Abstract
Tropomyosin isoforms contribute to generation of functionally divergent actin filaments. In the nematode Caenorhabditis elegans, multiple isoforms are produced from lev-11, the single tropomyosin gene, by combination of two separate promoters and alternative pre-mRNA splicing. In this study, we report that alternative splicing of lev-11 is regulated in a tissue-specific manner so that a particular tropomyosin isoform is expressed in each tissue. Reverse-transcription polymerase chain reaction analysis of lev-11 mRNAs confirms five previously reported isoforms (LEV-11A, LEV-11C, LEV-11D, LEV-11E, and LEV-11O) and identifies a new sixth isoform LEV-11T. Using transgenic alternative-splicing reporter minigenes, we find distinct patterns of preferential exon selections in the pharynx, body wall muscles, intestine, and neurons. The body wall muscles preferentially process splicing to produce high-molecular-weight isoforms, LEV-11A, LEV-11D, and LEV-11O. The pharynx specifically processes splicing to express a low-molecular-weight isoform LEV-11E, whereas the intestine and neurons process splicing to express another low-molecular-weight isoform LEV-11C. The splicing pattern of LEV-11T was not predominant in any of these tissues, suggesting that this is a minor isoform. Our results suggest that regulation of alternative splicing is an important mechanism to express proper tropomyosin isoforms in particular tissue and/or cell types in C. elegans.
Keywords: Actin, tropomyosin, isoforms, mRNA, alternative splicing
Introduction
Tropomyosin is a major actin-binding protein in fungi and metazoans, which binds along the side of actin filaments (Bailey 1946; Laki et al. 1962; Maruyama 1964) and is involved in numerous biological events (Gunning et al. 2008; Gunning et al. 2015; Holmes and Lehman 2008; Perry 2001). Tropomyosin directly alters the biophysical properties of actin filaments: tropomyosin slows down actin filament dynamics by reducing the rates of polymerization and depolymerization (Broschat 1990; Broschat et al. 1989; Hitchcock-DeGregori et al. 1988; Lal and Korn 1986) and increases the rigidity of actin filaments (Goldmann 2000; Greenberg et al. 2008). In addition, tropomyosin indirectly modulates the actin functions by recruiting or repelling other cytoskeletal proteins (Kuhn and Bamburg 2008; Wang and Coluccio 2010). Furthermore, multicellular organisms express a large number of tropomyosin isoforms to generate diverse cytoskeletal environments with different properties (Choi et al. 2012; Geeves et al. 2015; Gunning et al. 2015; Gunning et al. 2005; Lees-Miller and Helfman 1991; Lin et al. 1997; Pittenger et al. 1994; Schevzov et al. 2011). Changes in the expression patterns of tropomyosin isoforms are associated with pathological conditions such as heart disease (Karam et al. 2011; Rajan et al. 2010) and cancer cell transformation (Hendricks and Weintraub 1984; Lin et al. 1985; Matsumura et al. 1983; Pawlak et al. 2004). However, the regulatory mechanisms of tropomyosin isoform expression by alternative splicing have been characterized only in limited cases (Gooding and Smith 2008). Mammals have four tropomyosin genes producing more than 40 isoforms in total by alternative splicing (Geeves et al. 2015; Schevzov et al. 2011), yet further studies on the expression patterns of the tropomyosin isoforms will be necessary for understanding their isoform-specific functions.
The nematode Caenorhabditis elegans has a single tropomyosin gene lev-11 (also known as tmy-1), which gives rise to multiple isoforms (Kagawa et al. 1995). The lev-11 gene has two promoters and multiple alternatively spliced exons (Fig. 1). Five isoforms have been confirmed to date by cloning of full-length cDNAs, which include three high-molecular-weight isoforms (LEV-11A, LEV-11D, and LEV-11O) and two low-molecular-weight isoforms (LEV-11C and LEV-11E) (Anyanful et al. 2001; Barnes et al. 2018; Kagawa et al. 1995) (Fig. 1). Severe lev-11 mutants are paralyzed and arrested at an embryonic stage (Williams and Waterston 1994). Some of lev-11 mutants are resistant to levamisole, an agonist for acetylcholine receptor that induces muscle contraction (Lewis et al. 1980a; Lewis et al. 1980b), because slow muscle relaxation occurs while cholinergic neurons are active (Hwang et al. 2016). Splicing of two mutually exclusive exon 7s is differentially regulated in the head and body regions of the body wall muscle cells to express LEV-11O in the head muscles and LEV-11A in the rest of the body wall muscles, where these isoforms differentially regulate muscle contractility (Barnes et al. 2018). In addition, lev-11 is important for sarcomere organization (Ono and Ono 2002; Yu and Ono 2006), muscle arm development (Dixon and Roy 2005), ovulatory myoepithelial sheath contraction (Ono and Ono 2004), and contractility of male sex muscle (Gruninger et al. 2006). However, isoform-specific roles of C. elegans tropomyosins are largely unknown in these processes.
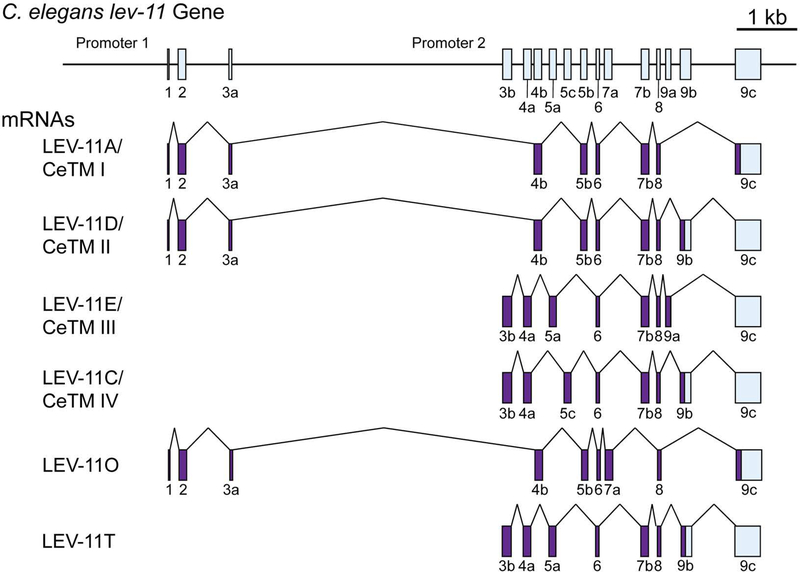
Figure 1. Structure of the C. elegans lev-11 gene.
The lev-11 gene structure is schematically shown on top with numbered boxes indicating exons. Below, structures of spliced mRNAs of five previously characterized isoforms (LEV-11A/CeTMI, LEV-11D/CeTMII, LEV-11E/CeTMIII, LEV-11C/CeTMIV, and LEV-11O) and a newly identified isoform (LEV-11T) are shown. Coding and non-coding regions are shown in purple and light blue, respectively.
