Abstract
Background
Cancer stem cells (CSCs) possess abilities of self-renewal and differentiation, have oncogenic potential and are regarded to be the source of cancer recurrence. However, the mechanism by which CSCs maintain their stemness remains largely unclear.
Methods
In this study, the cell line-derived ovarian CSCs (OCSCs), 3AO and Caov3, were enriched in serum-free medium (SFM). Differentially expressed proteins were compared between the OCSC subpopulation and parental cells using liquid chromatography (LC)-mass spectrometry (MS)/MS label-free quantitative proteomics. Sphere-forming ability assays, flow cytometry, quantitative real-time polymerase chain reaction (qPCR), western blotting, and in vivo xenograft experiments were performed to evaluate stemness. RNA-sequencing (RNA-seq) and pyrosequencing were used to reveal the mechanism by which STON2 negatively modulates the stem-like properties of ovarian cancer cells.
Results
Among the 74 most differentially expressed proteins, stonin 2 (STON2) was confirmed to be down-regulated in the OCSC subpopulation. We show that STON2 negatively modulates the stem-like properties of ovarian cancer cells, which are characterized by sphere formation, a CD44+CD24− ratio, and by CSC- and epithelial mesenchymal transition (EMT)-related markers. STON2 knockdown also accelerated tumorigenesis in NOD/SCID mice. Further investigation revealed a downstream target, mucin 1 (MUC1), as up-regulated upon the down regulation of STON2. A decrease in both DNA methyltransferase 1 (DNMT1) expression and methylation in the promoter region of MUC1 was associated with subsequently elevated MUC1 expression, as detected in STON2 knockdown in 3AO and Caov3 cells. Direct DNMT1 knockdown simultaneously elevated MUC1 expression. The functional significance of this STON2-DNMT1/MUC1 pathway is supported by the observation that STON2 overexpression suppresses MUC1-induced sphere formation of OCSCs. The paired expression of STON2 and MUC1 in ovarian cancer specimens was also detected revealing the prognostic value of STON2 expression to be highly dependent on MUC1 expression.
Conclusions
Our results imply that STON2 may negatively regulate stemness in ovarian cancer cells via DNMT1-MUC1 mediated epigenetic modification. STON2 is therefore involved in OCSC biology and may represent a therapeutic target for innovative treatments aimed at ovarian cancer eradication.
Electronic supplementary material
The online version of this article (10.1186/s13046-018-0977-y) contains supplementary material, which is available to authorized users.
Keywords: Ovarian cancer, Cancer stem cell, STON2, MUC1, DNMT1
Background
Ovarian cancer represents the most lethal of all gynecological neoplasms. The American Cancer Society predicts that 22,440 women will be diagnosed with ovarian cancer and 14,070 women will die from the disease in 2018. Cancer statistics gathered over the period from 2006 to 2012 suggest that the overall 5-year survival rate of ovarian cancer patients is 47%. However, in advanced stage patients this value drops to 29% [1]. Although 70% patients with ovarian cancer respond to first-line chemotherapy, most of them ultimately suffer from recurrence and metastasis [2]. It is noteworthy that these survival statistics have hardly changed over time, despite the introduction of platinum-based treatment more than 30 years ago [3]. Emerging evidence has confirmed that a small population of tumor cells with stemness properties, now known as CSCs or cancer stem-like cells (CSLCs), occur in cancer tissues [4]. These cells possess abilities of self-renewal and differentiation, have strong oncogenic potential and are considered as the source of cancer recurrence [5]. Thus, CSC-targeted therapy is expected to considerably improve the prognosis of patients with ovarian cancer [6]. Unfortunately, identification and eradication of CSCs has proved to be far from optimal [7]. Persistent exploration of CSC characteristics may be beneficial for the development of novel CSC-targeting drugs.
Since these cells were first isolated, cloned, and propagated in vitro, OCSCs have been widely recognized by their stemness or progenitor-like properties, which include sphere formation, self-renewal and tumorigenic abilities [8]. Previously, we have successfully enriched the OCSC subpopulation using a sphere formation model and observed that CD44+CD24− cells are highly tumorigenic [9]. Furthermore, we verified that cyclin D1 affects EMT in OCSLCs [10], whereas in other studies, NANOG and c-MYC were reported to be involved in OCSC regulation and acted as cancer stem related-markers [11, 12]. To gain deeper insight into the molecular basis for OCSCs, we used LC-MS/MS label-free quantitative proteomics and bioinformatic analysis to identify the key factors that are differentially down-regulated in the OCSC subpopulation. STON2, an endocytic sorting adaptor [13], was of particular interest. STON2 knockdown promoted OCSC stemness.
Recent analysis in the Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed that OCSC-specific gene expressions are enriched in the endocytosis pathway [14]. Many of these genes were noted to be involved in key steps of endocytosis related to the resurrection, multidrug resistance, stemness maintenance of CSCs [15, 16]. Here, we present novel observations, which indicate that STON2 is involved in modulating stemness in ovarian cancer cells.
The oncogenic MUC1, a member of the class of epigenetically controlled genes, is a transmembrane protein that is aberrantly overexpressed and confers poor prognosis in a variety of cancers, including pancreatic, colorectal, breast, lung and ovarian cancer [17]. Increasing evidence has shown that MUC1 is also associated with the stemness of lung cancer [18] and breast cancer [19, 20]. High expression levels of MUC1 are well documented as correlated with metastasis, chemoresistance, and the survival of ovarian cancer cells [21, 22]. However, the regulatory mechanisms of MUC1 in ovarian cancer remain elusive.
In this study, using RNA-seq and gene function experiments, we identify that MUC1 acts as a downstream target for STON2, and modulates stem-like properties. Interestingly, MUC1 levels are elevated by CpG demethylation in cancer cells, where promoter methylation plays an important role in determining MUC1 expression [23, 24]. We provide evidence that STON2-regulated MUC1 expression might be mediated by DNMT1-induced methylation in the promoter region of MUC1. The higher expression of STON2 together with lower expression of MUC1 is associated with a favorable prognosis. STON2, therefore, is involved in OCSC biology and may represent a therapeutic target for innovative treatments aimed at ovarian cancer eradication.
Methods
Cell culture and sphere-forming ability assay
The human epithelial ovarian cancer cell line, Caov3, was obtained from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). The human ovarian adenocarcinoma cell line 3AO was acquired from the Women’s Hospital, School of Medicine, Zhejiang University, where it was tested and authenticated. It was not cultured continuously for more than 3 months. Adherent Caov3 cells were cultured in their standard medium as recommended by ATCC. 3AO cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (BI, Kibbutz Beit-Haemek, Israel), supplemented with 10% fetal bovine serum (FBS) (Invitrogen, New York, USA), maintained at 37 °C in 5% CO2 and detached using trypsin/EDTA solution. The anchorage-independent spheroids were generated from Caov3 or 3AO cells after plating, 5 × 104 cells per well, in ultra-low attachment 6-well culture plates (Corning, New York, USA) and culturing in a SFM composed of Dulbecco’s modified Eagle’s medium (DMEM)/F12 (BI, Kibbutz Beit-Haemek, Israel), 10 ng/mL basic fibroblast growth factor, 20 ng/mL epidermal growth factor (PeproTech, Rocky Hill, USA), 1 mg/mL insulin (Sigma-Aldrich, Burlington, MA, USA), 10 μL/mL B27 additive (Life Technologies, Carlsbad, California, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin. Fresh medium was added after every two or three days and the cultures were maintained at 37 °C in 5% CO2 for 7 days. During the passaging of spheroids, the cells were centrifuged and digested with trypsin, and cultured in SFM.
After plating 400 cells per well in ultra-low attachment 96-well culture plates (Corning, New York, USA), Caov3 or 3AO cells were cultured in a SFM composed of DMEM/F12 (BI, Kibbutz Beit-Haemek, Israel), 10 ng/mL basic fibroblast growth factor, 20 ng/mL epidermal growth factor (PeproTech, Rocky Hill, USA), 1 mg/mL insulin (Sigma-Aldrich, Burlington, MA, USA), 10 μL/mL B27 additive (Life Technologies, Carlsbad, California, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin. Fresh medium was added after every two or three days and the cultures were maintained at 37 °C in 5% CO2 for 7 days.
LC-MS/MS label-free quantitative proteomics and bioinformatic analysis
Parental and spheroids of 3AO cells (three independent samples in each group) were cracked in 200 μl lysis buffer, disrupted with agitation, boiled for 10 min, and centrifuged at 12000 rpm for 10 min. The supernatant was then collected and digested overnight at 37 °C with trypsin (Promega, Madison, Wisconsin, USA). The peptide of each sample was desalted on C18 cartridges (Sigma, Burlington, MA, USA) and then resuspended in 0.1% trifluoroacetic acid after centrifugation. MS analysis was performed on a Finnigan LTQ VELOS MS (ThermoFisher, Waltham, MA, USA). Each scan cycle included one full MS1 scan in profile mode and 20 MS2 scans in centroid mode. The peptides were investigated automatically according to the MS/MS spectra in the International Protein Index (IPI) human protein database using the Bioworks Browser software suite (Thermo, Waltham, MA, USA). The quantification of peptides was performed using the DeCyder differential analysis software (GE Healthcare, Munich, Germany).
