The gold standard of cryptococcosis treatment consists of induction therapy with amphotericin B followed by lifelong maintenance therapy with fluconazole (FLC). However, prolonged exposure to FLC induces the emergence of clones heteroresistant to azoles. All the heteroresistant clones thus far analyzed have been shown to be aneuploids, but how the aneuploid is formed remains unclear. Aneuploidy in fungi and other eukaryotic cells is known to result most commonly from chromosome missegregation during cell division due to defects in any one of the multiple components and processes that are required for the formation of two genetically identical daughter cells. Although formation of multinucleated cells has been observed in cells exposed to FLC, evidence for the emergence of drug-resistant aneuploid populations directly from such cells has been lacking. We show the evidence that the aneuploid in fluconazole-heteroresistant clones of Cryptococcus neoformans is derived neither from multinucleated cells nor from chromosome missegregation.
KEYWORDS: fluconazole, heteroresistance, aneuploid, chromosomal endoduplication, nondisjunction
ABSTRACT
Heteroresistance to fluconazole (FLC) in Cryptococcus is a transient adaptive resistance which is lost upon release from the drug pressure. It is known that clones heteroresistant to FLC invariably contain disomic chromosomes, but how disomy is formed remains unclear. Previous reports suggested that the aneuploid heteroresistant colonies in Cryptococcus emerge from multinucleated cells, resembling the case in Candida albicans. Although a small number of cells containing multiple nuclei appear in a short time after FLC treatment, we provide evidence that the heteroresistant colonies in the presence of FLC arise from uninucleate cells without involving multinuclear/multimeric stages. We found that fidelity of chromosome segregation in mitosis plays an important role in regulation of FLC heteroresistance frequency in C. neoformans. Although FLC-resistant colonies occurred at a very low frequency, we were able to modulate the frequency of heteroresistance by overexpressing SMC1, which encodes a protein containing an SMC domain in chromosome segregation. Using time-lapse microscopy, we captured the entire process of colony formation from a single cell in the presence of FLC. All the multinucleated cells formed within a few hours of FLC exposure failed to multiply after a few cell divisions, and the cells able to proliferate to form colonies were all uninucleate without exception. Furthermore, no nuclear fusion event or asymmetric survival between mother and daughter cells, a hallmark of chromosome nondisjunction in haploid organisms, was observed. Therefore, the mechanisms of aneuploidy formation in C. neoformans appear different from most common categories of aneuploid formation known for yeasts.
INTRODUCTION
Cryptococcosis is one of the most devastating opportunistic fungal infections encountered by immunocompromised patients, especially those with HIV (1, 2). Globally, cryptococcal meningoencephalitis has been estimated to be responsible for 15% of AIDS-related deaths, which amounts to more than 180,000 a year (2). The patients diagnosed with cryptococcosis are most commonly treated by induction therapy consisting of amphotericin B with or without 5-fluorocytosine followed by lifelong maintenance therapy with fluconazole (FLC) (3). Due to its minimal side effects, FLC remains the most commonly used azole drug also for other yeast infections such as candidiasis. However, because FLC is fungistatic, prolonged exposure to the drug results in the emergence of cryptococcal populations that adapt to the drug stress and become resistant to FLC. It has been reported that every strain of Cryptococcus neoformans tested is innately heteroresistant to FLC (4). FLC heteroresistance is a transient adaptive resistance characterized by the emergence of a minor subpopulation within a single colony of the susceptible strain which can tolerate FLC concentrations higher than the strain’s MIC. By growing on drug-free medium, however, such acquired resistance is lost and the strain returns to its original MIC. The frequency of heteroresistant subpopulations is strain dependent and ranges from less than 1% to 10% (4). In the most commonly used reference strain, H99, all the heteroresistant clones thus far analyzed have been shown to be aneuploids containing a duplicated chromosome 1 (chr1). chr1 harbors both ERG11, the gene encoding the target of azoles, and AFR1, which encodes the major efflux pump for azoles (5). Heteroresistant clones can adapt to even higher levels of FLC, and such clones contain more duplicated chromosomes besides the disomic chr1 (5). In Candida albicans, aneuploidy is also associated with adaptation to severe FLC stress (6, 7). In clinical isolates of FLC-resistant C. albicans, aneuploidy is found to be most prevalent for chr5, which harbors ERG11 and the TAC1 gene, which encodes a transcription regulator of drug efflux pumps (6). In both yeasts, however, the precise mechanism by which FLC contributes to aneuploidy remains poorly understood (5, 8, 9).
Aneuploidy is known to occur commonly in fungi and other eukaryotic cells, and its occurrence can be spontaneous or as a consequence of genetic or environmental perturbations (10, 11). In either case, aneuploidy results most commonly from chromosome missegregation during cell division due to defects in any one of the multiple components and processes that are required for the formation of two genetically identical daughter cells (10–12). Chromosomal unequal segregation leading to the formation of aneuploid FLC-resistant progeny has been demonstrated in C. albicans (8). Prior to the formation of aneuploid progeny, FLC-exposed C. albicans cells first undergo a cell cycle delay followed by uncoupling of cytokinesis and nuclear division which leads to the formation of intermediate polyploid or multinucleated cells. Such an event is also common in the cells of aneuploid-rich solid tumors (13). Although formation of multimeric cells has been observed in C. neoformans exposed to FLC (9), evidence for the emergence of drug-resistant aneuploid populations directly from such cells has been lacking. The nuclear status of progenitor cells which yield aneuploid drug-resistant populations is critical to understand the mechanism of FLC heteroresistance. Our studies showed that C. neoformans cells exposed to FLC form multinucleated cells at a low frequency but the multinucleated cells fail to propagate to form colonies in the presence of FLC. Time-lapse imaging microscopy successfully captured the process of colony formation in the presence of FLC, and all such colonies were found to have originated from uninucleate progenitor cells. Furthermore, asymmetric survival between mother and daughter cells, a hallmark of chromosome missegregation in haploid cells, has not been detected. Our study revealed marked differences between C. albicans and C. neoformans in the formation of drug-associated aneuploidy.
RESULTS AND DISCUSSION
Ambiguity in flow cytometric determination of DNA content in fluconazole-treated C. neoformans cells stained by propidium iodide.