Profiling of their expression patterns is necessary to understand functional significance of tropomyosin isoforms in C. elegans. Previous studies have demonstrated tissue-specific activities of the two lev-11 promoters: the upstream promoter (promoter 1) drives transcription from exon 1 (E1) to express high-molecular-weight isoforms in the body wall muscle, vulva, and anal muscle, whereas the downstream promoter (promoter 2) drives from exon 3b (E3b) to express low-molecular-weight isoforms in the pharynx and intestine (Anokye-Danso et al. 2008; Anyanful et al. 2001; Kagawa et al. 1995). However, a pattern of alternative splicing has been characterized only for alternative exons 7a (E7a) and 7b (E7b) (Barnes et al. 2018). In this study, we determined in vivo selection patterns of alternative exons 4 (E4a and E4b), 5 (E5a, E5c, and E5b), and 9 (E9a, and E9b) in major C. elegans tissues using reverse transcription-PCR and transgenic alternative splicing reporters (Kuroyanagi et al. 2006; Kuroyanagi et al. 2010). We further identified a new sixth tropomyosin isoform LEV-11T. The results indicate that C. elegans has a mechanism to express different tropomyosin isoforms by tissue-specific alternative splicing, suggesting that evolution of tropomyosin isoforms is functionally relevant to tissue-specific functions of the actin cytoskeleton even in this relatively simple multicellular organism.
Results
Reverse transcription-PCR analysis of lev-11 reveals specific exon combinations and identifies a novel tropomyosin isoform
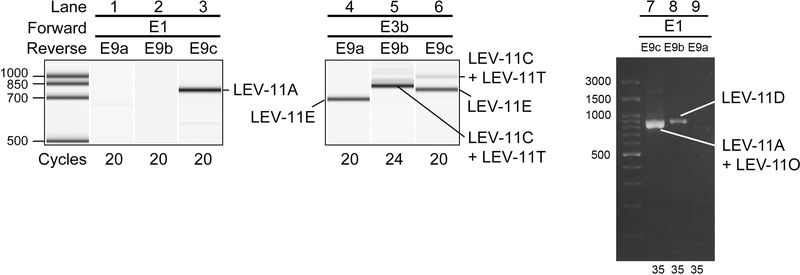
To analyze the processing patterns of the LEV-11 mRNAs, we performed reverse transcription (RT)-polymerase chain reaction (PCR) analyses using the L1-stage cDNAs with a forward primer on either E1 (the first exon for high-molecular-weight isoforms) or E3b (the first exon for low-molecular-weight isoforms) and a reverse primer on E9c, the most downstream exon that is included in all known transcripts either as coding or non-coding region (Fig. 1), to amplify full-length coding regions of cDNAs (Fig. 2). With the E1/E9c primer pair, we detected a single band (Fig. 2, lane 3), which contained LEV-11A/CeTMI (Fig. 1A) as the only detected isoform by direct sequencing. As the promoter upstream of E1 is most active in the body wall muscles, anal muscles and sex muscles (Kagawa et al. 1995), the representation of LEV-11A suggests that E4b, E5b, E7b, and E9c are mostly selected in these muscles. LEV-11O, which also contains E1 and E9c (Fig. 1), was not detected in this assay but was detected when it was specifically enriched and amplified (our unpublished observations), consistently with our recent observations that LEV-11O is a very minor isoform that is expressed in a subset of head muscle cells (Barnes et al. 2018). With the E3b/E9c primer pair, we detected a major lower band and a minor upper band (Fig. 2, lane 6). Cloning and sequencing of the RT-PCR products revealed that the major lower band was LEV-11E/CeTMIII, which includes E4a, E5a, E7b, and E9a, whereas the minor upper band was a mixture of LEV-11C/CeTMIV, which includes E4a, E5c, E7b, and E9b, (Fig. 1A) and a novel isoform LEV-11T that included E4a, E5a, E7b, and E9b (DDBJ accession number LC215398) (Fig. 1A). No cDNA containing E9c as a part of a coding region was detected when the E3b primer was used.
Figure 2. Analysis of lev-11 mRNAs by RT-PCR.
(Lanes 1–6) Total RNAs from synchronized wild-type L1 larvae were subjected to RT-PCR with indicated primer pairs (shown on top) and cycle numbers (shown on bottom), and the PCR products were analyzed with an on-chip capillary electrophoresis system, 2100 BioAnalyzer (Agilent). Results are shown in gel-like presentations. (Lanes 7–9) Total RNAs from mixed-stage wild-type worms were subjected to RT-PCR with indicated primer pairs (shown on top) and cycle numbers (shown on bottom), and the PCR products were analyzed with conventional agarose gel electrophoresis. Major cDNA species were cloned and sequenced, and identified isoforms are indicated. DNA size markers are shown on the left of lanes 1 and 7. The presented data are representative results from at least three separate experiments.
To further analyze the processing patterns of the LEV-11 mRNAs containing E9a and E9b, we utilized reverse primers on E9a and E9b. With the E1/E9a or E1/E9b primer pair, no detectable band was amplified from the L1-stage cDNAs (Fig. 2, lanes 1 and 2). However, when RT-PCR was performed using cDNAs from mixed-stage worms, a single band was amplified from the E1/E9b pair, which encoded LEV-11D/CeTMII (Fig. 2, lane 8), suggesting that LEV-11D is a developmentally regulated minor isoform. With the E3b/E9a primer pair, we detected a single band that included E4a, E5a and E7b (Fig. 2, lane 4), indicating that LEV-11E/CeTMIII is the major isoform in tissues where E9a is selected. With the E3b/E9b primer pair, a single band was detected (Fig. 2, lane 5) and it was a mixture of LEV-11C/CeTMIV and LEV-11T (Fig. 1A). Both the E3b/E9a and E3b/E9b RT-PCR products were consistent with the three E3b/E9c RT-PCR products including E9c as an untranslated region (Fig. 2, lane 6). Indistinguishable results were obtained with mixed-stage cDNAs for E3b/E9a, E3b/E9b, and E3b/E9c primer pairs (our unpublished observations).
In summary, our RT-PCR analyses demonstrated that: (1) the alternative exons of the lev-11 gene are selected in a limited number of combinations, and (2) a new splicing pattern produces a novel low-molecular-weight tropomyosin isoform LEV-11T.
