Abstract
This paper focuses on one of the most commonly encountered materials in our society, namely paper. Paper is an inherently complex material, yet its use provides for chemical analysis approaches that are elegant in their simplicity of execution. In the first half of the previous century, paper in scientific research was used mainly for filtration and chromatographic separation. While its use decreased with the rise of modern elution chromatography, paper remains a versatile substrate for low-cost analytical tests. Recently, we have seen renewed interest to work with paper in (bio)analytical science, a result of the growing demand for inexpensive, portable analysis. Dried blood spotting, paper microfluidics, and paper spray ionization are areas in which paper is (re)establishing itself as an important material. These research areas all exploit several properties of paper, including stable sample storage, passive fluid movement and manipulation, chromatographic separation/extraction, modifiable surface and/or volume, easily altered shape, easy transport, and low cost. We propose that the real, and to date underexploited, potential of paper lies in utilizing its combined characteristics to add new dimensions to paper-based (bio)chemical analysis, expanding its applicability. This article provides the reader with a short historical perspective on the scientific use of paper and the developments that led to the establishment of the aforementioned research areas. We review important characteristics of paper and place them in a scientific context in this descriptive, yet critical, assessment of the achieved and the achievable in paper-based analysis. The ultimate goal is the exploration of integrative approaches at the interface between the different fields in which paper is or can be used.
Paper is a unique material. One could say that the world as we know it may not have come to be, if not for paper.1 It has for millennia been the singlemost important information carrier, only now being surpassed by the digital format. Most of our scientific knowledge and cultural heritage has been preserved on paper. Before paper, we had animal skin (parchment), stone, wood, papyrus, and even silk to store and convey information, but these were variously bulky, heavy, expensive, not ubiquitously available, or difficult to inscribe. Not surprisingly, all of these materials became redundant after the introduction of paper roughly two millennia ago.1 There are several reasons underlying paper’s popularity for information management: (1) paper is lightweight and thin and thus easily stored and transported; (2) paper is cheap and easily produced and thus readily available; (3) paper has a porous structure and is therefore an excellent vessel for (long-term) storage of deposited material (such as ink).2−4
Another important application of paper is personal hygiene. Tissues, paper towels, and napkins are examples of items that are used on a daily basis by billions of people globally. Try to imagine what your day would be like without them. We can identify several reasons why we use paper in this context. First of all, paper is easily disposed of and recycled, as its main constituent is a biologically produced (and biodegradable) material. Second, paper is flexible and deformable and thus applicable to irregular surfaces. Third, the porosity and hydrophilicity of paper result in capillary action,5,6 so that paper can be used to absorb e.g. spilled liquids. Finally, paper can be specifically fabricated for different purposes1,7 (thickness, consistency, smoothness/softness, deformability) and modified to better suit the consumer’s needs (e.g., scented or moist tissues).
The applications of paper are vast, but we will conclude this introduction with one everyday example, namely the preparation of coffee, as a link to discussing the importance of paper in science. A common and easy way to make coffee is with a paper filter. The pores in paper can retain and thus separate solid particles from liquid, and as paper is nontoxic for humans, it can be used in the preparation of products for human consumption. Moreover, the paper filter nicely fits into the coffee machine, serving as an example of how easily paper can be shaped and combined with other materials to increase its utility.
In our consideration of the roles of paper in our everyday lives, we can distill out relevant properties for its use in science. These include availability, portability, affordability, biocompatibility, modifiability (structurally, physically, and chemically), application in sample/reagent storage, passive fluid propagation, filtration, separation, and combination with other materials. In this article, we will demonstrate that these properties can all contribute to the use of paper in analytical science and that approaches exploiting several properties simultaneously are the way to best leverage paper for (bio)analysis.
Molecular, Micro, and Macro Structure of Paper
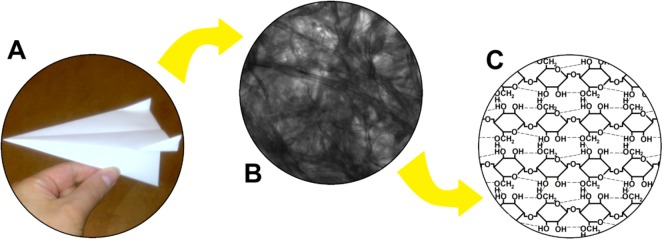
The word “paper” can refer to a range of materials. In ancient Egypt, the papyrus plant was used to make sheets for writing, and it is in fact this plant to which modern paper owes its name.1 However, papyrus is produced by laminating plant fibers, whereas modern-day paper is produced from a fiber-containing pulp (made from e.g. rags, straw, or wood), which undergoes a number of (chemical) processes (e.g., removal of lignin, bleaching) before it is pressed into sheets of paper and further processed.7,8 The different structural levels of paper in Figure 1 remind us of the properties of paper discussed above and help define “paper”. With respect to its macroscopic structure (Figure 1A), paper is a flexible sheet material, well below a millimeter in thickness, while having almost any desired dimension in width and length.
Figure 1.
(A) Macroscopic, (B) microscopic, and (C) molecular structures of paper.
When we look at paper through the microscope (Figure 1B), we see an intricate network of fibers of varying size and position. This network has a very open nature. The microstructure thus defines another important characteristic of paper, namely porosity, which is key to many of its applications.
The aforementioned fibers originate from plant material and are produced in plant cells to form the protective cell wall. The fibers consist of bundles of smaller fibers (microfibrils), which are made from sheets of molecular cellulose, a polymer of d-glucose (Figure 1C). Individual polymer chains are hydrophilic and interact with each other through hydrogen bonds between the hydroxyl and ether groups (dashed lines in Figure 1C). The molecular structure reveals the last defining characteristic of paper, namely, that it is made from cellulose or cellulose derivatives.9,10
History of Paper in Science
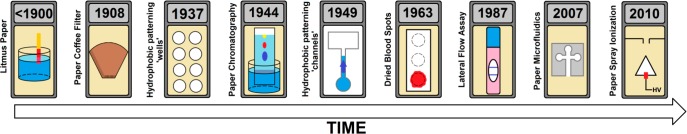
Paper has been used in science for many years (Figure 2). One of the oldest examples, with scientific reports going back to at least the early 19th century,11,12 involves the approximation of pH with litmus paper, a technique that is still used today. We can recognize the properties of paper that led to the conception of this product, namely disposability, storage capacity for the reagent, and passive fluid movement.
Figure 2.
A history of paper in science, highlighting important milestones.
The next milestone was the invention of the coffee filter by Melitta Bentz. The filtration process is crucial in any chemistry laboratory and exploits the disposability and deformability of paper, as well as its porosity and the possibility for integration in a rigid container.
In the 1940s, Martin and Synge investigated partition chromatography13−15 on paper.14 Although earlier examples of paper-chromatography-like approaches can be found in the literature,16 this work forms the basis of the method that paved the way for modern chromatography. Paper chromatography relies on passive fluid movement, differential retention, and the possibility of loading the pores with e.g. aluminum or silica gel to improve separation.17
The first example of a paper microfluidic structure was described in 1937 by Yagoda,18 who employed hydrophobic paraffin patterning to create hydrophilic “wells”. This was followed by a paraffin-patterned paper microfluidic channel reported by Müller et al. in 194919 for improving paper chromatography. Skipping a few decades, paper microfluidics regained attention in 2007, when the Whitesides group20 published their seminal paper on the subject, after a number of critical publications on the need for cheap diagnostics in low-resource settings.21,22 In this context, the aspects of affordability, portability, and disposability are important. Alternative processes for hydrophobic patterning are also being investigated for easier fabrication of devices.23−25
In 1963, paper was employed by Guthrie to test neonates for phenyl ketonuria (PKU),26 an illness which can lead to mental retardation if it goes unnoticed. A droplet of blood taken from a heel prick was applied to a piece of paper and dried. This dried blood spot (DBS) could be used for the diagnosis of the disease. A few decades later, with the rise of mass spectrometry (MS), the DBS technique has become increasingly popular due to the possibility of combining paper with the high-resolution, high-sensitivity analytical instrumentation.27,28 Various forms of paper are used for DBS for reasons of storage capability, disposability, and easy storage and transport.29 Now, DBS technology enables diagnostics for people in developing countries, as well as self-sampling patients who send their DBS to the laboratory by mail.
