Over the last decades, biofilms have gained more and more recognition as the common mode of growth microorganisms adapt in nature (1). Furthermore, many types of human infection have been found to progress with the involvement of biofilms or originate from biofilm-associated primary infections (2). Second only to the Gram-negative Pseudomonas aeruginosa, staphylococci have been in the focus of biofilm researchers. As common colonizers of the human skin, staphylococci are the most frequent sources of biofilm infections on surgically implanted indwelling medical devices. These include serious infections, including endocarditis and prosthetic joint infections (PJIs), and may lead to life-threatening conditions, such as sepsis (3–5).
For the longest time, biofilm research even in pathogenic bacteria has been characterized predominantly by in-vitro investigation, from which – despite the obvious limitations - researchers attempted to extrapolate to biofilm-associated infection. More recently, biofilm research has begun to integrate in-vivo infection models to obtain a better understanding of the mechanisms governing biofilm-associated infection. Infection models for P. aeruginosa - the biofilm-forming pathogen most intensely studied by in-vitro research - are extremely challenging to establish and perform (6). Thus, due to the possibility to perform biofilm-associated infection models that reflect human biofilm infection, staphylococcal biofilm research has gained considerably in relative importance. As an important further prerequisite to perform mechanistic biofilm research, methods to genetically manipulate staphylococci are continuously being improved.
In this chapter, the role of staphylococcal biofilms in the commensal state and during infection, our current knowledge of the molecular basis of staphylococcal biofilm formation and its regulation, and the role of biofilm formation in vivo as an important means of immune evasion will be discussed. Finally, potential strategies to develop therapeutics to treat biofilm-associated infections by staphylococci will be presented. There will be a focus on Staphylococcus aureus and Staphylococcus epidermidis, as we know most about biofilms in these two species.
STAPHYLOCOCCAL BIOFILMS IN THEIR NATURAL HABITAT AND DURING INFECTION
Many different definitions have been used for biofilms. Most define biofilm as a microbial community of cells that is embedded in a matrix of extracellular polymeric substances that they have produced. Some researchers restrict the term “biofilm” to aggregates attached to a surface, which can be biotic or abiotic. For others, free-floating bacterial aggregates also represent biofilms. Cells in a biofilm characteristically show a phenotype, with respect to metabolism, gene transcription, and protein production, that is different from that during planktonic growth. This has been shown for both S. aureus and S. epidermidis (7, 8).
Microbial aggregates on infected catheters or other indwelling medical devices obtained from patients resemble biofilms grown in in-vitro microtiter plates or biofilm reactors and there is no doubt that these fit the definition of a biofilm (9). Macro- or microscopic evaluation from wound infections also indicates that those infections, which often contain staphylococci among other pathogenic microbes, can be described as biofilm-associated (10). The situation becomes less clear when looking at the commensal colonization of the skin, nares or intestine, or disease-associated colonization, such as during atopic dermatitis. Some researchers would argue that the physiological situation of bacterial aggregates in those habitats may resemble that commonly found in biofilms, while others have raised doubt (11–13). Currently preventing a more detailed analysis of those potential “colonizing” biofilms is the lack of animal models that closely resemble human colonization. Therefore, in the following, only biofilms on devices and wound infections will be discussed.
Medical devices particularly prone to infection include contact lenses, peritoneal dialysis, urinary and central venous catheters (CVCs), endotracheal tubes, mechanical heart valves, pacemakers, and prosthetic joints (14). Staphylococci generally are the most frequent causes of infections on indwelling medical devices, with S. aureus and S. epidermidis representing the leading species (15). This is mostly due to their predominance in causing intravascular and prosthetic device-associated infections. However, on some specific devices, such as urinary catheters, other bacteria may dominate (16). Most commonly, indwelling medical devices are contaminated with the biofilm-forming bacteria during surgery. The source can be the skin or other colonized body sites of the patient or the health care personnel (14). In that regard, it is important that only ~ 20% of individuals are colonized by S. aureus – in contrast to many coagulase-negative staphylococci (CNS) including S. epidermidis, which are ubiquitous skin colonizers in humans (17, 18). Thus, the S. aureus carrier state of the patient is a risk factor for device infection, as it is for other types of S. aureus infection (19). Alternatively, already implanted devices can be infected via hematogenous seeding, although this is to be considered less frequent (20). Not surprisingly, immune- or otherwise compromised patients, such as AIDS patients, patients receiving immunosuppressive therapy, or premature newborns, are at increased risk of developing biofilm-associated infections on indwelling devices (14). These populations are also at increased risk regarding serious complications that may arise from infected devices. The most notable among those is blood infection (bacteremia), which may result in life-threatening sepsis. Central line (CVC)-associated bloodstream infections (CLABSIs) are among the most frequent and serious complications of medical device infection and cause thousands of deaths annually. In a 2004 study including 24,179 cases of nosocomial bloodstream infections (most of which stem from contaminated devices) in 49 US hospitals over a 7-year period from March 1995 through September 2002, CNS were responsible for 31%, and S. aureus for 20% of cases, far ahead any other microorganism (21).
Chronic wounds, such as chronic skin ulcers in diabetes patients, and acute burn wounds, are highly susceptible for infection by many pathogenic microorganisms. According to clinical observation, the infecting organisms, which are usually more than one, present as a biofilm (22). The species most often found in wound infections is S. aureus (23, 24). From a microbiological point of view, the multispecies biofilms in wounds are much more difficult to understand than infections of indwelling medical devices, which usually present as a monospecies infections. Several studies claim that bacterial interactions exist in wounds between S. aureus and, for example P. aeruginosa, but there is no commonly accepted view of their exact nature and role (25).
THE MOLECULAR BASIS OF BIOFILM FORMATION IN STAPHYLOCOCCI
Attachment phase
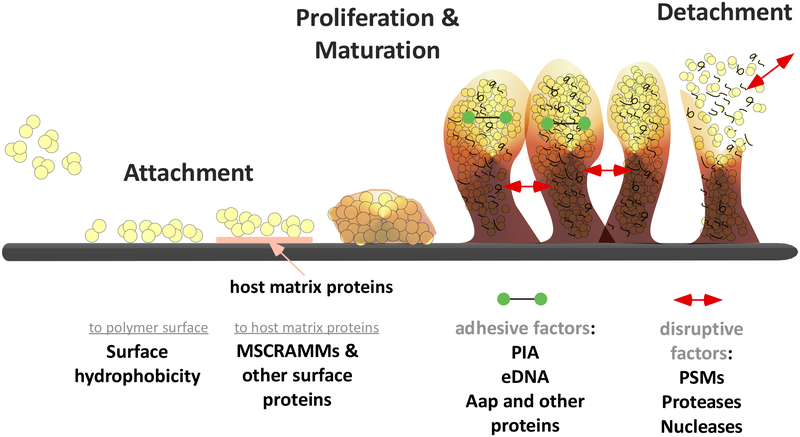
Biofilm development has been extensively studied in vitro using many different setups and microorganisms. Commonly, it is described as a process of 3 main stages: attachment, proliferation/maturation, and detachment (dispersal) (26) (Fig. 1).
Fig. 1. Staphylococcal biofilm development.
Attachment of cells to a surface (in the case of surface-attached biofilms) occurs via hydrophobic interactions to an abiotic surface, or via surface proteins that bind in a specific fashion to host matrix proteins covering an indwelling medical device. Growth of the biofilm in the proliferation/maturation stage is accompanied by the production of cell-cell-adhesive matrix components (such as, PIA, eDNA, and proteins) as well as disruptive factors (such as, PSMs and degradative secreted enzymes). Those disruptive factors can also cause detachment, a process of great importance for the initiation of complications of biofilm–associated infection, such as bacteremia.
Attachment to a surface is considered the first stage of biofilm development, although in “free-floating” biofilms, this stage is not relevant. In contrast to motile organisms, which may reach a surface in an active fashion, staphylococci gain contact with a surface passively. Staphylococci may directly attach to the abiotic surface of indwelling medical devices, or other abiotic surfaces in the environment (27). The hydrophobic character is the main factor enabling attachment to the usually hydrophobic material surface. However, specific staphylococcal molecules also have been implicated in surface attachment. These include the S. epidermidis major autolysin AtlE and its S. aureus homologue Atl (28, 29) and wall teichoic acids (WTAs) in S. aureus (30). These two may be linked mechanistically due to the fact that WTAs impact binding of Atl to the cell surface, controlling cell separation (31). Of note, primary attachment is often measured merely as a short-term variation of the frequently used simple microtiter-based biofilm assay. Whether results represent true primary attachment or initial phases of proliferation cannot be determined clearly using that assay.
