustrate how museum specimens are a powerful, albeit underutilized resource for documenting the Anthropocene—the emergence and spread of zoonotic diseases, environmental contamination, environmental stressors and climate change—among other assaults to our planet and its inhabitants. The continued collection of museum specimens will ensure detailed documentation of the Anthropocene and its myriad effects [145,146]. With the long time-scales they represent, museum specimens will play an even more important role in future studies of environmental change as preservation methods improve, novel technologies are developed, and creative thinking is applied to unlock their as yet unrealized potential.
Acknowledgements
We thank two anonymous reviewers for their constructive comments on the manuscript, and Gustavo Bravo for suggestions on figure 1.
Data accessibility
This article has no additional data.
Authors' contributions
C.J.S and S.V.E outlined the main scope of the review. C.J.S. led the writing of the paper; S.V.E. led the revision of the paper; and all authors contributed text to the final version and gave their final approval for publication.
Competing interests
The authors declare no competing interests related to the subject matter.
Funding
Fieldwork by S.V.E. and C.J.S. to collect avian specimens for deposit in the Museum of Comparative Zoology (MCZ) and other museums has been supported by Harvard University, the MCZ Putnam Fund, the Frank M. Chapman Memorial Award of the American Museum of Natural History, the Nuttall Ornithological Club, the Carl B. Koford and Louise Kellogg research awards of Museum of Vertebrate Zoology, UC Berkeley, grants from the National Geographic Society and NSF grants DEB 0077804, 0108249, 0815705, 0923088 and 1258784. Mammal specimens and associated parasites and pathogens were collected by J.A.C. on fieldwork supported by the US Geological Survey, USDA Forest Service, US Fish and Wildlife Service and NSF grants 0744025, 1057383, 1258010 and 1561342. Work by J.A.C. and S.V.E to integrate collections into undergraduate education was sponsored by NSF 0956129. K.Z.'s. fieldwork was funded by NSF grants DEB-1045960 and DEB-1120249.
References
- 1.Waters CN, et al. 2016. The Anthropocene is functionally and stratigraphically distinct from the Holocene. Science 351, aad2622 ( 10.1126/science.aad2622) [DOI] [PubMed] [Google Scholar]
- 2.Consortium PK. 2013. Continental-scale temperature variability during the past two millennia. Nat. Geosci. 6, 339–346. ( 10.1038/ngeo1797) [DOI] [Google Scholar]
- 3.Wanner H, Mercolli L, Grosjean M, Ritz SP. 2015. Holocene climate variability and change; a data-based review. J. Geol. Soc. 172, 254–263. ( 10.1144/jgs2013-101) [DOI] [Google Scholar]
- 4.Novakov T, Ramanathan V, Hansen JE, Kirchstetter TW, Sato M, Sinton JE, Sathaye JA. 2003. Large historical changes of fossil-fuel black carbon aerosols. Geophys. Res. Lett. 30, 1324 ( 10.1029/2002gl016345) [DOI] [Google Scholar]
- 5.Selin NE. 2009. Global biogeochemical cycling of mercury: a review. Ann. Rev. Environ. Res. 34, 43–63. ( 10.1146/annurev.environ.051308.084314) [DOI] [Google Scholar]
- 6.Lindberg S, Bullock R, Ebinghaus R, Engstrom D, Feng X, Fitzgerald W, Pirrone N, Prestbo E, Seigneur C. 2007. A synthesis of progress and uncertainties in attributing the sources of mercury in deposition. Ambio 36, 19–32. ( 10.1579/0044-7447(2007)36%5B19:ASOPAU%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 7.Hotez PJ. 2016. Neglected tropical diseases in the Anthropocene: the cases of Zika, Ebola, and other infections. PLoS Neglected Tropical Dis. 10, e0004648 ( 10.1371/journal.pntd.0004648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaffer HB, Fisher RN, Davidson C. 1998. The role of natural history collections in documenting species declines. Trends Ecol. Evol. 13, 27–30. ( 10.1016/S0169-5347(97)01177-4) [DOI] [PubMed] [Google Scholar]
- 9.Suarez AV, Tsutsui ND. 2004. The value of museum collections for research and society. BioScience 54, 66–74. ( 10.1641/0006-3568(2004)054%5B0066:TVOMCF%5D2.0.CO;2) [DOI] [Google Scholar]
- 10.Rocha LA, et al. 2014. Specimen collection: an essential tool. Science 344, 814–815. ( 10.1126/science.344.6186.814) [DOI] [PubMed] [Google Scholar]