At the end of the 1980s, the lateral flow (immuno)assay (LFA), or immunochromatography, was introduced,30 a format that became particularly popular after the introduction of the home pregnancy test. LFAs are formatted on nitrocellulose strips and make use of colored particles modified with antibodies.31 Particles are loaded onto a conjugate pad, resulting in the formation of antigen–antibody complexes when a sample is added. These complexes are immobilized further downstream by capture molecules, thus accumulating the detection-particle color (generally in a line).32 LFAs illustrate the merits of integrative paper-based approaches.
The most recent development in paper-based analysis has been the introduction by the Cooks group of paper spray ionization (PSI)33,34 for MS. The sample is deposited onto a sharp tip cut out of paper, which is then aligned in front of a mass spectrometer. When solvent and a high potential are applied, electrospray is generated at the tip, and the sample is introduced to the MS. In PSI, the sample generally does not require preprocessing, due to paper’s extractive properties. Since its introduction, more advanced PSI methods have been reported, which take advantage of a number of additional properties of paper.35−41
Building Function into Paper Devices
Capillary Action and Flow Control
The passive wicking behavior of fluids in paper is governed by capillary action, which was described in 1921 by Edward Washburn.6Equation 1, which was derived from the Poiseuille equation relating flow rate to pressure drop (assuming that the coefficient of slip at the capillary wall is zero), is used to describe capillary action in a capillary.
| 1 |
The linear fluid front velocity (dl/dt) is influenced by liquid properties (surface tension γ and viscosity η), capillary radius (r), and the contact angle between liquid and capillary (θ). It also depends on the gravitational acceleration on Earth (g, 9.8 m/s2) of a liquid having a density (ρ), that has traveled a certain distance through the capillary (ls) with the capillary positioned at an angle (ψ) to the horizontal axis. Furthermore, linear velocity decreases as ls increases. If we consider paper as a collection of parallel capillaries (admittedly somewhat of a simplification42), this equation still applies, but then the “radius” relates to the average pore size.6 Since eq 1 was derived for capillary flow, it does not take evaporation into account, which limits fluid propagation in an open system and fluid drag over the paper.42,43
Changing any of the variables in eq 1 will alter the linear velocity. For example, to obtain increased velocity, we can use a liquid with high surface tension (such as water), low viscosity (such as methanol), or (if flow runs upward) low density. Alternatively, lowering the surface energy of paper increases the contact angle of a liquid with high surface tension, and can slow down44−46 or stop19,20,23 capillary flow. Decreasing the surface energy of cellulose can be achieved by either physically depositing material into the pores19,20,47 or chemically bonding groups to hydroxyl groups on cellulose.23
Yet another way to influence flow velocity through paper is to change its geometric dimensions. Decreasing r leads to decreased flow velocity and can be achieved by compressing paper48 or by initially choosing a different type of paper. Deviation from a uniform rectangular paper strip affects flow rates as well.49−51 In lateral flow assays (LFAs), an absorbent pad is placed at the end of the strip to sustain flow.30,32 Alternatively, if a rectangular strip expands into a fanlike shape, the volumetric flow rate through the rectangular strip will shift from decreasing to quasi-stationary.49−51 The length of a strip of paper determines with which velocity a fluid reaches its end and proceeds to the next stage of the device. One can, for example, controllably time different solutions to arrive in a central chamber by choosing appropriate lengths for their respective supply wicks.49,52
We can also look into ways of controlling fluid movement over the surface of a wetted paper strip, instead of through the strip itself. In this case, wetted paper acts as a “wall” in a thin solution conduit. This can be achieved by e.g. scratching the paper in the direction of flow to create an open channel53 or by aligning the paper surface parallel to another paper or non-paper surface that is at least partially wettable by the liquid used.54,55 Solution is then wicked between the surface of the wetted paper and the second surface. This form of capillary transport has the advantage that it is far more rapid than flow through porous cellulose networks alone.
The Paper Microfluidic Toolbox
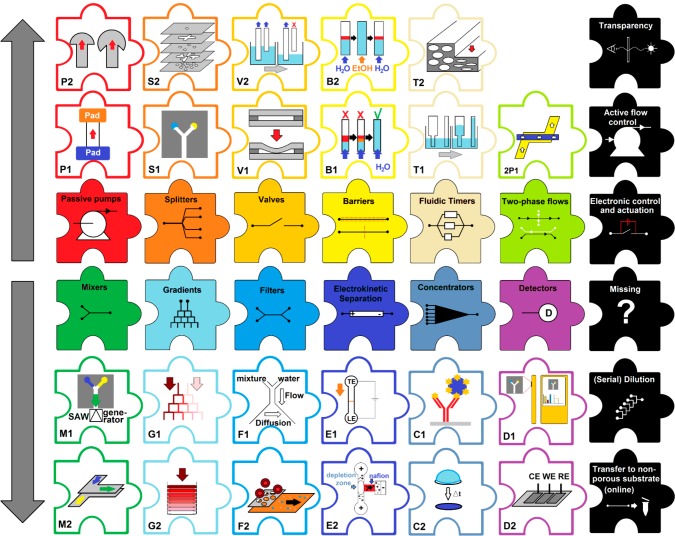
Early paper microfluidic devices (2007–2010) relied mainly on hydrophobic flow restrictions, specifically used for splitting up a flow into multiple streams.20,47,56−60 Since then, the field has evolved to introduce new ways to functionalize devices. In this section, we consider the paper microfluidic toolbox and identify which tools we have (Figure 3, color-coded) and which remain un(der)developed (Figure 3, black). We then look into possibilities for devices with higher levels of integration achieved by combining different tools.
Figure 3.
Paper microfluidic toolbox. The two central rows represent categories of tools, with color-coded examples in adjacent pieces (above or below). Black pieces represent tools that are still missing from the tool box. Pumps: absorbent pad32 (P1), fan51 (P2). Splitters: 2D20 (S1), 3D56 (S2). Valves: mechanical62 (V1), fluidic disconnects63 (V2). Barriers: dissolvable52 (B1), selectively permeable46 (B2). Fluidic timers: geometrical52 (T1), compressed48 (T2). Two-phase flows: countercurrent65 (2P1). Mixers: surface acoustic wave function66 (M1), geometrical61 (M2). Gradient: branched68 (G1), stacked69 (G2). Filters: Y-filter61 (F1), membrane70 (F2). Electrokinetic separation: isotachophoresis72 (E1), ion concentration polarization73 (E2). Concentrators: affinity based75 (C1), bulk76 (C2). Detectors: colorimetric/smartphone (D1), electrochemical77 (D2).
Basic paper microfluidic tools are passive pumping61 (see Capillary Action and Flow Control and Figure 3-P1/P2) and flow-splitting features (Figure 3-S1/S2) to guide the sample toward a number of different reagents simultaneously.20,59 For any paper device, the length of paper that can be used is limited, due to the decrease in flow rates as a function of distance traveled by the solvent front (eq 1).49 The number of times a channel can be split is equally limited (Figure 3-S1). However, when multiple layers of paper are stacked, a third physical dimension is added,56 in which we can selectively separate regions in different layers with hydrophobic layers having the thickness of a piece of tape, or one sheet of paper. This is well below the width of 2D patterns on paper. With the 3D approach, we can thus split a flow many more times within a given distance of porous material (Figure 3-S2).
One of the oldest paper microfluidic tools is the mechanical valve, operated by disconnecting and (re)connecting different paper regions (Figure 3-V1).23,62 Alternatively, wicks of a certain length can be suspended into a liquid, and the contact between those will be discontinued when the liquid level drops below a certain point (Figure 3-V2).54,63 We can also modify paper physically or chemically to manipulate flows. For instance, a temporary barrier can be formed through application of a dissolvable material52,64 (Figure 3-B1). When the applied material is not dissolvable, it alters the surface properties of paper and can be used to slow down or even stop flow.44,45 In the latter case, a solvent with different surface energy can be used to cross the barrier, thus achieving solvent-based on/off valving (Figure 3-B2).46 Another way to influence flow rates is to use fluidic timers (see Capillary Action and Flow Control and Figure 3-T1/T2)52,63 to ensure that compounds arrive at a common location at different times.