In vivo, direct interaction of staphylococcal cells with the abiotic surface is not considered relevant, as indwelling devices rapidly become covered by host matrix material, including fibronectin, fibrinogen, vitronectin, and other matrix molecules (27). Under those conditions, attachment to the device is facilitated by a series of specific staphylococcal surface proteins that interact with those human matrix proteins. The most important family of such surface-expressed staphylococcal binding proteins are the MSCRAMMs (microbial surface components recognizing adhesive matrix molecules). MSCRAMMs consist, from N- to C-terminus, of a signal peptide, a ligand-binding domain with characteristic repeat sequences, a cell wall-anchoring region, a membrane-spanning region, and a positively charged tail (32). At the C-terminus, the MSCRAMM contains a conserved LPXTG motif, which is covalently linked by the enzyme sortase to peptidoglycan (33). According to genome analysis, S. aureus has about 20 and S. epidermidis about 12 MSCRAMMs (34). Prominent members are the fibrinogen- and fibronectin proteins, which include clumping factors A and B (ClfA, ClfB), the serine/aspartate-rich (Sdr) protein family, and fibronectin-binding proteins A and B (FnBPA, FnBPB) (32). Non-covalently bound surface proteins may also mediate attachment. Among them are predominantly enzymes that have a primary catalytic role, such as the Atl-type autolysins. Furthermore, there is a loosely defined group of non-covalently surface-attached proteins collectively called SERAMs (secretable expanded repertoire adhesive molecules), which bind to the surface by not yet characterized mechanisms and usually show broad specificity in human matrix protein binding (35). The extracellular adherence protein Eap (also called “MHC class II analogous protein Map) and the extracellular matrix and plasma binding protein Emp of S. aureus belong to this group. Finally, the giant 1.1 MDa Embp protein of S. epidermidis and its S. aureus homologue Ebh (extracellular matrix binding protein homologues) are non-covalently surface-bound fibronectin-binding proteins (36, 37).
Proliferation and matrix formation
During the second stage of biofilm development, the microcolonies that have formed after attachment grow by proliferation. Additionally, cells secrete polymeric molecules to form the biofilm matrix. Among all aspects of biofilm development, the polymeric molecules that form the biofilm matrix have received most attention. Biofilm matrix polymers are of divergent chemical nature and include polysaccharides, proteins, and teichoic acids. In addition to those actively secreted molecules, polymeric substances from dead cells are also believed to contribute to the biofilm matrix. The most notable among those is DNA, which after release from dying cells is known as extracellular DNA (eDNA).
In many biofilm-forming bacteria, exopolysaccharides represent the matrix components that are most specifically implicated with biofilm formation. Staphylococci produce one main biofilm exopolysaccharide, which is called polysaccharide intercellular adhesin (PIA), or, according to its chemical composition, poly-N-acetylglucosamine (PNAG) (38) (Fig. 2). In contrast to chitin, another important N-acetylglucosamine homopolymer found in nature, the N-acetylglucosaminyl moieties in PIA are β−1–6 linked. One characteristic feature that distinguishes PIA from most other staphylococcal surface molecules, and also from many other bacterial biofilm exopolysacharides, is its cationic character, which is due to the enzyme-catalyzed removal of ~ 15 – 20% of N-acetyl groups after secretion. This feature may be important for electrostatic interaction with other surface polymers, and thus the formation of the sticky extracellular biofilm matrix (38, 39). There appears to be no specific “anchor” for PIA on the cell surface, and it has been shown directly that this function is not accomplished by wall teichoic acids (40).
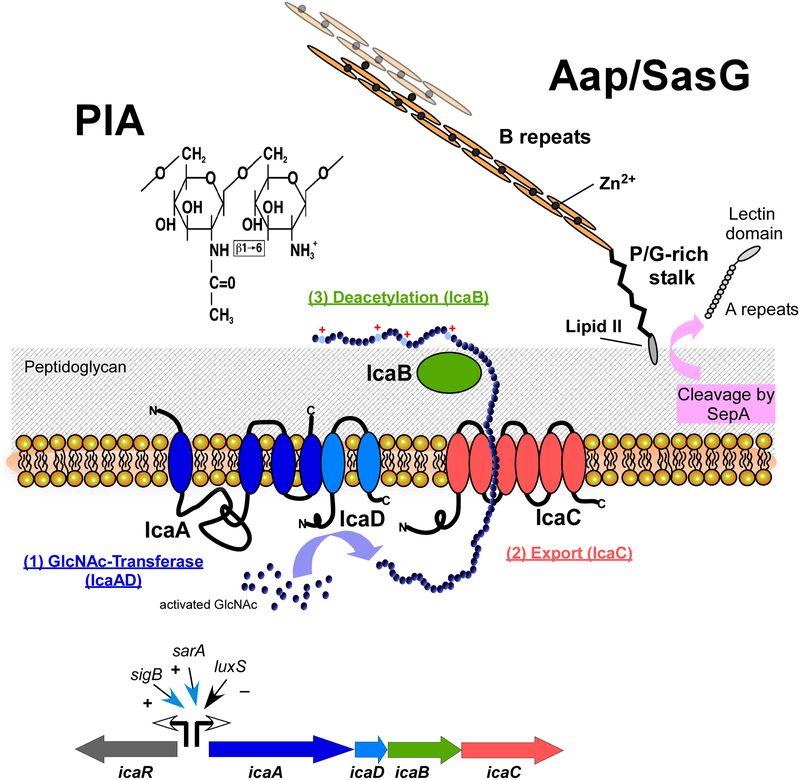
Fig. 2. Predominant staphylococcal biofilm matrix components.
The exopolysaccharide PIA (polysaccharide intercellular adhesin) (left), produced by many S. aureus and S. epidermidis isolates, is a β1–6-linked homopolymer of N-acetylglucosamine. It is synthesized in the cell by the combined activity of the membrane enzymes IcaA and IcaD, and likely exported by IcaC. The extracellular surface-bound enzyme IcaB removes a certain percentage (~ 15 – 20%) of N-acetyl moieties, which gives the otherwise neutral PIA molecule a positive net charge, anchoring PIA to the negatively charged cell surface. In addition to the ica biosynthetic genes icaADBC, the PIA biosynthesis locus also contains a regulatory gene, icaR. Several global regulators impact transcription from the icaR and icaADBC promoters.
The accumulation-associated protein (Aap), which is present in S. epidermidis and has a homologue in S. aureus called SasG, is produced as a 220-kD precursor protein, from which the secreted protease SepA cleaves off the N-terminal A-repeat and lectin domains. It is anchored to the cell wall via sortase-catalyzed covalent linkage to lipid II. Mature Aap forms extended fibrils out of B repeat domains, whose polymerization is dependent on Zn2+ ions. Zn2+ is also required for the interconnection of Aap/SasG proteins from different cells, which can happen in an interspecies manner.
PIA biosynthesis is accomplished by the products of the ica (intercellular adhesion) gene locus, which comprises the icaA, icaD, icaB, and icaC genes (41). IcaA is the N-acteylglucosamine transferase, which together with IcaD has been shown to produce an N-acetylglucosamine oligomer (42). Chain growth is dependent on the presence of IcaC, a membrane protein, assumed to be the PIA exporter. IcaB, which is located on the bacterial surface, is the PIA deacetylase (39). Deacetylation is crucial for the retention of PIA on the surface and the various functions PIA fulfills in staphylococcal physiology, which in addition to biofilm formation comprise increased resistance to antimicrobial peptides (AMPs) and neutrophil phagocytosis (39). Upstream of the ica operon is the icaR gene, which is transcribed in the opposite direction of the icaADBC operon. IcaR works as a repressor of the ica operon (43).