Plasmids, siRNA transfection, and lentivirus infection
FLAG-tagged full length open reading frames (ORFs) of human STON2 (NM_033104.3) were ligated into a pcDNA3.1 vector (BioVector, Beijing, China) after HindIII and XhoI digestion. HA-tagged human MUC1 (NM_001204286.1) was cloned into a pcDNA3.1 vector (BioVector, Beijing, China) after HindIII and EcoRI digestion. The sequences of all the constructs were verified by DNA sequencing. The primer sequences are listed in Additional file 1: Table S1. Transfections were performed using X-tremeGENE HP DNA transfection reagent (Roche, Basel, Switzerland) following the manufacturer’s protocols.
The short-hairpin RNAs (shRNAs) for STON2 and MUC1 oligonucleotides were cloned into hU6-MCS-CMV-puromycin lentivirus expression vectors between the AgeI and EcoRI sites and then transfected into adherent cells according to instructions. After 72 h of transfection, the cells were selected using 5 μg/ml puromycin for 4 days. All construct sequences were confirmed using DNA sequencing. The shRNA oligonucleotides are listed in Additional file 2: Table S2.
Small-interfering RNAs (siRNAs) against STON2 and DNMT1, along with RNAi negative controls, were purchased from Genepharma (Shanghai, China). Cells were transfected with the siRNAs (50 nM) using Lipofectamine™ RNAiMAX (ThermoFisher, Waltham, MA, USA) following the manufacturer’s protocols. The siRNA sequences are listed in Additional file 2: Table S2.
RNA extraction and qPCR
Total RNA was extracted using an RNA extraction kit (TaKaRa, Dalian, China) and reverse-transcribed into cDNA using the reverse transcription cDNA kit (TAKAR, Dalian, China). PCR reactions were performed using SYBR® Premix Ex Taq™ (TaKaRa, Dalian, China) and applied biosystems 7900HT fast real-time PCR system (Life Technologies, Carlsbad, California, USA). The relative mRNA expression was calculated using the 2-∆∆Ct method and normalized to GAPDH expression. Primer sequences are shown in Additional file 1: Table S1.
Western blot analysis
Cells were lysed using a radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) supplemented with PMSF inhibitor (Beyotime, Shanghai, China). Protein lysates were loaded and separated on a 10% sodium dodecyl sulfate polyacrylamide gel and transferred onto 0.22-μm polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, California, USA). The membranes were then blocked with Tris buffered saline Tween 20 (TBST) containing 5% non-fat milk for 1 h, and probed with primary antibodies overnight at 4 °C. They were then washed thrice with TBST for 10 min each and probed with secondary antibodies for 1 h, followed by washing three times in TBST for 10 min per wash. The bands were visualized using an enhanced chemiluminescence (ECL) kit (ThermoFisher, Waltham, MA, USA) in Image quant LAS400 mini (GE Healthcare, Munich, Germany). Primary antibodies against STON2 (Santa, Dallas, Texas), MUC1 (ThermoFisher, Waltham, MA, USA), NANOG (Sigma, Burlington, MA, USA), c-MYC (CST, Danvers, MA, USA), DNMT1 (Abcam, Cambridge, MA, USA), DNMT3A (Abcam, Cambridge, MA, USA), DNMT3B (Abcam, Cambridge, MA, USA), E-cadherin (Abcam, Cambridge, MA, USA), N-cadherin (CST, Danvers, MA, USA), and fibronectin (Abcam, Cambridge, MA, USA) were used. GAPDH (Abcam, Cambridge, MA, USA) was the loading control.
Flow cytometry analysis
Spheroids were centrifuged and digested with trypsin, after which 1 × 105 cells were counted and resuspended in 100 μl PBS, stained with either anti-CD44-FITC (eBiosciences, Vienna, Austria), anti-CD24-APC (eBiosciences, Vienna, Austria) antibodies or isotype control antibodies (eBiosciences, Vienna, Austria), and incubated for 30 min at room temperature following the manufacturer’s protocol. They were then detected using a FC 500 series flow cytometer (Beckman Coulter, Brea, CA, USA), and the data was subsequently analyzed using the CXP 2.1 software.
RNA-seq analysis
3AO cells transfected with shSTON2 lentivirus or control shRNA were cultured in a SFM for 7 days. Total RNA was prepared from spheroids using an RNeasy mini kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions. RNA quality was evaluated using a RNA Nano 6000 assay kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). High-quality RNA from shSTON2 and control groups was used for transcriptome sequencing (three independent samples for each group). The libraries were sequenced on an Illumina HiSeq platform using a 125 bp/150 bp paired-end sequencing strategy. The original image data generated by the sequencing machine was converted into sequence data via base calling using a TruSeq PE cluster kit v3-cBot-HS (Illumia, San Diego, CA, USA). These were then subjected to standard quality controls. Clean data was obtained by removing reads containing adapter, poly-N, and low quality reads from the raw data. Next, the Q20, Q30, and GC contents of the clean data were calculated. Paired-end clean reads were aligned to the reference genome using STAR. Read numbers mapped to each gene were counted by HTSeq v0.6.0. Differential expression analysis of two conditions was performed using the DESeq2 R package (1.10.1). The resulting P-values were adjusted using the benjamini and hochberg’s approach for controlling the false discovery rate (FDR). Genes with an adjusted P-value< 0.05, as identified by DESeq2, were considered to be differentially expressed.
DNA isolation, sodium bisulfite conversion, and pyrosequencing
Genomic DNA was extracted using a QIAamp DNA mini kit (Qiagen, Dusseldorf, Germany). One microgram of genomic DNA was bisulfite-modified using an EpiTect bisulfite kit (Qiagen, Dusseldorf, Germany) following the manufacturer’s protocol. The modified DNA was used as a template for subsequent PCR. Primers for PCR amplification and sequencing were designed using the pyromark assay design 2.0 software (Qiagen, Dusseldorf, Germany). A Hotstar Taq DNA polymerase PCR kit (Qiagen, Dusseldorf, Germany) was used for the experiments under the following conditions: 95 °C 3 min; 40 cycles of 94 °C 30 s, 52 °C 30 s, 72 °C for 1 min; 72 °C for 7 min. The acquisitions were assessed using gel electrophoresis and pyrosequenced with pyromark Q96 ID (Qiagen, Dusseldorf, Germany). The primers for pyrosequencing are shown in Additional file 1: Table S1.
Patient specimen selection and immunohistochemistry
With the permission of the patients and the ethical committee of the Women’s Hospital, School of Medicine, Zhejiang University, 165 formalin-fixed and paraffin-embedded tissue samples were obtained from patients that had been diagnosed with ovarian serous, mucinous, endometrioid, clear-cell and mixed carcinomas from January 2002 to December 2009. These were used for immunohistochemical analyses. All patients underwent primary surgery, followed by paclitaxel-based chemotherapy. The deadline of the follow-up was July 30, 2017. As 20 patients failed to re-contact, only the remaining 145 patients were used for the calculation of progression-free survival (PFS) and overall survival (OS). All pathological diagnosis was reconfirmed blindly by an expert pathologist. The section slides were counter-stained with haematoxylin, dehydrated and mounted. The details were described as previously [25]. Anti-STON2 (Abcam, Cambridge, MA, USA) (1:200) and anti-MUC1 (Abcam, Cambridge, MA, USA) (1:500) were used for IHC. Five microscope fields were picked for evaluating the semiquantification of STON2 and MUC1 staining. The intensity of immunostaining was graded as follows: 1+, weak; 2+, moderate; 3+, strong or 4+, very strong (Additional file 3: Figure S1). The area of positive cancer cells in was categorized as follows: 1+, 0 to 25%; 2+, 25 to 50%; 3+, 50 to 75% or 4+, 75 to 100%. The score for each microscopic field was obtained by multiplying the two parts of score, The sum was acquired by adding the scores of the five microscopic fields. The sum from 0 to 42 was assigned as “low expression”, from 43 to 80 was assigned as “high expression” [26].
In vivo xenograft experiments
All animal experiments were performed in accordance with Animal Research Reporting In Vivo Experiments (ARRIVE) guidelines for the use of laboratory animals and were approved by the ethics committee of Zhejiang University. NOD/SCID mice (female, 4–6-week-old) were purchased from the Chinese Academy of Medical Sciences (Beijing, China) and randomly assigned to each treatment group. For experiments, 1 × 105 or 1 × 104 3AO spheroids transfected with shSTON2 or shNC were resuspended in 100 μl PBS and injected into the right flank of mice, which were then monitored weekly for 4 weeks. The tumor volumes were calculated according to the formula V = a × b2 × 0.5, where a is the largest diameter and b is the smallest diameter of the tumor.
Statistical analysis
Differences in clinicopathological characteristics were assessed using the chi-squared test. Cumulative survival probabilities were calculated using the Kaplan-Meier method. Survival rates were compared using the log-rank test. Two-tailed Student’s t-tests were used to perform a statistical comparison between two groups unless otherwise indicated. Statistical tests were performed using the SPSS software, version 20.0 (SPSS Inc.) or with GraphPad Prism 6.0 (GraphPad Software, Inc.). The level of statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.001.