We can also consider introducing multiple streams of fluid with the express interest of allowing them to interact. We can, for example, use selective coatings to locally alter paper surface energies to allow countercurrent flow of two immiscible solvents (2P1).65 Moreover, we can mix miscible solvents on paper by applying an external force, such as acoustic wave functions (Figure 3-M1).66 Alternatively, providing a suitable path length and contact between two streams (e.g., by overlapping strips of paper) allows mixing through dispersion (Figure 3-M2).61 When mixing is incomplete, or when liquids are guided to interact through a branched network of channels, concentration gradients can be formed67,68 (Figure 3-G1). Concentration gradients can also be created by using 3D paper microfluidic stacks that are only supplied with a nutrient or gas from one side69 (Figure 3-G2). Diffusion across the interface between two side by side flows can also be used to separate compounds, as is achieved with the Y-filter (Figure 3-F1).61 Furthermore, filtration or separation can be achieved by tuning the pore size of the device. An example of this is the integration of a membrane that does not allow the passage of blood cells (Figure 3-F2)70 to extract serum from whole blood.
Another popular approach to achieve separations, and potentially orders of magnitude concentration increases, is to employ electric fields. Isotachophoresis (Figure 3-E1) can be used to focus the charged analyte in small bands on a paper substrate.71,72 The use of ion concentration polarization, in which ions are concentrated by application of an electric field over an ion-selective membrane/region, has been demonstrated as a possibility for on-paper concentration as well (Figure 3-E2).73 However, electrokinetic approaches require additional instrumentation and are only applicable to charged analytes. Therefore, the most common approach is affinity-based preconcentration of specific molecules on paper, as done in LFAs (Figure 3-C1).74,75 Instrument-free preconcentration of a bulk sample can be easily achieved by depositing a surplus of sample behind water-impermeable barriers, drying it, and eluting the concentrate with a stronger solvent76 (Figure 3-C2).
We have also identified paper microfluidic tools that are currently missing from the toolbox. Think here of on-paper serial dilution, and electronic control and actuation of paper microfluidic functions. The first steps toward their development have been reported. Simple dilution was demonstrated while proving the feasibility of a stacked mixer.61 A significant level of integration of electronic actuation has been shown in an example of pulsatile generation of colored bands by electrochemistry78 and electrokinetic separations.71,72 Also missing is the use of transparent materials to better visualize what is going on inside paper microfluidic devices. Such material can also be used to improve optical detection through background reduction and better penetration. Oils have been used to change refractive index and thus perform spectrophotometric and fluorescent detection in a paper format.79 However, transparent paper, which has already been developed,80 has not been implemented.
Other true challenges still lie ahead, especially the application of active flow control (rather than passive pumps) to alter flow rates during an experiment, and online off-paper extraction of sample or product. Advances in the application of more (active) flow control result from the fabrication of channel-like structures on paper53−55 to achieve higher flow rates, or the combination of paper-based devices with a digital microfluidic approach.81 Off-line extraction is achieved easily by cutting out a piece of the paper device and extracting its load,61 but this is a relatively irreproducible operation and results in substantial dilution. Both active flow control and online extraction may require the integration of the paper substrate in a container or casing to ensure that liquids cannot leave the paper via its pores and to be able to connect the paper strip to external components.
Now, imagine we need a point-of-care test to see if the level of a certain drug in a blood sample is within the therapeutic range. We can start constructing this from the building blocks in Figure 3. First, we need a user-friendly way to acquire the right sample volume. Paper’s passive pump will fill the substrate with sample, but we need valves to define a fixed amount. The sample matrix is complex; therefore, we install a mechanical filter to separate serum from the larger constituents of blood. Drug concentrations are often low, so that preconcentration of the filtrate is necessary. We then split the sample into different channels, where they are mixed with varying concentrations of the compound of interest for on-paper calibration. The choice of detection depends strongly on the assay itself, but will most likely be based on colorimetry (see Detection and Readout Platforms for Paper Devices). Furthermore, in order to make this setup easy to operate, it needs encasing (see Cartridge-Based Paper Analytics). By using this modular approach, we can easily select which tools are required to solve a problem as well as identify which tools still need to be developed.
Detection and Readout Platforms for Paper Devices
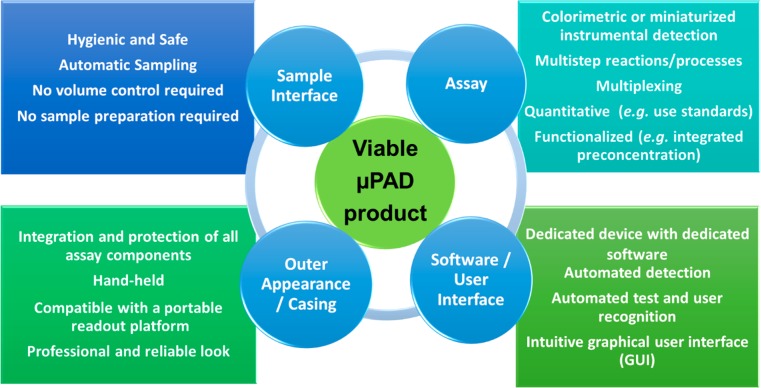
Detection is an important parameter in this discussion about integrated paper-based platforms. First and foremost, whatever detector is selected, it should be intuitive to operate but also portable and inexpensive, all of which is in line with the philosophy underlying on-site analyses (Figure 4). We can roughly distinguish three different approaches for paper-based analysis,32,82,83 namely, the unaided eye, digital colorimetry, and instrumental analysis.
Figure 4.
Required components of a fully functional μPAD from scientific and societal points of view.
A large number of paper-based assays, such as the pregnancy test, are interpreted simply by eye. Advantages of this approach are numerous, but only if we are interested in a qualitative answer, whereas many analytical tests require a quantitative outcome. Paper microfluidics has made a valuable contribution to readout with the unaided eye, through the implementation of easily interpretable and even legible results, such as an immunoassay that displays a patient’s blood type in patterned letters.84
The second, and very popular, approach is (digital) colorimetry. The reason for its popularity is very simple. To acquire results, it is enough to have a digital camera, which we find in every smartphone.82 Reactions giving a colorimetric response are numerous and include chemical reactions,20,85,86 enzymatic reactions,87 and immunoassays.88 Such approaches offer a sufficient detection limit for many medical and environmental purposes.83 However, the smartphone may not always be equally suitable as a detector, for the reasons discussed below.
The third category is instrumental analysis, a major representative of which is electrochemical detection.89−91 Electrochemistry offers sensitivity and can be used for many well-described medical and environmental tests. However, it requires a number of adaptations for use on paper, as well as dedicated instrumentation to carry out the test.82,83,92 PSI and many DBS analyses also fall into this category, since they rely on MS detection of sample on paper, via direct ionization or extraction, respectively. Since the biggest application fields for paper-based assays are point of care and environmental analysis, it has become clear that readout platforms should be portable and user friendly. Consequently, it seems paradoxical to develop paper-based tests which rely on laboratory-based instrumental analysis. Still, efforts in the miniaturization of readout platforms are making headway as well. A rather spectacular development in this respect, outside the scope of this article, is the field of miniaturized MS.93−95 Electrochemical detection is more easily miniaturized, as electrodes can be printed on paper and implemented using (trans)portable potentiostats.93−95
Recently, smartphone cameras, or adaptors with a built-in detector, have been applied for data acquisition, using a software app for interpretation.82 It is important to note the distinct difference between these two approaches, as smartphone cameras are used in this regard as the actual detection unit, whereas the adaptor approach employs the smartphone as a hand-held computer only. This “smart approach” contributes to the development of portable readout platforms for on-site assays but has numerous drawbacks as well. A crucial problem associated with smartphone-camera-based detection for diagnostic tests is that all smartphones produced globally would require certification (FDA-approved or similar), which is simply impossible. For tests which only give indicative, preliminary, or lifestyle information, however, there is more hope. Currently, it is possible to test yourself for alcohol, drugs, and allergies,96 and tests for environmental monitoring have also been reported.97
An additional concern for the use of smartphones comes from the contamination risk posed by the tested samples. There is a realistic danger of infecting/exposing the smartphone user, should his or her personal phone be used to test a potentially hazardous sample. Dedicated readout platforms, specifically developed for on-site analyses, give more freedom for the design and packaging of a device with special technical features. It is then probably easier to obtain the necessary approval for medical use. Furthermore, this reduces the threat of pathogen carryover from a sample to the user.