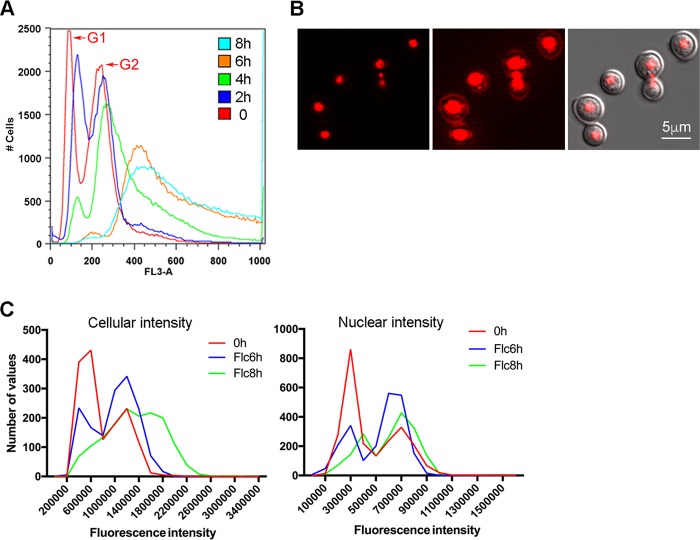
Fluconazole-heteroresistant strains of C. neoformans contain disomic chromosomes (4). It is possible that the disomic chromosomes were derived from polyploid cells in the presence of FLC. We used flow cytometry to monitor the DNA content of strain H99 cells exposed to FLC for different periods of time since flow cytometry has been commonly employed to monitor the DNA contents of FLC-treated cells and assessment of their ploidy status (8, 9). Flow cytometric analysis of the control log-phase cells stained with propidium iodide (PI) consistently showed two typical peaks corresponding to the G1 and G2 phases (Fig. 1A, left side, red line). Upon FLC treatment, both peaks shifted to the right with higher fluorescence intensity and the G1 peak gradually diminished in 8 h. The upshift of fluorescence intensity upon FLC treatment in C. neoformans was also reported in a previous publication (9), although the increase in fluorescence intensity in the previous report seemed to be less pronounced (see Fig. S1A in the supplemental material). The simplest explanation for the fluorescence intensity increase over time is, as postulated by Altamirano et al., that ploidy has changed in the analyzed population (9). However, this explanation is ambiguous. Because C. neoformans yeast cells are normally haploid, formation of multinucleate cells is the prerequisite for the formation of polyploid cells and hence multinucleated cell populations are expected to increase over time unless the ploidy changes occur by endogenous multiplications of the whole genome, bypassing the multinucleated stage. Since flow cytometry cannot differentiate PI-stained multinucleated cells from those of polyploid cells, we examined the PI-stained cells with a microscope to assess the effect of FLC treatment on the nuclear status. In the samples treated with FLC for up to 8 h, we very rarely detected multimeric cells or multimeras, which had been defined by Altamirano et al. as the formation of cells with two or more daughter cells attached (9). However, multinucleated cells were detectable in samples treated with FLC for 6 or 8 h and the frequency of such cells was less than 1% in the population. (Fig. S1, arrow). The multinucleated cells at this stage were most frequently found to contain three nuclei, one in the mother cells and the other two located near the isthmus between the mother and daughter cells (Fig. 1B). With such a low frequency, multinucleated cells could not have been the primary cause for the observed increase of fluorescence intensity in flow cytometry.
FIG 1.
FLC treatment affects flow cytometry patterns. (A) FLC treatment causes changes of fluorescence intensity in PI-stained cells. H99 cells grown to log phase were treated with 32 µg/ml of FLC for indicated times, stained with PI, and subjected to flow cytometry. The histogram of fluorescence intensity (FL3-A) is displayed. (B) FLC treatment causes formation of multinucleated cells. Micrograph on the left depicts the fluorescence image of PI-stained cells (red) treated with FLC for 6 h; in the middle, the same image was scaled to show the dim staining outside the nuclear area; on the right, merged image of DIC and fluorescence. (C) Nuclear fluorescence intensity is not altered by FLC treatment as determined by fluorescence microscopy. Cells were stained with PI and imaged by fluorescence microscopy. The intensity of 1,500 cells and nuclei was measured, and the histograms of fluorescence intensity of the cells (left panel) and nuclei (right panel) are displayed.
Fluconazole treatment affects cell size. (A) Flow cytometry patterns are similar in samples stained with different amounts of PI. Log-phase-grown H99 cells were treated with 32 µg/ml of FLC for indicated times and stained with indicated concentrations of PI. The left panel is the results of our standard method using 10 µg/ml of PI; the middle panel is the results of our standard method except without sonication; the right panel is the results using 0.5 µg/ml of PI and staining for 30 min as suggested and modified by L. Kozubowski (reference 9 and personal communication). The difference of the results between our standard method (left panel) and the method suggested by L. Kozubowski (right panel) could be due to difference of the intensity of PI-stained cells and settings of flow cytometer between the two methods. (B) Fluconazole treatment affects flow cytometry patterns. Cells were treated with 32 µg/ml of FLC for indicated times and analyzed by flow cytometry. The forward (x axis) and side (y axis) scatter patterns are increased in FLC-treated samples, indicating that treatment with FLC causes the increase of cell size and modification of cellular contents. (C) PI-stained cells were examined under a microscope and photographed. It is clear that cell size increases over time in FLC-treated samples. Arrows indicate the cells containing multiple nuclei. Download FIG S1, TIF file, 20.2 MB (20.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Another possibility for increase of fluorescence intensity could be the result of endogenous multiplication of the whole genome. To test this possibility, we used a fluorescence microscope to capture the images of PI-stained cells and measured the fluorescence intensity of each individual cell and nucleus separately. If endogenous multiplication of the whole genome had occurred, nuclear fluorescence intensity should have increased over time beyond 2N. When the fluorescence measurement was taken from whole cells, an upshift of fluorescence intensity was detected in FLC-treated samples (Fig. 1C, left panel). These data were similar to the flow cytometry results, although the upshift of fluorescence intensity was less pronounced in measurements derived from microscopy. In contrast, no clear change of fluorescence intensity patterns was observed in the FLC-treated samples when the fluorescence measurement was taken from only the nuclei (Fig. 1C, right panel). These results suggest that endogenous multiplication of the whole genome, if it ever occurred, would have been a rare event and should have had little or no contribution to the observed increase in fluorescence intensity.
We noted that the forward and side scatter intensity in the flow cytometry data increased over time in the FLC-treated samples (Fig. S1B), which indicated the cell size was increased and the cellular contents were modified upon treatment of FLC. The increase of cell size was further evident by microscopic examination (Fig. S1C). The increase of cell size upon FLC treatment has also been reported previously in C. neoformans and C. albicans (8, 9). It has been demonstrated that the fluorescence intensity of PI-stained samples in flow cytometry analysis is affected by changes in cell size or shape in Saccharomyces cerevisiae and the pattern of flow cytometry is affected irrespective of ploidy status (14). These results supported the notion that the upshift of fluorescence intensity in propidium iodide-stained samples was not contributed by the nuclear staining but was most likely due to the nonspecific extranuclear staining (Fig. 1B, middle panel). Thus, the increase of fluorescence intensity in flow cytometry cannot always be interpreted as the result of ploidy changes in PI-stained samples. We attempted to stain the H99 cells with Sybr green and Sytox green, two commonly used dyes recommended for better resolution than PI for flow cytometry (14, 15). However, both dyes showed nonspecific binding of C. neoformans cells treated with RNase A and yielded unreliable flow cytometric data (data not shown).
Inhibitors of ergosterol biosynthesis affect cell size and fluorescence intensity of PI-stained cells.