Several studies have shown that biofilm formation in vitro or in vivo can be accomplished by S. epidermidis or S. aureus isolates that do not harbor the ica locus (44–46). Although, the extension, structuring, and robustness of PIA-negative biofilms appears to be lower than those formed by PIA, at least in S. epidermidis (47, 48). In S. aureus isolates, biofilm formation appears to be predominantly protein-dependent (49). Earlier studies reported differences between methicillin-resistant (MRSA)and –sensitive (MSSA) isolates in that regard, while a more recent study indicates that both MRSA and MSSA form almost exclusively protein-dependent biofilms (49, 50). In contrast, S. epidermidis more frequently relies on PIA to form biofilms, with an additional protein-mediated contribution (46). Replacing PIA in ica-negative isolates, or adding to its function in ica-positive isolates, are a series of macromolecules that include polymeric proteins and non-protein polymers. Among the many surface-located proteins implicated in staphylococcal biofilm formation, the S. epidermidis accumulation-associated protein (Aap) and its S. aureus homologue, SasG (51), stand out due their capacity to self-polymerize and form fibrils that interconnect cells (52–54) (Fig. 2). Aap is a 220-kD protein that is cleaved by the SepA protease to an active, mature form (55–57). At the N-terminus, the A domain of Aap contains repeats that may initiate biofilm formation in some strains and also bind to human corneocytes (58). It is followed by a lectin domain. These two domains are not present in mature Aap, which contains a large stretch of B repeats in front of a C-terminal proline/glycine-rich region and the cell wall-anchoring domain. The B repeats are responsible for self- and intercellular adhesion in a Zn2+-dependent manner, assisted by cooperative folding (53, 59–61). Recently, the proline/glycine-rich region was shown to form an extended stalk that pushes the B repeat binding domain away from the cell surface, likely to enable better interaction with Aap from other cells, promoting biofilm formation (62). Notably, it has been shown that the homologous proteins SasG and Aap can interact, enabling interspecies biofilm connections (63). Several other surface proteins have also been implicated in intercellular adhesion, but in many of those cases it is difficult to distinguish between a surface/tissue attachment and genuine intercellular adhesion function. These proteins contain many surface binding proteins of the MSCRAMM family in addition to the biofilm-associated protein (Bap), found in S. aureus mastitis isolates, of which a homologue (Bhp) is present in S. epidermidis (64, 65). In support of a specific involvement in matrix formation, Bap has recently been reported to assemble into amyloid-like matrices to promote biofilm assembly (66), reminiscent of the function of Aap.
Non-protein extracellular polymers implicated in biofilm formation include teichoic acids and eDNA. Teichoic acids occur in two forms: wall teichoic acid (WTA), which is covalently linked to the cell wall, and lipoteichoic acid (LTA), which is surface-anchored via a lipid moiety that intercalates in the cytoplasmic membrane lipid bilayer (67). A specific contribution of the teichoic acid surface polymers to the biofilm matrix is likely, but has only been shown for WTA of S. epidermidis (68). Teichoic acids devoid of D-alanylation, which alters their net charge, show diminished biofilm formation in S. aureus (30). Teichoic acids are also implicated in receptor-mediated adhesion to the nasal epithelium (69). Finally, eDNA contributes to biofilm formation in vitro (70). Likely due the highly polymeric and anionic character of DNA, it is extremely “sticky” and thus, eDNA is prone to interact with many other surface molecules, contributing to the formation of the extracellular biofilm matrix network.
Structuring and detachment
Biofilms do not grow as undifferentiated “bricks”, but have a characteristic three-dimensional structure, which contains towers, often described as “mushroom”-like, as well as fluid-filled channels between those towers (26). The second (maturation) stage of biofilm development comprises disruptive forces to create those structures in addition to the adhesive mechanisms described above. These are mediated by enzymes that degrade biofilm polymers, such as nucleases and proteases, and surfactant-like molecules, like the staphylococcal phenol-soluble modulins (PSMs), which disrupt non-covalent interactions (71, 72). The same forces can also ultimately cause biofilm detachment, which facilitates systemic dissemination and subsequent complications during device-associated infection. Both biofilm structuring and detachment/dispersal effects have been described for the PSMs, while research on degradative enzymes has been mostly limited to detachment/dispersal.
PSMs are a family of alpha-helical peptides with pronounced amphipathy, thus exhibiting characteristics that give them surfactant-like properties (73). Members of the PSM family are produced in S. aureus, S. epidermidis, and most likely virtually all staphylococci (73, 74). PSMs can be grouped in α-type peptides (~20–25 amino acids in length), which in S. aureus comprise the PSMα and δ-toxin peptides, and the longer β-type PSMs (~44–45 amino acids) (73). Confocal-laser scanning microscopy, a method often used to visualize biofilm structure, revealed that the characteristic channel-containing biofilm structure is not present in psm deletion mutants, which grow in an undifferentiated and extended fashion (48, 75). In contrast to biofilm polymer-degrading enzymes, PSMs structure PIA -dependent and –independent biofilms (48). PSMs work on growing biofilms; when biofilms are formed, external addition of PSMs has only limited biofilm-dispersing activity. Somewhat contrastingly, PSMs have also been described to form amyloid structures that prevent the enzymatic degradation of pre-formed biofilms (76). A double mutant in the psmα and psmβ loci was impaired in preventing degradation, while the δ-toxin was not specifically addressed in that study. While there appears to be agreement on the in-vitro capacity of PSMs to form amyloid-like structures, the role of PSM amyloids in biofilms remains controversial (77).
Among biofilm-degrading enzymes, proteases are the most important (72), while enzymes degrading PIA are not produced by staphylococci. S. aureus and S. epidermidis produce a series of exoproteases. S. aureus secretes ten proteases, including seven serine proteases (SspA and SplA-F), two cysteine proteases (SspB and ScpA), and one metalloprotease (Aur) (78). S. epidermidis produces at least three major proteases, a homologue of the staphopain cysteine protease SspB, a metalloprotease called SepA, and a homologue of the S. aureus SspA (V8) serine protease (17). Mutations in specific S. aureus protease genes, or addition of protease inhibitors, impair biofilm formation or lead to biofilm degradation, respectively, in biofilms that are protein-dependent (49, 79). While specific matrix protein targets of several of the S. aureus proteases have been identified (80, 81), they overall show relatively low target sequence specificity (82). The S. epidermidis protease SspA (Esp) has been shown to be able to degrade protein-dependent S. aureus biofilms (83). As biofilm formation in S. epidermidis is often strongly PIA-dependent, research on the role of S. epidermidis proteases in biofilm development has been limited.
The biofilm matrix component eDNA is subject to degradation by nucleases, which originally has been shown using human DNase I (70), which is abundant in human serum. Both S. aureus and S. epidermidis secrete nuclease activity. S. aureus is known to produce two secreted nucleases, Nuc1 and Nuc2 (84). Expression of Nuc1 reduces biofilms and a nuc1 mutation in strain USA300 shows increased in-vitro biofilm formation (85), as would be expected from abolished eDNA degradation. However, isogenic nuc1 or nuc2, or combined nuc12 mutants in a different strain (UAMS-1) did not exhibit enhanced in-vitro or, notably, in-vivo biofilm formation (86).
REGULATION OF STAPHYLOCOCCAL BIOFILM FORMATION
Biofilms have been recognized as the common form of microbial growth. It is thus not surprising that many regulators that adapt staphylococcal gene expression to changing environmental conditions have an impact on biofilm formation. Here, there will be a focus on regulatory effects on the main biofilm-forming factors and on the biofilm effects of the major global regulators, the quorum-sensing system Agr, the Sar family of regulators, and the alternative sigma factor, SigB.
In 1998, Davies et al. reported that a quorum-sensing regulator in P. aeruginosa positively impacted biofilm formation (87), a result soon generalized regarding the relationship of quorum-sensing control and biofilm formation in bacteria. Soon after, Vuong et al. reported increased biofilm formation in isogenic mutants of the Agr quorum-sensing system in S. aureus (88), in clear contrast to the generalized P. aeruginosa finding. Ever since, mechanistic investigation has shed more light on the impact quorum-sensing systems have on biofilm formation in several different bacteria. As a common theme, quorum-sensing systems appear to frequently control biofilm structuring processes, which may result in more or less extended biofilms depending on the specific conditions (71). Agr in staphylococci controls two main biofilm structuring and dispersal factors described above, PSMs and proteases, with the control of psm genes being exceptionally direct and tight (8, 89, 90). As a result, biofilms of agr mutants grow in an unstructured and extended fashion, as shown for both S. aureus and S. epidermidis (75, 91), and applying the extracellular autoinducing peptide (AIP) signal of the Agr quorum-sensing system results in biofilm dispersal (79). However, it remains poorly understood which mechanisms lead to the spatial and temporal heterogeneity of Agr expression that is observed in biofilms (75) and which are a theoretical prerequisite for the Agr-mediated biofilm structuring effect. Finally, the autoinducer-2 quorum-sensing system, which remains less well investigated in staphylococci, represses biofilm formation in an ica-dependent manner (92, 93). Both quorum-sensing systems thus affect biofilm formation negatively, but using different mechanisms.
The prototype of the Sar paralogue family of regulators, SarA, impacts biofilm formation via regulation of Agr, as well as in an Agr-independent manner (94–97). The strong regulatory impact of SarA on protease production appears to play a preeminent role in this effect (98), and this, historically, was how the biofilm effect of S. aureus proteases was discovered. Furthermore, SarA positively controls ica transcription in both S. aureus and S. epidermidis (95, 97), while its paralogue SarZ appears to have opposite effects in the two species – positive in S. epidermidis and negative in S. aureus (99, 100). Generally, which environmental signals stimulate Sar expression is not known, and therefore the biological context of biofilm regulation by Sar paralogues remains poorly defined.