Cartridge-Based Paper Analytics
In commercial products, the test is often enclosed in a cartridge. Fitting a paper-based test into a cartridge can serve many purposes that enhance the end-user experience, test functionality, or both. Use of a cartridge to enhance end-user experience is exemplified in such well-known examples as the pregnancy test. Here, the cartridge serves as protection, integrative casing, and easy-to-use interface, in addition to having a professional and reliable appearance. The cartridge is paramount for both a successful experiment in terms of reliability and reproducibility and a potential future product in terms of operational simplicity and appeal. The cartridge is also used to add functionality to the test by, for instance, imparting additional control over fluid flows by capillary action between paper and the surface of the cartridge.54 Movable features36 may be integrated, as can mechanical support, to achieve functional alignment of paper and non-paper components. It is also possible to integrate other materials and objects into the assay (e.g., electrodes).98
In the case of paper-based tests, integration into a cartridge is often a logical step in the development process. When we encounter a practical problem and try to solve it in the laboratory, we often envision a slick device that is ready for the market. In reality, however, we often end up building a setup “McGyver-style” which cannot be used by anyone except the inventors. The design and fabrication of a cartridge by most processes can be a costly and time-consuming affair. Furthermore, it limits the number of changes that can be made to the device later on. Many researchers have therefore started to rely on rapid prototyping approaches,99−102 the newly crowned king of which is 3D printing. This technology is very affordable nowadays, and even low-end printers can be used for producing casings and cartridges. The added benefit of 3D-printing approaches is that the entire development and production process is kept in house, thus drastically reducing production time.
Applications of Paper in (Bio-)Chemical Analysis
Biochemistry on Paper
Paper-based analytical devices (μPADs)20 based on biochemical reactions are of particular interest for paper-based diagnostic83,103,104 and environmental assays.105 There has been a tendency to omit LFAs from this μPAD category. To the best of our knowledge, however, no formal definition exists for paper microfluidics that would justify this omission. An obvious and historically justified definition (History of Paper in Science and Figure 2) could be “the controlled manipulation of small quantities of liquid in and through a porous (cellulose) network”. Seen in this context, one could argue that LFAs are simply one of the many possible embodiments of paper microfluidics. In this section, we broadly discuss paper-based biochemical assays and, in doing so, present a comparison of established LFAs and the newer μPADs (Table 1).
Table 1. Comparison of LFAs with μPADs, on the Basis of References Throughout the Text and Conclusions Drawn from Those.
| lateral flow assay | μPAD | |
|---|---|---|
| type of chemistry | immunological, colorimetric | immunological, enzymatic, colorimetric, electrochemical |
| reaction/process complexity | mostly two binding steps | mainly single-step chemistry |
| on-paper flow restriction | no | usually |
| type of readout | unaided eye, dedicated analyzer | unaided eye, smartphone, standard laboratory equipment |
| type of results | mainly qualitativea | qualitative or quantitative |
| uniform format | yes | no |
| integrated control assay | yes | rare |
| external equipment needed | no | sometimes |
| multiplexing | possible | possible |
| stability of a test | stable | often not yet established |
| casing | yes | rare |
| size | hand-held (∼10 cm) | ∼1–5 cm |
| social acceptance | known worldwide and accepted | lack of end-user awareness |
Dedicated read-out devices can provide quantitative data (http://idetekt.com/reader-systems/).
Biochemistry is the chemistry of biologically active molecules, such as enzymes and antibodies, which are generally more prone to degradation in comparison to small molecules. Biochemical analysis generally utilizes these biomolecules in multistep processes including immobilization and binding steps, which require devices with integrated multifunctionality to carry out. The LFA is specifically designed and optimized for this purpose, whereas the μPAD is still often based on one-step reactions, with additional operational steps for the user.103 In both LFAs and μPADs, colorimetry is mostly employed as a readout. However, where LFAs mostly rely on qualitative interpretation by the user, detection in most μPADs is of a quantitative nature, and the range of applied analytical techniques is vast (see Detection and Readout Platforms for Paper Devices).
With respect to the performance of LFAs and μPADs, one could contend that LFAs have a more intricate and complete design in comparison to many μPADs. However, more recently we see that the field of paper microfluidics has begun to implement additional functionality and complexity.83,92,103,105−108 Recent advances in LFA development include dedicated readout platforms (for example: http://www.lateralflowreader.com/ and http://www.alere.com/en/home.html) and multiplexing (http://symbolicsdx.com/). However, the main strategy behind present-day LFA development appears to be their successful implementation for new applications. While most individual μPADs might not be as sophisticated as the LFA, the total scope of their applicability is immense and covers many areas of analytical science.
With respect to user friendliness, LFAs surpass the majority of μPADs, because they are straightforward to operate, are hand-held, and yield unambiguous results. Arguably though, μPADs to date have been mostly proof-of-concept devices. Current drawbacks of μPADs in this regard are that they require more input from the user, and that their size can range from something as small as a postage stamp to a complete, laboratory-based setup.
Before μPADs can achieve market or societal success comparable to that of LFAs, they must obtain an edge over these in terms of e.g. user friendliness, integrated functionality, and cost. A higher level of integration is achievable by the implementation of still un(der)exploited functionalities (see The Paper Microfluidic Toolbox). Moreover, the fact that a quantitative answer is desired85,109 in many applications offers opportunities for μPADs. Figure 4 describes the ideal μPAD, which integrates the flexibility of μPADs with lessons learned from LFAs.
One of the great values of paper-based devices is the possibility of taking a standard laboratory test and performing it on site.20,109 A great example from biochemistry in this regard is the paper-based polymerase chain reaction (PCR), which can be a game-changing approach for diagnosis and pathogen identification in low-resource settings. The first success reported was the use of a paper device to identify Ebola RNA using isothermal amplification, namely reverse transcription recombinase polymerase amplification.110 This work amply proves the possibility of efficient application of PCR in a μPAD format and its contribution to overcoming contemporary societal problems such as outbreaks of Ebola.
Paper Spray Ionization: Mass Spectrometric Detection
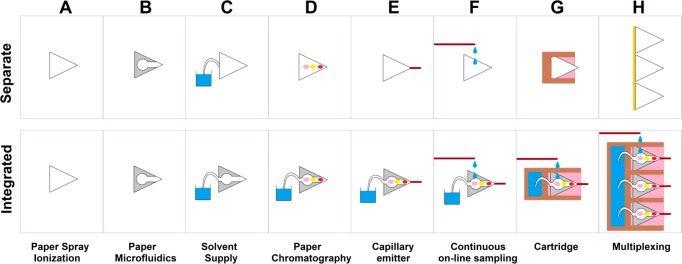
Since its conception in 2010, PSI has experienced a rapid increase in the number of reported applications.93,111 Most of this work focuses on the employment of this ambient ionization method to applications using unmodified paper tips (Figure 5A). Additionally, attention has been devoted to the improvement and functionalization of the method (Figure 5B–H). In this section, we discuss a number of these improvements (Figure 5, top panel) and consider them from an integrative point of view (Figure 5, bottom panel), extrapolating future directions for PSI, and paper-based analytics in general.
Figure 5.
Functionalization of PSI. The top row shows individual improvements to the method. The second row shows how individual developments may be combined for more integrated functionality.
One obvious choice for the functionalization of PSI tips is the integration of paper microfluidics to create “smart tips”. However, only recently have we seen the first reports that demonstrate such a union.35,112 Up to now, we have seen PSI tips with straight channels (Figure 5B), which help to reduce lateral diffusion as well as increase the electrical field strength, allowing the use of lower potentials. Furthermore, we have seen the introduction of selectively permeable valves on paper tips to preconcentrate the sample.76 The PSI–paper microfluidics marriage yields many interesting options for future exploration, which can be viewed as “functionalization of PSI tips”, or in broader terms as “MS detection for paper microfluidics”. In principle, any paper microfluidic structure could be patterned on a PSI tip, functionalizing it e.g. for sampling, chemical derivatization, or improved separation.
Spray time, which is conventionally limited to approximately 1 min, can be significantly extended by passively supplying solvent from a reservoir via a hydrophilic wick54,113 or a supply channel114,115 (Figure 5C). Once a continuous solvent supply is ensured, we can perform an online paper chromatographic separation with MS detection54,116 (Figure 5D).