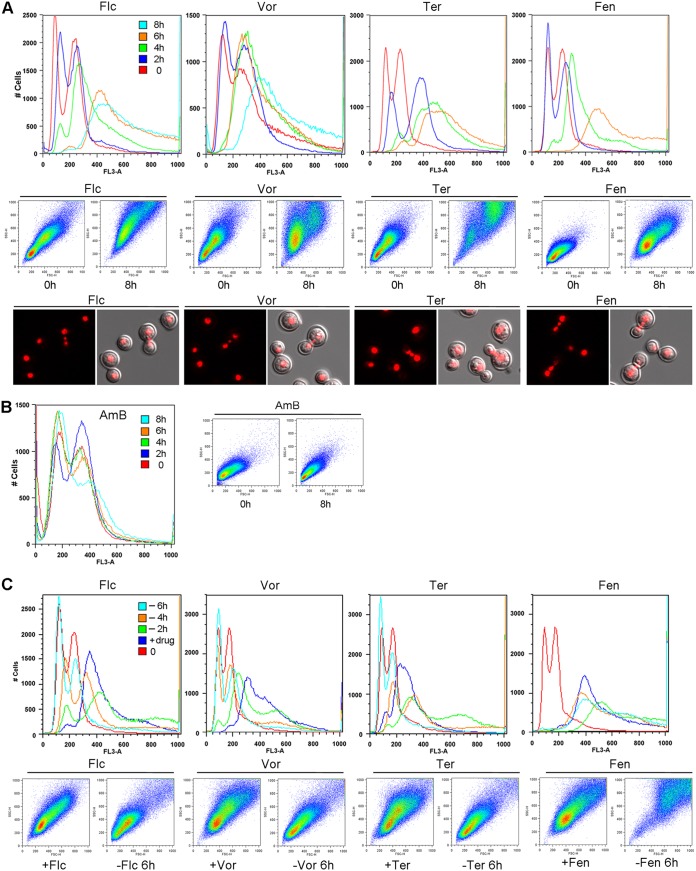
The observed changes of flow cytometry patterns in FLC-treated cells are likely the result of perturbation in ergosterol biosynthesis because FLC targets Erg11, a lanosterol 14α-demethylase, which is an essential protein for ergosterol biosynthesis. We postulated that drugs interfering with different steps in ergosterol biosynthesis might produce similar changes in flow cytometry patterns as FLC. We examined the effect of three other antifungal drugs: voriconazole, a triazole which also targets Erg11; fenpropimorph, which targets Erg2 and Erg24 (16); and terbinafine, which inhibits squalene epoxidase (17). All three drugs caused an increase of PI fluorescence intensity and forward-side scattering patterns similar to the FLC-treated cells (Fig. 2A, upper two rows). Furthermore, multinucleated cells containing three nuclei near the isthmus could be detected in samples treated with the three drugs for 8 h (Fig. 2A). In contrast, no changes in PI fluorescence intensity or forward-side scattering patterns were observed when cells were treated with amphotericin B, an antifungal drug which does not target ergosterol biosynthesis (Fig. 2B). These results indicated that treatment with drugs that inhibit ergosterol biosynthesis affects cell size and cellular content, thereby increasing fluorescence intensity.
FIG 2.
Treatment with drugs inhibiting ergosterol biosynthesis affects flow cytometry patterns. Log-phase-grown H99 cells were treated with indicated drugs for various times. PI-stained cells were analyzed by flow cytometry. (A) Top row shows the histograms of fluorescence intensity (FL3-A) of cells treated with indicated drugs. Middle row shows the forward (x axis) and side (y axis) scatter patterns. Bottom row shows the micrographs of PI-stained cells. (B) Histogram of fluorescence intensity (left panel) and forward-side scatter patterns (right panel) of cells treated with amphotericin B for various times. (C) The effects of azole drugs are reversible. Log-phase YPD-grown cells (0) were treated with indicated drugs for 4 h (+drug). Cells were washed and transferred to YPD without drug for 2, 4, and 6 h (−2h, −4h, −6h). The PI-stained cells were analyzed by flow cytometry. Top row shows the histograms of fluorescence intensity. Bottom row shows the forward-side scatter patterns. Flc, 32 µg/ml of fluconazole; Vor, 4 µg/ml voriconazole; Ter, 1 µg/ml terbinafine; Fen, 3 µg/ml fenpropimorph.
It is noteworthy that the pattern of increased fluorescence intensity was reversible when the cells were treated with FLC for 4 h and then transferred into drug-free medium for different periods of time. The peaks of fluorescence intensity increased slightly 2 h after the transfer, which could have been due to the effect of residual FLC (Fig. 2C). Upon longer incubation in the drug-free medium, however, the peaks of fluorescence intensity reverted to the patterns resembling FLC-untreated control cells (Fig. 2C, 6 h, cyan color). In addition, the forward-side scattering patterns in samples transferred to drug-free medium for 6 h also reverted to the patterns similar to that of the cells never treated with FLC (Fig. 2C), indicating that the cell size and cellular content were back to normal. These results further suggested that a massive change of ploidy likely did not occur because resolution of polyploidy seems unlikely to occur within this short time frame. A similar reversible effect was observed in cells pretreated with voriconazole and terbinafine, but the fluorescence intensity and forward-side scattering patterns in the cells pretreated with fenpropimorph did not revert in drug-free medium (Fig. 2C). This suggests that fenpropimorph might have a long-lasting inhibitory effect on ergosterol biosynthesis.
The increase of fluorescence intensity and change in forward-side scattering patterns were also observed in C. albicans strain B-4497 treated with FLC as well as with the other three ergosterol biosynthesis inhibitors (Fig. S2A). However, the frequency of multinucleated cells was significantly higher in FLC-treated C. albicans than in FLC-treated H99, and the location of the multinuclei in C. albicans was not restricted to the site near the isthmus between mother and daughter cells. When S. cerevisiae strain S288C was treated with the four drugs, a slight change of fluorescence intensity and forward-side scattering patterns was observed, although the changes was not as obvious as in C. albicans and C. neoformans (Fig. S2B). In contrast, no clear changes in fluorescence intensity or forward-side scattering patterns were observed in the C. glabrata strain 1660 treated with the same four drugs (Fig. S2C). These results indicate that the inhibitors of ergosterol biosynthesis do not exert the same influence in forward-side scattering patterns and fluorescence intensity of PI-stained cells among different species of yeast. It is not clear how inhibition of ergosterol synthesis causes the morphological changes in fungi.
Effect of different drugs on the patterns of fluorescence intensity in different yeast strains. Log-phase-grown cells were treated with indicated drugs for 6 hours, and PI-stained cells were analyzed by flow cytometry. Figures show histogram of fluorescence intensity (FL3-A) and forward (x axis) and side (y axis) scatter patterns. The morphology of PI-stained C. albicans treated with different drugs is also shown (A, bottom row). The amounts of each drug used were adjusted according to each strain’s MIC. (A) C. albicans strain B-4497. Flc, 32 µg/ml of fluconazole; Vor, 1 µg/ml voriconazole; Ter, 32 µg/ml terbinafine; Fen, 1 µg/ml fenpropimorph. (B) S. cerevisiae strain S288C. Flc, 256 µg/ml of fluconazole; Vor, 16 µg/ml voriconazole; Ter, 16 µg/ml terbinafine; Fen, 16 µg/ml fenpropimorph. (C) C. glabrata strain 1660. Flc, 256 µg/ml of fluconazole; Vor, 8 µg/ml voriconazole; Ter, 16 µg/ml terbinafine; Fen, 16 µg/ml fenpropimorph. Download FIG S2, TIF file, 20.2 MB (20.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Multinucleated cell populations do not yield a higher number of FLC-heteroresistant colonies.
