Abstract
Objective
To identify novel genes involved in the etiology of intracranial aneurysms (IAs) or subarachnoid hemorrhages (SAHs) using whole-exome sequencing.
Methods
We performed whole-exome sequencing in 13 individuals from 3 families with an autosomal dominant IA/SAH inheritance pattern to look for candidate genes for disease. In addition, we sequenced PCNT exon 38 in a further 161 idiopathic patients with IA/SAH to find additional carriers of potential pathogenic variants.
Results
We identified 2 different variants in exon 38 from the PCNT gene shared between affected members from 2 different families with either IA or SAH (p.R2728C and p.V2811L). One hundred sixty-four samples with either SAH or IA were Sanger sequenced for the PCNT exon 38. Five additional missense mutations were identified. We also found a second p.V2811L carrier in a family with a history of neurovascular diseases.
Conclusion
The PCNT gene encodes a protein that is involved in the process of microtubule nucleation and organization in interphase and mitosis. Biallelic loss-of-function mutations in PCNT cause a form of primordial dwarfism (microcephalic osteodysplastic primordial dwarfism type II), and ≈50% of these patients will develop neurovascular abnormalities, including IAs and SAHs. In addition, a complete Pcnt knockout mouse model (Pcnt−/−) published previously showed general vascular abnormalities, including intracranial hemorrhage. The variants in our families lie in the highly conserved PCNT protein-protein interaction domain, making PCNT a highly plausible candidate gene in cerebrovascular disease.
Intracranial aneurysms (IAs) are acquired vascular lesions responsible for ≈80% of all nontraumatic subarachnoid hemorrhages (SAHs).1 SAHs secondary to ruptured IA have an overall incidence of 6 to 7 per 100,000 persons per year in most populations.2 The prevalence of unruptured IA in populations >30 years of age ranges from 3.6% to 6.5%, depending on ethnicity and several risk factors.3 Therefore, the low incidence of SAH in comparison to unruptured IA suggests that most IAs do not rupture.4,5 Neurological disorders such as stroke and SAH are a leading cause of disability and death.6 New strategies and preventive measures are needed to ultimately alleviate this burden.7
The prevalence of IA can increase up to 9.8% if a family history of cerebrovascular disease is present (relative risk 4.0 compared to the healthy population),8 suggesting that genetic variation can contribute to susceptibility. Furthermore, population-based genome-wide association studies have found and replicated several chromosomal regions that are associated with increased risk of developing IA.9–12 Recently, a rare coding variant in the ANGPTL6 gene (c.1378A>T) was identified in a family with IA and was present in all affected individuals in addition to 5 of 22 unaffected family members.13
Given the clinical heterogeneity and environmental influences in IA and cerebrovascular disease, large families with mendelian patterns of disease inheritance are uncommon. In the current study, we performed exome sequencing in 3 families with a likely autosomal dominant IA inheritance pattern to look for candidate mutations and novel disease genes.
Methods
Study design
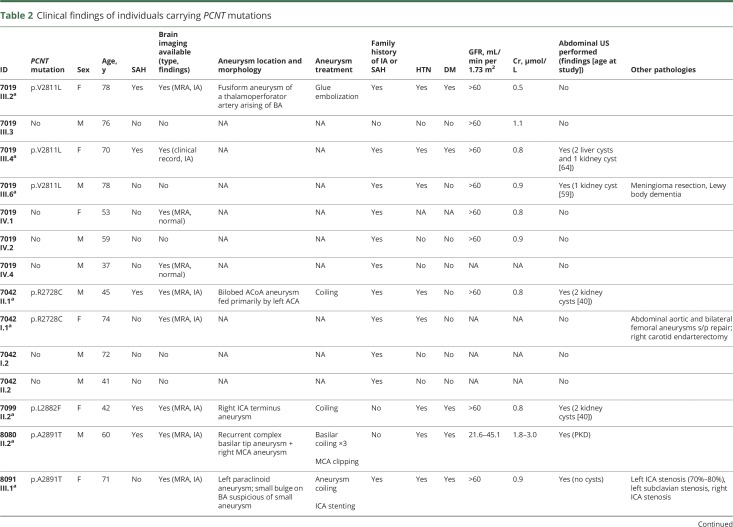
Three multigenerational families with either nontraumatic SAH or unruptured IA family history were recruited from the inpatient, consultation, and outpatient services at the Mayo Clinic (Jacksonville, FL) as part of the ongoing prospective Mayo Clinic Familial Cerebrovascular Diseases Registry (MCFCDR; figure 1A). SAH was diagnosed with axial CT or MRI of the head, and IA was diagnosed by CT angiography (CTA), magnetic resonance angiography (MRA), or digital subtraction angiography. In addition, 161 probands from nonrelated independent families from the MCFCDR with either SAH or unruptured IA were studied in follow-up analyses. The entire series consists of 126 whites, 26 blacks, 9 Hispanics or Latinos, and 3 patients with admixed ethnicity. We grouped all ethnicities other than white in a single cohort because of the small sample size (table 1).