Sigma factors are transcription initiation factors that provide promoter recognition specificity to RNA polymerase (101). The “alternative” sigma factor SigB is expressed during stationary growth phase and environmental stress and is involved in controlling manifold aspects of staphylococcal physiology and virulence. SigB regulation of biofilm formation in S. epidermidis biofilms was reported to occur by an impact on ica transcription via SigB dependence of the icaR promoter (102), and in S. aureus possibly via SarA-dependent protease regulation (103, 104). However, other studies found no impact of SigB on biofilm formation in S. aureus and S. epidermidis (97, 105).
IN VIVO ROLES OF STAPHYLOCOCCAL BIOFILM FACTORS
The recognition that microbes grow in biofilms in nature as well as during many types of infection has revolutionized microbiology in the second half of the previous century. However, in comparison to the wealth of in-vitro biofilm studies, biofilm microbiology has not yet widely included in-vivo experimentation to understand mechanisms of biofilm-associated infection and determine whether general principles established in-vitro bear in-vivo relevance. In part, this is due to the problem that animal models for biofilm-associated lung infection, by most biofilm researchers’ favorite model organism, P. aeruginosa, are extremely challenging. The situation is somewhat better for staphylococcal biofilm-associated infection, as catheter infections can quite easily be mimicked by placing a plastic device under the skin of test animals, such as mice or rabbits. Furthermore, more sophisticated models of biofilm-associated staphylococcal endocarditis are available.
Staphylococcal biofilm infection has originally been associated mainly with S. epidermidis. Biochemical characterization of PIA and genetic approaches to find biofilm-related factors using transposon mutagenesis of S. epidermidis then resulted in the identification of the ica and atlE loci as important for matrix formation or initial adherence, respectively, during in-vitro biofilm formation (29, 41). For that reason, these were the factors first assayed in animal models of biofilm-associated infection. An isogenic transposon mutant in the ica genes (S. epidermidis M10) was tested for the impact of PIA production on infection outcome in a mouse model of device-associated infection, using a subcutaneously implanted catheter, and a rat CVC-associated infection model (106, 107). In the mouse subcutaneous catheter model, the wild-type produced significantly more biofilm on the catheter and a more pronounced abscess (106). In the rat model, more animals challenged with the wild-type strain developed CVC-associated infection than did those challenged with the ica-negative M10 strain (107). The impact of PIA and AtlE were also assayed in a different strain background in the rat CVC-associated model. Both PIA and AtlE had a significant impact on biofilm formation on the device as well as on associated bacteremia (108). Interestingly, later it was also shown that the deacetylation of PIA by IcaB is crucial for the development of biofilm-associated infection using a mouse subcutaneous catheter model (39). As for S. aureus, there is no direct evidence from animal infection models suggesting a key role of PIA in biofilm-associated infection. Furthermore, one study showed that the S. aureus homologue of AtlE, the major autolysin Atl, does not impact biofilm-associated infection (109). These results reflect in-vitro findings indicating that biofilm formation in S. aureus is mainly dependent on other factors.
Biofilm formation in S. aureus is believed to depend on surface proteins in a majority of strains. However, only for very few surface proteins, has an impact on biofilm-associated infection been directly demonstrated using biofilm-associated infection models and deletion mutants. These include S. aureus Bap and protein A (Spa), for which a significant impact on pathogenesis in mouse catheter implant infection models was found (64, 110). For some other surface proteins, there is some circumstantial evidence for roles in in-vivo biofilm formation that is derived from using antibodies. For example, human IgG recognizing ClfA was protective in a rabbit endocarditis model (111). As for S. epidermidis, the difficulty in producing isogenic deletion mutants has hampered research along those lines, but circumstantial evidence obtained by heterologous expression has been obtained indicating a role of the surface protein SdrF, which is produced by a subset of S. epidermidis strains (112), in the initiation of experimental device driveline-related infection in mice (113). In general, the paucity of experimental in-vivo evidence underlining the importance of surface proteins in biofilm-associated infection may be due to their pronounced functional redundancy. More recently, an ex-vivo approach was taken to investigate biofilm formation during PJI. Dastgheyb et al. used synovial fluid obtained during surgery from uninfected human joints. Using the Nebraska transposon bank, which contains a copy of every non-essential gene in S. aureus, they identified ClfA, ClfB, FnBPA and FnBPB, as well as characteristically low Agr activity and PSM concentration in synovial fluid as driving forces of the extensive formation of antibiotic-resistant biofilms seen in PJI (114–116).
In-vivo information on biofilm structuring factors is available for the PSMβ peptides of S. epidermidis, which have been shown to drive dispersal and systemic dissemination from an implanted device in a mouse model (48). Mutants in the psmα, psmβ, or hld (encoding δ-toxin, now classified as a PSM) loci of S. aureus show the same effect, with that of a triple psmαβhld mutant being comparable to that of an agr mutant (75). However, at least in the case of α-type PSMs (PSMα peptides, δ-toxin), these outcomes may be influenced also by the cytolysis-mediated role of PSMs in immune evasion (117). Which role proteases play in biofilm-associated infection remains largely undefined. With proteases presumably having multiple roles in infection, their specific role in in-vivo biofilms is difficult to define. In a recent study using the USA300 strain LAC, a deletion mutant in all ten protease genes did not reveal a significantly changed bacterial load on the device in experimental implanted device infection in mice (118). Similarly, S. aureus nuc1 and nuc2 mutants do not have a significant in-vivo effect during biofilm-associated infection (86), casting some doubt also on the role of eDNA in in-vivo biofilms.
STAPHYLOCOCCAL BIOFILM FORMATION AS AN IMMUNE EVASION AND ANTIBIOTIC RESISTANCE MECHANISM
In-vivo biofilm formation is subject to several factors that are specifically due to the in-vivo environment, such as, most notably, interaction with innate host defenses, in addition to interaction with antibiotics during antibiotic therapy. Innate host defenses consist primarily of phagocytes, such as neutrophils and macrophages, and antimicrobial peptides (AMPs), which are a part of the intracellular killing mechanisms phagocytes employ (119). AMPs are also produced in the skin, where they are secreted to control skin-colonizing or –infecting bacteria (120). Secreted AMPs may thus also form part of the host’s defenses on subcutaneously placed catheters (Fig. 3).
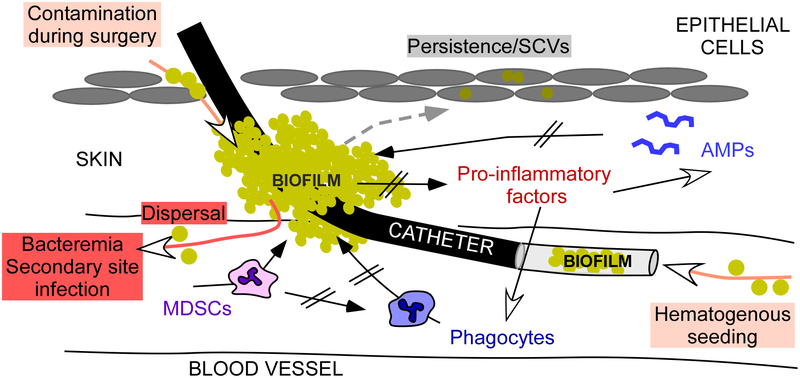
Fig. 3. Staphylococcal biofilm-associated infection on medical devices.
Mechanisms underlying staphylococcal device-associated infection are depicted in an exemplary fashion for an intravascular catheter-associated biofilm. Many of those are still hypothetical. Biofilm formation on a catheter may originate from bacteria introduced as a contamination during surgery/catheter insertion; those are believed to initiate biofilms on the catheter outside. Alternatively, biofilms in the lumen of the catheter can originate from bacteremia and other infection sites due to hematogenous seeding from those sites. Compared to planktonic growth, biofilms secrete less pro-inflammatory factors, which normally cause considerable responses by innate host defenses, such as phagocyte influx and AMP production. In addition to eliciting less such responses, the biofilm matrix provides a shelter from AMPs and phagocyte intrusion. S. aureus biofilms also attract myeloid-derived suppressor cells (MDSCs), which add to decreasing inflammatory responses, particularly phagocyte influx. Finally, internalization of bacteria for example by epithelial cells can produce SCVs, prone to persist and cause recurrent infections.