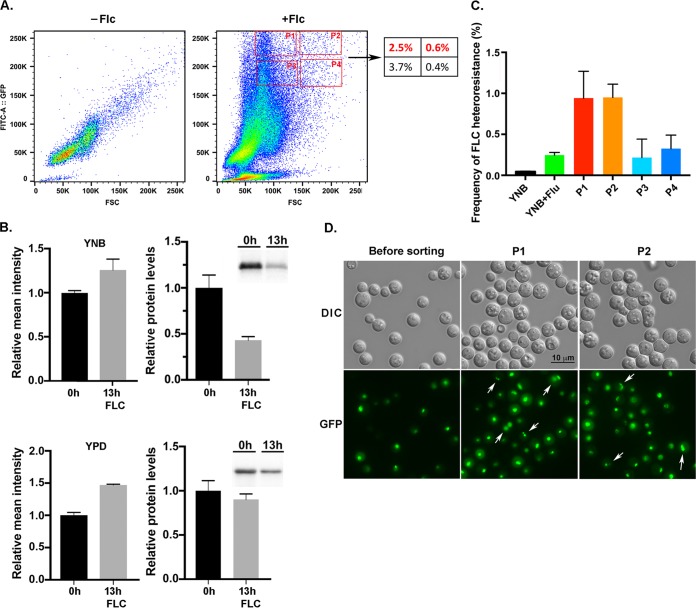
Although there was no massive ploidy change after FLC treatment, it was possible that the multinucleated cells or multimeric cells (9) detected in FLC-treated samples could have been the primary origin of FLC-heteroresistant colonies. This hypothesis requires ascertaining the fate of the multinucleated cells that emerge after FLC treatment. To isolate multinucleated cells and determine if these cells could produce FLC-heteroresistant colonies, we tagged the histone 2B with GFP in H99 (H2B-GFP) to track the status of nuclear content. The H2B-GFP cells were treated with FLC for 13 h to induce the multimeric cells (9). As expected, treatment of FLC increased the forward scatter intensity, but unexpectedly there was a massive upshift of fluorescence intensity when the cells were treated with FLC for 13 h in YNB or YPD medium (Fig. 3A and data not shown). If the GFP fluorescence intensity had truly reflected the nuclear content, this observation was not consistent with our model that FLC treatment did not cause massive ploidy change. We quantitated the amounts of H2B-GFP in the total protein extract by western blot analysis. Although the relative mean intensity of GFP was higher, no increase was observed in the amounts of H2B-GFP protein from the cells grown in YNB or YPD medium with FLC (Fig. 3B). The increase of the H2B-GFP fluorescence intensity was also visible by microscopy of FLC-treated samples (Movie S1, 0 h versus 13 h, see below). It was not clear why FLC treatment caused the increase in fluorescence intensity of H2B-GFP without causing the increase in the amount of H2B-GFP protein. It is conceivable that FLC treatment may affect the crowding of histone-GFP in the nucleosomes and thus alter the fluorescence intensity signal per GFP molecule. Such a possibility is not in the scope of this work and remains to be further investigated. Despite the unanticipated complications, we assumed that the cells in the population with the high GFP intensity and high forward scatter should contain more multinucleated cells and thus performed the cell sorting experiments. We used flow cytometry and cell sorting to collect four subpopulations of the cells that had high fluorescence intensity and high forward scatter to determine if cells in these subpopulations had altered frequency of FLC-heteroresistant colony formation (Fig. 3A, P1 to P4). The sorted cells from P1 to P4 subpopulations were plated on media with or without 32 µg/ml of FLC, and the frequency of FLC-heteroresistant colony formation was determined for each subpopulation. The subpopulations which had the highest fluorescence intensity, P1 and P2, produced close to a 4-fold-higher frequency of FLC heteroresistant colonies than the unsorted control cells (Fig. 3C). In contrast, the sorted cells from the subpopulations with slightly lower fluorescence intensity, P3 and P4, produced a similar frequency of FLC-resistant colonies as the unsorted control cells. It was evident that the cell size was larger and the GFP fluorescence intensity was higher in the cells of the P1 and P2 subpopulations (Fig. 3D). In addition, the P1 and P2 populations contained multinucleated cells which were infrequently detected in the unsorted cell samples. Furthermore, the morphology of multinucleated cells was different from the multinucleated cells of samples treated with FLC for 6 or 8 h described above, and it was also different from the multimeric cells described in the previous report (9). It is possible that the cell morphology was modified during the washing and sorting process without affecting the multinucleated status. The fraction of multinucleated cells was about 9% in P1 (4/43) and P2 (3/32) populations, but the frequency of FLC-heteroresistant colonies derived from these subpopulations was only about 1%. These results suggested that if all the FLC-heteroresistant colonies were supposed to have been derived from the multinucleated cells in the P1 and P2 subpopulations, the majority of multinucleated cells from the sorted population must have failed to form FLC-heteroresistant colonies.
FIG 3.
Cell sorting of FLC-treated population. (A) FLC treatment causes the increase of H2B-GFP fluorescence. Log-phase YNB-grown cells (−FLC) were treated with 32 µg/ml of FLC for 13 h (+FLC), and the fluorescence patterns were determined by flow cytometry. Figures are a representative of three independent experiments. The number in the box on the right panel indicates the percentage of the P1 to P4 subpopulations in the analyzed sample. (B) The amounts of H2B-GFP do not increase in FLC-treated samples. Total protein extracts were isolated from YNB- or YPD-grown cells treated or untreated with 32 µg/ml of FLC for 13 h. The amount of H2B-GFP was quantitated by western blot analysis. The error bars represent the standard deviation from three biological repeats. The insets on the right show the H2B-GFP band intensity. (C) Frequency of FLC heteroresistance among the sorted populations. Cells from the P1 to P4 sorted populations were plated on YPD with or without 32 µg/ml of FLC, and the frequency of FLC heteroresistance was determined. The error bars represent the standard deviation from three biological repeats. (D) Micrographs of H2B-GFP-tagged H99 cells before and after sorting. Arrows indicate the multinucleated cells.
Time-lapse images of the formation of FLC-resistant colony. Red circle indicates the progenitor mother cell. Yellow circles indicate the origin of cells producing multimeric/multinucleated progeny. Download Movie S1, MOV file, 8.8 MB (9MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Approaches to trace the progenitor of FLC-heteroresistant colonies.
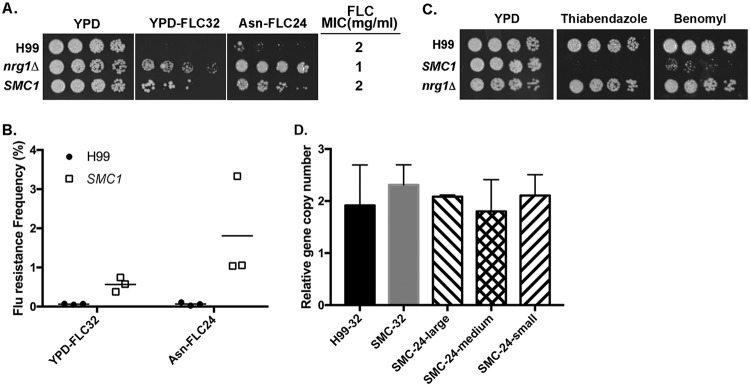
In order to identify the progenitor cells from which the FLC-resistant colonies emerged, we attempted to use a time-lapse microscopy approach to monitor the cellular events that take place from the naive single cells to the formation of visible colonies on FLC medium. Since the frequency of FLC-resistant population at 32 µg/ml of FLC in strain H99 is less than 0.5% (4), it was impractical to attempt capturing the progenitor of the resistant population by time-lapse imaging. To achieve substantial increase in the frequency of FLC heteroresistance, we first screened 9 compounds that were known to affect one or several components of the mitotic apparatus, including ansamitocin P-3, benomyl, cytochalasins, jasplakinolide, monastrol, nocodazole, rhizoxin, thiabendazole, and tubulysin A. We found the incorporation of monastrol, nocodazole, rhizoxin, and thiabendazole in the culture media substantially increased the frequency of FLC-heteroresistant colonies (Fig. S3). However, considering the possibility that these chemicals could have nonspecific effects and interfere with normal cellular behavior, this approach was not pursued further. We next screened a library containing a collection of deletion mutants (18) to identify strains that displayed increased frequency of FLC heteroresistance. Five strains containing deletions of CHL1 (CNAG_04026), GCN5 (CNAG_00375), NRG1 (CNAG_05222), NSR1 (CNAG02130), or SSN8 (CNAG_00440) showed substantially increased frequency of FLC-resistant clone formation on 32 µg/ml FLC medium. One of the genes, NRG1, encodes a transcription factor, and the gene expression profile of the nrg1Δ strain had been studied in C. neoformans serotype D strain JEC21 (19). Since we discovered that some of the compounds known to affect the function of the mitotic apparatus also influenced the frequency of FLC heteroresistance, we hypothesized that deletion of NRG1 might affect the expression of genes associated with the function of mitotic apparatus. We found two genes with annotation related to mitotic apparatus function which were upregulated at least 2.5-fold among the 71 genes whose expression has been affected in the nrg1Δ mutant. One is CNI03680, which contains a chromosome segregation protein SMC domain, and the other is CNA01600, which contains a myosin and kinesin motor domain. The orthologous genes of CNI03680 and CNA01600 also exist in H99 (CNAG_04227 and CNAG_00172, respectively). We overexpressed the two orthologous genes in H99 separately and found that overexpression of CNAG_04227, which we designated SMC1, increased the FLC-heteroresistant frequency approximately 9-fold, although the strain overexpressing SMC1 had the same FLC MIC as the wild-type control (Fig. 4A and B). In addition, the SMC1-overexpressing strain produced an even higher frequency (about 3-fold) of FLC-heteroresistant colonies when the FLC concentration was reduced from 32 to 24 µg/ml. These results indicated that we could increase the FLC heteroresistance frequency to nearly 27-fold by modulating the SMC1 expression level and the FLC concentration. Figure 4C shows that the SMC1-overexpressing strain was hypersensitive to microtubule destabilizers benomyl and thiabendazole, suggesting that this strain may have altered microtubule-associated functions.