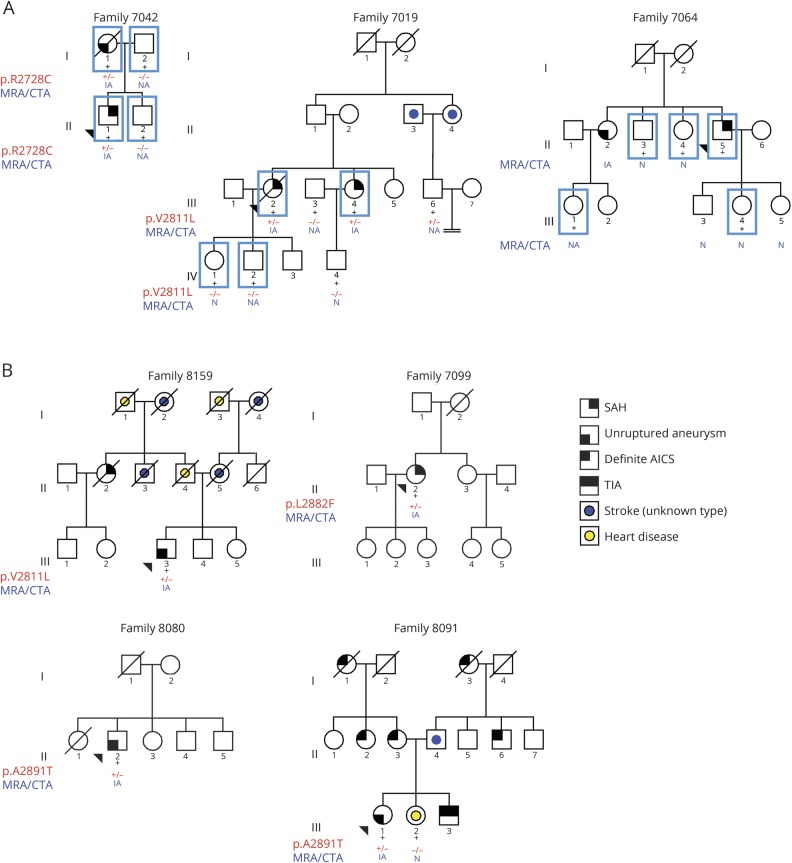
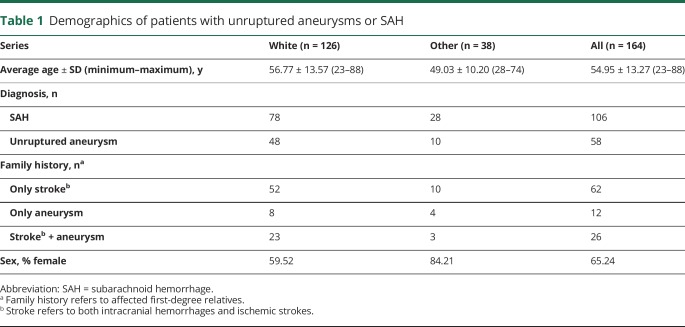
Figure 1. Pedigrees of the families analyzed in this study.
(A) Simplified pedigrees of the families who underwent exome analysis. Black + = individuals for which DNA is available; blue square = individuals for whom exome data is available. (B) Pedigrees of additional families carrying missense variants in the PCNT gene. Simplified pedigrees of the families carrying PCNT missense variants. Squares = men; circles = women; black + = individuals for whom DNA is available who were Sanger sequenced; red + = mutated allele; red − = wild-type allele; forward black slash = deceased individual; arrowhead = proband of each family. Families 7019 and 8159 carry the same PCNT p.V2811L missense mutation. Families 8080 and 8091 carry the same PCNT p.A2891T missense mutation. AICS = acute ischemic cerebrovascular syndrome; CTA = computed tomography angiography; IA = intracranial aneurysm; MRA = magnetic resonance angiography; N = normal; NA = not available; SAH = subarachnoid hemorrhage.
Table 1.
Demographics of patients with unruptured aneurysms or SAH
Standard protocol approvals, registrations, and patient consents
The Mayo Clinic Institutional Review Board approved this study (No. 08-003878), and written informed consent was obtained from all individuals before their participation in the study.
Exome sequencing
Genomic DNA was isolated from available individuals from peripheral blood. Exome sequencing was performed with the Sure Select V4 + UTR exome capture kit (Agilent Technologies, Santa Clara, CA) using 2.1 µg genomic DNA following the manufacturer's standard protocol. The Mayo Clinic Bioinformatics Core (Rochester, MN) performed sequencing on a HiSeq 2000 sequencer (Illumina, San Diego, CA). Alignment and base calling were performed by the Mayo Clinic Bioinformatics Core using custom-built pipelines. Single nucleotide polymorphism (SNP) annotation was performed with SNP & Variation Suite 8.4.2 (Golden Helix Inc, Bozeman, MT).14 Variants were numbered according to standard nomenclature (hgvs.org/mutnomen/)15 based on RefSeq NM_006031.5 and NP_006022.3 accession numbers.
Variant selection criteria
Analysis and selection criteria were applied with SNP & Variation Suite version 8.4.2 using the combined vcf files from each family. Low-quality variants were excluded from the analysis by converting them into unknown genotypes if the sample genotype quality was <20 or the read depth was <10. SNPs with a minor allele frequency (MAF) >1% in the Exome Sequencing Project 6500, in the 1,000 Genomes database, or in the non-Finnish European population from the Genome Aggregation Database (gnomAD) were removed from further analysis. Variants present in affected patients and absent in familial healthy individuals were selected for further follow-up. Variants not fulfilling the following criteria were removed from the analysis: missense mutations, coding deletions or insertions, frameshift mutations, or intronic and synonymous variants near splice sites (≤5 nucleotides).
Candidate variant sequencing and cosegregation
Candidate variants were validated in all individuals from both families by direct Sanger sequencing to confirm a true call and to check cosegregation with disease in the family. MAFs for non-Finnish European population from the gnomAD are provided for candidate variants (data available from Dryad, tables e-1 and e-2, doi.org/10.5061/dryad.8br9852). Bidirectional sequencing of PCNT exon 38 was performed in the 164 samples from our cohort with specific primers designed with Primer316 (data available from Dryad, table e-3), and sequences were aligned and analyzed with SeqScape version 2.5 software (Applied Biosystems, Foster City, CA).