Biofilm formation per se has long been known to provide resistance to attacks by phagocytes, which are impaired in penetrating through the biofilm matrix (121, 122). Similarly, the biofilm matrix may be impermeable to certain AMPs and antibiotics. On the other hand, some can readily penetrate, which is likely dependent on the AMP’s or antibiotic’s chemical features (123). It has been estimated that biofilms provide 10 – 1000 times increased tolerance to antibiotic effects (124). One antibiotic that has been shown to be exceptionally active against biofilms and is often proposed to treat biofilm infections is rifampicin (rifampin) (125). However, resistance to rifampicin can develop quickly during treatment (126). Interestingly, recent in-vivo research indicates that specific other antibiotics, such as ceftaroline, are more active against S. aureus biofilms than those commonly used in the clinical setting (127). Decreased anti-biofilm efficacy of antibiotics whose penetration is not decreased is due the specific physiology of cells in a biofilm. For example, the reduced proliferative and metabolic activity of biofilm cells makes them more tolerant to antibiotics that target those processes, such as cell wall-targeting antibiotics (e.g., beta-lactams) or antibiotics that target DNA or protein synthesis (128). Finally, biofilms have been shown to contain more so-called “persisters”, dormant cells which are indifferent to antibiotics, as compared to the majority of the bacterial population, an effect that enables this subset of cells to withstand antibiotic concentrations far above the MIC (129).
In addition to those general effects, staphylococcal biofilms show specific alterations in physiology, as compared to planktonic cells, that dampen inflammation and the innate immune response. First, Agr as a regulator of many toxins, which have pro-inflammatory effects, shows limited expression in biofilms (75, 91). However, whether this effect has in-vivo consequences remains to be shown directly. In support of that notion, many S. aureus and S. epidermidis isolates obtained from chronic and device-associated infections are functionally Agr-negative (91, 130). It appears as if phenotypically or genotypically Agr-negative isolates have selective advantages during chronic infection, of which the mechanistic underpinnings still need to be elucidated.
Second, S. aureus biofilms have been shown to reduce inflammatory effects, such as phagocyte influx. Specifically, S. aureus biofilms in murine biofilm-associated infection appear to skew the immune response away from a microbicidal phenotype by circumventing locus specific pathogen recognition pathways (131) – a finding in line with reduced Agr activity. Furthermore, during experimental PJI, S. aureus biofilms stimulate the influx of myeloid-derived suppressor cells (MDSCs), resulting in reduced phagocyte influx (132), and human PJIs are associated with MDSCs (133). Finally, cyclic di-AMP, released by cell lysis, has been shown to stimulate a type I interferon-mediated anti-inflammatory effect in biofilms (134). However, it is not clear whether these immune evasion mechanisms are specific to a biofilm infection as opposed to other S. aureus infection types, or, in the case of cyclic di-AMP release, occur in vivo. Somewhat in contrast, it has also been shown that in a long-lasting murine biofilm infection, downregulated responses in regulatory T-cells (Tregs) during later stages of the infection provide for an unrestrained inflammatory response (135).
Some prominent biofilm matrix molecules have been described to promote immune evasion mechanisms independently of their role in matrix formation. PIA in its de-acetylated form provides resistance to neutrophil phagocytosis and killing by AMPs (39). Possibly, this activity is responsible for the effects PIA has shown in non-biofilm associated experimental infection (136). The S. aureus Bap protein, in addition to promoting adhesion, prevents S. aureus internalization by epithelial cells (137). This contrasts with fibronectin-binding proteins, which have long been known to facilitate such internalization (138). Which role internalization and potential intercellular persistence in non-professional phagocytic cells has in biofilm-associated infection still needs to be investigated in more depth. There is evidence indicating that the formation of small-colony variants (SCVs), cells with severely reduced metabolism, contributes considerably to such persistence (139).
THERAPEUTIC STRATEGIES FOR STAPHYLOCOCCAL BIOFILM-ASSOCIATED INFECTIONS
Biofilm infections remain extremely resistant to antibiotic therapy. Consequently, there are currently no alternatives, and excision and replacement of an infected device are often the only remedy. In the following, some strategies to develop alternative biofilm therapeutics will be presented. Most of them are still in the pre-clinical stage.
Antimicrobial peptides
With most antibiotics not being efficacious against biofilm cells due to their mode of action that only targets active cell processes, one strategy focuses on developing bactericidal antibacterial compounds. In the center of this approach are AMPs, most of which work by forming pores in the bacterial membrane (140). However, AMPs are subject to a series of resistance mechanisms that are based on the barrier function of the biofilm matrix in addition to more specific mechanisms, such as efflux pumps and AMP repulsion. Furthermore, staphylococci have learned to sense the presence of AMPs during their co-evolution with humans and react with efficient countermeasures (141, 142).
Material alterations
Many attempts have been made to deal with the adhesion of bacteria to medical devices by altering the surface so as to lower adhesive features, or by coating the device surface with antibacterial compounds (143). The latter mostly comprise antibacterial metals, such as silver or copper, in addition to antibiotics or AMPs. Such optimized devices, mainly catheters, are in clinical use and have had some success; however, biofilm formation still happens and it appears impossible to completely inhibit adhesion by altering device surface composition. Unfortunately, once adhesion is accomplished, altered device surfaces do not impact further biofilm growth. One reason for the limited success of altering device surfaces is the abovementioned fact that they are prone to being covered by human matrix proteins, to which bacteria attach independently of the device surface.
Vaccines
So far, there is no working vaccine for S. aureus, even for non-biofilm associated infections; and many have argued that development of an S. aureus vaccine is inherently difficult (144). It has been proposed to design vaccine approaches for biofilm infections depending on which antigens are present in the biofilm mode of growth (145), but no such attempts have been actively pursued. There appears to be some pre-clinical success with anti-PIA antibodies, as shown in an S. aureus periprosthetic osteomyelitis rat model (146). As for S. epidermidis, there has been success using immunization with the surface protein SesC in a subcutaneous foreign body in a rat model (147). However, vaccination against S. epidermidis as a beneficial part of the skin microbiota is generally debatable. Furthermore, the many positive results from anti-staphylococcal vaccine experiments in animals have so far never resulted in a vaccine that works in humans.
Bacteriophage and lysins
Bacteriophage therapy is controversial for several reasons, but it has the advantage of having a bactericidal mechanism, and thus not being subject to the efficacy-lowering bacterial physiology of biofilms or persister cells (148). Then again, cell surface-located bacteriophage receptor molecules may not be accessible in the biofilm matrix. However, in several cases bacteriophages have proven efficacious against in-vitro staphylococcal biofilms. For example, bacteriophages ϕIPLA-RODI and ϕIPLA-C1C reduced biofilms of S. aureus and S. epidermidis in vitro (149). Bacteriophage K is also often proposed as an anti-staphylococcal biofilm agent (150, 151). Furthermore, bacteriophage lysins may represent a means to enzymatically disrupt biofilms, such as bacteriophage lysin CF-301 (152, 153). In-vivo efficacy of bacteriophages or bacteriophage lysins against Staphylococcus biofilm-associated infection still needs to be demonstrated.
Biofilm-degrading enzymes
The idea to enzymatically disrupt staphylococci in biofilms for treatment is not limited to bacteriophage-derived lysins. Lysostaphin, a lysin produced by S. staphylolyticus, is used in the laboratory to digest staphylococcal cells and has been investigated intensely as an anti-staphylococcal agent (154). It has also been shown to work against in-vitro S. aureus biofilms, an effect that could be enhanced when used together with antibiotics (155–157). Notably, lysostaphin eradicated S. aureus in a catheter-associated infection model in mice when applied through the catheters (158). S. epidermidis shows increased resistance to lysostaphin due to the different composition of the pentapeptide bridge in peptidoglycan, the target of lysostaphin (159). Another enzyme-based potential therapeutic strategy consists of applying matrix-degrading enzymes. Dispersin B is a PIAse produced by Actinobacillus actinomycetemcomitans (160). It has been shown to degrade staphylococcal biofilms in vitro that are dependent on PIA, thus, predominantly S. epidermidis biofilms (161, 162). Other matrix-degrading enzymes, such as proteases, have also been proposed to treat biofilm infections (79, 155), a strategy obviously also dependent on the mode of biofilm matrix formation. In-vivo efficacy of biofilm matrix-degrading enzymes still needs to be shown.
Quorum-sensing blockers
Finally, quorum-sensing blockers are often proposed for biofilm infections; however, due to the biofilm-structuring and –dispersing effect of the Agr quorum-sensing system, this approach appears counterproductive in staphylococci. Nevertheless, one needs to consider that severe and life-threatening conditions, such as bloodstream infections, often only arise after dispersal from a biofilm. For those, quorum-sensing blockers or other approaches targeting aggressive virulence factors, such as toxins, may be invaluable. Anti-toxin antibodies show great promise in that regard (163).