FIG 4.
Overexpression of SMC1 increases frequency of FLC heteroresistance. (A) Spot assays. Threefold serial dilutions of each strain were spotted on YPD medium or YPD medium containing 32 µg/ml of FLC (YPD-FLC32) or Asn medium containing 24 µg/ml of FLC (Asn-FLC24), incubated at 30°C for 5 days, and photographed. SMC1 represents the strain overexpressing the SMC1 gene. The FLC MIC of each strain is listed on the right. (B) Frequency of FLC heteroresistance. Cells from H99 and SMC1-overexpressing strains were plated on indicated media, and the frequency of FLC heteroresistance was determined. (C) The SMC1-overexpressing strain is hypersensitive to thiabendazole (5 µg/ml) and benomyl (2 µg/ml). (D) The chr1 gene in FLC-resistant clones derived from the SMC1-overexpressing strain is duplicated. Randomly selected colonies derived from H99 and SMC1-overexpresseing strains resistant to 32 µg/ml of FLC (H99-32, SMC-32) as well as colonies with small, medium, or large size derived from the SMC1-overexpressing strain resistant to 24 µg/ml of FLC (SMC-24-large, SMC-24-medium, SMC-24-small) were used for quantitative PCR. Probes specific for different chromosomes were chosen to assess the changes in genomic copy number. The PCR results of the chr1-specific probes were compared to that of chr5, which served as unduplicated internal control in the indicated strains. Values represent the means ± standard deviations from three independent FLC-resistant clones of each strain.
(A) Plate assay showing drugs that increase the frequency of FLC heteroresistance. Approximately 5,000 H99 cells were plated on YPD, YPD with 32 µg/ml FLC (FLC), or YPD with 32 µg/ml FLC plus indicated amounts of each drugs (in µg/ml). Plates were incubated at 30°C for 5 days and photographed. Monastrol (Mon), nocodazole (Noc), rhizoxin (Rhiz), and thiabendazole (Thb). (B) Fluctuation test. The FLC mutation rates of 15 different individual subcultures of H99 and the control population were determined. Briefly, two independent colonies of H99 from YPD plate were separately inoculated in 20 ml of YPD broth and placed in the 30°C shaker incubator overnight to reach OD600 0.4. Approximately 100 yeast cells from each culture were suspended in 3 ml of YPD broth in each of 15 test tubes. The same overnight cultures were diluted 10-fold in a sixteenth test tube containing 3 ml of YPD broth to serve as controls. All tubes were allowed to incubate for 2 days in the 30°C shaker incubator. After the incubation, OD600 of each tube was determined and adequate dilution from each tube was plated on YPD agar to determine the final number of cells in each tube. In addition, approximately 20,000 cells from each of the 15 tubes were plated on YPD agar containing 32 μg/ml FLC and incubated at 30°C. Fifteen aliquots from the sixteenth tube which served as controls were similarly plated. The colony numbers appearing on plates of YPD and YPD containing 32 μg/ml FLC were determined after 3 and 7 days of incubation, respectively. A web tool based on the Ma-Sandri-Sarkar maximum likelihood estimator method was used to evaluate the mutation rate (29). The experiments were repeated four times, and error bars represent the 95% confidence intervals of each experiment. No statistically significant difference was found in each experiment. Download FIG S3, TIF file, 6.7 MB (6.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
It has been shown that the H99 cells heteroresistant to 32 µg/ml FLC contain two copies of chr1 (4). To confirm that FLC-resistant clones derived from the SMC1-overexpressing strain also contained disomic chr1, we indirectly assessed the copy number of chromosome by using single-colony quantitative PCR for dosage determination of the genes located on chr1 (4). The relative copy number of the chr1 gene was close to 2 in randomly selected colonies resistant to 32 µg/ml FLC derived from H99 as well as the SMC1-overexpressing strains (Fig. 4D). Additionally, the relative copy number of chr1 was also close to 2 in colonies resistant to 24 µg/ml FLC derived from the SMC1-overexpressing strain, and the relative copy number was independent of the size of heteroresistant colonies. This result suggests that chr1 was duplicated in the FLC-resistant clones derived from the SMC1-overexpressing strain, as was the case with the wild type.
Detection of the progenitor cells that form FLC-resistant colonies by time-lapse microscopic imaging.
To monitor the nuclear status during time-lapse imaging experiments, the SMC1-overexpressing strain was tagged with GFP at the C terminus of histone 2B. The GFP-tagged cells were placed in the YC-1 flow chamber which was sandwiched between a cellulose membrane and a polydimethylsiloxane layer. This design prevents cell locomotion during the experiment and provides a single focal plane for imaging by restricting cell proliferation in two dimensions. Growth medium containing 24 µg/ml FLC was infused into the chamber at a flow rate of 0.8 ml/h during 3 days of the experiment. We successfully captured and recorded 29 different time-lapse images covering the whole process of colony formation starting from single naive cells in the presence of FLC. A representative time-lapse image of the colony formation is shown in Movie S1. It was clear that the vast majority of the cells loaded in the chamber failed to multiply after a few cell divisions and the GFP fluorescence diminished by 30 h. No multinucleated cells were detected before 13 h, which was consistent with the aforementioned results that multinucleated cells were a rare event in samples treated with FLC for 6 to 8 h. In the movie, we marked five cells with yellow circles among the initial input population in which the five cells produced multimeric cells after 14 h or later. Close monitoring of the multimeric cells indicated that these multinucleated cells exhibited a cytokinesis defect in which cell separation did not occur while nuclear division continued. This observation corroborates previous studies showing pleiotropic effects of FLC on growth of C. neoformans (9). However, all the multimeric progeny originating from the five different cells failed to proliferate further after a few nuclear divisions. Furthermore, all the multimeric cells among the more than 870 recorded time-lapse images eventually ceased to divide, and some of the multinucleated cells ruptured and expelled their cellular contents (Movie S2).
Time-lapse images of the rupturing of multinucleated cell. Download Movie S2, MOV file, 1.9 MB (2MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
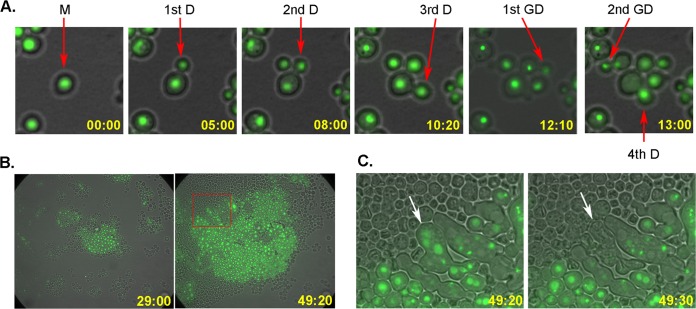
Most importantly, the cells in the developed colony were derived from a uninucleated mother cell which continued to divide for 18 times (red circle in Movie S1 pointing to the original mother cell). Figure 5 shows the events of the colony-forming process from selected frames of Movie S1. Furthermore, all the progenitors of 29 FLC-resistant colonies filmed by time-lapse imaging microscopy were clearly identified as uninucleated cells.