Since paper is a complex network of cellulose fibers, the microstructure of a macroscopic tip is difficult to reproduce properly, which might cause differences in spray intensity and stability during and between experiments. One approach to solve this problem is to attach a capillary to the tip to serve as the emitter117 (Figure 5E). This way, we can use paper with its useful characteristics (separation, passive fluid propagation), while relying on a capillary emitter for reproducible spray stability.
Sample application to the paper tip generally consists of a single sampling moment but might require sampling at a certain frequency. Periodic sampling from a system that requires continuous monitoring can be achieved by slowly pumping the solution through a capillary, which then drips the sample onto the paper tip38 (Figure 5F). If a steady supply of solvent and a stable spray is ensured, sample can be directly “injected” into the flow.
Proof-of-concept setups are often bulky, not very reproducible, and certainly not user friendly. All of these problems can be solved using a single solution, namely putting the paper tip into a disposable cartridge36,40,93,115,118 (Figure 5G). The cartridge not only protects the tip but provides physical support, allows easy positioning in a complementary holder, and can integrate features such as solvent reservoirs and sample delivery systems (see also section 2.4).
As we all want to do as many analyses as possible in the least amount of time, multiplexing is imperative. So-called “high-throughput” PSI was reported by mounting a number of paper tips next to each other and moving them along past the mass spectrometer orifice39 (Figure 5H).
As a result of integration, we can end up with a system which exhibits improved sensitivity due to hydrophobic patterning, improved spray stability due to the capillary emitter, longer spray time due to the hydrophilic wick in a solvent reservoir, and improved integration, protection, and stability by incorporation into a cartridge. All of these features improve PSI, without putting additional demands upon the user. If anything, it makes PSI easier to use. It is therefore rather surprising that most PSI applications published to date do not implement this type of development. Moreover, there are optional functions such as multiplexing and periodic sampling which, though definitely not needed in every application, can greatly contribute in some.
Paper-Based Cell Cultivation Systems
In the past decade, microfluidic systems for cell cultures have been increasingly employed to bridge the gap between well-plate cultures and animal studies, by striving for in vivo-like conditions in specially designed in vitro systems.119−124 However, these achievements often come with expensive device fabrication techniques and relatively complicated system operation. Recently, paper has been introduced as a viable candidate for the simplification of various cell-culture platforms.69,124,125 The most relevant question for the development of paper-based cell cultivation, as is the case for any new cell culture platform, is whether it better resembles in vivo conditions in comparison to conventional cell culture platforms. This section addresses that question.
When paper is used for cell cultivation, the pores of paper serve as a 3D mesh to contain them, while the same paper can simultaneously contain integrated sensors. Paper can support cell cultures either directly by providing a surface for cell attachment69 or indirectly as a scaffold for deposition of cells in hydrogel.126 Moreover, paper can be easily modified and prepared for different cell types, ranging from immortalized cell lines to sensitive primary cell cultures, including stem cells.127 Other benefits are the low price of the material, biocompatibility, and accessibility.
The passive wicking of liquids through paper can be employed to regulate continuous nutrient delivery, whereas the open architecture allows unhindered gas exchange with the direct environment. In addition, paper-based cell cultivation can benefit from the ease with which new features can be integrated into a device,69,126,128 especially for 3D cell cultures. For example, stacking sheets of paper with cultivated cells results in a 3D construct that can be used for monitoring cell–cell interaction and nutrient distribution.69,124,125,127 Furthermore, this allows for disassembly of the layers and separate analysis of different cell types in those layers. Stacking layers with different chemical content, or with different distances to a source of nutrients, results in the formation of gradients for the study of e.g. nutrient/waste transport or the oxygenation level of a culture.69
Another elegant demonstration of paper for cell cultures was the cultivation of neurons, which formed functional connections through the paper and could transfer electrical impulses.129 The features of paper primarily contributing to this work were porosity, which allowed the neurons to develop in 3D, the structural modifiability of paper, which allowed directional growth of the neurons, and the low electrical conductivity of the material.
Paper is a good matrix as well for the cultivation and identification of bacteria and tests of their antibiotic resistance.130 This assay was inspired by standard Petri-dish cultures and therefore successfully exemplifies how one can adapt existing tests and make them broadly applicable, also outside the microbiological laboratories. Tests for antimicrobial resistance on paper can be easily multiplexed in terms of identifying multiple pathogens and evaluating the resistance to different antibiotics.
Some approaches in cell culture could actually be easier to achieve with paper than with standard labware. For example, the printing and deposition of cells (in medium or hydrogel) on cultivation devices should be simple, because cells and their matrix will be directly retained within the porous network. This would have an additional benefit in heterogeneous cell cultures where different cell types have to be spatially separated or arranged in a fashion which helps to emulate their function (e.g., smooth muscle cells below endothelial cells).
The most popular method for cell inspection is microscopy, which is not fully compatible with paper-based cell cultures, due to the opacity of paper. This obstacle can potentially be overcome with the application of transparent paper80 for paper-based cell culture systems. Transparent paper differs from opaque paper, because the cellulose fibers have a smaller diameter and their density in the sheet is higher.
Perspectives
There are a number of challenges ahead if paper-based assays are to provide society with the tools to perform the on-site analyses we keep talking about. Below are our conclusions, drawn from our observation of the field, which can be taken into consideration as recommendations for future paper-based research.
(1) Learn from previous successes. For example, LFAs are viable, paper-based, commercial products that address societal demand for fast, easy-to-use diagnostics. While LFAs are not as diverse as paper microfluidics, they are great examples of optimized devices for on-site analysis, to the point that they can literally be used by anyone. This is highly important for paper microfluidics as well, yet has not always been taken into account. For instance, researchers still use pipets for sample application and complicated or insufficient readout systems for paper microfluidics.
(2) Look beyond the piece of paper. Conventional microfluidics (and other fields) can inspire increased functionality in paper-based devices. Cartridges can be applied for integrating various functional elements, adding functionality, and if nothing else, making paper devices easier to handle and appealing. Integration of electronics on paper or applying hybrid patterning strategies can further extend the possibilities.
(3) Think top-down, rather than bottom-up. It is fair to say that academic research does not need to be immediately translatable into products, but this is exactly what justifies paper microfluidics. We should identify an analytical problem and design the solution, rather than defining a problem that suits the solution. We can then start working our way down to focus on the individual components that are required.
(4) Focus on expanding the paper microfluidic toolbox. Some tools are still completely absent. Others have been invented but are still in an early stage of development. Integration is important, but in order to develop complete assays, we need to keep working on the components. Paper has many different favorable characteristics, each of which can be viewed as an additional dimension to operate in, an additional level of complexity for the device.
(5) Re-evaluate smartphone detection. It is impossible to calibrate and certify each and every smartphone for diagnostic readout. However, it is possible to do this for dedicated readout devices, which would also be more affordable than a smartphone. Furthermore, the possible contamination of a smartphone with samples bears health risks for the owner. Having said that, smartphones can be attractive detectors for assays with a less regulated result, such as home tests for certain lifestyle markers or scanning tests to identify epidemiological trends or samples that should be submitted for FDA-approved testing.
(6) Identify patterns in patterning strategies. Many patterning approaches have been described, each differing in one way or the other. It is important that we characterize and categorize the different types of barriers, so that we can make educated decisions as to which barrier to use for a given application. Most barriers can confine aqueous solutions, but little effort has been dedicated to testing them with more challenging solutions, containing e.g. detergents or alcohols.
(7) Customer demand is as important as the technology. In the case of diagnostics, especially in low-resource settings, we cannot create new demand but should rather recognize it and offer a solution which addresses as many objectives as possible. Therefore, devices that can answer more questions simultaneously in a reliable and accurate fashion win out against solutions offering less information. Equally true is that the less effort or input which is required to perform the test, the more likely it is that it will be adopted by end users. Importantly, we need to consider the dimensions and shape of the assay, which preferably can be hand-held yet is not microscopic in size, which could make it difficult to operate and easy to contaminate. By applying a unique shape (or color), we can help the user recognize a specific test and provide lock-key fitting into the readout device.
(8) Researchers often proudly announce that their μPAD costs less than $0.01 per piece. However, this price is usually solely based on the cost of the materials and reagents, and the price does not include manpower or even the costs for the usage of laboratory space and equipment. It is true that paper-based assays can be cheaper than any other approach, but to this price we must add the cost of setting up a production line ($150k–500k103), costs related to logistics, and profits for those involved in μPAD development and production.