Family 7019 and 8159 haplotype analysis
Haplotype analysis was performed by analyzing 5 polymorphic microsatellites covering a 4.72-cM interval (data available from Dryad, table e-4, doi.org/10.5061/dryad.8br9852) at chromosome 21q22.3, in which the PCNT gene is located, to check whether the p.V2811L variant shared by families 7019 and 8159 had a common ancestor. One of the microsatellites (PCNT) was designed by us within a PCNT gene repeat region. To estimate the allele frequencies of this custom microsatellite in the general population, we screened a series of 80 white samples (data available from Dryad, table e-5). Microsatellite electropherograms were analyzed with GeneMapper version 4.0 (Applied Biosystems). Further analysis was performed with Pedcheck software17 to remove mendelian inconsistencies and genotyping errors and with Simwalk2 v.2.9118 to reconstruct haplotypes based on the microsatellite data.
Neuroimaging protocol
Imaging modalities used to identify aneurysms included cerebral angiography, MRA, and CTA. MRA was performed both with and without gadolinium contrast enhancement. All probands were diagnosed with cerebral angiography. Consenting relatives of the index patients were screened with MRA or CTA. The majority of studies were performed at our institution. Those studies were initially interpreted by a neuroradiologist as part of a clinical workup and then reviewed independently by a second neuroradiologist. Some patients were imaged at outside institutions. For those imaging studies, the outside report was evaluated when available, and the images were independently reviewed by an experienced neuroradiologist at our institution.
All MRAs were obtained on 1.5T (or occasionally 3T) Siemens MRI scanners (Siemens Medical Solutions, Malvern, PA). The algorithms used were the standard algorithms used for clinical imaging. When feasible, MRA with contrast directed at the circle of Willis was performed. Patients with contraindications to gadolinium administration received noncontrast MRA studies. In these cases, 3-dimensional time-of-flight technique was used in lieu of a contrast study. Both source images and maximum-intensity angiographic projections of the MRA images were generated in all cases. The maximum-intensity projection images were generated with the standard reconstruction algorithms available on the specific machine that performed the study. Outside MRA examinations were mostly noncontrast MRAs, with a few contrast-enhanced examinations.
All CTAs obtained at our institution were performed on the Siemens FLASH high-speed CT scanners. The images were processed with the standard clinical software available on each machine. Most CTAs were performed with 100 mL iodinated contrast, although the protocol did allow a decreased dose in the setting of renal insufficiency. Three-dimensional reconstructions of the data were performed by using preset volume-rendered and maximum-intensity projection display algorithms. In addition to the clinical interpretation, the source images and multiplanar reformatted images were reviewed independently by an experienced neuroradiologist.
Kidney function and morphology
Kidney function was assessed by serum creatinine concentration and glomerular filtration rate estimated with the Modification of Diet in Renal Disease equation, which, in addition to creatinine, uses age, race, sex, serum concentrations of urea nitrogen, and albumin.19 All available CT or ultrasonographic images of the kidneys obtained as part of routine clinical care were reviewed for the presence of cysts.
Data availability
Deidentified exome sequencing data from the studied families will be made available for other researchers on request and deposited in the Database of Genotypes and Phenotypes. Variants identified of interest will be submitted to ClinVar and appropriate variant databases.
Results
Exome sequencing of candidate families
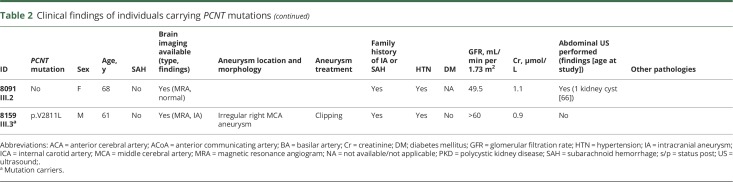
Through the MCFDR, we identified 3 families with a history of IA or SAH (figure 1A). Table 2 provides detailed clinical phenotypes for each family. Four individuals from family 7019 were sent for exome sequencing: 2 sisters (III.2 and III.4) who had an SAH at the age of 78 and 55 years and 2 healthy children (7019 IV.1 and 7019 IV.2) from sister 7019 III.2. Family 7042 consists of 4 individuals (both parents and 2 children). The mother (7042 I.1) carried an unruptured IA and had multiple vascular abnormalities. Her son (7042 II.1) had a massive SAH at the age of 40 years secondary to the rupture of an IA. Exome sequencing was performed in 5 individuals from family 7064: 1 male patient with an SAH (7064 II.5), his healthy daughter (7064 III.4), 2 healthy sisters (7064 II.3 and II.4), and a niece (7064 III.1), whose mother (7064 II.2) had been diagnosed with an unruptured IA. Unfortunately, DNA from 7064 II.2 was not available for analysis.
Table 2.
Clinical findings of individuals carrying PCNT mutations
Variants fulfilling selection criteria in the 3 families were further analyzed for validation and cosegregation (data available from Dryad, tables e-1 and e-2, doi.org/10.5061/dryad.8br9852). The mother (I.1) and her affected son (II.1) from family 7042 shared 118 variants, but taking into account the variants present in the father (individual I.2) and in the healthy son (individual II.2), the number of variants could be reduced to 55. After Sanger sequencing, we ended up with 53 confirmed variants that were shared by affected members of the family (data available from Dryad, table e-1). The exome sequencing analysis in family 7019 revealed that the 2 sisters (III.2 and III.4) shared 83 variants, thus meeting selection criteria. This number could be reduced to 20 variants when healthy individuals IV.1 and IV.2 were added to the analysis to rule out pathogenic variants (data available from Dryad, table e-2). DNA was also available for an additional paternal cousin from the 2 affected siblings. Variants shared by patients of families 7042 and 7019 but present in healthy individuals from these families are also provided (data available from Dryad, tables e-7 and e-8, respectively). Cosegregation analyses in family 7064 were unable to be assessed because we had only a single patient (II.5) with a definite diagnosis of SAH. This patient carried 347 candidate variants that met the variant filtering criteria. No rare variants were identified in the ANGPTL6 gene in any of the studied families.