Results
STON2 expression is down-regulated in an OCSC-enriched population
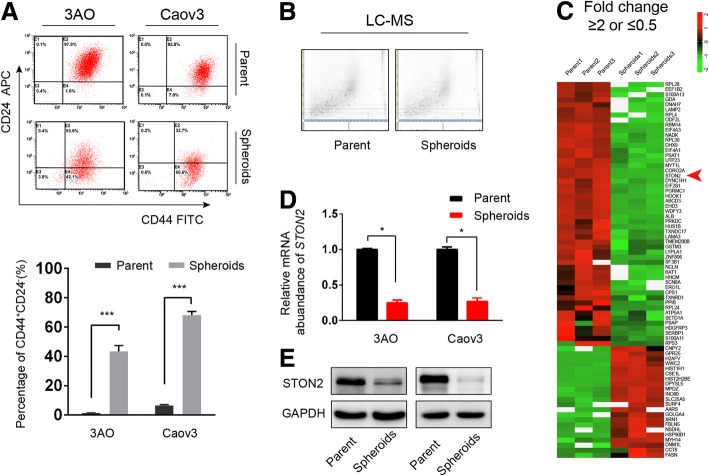
The ovarian cancer cells were cultured in suspension culture conditions to enrich the spheroids [9]. As shown in Fig. 1a, spheroids with considerably increased CD44+CD24− subpopulation are characterized as CSCs and referred to as OCSCs for the subsequent experiments. Using proteomics, we analyzed the differential protein expression between parental cells and spheroids from 3AO cells. The 2-dimensional image generated by LC-MS/MS analysis (Fig. 1b) revealed that 685 peptides and 187 nonredundant peptides had been differentially expressed between parental cells and spheroids (FDR < 0.05, Additional file 4: Table S3). The heat map of Fig. 1c shows 74 of the most differentially expressed proteins in DeCyder differential analysis (FDR < 0.05, fold change ≥2 or ≤ 0.5). Notably, qPCR revealed a lesser STON2 expression in the spheroids than in the parental cells (Fig. 1d). Western blotting further validated the significantly lower expression of STON2 protein in the spheroids than in parental 3AO and Caov3 cells (Fig. 1e).
Fig. 1.
STON2 expression is down-regulated in an OCSC-enriched population. a 3AO and Caov3 cells were cultured in spheroid culture conditions to enrich OCSCs. The CD44+CD24− cell population were analyzed by FCM. The parental cells in normal culture were used as the control. b 2D feature maps of LC-MS/MS data from the analysis between parental cells and spheroids of 3AO cells. The horizontal axis shows retention time, the vertical axis shows mass-to-charge ratio, and the gray color level indicates the intensity value. c Protein cluster map generated by Cluster software (FDR < 0.05, fold change ≥2 or ≤ 0.5). Proteins up-regulated in spheroids are shown in red, and the down-regulated proteins in green. The intensity of the color green or red corresponds to the degree of alteration, according to the color strip at the right of the figure. The arrow indicates STON2. d qPCR analysis of STON2 expression in parental cells and spheroids. e Immunoblot analysis of STON2 expression in parental cells and spheroids. Data represents the mean ± S.E. of three independent experiments. The level of significance is indicated by *P < 0.05, ***P < 0.001
STON2 negatively modulates stem-like properties in ovarian cancer cells
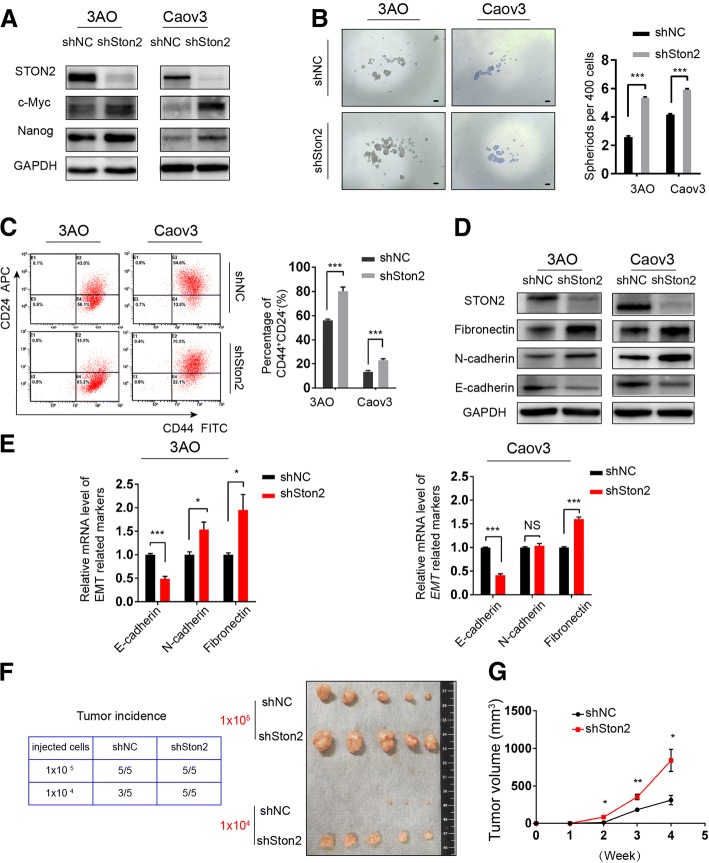
To explore the function of STON2 in ovarian cancer, a shRNA was used to suppress STON2 expression in 3AO and Caov3 cells cultured in SFM for 7 days. STON2 protein levels were successfully inhibited by specific shRNA (Fig. 2a). We observed that STON2 knockdown up-regulated the expression of stem cell related-markers such as NANOG and c-MYC [27] (Fig. 2a) and that spheroid numbers were significantly higher in OCSCs than in shNC cells (Fig. 2b). Flow cytometry analysis showed that the proportion of CD44+CD24− cells had also increased (Fig. 2c). We quantified both the protein and mRNA levels of EMT-related key factors, including E-cadherin, N-cadherin, and fibronectin, all of which revealed significant differences in expressions between shSTON2 and shNC cells, except the N-cadherin mRNA expression in Caov3 line (Fig. 2d, e).
Fig. 2.
STON2 knockdown promotes stem-like properties of ovarian cancer cells. a 3AO and Caov3 cells were transfected with a STON2-specific shRNA for 48 h, and cultured in spheroid culture conditions for 7 days. The expressions of STON2 and the CSC-related markers NANOG and c-MYC were detected by immunoblot analysis. b Quantitation and representative images of sphere-formation (sphere > 50 μm), Scale bars, 50 μm. c The effect of STON2 knockdown on the CD44+CD24− phenotype was analyzed using FCM. d Immunoblot analysis of EMT-related markers (E-cadherin, N-cadherin, and fibronectin). e qPCR analysis of EMT-related markers (E-cadherin, N-cadherin, and fibronectin). f, g 3AO cells were transfected with shNC or shSTON2 for 48 h, cultured in spheroid culture conditions for 7 days, and then injected into NOD/SCID mice (n = 5 in each group). All mice were sacrificed at week 4 and the tumor incidence evaluated (left panel). Subcutaneous tumors are shown (right panel) (f).Tumor diameters of xenografts from mice injected with 1 × 105 cells were measured at a regular intervals of 1 week for up to 4 weeks and the tumor volume was calculated (g). Data represents mean ± S.E. of three independent experiments. The level of significance is indicated by *P < 0.05, **P < 0.01, ***P < 0.001
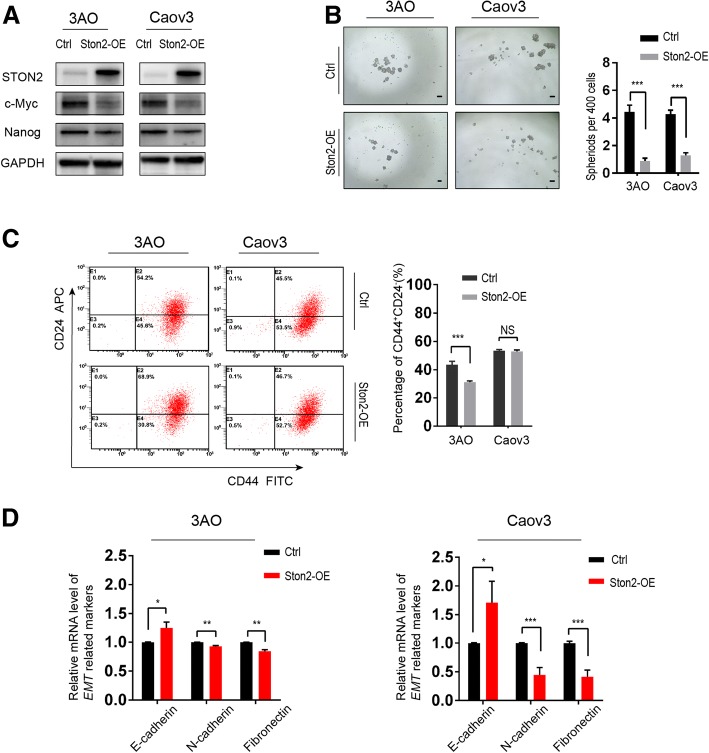
Next, we considered whether STON2 overexpression negatively affects OCSCs. 3AO and Caov3 cells were transfected with STON2 plasmids and cultured in SFM for 7 days. Cells transfected with the empty vectors acted as controls. Analysis of the expression of CSC-related proteins showed that their expression levels were reduced in cells showing STON2 overexpression (Fig. 3a). STON2 overexpression also significantly inhibited spheroid number (Figs. 3b) and the proportion of cells with the CD44+CD24−phenotype in the 3AO cell line, but not in the Caov3 cell line (Fig. 3c). It also altered the expression of EMT-related key factors (Fig. 3d), as compared to those of the control groups.