CONCLUDING REMARKS
Biofilm formation in staphylococci remains a serious clinical problem due to the enormous and continued difficulties in treating staphylococcal biofilm-associated infections. Therefore, mechanisms of staphylococcal biofilm formation have received much attention over the last decades; and many important results, for example on staphylococcal biofilm genetics and regulation, have been achieved using in-vitro setups of biofilm development. However, it has become clear that biofilm-related in-vitro observations are often not transferable to the in-vivo level, with the in-vivo situation differing in substantial aspects, such as the interaction with host matrix proteins and immune defenses. With the interest in staphylococcal biofilm formation clearly stemming from the involvement of staphylococci in infections, there should be a more pronounced focus on the in-depth evaluation of biofilm formation in an in-vivo context in the future. This is equally imperative in pre-clinical research on potential anti-biofilm therapeutics, which currently often lacks this crucial aspect. Such approaches should contain more animal models, which still can be optimized to better reflect human biofilm-associated infection, and ex-vivo research using human cells, particularly immune cells. The increasing recent interest in the genetics and immunology underlying the asymptomatic colonization of human skin and mucous surfaces, which can be seen as proceeding in a biofilm-like fashion, will also be of great value to gain a comprehensive understanding of staphylococcal biofilm physiology in the commensal and infectious states.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, and Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–45. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Stewart PS, and Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–22. [DOI] [PubMed] [Google Scholar]
- 3.Raad I 1998. Intravascular-catheter-related infections. Lancet 351:893–8. [DOI] [PubMed] [Google Scholar]
- 4.Rupp ME 1997. Coagulase-negative staphylococcal infections: an update regarding recognition and management. Curr Clin Top Infect Dis 17:51–87. [PubMed] [Google Scholar]
- 5.Dougherty SH 1988. Pathobiology of infection in prosthetic devices. Rev Infect Dis 10:1102–17. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann N, Rasmussen TB, Jensen PO, Stub C, Hentzer M, Molin S, Ciofu O, Givskov M, Johansen HK, and Hoiby N. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect Immun 73:2504–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resch A, Rosenstein R, Nerz C, and Götz F. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol 71:2663–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y, Sturdevant DE, and Otto M. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis 191:289–98. [DOI] [PubMed] [Google Scholar]
- 9.Donlan RM 2001. Biofilms and device-associated infections. Emerging Infectious Diseases 7:277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metcalf DG, and Bowler PG. 2013. Biofilm delays wound healing: A review of the evidence. Burns Trauma 1:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krismer B, and Peschel A. 2011. Does Staphylococcus aureus nasal colonization involve biofilm formation? Future Microbiol 6:489–93. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez T, Biagini Myers JM, Herr AB, and Khurana Hershey GK. 2017. Staphylococcal Biofilms in Atopic Dermatitis. Curr Allergy Asthma Rep 17:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto M 2014. Physical stress and bacterial colonization. FEMS Microbiol Rev 38:1250–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donlan RM 2001. Biofilms and device-associated infections. Emerg Infect Dis 7:277–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rupp ME 2014. Clinical characteristics of infections in humans due to Staphylococcus epidermidis. Methods Mol Biol 1106:1–16. [DOI] [PubMed] [Google Scholar]
- 16.Delcaru C, Alexandru I, Podgoreanu P, Grosu M, Stavropoulos E, Chifiriuc MC, and Lazar V. 2016. Microbial Biofilms in Urinary Tract Infections and Prostatitis: Etiology, Pathogenicity, and Combating strategies. Pathogens 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otto M 2009. Staphylococcus epidermidis--the ‘accidental’ pathogen. Nat Rev Microbiol 7:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, and Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–62. [DOI] [PubMed] [Google Scholar]
- 19.Datta R, and Huang SS. 2008. Risk of infection and death due to methicillin-resistant Staphylococcus aureus in long-term carriers. Clin Infect Dis 47:176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murdoch DR, Roberts SA, Fowler VG Jr., Shah MA, Taylor SL, Morris AJ, and Corey GR. 2001. Infection of orthopedic prostheses after Staphylococcus aureus bacteremia. Clin Infect Dis 32:647–9. [DOI] [PubMed] [Google Scholar]
- 21.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, and Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–17. [DOI] [PubMed] [Google Scholar]
- 22.Hurlow J, and Bowler PG. 2009. Clinical experience with wound biofilm and management: a case series. Ostomy Wound Manage 55:38–49. [PubMed] [Google Scholar]
- 23.Hansson C, Hoborn J, Moller A, and Swanbeck G. 1995. The microbial flora in venous leg ulcers without clinical signs of infection. Repeated culture using a validated standardised microbiological technique. Acta Derm Venereol 75:24–30. [DOI] [PubMed] [Google Scholar]
- 24.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, and Krogfelt KA. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 3:225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alves PM, Al-Badi E, Withycombe C, Jones PM, Purdy KJ, and Maddocks SE. 2018. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed-biofilm. Pathog Dis doi:10.1093/femspd/fty003. [DOI] [PubMed] [Google Scholar]
- 26.O’Toole G, Kaplan HB, and Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. [DOI] [PubMed] [Google Scholar]
- 27.Heilmann C 2011. Adhesion mechanisms of staphylococci. Adv Exp Med Biol 715:105–23. [DOI] [PubMed] [Google Scholar]
- 28.Bose JL, Lehman MK, Fey PD, and Bayles KW. 2012. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7:e42244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heilmann C, Hussain M, Peters G, and Gotz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24:1013–24. [DOI] [PubMed] [Google Scholar]
- 30.Gross M, Cramton SE, Gotz F, and Peschel A. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun 69:3423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, and Gotz F. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75:864–73. [DOI] [PubMed] [Google Scholar]
- 32.Clarke SR, and Foster SJ. 2006. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol 51:187–224. [DOI] [PubMed] [Google Scholar]
- 33.Mazmanian SK, Liu G, Ton-That H, and Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–3. [DOI] [PubMed] [Google Scholar]
- 34.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, and Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chavakis T, Wiechmann K, Preissner KT, and Herrmann M. 2005. Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thromb Haemost 94:278–85. [DOI] [PubMed] [Google Scholar]
- 36.Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, Pehle P, Kroll G, Schulze C, Buck F, Mack D, Aepfelbacher M, and Rohde H. 2010. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol 75:187–207. [DOI] [PubMed] [Google Scholar]
- 37.Clarke SR, Harris LG, Richards RG, and Foster SJ. 2002. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect Immun 70:6680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, and Laufs R. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol 178:175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, and Otto M. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279:54881–6. [DOI] [PubMed] [Google Scholar]
- 40.Vergara-Irigaray M, Maira-Litran T, Merino N, Pier GB, Penades JR, and Lasa I. 2008. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology 154:865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, and Gotz F. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol 20:1083–91. [DOI] [PubMed] [Google Scholar]
- 42.Gerke C, Kraft A, Süssmuth R, Schweitzer O, and Götz F. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273:18586–93. [DOI] [PubMed] [Google Scholar]
- 43.Conlon KM, Humphreys H, and O’Gara JP. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol 184:4400–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzpatrick F, Humphreys H, and O’Gara JP. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol 43:1973–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kogan G, Sadovskaya I, Chaignon P, Chokr A, and Jabbouri S. 2006. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett 255:11–6. [DOI] [PubMed] [Google Scholar]
- 46.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JK, Ragunath C, Kaplan JB, and Mack D. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28:1711–20. [DOI] [PubMed] [Google Scholar]
- 47.Schommer NN, Christner M, Hentschke M, Ruckdeschel K, Aepfelbacher M, and Rohde H. 2011. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect Immun 79:2267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, and Otto M. 2011. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest 121:238–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loughran AJ, Atwood DN, Anthony AC, Harik NS, Spencer HJ, Beenken KE, and Smeltzer MS. 2014. Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. Microbiologyopen 3:897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, and O’Gara JP. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8:e1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corrigan RM, Rigby D, Handley P, and Foster TJ. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435–46. [DOI] [PubMed] [Google Scholar]
- 52.Banner MA, Cunniffe JG, Macintosh RL, Foster TJ, Rohde H, Mack D, Hoyes E, Derrick J, Upton M, and Handley PS. 2007. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J Bacteriol 189:2793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, and Herr AB. 2008. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A 105:19456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O’Gara JP, Potts JR, and Foster TJ. 2010. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol 192:5663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, and Peters G. 1997. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun 65:519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, and Mack D. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol 55:1883–95. [DOI] [PubMed] [Google Scholar]