- 11.Rainbow PS. 2009. Marine biological collections in the 21st century. Zool. Scr. 38, 33–40. ( 10.1111/j.1463-6409.2007.00313.x) [DOI] [Google Scholar]
- 12.Holmes MW, et al. 2016. Natural history collections as windows on evolutionary processes. Mol. Ecol. 25, 864–881. ( 10.1111/mec.13529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward DF, Leschen RAB, Buckley TR. 2015. More from ecologists to support natural history museums. Trends Ecol. Evol. 30, 373–374. ( 10.1016/j.tree.2015.04.015) [DOI] [PubMed] [Google Scholar]
- 14.Brooks SJ, et al. 2011. Natural history collections as sources of long-term datasets. Trends Ecol. Evol. 26, 153–154. ( 10.1016/j.tree.2010.12.009) [DOI] [PubMed] [Google Scholar]
- 15.Joseph L. 2011. Museum collections in ornithology: today's record of avian biodiversity for tomorrow's world. Emu Austral Ornithol. 111, i–xii. ( 10.1071/MU10050) [DOI] [Google Scholar]
- 16.Winker K. 2004. Natural history museums in a postbiodiversity era. BioScience 54, 455–459. ( 10.1641/0006-3568(2004)054%5B0455:NHMIAP%5D2.0.CO;2) [DOI] [Google Scholar]
- 17.Edwards SV, Birks S, Brumfield RT, Hanner R. 2005. Avian genetic resources collections: archives of evolutionary and environmental history. Auk 122, 979–984. ( 10.1642/0004-8038(2005)122%5B0979:FOAGRC%5D2.0.CO;2) [DOI] [Google Scholar]
- 18.Stephens PR, et al. 2016. The macroecology of infectious diseases: a new perspective on global-scale drivers of pathogen distributions and impacts. Ecol. Lett. 19, 1159–1171. ( 10.1111/ele.12644) [DOI] [PubMed] [Google Scholar]
- 19.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. ( 10.1038/nature10947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daszak P, Cunningham AA, Hyatt AD. 2000. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science 287, 443–449. ( 10.1126/science.287.5452.443) [DOI] [PubMed] [Google Scholar]
- 21.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451, 990–993. ( 10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yates TL, et al. 2002. The ecology and evolutionary history of an emergent disease: hantavirus pulmonary syndrome. Bioscience 52, 989–998. ( 10.1641/0006-3568(2002)052%5B0989:TEAEHO%5D2.0.CO;2) [DOI] [Google Scholar]
- 23.Lopez N, Padula P, Rossi C, Lazaro ME, Franze-Fernandez MT. 1996. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology 220, 223–226. ( 10.1006/viro.1996.0305) [DOI] [PubMed] [Google Scholar]
- 24.Nelson R, Canate R, Pascale JM, Dragoo JW, Armien B, Armien AG, Koster F. 2010. Confirmation of Choclo virus as the cause of hantavirus cardiopulmonary syndrome and high serum antibody prevalence in Panama. J. Med. Virol. 82, 1586–1593. ( 10.1002/jmv.21864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cook JA, et al. 2016. Transformational principles for NEON sampling of mammalian parasites and pathogens: a response to Springer and colleagues. BioScience 66, 917–919. ( 10.1093/biosci/biw123) [DOI] [Google Scholar]
- 26.Rodriguez D, Becker CG, Pupin NC, Haddad CFB, Zamudio KR. 2013. Long-term endemism of two highly divergent lineages of the amphibian-killing fungus in the Atlantic Forest of Brazil. Mol. Ecol. 23, 774–787. ( 10.1111/mec.12615) [DOI] [PubMed] [Google Scholar]
- 27.Cheng TL, Rovito SM, Wake DB, Vredenburg VT. 2011. Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc. Natl Acad. Sci. USA 108, 9502–9507. ( 10.1073/pnas.1105538108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho T, Becker CG, Toledo LF. 2017. Historical amphibian declines and extinctions in Brazil linked to chytridiomycosis. Proc. R. Soc. B 284, 20162254 ( 10.1098/rspb.2016.2254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campana MG, Kurata NP, Foster JT, Helgen LE, Reeder DM, Fleischer RC, Helgen KM. 2017. White-nose syndrome fungus in a 1918 bat specimen from France. Emerg. Infect. Dis. J. 23, 1611–1612. ( 10.3201/eid2309.170875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass GE, Shields TM, Parmenter RR, Goade D, Mills JN, Cheek J, Cook JA, Yates TL. 2006. Predicted hantavirus risk in 2006 for the Southwestern U.S. Occasional Papers, Musuem Texas Tech Univ. 225, 1–16. [Google Scholar]