When analyzing the 3 families, we observed only 2 genes with a variant shared by patients in >1 family. A variant in the KIF20B gene was shared by patients in family 7042 (p.I1121M), and another variant in the same gene was present in patient 7064 II.5 (p.S215N). The KIF20B gene is a kinesin-related gene involved in regulating the polarization of migrating neurons.20 Two different rare variants in the PCNT gene were also shared between affected members in families 7019 and 7042. Deletions and frameshift mutations in the PCNT gene cause microcephalic osteodysplastic primordial dwarfism type II (MOPD-II, Mendelian Inheritance in Man No. 605925) in an autosomal recessive manner. Up to 20% of patients with this disorder develop single or multiple IAs. Therefore, the PCNT gene is a plausible candidate gene from a biological perspective. Family 7042 patients I.1 and II.1 carry the PCNT p.R2728C variant. This mutation was absent in healthy individuals I.2 and II.2. Family 7019 patients III.2, III.4, and III.6 shared a missense PCNT p.V2811L variant in the exon 38 of the gene. This mutation was absent in healthy individuals IV.1 and IV.2. Healthy individual IV.1 underwent screening for IA with MRA and screening for renal and hepatic cysts with abdominal ultrasound and showed no IAs. Individual IV.2 refused any imaging studies. These 2 families carry a different missense variant in exon 38 of the PCNT gene. The mean coverage for all exonic regions for the PCNT gene was 44.93 reads, and the mean coverage for exon 38 was 31.71 reads. Details about the coverage for each single sample for the entire gene and for exon 38 are provided in table e-6 available from Dryad (doi.org/10.5061/dryad.8br9852).
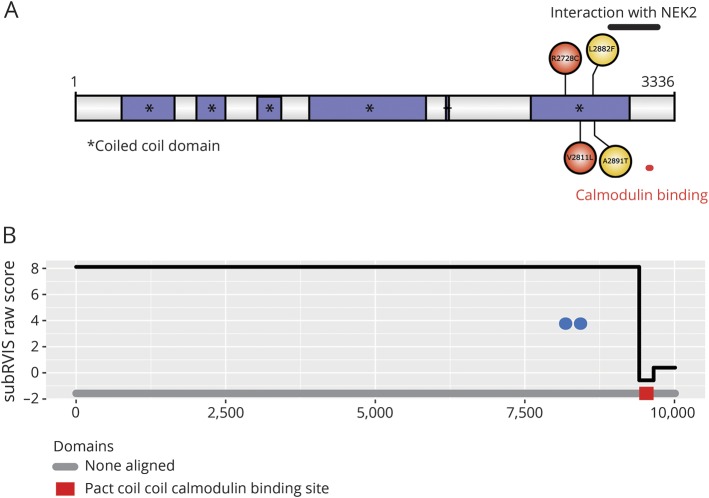
The PCNT p.R2728C and p.V2811L substitutions have an MAF of 0.00009497 and 0.0007343, respectively, in the non-Finnish European population from the gnomAD (table 3), which suggests that they are rare variants in the general population. In addition, both variants are located in exon 38, which encodes part of the protein-protein interaction domain of the PCNT protein (figure 2A). Both mutations are located in the coiled coil domain that forms part of the highly conserved pericentrin-AKAP-450 centrosomal targeting (PACT) domain of the protein, which serves to bind the PCNT protein to the centrosome/spindle pole (in mitosis).21 The subregion residual variation intolerance score is an algorithm that calculates the degree of intolerance (the lower the score, the more intolerant) of a certain gene subregion (either an exon or protein domain) in which a variant is located.22 According to the subregion residual variation intolerance score, any variant located in the calmodulin binding site of the PACT domain (−0.57) would likely be pathogenic (figure 2B). However, the protein-protein interaction domain extends outside this calmodulin binding site (see NEK2 interaction region in figure 2A), and it is possible that mutations in the entire protein-protein interaction domain of PCNT could be potentially pathogenic.
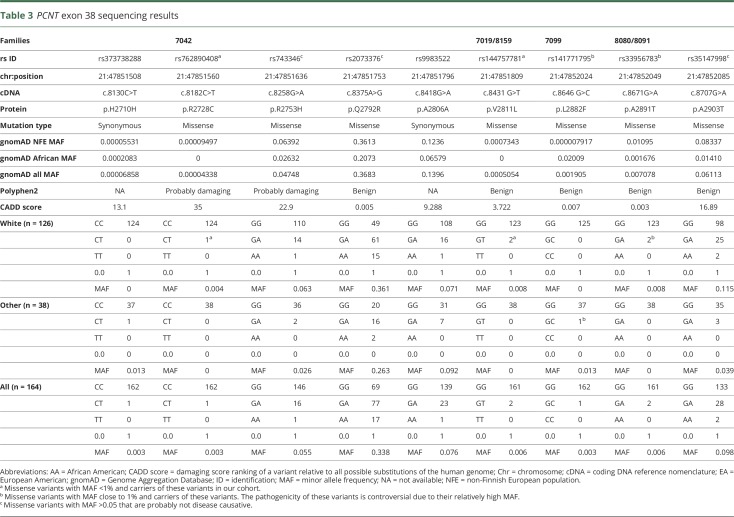
Table 3.
PCNT exon 38 sequencing results
Figure 2. PCNT protein structure.