This perspective on paper describes a number of scientific fields that could benefit from choosing a more integrative approach toward research. All of us doing research in one of these fields share a common belief that paper is in fact the material that will help us overcome the obstacles to successful on-site analysis in many application fields. Such research could prove important for diagnosing those who currently cannot be diagnosed due to financial, geographic, or cultural restrictions. Paper has the potential to help us reach that goal, if we continue to reinvent its application for analytical chemistry.
The authors declare no competing financial interest.
References
- Basbanes N. A.On Paper - The Everything of its two-thousand-year history, 1st ed.; Vintage Books: New York, 2014. [Google Scholar]
- Behets F.; Kashamuka M.; Pappaioanou M.; Green T.; Ryder R.; Batter V.; George J.; Hannon W.; Quinn T. J. Clin. Microbiol. 1992, 30, 1179–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfazil A.; Anderson R. J. Anal. Toxicol. 2008, 32, 511–515. 10.1093/jat/32.7.511. [DOI] [PubMed] [Google Scholar]
- Davis G.; Poholek R. Clin. Chem. 1979, 25, 24–25. [PubMed] [Google Scholar]
- Darcy H.Les fontaines publiques de la ville de Dijon; 1856. [DOI] [PubMed]
- Washburn E. Phys. Rev. 1921, 17, 273–283. 10.1103/PhysRev.17.273. [DOI] [Google Scholar]
- Sawyer F.; Beals C.; Neubauer A. Ind. Eng. Chem. 1950, 42, 1007–1020. 10.1021/ie50486a012. [DOI] [Google Scholar]
- Hubbe M.; Venditti R.; Rojas O. BioResources 2007, 2, 739–788. [Google Scholar]
- Poletto M.; Pistor V.; Zattera A. J. Cellul.-Fundam. Asp. 2013, 45–68. 10.5772/50452. [DOI] [Google Scholar]
- Hon D. N. S. Cellulose 1994, 1, 1–25. 10.1007/BF00818796. [DOI] [Google Scholar]
- Davy J. Philos. Trans. R. Soc. London 1812, 102, 352–369. 10.1098/rstl.1812.0020. [DOI] [Google Scholar]
- Gregor W. Abstr. Pap. Print. Philos. Trans. R. Soc. London 1814, 1, 209–210. [Google Scholar]
- Gordon A. H.; Martin A. J. P.; Synge R. L. M. Biochem. J. 1943, 37, 79–86. 10.1042/bj0370079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consden R.; Gordon A. H.; Martin A. J. P. Biochem. J. 1944, 38, 224–232. 10.1042/bj0380224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. H.; Martin A. J. P.; Synge R. L. M. Biochem. J. 1944, 38, 65–68. 10.1042/bj0380065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. D.; Wilson I. D. Chromatographia 2004, 60, 135–136. 10.1365/s10337-004-0316-7. [DOI] [Google Scholar]
- Valadon L.; Mummery R. Phytochemistry 1972, 11, 413–414. 10.1016/S0031-9422(00)90028-1. [DOI] [Google Scholar]
- Yagoda H.Test Paper. U.S. Patent US2129754, 1937.
- Müller R. H.; Clegg D. L. Anal. Chem. 1949, 21, 1123–1125. 10.1021/ac60033a032. [DOI] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Butte M. J.; Whitesides G. M. Angew. Chem., Int. Ed. 2007, 46, 1318–1320. 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. D.; Linder V.; Sia S. K. Lab Chip 2007, 7, 41–57. 10.1039/B611455E. [DOI] [PubMed] [Google Scholar]
- Yager P.; Edwards T.; Fu E.; Helton K.; Nelson K.; Tam M. R.; Weigl B. H. Nature 2006, 442, 412–418. 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- Li X.; Tian J.; Nguyen T.; Shen W. Anal. Chem. 2008, 80, 9131–9134. 10.1021/ac801729t. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Lin B.; Qin J. Anal. Chem. 2011, 83, 1830–1835. 10.1021/ac102577n. [DOI] [PubMed] [Google Scholar]
- Olkkonen J.; Lehtinen K.; Erho T. Anal. Chem. 2010, 82, 10246–10250. 10.1021/ac1027066. [DOI] [PubMed] [Google Scholar]
- Guthrie R.; Susi A. Pediatrics 1963, 32, 338–343. [PubMed] [Google Scholar]
- Li W.; Tse F. Biomed. Chromatogr. 2010, 24, 49–65. 10.1002/bmc.1367. [DOI] [PubMed] [Google Scholar]
- Espy R. D.; Manicke N. E.; Ouyang Z.; Cooks R. G. Analyst 2012, 137, 2344–2349. 10.1039/c2an35082c. [DOI] [PubMed] [Google Scholar]
- McDade T.; Williams S.; Snodgrass J. Demography 2007, 44, 899–925. 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- Campbell R. L.; Wagner D. B.; O’Connell J. P.. Solid Phase Assay with Visual Readout. U.S. Patent US4703017, 1987.
- Litman D. J.; Hanlon T. M.; Ullman E. F. Anal. Biochem. 1980, 106, 223–229. 10.1016/0003-2697(80)90141-4. [DOI] [PubMed] [Google Scholar]
- Posthuma-Trumpie G. A.; Korf J.; van Amerongen A. Anal. Bioanal. Chem. 2009, 393, 569–582. 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- Wang H.; Liu J.; Cooks R. G.; Ouyang Z. Angew. Chem., Int. Ed. 2010, 49, 877–880. 10.1002/anie.200906314. [DOI] [PubMed] [Google Scholar]
- Liu J.; Wang H.; Manicke N. E.; Lin J.-M.; Cooks R. G.; Ouyang Z. Anal. Chem. 2010, 82, 2463–2471. 10.1021/ac902854g. [DOI] [PubMed] [Google Scholar]
- Colletes T. C.; Garcia P. T.; Campanha R. B.; Abdelnur P. V.; Romão W.; Coltro W. K. T.; Vaz B. G. Analyst 2016, 141, 1707–1713. 10.1039/C5AN01954K. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Manicke N. E. Anal. Chem. 2015, 87, 6212–6219. 10.1021/acs.analchem.5b00884. [DOI] [PubMed] [Google Scholar]
- Narayanan R.; Sarkar D.; Cooks R. G.; Pradeep T. Angew. Chem., Int. Ed. 2014, 53, 5936–5940. 10.1002/anie.201311053. [DOI] [PubMed] [Google Scholar]
- Liu W.; Wang N.; Lin X.; Ma Y.; Lin J.-M. Anal. Chem. 2014, 86, 7128–7134. 10.1021/ac501678q. [DOI] [PubMed] [Google Scholar]
- Shen L.; Zhang J.; Yang Q.; Manicke N. E.; Ouyang Z. Clin. Chim. Acta 2013, 420, 28–33. 10.1016/j.cca.2012.10.025. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Wang H.; Maas J. D.; Chappell W. J.; Manicke N. E.; Cooks R. G.; Ouyang Z. Int. J. Mass Spectrom. 2012, 312, 201–207. 10.1016/j.ijms.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Xu W.; Manicke N. E.; Cooks R. G.; Ouyang Z. Anal. Chem. 2012, 84, 931–938. 10.1021/ac202058w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaar D.; Blokhuis G. Appl. Sci. Res. 1950, 2, 125–141. 10.1007/BF00411977. [DOI] [Google Scholar]
- Fujita H. J. Phys. Chem. 1952, 56, 625–629. 10.1021/j150497a015. [DOI] [Google Scholar]
- Noh H.; Phillips S. T. Anal. Chem. 2010, 82, 8071–8078. 10.1021/ac1005537. [DOI] [PubMed] [Google Scholar]
- Noh H.; Phillips S. T. Anal. Chem. 2010, 82, 4181–4187. 10.1021/ac100431y. [DOI] [PubMed] [Google Scholar]
- Salentijn G. IJ.; Hamidon N. N.; Verpoorte E. Lab Chip 2016, 16, 1013–1021. 10.1039/C5LC01355K. [DOI] [PubMed] [Google Scholar]