- 57.Paharik AE, Kotasinska M, Both A, Hoang TN, Buttner H, Roy P, Fey PD, Horswill AR, and Rohde H. 2017. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol Microbiol 103:860–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Macintosh RL, Brittan JL, Bhattacharya R, Jenkinson HF, Derrick J, Upton M, and Handley PS. 2009. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J Bacteriol 191:7007–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conrady DG, Wilson JJ, and Herr AB. 2013. Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A 110:E202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruszka DT, Mendonca CA, Paci E, Whelan F, Hawkhead J, Potts JR, and Clarke J. 2016. Disorder drives cooperative folding in a multidomain protein. Proc Natl Acad Sci U S A 113:11841–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, and Clarke J. 2015. Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein. Nat Commun 6:7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yarawsky AE, English LR, Whitten ST, and Herr AB. 2017. The Proline/Glycine-Rich Region of the Biofilm Adhesion Protein Aap Forms an Extended Stalk that Resists Compaction. J Mol Biol 429:261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Formosa-Dague C, Speziale P, Foster TJ, Geoghegan JA, and Dufrene YF. 2016. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc Natl Acad Sci U S A 113:410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, and Penades JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol 183:2888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, and Hook M. 2005. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151:1453–64. [DOI] [PubMed] [Google Scholar]
- 66.Taglialegna A, Navarro S, Ventura S, Garnett JA, Matthews S, Penades JR, Lasa I, and Valle J. 2016. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLoS Pathog 12:e1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajagopal M, and Walker S. 2017. Envelope Structures of Gram-Positive Bacteria. Curr Top Microbiol Immunol 404:1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holland LM, Conlon B, and O’Gara JP. 2011. Mutation of tagO reveals an essential role for wall teichoic acids in Staphylococcus epidermidis biofilm development. Microbiology 157:408–18. [DOI] [PubMed] [Google Scholar]
- 69.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, and Peschel A. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med 10:243–5. [DOI] [PubMed] [Google Scholar]
- 70.Whitchurch CB, Tolker-Nielsen T, Ragas PC, and Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 71.Otto M 2013. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64:175–88. [DOI] [PubMed] [Google Scholar]
- 72.Boles BR, and Horswill AR. 2011. Staphylococcal biofilm disassembly. Trends Microbiol 19:449–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheung GY, Joo HS, Chatterjee SS, and Otto M. 2014. Phenol-soluble modulins--critical determinants of staphylococcal virulence. FEMS Microbiol Rev 38:698–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rautenberg M, Joo HS, Otto M, and Peschel A. 2011. Neutrophil responses to staphylococcal pathogens and commensals via the formyl peptide receptor 2 relates to phenol-soluble modulin release and virulence. Faseb j 25:1254–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, and Otto M. 2012. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A 109:1281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwartz K, Syed AK, Stephenson RE, Rickard AH, and Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog 8:e1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng Y, Joo HS, Nair V, Le KY, and Otto M. 2017. Do amyloid structures formed by Staphylococcus aureus phenol-soluble modulins have a biological function? Int J Med Microbiol doi:10.1016/j.ijmm.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shaw L, Golonka E, Potempa J, and Foster SJ. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150:217–28. [DOI] [PubMed] [Google Scholar]
- 79.Boles BR, and Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abraham NM, and Jefferson KK. 2012. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology 158:1504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGavin MJ, Zahradka C, Rice K, and Scott JE. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun 65:2621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dubin G 2002. Extracellular proteases of Staphylococcus spp. Biol Chem 383:1075–86. [DOI] [PubMed] [Google Scholar]
- 83.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, and Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–9. [DOI] [PubMed] [Google Scholar]
- 84.Tang J, Zhou R, Shi X, Kang M, Wang H, and Chen H. 2008. Two thermostable nucleases coexisted in Staphylococcus aureus: evidence from mutagenesis and in vitro expression. FEMS Microbiol Lett 284:176–83. [DOI] [PubMed] [Google Scholar]
- 85.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, and Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beenken KE, Spencer H, Griffin LM, and Smeltzer MS. 2012. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect Immun 80:1634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, and Greenberg EP. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–8. [DOI] [PubMed] [Google Scholar]
- 88.Vuong C, Saenz HL, Gotz F, and Otto M. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis 182:1688–93. [DOI] [PubMed] [Google Scholar]
- 89.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, and Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, and Otto M. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vuong C, Kocianova S, Yao Y, Carmody AB, and Otto M. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis 190:1498–505. [DOI] [PubMed] [Google Scholar]
- 92.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, and Gao Q. 2006. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun 74:488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu D, Zhao L, Xue T, and Sun B. 2012. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol 12:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, Rice KC, Horswill AR, Bayles KW, and Smeltzer MS. 2010. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One 5:e10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tormo MA, Marti M, Valle J, Manna AC, Cheung AL, Lasa I, and Penades JR. 2005. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J Bacteriol 187:2348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beenken KE, Blevins JS, and Smeltzer MS. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun 71:4206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, and Lasa I. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol 48:1075–87. [DOI] [PubMed] [Google Scholar]
- 98.Mrak LN, Zielinska AK, Beenken KE, Mrak IN, Atwood DN, Griffin LM, Lee CY, and Smeltzer MS. 2012. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS One 7:e38453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tamber S, and Cheung AL. 2009. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect Immun 77:419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Li M, Dong D, Bach TH, Sturdevant DE, Vuong C, Otto M, and Gao Q. 2008. SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J Infect Dis 197:1254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kazmierczak MJ, Wiedmann M, and Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev 69:527–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Handke LD, Slater SR, Conlon KM, O’Donnell ST, Olson ME, Bryant KA, Rupp ME, O’Gara JP, and Fey PD. 2007. SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can J Microbiol 53:82–91. [DOI] [PubMed] [Google Scholar]
- 103.Rom JS, Atwood DN, Beenken KE, Meeker DG, Loughran AJ, Spencer HJ, Lantz TL, and Smeltzer MS. 2017. Impact of Staphylococcus aureus regulatory mutations that modulate biofilm formation in the USA300 strain LAC on virulence in a murine bacteremia model. Virulence 8:1776–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Atwood DN, Loughran AJ, Courtney AP, Anthony AC, Meeker DG, Spencer HJ, Gupta RK, Lee CY, Beenken KE, and Smeltzer MS. 2015. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen 4:436–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kies S, Otto M, Vuong C, and Gotz F. 2001. Identification of the sigB operon in Staphylococcus epidermidis: construction and characterization of a sigB deletion mutant. Infect Immun 69:7933–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rupp ME, Ulphani JS, Fey PD, Bartscht K, and Mack D. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun 67:2627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rupp ME, Ulphani JS, Fey PD, and Mack D. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun 67:2656–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rupp ME, Fey PD, Heilmann C, and Gotz F. 2001. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis 183:1038–42. [DOI] [PubMed] [Google Scholar]
- 109.McCarthy H, Waters EM, Bose JL, Foster S, Bayles KW, O’Neill E, Fey PD, and O’Gara JP. 2016. The major autolysin is redundant for Staphylococcus aureus USA300 LAC JE2 virulence in a murine device-related infection model. FEMS Microbiol Lett 363. [DOI] [PubMed] [Google Scholar]
- 110.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penades JR, and Lasa I. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol 191:832–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vernachio J, Bayer AS, Le T, Chai YL, Prater B, Schneider A, Ames B, Syribeys P, Robbins J, and Patti JM. 2003. Anti-clumping factor A immunoglobulin reduces the duration of methicillin-resistant Staphylococcus aureus bacteremia in an experimental model of infective endocarditis. Antimicrob Agents Chemother 47:3400–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McCrea KW, Hartford O, Davis S, Eidhin DN, Lina G, Speziale P, Foster TJ, and Hook M. 2000. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146 (Pt 7):1535–46. [DOI] [PubMed] [Google Scholar]