Alternative exons of the lev-11 gene are selected mostly in a tissue-type-specific manner
To examine in vivo selection patterns of the lev-11 alternative exons, we utilized fluorescence reporter systems (Kuroyanagi et al. 2006). A choice of E1/E2/E3a or E3b was not examined here because these are determined by two alternative promoters (Kagawa et al. 1995). To examine selection patterns of E4a/E4b and E5a/E5c/E5b in vivo, we constructed a pair of fluorescence reporter minigenes (Fig. 3A and B). In the enhanced green fluorescent protein (EGFP) minigene (E3b-E6-EGFP), a lev-11 genomic fragment spanning from E3b through E6 was connected in-frame with the EGFP cDNA so that the cells were forced to use E3b (Fig. 3A). In the monomeric red fluorescent protein (mRFP) minigene (E4b-E6-mRFP), a lev-11 genomic fragment spanning from E4b through E6 was connected in-frame to mRFP1 cDNA so that the cells were forced to use E4b (Fig. 3B). As each tissue can select any E4 (in the EGFP minigene) and any E5 (in both of the minigenes) without constraint, we anticipated that both EGFP and mRFP would be uniformly detected in all the tissues when the minigenes were ubiquitously expressed by the eef-1A.1 promoter. However, expression of the fluorescent proteins in the transgenic reporter worms exhibited striking tissue specificity: the pharyngeal muscle preferentially expressed EGFP (Fig. S1A), whereas the body wall muscle predominantly expressed mRFP (Fig. S1B). Although these and other reporter constructs contain small portions of the LEV-11 proteins fused with the fluorescent proteins, we did not observe any dominant phenotypes due to expression of reporters in terms of morphology of tissues and cells and locomotive activity of the transgenic worms.
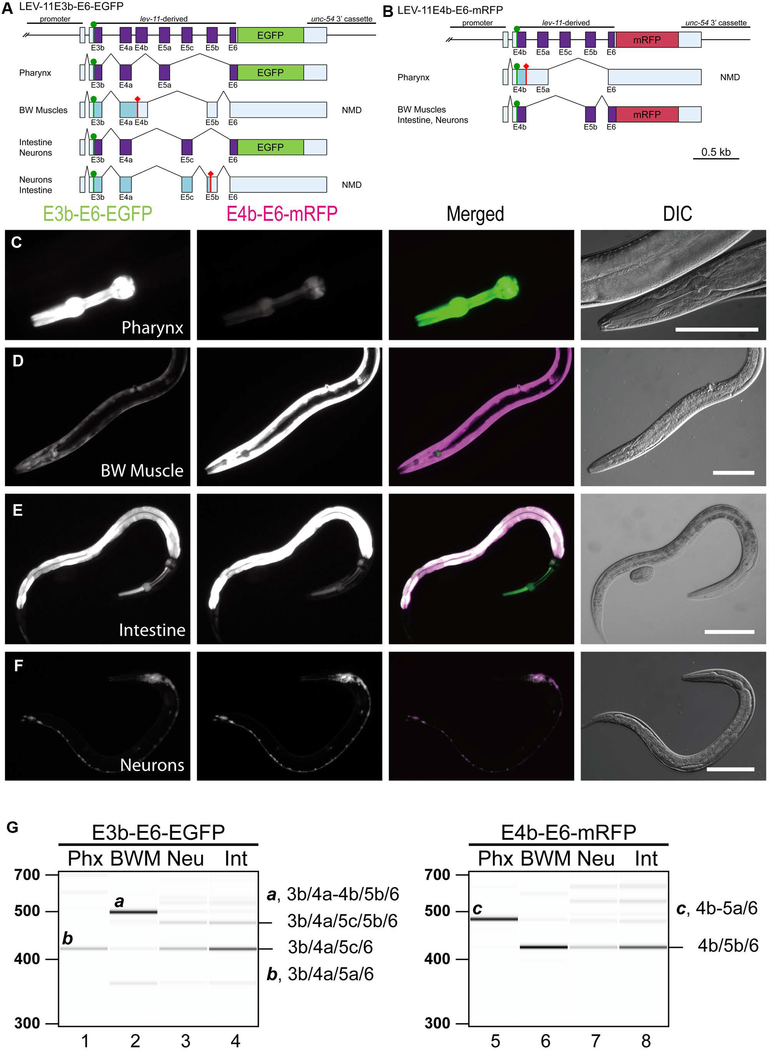
Figure 3. Fluorescence splicing reporter analysis for lev-11 E4s and E5s.
(A, B) Schematic illustration of lev-11 reporter minigenes, E3b-E6-EGFP (A) and E4b-E6-mRFP (B). Structures of major mRNA isoforms expressed in the pharynx, body wall muscles (BW Muscles), intestine, and neurons are indicated below each minigene. The cDNA cassettes and the predicted open reading frames (ORFs) for EGFP and mRFP are colored in green and magenta, respectively. Frames shown in cyan are not in-frame with the fluorescent proteins and such mRNAs are likely degraded by nonsense-mediated mRNA decay (NMD). Green circles indicate artificially-introduced initiation codons. (C-F) Fluorescence images showing expression of E3b-E6-EGFP and E4b-E6-mRFP in L4 larvae or adults under the control of the myo-2 (pharynx) (C), myo-3 (body wall muscle) (D), gst-42 (intestine) (E), or rgef-1 (neurons) (F) promoter. Micrographs were taken with the same exposure settings for the four strains. Merged images of EGFP in green and mRFP in magenta and DIC images are also shown. Bars, 100 μm. (G) RT-PCR analysis of polyadenylated mRNAs from E3b-E6-EGFP (lanes 1–4) and E4b-E6-mRFP (lanes 5–8). Major cDNA species were cloned, sequenced, and identified exon combinations are indicated (also shown schematically in A and B). These reporters were analyzed in NMD-deficient smg-2 mutant background. DNA size markers are shown on the left of lanes 1 and 5. The presented results were consistent in two independent transgenic strains.