- 31.Taylor LH, Latham SM, Woolhouse MEJ. 2001. Risk factors for human disease emergence. Phil. Trans. R. Soc. B 356, 983–989. ( 10.1098/rstb.2001.0888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiEuliis D, Johnson KR, Morse SS, Schindel DE. 2016. Specimen collections should have a much bigger role in infectious disease research and response. Proc. Natl Acad. Sci. USA 113, 4–7. ( 10.1073/pnas.1522680112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger L, et al. 1998. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc. Natl Acad. Sci. USA 95, 9031–9036. ( 10.1073/pnas.95.15.9031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaustein AR, Wake DB. 1990. Declining amphibian populations: a global phenomenon? Trends Ecol. Evol. 5, 203–204. ( 10.1016/0169-5347(90)90129-2) [DOI] [Google Scholar]
- 35.Campos PF, Gilbert TMP. 2012. Extraction from formalin-fixed material. In Ancient DNA: methods and protocols (eds Shapiro B, Hofreiter M), pp. 81–85. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 36.McGuire JA, et al. 2018. Squeezing water from a stone: high-throughput sequencing from a 145-year old holotype resolves (barely) a cryptic species problem in flying lizards. PeerJ 6, e4470 ( 10.7717/peerj.4470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hykin SM, Bi K, McGuire JA. 2015. Fixing formalin: a method to recover genomic-scale DNA sequence data from formalin-fixed museum specimens using high-throughput sequencing. PLoS ONE 10, e0141579 ( 10.1371/journal.pone.0141579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruane S, Austin Christopher C. 2017. Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Mol. Ecol. Res. 17, 1003–1008. ( 10.1111/1755-0998.12655) [DOI] [PubMed] [Google Scholar]
- 39.Morehouse EA, James TY, Ganley AR, Vilgalys R, Berger L, Murphy PJ, Longcore JE. 2003. Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Mol. Ecol. 12, 395–403. ( 10.1046/j.1365-294X.2003.01732.x) [DOI] [PubMed] [Google Scholar]
- 40.James TY, et al. 2009. Rapid global expansion of the fungal disease Chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 5, e1000458 ( 10.1371/journal.ppat.1000458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyle DG, Boyle DB, Olsen V, Morgan JA, Hyatt AD. 2004. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60, 141–148. ( 10.3354/dao060141) [DOI] [PubMed] [Google Scholar]
- 42.Rosenblum EB, et al. 2013. Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc. Natl Acad. Sci. USA 110, 9385–9390. ( 10.1073/pnas.1300130110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Hanlon SJ, et al. 2018. Recent Asian origin of chytrid fungi causing global amphibian declines. Science 360, 621–627. ( 10.1126/science.aar1965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fong JJ, Cheng TL, Bataille A, Pessier AP, Waldman B, Vredenburg VT. 2015. Early 1900s detection of Batrachochytrium dendrobatidis in Korean Amphibians. PLoS ONE 10, e0115656 ( 10.1371/journal.pone.0115656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yap TA, Koo MS, Ambrose RF, Vredenburg VT. 2018. Introduced bullfrog facilitates pathogen invasion in the western United States. PLoS ONE 13, e0188384 ( 10.1371/journal.pone.0188384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burrowes PA, De la Riva I. 2017. Unraveling the historical prevalence of the invasive chytrid fungus in the Bolivian Andes: implications in recent amphibian declines. Biol. Invasions 19, 1781–1794. ( 10.1007/s10530-017-1390-8) [DOI] [Google Scholar]
- 47.Martel A, et al. 2013. Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc. Natl Acad. Sci. USA 110, 15 325–15 329. ( 10.1073/pnas.1307356110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martel A, et al. 2014. Recent introduction of a chytrid fungus endangers Western Palearctic salamanders. Science 346, 630–631. ( 10.1126/science.1258268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu W, et al. 2014. A survey for Batrachochytrium salamandrivorans in Chinese amphibians. Cur. Zool. 60, 729–735. ( 10.1093/czoolo/60.6.729) [DOI] [Google Scholar]
- 50.Nichol ST, et al. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262, 914–917. ( 10.1126/science.8235615) [DOI] [PubMed] [Google Scholar]
- 51.Horgan J. 1993. Were four corners victims Biowar casualties? Sci. Am. 269, 16. [DOI] [PubMed] [Google Scholar]