Fig. 3.
STON2 overexpression inhibits stem-like properties of ovarian cancer cells. a 3AO and Caov3 cells were transfected with STON2 overexpressing plasmid for 48 h and cultured in spheroid culture conditions for 7 days. Expression of STON2 and the CSC-related markers NANOG and c-MYC were determined by immunoblot analysis. b Quantitation and representative images of sphere-formation (sphere > 50 μm), Scale bars, 50 μm. c Effect of STON2 overexpression on the CD44+CD24− phenotype was analyzed using FCM. d qPCR analysis of EMT-related markers (E-cadherin, N-cadherin, and fibronectin). Data represents the mean ± S.E. of three independent experiments. The level of significance is indicated by *P < 0.05, **P < 0.01, ***P < 0.001, NS indicates P > 0.05
We then investigated the effect of STON2 knockdown on ovarian tumorigenicity. We performed xenograft assays using different gradients of CSC subpopulations enriched from 3AO-shNC or shSTON2 cells. As shown in Fig. 2f, by 4 weeks after injection, 1 × 104 shSTON2 cells had efficiently generated large tumors in all mice (n = 5), but equal numbers of shNC cells had produced only three tumors among five injected NOD/SCID mice. These data indicated that STON2 knockdown enhanced the CSC subpopulation-induced tumorigenicity in vivo. In addition, the tumor diameters of mice injected with 1 × 105 cells were measured weekly for 4 weeks after implantation. As shown in Fig. 2g, the volume of tumors in the shSTON2 groups was larger than those in the shNC groups.
MUC1 acts as a downstream target for STON2
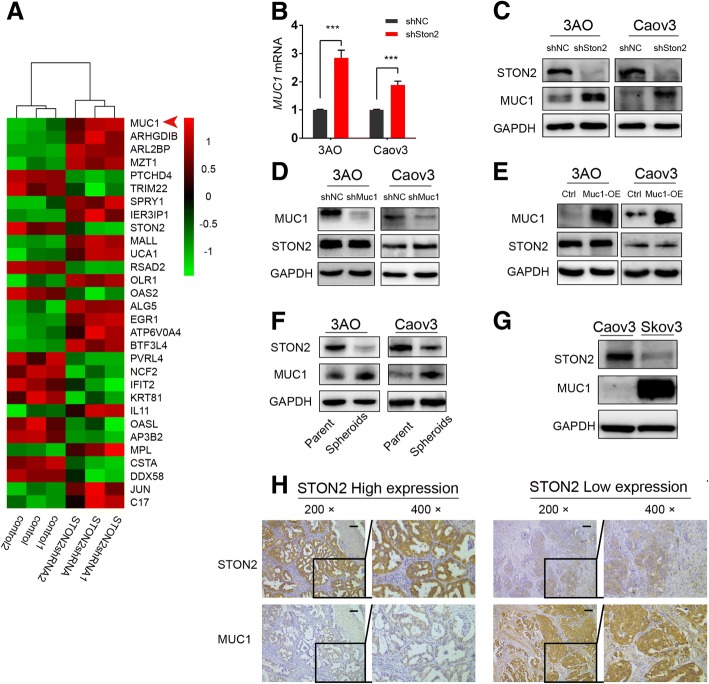
As STON2 modulates stem-like properties in ovarian cancer cells, we attempted to determine the downstream targets of STON2. Total RNA was extracted from spheroid cells treated with shNC or shSTON2, and the differential mRNA expression was detected using RNA-seq (three independent samples in each group). DESeq2 software analysis showed that a series of genes was differentially expressed (< 0.05-Padj) (Fig. 4a). We focused on MUC1, which was shown to be particularly up-regulated (− 0.0000446-Padj) and has been demonstrated to be related to stemness in breast cancer [28] and pancreatic cancer [29].
Fig. 4.
MUC1 acts as a downstream target for STON2. a RNA-seq data from the analysis between shNC and shSTON2 of 3AO cells derived stem cells. The cluster map generated by DESeq2 software analysis shows a series of differentially expressed genes (Padj < 0.05). Genes up-regulated in shSTON2 are shown in red, and down-regulated genes in green. The intensity of the color green or red corresponds to the degree of alteration, according to the color strip at the right of the figure. The arrow indicates MUC1. b qPCR analysis of MUC1 mRNA expression regulated by STON2 in 3AO and Caov3. c Immunoblot analysis of MUC1 protein level regulated by STON2 in 3AO and Caov3. d STON2 protein level after MUC1 knockdown in 3AO and Caov3. e STON2 protein level after MUC1 overexpression in 3AO and Caov3. f Immunoblot analysis of STON2 and MUC1 expression in parental cells and spheroids. g Immunoblot analysis of STON2 and MUC1 expression in Caov3 and Skov3 cells. h Specimen sections from the paraffin-embedded block from ovarian cancer patients were used for detection using anti-STON2 and anti-MUC1 antibodies, respectively. Paired STON2 and MUC1 staining is shown at 200× and 400× magnifications. Data represents the mean ± S.E. of three independent experiments. The level of significance is indicated by ***P < 0.001
We determined MUC1 expression in 3AO and Caov3 cells using qPCR and western blotting. Results showed that the expression levels of MUC1 mRNA and protein were elevated after STON2 knockdown (Fig. 4b, c), whereas, STON2 expression remained unaltered upon MUC1 knockdown or over-expression (Fig. 4d, e). This suggests that MUC1 acts downstream of STON2 in ovarian cancer cells.
We also analyzed the expression of MUC1 in parental as well as in cancer cell derived stem cells. Western blotting showed a higher expression of MUC1 associated with a lower expression of STON2 in spheroids, as compared to those in parental cells (Fig. 4f). We also employed a highly aggressive ovarian cancer cell line Skov3 [30] to examine MUC1 expression. As expected, a much higher expression of MUC1 was associated with a lower expression of STON2 in the Skov3 cells, as compared to those in Caov3 cells (Fig. 4g).
Further random examinations of specimen sections from the paraffin-embedded blocks of ovarian cancer patients confirmed that STON2 and MUC1 expression correlated negatively (Fig. 4h).
DNMT1-MUC1 mediated epigenetic modification contributes to STON2 regulation in ovarian cancer cells
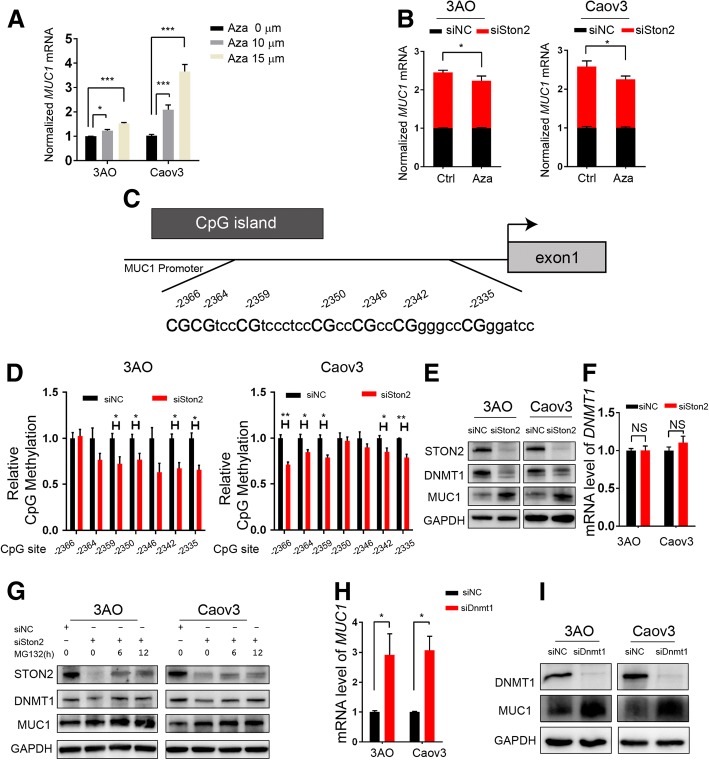
Previous studies have shown that MUC1 expression is regulated by DNA methylation [24, 31]. Hence, we treated 3AO and Caov3 cells with different concentrations of DNA methylation inhibitor azacitidine (Aza) for 48 h. An Aza dosage-dependent increase in MUC1 expression was observed (Fig. 5a). We then transfected a STON2-specific siRNA or control siRNA in 3AO and Caov3 cells and incubated them for 24 h. This was followed by treatment with Aza or control for 48 h. Results showed that Aza was able to partially block MUC1 up-regulation post-STON2 knockdown (Fig. 5b), suggesting that STON2 knockdown-induced MUC1 up-regulation might occur via a reduction in MUC1 promoter methylation. We further identified one CpG-rich region in the MUC1 promoter (Fig. 5c) and designed primers to analyze the CpG-rich region using a PyroMark assay design. 3AO and Caov3 cells were treated with Aza for 24 h after transfection with the STON2-specific siRNA or control siRNA. The DNA methylation levels of the specific CpG nucleotides were then detected using pyrosequencing. Results showed that the extent of DNA methylation at multiple sites in the siSTON2 group was less than that of the control group (Fig. 5d and Additional file 5: Figure S2).
Fig. 5.