To obtain further insight into the tissue-specific splicing patterns, we generated transgenic reporter worms expressing these reporter minigenes in four major tissues. Pharynx, body wall muscles and intestine are the tissues in which activity of the lev-11 promoters have been detected (Anyanful et al. 2001; Kagawa et al. 1995). Neurons were also analyzed because they constitute >30% of the somatic cell population at the adult stage. We then analyzed fluorescent protein expression by fluorescence microscopy (Fig. 3C-F) and major mRNA species by RT-PCR (Fig. 3G). From the E3b-E6-EGFP reporter, EGFP was strongly expressed in the pharynx (Fig. 3C), and intestine (Fig. 3E), and moderately in neurons (Fig. 3F). Sequencing of the RT-PCR products revealed that, in the pharynx, a single band containing E3b/E4a/E5a/E6 (LEV-11E/T type) was predominantly produced (Fig. 3G, lane 1), whereas, in the intestine and neurons, two bands were produced (Fig. 3G, lanes 3 and 4). The lower bands in the intestine and neurons contained E3b/E4a/E5c/E6 isoform (LEV-11C type) (Fig. 3A and G, lanes 3 and 4), but the upper bands contained E3b/E4a/E5c/E5b/E6, which includes a premature stop codon to prevent translation of EGFP (Fig. 3A and G). The body wall muscles, however, processed the mRNAs in an unexpected manner; E4b and E5b are specifically selected as expected from the LEV-11A/O-type splicing, yet E4b is fused with E4a with a short intron between them retained in the mature mRNA (E3b/E4a-E4b/E5b/E6 isoform) (Fig. 3A and G, lane 2). This mRNA also includes a premature stop codon to prevent translation of EGFP (Fig. 3A). Nonetheless, weak expression of EGFP was detected in the body wall muscles (Fig. 3D), and an exon combination for this mRNA remains unknown. The intron between E4a and E4b could not be excised probably because it is too short (36 nt) for the spliceosome (Lander et al. 2001). From the E4b-E6-mRFP reporter, mRFP was strongly expressed in the body wall muscle (Fig. 3D), intestine (Fig. 3E), and neurons (Fig. 3F). RT-PCR analysis showed that these tissues primarily expressed E4b/E5b/E6 isoform (LEV-11A/O type) (Fig. 3B and G, lanes 6–8). The pharynx predominantly selected E5a, yet the intron between E4b and E5a was retained in the mature mRNAs (E4b-E5a/E6 isoform) (Fig. 3B and G, lane 5) although the intron (123 nt) appears to be long enough for the spliceosome. These results suggested that (1) each E5 is selected in mutually exclusive and tissue-specific manners, (2) E4b can be properly recognized only in the body wall muscles, and (3) intron 3b (between E3b and E4a) can be excised in any tissues if exon 3b is forced to be included.
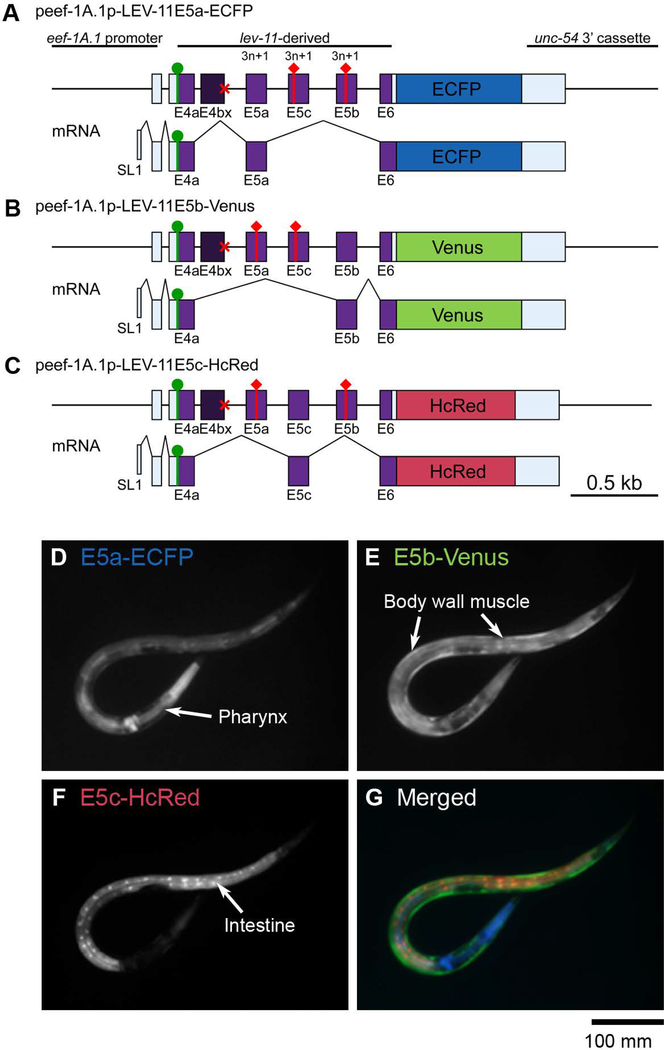
To further determine the selection patterns of E5s in vivo, we constructed a trio of reporter minigenes in which a lev-11 genomic fragment spanning from E4a through E6 is connected in-frame to one of three fluorescent protein cDNAs (Fig. 4A-C). To focus on the mutually exclusive selection from three E5s, we disrupted the 5’-splice site of E4b (Fig. 4A-C, red crosses) so that all the tissues are forced to use the 5’-splice site of E4a. Artificial termination codons were then introduced into two of the three E5s in each of the minigenes (Fig. 4A-C, red diamonds) so that each fluorescent protein is expressed upon selection of a specific E5 (Kuroyanagi et al. 2013a). When these minigenes were expressed under the control of the common, ubiquitous eef-1A.1 promoter, we detected preferential expression of one fluorescent protein in each of the three major tissues: the pharynx predominantly expressed E5a-enhanced cyan fluorescent protein (ECFP) (Fig. 4D), the body wall muscles expressed E5b-Venus (Fig. 4E), and the intestine predominantly expressed E5c-HcRed (Fig. 4F). None of the reporters were detected in the neurons for an unknown reason. As these expression patterns were consistent with the RT-PCR analyses (Fig. 3G), we concluded that E5a, E5c and E5b are preferentially selected in the pharynx, the intestine, and the body wall muscles, respectively.
Figure 4. Fluorescence splicing reporter analysis for lev-11 E5s.
(A-C) Schematic illustration of reporter minigenes for lev-11 E5a (E5a-ECFP) (A), E5b (E5b-Venus) (B), and E5c (E5c-HcRed) (C). These reporters were expressed under the control of the ubiquitous eef-1A.1 promoter. Structures of mRNA isoforms that encode the fluorescent proteins are indicated below each minigene. The cDNA cassettes and the predicted ORFs for ECFP, Venus and HcRed are colored in cyan, green and magenta, respectively. Green circles and red diamonds indicate artificially-introduced initiation and termination codons, respectively. Red crosses indicate disruption of the 5’-splice sites for E4b. (D-G) Fluorescence images showing expression of E5a-ECFP (D), E5b-Venus (E) and E5c-HcRed (F). A merged image with ECFP in blue, Venus in green, and HcRed in red, is shown in G. Bar, 100 μm.