- 52.Yanagihara R, Gu SH, Arai S, Kang HJ, Song JW. 2014. Hantaviruses: rediscovery and new beginnings. Virus Res. 187, 6–14. ( 10.1016/j.virusres.2013.12.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmaljohn C, Hjelle B. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3, 95–104. ( 10.3201/eid0302.970202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters CJ, Mills JN, Spiropoulou CF, Zaki SR, Rollin PE. 1999. Hantaviruses. In Tropical infectious diseases: principles, pathogens, and practice (eds Guerrant RL, Walker DH, Weller PF), pp. 1214–1235. New York, NY: W. B. Sanders. [Google Scholar]
- 55.Holmes EC, Zhang YZ. 2015. The evolution and emergence of hantaviruses. Curr. Opin. Virol. 10, 27–33. ( 10.1016/j.coviro.2014.12.007) [DOI] [PubMed] [Google Scholar]
- 56.Song JW, et al. 2009. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura iasiura). J. Virol. 83, 6184–6191. ( 10.1128/JVI.00371-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sumibcay L, Kadjo B, Gu SH, Kang HJ, Lim BK, Cook JA, Song JW, Yanagihara R. 2012. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Cote d'Ivoire. Virol. J. 9, 34 ( 10.1186/1743-422X-9-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milholland MT, Castro-Arellano I, Suzán G, Garcia-Peña GE, Lee TE, Rohde RE, Alonso Aguirre A, Mills JN. 2018. Global diversity and distribution of Hantaviruses and their hosts. EcoHealth 15, 163–208. ( 10.1007/s10393-017-1305-2) [DOI] [PubMed] [Google Scholar]
- 59.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19, 531–545. ( 10.1128/CMR.00017-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brook CE, Dobson AP. 2015. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 23, 172–180. ( 10.1016/j.tim.2014.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yanagihara R, Gu SH, Song J-W. 2015. Expanded host diversity and global distribution of hantaviruses: implications for identifying and investigating previously unrecognized hantaviral diseases. In Global virology I - identifying and investigating viral diseases (eds Shapshak P. et al), pp. 161–198. New York, NY: Springer New York: [Google Scholar]
- 62.Arai S, et al. 2016. Molecular phylogeny of a genetically divergent hantavirus harbored by the Geoffroy's rousette (Rousettus amplexicaudatus), a frugivorous bat species in the Philippines. Infect. Genet. Evol. 45, 26–32. ( 10.1016/j.meegid.2016.08.008) [DOI] [PubMed] [Google Scholar]
- 63.Dunnum JL, Yanagihara R, Johnson KM, Armien B, Batsaikhan N, Morgan L, Cook JA. 2017. Biospecimen repositories and integrated databases as critical infrastructure for pathogen discovery and pathobiology research. PLoS Neglected Tropical Dis. 11, e0005133 ( 10.1371/journal.pntd.0005133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morse SS, Mazet JAK, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet 380, 1956–1965. ( 10.1016/S0140-6736(12)61684-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller GE, Grant PM, Kishore R, Steinkruger FJ, Rowland FS, Guinn VP. 1972. Mercury concentrations in museum specimens of tuna and swordfish. Science 175, 1121 ( 10.1126/science.175.4026.1121) [DOI] [PubMed] [Google Scholar]
- 66.Palmer PT, Martin M, Wentworth G, Caldararo N, Davis L, Kane S, Hostler D. 2003. Analysis of pesticide residues on museum objects repatriated to the Hupa Tribe of California. Environ. Sci. Technol. 37, 1083–1088. ( 10.1021/es026235n) [DOI] [PubMed] [Google Scholar]
- 67.Thompson DR, Furness RW, Walsh PM. 1992. Historical changes in mercury concentrations in the marine ecosystem of the north and north-east Atlantic Ocean as indicated by seabird feathers. J. Appl. Ecol. 29, 79–84. ( 10.2307/2404350) [DOI] [Google Scholar]
- 68.Thompson DR, Furness RW, Lewis SA. 1993. Temporal and spatial variation in mercury concentrations in some albatrosses and petrels from the sub-Antarctic. Polar Biol. 13, 239–244. ( 10.1007/BF00238759) [DOI] [Google Scholar]
- 69.Thompson DR, Furness RW. 1989. Comparison of the levels of total and organic mercury in seabird feathers. Mar. Pollut. Bull. 20, 577–579. ( 10.1016/0025-326X(89)90361-5) [DOI] [Google Scholar]
- 70.Thompson DR, Becker PH, Furness RW. 1993. Long-term changes in mercury concentrations in herring gulls Larus argentatus and common terns Sterna hirundo from the German North sea coast. J. Appl. Ecol. 30, 316–320. ( 10.2307/2404633) [DOI] [Google Scholar]