(A) Graphic representation of the PCNT protein with its multiple coiled coil domains and the location of the 4 missense variants identified in the present study. Red circles = mutations shown to cosegregate within the families; yellow circles = mutations found in sporadic patients or cosegregation analysis was not performed due to lack of DNA of affected relatives. This figure was designed with the Uniprot database.39 (B) Distribution of the 2 variants in the PCNT gene that cosegregated within the families. The y-axis represents the domain subregion residual variation intolerance scores. Each variant is depicted with a blue circle. NEK = NIMA (never in mitosis gene A)–related kinase; Pact = pericentrin-AKAP-450 centrosomal targeting; subRVIS = subregion residual variation intolerance score.
The imaging studies available in the 2 family members showed that affected patients had both IA/SAH and multiple kidney cysts (table 2). Family 7019 patient III.2 was a woman who presented at the age of 78 years with an SAH secondary to a 4.7-mm ruptured aneurysm in a thalamoperforator branch arising from the right posterior cerebral artery (data available from Dryad, figure e-1, doi.org/10.5061/dryad.8br9852). She died 2 weeks later of several complications, including severe hydrocephalus and perioperative stroke after glue embolization of the aneurysm. No abdominal imaging was available on her for evaluation of the presence of cysts in abdominal organs. Her sister (patient III.4) also had an SAH at the age of 55 years. In addition, she had a history of renal and hepatic cysts identified incidentally on abdominal CT when she was 64 years old. Family 7042 patient II.1, a white man, presented at the age of 40 with a gradually worsening headache 2 days after sustaining mild head trauma. Brain imaging showed a massive SAH (data available from Dryad, figure e-2A) secondary to the rupture of a 4.5-mm aneurysm in the anterior communicating artery (data available from Dryad, figure e-2B and e-2C), which was subsequently treated with endovascular coiling. In addition, he had 2 cysts in the upper and lower poles of the left kidney (data available from Dryad, figure e-2D–e-2F). His mother, patient I.1, had a history of a small unruptured IA. In addition, she had undergone surgical repair of an abdominal aortic aneurysm and bilateral femoral aneurysms in her 50s. Further information on the presence of any cystic lesions in her kidneys or liver was not available.
PCNT exon 38 sanger sequencing
Because of the presence of 2 different missense mutations (p.R2728C and p.V2811L) in PCNT exon 38 that were shared between affected members in 2 different families with SAH, we decided to sequence this exon, which encodes the protein-protein interaction domain of PCNT, in the complete series of patients harboring an IA or with history of SAH from the MCFCDR.
One hundred sixty-four samples with either SAH or IA, including the probands from the 3 families, were Sanger sequenced for PCNT exon 38. We found 9 coding mutations, 7 of which were missense mutations and 2 that were synonymous mutations. From the missense mutations, 3 had a high MAF (>5%; p.R2753H, p.Q2792R, and p.A2903T), 2 had an MAF close to 1% (p.L2882F and p.A2891T), and 2 others had an MAF <1% (p.R2728C and p.V2811L) according to gnomAD MAF when considering the ethnicities of the families carrying the variants (table 3). Sanger sequencing confirmed that variant p.R2728C was carried by family 7042 patients and that variant p.V2811L was carried by family 7019 patients. In addition, we found a second carrier of variant p.V2811L in a family (8159) with a large family history of both ischemic and hemorrhagic stroke (figure 1B).
We were not able to establish the potential pathogenicity of the 2 variants with MAF close to 1% because 2 of the patients carrying these variants were sporadic cases (patients 7099:II.2 and 8080:II.2), and we had no DNA on additional patients with SAH or IA carriers from family 8091. Furthermore, the MAF of these mutations (≈1%) makes them less likely to be pathogenic.
Clinical description of additional mutation carriers
We found 4 additional probands carrying at least 1 PCNT missense mutation (see data available from Dryad material for detailed clinical data on these families, doi.org/10.5061/dryad.8br9852). Patient II:2 from family 7099 (figure 1B) is a 42-year-old black female who had a diffuse SAH secondary to the rupture of a 3.4-mm right internal carotid artery terminus aneurysm (data available from Dryad, figure e-3). She carries a PCNT p.L2882F substitution, and exome sequencing identified 2 additional rare missense variants in the PCNT gene: resulting in p.D859N and p.S1985F substitutions (data available from Dryad, table e-9), but additional DNA on relatives was not available, so it was not possible to perform segregation analyses in this family for the 3 PCNT variants. Patient II:2 from family 8080 (figure 1B) is a 60-year-old white man diagnosed with a large wide-neck basilar tip aneurysm that required multiple interventions (data available from Dryad, figure e-4) and a right middle cerebral artery bifurcation aneurysm. This patient carries the PCNT p.A2891T substitution, which has an MAF of ≈1% in general population. Patient III:1 from family 8091 (figure 1B) is a 71-year-old woman with a 7-mm left paraclinoid aneurysm (data available from Dryad, figure e-5), a left internal carotid artery stenosis, a left subclavian stenosis, a right internal carotid artery stenosis, and a small bulge on the basilar tip suggestive of a small aneurysm. This patient carries the PCNT p.A2891T substitution that was also present in family 8080, which has an MAF of ≈1% in general population. Patient III-3 from family 8159 (figure 1B) is a 61-year-old white man with a right middle cerebral artery aneurysm that was 8 to 9 mm in its greatest diameter with surface irregularities (data available from Dryad, figure e-6) and some calcifications and mild stenosis of his carotid arteries in the cervical region. This patient carries the same missense PCNT p.V2811L substitution that is shared by the 2 affected members from family 7019. The haplotype reconstruction between the 2 patients of family 7019 and the proband of family 8159 (data available from Dryad, figure e-7) showed a potential common ancestor with allele sharing of 4 microsatellites surrounding and within the PCNT gene (D21S1903, D21S1897, PCNT, and D21S1446).