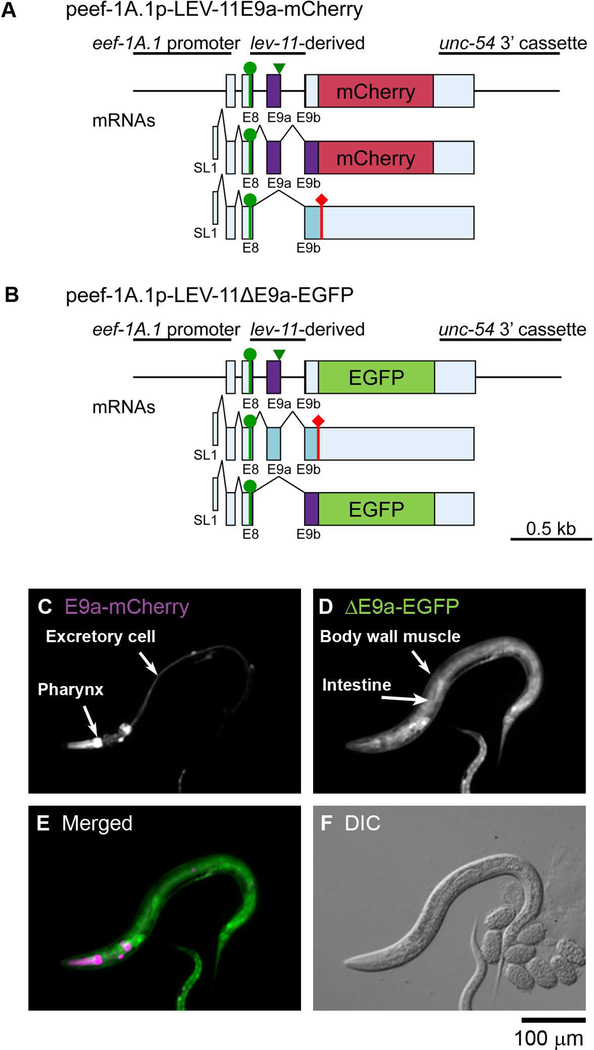
As determined by the RT-PCR analysis (Fig. 2), alternative splicing of E9s is somewhat complex. Although each of E9a, E9b, or E9c is selected as a translated region, E9c is also used as an untranslated region when E9a or E9b is selected (Fig. 1). Thus, E9a and E9b are classified as tandem cassette exons, which are either included or skipped. We examined patterns of inclusion or exclusion of E9a in vivo using a pair of reporter minigenes in which a lev-11 genomic fragment spanning from E8 through E9b is connected in-frame to one of two fluorescent protein cDNAs (Fig. 5A and B) to detect inclusion of E9a (E9a-mCherry) or exclusion of E9a (ΔE9a-EGFP). When these minigenes were ubiquitously expressed, only the pharynx and the excretory cells expressed E9a-mCherry (Fig. 5C), whereas the other tissues expressed ΔE9a-EGFP (Fig. 5D). Since LEV-11E is the only known isoform containing E9a (Fig. 1), these results suggest that this isoform is expressed specifically in these limited tissues.
Figure 5. Fluorescence splicing reporter analysis for lev-11 E9s.
(A, B) Schematic illustration of reporter minigenes for inclusion (E9a-mCherry) (A) and exclusion (ΔE9a-EGFP) (B) of lev-11 E9a. These reporters were expressed under the control of the ubiquitous eef-1A.1 promoter. Structures of mRNA isoforms are indicated below the minigenes. The cDNA cassettes and the predicted ORFs for mCherry and EGFP are colored in magenta and green, respectively. Green circles indicate the artificially-introduced initiation codons. Green arrowheads indicate nucleotide substitutions for eliminating a termination codon. (C-F) Fluorescence images showing expression of E9a-mCherry (C) and ΔE9a-EGFP (D). A merged image with mCherry in magenta and EGFP in green is shown in (E). A DIC image is shown in (F). Bar, 100 μm.
Tropomyosin isoforms are expressed in a tissue-specific manner by alternative splicing
Based on the splicing patterns, tissue-specific expression of LEV-11 isoforms are summarized in Table 1. Most of the body wall muscle cells express the high-molecular-weight isoforms LEV-11A or LEV-11D, which includes E4b (Fig. 3A), E5b (Fig. 4E), and E7b (Barnes et al. 2018) but excludes E9a (Fig. 5D). A subset of head muscle cells predominantly express LEV-11O that includes E7a (Barnes et al. 2018). The pharynx expresses LEV-11E that includes E4a (Fig. 3A), E5a (Fig. 4D), E7b (Barnes et al. 2018), and E9a (Fig. 5C). The intestine and neurons express LEV-11C, which includes E5c (Fig. 4F) and E7b (Barnes et al. 2018), and excludes E9a (Fig. 5D). The excretory cells are likely to express LEV-11E, since this is the only known isoform containing E9a (Fig. 5C). Expression pattern of LEV-11T remains unclear because selection of E5a and E9b is to be simultaneously elucidated. Additional analysis using the reporters under the control of tissue-specific promoters will be required to determine lev-11 splicing patterns in the excretory cell as well as in other minor tissues.
Table 1.
Summary of LEV-11 tropomyosin isoforms
| Isoform | Other name | Exons | Length (amino acids) | Expression |
|---|---|---|---|---|
| LEV-11A | CeTMI | E1/E2/E3a/E4b/E5b/E6/E7b/E8/E9c | 284 | Body wall muscle (main body) |
| LEV-11D | CeTMII | E1/E2/E3a/E4b/E5b/E6/E7b/E8/E9b/(E9c)1 | 284 | Body wall muscle (main body) |
| LEV-11E | CeTMIII | E3b/E4a/E5a/E6/E7b/E8/E9a/(E9c) | 256 | Pharynx, excretory cells |
| LEV-11C | CeTMIV | E3b/E4a/E5c/E6/E7b/E8/E9b/(E9c) | 256 | Intestine, neurons |
| LEV-11O | - | E1/E2/E3a/E4b/E5b/E6/E7a/E8/E9c | 284 | Body wall muscle (head) |
| LEV-11T | - | E3b/E4a/E5a/E6/E7b/E8/E9b/(E9c) | 256 | Unknown |
E9c in parentheses only contains a 3’-untranslated region.
Discussion
In the present and previous studies, we demonstrated that selection patterns of alternative exons of the lev-11 gene are regulated specifically in certain tissues and cell types by utilizing fluorescence splicing reporters (Barnes et al. 2018). It should be noted that there are technical limitations to detect low-level expression of the splicing reporters by fluorescence microscopy. It is also likely that minor fluorescent signals are false-positives due to artificial structure or intense promoter activity of the reporter minigene. Therefore, this technique is highly effective in characterizing major splicing patterns, but occurrence of minor splicing events remains ambiguous. Nevertheless, our study showed remarkable preference in tropomyosin isoform expression in major C. elegans tissues. To date, a splicing factor for the lev-11 gene has not been identified. For several other alternatively spliced genes in C. elegans, combinatorial regulation by multiple factors such as ASD-1, FOX-1, SUP-12, ASD-2, UNC-75 and PTB-1 have been characterized as the basis of tissue- and cell-type-specific alternative splicing (Anyanful et al. 2004; Kuroyanagi et al. 2006; Kuroyanagi et al. 2007; Kuroyanagi et al. 2010; Kuroyanagi et al. 2013a; Ohno et al. 2008; Ohno et al. 2012; Tomioka et al. 2016). The fluorescence splicing reporter worms generated in this study should be useful tools for future genetic screening and characterization of splicing regulators in vivo, as well as determination of cis-elements critical for the tight regulation of tropomyosin isoform expression in nematodes.