- 113.Arrecubieta C, Toba FA, von Bayern M, Akashi H, Deng MC, Naka Y, and Lowy FD. 2009. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS Pathog 5:e1000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dastgheyb SS, Villaruz AE, Le KY, Tan VY, Duong AC, Chatterjee SS, Cheung GY, Joo HS, Hickok NJ, and Otto M. 2015. Role of Phenol-Soluble Modulins in Formation of Staphylococcus aureus Biofilms in Synovial Fluid. Infect Immun 83:2966–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dastgheyb SS, Hammoud S, Ketonis C, Liu AY, Fitzgerald K, Parvizi J, Purtill J, Ciccotti M, Shapiro IM, Otto M, and Hickok NJ. 2015. Staphylococcal persistence due to biofilm formation in synovial fluid containing prophylactic cefazolin. Antimicrob Agents Chemother 59:2122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dastgheyb S, Parvizi J, Shapiro IM, Hickok NJ, and Otto M. 2015. Effect of biofilms on recalcitrance of staphylococcal joint infection to antibiotic treatment. J Infect Dis 211:641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peschel A, and Otto M. 2013. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol 11:667–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, and Smeltzer MS. 2012. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol 86:1183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rigby KM, and DeLeo FR. 2012. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol 34:237–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wiesner J, and Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–64. [DOI] [PubMed] [Google Scholar]
- 121.Heinzelmann M, Herzig DO, Swain B, Mercer-Jones MA, Bergamini TM, and Polk HC Jr. 1997. Phagocytosis and oxidative-burst response of planktonic Staphylococcus epidermidis RP62A and its non-slime-producing variant in human neutrophils. Clin Diagn Lab Immunol 4:705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Donlan RM, and Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singh R, Ray P, Das A, and Sharma M. 2010. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother 65:1955–8. [DOI] [PubMed] [Google Scholar]
- 124.Mah TF, and O’Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–9. [DOI] [PubMed] [Google Scholar]
- 125.Trampuz A, and Zimmerli W. 2006. Antimicrobial agents in orthopaedic surgery: Prophylaxis and treatment. Drugs 66:1089–105. [DOI] [PubMed] [Google Scholar]
- 126.Croes S, Beisser PS, Neef C, Bruggeman CA, and Stobberingh EE. 2010. Unpredictable effects of rifampin as an adjunctive agent in elimination of rifampin-susceptible and -resistant Staphylococcus aureus strains grown in biofilms. Antimicrob Agents Chemother 54:3907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meeker DG, Beenken KE, Mills WB, Loughran AJ, Spencer HJ, Lynn WB, and Smeltzer MS. 2016. Evaluation of Antibiotics Active against Methicillin-Resistant Staphylococcus aureus Based on Activity in an Established Biofilm. Antimicrob Agents Chemother 60:5688–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hogan D, and Kolter R. 2002. Why are bacteria refractory to antimicrobials? Curr Opin Microbiol 5:472–7. [DOI] [PubMed] [Google Scholar]
- 129.Conlon BP, Rowe SE, and Lewis K. 2015. Persister cells in biofilm associated infections. Adv Exp Med Biol 831:1–9. [DOI] [PubMed] [Google Scholar]
- 130.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, and Novick RP. 2008. agr function in clinical Staphylococcus aureus isolates. Microbiology 154:2265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, and Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol 186:6585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heim CE, Vidlak D, Scherr TD, Kozel JA, Holzapfel M, Muirhead DE, and Kielian T. 2014. Myeloid-derived suppressor cells contribute to Staphylococcus aureus orthopedic biofilm infection. J Immunol 192:3778–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heim CE, Vidlak D, Odvody J, Hartman CW, Garvin KL, and Kielian T. 2017. Human prosthetic joint infections are associated with myeloid-derived suppressor cells (MDSCs): Implications for infection persistence. J Orthop Res doi:10.1002/jor.23806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gries CM, Bruger EL, Moormeier DE, Scherr TD, Waters CM, and Kielian T. 2016. Cyclic di-AMP Released from Staphylococcus aureus Biofilm Induces a Macrophage Type I Interferon Response. Infect Immun 84:3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Prabhakara R, Harro JM, Leid JG, Harris M, and Shirtliff ME. 2011. Murine immune response to a chronic Staphylococcus aureus biofilm infection. Infect Immun 79:1789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kropec A, Maira-Litran T, Jefferson KK, Grout M, Cramton SE, Gotz F, Goldmann DA, and Pier GB. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun 73:6868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Valle J, Latasa C, Gil C, Toledo-Arana A, Solano C, Penades JR, and Lasa I. 2012. Bap, a biofilm matrix protein of Staphylococcus aureus prevents cellular internalization through binding to GP96 host receptor. PLoS Pathog 8:e1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dziewanowska K, Patti JM, Deobald CF, Bayles KW, Trumble WR, and Bohach GA. 1999. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect Immun 67:4673–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Proctor RA, Kriegeskorte A, Kahl BC, Becker K, Loffler B, and Peters G. 2014. Staphylococcus aureus Small Colony Variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front Cell Infect Microbiol 4:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pletzer D, Coleman SR, and Hancock RE. 2016. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr Opin Microbiol 33:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peschel A, and Sahl HG. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol 4:529–36. [DOI] [PubMed] [Google Scholar]
- 142.Joo HS, and Otto M. 2015. Mechanisms of resistance to antimicrobial peptides in staphylococci. Biochim Biophys Acta 1848:3055–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Romano CL, Scarponi S, Gallazzi E, Romano D, and Drago L. 2015. Antibacterial coating of implants in orthopaedics and trauma: a classification proposal in an evolving panorama. J Orthop Surg Res 10:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schaffer AC, and Lee JC. 2009. Staphylococcal vaccines and immunotherapies. Infect Dis Clin North Am 23:153–71. [DOI] [PubMed] [Google Scholar]
- 145.Harro JM, Peters BM, O’May GA, Archer N, Kerns P, Prabhakara R, and Shirtliff ME. 2010. Vaccine development in Staphylococcus aureus: taking the biofilm phenotype into consideration. FEMS Immunol Med Microbiol 59:306–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Soe NH, Jensen NV, Jensen AL, Koch J, Poulsen SS, Pier GB, and Johansen HK. 2017. Active and Passive Immunization Against Staphylococcus aureus Periprosthetic Osteomyelitis in Rats. In Vivo 31:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shahrooei M, Hira V, Khodaparast L, Stijlemans B, Kucharíková S, Burghout P, Hermans PW, and Van Eldere J. 2012. Vaccination with SesC decreases Staphylococcus epidermidis biofilm formation. Infect Immun 80:3660–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wittebole X, De Roock S, and Opal SM. 2014. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gutierrez D, Vandenheuvel D, Martinez B, Rodriguez A, Lavigne R, and Garcia P. 2015. Two Phages, phiIPLA-RODI and phiIPLA-C1C, Lyse Mono- and Dual-Species Staphylococcal Biofilms. Appl Environ Microbiol 81:3336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lungren MP, Christensen D, Kankotia R, Falk I, Paxton BE, and Kim CY. 2013. Bacteriophage K for reduction of Staphylococcus aureusbiofilm on central venous catheter material. Bacteriophage 3:e26825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cerca N, Oliveira R, and Azeredo J. 2007. Susceptibility of Staphylococcus epidermidis planktonic cells and biofilms to the lytic action of Staphylococcus bacteriophage K. Lett Appl Microbiol 45:313–7. [DOI] [PubMed] [Google Scholar]
- 152.Fischetti VA 2017. Lysin Therapy for Staphylococcus aureus and Other Bacterial Pathogens. Curr Top Microbiol Immunol 409:529–540. [DOI] [PubMed] [Google Scholar]
- 153.Schuch R, Khan BK, Raz A, Rotolo JA, and Wittekind M. 2017. Bacteriophage Lysin CF-301, a Potent Antistaphylococcal Biofilm Agent. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kokai-Kun JF, Chanturiya T, and Mond JJ. 2007. Lysostaphin as a treatment for systemic Staphylococcus aureus infection in a mouse model. J Antimicrob Chemother 60:1051–9. [DOI] [PubMed] [Google Scholar]
- 155.Hogan S, Zapotoczna M, Stevens NT, Humphreys H, O’Gara JP, and O’Neill E. 2017. Potential use of targeted enzymatic agents in the treatment of Staphylococcus aureus biofilm-related infections. J Hosp Infect 96:177–182. [DOI] [PubMed] [Google Scholar]
- 156.Aguinaga A, Frances ML, Del Pozo JL, Alonso M, Serrera A, Lasa I, and Leiva J. 2011. Lysostaphin and clarithromycin: a promising combination for the eradication of Staphylococcus aureus biofilms. Int J Antimicrob Agents 37:585–7. [DOI] [PubMed] [Google Scholar]
- 157.Wu JA, Kusuma C, Mond JJ, and Kokai-Kun JF. 2003. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob Agents Chemother 47:3407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kokai-Kun JF, Chanturiya T, and Mond JJ. 2009. Lysostaphin eradicates established Staphylococcus aureus biofilms in jugular vein catheterized mice. J Antimicrob Chemother 64:94–100. [DOI] [PubMed] [Google Scholar]
- 159.Donegan EA, and Riggs HG Jr. 1974. In vitro incorporation of serine into the staphylococcal cell wall. Infect Immun 10:264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ramasubbu N, Thomas LM, Ragunath C, and Kaplan JB. 2005. Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J Mol Biol 349:475–86. [DOI] [PubMed] [Google Scholar]
- 161.Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, and Jabbouri S. 2007. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl Microbiol Biotechnol 75:125–32. [DOI] [PubMed] [Google Scholar]
- 162.Izano EA, Amarante MA, Kher WB, and Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol 74:470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dickey SW, Cheung GYC, and Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. [DOI] [PubMed] [Google Scholar]