Discussion
It is becoming clearer that mutations in the same genes that cause rare young-onset disorders can predispose individuals to more common disorders usually with milder phenotypes and later presentations in adults.23,24 It is also established that heterozygous carriers of mutations in recessive disorders may be at increased risk of disease or complex diseases phenotypes. Biallelic PCNT deletion and frameshift mutations cause MOPD-II, an autosomal recessive disorder characterized by severe prenatal and postnatal growth retardation, marked microcephaly, a characteristic skeletal dysplasia, craniofacial dysmorphism, and small teeth.25 Approximately 20% to 50% of these patients develop cerebral neurovascular abnormalities, including moyamoya angiopathy or IAs; aneurysmal SAH is a significant cause of death in patients with MOPD-II.26–28
In the present study, we exome sequenced 3 multigenerational families with either SAH or IA and found 2 different substitutions (p.R2728C and p.V2811L) in the PCNT gene (exon 38) that were shared between affected members in 2 independent families. Sanger sequencing of PCNT exon 38 in 161 additional patients with either SAH or IA revealed the presence of p.V2811L mutation in a patient with familial IA. Furthermore, we identified 2 additional missense mutations (resulting in p.L2882F and p.A2891T substitutions) in PCNT exon 38 (MAF of ≈1% in ethnicity-matched control population) in 3 patients with sporadic SAH/IA (figure 1B and table 3). We observed rare variants in our families, and it is worth noting that although several genome-wide association studies have linked specific chromosomal loci with an increased risk of developing IA,9–12 common variation in the PCNT gene has not been nominated to date.
The PCNT gene encodes multiple processed transcripts and at least 2 different coding proteins (data available from Dryad, figure e-8, doi.org/10.5061/dryad.8br9852): a fully functional large isoform (PCNT-001; ENST00000359568) encoded by 47 exons and a small protein (PCNT-007; ENST00000418394) with unknown functionality encoded by PCNT exons 40 to 44 and 47. Both isoforms contain the PF-10495 domain, which is a coiled coil region close to the C-terminal part of the protein involved in the recruitment of AKAP-450 and PCNT itself to the centrosome. Thus, even if a copy of the full-length functional protein is damaged, it is still possible that PCNT-007 could rescue some of the centrosome-related functions of PCNT-001 protein isoform. This hypothesis is supported by studies in mice showing that there are multiple PCNT transcripts, including even kidney-specific PCNT isoforms.29
Similar findings have been reported in human cells. In fact, a recent study showed that several human cell cultures showed different expression levels of different PCNT isoforms, suggesting unique PCNT expression patterns for every human tissue.30 This would support the hypothesis that aneurysm development and severity could depend on the type and location of PCNT mutations. The variants in our families lie in the coiled coil domain that constitutes part of the PCNT protein-protein interaction domain. This PACT domain, which is highly conserved across species (data available from Dryad, figures e-9 and e-10, doi.org/10.5061/dryad.8br9852),31 is located in the C-terminus of PCNT and the final 22 residues of this domain seem to be a specific calmodulin-binding site (figure 2B).21 While the mutations that we have found are located in the interaction domain of PCNT, it remains to be elucidated whether variants in other domains of the protein could disrupt the protein functionality and cause IA.
The PCNT protein is essential for centrosome maturation, the process of building centrosomes/spindle poles. It is directly involved in microtubule formation and organization throughout the cell cycle and is essential for spindle formation, centrosome assembly/function, and cell division. PCNT binds the cilia protein PKD2 and is essential for the formation of the primary cilium.32 Moreover, a specific set of essential centrosomal proteins (CEP215, ninein, and centriolin) have an absolute requirement for PCNT for their recruitment to centrosomes/spindle poles and for orientation of the mitotic spindle.33,34 Recently, a group developed a complete Pcnt knockout mouse model (Pcnt−/−)33 to study MOPD-II abnormalities in an animal model. The Pcnt−/− model showed not only a smaller size than control mice, as expected, but also general vascular abnormalities, including intracranial hemorrhage, and cystic and duplicated kidneys.33,35
It is worthwhile noting that several patients in our families also have kidney cysts, but there is not enough evidence to suggest that PCNT mutations are involved in the development of kidney cysts, and this finding may be coincidental. Moreover, although cystic kidneys have been seen in the Pcnt−/− mouse model, they are not a common feature of patients with MOPD-II. Autosomal dominant polycystic kidney disease (ADPKD), due to mutations in the PKD2 gene, is characterized by severe kidney dysfunction and an increased risk of developing IA.36 According to a recent meta-analysis, patients with ADPKD have a prevalence ratio of 6.9 compared with general population of developing IAs.3 Several affected members having SAH or unruptured aneurysms also carried kidney cysts (7019 III.4, 7019 III.6, 7042 II.1, 7099 II.2, 8080 II.2, and 8091 III.2). Three patients (7099 II.2, 8080 II.2, and 8091 III.1) who were sequenced for PCNT exon 38 also underwent exome sequencing to rule out variants in other genes causing polycystic kidney disease (PKD) because the PCNT missense mutations they carried had an MAF in their respective ethnic specific populations of 1%. In fact, patient 8080 II.2 had a diagnosis of PKD, and by exome sequencing, we identified a PKD1 mutation (p.Q4004*), which has been described before to cause ADPKD in a Chinese family.37 In addition, patient 8091 III.1 carried the PKD1 p.S1352N missense variant, a variant of uncertain significance. The presence of these PKD1 mutations makes the interpretation of the renal cysts in our patients even more difficult. Therefore, the relation between PCNT mutations and cysts development is only suggestive. However, given the fact that patients 8080 II.2 (data available from Dryad, figure e-4, doi.org/10.5061/dryad.8br9852) and 8091 III.1 (data available from Dryad, figure e-5) had severe clinical phenotypes, an additive effect of carrying mutations in both the PCNT and PKD1 genes cannot be ruled out.