- Bruzewicz D. A.; Reches M.; Whitesides G. M. Anal. Chem. 2008, 80, 3387–3392. 10.1021/ac702605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. H.; Park J.; Kim S. H.; Park J. K. Biomicrofluidics 2014, 8, 054121. 10.1063/1.4899773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu E.; Ramsey S. A.; Kauffman P.; Lutz B.; Yager P. Microfluid. Nanofluid. 2011, 10, 29–35. 10.1007/s10404-010-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina A.; Pérez-Rosales C.; Pineda A.; Higuera F. J. Rev. Mex. Fis. 2001, 47, 537–541. [Google Scholar]
- Mendez S.; Fenton E. M.; Gallegos G. R.; Petsev D. N.; Sibbett S. S.; Stone H. A.; Zhang Y.; López G. P. Langmuir 2010, 26, 1380–1385. 10.1021/la902470b. [DOI] [PubMed] [Google Scholar]
- Fu E.; Lutz B.; Kauffman P.; Yager P. Lab Chip 2010, 10, 918–920. 10.1039/b919614e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giokas D. L.; Tsogas G. Z.; Vlessidis A. G. Anal. Chem. 2014, 86, 6202–6207. 10.1021/ac501273v. [DOI] [PubMed] [Google Scholar]
- Salentijn G. IJ.; Permentier H. P.; Verpoorte E. Anal. Chem. 2014, 86, 11657–11665. 10.1021/ac502785j. [DOI] [PubMed] [Google Scholar]
- Channon R. B.; Nguyen M. P.; Scorzelli A. G.; Henry E. M.; Volckens J.; Dandy D. S.; Henry C. S. Lab Chip 2018, 18, 793–802. 10.1039/C7LC01300K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Whitesides G. M. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 19606–19611. 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Carrilho E.; Thomas S. W.; Sindi H.; Whitesides G. M. Anal. Chem. 2008, 80, 3699–3707. 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; Shi W.; Jiang L.; Qin J.; Lin B. Electrophoresis 2009, 30, 1497–1500. 10.1002/elps.200800563. [DOI] [PubMed] [Google Scholar]
- Fenton E. M.; Mascarenas M. R.; López G. P.; Sibbett S. S. ACS Appl. Mater. Interfaces 2009, 1, 124–129. 10.1021/am800043z. [DOI] [PubMed] [Google Scholar]
- Dungchai W.; Chailapakul O.; Henry C. S. Anal. Chim. Acta 2010, 674, 227–233. 10.1016/j.aca.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Osborn J. L.; Lutz B.; Fu E.; Kauffman P.; Stevens D. Y.; Yager P. Lab Chip 2010, 10, 2659–2665. 10.1039/c004821f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Nie Z.; Cheng C.-M.; Carrilho E.; Wiley B. J.; Whitesides G. M. Lab Chip 2010, 10, 2499–2504. 10.1039/c0lc00021c. [DOI] [PubMed] [Google Scholar]
- Lutz B. R.; Trinh P.; Ball C.; Fu E.; Yager P. Lab Chip 2011, 11, 4274–4278. 10.1039/c1lc20758j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley G. E.; Le H. Q.; Fu E.; Yager P. Lab Chip 2012, 12, 4321–4327. 10.1039/c2lc40785j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentijn G. IJ.; Grajewski M.; Verpoorte E. Lab Chip 2017, 17, 3401–3404. 10.1039/C7LC00770A. [DOI] [PubMed] [Google Scholar]
- Rezk A. R.; Qi A.; Friend J. R.; Li W. H.; Yeo L. Y. Lab Chip 2012, 12, 773–779. 10.1039/C2LC21065G. [DOI] [PubMed] [Google Scholar]
- Hong B.; Xue P.; Wu Y.; Bao J.; Chuah Y. J.; Kang Y. Biomed. Microdevices 2016, 18, 1–8. 10.1007/s10544-016-0054-2. [DOI] [PubMed] [Google Scholar]
- Schaumburg F.; Urteaga R.; Kler P. A.; Berli C. L. A. J. Chromatogr. A 2018, 1561, 83–91. 10.1016/j.chroma.2018.05.040. [DOI] [PubMed] [Google Scholar]
- Derda R.; Laromaine A.; Mammoto A.; Tang S. K. Y.; Mammoto T.; Ingber D. E.; Whitesides G. M. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 18457–18462. 10.1073/pnas.0910666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songjaroen T.; Dungchai W.; Chailapakul O.; Henry C. S.; Laiwattanapaisal W. Lab Chip 2012, 12, 3392–3398. 10.1039/c2lc21299d. [DOI] [PubMed] [Google Scholar]
- Rosenfeld T.; Bercovici M. Lab Chip 2014, 14, 4465–4474. 10.1039/C4LC00734D. [DOI] [PubMed] [Google Scholar]
- Moghadam B. Y.; Connelly K. T.; Posner J. D. Anal. Chem. 2015, 87, 1009–1017. 10.1021/ac504552r. [DOI] [PubMed] [Google Scholar]
- Phan D. T.; Shaegh S. A. M.; Yang C.; Nguyen N. T. Sens. Actuators, B 2016, 222, 735–740. 10.1016/j.snb.2015.08.127. [DOI] [Google Scholar]
- Abe K.; Kotera K.; Suzuki K.; Citterio D. Anal. Bioanal. Chem. 2010, 398, 885–893. 10.1007/s00216-010-4011-2. [DOI] [PubMed] [Google Scholar]
- Fu E.; Kauffman P.; Lutz B.; Yager P. Sens. Actuators, B 2010, 149, 325–328. 10.1016/j.snb.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidon N. N.; Hong Y.; Salentijn G. IJ.; Verpoorte E. Anal. Chim. Acta 2018, 1000, 180–190. 10.1016/j.aca.2017.10.040. [DOI] [PubMed] [Google Scholar]
- Dungchai W.; Chailapakul O.; Henry C. S. Anal. Chem. 2009, 81, 5821–5826. 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- Kauffman P.; Fu E.; Lutz B.; Yager P. Lab Chip 2010, 10, 2614–2617. 10.1039/c004766j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrilho E.; Phillips S. T.; Vella S. J.; Martinez A. W.; Whitesides G. M. Anal. Chem. 2009, 81, 5990–5998. 10.1021/ac900847g. [DOI] [PubMed] [Google Scholar]
- Nogi M.; Iwamoto S.; Nakagaito A. N.; Yano H. Adv. Mater. 2009, 21, 1595–1598. 10.1002/adma.200803174. [DOI] [Google Scholar]
- Abadian A.; Sepehri Manesh S.; Jafarabadi Ashtiani S. Microfluid. Nanofluid. 2017, 21, 1–11. 10.1007/s10404-017-1899-2. [DOI] [Google Scholar]
- Yang K.; Peretz-Soroka H.; Liu Y.; Lin F. Lab Chip 2016, 16, 943–958. 10.1039/C5LC01524C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate D. M.; Adkins J. A.; Mettakoonpitak J.; Henry C. S. Anal. Chem. 2015, 87, 19–41. 10.1021/ac503968p. [DOI] [PubMed] [Google Scholar]
- Li M.; Tian J.; Al-Tamimi M.; Shen W. Angew. Chem., Int. Ed. 2012, 51, 5497–5501. 10.1002/anie.201201822. [DOI] [PubMed] [Google Scholar]
- Ellerbee A. K.; Phillips S. T.; Siegel A. C.; Mirica K. A.; Martinez A. W.; Striehl P.; Jain N.; Prentiss M.; Whitesides G. M. Anal. Chem. 2009, 81, 8447–8452. 10.1021/ac901307q. [DOI] [PubMed] [Google Scholar]
- Bhakta S. A.; Borba R.; Taba M. Jr; Garcia C. D.; Carrilho E. Anal. Chim. Acta 2014, 809, 117–122. 10.1016/j.aca.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W. J.; Feng D. Q.; Chen M.; Chen Z. D.; Zhu R.; Fang H. L.; Wang W. Sens. Actuators, B 2014, 190, 414–418. 10.1016/j.snb.2013.09.007. [DOI] [Google Scholar]
- Bagherbaigi S.; Corcoles E. P.; Wicaksono D. H. B. Anal. Methods 2014, 6, 7175–7180. 10.1039/C4AY01071J. [DOI] [Google Scholar]
- Nie Z.; Deiss F.; Liu X.; Akbulut O.; Whitesides G. M. Lab Chip 2010, 10, 3163–3169. 10.1039/c0lc00237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiphung J.; Songjaroen T.; Dungchai W.; Henry C. S.; Chailapakul O.; Laiwattanapaisal W. Anal. Chim. Acta 2013, 788, 39–45. 10.1016/j.aca.2013.06.021. [DOI] [PubMed] [Google Scholar]