FIG 5.
Time-lapse imaging for the formation of FLC-resistant colony. (A) Cellular events starting from a progenitor mother cell. Pictures show that the mother cell as well as the first two daughter cells continued to divide, and no asymmetrical survival was observed. Picture frames were obtained from Movie S1. The chronological appearance of the first few cells is indicated on the lower right corner of each frame. D, daughter cell; GD, granddaughter cell; M, mother cell. (B) Colony formation at 29:00 and 40:30 h in the presence of FLC. The majority of the initial input cells had stopped dividing and lost GFP signal. Only the progeny from the progenitor mother cell continued to divide. (C) The inset in the right panel of panel B is magnified to show some abnormally elongated multinucleated cells. Between two movie frames, collapse of one of the multinucleated cells and loss of its GFP signal were observed (arrow).
We have confirmed that all the FLC-heteroresistant colonies examined contained a disomy of chr1 (Fig. 4D). One of the most common mechanisms for aneuploidy formation is via chromosome missegregation during cell division (for review, see references 10 to 12). Because C. neoformans is a haploid yeast, if chr1 disomy has resulted from chromosome missegregation during mitotic division, chr1 would be missing in one of the daughter cells and would not have survived (asymmetric survival). This is because chr1 is the largest chromosome, consisting of more than 10% of the genome, and contains several essential genes such as ERG11. However, when we traced the fate of the first three daughters derived from the original mother cell and the first granddaughters budded out from the first three daughters, all of the traced cells continued to divide at least four times (Fig. 5A and Movie S1). Furthermore, we did not detect any asymmetric survival during mitosis in all the 29 colonies captured by time-lapse experiments. Therefore, our data suggested that chromosome missegregation is the unlikely cause of chr1 disomy in the FLC-heteroresistant colonies.
There are two other possible causes for the formation of FLC-resistant aneuploid cells. One is the possibility of preexistence of aneuploid cells in the naive H99 population and exposure to FLC selecting for those chr1 disomic cells. The other possibility is endoduplication of chr1 in response to FLC pressure. In this study, we have identified that several chemicals which are known to inhibit functions of the mitotic apparatus also affect the frequency of FLC heteroresistance. In addition, overexpression of the SMC1 gene, which encodes a protein containing a chromosome segregation SMC domain, increased the frequency of FLC heteroresistance and the strain overexpressing SMC1 had altered microtubule-associated functions. These findings suggest that fidelity of chromosome segregation in mitosis may play important roles in FLC heteroresistance. Since aneuploidy can be produced by loss of fidelity of chromosome segregation in mitosis, one explanation for the time-lapse imaging is that the progenitor is a preexisting aneuploid with disomic chr1 that is present in the population overexpressing SMC1. However, we did not observe a difference in the timing of appearance of FLC-heteroresistant colonies between the wild type and the SMC1-overexpressing strain (data not shown). To determine if there are preexisting FLC-resistant cells among the naive cell population, we carried out the fluctuation analysis originally described by Luria and Delbruck (20). We found there were no significant variations in the rate of heteroresistant clones between individual cultures and to the pooled control cultures (Fig. S3B). However, the fluctuation test would lose its validity if the nature of the mutation was unstable. For example, if the preexisting aneuploid cells subsisted very transiently in a naive population, the fluctuation test would have been invalid. In addition, it has been observed that the growth rate of the aneuploid cells was reduced in the FLC-free medium (5), which rendered the fluctuation test unreliable for testing the preexisting FLC-resistant cells.
In S. cerevisiae, aneuploidy occurs spontaneously once every 5 × 105 cell divisions (21), although this rate can increase more than 100-fold when the cells are under certain stressful environmental conditions (22). Our previous screening of more than 100 C. neoformans strains showed that the frequency of the FLC-heteroresistant population was strain dependent and varied from 0.03% to 10% (4). If the FLC-heteroresistant colonies were derived from preexisting spontaneous aneuploidy, such high frequency and wide variation of spontaneous aneuploidy have never been reported in haploid yeasts. Therefore, preexisting transient aneuploidy is an unlikely cause for FLC heteroresistance. Consequently, these results raised the possibility that disomy of chr1 was through endoduplication under FLC stress. The terms endoduplication and de novo formation of aneuploidy have been frequently used in the field of cancer biology (for a review, see reference 23). However, there is little experimental evidence of aneuploidy formation directly by endoduplication of chromosomes. One possible approach to determine if the chr1 disomy is through chromosomal endoduplication or preexisting aneuploidy is to use two different fluorescent proteins to separately tag chr1 and another chromosome which is not known to be duplicated under FLC stress in the naive strain to monitor the intensity ratio between the two fluorophores in the presence of FLC with time-lapse imaging. We used GFP and RFP to separately tag two chromosomes with Lac and Lex operator arrays (for reviews, see references 24 and 25). We have modulated the expression levels of fluorescently tagged proteins by using different strengths of promoters and different types of nuclear location signals. We have also increased the number of operators to as high as 192 copies. However, with all the modifications in our various constructs, the signal intensity and signal-to-noise ratio were unacceptably low and prevented us from pursuing the time-lapse imaging experiment. Perhaps with the ever-improving technology in microscopy and protein tagging, similar approaches can be revisited in the future as the suitable technology becomes available.
Interestingly, some of the progeny cells in Movie S1 were elongated with aberrant morphology and contained multinuclei (Fig. 4C). Formation of the morphologically abnormal elongated cells was also observed when the time-lapse experiments were carried out by using cells directly isolated from colonies resistant to FLC and observed microscopically (Movie S3). These results suggested that although the FLC-resistant cells were able to survive by the formation of disomic chromosomes, FLC continued to exert its detrimental impact on the process of cytokinesis.
Time-lapse image of the fate of cells directly isolated from colony resistant to FLC. Multinucleated cells are formed in the images. Download Movie S3, MOV file, 2.0 MB (2.1MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
FLC exposure in C. albicans causes a cytokinesis defect and quickly leads to so-called “trimeras,” cells which are composed of three connected cells of a mother, daughter, and granddaughter (8). The trimeras undergo coordinated nuclear division and form tetraploid cells which give rise to aneuploid FLC-resistant populations via chromosome missegregation. Interestingly, no trimera formation was detected in an FLC-resistant clinical isolate of C. albicans. These results are clearly different from our observations indicating the fundamental differences in handling FLC stress between these two phylogenetically distant species. Although the mechanism primarily contributing to heteroresistance in C. neoformans remains to be established, it is clear that it deviates from the most common categories of aneuploid formation reported for yeasts. We have identified five genes in this study and additional transcription factors that play a role in determination/regulation of FLC heteroresistance frequency (data not shown). These findings pave the way for future studies to decipher not only the mechanisms of FLC heteroresistance in C. neoformans but also the mechanism of chromosome duplication in general.
MATERIALS AND METHODS
Strains.