DNMT1-MUC1 mediated epigenetic modification contributes to STON2 regulation in ovarian cancer cells. a 3AO and Caov3 cells were treated with 0 μM, 10 μM, and 15 μM Aza (S1782, Selleck, USA) for 48 h, and MUC1 expression was detected using qPCR analysis. b 3AO and Caov3 cells were transfected with a STON2-specific siRNA or siNC for 24 h, followed by treatment with Aza (15 μM for 48 h). MUC1 expression was detected using qPCR analysis. c Schematic illustration of the MUC1 promoter embedded in one CpG-rich region (− 2366 to − 2327 bp). d 3AO and Caov3 cells were transfected with a STON2-specific siRNA or control for 24 h, followed by pyrosequencing to assess the DNA methylation status of MUC1 (three coupled independent samples). Each column represents the relative average DNA methylation level at one CpG site compared to the control group (see also raw pyrograms of representative experiments in Additional file 5: Figure S2). e-g 3AO and Caov3 cells were transfected with a STON2-specific siRNA for 72 h. DNMT1 and MUC1 expression were detected by immunoblot analysis (e). DNMT1 expression was determined by qPCR analysis (f). DNMT1 and MUC1 expression was assayed by immunoblotting in the presence of MG132 (10 μM for 0 h, 6 h, 12 h) g. (h-i) 3AO and Caov3 cells were transfected with a DNMT1-specific siRNA for 72 h. MUC1 expression was detected using qPCR analysis (h). MUC1 expression was detected using immunoblot analysis (i). Data represents the mean ± S.E. of three independent experiments. The level of significance is indicated by *P < 0.05, **P < 0.01, ***P < 0.001, NS indicates P > 0.05
DNMT1, a key member of the DNA methyltransferase family [32], is known to maintain methylation patterns and suppress transcription [33]. Our results showed that STON2 knockdown decreased DNMT1 but increased MUC1 expression (Fig. 5e). We also observed that STON2 knockdown did not alter DNMT1 mRNA levels but changed DNMT1 protein levels (Fig. 5e, f). Growing evidence suggests that the stability of DNMT1 is regulated by post-translational modifications such as phosphorylation, acetylation, methylation, and ubiquitination [34–36]. Using specific siRNAs, we confirmed that the STON2 knockdown-induced reduction in DNMT1 could be partially rescued by the proteasome inhibitor MG132, and that a time-dependent increase in MUC1 expression was observed (Fig. 5g). In addition, both the mRNA expression and protein level of MUC1 were increased by DNMT1 knockdown (Fig. 5h, i). Correspondingly, the MUC1 promoter DNA methylation levels of specific CpG nucleotides at multiple sites in the siDNMT1 group were lower than those in the control group (Additional file 6: Figure S3). These results suggest that, for cancer stem cells, STON2 functions at the post-transcriptional level via the DNMT1/MUC1 pathway.
MUC1 participates in the STON2-mediated modulation of stem-like properties in ovarian cancer cells
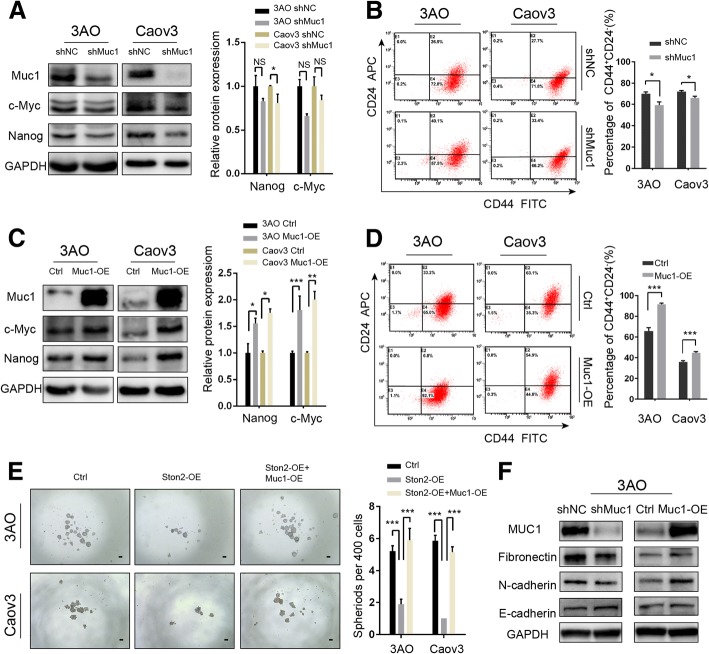
Finally, we constructed shRNA to suppress MUC1 in 3AO and Caov3 cells cultured in SFM. We analyzed the relative expression levels of CSC-related proteins, NANOG and c-MYC. The results showed that MUC1 knockdown significantly reduced the expression level of NANOG in Caov3 (p < 0.05), but not in 3AO (Fig. 6a). The reduction of c-MYC expression was not significant in 3AO or Caov3 cells (Fig. 6a). MUC1 knockdown also significantly reduced CD44+CD24−phenotypes (Fig. 6b). Conversely, MUC1 overexpression significantly up-regulated both NANOG (p < 0.05) and c-MYC (p < 0.01) (Fig. 6c) and increased the proportion of cells with the CD44+CD24− phenotype (Fig. 6d). Furthermore, we observed that the STON2 overexpression-induced reduction of sphere-forming ability was partially reversed by MUC1 overexpression (Fig. 6e).
Fig. 6.
MUC1 participates in the STON2-mediated modulation of stem-like properties in ovarian cancer cells. a, b 3AO and Caov3 cells were transfected with a MUC1-specific shRNA for 48 h and cultured in spheroid culture conditions for 7 days. Representative images and the relative expression levels of CSC-related markers NANOG and c-MYC were detected by immunoblot analysis (a), and the CD44+CD24− population was detected by FCM (b). c, d 3AO and Caov3 cells were transfected with MUC1 overexpressing plasmid for 48 h, and cultured in spheroid culture conditions for 7 days. Representative images and the relative expression levels of CSC-related markers NANOG and c-MYC were detected by immunoblot analysis (c), and the CD44+CD24− population was detected using FCM (d). e 3AO and Caov3 cells were transfected with a control vector, STON2 overexpression plasmid, or STON2 overexpression plasmid plus MUC1 overexpression plasmid for 48 h and cultured in spheroid culture conditions for 7 days. Sphere-formation (sphere> 50 μm) was assessed. Scale bars, 50 μm. f 3AO cells were transfected with a MUC1-specific shRNA, or MUC1 overexpressing plasmid for 48 h, and cultured in spheroid culture conditions for 7 days. EMT-related markers (E-cadherin, N-cadherin, and fibronectin) were detected using immunoblot analysis. Data represents the mean ± S.E. of three independent experiments. The level of significance is indicated by *P < 0.05, ***P < 0.001
Using MUC1 knockdown or overexpression in the 3AO cell line, we detected the expression levels of EMT-related key factors by western blotting. As expected, the results showed that the expression of E-cadherin increased, while N-cadherin and fibronectin decreased in shMUC1 cells, as compared to those of the shNC cells (Fig. 6f). MUC1 overexpression up-regulated the expression of N-cadherin and fibronectin, but did not significantly change E-cadherin, as compared to those of the control groups (Fig. 6f). However, we failed to co-express the shSTON2 and shMUC1 in 3AO cells. Based on previous results that the EMT-related protein expression was regulated by shSTON2 (Fig. 2d), it is conceivable that MUC1 participates in STON2 induced changes in EMT-related markers.
The clinical significance of MUC1 and STON2 expression in ovarian cancer patients
We firstly investigated the correlation between STON2 expression and clinical parameters based on immunohistological examination of tissue samples from patients. Kaplan–Meier survival analysis failed to reveal any correlation of STON2 expression with progression-free survival (PFS) or overall survival (OS) in our patient cohort (Additional file 7: Figure S5A). However, significantly lower STON2 immunoreactivity was detected in poorly differentiated ovarian cancer (P = 0.000, Additional file 8: Table S4). We also analyzed the correlation of MUC1 expression with clinicopathological characteristics, and prognosis of ovarian cancer. Results showed that significantly higher expression of MUC1 was associated with FIGO stage (P = 0.004), tumor grade (P = 0.01), serum CA125 (P = 0.005), and PFS (P = 0.006), OS (P = 0.019) (Additional file 9: Table S5 and Fig. 7a).
Fig. 7.