The fact that biallelic deletion and frameshift mutations in the PCNT gene have already been involved in the development of IA makes it plausible that point mutations in this gene could play a role in the development of IA, similar to patients with MOPD-II. To assess whether the relatives of patients with MOPD-II have an increased risk of carrying IA or having SAH, we reviewed the clinical features of the relatives of one of the largest clinical cohorts of patients with MOPD-II.38 However, these individuals had not been screened for IA or asked specifically for the occurrence of SAH. The cohort includes 42 patients with a clinical diagnosis of MOPD-II; 35 of those individuals have a molecular diagnosis. Family history is available on 26 parents and 48 grandparents from 17 molecularly confirmed patients. We identified 3 relatives with history of heart attack, 4 relatives with history of stroke of unknown etiology, 3 relatives with congenital heart disease, and 1 individual with an anomalous pulmonary artery.
The identification of PCNT mutations in familial IA is intriguing, and further study is necessary to examine its role in neurovascular disease. Future association studies with large sample sizes will have to evaluate whether other mutations in the PCNT gene also increase the risk of developing either IA or SAH. A potential caveat of our study is that we were not able to rule out the KIF20B variants (p.I1121M and p.S215N) present in family 7042 and the proband of family 7064, respectively. Another potential caveat of our methodology is that we included the healthy brother II.2 from family 7042 in the analysis to rule out potential pathogenic variants, even though he can be still at risk of developing an IA. Ultimately, the possibility of identifying and screening for putative pathogenic variants in this gene and others would allow clinicians to follow up these patients more closely so that an IA can be detected in the early stages with periodic neurovascular imaging studies. If these patients are diagnosed before IA rupture, their life expectancy could be increased through earlier intervention, thus diminishing the risk of an acute presentation.
Acknowledgment
The authors thank the patients and families who donated DNA samples for this work, including those participants of the MCFCDR.
Glossary
- ADPKD
autosomal dominant polycystic kidney disease
- CTA
CT angiography
- gnomAD
Genome Aggregation Database
- IA
intracranial aneurysm
- MAF
minor allele frequency
- MCFCDR
Mayo Clinic Familial Cerebrovascular Diseases Registry
- MOPD-II
microcephalic osteodysplastic primordial dwarfism type II
- MRA
magnetic resonance angiography
- PACT
pericentrin-AKAP-450 centrosomal targeting
- SAH
subarachnoid hemorrhage
- SNP
single nucleotide polymorphism
Author contributions
Dr. Oswaldo Lorenzo-Betancor: study concept and design, analysis and interpretation, drafting and critical revision of the manuscript for important intellectual content. Dr. Patrick R. Blackburn: analysis and interpretation, drafting and critical revision of the manuscript for important intellectual content. Emily Edwards and Rocío Vázquez-do-Campo: acquisition of data. Eric W. Klee and Dr. Catherine Labbé: critical revision of the manuscript for important intellectual content. Kyndall Hodges and Patrick Glover: analysis and interpretation. Ashley N. Sigafoos: acquisition of data. Alexandra I. Soto and Ronald L. Walton: analysis and interpretation. Dr. Stephen Doxsey and Dr. Michael B. Bober: critical revision of the manuscript for important intellectual content. Sarah Jennings: analysis and interpretation. Dr. Karl J. Clark: critical revision of the manuscript for important intellectual content. Dr. Yan Asmann: analysis and interpretation. David Miller and William D. Freeman: acquisition of data and critical revision of the manuscript for important intellectual content. James Meschia: acquisition of data, critical revision of the manuscript for important intellectual content, and study supervision. Dr. Owen A. Ross: study concept and design, acquisition of data, critical revision of the manuscript for important intellectual content, and study supervision.
Study funding
The MCFCDR received funds from the Mayo Foundation for Medical Education and Research, and work was supported in part by the Randall S. and Friedgard D. Acree, James and Esther King Biomedical Research Program, American Heart Association, Mayo Clinic Office of Health Disparities Research, Mayo Clinic Florida Neuroscience Focused Research Team and Center for Individualized Medicine, Myron and Jane Hanley Award for Stroke Research, and Joe Niekro Foundation. The Primordial Dwarfism Registry at the A.I. duPont Hospital for Children was supported by the Potentials Foundation and the Walking With Giants Foundation.
Disclosure
O. Lorenzo-Betancor was supported in part by a postdoctoral fellowship award from the Department of Veterans Affairs. P. Blackburn, E. Edwards, R. Vázquez-do-Campo, E. Klee, C. Labbé, K. Hodges, P. Glover, A. Sigafoos, A. Soto, R. Walton, S. Doxsey, M. Bober, S. Jennings, K. Clark, Y. Asmann, D. Miller, W. Freeman, and J. Meschia report no disclosures relevant to the manuscript. O. Ross received support from R01-NS078086, P50-NS072187 and U54 NS100693 and the Michael J. Fox Foundation. O.A.R. is an editorial board member of American Journal of Neurodegenerative Disease and Molecular Neurodegeneration. Go to Neurology.org/N for full disclosures.