- 71.Monteiro LR, Furness RW. 2009. Accelerated increase in mercury contamination in north Atlantic mesopelagic food chains as indicated by time series of seabird feathers. Environ. Toxicol. Chem. 16, 2489–2493. ( 10.1002/etc.5620161208) [DOI] [Google Scholar]
- 72.Rocque DA, Winker K, Zink RM. 2005. Use of bird collections in contaminant and stable-isotope studies. Auk 122, 990–994. ( 10.1642/0004-8038(2005)122%5B0990:UOBCIC%5D2.0.CO;2) [DOI] [Google Scholar]
- 73.Dosch JJ. 2007. Jerald J. Dosch on dead birds' tales: museum specimen feathers as historical archives of environmental pollutants. Environmental History 12, 661–665. ( 10.1093/envhis/12.3.661) [DOI] [Google Scholar]
- 74.Movalli P, Bode P, Dekker R, Fornasari L, van der Mije S, Yosef R. 2017. Retrospective biomonitoring of mercury and other elements in museum feathers of common kestrel Falco tinnunculus using instrumental neutron activation analysis (INAA). Environ. Sci. Pollut. Res. 24, 25 986–26 005. ( 10.1007/s11356-017-0157-1) [DOI] [PubMed] [Google Scholar]
- 75.DuBay SG, Fuldner CC. 2017. Bird specimens track 135 years of atmospheric black carbon and environmental policy. Proc. Natl Acad. Sci. USA 114, 11 321–11 326. ( 10.1073/pnas.1710239114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vo AT, Bank MS, Shine JP, Edwards SV. 2011. Temporal increase in organic mercury in an endangered pelagic seabird assessed by century-old museum specimens. Proc. Natl Acad. Sci. USA 108, 7466–7471. ( 10.1073/pnas.1013865108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bond TC, et al. 2013. Bounding the role of black carbon in the climate system: a scientific assessment. J. Geophys. Res. Atmos. 118, 5380–5552. ( 10.1002/jgrd.50171) [DOI] [Google Scholar]
- 78.Bond TC, Bhardwaj E, Dong R, Jogani R, Jung S, Roden C, Streets DG, Trautmann NM. 2007. Historical emissions of black and organic carbon aerosol from energy-related combustion, 1850–2000. Global Biogeochem. Cycles 21, GB2018 ( 10.1029/2006gb002840) [DOI] [Google Scholar]
- 79.Ramanathan V, Carmichael G. 2008. Global and regional climate changes due to black carbon. Nat. Geosci. 1, 221–227. ( 10.1038/ngeo156) [DOI] [Google Scholar]
- 80.Stern AC, Professor E. 1982. History of air pollution legislation in the United States. J. Air Pollut. Control Assoc. 32, 44–61. ( 10.1080/00022470.1982.10465369) [DOI] [PubMed] [Google Scholar]
- 81.Shindell D, et al. 2012. Simultaneously mitigating near-term climate change and improving human health and food security. Science 335, 183–189. ( 10.1126/science.1210026) [DOI] [PubMed] [Google Scholar]
- 82.Compeau GC, Bartha R. 1985. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 50, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crewther WG, Fraser RDB, Lennox FG, Lindley H. 1965. The chemistry of keratins. In Advances in protein chemistry (ed Anfinsen CB, et al.), pp. 191–346. Academic Press. [DOI] [PubMed] [Google Scholar]
- 84.Furness RW, Muirhead SJ, Woodburn M. 1986. Using bird feathers to measure mercury in the environment: relationships between mercury content and moult. Mar. Pollut. Bull. 17, 27–30. ( 10.1016/0025-326X(86)90801-5) [DOI] [Google Scholar]
- 85.Chen C, Amirbahman A, Fisher N, Harding G, Lamborg C, Nacci D, Taylor D. 2008. Methylmercury in marine ecosystems: spatial patterns and processes of production, bioaccumulation, and biomagnification. Ecohealth 5, 399–408. ( 10.1007/s10393-008-0201-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bond AL, Hobson KA, Branfireun BA. 2015. Rapidly increasing methyl mercury in endangered ivory gull Pagophila eburnea feathers over a 130 year record. Proc. R. Soc. B 282, 20150032 ( 10.1098/rspb.2015.0032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar A, Divoll TJ, Ganguli PM, Trama FA, Lamborg CH. 2018. Presence of artisanal gold mining predicts mercury bioaccumulation in five genera of bats (Chiroptera). Environ. Pollut. 236, 862–870. ( 10.1016/j.envpol.2018.01.109) [DOI] [PubMed] [Google Scholar]
- 88.IUCN. 2018. IUCN 2013. IUCN Red List of Threatened Species. Version 2018.1. See http://www.redlist.org/ (accessed 30 August 2018).