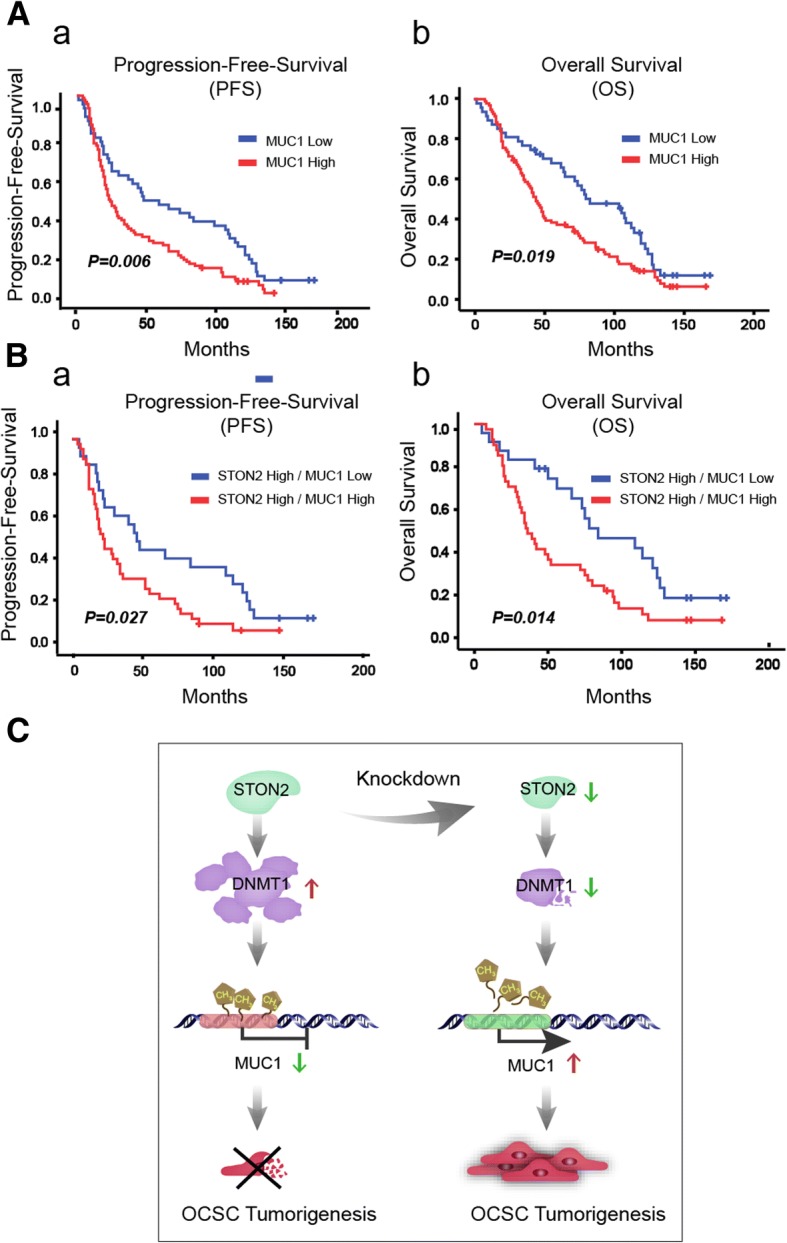
The clinical significance of MUC1 and STON2 expression in ovarian cancer patients. a, b Cumulative survival probabilities (a, PFS and b, OS) were calculated using the Kaplan–Meier method (n = 145) based on STON2 and MUC1 expression. Survival rates were compared using log-rank test. Patients expressing low or high levels of MUC1 (a). Patients expressing high levels of STON2 combined with low MUC1 expression or high MUC1 expression (b). c A schematic model of the role of STON2 in regulating CSCs traits
IHC analysis of the co-expression relationships between STON2 and MUC1 revealed that the prognostic value of STON2 expression was highly dependent on MUC1 expression, in which the high expression of STON2 twinned with a low expression of MUC1 was associated with a favorable prognosis (Fig. 7b). A trend was also revealed in that patients expressing high STON2 and low MUC1 were associated with a best survival of all the remaining patients (PFS, P = 0.095, and OS, P = 0.06). However, with a low expression of STON2, there was no prognostic value of MUC1 expression (Additional file 7: Figure S5B). Taken together, these observations provide evidence that STON2 acts as a negative modulator via the MUC1-mediated pathway in ovarian cancer.
Discussion
We have shown that the OCSC-enriched population displays a dramatically decreased STON2 expression. STON2 knockdown promotes stem-like properties in ovarian cancer cells and its overexpression suppresses MUC1-induced sphere formation in OCSCs, in which DNMT1-mediated methylation in the promoter region of MUC1 has been confirmed to be modulated by STON2. Our findings suggest that STON2 depression up-regulates MUC1 expression, along with inhibition of DNMT1, in a process that contributes to the maintenance of stem-like properties in ovarian cancer cells (Fig. 7c).
In our study, STON2 knockdown increased both MUC1 mRNA and protein levels. This suggests that the regulation of MUC1 by STON2 occurs at the transcriptional level. MUC1 expression has been reported to be associated with DNA methylation [23, 24, 37]. MassARRAY assays have been used to detect high methylation levels in the MUC1 promoter region in MDA-MB-453, a MUC1-negative cancer cell line. MUC1 mRNA expression in MUC1-negative cells has been shown to be restored by treatment with a DNA methylation inhibitor. MCF-7, a MUC1-positive cell line, has been associated with a low methylation level in the MUC1 promoter region [24].
DNA methylation is an important genome modification that occurs at the cytosine residues within CpG dinucleotides. There are three enzymatically active mammalian methyltransferases: DNMT1, DNMT3A, and DNMT3B [38]. DNMT1 is responsible for maintaining methylation patterns in CpG dinucleotide-rich regions as well as for transcriptional repression [39], whereas DNMT3A and DNMT3B are involved in de novo methylation [40]. In the present study, we observed that only DNMT1, but not DNMT3A or DNMT3B, responded to STON2 knockdown, (Additional file 10: Figure S4). Subsequently, STON2 knockdown suppressed DNMT1 and increased MUC1 expression. Pyrosequencing data also revealed that the methylation level was reduced in the MUC1 promoter region in STON2 or DNMT1 knocked down ovarian cancer cells. Therefore, we are the first to demonstrate that STON2 is involved in the DNMT1-mediated epigenetic modification of MUC1 promoter methylation in OCSCs.
Compared to other tumor types, for which there is a more generalized consensus on the CSC-related markers (such as for CD44+CD24− in breast carcinomas, CD133+ in glioblastomas, and LGR5+ in colon carcinomas, etc.), it is striking to see how heterogeneous the sets of putative OCSC markers are [41], particularly, CD44, CD133, CD117, ALDH1 [42], and the combinations CD44+/CD24−, CD44+/CD117+ [43]. Isolated OCSCs are often characterized by the expression of stemness-associated genes such as NANOG [44] and c-MYC [11], both of which play key roles in maintaining the pluripotency of embryonic stem cells [45, 46]. Similarly, CSCs isolated from ascites derived from ovarian cancer patients showed elevated NANOG expressions [47]. NANOG has been widely used as a stem cell marker [48], is associated with stemness maintenance [12] and EMT acceleration [44], and is taken as an indicator of a poorer prognosis in ovarian cancers [49]. In addition, c-MYC expression has been most often proposed as a general feature of the CSCs of various cancers including breast [50], prostate [51], esophagus [52] and tongue [53], and the expression of c-MYC together with NANOG has been previously noted as a feature in a population of ovarian cancer cells [27]. In our previous study [9], we successfully enriched and characterized OCSCs with CD44+CD24−. Interestingly, here we showed that NANOG and c-MYC expression, as stemness-associated factors, were detected in these OCSCs and that their cancer stem-like properties were negatively regulated by STON2 expression.
In the present study, we found that analysis of STON2 in isolation did not have any prognostic value in ovarian cancer. This provides a contrast to a previously published finding that STON2 overexpression is correlated with unfavorable prognosis in 89 ovarian cancer patients [54]. The reason for such an inconsistency is unclear, but our paper certainly represents a fairly comprehensively exploration of the prognostic value of STON2 expression and MUC1 expression in ovarian cancer. Conversely, we found that the prognostic value of high STON2 expression was highly dependent on MUC1 expression, with a trend that if a high expression of STON2 occurred together with a low expression of MUC1, then this was positively associated with a favorable prognosis. A clinical study based on larger patient samples is required for further confirmation.
Conclusions
In summary, we observed for the first time that STON2 acts as a negative modulator in ovarian cancer cells via DNMT1/MUC1-mediated epigenetic mechanisms. STON2, is therefore involved in OCSC biology and may represent a therapeutic target for innovative treatments aimed at ovarian cancer eradication.
Additional files
Table S1. Primer sequences used in this study (DOCX 18 kb)
Table S2. Target sequences in this study. (DOCX 18 kb)
Figure S1. Intensity of STON2 and MUC1 immunohistochemical staining in ovarian cancer tissues. Representative images of STON2 and MUC1 staining in ovarian cancer tissues are shown. Scale bars represent approximately 50 μm. Staining intensity is graded as follows: 1+, weak; 2+, moderate; 3+, strong; 4+, very strong. (PNG 4594 kb)
Table S3. Peptide information from the LC-MS/MS data after analyzing parental cells and spheroids of 3AO cells (FDR < 0.05). Av. Ratio (Lin): the ratio of spheroids to parental cells. (XLS 154 kb)
Figure S2. (Related to pyrosequencing data shown in Fig. 5d) Representative pyrogramms of MUC1 promoter in 3AO and Caov3 cells (siNC or siSTON2). (TIF 760 kb)
Figure S3. 3AO and Caov3 cells were transfected with a DNMT1-specific siRNA or controls for 24 h, and then subjected to pyrosequencing to assess the DNA methylation status of MUC1 (three coupled independent samples) (A-B). Each column represents the relative average DNA methylation level at one CpG site compared to the control group (A). Raw pyrograms of representative experiments (B). (TIF 1569 kb)
Figure S5. (A-B) Cumulative survival probabilities (a, PFS and b, OS) were calculated using the Kaplan–Meier method (n = 145) based on STON2 and MUC1 expression. Survival rates were compared using a log-rank test. Patients expressing low or high levels of STON2 (A). Patients expressing low levels of STON2 combined with low MUC1 expression or high MUC1 expression (B). (TIF 912 kb)
Table S4. Association of STON2 expression with clinicopathological characteristics in patients with ovarian cancer (n = 165). (XLSX 11 kb)
Table S5. Association of MUC1 expression with clinicopathological characteristics in patients with ovarian cancer (n = 165). (XLSX 11 kb)
Figure S4. 3AO and Caov3 cells were transfected with a STON2-specific siRNA for 72 h, and the expression of DNMT1 (A), DNMT3A (B), or DNMT3B (C) was assayed using immunoblotting analysis. (TIF 808 kb)
Acknowledgements
We thank our colleagues for valuable assistance and helpful discussions. We would like to thank prof. Chris Wood and Yongmei Xi from Zhejiang University for improvement of English writing of the manuscript. We also would like to thank the experts who were careful reading of our manuscript and the editors who gave our article a chance to be published.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No.81772771 and No. 81572550).