Strains relevant to this study are listed in Table S1 in the supplemental material. Asn medium was yeast nitrogen base without amino acids and ammonium sulfate but contained 10 mM asparagine as the sole nitrogen source. To tag histone H2B with GFP, cDNA of CNAG_06746 was placed under the promoter control of the glyceraldehyde-3-phosphate dehydrogenase gene (CNAG_06699) and fused with mNG at its C terminus. The resulting construct was target integrated into chr3 as described previously (5) to yield C1746. To overexpress SMC1, the CNAG_04227 gene was placed under the promoter control of the 60S ribosomal protein RPL2 gene (CNAG_05232) and was target integrated at the CNAG_04227 locus to yield C1352. The GFP-tagged histone H2B was target integrated into chr3 of the SMC1-overexpressing strain C1352 to yield C1753. The reference method CLSI M27-A3 was used to determine the MIC of FLC for the indicated strains (26).
PCR primers relevant to the study. Download Table S1, DOCX file, 0.01 MB (13KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Flow cytometry analysis.
FACS analysis was performed as described previously (27). Briefly, cells were fixed in ice-cold 70% ethanol overnight at 4°C. The cells were washed, resuspended in NS buffer (10 mM Tris-HCl [pH 7.2], 0.25 M sucrose, 1 mM EDTA, 1 mM MgCl2, 0.1 mM ZnCl2, 0.4 mM phenylmethylsulfonyl fluoride, 7 mM β-mercaptoethanol), digested with 0.5 mg/ml of RNase A for 2 h, and stained with propidium iodide (10 μg/ml) for an additional 2 h at 37°C in the dark. The cells were sonicated for 30 s before FACS analysis. Fluorescence was measured with a FACSCalibur flow cytometer (Becton, Dickinson, San Jose, CA), and data were acquired using CellQuest and analyzed with FlowJo software. At least 30,000 cells of each strain were analyzed for each sample.
Identification of mutants showing increased frequency of fluconazole heteroresistance.
We screened the signature tag mutagenesis (STM) deletion collection of C. neoformans (18). The STM strains were first replica-spotted to YPD agar medium containing 32 µg/ml FLC. Six mutants that showed higher frequency of heteroresistance than H99 were identified. The wild-type genes of each mutant were amplified by PCR, cloned, and sequenced. The plasmid containing the correct sequence was transformed into the respective mutants. The resulting transformants were retested for their levels of FLC resistance. We confirmed that five genes including CHL1 (CNAG_04026), GCN5 (CNAG_00375), NRG1 (CNAG_05222), NSR1 (CNAG02130), and SSN8 (CNAG_00440) were able to complement the mutant phenotype and showed a clear reduction of the resistance frequency to levels similar to the wild type, indicating that the deletion of the five genes caused the change of FLC resistance frequency.
Microscopy and time-lapse imaging.
Cells were stained with PI as described above and imaged by a Zeiss Axiovert fluorescence microscope equipped with an AxioCam MRm digital camera. The image was captured with AxioVision (version 4.0). We made certain that all the signal intensities used for the quantification were not saturated. We employed the Surface function from the Imaris software (Bitplane Scientific Software, USA) for the quantification of PI fluorescence intensity, which allows image segmentation based on highly fluorescent spots. To determine the fluorescence intensity of the whole cell, the spot was set with a predefined area sized to select the entire cell. Thus, the quantitation represents the fluorescence intensity of each cell (nucleus + nonnuclear region) but not any neighboring or attached cells (including daughters). The fluorescence intensity of the nuclei was also similarly determined except that only the areas of the nuclei were measured. The fluorescence intensity of 1,500 cells and nuclei from each treatment was measured. The intensity sum of each data point was imported into Prism 7, and the histogram of fluorescence intensity distribution was analyzed.
For time-lapse imaging, log-phase-grown cells were placed in a YC-1 flow chamber (Warner Instruments). A constant flow of Asn medium containing 24 µg/ml FLC was infused into the chamber at a flow rate of 0.8 ml/h using an Instech P720 peristaltic pump. The chamber was mounted on a Leica DMI6000 microscope equipped with a 63×/1.4-NA oil immersion objective, an adaptive focus control, a motorized XY-stage, and a GFP filter cube, which was kept in an environmental chamber maintained at 30°C. The image was captured using a PCO Edge 5.5 camera and Leica LAS X software (Leica Microsystems, Wetzlar, Germany). The initial number of cells in each imaged position was approximately 50 to 200 per field of view. Images were recorded every 10 min in each position for 3 days. We recorded greater than 870 positions, and there were more than 29 colonies formed in the recorded positions after 3 days of FLC treatment, but we were not able to trace the origin of progenitor cells in a portion of the colonies due either to the location of the FLC-resistant colonies in the field of view or shifting of the positions of the cells during the analysis, which was an inherent issue of the YC-1 chamber. However, we successfully imaged 29 colonies in which the progenitor cell of colonies was clearly traceable after 3 days of FLC treatment.
Protein extraction and immunoblot analysis.
To extract total protein, cells were spun down, washed in ice-cold water, lyophilized, disrupted with a bead using FastPrep-24 (MP Biomedicals, CA), and resuspended in cold lysis buffer containing 0.269 N NaOH and 1% 2-mercaptoethanol. Cell lysate was neutralized with 10 M cold acetic acid. The solution was immediately adjusted to 10 mM Tris (pH 7.5), 1 mM EDTA, 1 mM PMSF, and 1× proteinase inhibitor cocktail (Roche). An equal amount of protein (20 μg) was loaded and run on the Any kD Criterion TGX stain-free gel (Bio-Rad, Richmond, CA), and proteins were transferred to a PVDF membrane. The western blot was incubated with anti-FLAG antibody and developed using the Clarity Western ECL (Bio-Rad, Richmond, CA). The signal was quantitated using the ChemiDoc MP imaging system (Bio-Rad, Richmond, CA), and band intensities were normalized to stain-free blots to control for loading as described previously (28).
Oligonucleotides used for qPCR assays. Download Table S2, DOC file, 0.03 MB (29KB, doc) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of strains relevant to this study. Download Table S3, DOCX file, 0.02 MB (18.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research (DIR), NIAID, NIH.
We thank M. Smelkinson and J. Kabat at the Biological Imaging Facility, NIAID, for their valuable assistance in microscopy. We also thank A. Varma and M. Davis for critical reading of the manuscript.
Footnotes
Citation Chang YC, Khanal Lamichhane A, Kwon-Chung KJ. 2018. Cryptococcus neoformans, unlike Candida albicans, forms aneuploid clones directly from uninucleated cells under fluconazole stress. mBio 9:e01290-18. https://doi.org/10.1128/mBio.01290-18.