- 89.Wiley AE, James HF, Ostrom PH. 2017. Emerging techniques for isotope studies of avian ecology. In The extended specimen: emerging frontiers in collections-based ornithological research (ed. Webster MS.), pp. 89–109. Boca Raton, FL: CRC Press. [Google Scholar]
- 90.Edwards MS, Turner TF, Sharp ZD, Montgomery WL. 2002. Short- and long-term effects of fixation and preservation on stable isotope values (δ13C, δ15N, δ34S) of fluid-preserved museum specimens. Copeia 2002, 1106–1112. ( 10.1643/0045-8511(2002)002%5B1106:SALTEO%5D2.0.CO;2) [DOI] [Google Scholar]
- 91.Bortolotti GR, Marchant TA, Blas J, German T. 2008. Corticosterone in feathers is a long-term, integrated measure of avian stress physiology. Funct. Ecol. 22, 494–500. ( 10.1111/j.1365-2435.2008.01387.x) [DOI] [Google Scholar]
- 92.Bortolotti GR, Marchant T, Blas J, Cabezas S. 2009. Tracking stress: localisation, deposition and stability of corticosterone in feathers. J. Exp. Biol. 212, 1477–1482. ( 10.1242/jeb.022152) [DOI] [PubMed] [Google Scholar]
- 93.van Rijssel JC, Hecky RE, Kishe-Machumu MA, Witte F. 2017. Changing ecology of Lake Victoria cichlids and their environment: evidence from C13 and N15 analyses. Hydrobiologia 791, 175–191. ( 10.1007/s10750-016-2790-y) [DOI] [Google Scholar]
- 94.Walker JL, Macko SA. 2006. Dietary studies of marine mammals using stable carbon and nitrogen isotopic ratios of teeth. Marine Mammal Sci. 15, 314–334. ( 10.1111/j.1748-7692.1999.tb00804.x) [DOI] [Google Scholar]
- 95.Hobson KA, Sease JL. 2006. Stable isotope analyses of tooth annuli reveal temporal dietary records: an example using steller sea lions. Marine Mammal Sci. 14, 116–129. ( 10.1111/j.1748-7692.1998.tb00694.x) [DOI] [Google Scholar]
- 96.Fleming TH, Nuñez RA, Sternberg LD. 1993. Seasonal changes in the diets of migrant and non-migrant nectarivorous bats as revealed by carbon stable isotope analysis. Oecologia 94, 72–75. ( 10.1007/BF00317304) [DOI] [PubMed] [Google Scholar]
- 97.Negrete P, Sallaberry M, Barceló G, Maldonado K, Perona F, McGill RAR, Quillfeldt P, Sabat P. 2017. Temporal variation in isotopic composition of Pygoscelis penguins at Ardley Island, Antarctic: are foraging habits impacted by environmental change? Polar Biol. 40, 903–916. ( 10.1007/s00300-016-2017-8) [DOI] [Google Scholar]
- 98.Fairhurst GD, Bond AL, Hobson KA, Ronconi RA. 2014. Feather-based measures of stable isotopes and corticosterone reveal a relationship between trophic position and physiology in a pelagic seabird over a 153-year period. Ibis 157, 273–283. ( 10.1111/ibi.12232) [DOI] [Google Scholar]
- 99.English PA, Green DJ, Nocera JJ. 2018. Stable isotopes from museum specimens may provide evidence of long-term change in the trophic ecology of a migratory aerial insectivore. Front. Ecol. Evol. 6, 14 ( 10.3389/fevo.2018.00014) [DOI] [Google Scholar]
- 100.Hobson KA, Greenberg R, Wilgenburg SLV, Mettke-Hofmann C. 2010. Migratory connectivity in the rusty blackbird: isotopic evidence from feathers of historical and contemporary specimens. Condor 112, 778–788. ( 10.1525/cond.2010.100146) [DOI] [Google Scholar]
- 101.Gadek C, Newsome S, Beckman E, Chavez A, Galen S, Bautista E, Witt C. 2017. Why are tropical mountain passes ‘low’ for some species? Genetic and stable-isotope tests for differentiation, migration, and expansion in elevational generalist songbirds. J. Anim. Ecol. 87, 741–753. ( 10.1111/1365-2656.12779) [DOI] [PubMed] [Google Scholar]
- 102.Pergams ORW, Lawler JJ. 2009. Recent and widespread rapid morphological change in rodents. PLoS ONE 4, e6452 ( 10.1371/journal.pone.0006452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Desrochers A. 2010. Morphological response of songbirds to 100 years of landscape change in North America. Ecology 91, 1577–1582. ( 10.1890/09-2202.1) [DOI] [PubMed] [Google Scholar]
- 104.Yom-Tov Y, Geffen E. 2010. Recent spatial and temporal changes in body size of terrestrial vertebrates: probable causes and pitfalls. Biol. Rev. 86, 531–541. ( 10.1111/j.1469-185X.2010.00168.x) [DOI] [PubMed] [Google Scholar]
- 105.Parmesan C. 2006. Ecological and evolutionary responses to recent climate change. Ann. Rev. Ecol. Evol. Syst. 37, 637–669. ( 10.1146/annurev.ecolsys.37.091305.110100) [DOI] [Google Scholar]
- 106.Rosenzweig C, et al. 2008. Attributing physical and biological impacts to anthropogenic climate change. Nature 453, 353–357. ( 10.1038/nature06937) [DOI] [PubMed] [Google Scholar]
- 107.Walther G-R, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC, Fromentin J-M, Hoegh-Guldberg O, Bairlein F. 2002. Ecological responses to recent climate change. Nature 416, 389–395. ( 10.1038/416389a) [DOI] [PubMed] [Google Scholar]
- 108.Parmesan C, Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42. ( 10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 109.Moritz C, Patton JL, Conroy CJ, Parra JL, White GC, Beissinger SR. 2008. Impact of a century of climate change on small-mammal communities in Yosemite National Park, USA. Science 322, 261–264. ( 10.1126/science.1163428) [DOI] [PubMed] [Google Scholar]