Availability of data and materials
All data are fully available without restriction, and the RNA-seq data had been deposited in the SRA database (SRP135729) and wil be accessible without restriction from the date of publication.
Abbreviations
- ARRIVE
Animal research reporting in vivo experiments
- ATCC
American type culture collection
- Aza
azacitidine
- CSCs
Cancer stem cells
- CSLCs
Cancer stem-like cells
- DMEM
Dulbecco’s modified eagle’s medium
- DNMT1
Dna methyltransferase 1
- ECL
Enhanced chemiluminescence
- EMT
Epithelial mesenchymal transition
- FBS
Fetal bovine serum
- FCM
Flow cytometry analysis
- FDR
False discovery rate
- FIGO
Federation international of gynecology and obstetrics
- IHC
Immunohistochemistry
- IPI
International protein index
- KEGG
Kyoto encyclopedia of genes and genomes
- LC
Liquid chromatography
- MS
Mass spectrometry
- MUC1
Mucin1
- OCSCs
Ovarian cancer stem cells
- OS
Overall survival
- PFS
Progression-free survival
- PVDF
Polyvinylidene fluoride
- qPCR
Quantitative real-time polymerase chain reaction
- RIPA
Radioimmunoprecipitation assay
- RNA-Seq
RNA-sequencing
- RPMI
Roswell park memorial institute
- SFM
Serum-free medium
- shRNA
Short-hairpin RNA
- siRNAs
Small-interfering RNAs
- STON2
Stonin2
- TBST
Tris buffered saline Tween 20
- TCGA
The cancer genome atlas
Authors’ contributions
WL and XW conceptualized and designed the study, revised the manuscript, supervised and administrated the study. SX designed the study, performed most of the experiments, analyzed the data, and wrote the manuscript. YY performed the experiments, analyzed the data, and wrote the manuscript. SZ performed most of the vivo xenograft experiments. CZ performed the immunohistochemistry and analyzed the data. XC and XX contributed experiments and provided technical and theoretical support. All of the authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were carried out in accordance with Animal Research: Reporting In Vivo Experiments (ARRIVE) guidelines and use of Laboratory animals and were approved by the ethics committee of Zhejiang University. The immunohistochemistry study of patient tissues was approved by the ethical committee of the Women’s Hospital, School of Medicine, Zhejiang University.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shanshan Xu, Email: qingwu@zju.edu.cn.
Yongfang Yue, Email: 11518242@zju.edu.cn.
Songfa Zhang, Email: zhangsongfa@zju.edu.cn.
Caiyun Zhou, Email: zhou-caiyun@163.com.
Xiaodong Cheng, Email: chengxd@zju.edu.cn.
Xing Xie, Email: xiex@zju.edu.cn.
Xinyu Wang, Phone: 0086-571-87061501, Email: wangxy@zju.edu.cn.
Weiguo Lu, Phone: 0086-571-87061501, Email: lbwg@zju.edu.cn.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaughan Sebastian, Coward Jermaine I., Bast Robert C., Berchuck Andy, Berek Jonathan S., Brenton James D., Coukos George, Crum Christopher C., Drapkin Ronny, Etemadmoghadam Dariush, Friedlander Michael, Gabra Hani, Kaye Stan B., Lord Chris J., Lengyel Ernst, Levine Douglas A., McNeish Iain A., Menon Usha, Mills Gordon B., Nephew Kenneth P., Oza Amit M., Sood Anil K., Stronach Euan A., Walczak Henning, Bowtell David D., Balkwill Frances R. Rethinking ovarian cancer: recommendations for improving outcomes. Nature Reviews Cancer. 2011;11(10):719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 5.Ffrench B, Gasch C, O'Leary JJ, Gallagher MF. Developing ovarian cancer stem cell models: laying the pipeline from discovery to clinical intervention. Mol Cancer. 2014;13:262. doi: 10.1186/1476-4598-13-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nassar D, Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- 7.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 8.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 9.Shi MF, Jiao J, Lu WG, Ye F, Ma D, Dong QG, Xie X. Identification of cancer stem cell-like cells from human epithelial ovarian carcinoma cell line. Cell Mol Life Sci. 2010;67:3915–3925. doi: 10.1007/s00018-010-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao J, Huang L, Ye F, Shi M, Cheng X, Wang X, Hu D, Xie X, Lu W. Cyclin D1 affects epithelial-mesenchymal transition in epithelial ovarian cancer stem cell-like cells. Onco Targets Ther. 2013;6:667–677. doi: 10.2147/OTT.S44177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motohara T, Masuko S, Ishimoto T, Yae T, Onishi N, Muraguchi T, Hirao A, Matsuzaki Y, Tashiro H, Katabuchi H, et al. Transient depletion of p53 followed by transduction of c-Myc and K-Ras converts ovarian stem-like cells into tumor-initiating cells. Carcinogenesis. 2011;32:1597–1606. doi: 10.1093/carcin/bgr183. [DOI] [PubMed] [Google Scholar]
- 12.Xu CX, Xu M, Tan L, Yang H, Permuth-Wey J, Kruk PA, Wenham RM, Nicosia SV, Lancaster JM, Sellers TA, Cheng JQ. MicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/Nanog. J Biol Chem. 2012;287:34970–34978. doi: 10.1074/jbc.M112.374611. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Jung N, Wienisch M, Gu M, Rand JB, Muller SL, Krause G, Jorgensen EM, Klingauf J, Haucke V. Molecular basis of synaptic vesicle cargo recognition by the endocytic sorting adaptor stonin 2. J Cell Biol. 2007;179:1497–1510. doi: 10.1083/jcb.200708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Ju B, Tian J, Liu F, Yu H, Xiao H, Liu X, Liu W, Yao Z, Hao Q. Ovarian cancer stem cell-specific gene expression profiling and targeted drug prescreening. Oncol Rep. 2014;31:1235–1248. doi: 10.3892/or.2014.2976. [DOI] [PubMed] [Google Scholar]
- 15.Jinesh GG, Kamat AM. Endocytosis and serpentine filopodia drive blebbishield-mediated resurrection of apoptotic cancer stem cells. Cell Death Discov. 2016;2. [DOI] [PMC free article] [PubMed]
- 16.Garcia-Heredia JM, Verdugo Sivianes EM, Lucena-Cacace A, Molina-Pinelo S, Carnero A. Numb-like (NumbL) downregulation increases tumorigenicity, cancer stem cell-like properties and resistance to chemotherapy. Oncotarget. 2016;7:63611–63628. doi: 10.18632/oncotarget.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alam M, Ahmad R, Rajabi H, Kufe D. MUC1-C induces the LIN28B-->LET-7-->HMGA2 Axis to regulate self-renewal in NSCLC. Mol Cancer Res. 2015;13:449–460. doi: 10.1158/1541-7786.MCR-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget. 2014;5:2622–2634. doi: 10.18632/oncotarget.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiraki M, Maeda T, Bouillez A, Alam M, Tagde A, Hinohara K, Suzuki Y, Markert T, Miyo M, Komura K, et al. MUC1-C activates BMI1 in human cancer cells. Oncogene. 2017;36:2791–2801. doi: 10.1038/onc.2016.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng J, Wang L, Chen H, Li L, Ma Y, Ni J, Li Y. The role of tumour-associated MUC1 in epithelial ovarian cancer metastasis and progression. Cancer Metastasis Rev. 2013;32:535–551. doi: 10.1007/s10555-013-9423-y. [DOI] [PubMed] [Google Scholar]
- 22.Budiu RA, Mantia-Smaldone G, Elishaev E, Chu T, Thaller J, McCabe K, Lenzner D, Edwards RP, Vlad AM. Soluble MUC1 and serum MUC1-specific antibodies are potential prognostic biomarkers for platinum-resistant ovarian cancer. Cancer Immunol Immunother. 2011;60:975–984. doi: 10.1007/s00262-011-1010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zrihan-Licht S, Weiss M, Keydar I, Wreschner DH. DNA methylation status of the MUC1 gene coding for a breast-cancer-associated protein. Int J Cancer. 1995;62:245–251. doi: 10.1002/ijc.2910620303. [DOI] [PubMed] [Google Scholar]
- 24.Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S. MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res. 2008;68:2708–2716. doi: 10.1158/0008-5472.CAN-07-6844. [DOI] [PubMed] [Google Scholar]
- 25.Li YL, Ye F, Cheng XD, Hu Y, Zhou CY, Lu WG, Xie X. Identification of glia maturation factor beta as an independent prognostic predictor for serous ovarian cancer. Eur J Cancer. 2010;46:2104–2118. doi: 10.1016/j.ejca.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Zhang SF, Wang XY, Fu ZQ, Peng QH, Zhang JY, Ye F, Fu YF, Zhou CY, Lu WG, Cheng XD, Xie X. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy. 2015;11:225–238. doi: 10.1080/15548627.2014.998931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di J, Duiveman-de Boer T, Zusterzeel PL, Figdor CG, Massuger LF, Torensma R. The stem cell markers Oct4A, Nanog and c-Myc are expressed in ascites cells and tumor tissue of ovarian cancer patients. Cell Oncol (Dordr) 2013;36:363–374. doi: 10.1007/s13402-013-0142-8. [DOI] [PubMed] [Google Scholar]