- Santhiago M.; Kubota L. T. Sens. Actuators, B 2013, 177, 224–230. 10.1016/j.snb.2012.11.002. [DOI] [Google Scholar]
- Li X.; Ballerini D. R.; Shen W. Biomicrofluidics 2012, 6, 011301. 10.1063/1.3687398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder D. T.; Pulliam C. J.; Ouyang Z.; Cooks R. G. Anal. Chem. 2016, 88, 2–29. 10.1021/acs.analchem.5b03070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Manicke N. E.; Cooks R. G.; Ouyang Z. JALA 2010, 15, 433–439. 10.1016/j.jala.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks R. G.; Manicke N. E.; Dill A. L.; Ifa D. R.; Eberlin L. S.; Costa A. B.; Wang H.; Huang G.; Ouyang Z. Faraday Discuss. 2011, 149, 247. 10.1039/C005327A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun A. F.; Wong J.; Khodadadi D.; Nagi R.; Tey A.; Ozcan A. Lab Chip 2013, 13, 636–640. 10.1039/C2LC41152K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q.; Nagi R.; Sadeghi K.; Feng S.; Yan E.; Ki S. J.; Caire R. ACS Nano 2014, 8, 1121–1129. 10.1021/nn406571t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentijn G. IJ.; Oleschuk R. D.; Verpoorte E. Anal. Chem. 2017, 89, 11419–11426. 10.1021/acs.analchem.7b02490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femmer T.; Jans A.; Eswein R.; Anwar N.; Moeller M.; Wessling M.; Kuehne A. J. C. ACS Appl. Mater. Interfaces 2015, 7, 12635–12638. 10.1021/acsami.5b03969. [DOI] [PubMed] [Google Scholar]
- Bonyár A.; Sántha H.; Varga M.; Ring B.; Vitéz A.; Harsányi G. Int. J. Mater. Form. 2014, 7, 189–196. 10.1007/s12289-012-1119-2. [DOI] [Google Scholar]
- O’Neill P. F.; Ben Azouz A.; Vázquez M.; Liu J.; Marczak S.; Slouka Z.; Chang H. C.; Diamond D.; Brabazon D. Biomicrofluidics 2014, 8, 052112. 10.1063/1.4898632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. D. Nat. Chem. 2012, 4, 338–339. 10.1038/nchem.1333. [DOI] [PubMed] [Google Scholar]
- Yetisen A. K.; Akram M. S.; Lowe C. R. Lab Chip 2013, 13, 2210–2251. 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- Liu B.; Du D.; Hua X.; Yu X. Y.; Lin Y. Electroanalysis 2014, 26, 1214–1223. 10.1002/elan.201400036. [DOI] [Google Scholar]
- Meredith N.; Quinn C.; Cate D.; Reilly T.; Volckens J.; Henry C. S. Analyst 2016, 141, 1874–1887. 10.1039/C5AN02572A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins J.; Boehle K.; Henry C. S. Electrophoresis 2015, 36, 1811–1824. 10.1002/elps.201500084. [DOI] [PubMed] [Google Scholar]
- Ahmed S.; Bui M. P. N.; Abbas A. Biosens. Bioelectron. 2016, 77, 249–263. 10.1016/j.bios.2015.09.038. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Si J.; Li Z. Biosens. Bioelectron. 2016, 77, 774–789. 10.1016/j.bios.2015.10.032. [DOI] [PubMed] [Google Scholar]
- Jeong S. G.; Kim J.; Nam J. O.; Song Y. S.; Lee C. S. Int. Neurourol. J. 2013, 17, 155–161. 10.5213/inj.2013.17.4.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro L.; Jacquelin B.; Escadafal C.; Garneret P.; Kwasiborski A.; Manuguerra J. C.; Monti F.; Sakuntabhai A.; Vanhomwegen J.; Lafaye P.; Tabeling P. Sci. Rep. 2017, 7, 1–9. 10.1038/s41598-017-00758-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira C. R.; Yannell K. E.; Jarmusch A. K.; Pirro V.; Ouyang Z.; Cooks R. G. Clin. Chem. 2016, 62, 99–110. 10.1373/clinchem.2014.237164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon D. D.; Maher Y. S.; Yin M.; Jjunju F.; Young I. S.; Taylor S.; Maher S.; Badu-Tawiah A. K. Analyst 2016, 141, 3866–3873. 10.1039/C6AN00168H. [DOI] [PubMed] [Google Scholar]
- Manicke N. E.; Yang Q.; Wang H.; Oradu S.; Ouyang Z.; Cooks R. G. Int. J. Mass Spectrom. 2011, 300, 123–129. 10.1016/j.ijms.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.; Jhang C.-S.; Liu J.-T.; Lin C.-H. J. Sep. Sci. 2012, 35, 2822–2825. 10.1002/jssc.201200480. [DOI] [PubMed] [Google Scholar]
- Duarte L. C.; de Carvalho T. C.; Lobo-Júnior E. O.; Abdelnur P. V.; Vaz B. G.; Coltro W. K. T. Anal. Methods 2016, 8, 496–503. 10.1039/C5AY03074A. [DOI] [Google Scholar]
- Ren Y.; Wang H.; Liu J.; Zhang Z.; McLuckey M. N.; Ouyang Z. Chromatographia 2013, 76, 1339–1346. 10.1007/s10337-013-2458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y.; Chiang S.; Zhang W.; Wang X.; Lin Z.; Ouyang Z. Anal. Bioanal. Chem. 2016, 408, 1385–1390. 10.1007/s00216-015-9129-9. [DOI] [PubMed] [Google Scholar]
- Bare R. O.; Miller R. D.; Pacala T. J.; Payne T. J.; Hertig J. C.; Manicke N. E.; Sistiabudi R.; Yang Q.; Ouyang Z.. Cassettes, systems, and methods for ion generation using wetted porous materials. WO patent WO2012/170301 A1, 2012.
- Bhatia S. N.; Ingber D. E. Nat. Biotechnol. 2014, 32, 760–772. 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Sackmann E. K.; Fulton A. L.; Beebe D. J. Nature 2014, 507, 181–189. 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- Meyvantsson I.; Beebe D. J. Annu. Rev. Anal. Chem. 2008, 1, 423–449. 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- El-Ali J.; Sorger P. K.; Jensen K. F. Nature 2006, 442, 403–411. 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- Kim L.; Toh Y.-C.; Voldman J.; Yu H. Lab Chip 2007, 7, 681–694. 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- Kenney R. M.; Lloyd C. C.; Whitman N. A.; Lockett M. R. Chem. Commun. 2017, 53, 7194–7210. 10.1039/C7CC02357J. [DOI] [PubMed] [Google Scholar]
- Rodenhizer D.; Cojocari D.; Wouters B. G.; McGuigan A. P. Biofabrication 2016, 8, 045008. 10.1088/1758-5090/8/4/045008. [DOI] [PubMed] [Google Scholar]
- Tao F. F.; Xiao X.; Lei K. F.; Lee I.-C. BioChip J. 2015, 9, 97–104. 10.1007/s13206-015-9202-7. [DOI] [Google Scholar]
- Ng K.; Gao B.; Yong K. W.; Li Y.; Shi M.; Zhao X.; Li Z.; Zhang X. H.; Pingguan-Murphy B.; Yang H.; Xu F. Mater. Today 2017, 20, 32–44. 10.1016/j.mattod.2016.07.001. [DOI] [Google Scholar]
- Derda R.; Tang S. K. Y.; Laromaine A.; Mosadegh B.; Hong E.; Mwangi M.; Mammoto A.; Ingber D. E.; Whitesides G. M. PLoS One 2011, 6, e18940. 10.1371/journal.pone.0018940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Steckel G.; Dermutz H.; de Lange V.; Demkó L.; Vörös J. In Proceedings of the 19th International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS 2015); October 25−29, 2015; Gyeongju, Korea; 2015; pp 41–43.
- Deiss F.; Funes-Huacca M. E.; Bal J.; Tjhung K. F.; Derda R. Lab Chip 2014, 14, 167–171. 10.1039/C3LC50887K. [DOI] [PubMed] [Google Scholar]