References
- 1.Brown RD. Unruptured intracranial aneurysms. Semin Neurol 2010;30:537–544. [DOI] [PubMed] [Google Scholar]
- 2.van Gijn J, Kerr RS, Rinkel GJ. Subarachnoid haemorrhage. Lancet 2007;369:306–318. [DOI] [PubMed] [Google Scholar]
- 3.Vlak MH, Algra A, Brandenburg R, Rinkel GJ. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol 2011;10:626–636. [DOI] [PubMed] [Google Scholar]
- 4.Vernooij MW, Ikram MA, Tanghe HL, et al. Incidental findings on brain MRI in the general population. N Engl J Med 2007;357:1821–1828. [DOI] [PubMed] [Google Scholar]
- 5.Mackey J, Brown RD Jr, Moomaw CJ, et al. Unruptured intracranial aneurysms in the Familial Intracranial Aneurysm and International Study of Unruptured Intracranial Aneurysms cohorts: differences in multiplicity and location. J Neurosurg 2012;117:60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Global, regional, and national burden of neurological disorders during 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol 2017;16:877–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodel R, Winter Y, Ringel F, et al. Cost of illness in subarachnoid hemorrhage: a German Longitudinal Study. Stroke 2010;41:2918–2923. [DOI] [PubMed] [Google Scholar]
- 8.Rinkel GJ, Djibuti M, Algra A, van Gijn J. Prevalence and risk of rupture of intracranial aneurysms: a systematic review. Stroke 1998;29:251–256. [DOI] [PubMed] [Google Scholar]
- 9.Foroud T, Koller DL, Lai D, et al. Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke 2012;43:2846–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foroud T, Lai D, Koller D, et al. Genome-wide association study of intracranial aneurysm identifies a new association on chromosome 7. Stroke 2014;45:3194–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foroud T, Sauerbeck L, Brown R, et al. Genome screen in familial intracranial aneurysm. BMC Med Genet 2009;10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foroud T, Sauerbeck L, Brown R, et al. Genome screen to detect linkage to intracranial aneurysm susceptibility genes: the Familial Intracranial Aneurysm (FIA) Study. Stroke 2008;39:1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourcier R, Le Scouarnec S, Bonnaud S, et al. Rare coding variants in ANGPTL6 are associated with familial forms of intracranial aneurysm. Am J Hum Genet 2018;102:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SNP & Variation Suite (Version 8.4.2) [Software]. Bozeman, MT: Golden Helix, Inc. Available from http://www.goldenhelix.com. [Google Scholar]
- 15.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet 2001;109:121–124. [DOI] [PubMed] [Google Scholar]
- 16.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000;132:365–386. [DOI] [PubMed] [Google Scholar]
- 17.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 19.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation: Modification of Diet in Renal Disease Study Group. Ann Intern Med 1999;130:461–470. [DOI] [PubMed] [Google Scholar]
- 20.Sapir T, Levy T, Sakakibara A, Rabinkov A, Miyata T, Reiner O. Shootin1 acts in concert with KIF20B to promote polarization of migrating neurons. J Neurosci 2013;33:11932–11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep 2000;1:524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol 2016;17:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet 2001;69:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010;11:415–425. [DOI] [PubMed] [Google Scholar]
- 25.Bober MB, Jackson AP. Microcephalic osteodysplastic primordial dwarfism, type II: a clinical review. Curr Osteoporos Rep 2017;15:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bober MB, Khan N, Kaplan J, et al. Majewski osteodysplastic primordial dwarfism type II (MOPD II): expanding the vascular phenotype. Am J Med Genet A 2010;152A:960–965. [DOI] [PubMed] [Google Scholar]
- 27.Brancati F, Castori M, Mingarelli R, Dallapiccola B. Majewski osteodysplastic primordial dwarfism type II (MOPD II) complicated by stroke: clinical report and review of cerebral vascular anomalies. Am J Med Genet A 2005;139:212–215. [DOI] [PubMed] [Google Scholar]
- 28.D'Angelo VA, Ceddia AM, Zelante L, Florio FP. Multiple intracranial aneurysms in a patient with Seckel syndrome. Childs Nerv Syst 1998;14:82–84. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi K, Asanuma M, Miyazaki I, Matsuzaki S, Tohyama M, Ogawa N. Characterization of pericentrin isoforms in vivo. Biochem Biophys Res Commun 2006;351:745–749. [DOI] [PubMed] [Google Scholar]
- 30.Muhlhans J, Giessl A. Pericentrin in health and disease: exploring the patchwork of Pericentrin splice variants. Commun Integr Biol 2012;5:304–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 1994;76:639–650. [DOI] [PubMed] [Google Scholar]
- 32.Jurczyk A, Gromley A, Redick S, et al. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol 2004;166:637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CT, Hehnly H, Yu Q, et al. A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr Biol 2014;24:2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S, Rhee K. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One 2014;9:e87016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang SJ, Yang Q, Meigs JB, Pearce EN, Fox CS. A genome-wide association for kidney function and endocrine-related traits in the NHLBI's Framingham Heart Study. BMC Med Genet 2007;8(suppl 1):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrone RD, Malek AM, Watnick T. Vascular complications in autosomal dominant polycystic kidney disease. Nat Rev Nephrol 2015;11:589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao DX, Cao QW, Ding KJ, et al. An analysis for the phenotype and genotype of autosomal dominant polycystic kidney disease from two Chinese families [in Chinese]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2006;23:23–26. [PubMed] [Google Scholar]
- 38.Bober MB, Niiler T, Duker AL, et al. Growth in individuals with Majewski osteodysplastic primordial dwarfism type II caused by pericentrin mutations. Am J Med Genet A 2012;158A:2719–2725. [DOI] [PubMed] [Google Scholar]
- 39.UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017;45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Deidentified exome sequencing data from the studied families will be made available for other researchers on request and deposited in the Database of Genotypes and Phenotypes. Variants identified of interest will be submitted to ClinVar and appropriate variant databases.