- 28.Engelmann K, Shen H, Finn OJ. MCF7 side population cells with characteristics of cancer stem/progenitor cells express the tumor antigen MUC1. Cancer Res. 2008;68:2419–2426. doi: 10.1158/0008-5472.CAN-07-2249. [DOI] [PubMed] [Google Scholar]
- 29.Curry JM, Thompson KJ, Rao SG, Besmer DM, Murphy AM, Grdzelishvili VZ, Ahrens WA, McKillop IH, Sindram D, Iannitti DA, et al. The use of a novel MUC1 antibody to identify cancer stem cells and circulating MUC1 in mice and patients with pancreatic cancer. J Surg Oncol. 2013;107:713–722. doi: 10.1002/jso.23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez L, Kim MK, Lyle LT, Bunch KP, House CD, Ning F, Noonan AM, Annunziata CM. Characterization of ovarian cancer cell lines as in vivo models for preclinical studies. Gynecol Oncol. 2016;142:332–340. doi: 10.1016/j.ygyno.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thai P, Loukoianov A, Wachi S, Wu R. Regulation of airway mucin gene expression. Annu Rev Physiol. 2008;70:405–429. doi: 10.1146/annurev.physiol.70.113006.100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2017. [DOI] [PubMed]
- 33.Maslov AY, Lee M, Gundry M, Gravina S, Strogonova N, Tazearslan C, Bendebury A, Suh Y, Vijg J. 5-aza-2′-deoxycytidine-induced genome rearrangements are mediated by DNMT1. Oncogene. 2012;31:5172–5179. doi: 10.1038/onc.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Signal. 2011;4:pe3. doi: 10.1126/scisignal.2001764. [DOI] [PubMed] [Google Scholar]
- 35.Kinney SR, Pradhan S. Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells. Prog Mol Biol Transl Sci. 2011;101:311–333. doi: 10.1016/B978-0-12-387685-0.00009-3. [DOI] [PubMed] [Google Scholar]
- 36.Cheng J, Yang H, Fang J, Ma L, Gong R, Wang P, Li Z, Xu Y. Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation. Nat Commun. 2015;6:7023. doi: 10.1038/ncomms8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama S, Higashi M, Kitamoto S, Oeldorf M, Knippschild U, Kornmann M, Maemura K, Kurahara H, Wiest E, Hamada T, et al. Aberrant methylation of MUC1 and MUC4 promoters are potential prognostic biomarkers for pancreatic ductal adenocarcinomas. Oncotarget. 2016;7:42553–42565. doi: 10.18632/oncotarget.9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto Y, Zumwalt TJ, Goel A. DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer. Epigenomics. 2016;8:685–703. doi: 10.2217/epi-2015-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bigey P, Ramchandani S, Theberge J, Araujo FD, Szyf M. Transcriptional regulation of the human DNA methyltransferase (dnmt1) gene. Gene. 2000;242:407–418. doi: 10.1016/S0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- 40.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 41.Lupia M, Cavallaro U. Ovarian cancer stem cells: still an elusive entity? Mol Cancer. 2017;16:64. doi: 10.1186/s12943-017-0638-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed N, Abubaker K, Findlay JK. Ovarian cancer stem cells: molecular concepts and relevance as therapeutic targets. Mol Asp Med. 2014;39:110–125. doi: 10.1016/j.mam.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhang R, Zhang P, Wang H, Hou D, Li W, Xiao G, Li C. Inhibitory effects of metformin at low concentration on epithelial-mesenchymal transition of CD44(+)CD117(+) ovarian cancer stem cells. Stem Cell Res Ther. 2015;6:262. doi: 10.1186/s13287-015-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iv Santaliz-Ruiz LE, Xie X, Old M, Teknos TN, Pan Q. Emerging role of nanog in tumorigenesis and cancer stem cells. Int J Cancer. 2014;135:2741–2748. doi: 10.1002/ijc.28690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullin NP, Yates A, Rowe AJ, Nijmeijer B, Colby D, Barlow PN, Walkinshaw MD, Chambers I. The pluripotency rheostat Nanog functions as a dimer. Biochem J. 2008;411:227–231. doi: 10.1042/BJ20080134. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 47.Hu L, McArthur C, Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br J Cancer. 2010;102:1276–1283. doi: 10.1038/sj.bjc.6605626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobello N, Biamonte F, Pisanu ME, Faniello MC, Jakopin Z, Chiarella E, Giovannone ED, Mancini R, Ciliberto G, Cuda G, Costanzo F. Ferritin heavy chain is a negative regulator of ovarian cancer stem cell expansion and epithelial to mesenchymal transition. Oncotarget. 2016;7:62019–62033. doi: 10.18632/oncotarget.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siu MK, Wong ES, Kong DS, Chan HY, Jiang L, Wong OG, Lam EW, Chan KK, Ngan HY, Le XF, Cheung AN. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene. 2013;32:3500–3509. doi: 10.1038/onc.2012.363. [DOI] [PubMed] [Google Scholar]
- 50.Nair R, Roden DL, Teo WS, McFarland A, Junankar S, Ye S, Nguyen A, Yang J, Nikolic I, Hui M, et al. C-Myc and Her2 cooperate to drive a stem-like phenotype with poor prognosis in breast cancer. Oncogene. 2014;33:3992–4002. doi: 10.1038/onc.2013.368. [DOI] [PubMed] [Google Scholar]
- 51.Li Q, Ye L, Guo W, Wang M, Huang S, Peng X. PHF21B overexpression promotes cancer stem cell-like traits in prostate cancer cells by activating the Wnt/beta-catenin signaling pathway. J Exp Clin Cancer Res. 2017;36:85. doi: 10.1186/s13046-017-0560-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang HF, Wu C, Alshareef A, Gupta N, Zhao Q, Xu XE, Jiao JW, Li EM, Xu LY, Lai R. The PI3K/AKT/c-MYC Axis promotes the Acquisition of Cancer Stem-like Features in esophageal squamous cell carcinoma. Stem Cells. 2016;34:2040–2051. doi: 10.1002/stem.2395. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, He Q, Ding X, Zhao T, Zhao L, Wang A. SOD2 is a C-myc target gene that promotes the migration and invasion of tongue squamous cell carcinoma involving cancer stem-like cells. Int J Biochem Cell Biol. 2015;60:139–146. doi: 10.1016/j.biocel.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 54.Sun X, Zhang W, Li H, Niu C, Ou Y, Song L, Zhang Y. Stonin 2 overexpression is correlated with unfavorable prognosis and tumor invasion in epithelial ovarian Cancer. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences used in this study (DOCX 18 kb)
Table S2. Target sequences in this study. (DOCX 18 kb)
Figure S1. Intensity of STON2 and MUC1 immunohistochemical staining in ovarian cancer tissues. Representative images of STON2 and MUC1 staining in ovarian cancer tissues are shown. Scale bars represent approximately 50 μm. Staining intensity is graded as follows: 1+, weak; 2+, moderate; 3+, strong; 4+, very strong. (PNG 4594 kb)
Table S3. Peptide information from the LC-MS/MS data after analyzing parental cells and spheroids of 3AO cells (FDR < 0.05). Av. Ratio (Lin): the ratio of spheroids to parental cells. (XLS 154 kb)
Figure S2. (Related to pyrosequencing data shown in Fig. 5d) Representative pyrogramms of MUC1 promoter in 3AO and Caov3 cells (siNC or siSTON2). (TIF 760 kb)
Figure S3. 3AO and Caov3 cells were transfected with a DNMT1-specific siRNA or controls for 24 h, and then subjected to pyrosequencing to assess the DNA methylation status of MUC1 (three coupled independent samples) (A-B). Each column represents the relative average DNA methylation level at one CpG site compared to the control group (A). Raw pyrograms of representative experiments (B). (TIF 1569 kb)
Figure S5. (A-B) Cumulative survival probabilities (a, PFS and b, OS) were calculated using the Kaplan–Meier method (n = 145) based on STON2 and MUC1 expression. Survival rates were compared using a log-rank test. Patients expressing low or high levels of STON2 (A). Patients expressing low levels of STON2 combined with low MUC1 expression or high MUC1 expression (B). (TIF 912 kb)
Table S4. Association of STON2 expression with clinicopathological characteristics in patients with ovarian cancer (n = 165). (XLSX 11 kb)
Table S5. Association of MUC1 expression with clinicopathological characteristics in patients with ovarian cancer (n = 165). (XLSX 11 kb)
Figure S4. 3AO and Caov3 cells were transfected with a STON2-specific siRNA for 72 h, and the expression of DNMT1 (A), DNMT3A (B), or DNMT3B (C) was assayed using immunoblotting analysis. (TIF 808 kb)
Data Availability Statement
All data are fully available without restriction, and the RNA-seq data had been deposited in the SRA database (SRP135729) and wil be accessible without restriction from the date of publication.