Organized regulation of multiple alternatively spliced exons can potentially contribute to production of a number of tropomyosin isoforms. High- and low-molecular-weight lev-11 tropomyosin isoforms are encoded by nine and seven exons, respectively, and only E6 and E8 are used constitutively as protein-coding regions in all known isoforms (Fig. 1). With the two promoters and alternative choices of exons 4, 5, 7, and 9, up to 72 tropomyosin isoforms could be theoretically produced. This indicates that the C. elegans lev-11 gene is more extensively alternatively spliced than any mammalian tropomyosin genes: human tropomyosin isoforms are similarly encoded by nine or seven exons, and each of the four tropomyosin genes has five constitutively used exons (Geeves et al. 2015; Schevzov et al. 2011). Therefore, extensive alternative splicing of the lev-11 gene might be an adaptation to produce many tropomyosin isoforms from a single gene to fulfill a variety of tropomyosin functions.
Expression of different tropomyosin isoforms were detected in distinct types of muscles, which might be related to the major function of tropomyosin to regulate muscle contraction by mediating calcium-dependent conformational changes of troponin to actin filaments (Ebashi 1984). In C. elegans, multiple isoforms of troponin components are expressed in both striated and non-striated muscles (Barnes et al. 2016; Burkeen et al. 2004; Myers et al. 1996; Obinata et al. 2010; Ruksana et al. 2005; Terami et al. 1999), and there may be isoform-specific interactions among tropomyosin and troponin components. The body wall muscles are obliquely striated and have similarities to vertebrate cross-striated muscles (Ono 2014). A C. elegans hermaphrodite has total 95 mononucleated body wall muscle cells, which contain indistinguishable sarcomere structures. However, eight muscle cells in the head region (head muscles) exhibit a distinct pattern of gene expression from other posterior muscle cells (Cao et al. 2017). Alternative splicing of lev-11 exon 7s results in expression of LEV-11O in the head muscles and LEV-11A in the other body wall muscle cells (Barnes et al. 2018). This pattern is correlated with the expression patterns of two of the four troponin I isoforms: TNI-3 is enriched in the head muscles, whereas UNC-27 is expressed in the other body wall muscle cells (Burkeen et al. 2004; Ruksana et al. 2005). A previous study suggests that LEV-11A and UNC-27 are a functional isoform combination of tropomyosin and troponin I (Barnes et al. 2018), but compatibility among other isoforms remains unknown. The pharyngeal muscles represent a unique type of muscle in which only a low-molecular-weight tropomyosin, LEV-11E, is expressed together with distinct isoforms of troponin components (Amin et al. 2007; Burkeen et al. 2004; Ruksana et al. 2005). Typically, high-molecular-weight tropomyosins play a major role in the regulation of muscle contraction. Therefore, this unique set of tropomyosin and troponin may have a specific function in the pharyngeal muscle.
Tropomyosins are expressed in both muscle and non-muscle cells in animals (Gunning et al. 2015). In C. elegans, expression of LEV-11 isoforms have been characterized in major tissues, including the body wall muscle, pharynx, and intestine (Anyanful et al. 2001; Barnes et al. 2018; Kagawa et al. 1995; Ono and Ono 2002). However, the expression and function of most of non-muscle LEV-11 tropomyosins have not been determined. The previous promoter-reporter analyses reported that the lev-11 gene has two promoters, which are active only in limited tissues (Anyanful et al. 2001; Kagawa et al. 1995) and are unlikely to represent all tropomyosin-expressing tissues. For example, LEV-11 is expressed in the myoepithelial sheath and spermatheca in the somatic gonad and essential for ovulation (Ono and Ono 2004; Ono et al. 2007), but the promoter activity has not been detected in these tissues, suggesting that the examined promoter regions might not have included all tissue-specific cis-acting elements. Using heterologous promoters, we demonstrated that neurons and excretory cell processed specific patterns of alternative splicing in the lev-11 minigene constructs. Additional profiling of lev-11 splicing using other tissue/cell-type specific promoters should help to determine distribution of tropomyosin isoforms in C. elegans.
Among 16 exons in the lev-11 gene, only E6, E8, and E9c are constitutively selected. We therefore constructed atypical reporter minigenes in this study: alternatively spliced exons were forced to be a part of constitutive exons (Fig. 3A-B, Fig. 4A-C, and Fig. 5A-B). These experiments revealed some unexpected results. For instance, intron 3b in the LEV-11E3b-E6-EGFP minigene was constitutively excised, and E4a was included even in body wall muscles where only E4b should be selected (Fig. 3A and G). E5b in the LEV-11E4b-E6-mRFP minigene was preferentially selected even in intestine and neurons (Fig. 3B and G) that prefer E5c in other minigenes (Fig. 3A, 4F). These results imply that context-dependent mechanisms play substantial roles in the tissue-specific alternative splicing of lev-11 pre-mRNA. This may be the reason why we did not detect any of the fluorescent proteins in the nervous system with the three-color E5 reporter.
In conclusion, tropomyosin isoforms have evolved in the nematode C. elegans by increasing the complexity of tissue/cell-specific alternative splicing of the single gene, which must have accompanied functional differentiation of tropomyosin isoforms that support various actin-based cytoskeletal structures. Further correlative studies on tropomyosin isoforms in C. elegans using genetics, cell biology, and biochemistry should be useful in the future to understand the basis of cytoskeletal differentiation in multicellular organisms.
Materials and methods
Worm culture
Worms were cultured following standard methods (Stiernagle 2006). Wild-type C. elegans strain N2 was obtained from the Caenorhabditis Genetics Center (Minneapolis, MN) and used in this study. Transgenic worms were generated as described previously (Kuroyanagi et al. 2010). At least two independent lines with extrachromosomal arrays were established for each combination of the lev-11 reporter minigenes. The expression patterns of the fluorescent proteins were reproducible and consistent within the lines as well as among the independent lines as we have already reported for other reporters (Kuroyanagi et al. 2010). Genotypes of the strains analyzed in the Figures are listed in Supplemental Table S1.
RT-PCR and sequencing
Total RNAs from synchronized L1 worms were extracted as described previously (Kuroyanagi et al. 2010; Kuroyanagi et al. 2013b). Total RNAs from mixed-stage worms were extracted using TRI Reagent® (Sigma-Aldrich). RT-PCR was performed essentially as described previously (Kuroyanagi et al. 2010; Kuroyanagi et al. 2013b). Sequences of the primers used in the RT-PCR experiments are available in Supplemental Table S2. RT-PCR products were analyzed by using 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA) or conventional agarose-gel electrophoresis and ethidium bromide staining. Sequences of the RT-PCR products were confirmed by direct sequencing or by cloning and sequencing.