- 110.Lenoir J, Gégout JC, Marquet PA, de Ruffray P, Brisse H. 2008. A significant upward shift in plant species optimum elevation during the 20th century. Science 320, 1768–1771. ( 10.1126/science.1156831) [DOI] [PubMed] [Google Scholar]
- 111.Thomas CD, et al. 2004. Extinction risk from climate change. Nature 427, 145–148. ( 10.1038/nature02121) [DOI] [PubMed] [Google Scholar]
- 112.Williams JW, Jackson ST. 2007. Novel climates, no-analog communities, and ecological surprises. Front. Ecol. Environ. 5, 475–482. ( 10.1890/070037) [DOI] [Google Scholar]
- 113.Faurby S, Araújo MB. 2018. Anthropogenic range contractions bias species climate change forecasts. Nat. Clim. Change 8, 252–256. ( 10.1038/s41558-018-0089-x) [DOI] [Google Scholar]
- 114.Beissinger SR, Peery MZ. 2007. Reconstructing the historic demography of an endangered seabird. Ecology 88, 296–305. ( 10.1890/06-0869) [DOI] [PubMed] [Google Scholar]
- 115.Peery MZ. et al. 2010. Genetic analyses of historic and modern marbled murrelets suggest decoupling of migration and gene flow after habitat fragmentation. Proc. R. Soc. B 277, 697–706. ( 10.1098/rspb.2009.1666) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Post E. 2013. Ecology of climate change: the importance of biotic interactions. Princeton, NJ: Princeton University Press. [Google Scholar]
- 117.Bi K, Linderoth T, Vanderpool D, Good Jeffrey M, Nielsen R, Moritz C. 2013. Unlocking the vault: next-generation museum population genomics. Mol. Ecol. 22, 6018–6032. ( 10.1111/mec.12516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Santos MJ, Smith AB, Thorne JH, Moritz C. 2017. The relative influence of change in habitat and climate on elevation range limits in small mammals in Yosemite National Park, California. U.S.A. Clim. Change Responses 4, 7 ( 10.1186/s40665-017-0035-6) [DOI] [Google Scholar]
- 119.Tingley MW, Beissinger SR. 2013. Cryptic loss of montane avian richness and high community turnover over 100 years. Ecology 94, 598–609. ( 10.1890/12-0928.1) [DOI] [PubMed] [Google Scholar]
- 120.Tingley MW, Monahan WB, Beissinger SR, Moritz C. 2009. Birds track their Grinnellian niche through a century of climate change. Proc. Natl Acad. Sci. USA 106, 19 637–19 643. ( 10.1073/pnas.0901562106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stewart JAE, Wright DH, Heckman KA. 2017. Apparent climate-mediated loss and fragmentation of core habitat of the American Pika in the Northern Sierra Nevada, California, USA. PLoS ONE 12, e0181834 ( 10.1371/journal.pone.0181834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tingley MW, Beissinger SR. 2009. Detecting range shifts from historical species occurrences: new perspectives on old data. Trends Ecol. Evol. 24, 625–633. ( 10.1016/j.tree.2009.05.009) [DOI] [PubMed] [Google Scholar]
- 123.McCormack JE, Rodríguez-Gómez F, Tsai WL, Faircloth BC. 2017. Transforming museum specimens into genomic resources. In The extended specimen: emerging frontiers in collections based ornithological research (ed. Webster MS.), pp. 143–156. Boca Raton, FL: CRC Press. [Google Scholar]
- 124.Galbreath KE, Hafner DJ, Zamudio KR. 2009. When cold is better: climate-driven elevation shifts yield complex patterns of diversification and demography in an alpine specialist (American Pika, Ochotona princep). Evolution 63, 2848–2863. ( 10.1111/j.1558-5646.2009.00803.x) [DOI] [PubMed] [Google Scholar]
- 125.Minteer BA, Collins JP, Love KE, Puschendorf R. 2014. Avoiding (Re)extinction. Science 344, 260–261. ( 10.1126/science.1250953) [DOI] [PubMed] [Google Scholar]
- 126.Malaney JL, Cook JA. 2018. A perfect storm for mammalogy: declining sample availability in a period of rapid environmental degradation. J. Mammalogy 99, 773–788. ( 10.1093/jmammal/gyy082) [DOI] [Google Scholar]
- 127.Hoekman D, et al. 2016. Design for mosquito abundance, diversity, and phenology sampling within the National Ecological Observatory Network. Ecosphere 7, e01320 ( 10.1002/ecs2.1320) [DOI] [Google Scholar]
- 128.Hoekman D, et al. 2017. Design for ground beetle abundance and diversity sampling within the National Ecological Observatory Network. Ecosphere 8, e01744 ( 10.1002/ecs2.1744) [DOI] [Google Scholar]
- 129.Springer YP, et al. 2016. Tick-, mosquito-, and rodent-borne parasite sampling designs for the National Ecological Observatory Network. Ecosphere 7, 40 ( 10.1002/ecs2.1271) [DOI] [Google Scholar]
- 130.Webster MS. 2017. The extended specimen. In The extended specimen: emerging frontiers in collections-based ornithological research (ed. Webster MS.), pp. 1–9. Boca Raton, FL: CRC Press. [Google Scholar]
- 131.Global Genome Initiative. Smithsonian National Museum of Natural History. https://ggi.si.edu (accessed 16 October 2018).