REFERENCES
- 1.Kwon-Chung KJ, Fraser JA, Doering TL, Wang Z, Janbon G, Idnurm A, Bahn YS. 2014. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med 4:a019760. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sionov E, Chang YC, Garraffo HM, Kwon-Chung KJ. 2009. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob Agents Chemother 53:2804–2815. doi: 10.1128/AAC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog 6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman J. 2016. Ploidy plasticity: a rapid and reversible strategy for adaptation to stress. FEMS Yeast Res 16:fow020. doi: 10.1093/femsyr/fow020. [DOI] [PubMed] [Google Scholar]
- 8.Harrison BD, Hashemi J, Bibi M, Pulver R, Bavli D, Nahmias Y, Wellington M, Sapiro G, Berman J. 2014. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol 12:e1001815. doi: 10.1371/journal.pbio.1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altamirano S, Fang D, Simmons C, Sridhar S, Wu P, Sanyal K, Kozubowski L. 2017. Fluconazole-induced ploidy change in Cryptococcus neoformans results from the uncoupling of cell growth and nuclear division. mSphere 2:e00205-17. doi: 10.1128/mSphere.00205-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres EM, Williams BR, Amon A. 2008. Aneuploidy: cells losing their balance. Genetics 179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon-Chung KJ, Chang YC. 2012. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog 8:e1003022. doi: 10.1371/journal.ppat.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulla W, Zhu J, Li R. 2014. Yeast: a simple model system to study complex phenomena of aneuploidy. FEMS Microbiol Rev 38:201–212. doi: 10.1111/1574-6976.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albertson DG, Collins C, McCormick F, Gray JW. 2003. Chromosome aberrations in solid tumors. Nat Genet 34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 14.Haase SB, Reed SI. 2002. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1:132–136. [PubMed] [Google Scholar]
- 15.Delobel P, Tesniere C. 2014. A simple FCM method to avoid misinterpretation in Saccharomyces cerevisiae cell cycle assessment between G0 and sub-G1. PLoS One 9:e84645. doi: 10.1371/journal.pone.0084645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11:382–402. doi: 10.1128/CMR.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryder NS. 1992. Terbinafine: mode of action and properties of the squalene epoxidase inhibition. Br J Dermatol 126(Suppl 39):2–7. doi: 10.1111/j.1365-2133.1992.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cramer KL, Gerrald QD, Nichols CB, Price MS, Alspaugh JA. 2006. Transcription factor Nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryot Cell 5:1147–1156. doi: 10.1128/EC.00145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luria SE, Delbruck M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwell LH, Dutcher SK, Wood JS, Garvik B. 1982. The fidelity of mitotic chromosome reproduction in S. cerevisiae. Rec Adv Yeast Mol Biol 1:28–38. [Google Scholar]
- 22.Chen G, Bradford WD, Seidel CW, Li R. 2012. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandrioli D, Belpoggi F, Silbergeld EK, Perry MJ. 2016. Aneuploidy: a common and early evidence-based biomarker for carcinogens and reproductive toxicants. Environ Health 15:97. doi: 10.1186/s12940-016-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meister P, Gehlen LR, Varela E, Kalck V, Gasser SM. 2010. Visualizing yeast chromosomes and nuclear architecture. Methods Enzymol 470:535–567. doi: 10.1016/S0076-6879(10)70021-5. [DOI] [PubMed] [Google Scholar]
- 25.Zuo J, Niu QW, Chua NH. 2000. Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 26.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts—3rd ed, approved standard M27-A3. CLSI, Wayne, PA. [Google Scholar]
- 27.Tanaka R, Taguchi H, Takeo K, Miyaji M, Nishimura K. 1996. Determination of ploidy in Cryptococcus neoformans by flow cytometry. J Med Vet Mycol 34:299–301. doi: 10.1080/02681219680000521. [DOI] [PubMed] [Google Scholar]
- 28.Colella AD, Chegenii N, Tea MN, Gibbins IL, Williams KA, Chataway TK. 2012. Comparison of stain-free gels with traditional immunoblot loading control methodology. Anal Biochem 430:108–110. doi: 10.1016/j.ab.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Hall BM, Ma CX, Liang P, Singh KK. 2009. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluconazole treatment affects cell size. (A) Flow cytometry patterns are similar in samples stained with different amounts of PI. Log-phase-grown H99 cells were treated with 32 µg/ml of FLC for indicated times and stained with indicated concentrations of PI. The left panel is the results of our standard method using 10 µg/ml of PI; the middle panel is the results of our standard method except without sonication; the right panel is the results using 0.5 µg/ml of PI and staining for 30 min as suggested and modified by L. Kozubowski (reference 9 and personal communication). The difference of the results between our standard method (left panel) and the method suggested by L. Kozubowski (right panel) could be due to difference of the intensity of PI-stained cells and settings of flow cytometer between the two methods. (B) Fluconazole treatment affects flow cytometry patterns. Cells were treated with 32 µg/ml of FLC for indicated times and analyzed by flow cytometry. The forward (x axis) and side (y axis) scatter patterns are increased in FLC-treated samples, indicating that treatment with FLC causes the increase of cell size and modification of cellular contents. (C) PI-stained cells were examined under a microscope and photographed. It is clear that cell size increases over time in FLC-treated samples. Arrows indicate the cells containing multiple nuclei. Download FIG S1, TIF file, 20.2 MB (20.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of different drugs on the patterns of fluorescence intensity in different yeast strains. Log-phase-grown cells were treated with indicated drugs for 6 hours, and PI-stained cells were analyzed by flow cytometry. Figures show histogram of fluorescence intensity (FL3-A) and forward (x axis) and side (y axis) scatter patterns. The morphology of PI-stained C. albicans treated with different drugs is also shown (A, bottom row). The amounts of each drug used were adjusted according to each strain’s MIC. (A) C. albicans strain B-4497. Flc, 32 µg/ml of fluconazole; Vor, 1 µg/ml voriconazole; Ter, 32 µg/ml terbinafine; Fen, 1 µg/ml fenpropimorph. (B) S. cerevisiae strain S288C. Flc, 256 µg/ml of fluconazole; Vor, 16 µg/ml voriconazole; Ter, 16 µg/ml terbinafine; Fen, 16 µg/ml fenpropimorph. (C) C. glabrata strain 1660. Flc, 256 µg/ml of fluconazole; Vor, 8 µg/ml voriconazole; Ter, 16 µg/ml terbinafine; Fen, 16 µg/ml fenpropimorph. Download FIG S2, TIF file, 20.2 MB (20.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Time-lapse images of the formation of FLC-resistant colony. Red circle indicates the progenitor mother cell. Yellow circles indicate the origin of cells producing multimeric/multinucleated progeny. Download Movie S1, MOV file, 8.8 MB (9MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
(A) Plate assay showing drugs that increase the frequency of FLC heteroresistance. Approximately 5,000 H99 cells were plated on YPD, YPD with 32 µg/ml FLC (FLC), or YPD with 32 µg/ml FLC plus indicated amounts of each drugs (in µg/ml). Plates were incubated at 30°C for 5 days and photographed. Monastrol (Mon), nocodazole (Noc), rhizoxin (Rhiz), and thiabendazole (Thb). (B) Fluctuation test. The FLC mutation rates of 15 different individual subcultures of H99 and the control population were determined. Briefly, two independent colonies of H99 from YPD plate were separately inoculated in 20 ml of YPD broth and placed in the 30°C shaker incubator overnight to reach OD600 0.4. Approximately 100 yeast cells from each culture were suspended in 3 ml of YPD broth in each of 15 test tubes. The same overnight cultures were diluted 10-fold in a sixteenth test tube containing 3 ml of YPD broth to serve as controls. All tubes were allowed to incubate for 2 days in the 30°C shaker incubator. After the incubation, OD600 of each tube was determined and adequate dilution from each tube was plated on YPD agar to determine the final number of cells in each tube. In addition, approximately 20,000 cells from each of the 15 tubes were plated on YPD agar containing 32 μg/ml FLC and incubated at 30°C. Fifteen aliquots from the sixteenth tube which served as controls were similarly plated. The colony numbers appearing on plates of YPD and YPD containing 32 μg/ml FLC were determined after 3 and 7 days of incubation, respectively. A web tool based on the Ma-Sandri-Sarkar maximum likelihood estimator method was used to evaluate the mutation rate (29). The experiments were repeated four times, and error bars represent the 95% confidence intervals of each experiment. No statistically significant difference was found in each experiment. Download FIG S3, TIF file, 6.7 MB (6.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Time-lapse images of the rupturing of multinucleated cell. Download Movie S2, MOV file, 1.9 MB (2MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Time-lapse image of the fate of cells directly isolated from colony resistant to FLC. Multinucleated cells are formed in the images. Download Movie S3, MOV file, 2.0 MB (2.1MB, mov) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
PCR primers relevant to the study. Download Table S1, DOCX file, 0.01 MB (13KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Oligonucleotides used for qPCR assays. Download Table S2, DOC file, 0.03 MB (29KB, doc) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
List of strains relevant to this study. Download Table S3, DOCX file, 0.02 MB (18.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.