Construction of vectors for splicing reporters
Fluorescence lev-11 splicing reporter minigenes were constructed essentially as described previously (Kuroyanagi et al. 2010; Kuroyanagi et al. 2013a). Briefly, the lev-11 genomic DNA fragment was cloned into a Gateway pENTR-L1/R5 vector (Life Technologies, Carlsbad, CA). Mutagenesis in the exons were performed by utilizing PrimeStar GXL DNA polymerase (Takara Bio, Inc., Shiga, Japan). Expression vectors were constructed by homologous recombination between the genomic DNA cassette and a fluorescent protein cassette in the pENTR-L1/R5 vector and a destination vector pDEST-eef-1A.1p, pDEST-myo-2p, pDEST-myo-3p, pDEST-gst-42p, or pDEST-rgef-1p (Kuroyanagi et al. 2006; Kuroyanagi et al. 2010) by utilizing LR Clonase II Plus (Life Technologies, Carlsbad, CA). Sequences of the primers used in the plasmid construction and mutagenesis are available in Supplemental Table S3. Genotypes of the generated strains are listed in Supplemental Table S1. Sequence information of the reporter minigenes are available upon request to HK.
Microscopy
We observed the expression patterns of the fluorescent proteins in at least 100 worms from each strain using high-magnification epifluorescence stereomicroscopes (MZ16FA and M205FA with Fluo Combi III, Leica Microsystems, Wetzlar, Germany) and confirmed consistent patterns in nearly all observed worms. Then, images of representative fluorescence reporter worms were captured using a fluorescence compound microscope (DM6000B, Leica Microsystems, Wetzlar, Germany) equipped with a color cooled CCD camera (DFC310FX, Leica Microsystems, Wetzlar, Germany). Images were processed with Photoshop CS2 (Adobe, San Jose, CA).
Supplementary Material
Acknowledgements
The authors declare no conflict of interest. We thank Masayuki Hagiwara, Hiroaki Sakane, Hajime Ito, Hiroyuki Maruoka, Chenxi Zhao, Hiroshi Kurokawa, Yohei Watanabe and Marina Togo-Ohno for technical assistance. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by KAKENHI from Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) or the Japan Society for the Promotion of Science (JSPS) [Grant numbers JP20112004, JP25118506, JP26291003, JP15H01350, JP15H01467, JP15KK0252, JP17H03633 and JP17H05596] to H. K., Precursory Research for Embryonic Science and Technology (PRESTO) from Japan Science and Technology Agency (JST) to H. K., and a grant from the National Institutes of Health (AR048615) to S. O.
References
- Amin MZ, Bando T, Ruksana R, Anokye-Danso F, Takashima Y, Sakube Y, Kagawa H. 2007. Tissue-specific interactions of TNI isoforms with other TN subunits and tropomyosins in C. elegans: the role of the C- and N-terminal extensions. Biochim Biophys Acta 1774:456–465. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Anyanful A, Sakube Y, Kagawa H. 2008. Transcription factors GATA/ELT-2 and forkhead/HNF-3/PHA-4 regulate the tropomyosin gene expression in the pharynx and intestine of Caenorhabditis elegans. J Mol Biol 379:201–211. [DOI] [PubMed] [Google Scholar]
- Anyanful A, Ono K, Johnsen RC, Ly H, Jensen V, Baillie DL, Ono S. 2004. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J Cell Biol 167:639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyanful A, Sakube Y, Takuwa K, Kagawa H. 2001. The third and fourth tropomyosin isoforms of Caenorhabditis elegans are expressed in the pharynx and intestines and are essential for development and morphology. J Mol Biol 313:525–537. [DOI] [PubMed] [Google Scholar]
- Bailey K 1946. Tropomyosin: a new asymmetric protein component of muscle. Nature 157:368. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Hwang H, Ono K, Lu H, Ono S. 2016. Molecular evolution of troponin I and a role of its N-terminal extension in nematode locomotion. Cytoskeleton 73:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Watabe E, Ono K, Kwak E, Kuroyanagi H, Ono S. 2018. Tropomyosin isoforms differentially affect muscle contractility in the head and body regions of the nematode Caenorhabditis elegans. Mol Biol Cell 29:1075–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broschat KO. 1990. Tropomyosin prevents depolymerization of actin filaments from the pointed end. J Biol Chem 265:21323–21329. [PubMed] [Google Scholar]
- Broschat KO, Weber A, Burgess DR. 1989. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry 28:8501–8506. [DOI] [PubMed] [Google Scholar]
- Burkeen AK, Maday SL, Rybicka KK, Sulcove JA, Ward J, Huang MM, Barstead R, Franzini-Armstrong C, Allen TS. 2004. Disruption of Caenorhabditis elegans muscle structure and function caused by mutation of troponin I. Biophys J 86:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ and others. 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Kim D, Kim S, Jeong S, Song E, Helfman DM. 2012. From skeletal muscle to cancer: insights learned elucidating the function of tropomyosin. J Struct Biol 177:63–69. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Roy PJ. 2005. Muscle arm development in Caenorhabditis elegans. Development 132:3079–3092. [DOI] [PubMed] [Google Scholar]
- Ebashi S 1984. Ca2+ and the contractile proteins. J Mol Cell Cardiol 16:129–136. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Hitchcock-DeGregori SE, Gunning PW. 2015. A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil 36:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann WH. 2000. Binding of tropomyosin-troponin to actin increases filament bending stiffness. Biochem Biophys Res Commun 276:1225–1228. [DOI] [PubMed] [Google Scholar]
- Gooding C, Smith CW. 2008. Tropomyosin exons as models for alternative splicing. Adv Exp Med Biol 644:27–42. [DOI] [PubMed] [Google Scholar]
- Greenberg MJ, Wang CL, Lehman W, Moore JR. 2008. Modulation of actin mechanics by caldesmon and tropomyosin. Cell Motil Cytoskeleton 65:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruninger TR, Gualberto DG, LeBoeuf B, Garcia LR. 2006. Integration of male mating and feeding behaviors in Caenorhabditis elegans. J Neurosci 26:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, O’Neill G, Hardeman E. 2008. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev 88:1–35. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. 2015. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J Cell Sci 128:2965–2974. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. 2005. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol 15:333–341. [DOI] [PubMed] [Google Scholar]
- Hendricks M, Weintraub H. 1984. Multiple tropomyosin polypeptides in chicken embryo fibroblasts: differential repression of transcription by Rous sarcoma virus transformation. Mol Cell Biol 4:1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Sampath P, Pollard TD. 1988. Tropomyosin inhibits the rate of actin polymerization by stabilizing actin filaments. Biochemistry 27:9182–9185. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Lehman W. 2008. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil 29:213–219. [DOI] [PubMed] [Google Scholar]