- 132.Grayson P, Sin SYW, Sackton TB, Edwards SV. 2017. Comparative genomics as a foundation for evo-devo studies in birds. In Methods in molecular biology: avian and reptilian developmental biology (ed. Shen G.), pp. 11–46. New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- 133.Claramunt S, Wright NA. 2017. Using museum specimens to study flight and dispersal. In The extended specimen: emerging frontiers in collections-based ornithological research (ed. Webster MS.), pp. 127–141. Boca Raton, FL: CRC Press. [Google Scholar]
- 134.Li Z, Clarke JA. 2016. The craniolingual morphology of waterfowl (Aves, Anseriformes) and its relationship with feeding mode revealed through contrast-enhanced X-ray computed tomography and 2D morphometrics. Evol. Biol. 43, 12–25. ( 10.1007/s11692-015-9345-4) [DOI] [Google Scholar]
- 135.Dickson BV, Sherratt E, Losos JB, Pierce SE. 2017. Semicircular canals in Anolis lizards: ecomorphological convergence and ecomorph affinities of fossil species. R. Soc. open sci. 4, 170058 ( 10.1098/rsos.170058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lacey EA, et al. 2017. Climate change, collections and the classroom: using big data to tackle big problems. Evol. Edu. Outreach 10, 2 ( 10.1186/s12052-017-0065-3) [DOI] [Google Scholar]
- 137.Movalli P, Dekker R, Koschorreck J, Treu G. 2017. Bringing together raptor collections in Europe for contaminant research and monitoring in relation to chemicals regulations. Environ. Sci. Pollut Res. Int. 24, 24 057–24 060. ( 10.1007/s11356-017-0096-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wehi PM, Whaanga H, Trewick SA. 2012. Artefacts, biology and bias in museum collection research. Mol. Ecol. 21, 3103–3109. ( 10.1111/j.1365-294X.2012.05589.x) [DOI] [PubMed] [Google Scholar]
- 139.Johnson KW. 2018. The ornithologist the internet called a murderer. The New York Times, 16 June 2018, p. SR1. New York, NY: The New York Times Company. [Google Scholar]
- 140.Hope AG, Sandercock BK, Malaney JL. 2018. Collection of scientific specimens: benefits for biodiversity sciences and limited impacts on communities of small mammals. BioScience 68, 35–42. ( 10.1093/biosci/bix141) [DOI] [Google Scholar]
- 141.Remsen JV., Jr 1995. The importance of continued collecting of bird specimens to ornithology and bird conservation. Bird Conserv. Int. 5, 145–180. ( 10.1017/S095927090000099X) [DOI] [Google Scholar]
- 142.Roger E, Klistorner S. 2016. BioBlitzes help science communicators engage local communities in environmental research. Jcom-J. Sci. Commun. 15, 18 ( 10.22323/2.15030206) [DOI] [Google Scholar]
- 143.Hiller AE, Cicero C, Albe MJ, Barclay TLW, Spencer CL, Koo MS, Bowie RCK, Lacey EA. 2017. Mutualism in museums: a model for engaging undergraduates in biodiversity science. PLoS Biol. 15, e2003318 ( 10.1371/journal.pbio.2003318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cook JA, et al. 2014. Natural history collections as emerging resources for innovative education. Bioscience 64, 725–734. ( 10.1093/biosci/biu096) [DOI] [Google Scholar]
- 145.Morrison SA, Sillett TS, Funk WC, Ghalambor CK, Rick TC. 2017. Equipping the 22nd-century historical ecologist. Trends Ecol. Evol. 32, 578–588. ( 10.1016/j.tree.2017.05.006) [DOI] [PubMed] [Google Scholar]
- 146.Funk VA. 2018. Collections-based science in the 21st century. J. Syst. Evol. 56, 175–193. ( 10.1111/jse.12315) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.