- Hwang H, Barnes DE, Matsunaga Y, Benian GM, Ono S, Lu H. 2016. Muscle contraction phenotypic analysis enabled by optogenetics reveals functional relationships of sarcomere components in Caenorhabditis elegans. Sci Rep 6:19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H, Sugimoto K, Matsumoto H, Inoue T, Imadzu H, Takuwa K, Sakube Y. 1995. Genome structure, mapping and expression of the tropomyosin gene tmy-1 of Caenorhabditis elegans. J Mol Biol 251:603–613. [DOI] [PubMed] [Google Scholar]
- Karam CN, Warren CM, Rajan S, de Tombe PP, Wieczorek DF, Solaro RJ. 2011. Expression of tropomyosin-κ induces dilated cardiomyopathy and depresses cardiac myofilament tension by mechanisms involving cross-bridge dependent activation and altered tropomyosin phosphorylation. J Muscle Res Cell Motil 31:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn TB, Bamburg JR. 2008. Tropomyosin and ADF/cofilin as collaborators and competitors. Adv Exp Med Biol 644:232–249. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. 2006. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods 3:909–915. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Mitani S, Hagiwara M. 2007. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol 27:8612–8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M. 2010. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat Protoc 5:1495–1517. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Hagiwara M. 2013a. CELF family RNA-binding protein UNC-75 regulates two sets of mutually exclusive exons of the unc-32 gene in neuron-specific manners in Caenorhabditis elegans. PLoS Genet 9:e1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Suzuki Y, Hagiwara M. 2013b. Position-dependent and neuron-specific splicing regulation by the CELF family RNA-binding protein UNC-75 in Caenorhabditis elegans. Nucleic Acids Res 41:4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laki K, Maruyama K, Kominz DR. 1962. Evidence for the interaction between tropomyosin and actin. Arch Biochem Biophys 98:323–330. [DOI] [PubMed] [Google Scholar]
- Lal AA, Korn ED. 1986. Effect of muscle tropomyosin on the kinetics of polymerization of muscle actin. Biochemistry 25:1154–1158. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W and others. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Lees-Miller JP, Helfman DM. 1991. The molecular basis for tropomyosin isoform diversity. Bioessays 13:429–437. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Wu CH, Berg H, Levine JH. 1980a. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics 95:905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Wu CH, Levine JH, Berg H. 1980b. Levamisole-resistant mutants of the nematode Caenorhabditis elegans appear to lack pharmacological acetylcholine receptors. Neuroscience 5:967–989. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Helfman DM, Hughes SH, Chou CS. 1985. Tropomyosin isoforms in chicken embryo fibroblasts: purification, characterization, and changes in Rous sarcoma virus-transformed cells. J Cell Biol 100:692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Warren KS, Wamboldt DD, Wang T, Lin JL. 1997. Tropomyosin isoforms in nonmuscle cells. Int Rev Cytol 170:1–38. [DOI] [PubMed] [Google Scholar]
- Maruyama K 1964. Interaction of tropomyosin with actin. A flow birefringence study. Arch Biochem Biophys 105:142–150. [DOI] [PubMed] [Google Scholar]
- Matsumura F, Lin JJ, Yamashiro-Matsumura S, Thomas GP, Topp WC. 1983. Differential expression of tropomyosin forms in the microfilaments isolated from normal and transformed rat cultured cells. J Biol Chem 258:13954–13964. [PubMed] [Google Scholar]
- Myers CD, Goh PY, Allen TS, Bucher EA, Bogaert T. 1996. Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans. J Cell Biol 132:1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinata T, Ono K, Ono S. 2010. Troponin I controls ovulatory contraction of non-striated actomyosin networks in the C. elegans somatic gonad. J Cell Sci 123:1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno G, Hagiwara M, Kuroyanagi H. 2008. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev 22:360–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno G, Ono K, Togo M, Watanabe Y, Ono S, Hagiwara M, Kuroyanagi H. 2012. Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans. PLoS Genet 8:e1002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ono S. 2004. Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol Biol Cell 15:2782–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yu R, Ono S. 2007. Structural components of the nonstriated contractile apparatuses in the Caenorhabditis elegans gonadal myoepithelial sheath and their essential roles for ovulation. Dev Dyn 236:1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S 2014. Regulation of structure and function of sarcomeric actin filaments in striated muscle of the nematode Caenorhabditis elegans. Anat Rec (Hoboken) 297:1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. 2002. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol 156:1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak G, McGarvey TW, Nguyen TB, Tomaszewski JE, Puthiyaveettil R, Malkowicz SB, Helfman DM. 2004. Alterations in tropomyosin isoform expression in human transitional cell carcinoma of the urinary bladder. Int J Cancer 110:368–373. [DOI] [PubMed] [Google Scholar]
- Perry SV. 2001. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil 22:5–49. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Kazzaz JA, Helfman DM. 1994. Functional properties of non-muscle tropomyosin isoforms. Curr Opin Cell Biol 6:96–104. [DOI] [PubMed] [Google Scholar]
- Rajan S, Jagatheesan G, Karam CN, Alves ML, Bodi I, Schwartz A, Bulcao CF, D’Souza KM, Akhter SA, Boivin GP, Dube DK, Petrashevskaya N, Herr AB, Hullin R, Liggett SB, Wolska BM, Solaro RJ, Wieczorek DF. 2010. Molecular and functional characterization of a novel cardiac-specific human tropomyosin isoform. Circulation 121:410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruksana R, Kuroda K, Terami H, Bando T, Kitaoka S, Takaya T, Sakube Y, Kagawa H. 2005. Tissue expression of four troponin I genes and their molecular interactions with two troponin C isoforms in Caenorhabditis elegans. Genes Cells 10:261–276. [DOI] [PubMed] [Google Scholar]
- Schevzov G, Whittaker SP, Fath T, Lin JJ, Gunning PW. 2011. Tropomyosin isoforms and reagents. BioArchitecture 1:135–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T 2006. Maintenance of C. elegans. WormBook:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terami H, Williams BD, Kitamura S, Sakube Y, Matsumoto S, Doi S, Obinata T, Kagawa H. 1999. Genomic organization, expression, and analysis of the troponin C gene pat-10 of Caenorhabditis elegans. J Cell Biol 146:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Naito Y, Kuroyanagi H, Iino Y. 2016. Splicing factors control C. elegans behavioural learning in a single neuron by producing DAF-2c receptor. Nat Commun 7:11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Coluccio LM. 2010. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int Rev Cell Mol Biol 281:91–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol 124:475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Ono S. 2006. Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton 63:659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.