Abstract
Background
In view of the limited knowledge of plasma biomarkers relating to cancer resistance to radiotherapy, we have set up screening, training and testing stages to investigate the microRNAs (miRNAs) expression profile in plasma to predict between the poor responsive and responsive groups after 6 months of radiotherapy.
Methods
Plasma was collected prior to and after radiotherapy, and the microRNA profiles were analyzed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) arrays. Candidate miRNAs were validated by single qRT-PCR assays from the training and testing set. The classifier for ancillary prognosis was developed by multiple logistic regression analysis to correlate the ratios of miRNAs expression levels with clinical data.
Results
We revealed that eight miRNAs expressions had significant changes after radiotherapy and the expression levels of miR-374a-5p, miR-342-5p and miR-519d-3p showed significant differences between the responsive and poor responsive groups in the pre-radiotherapy samples. The Kaplan–Meier curve analysis also showed that low miR-342-5p and miR-519d-3p expressions were associated with worse prognosis. Our results revealed two miRNA classifiers from the pre- and post-radiotherapy samples to predict radiotherapy response with area under curve values of 0.8923 and 0.9405.
Conclusions
The expression levels of miR-374a-5p, miR-342-5p and miR-519d-3p in plasma are associated with radiotherapy responses. Two miRNA classifiers could be developed as a potential non-invasive ancillary tool for predicting patient response to radiotherapy.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1711-4) contains supplementary material, which is available to authorized users.
Keywords: microRNAs, Radiotherapy, Biomarkers, Prognosis, Cancer
Background
With more than half of the cancer patients will receive radiation therapy as part of treatment in head and neck and rectal cancer, recurrence is still a major cause of treatment failure despite the advances of combination chemo-irradiation and preoperative radiotherapy [1]. Hence many studies have investigated the tumor radioresistance and signaling pathway [2–4]. Interestingly, it has been reported that the IGF1R, MAPK, PI3K and DNA repair signaling pathways are associated with radioresistance in several cancers [5–8]. However, the lack of a sensitive biomarker for radiotherapy and an understanding of mechanisms of related radioresistance hinder the success of radiation as a treatment for many patients [9–12].
The circulating miRNA profile is believed to be a molecular tool as disease biomarkers to predict or differentiate different types of disease [13–15]. The expression level of circulating miRNAs was related to the progression and development of cancers [13, 16]. In addition, circulating miRNA can be packaged in exosomes, the microvasculature or innate structures, enhancing its stability and avoiding degradation in biofluids [17, 18].
A large number of studies have examined the general and specific effects of miRNA perturbation in radiation-exposed cells [19–25]. However, cell line-based studies do not always correlate well with the results from clinical studies and no reliable and predictive biomarker could be applied in clinical for radiotherapy. Therefore, investigations on the non-invasive way to assess miRNA expression patterns to predict radiotherapy response are our primary interest. In the present work, we aimed to study the effects of radiotherapy on the expression levels of miRNAs in plasma. We further used these miRNA signatures to develop prediction classifiers for samples with an unknown radiotherapy status.
Materials and methods
Patients and samples
A total of 62 patients, including 26 and 36 patients with non-metastatic rectal cancer and head and neck cancers, respectively, were enrolled from December 2012–2015 (LSH-IRB-12-15). All patients were treated with radiation as part of curative treatment using a linear accelerator (6 MV, 10 MV) with standard dose fraction (2 Gy per day). Treatment response was evaluated 3 to 6 months after treatment including computed tomography (CT) imaging, magnetic resonance (MR) imaging and positron emission tomography (PET) imaging. Primary tumor with complete and partial response was defined as responsive group and the other as poor responsive. Response assessed with the use of response evaluation criteria in solid tumors (RECIST), version 1.1 [26]. In all, 15 poor responsive and 47 responsive patients were compared in this study.
Peripheral blood samples were collected from patients after obtaining informed consent. The samples were collected within 5 days before and after conclusion of radiotherapy. Samples were centrifuged and separated into plasma and carefully stored at − 80 °C.
RNA isolation from plasma samples
Total RNA from 0.5 ml of plasma was extracted by using TRIzol® LS Reagent and a mirVana™ miRNA Isolation Kit according to the standard protocol. We used Syn-cel-miR-39 as spiked-in control for some of the technical variability of plasma RNA extraction. The median of the syn-cel-miR-39 CT value obtained from all the samples was calculated. The RNA quality from the plasma was detected by a spectrophotometer (BioTek). All the RNA samples were carefully stored at − 80 °C.
Reverse transcription
cDNAs were reverse transcribed from miRNAs using a TaqMan™ MicroRNA Reverse Transcription Kit (Applied Biosystems) with 600 ng of total RNA and miRNA specific stem loop primers including miRNA PCR array A (Megaplex RT primers for Human Pool A) and the miRNA candidate pool. The conditions for reverse transcription were in accordance with the standard protocol. cDNA was generated using TaqMan® 2× Universal PCR master mix without UNG and TaqMan® Array Human MicroRNA Cards A or TaqMan® miRNA single assays.
MiRNA profiling and individual miRNA quantification by RT-PCR
miRNA PCR profiling in plasma samples were carried out using TaqMan® Array Human MicroRNA Cards (Applied Biosystems). To quantify individual miRNA levels, we used TaqMan® miRNA single assays as the main detection method as described before [27]. The expression of miRNAs was determined using the 2−ΔCT method relative to U6. The raw data of miRNA expressions was transformed to log10 form since the data with log10 form was in accordance with the normal distribution. In our analysis, the value of no detection miRNAs expression was replaced into − 4.5 value at log10 form.
Survival curve analysis
A public website of a smRNA-seq analysis of the clinical specimens was compared to survival status at YM500v3: a database for small RNA sequencing in human cancer research (http://driverdb.tms.cmu.edu.tw/ym500v3/index.php) [28]. Then, miR-374a-5p, miR-342-5p and miR-519d-3p expression values from clinical specimens were used to perform Kaplan–Meier survival curve analysis according to the clinical parameter provided in the same dataset. High and low expression groups were created by using the quantile and median value, respectively as a cutoff.
Data statistical analysis
Clinical characteristics between poor responsive and responsive patients were evaluated by using Pearson’s Chi squared test for categorical variables. Normality and Student’s t test were used for unpaired comparisons of two groups. All tests were two-tailed and were assessed by Levene’s test. All the statistical analyses were completed with GraphPad Prism software. The logistic regression of miRNA ratios combination were completed with SigmaPlot software.
Results
Identification of differentially expressed candidate miRNAs in plasma between poor responsive and responsive groups of radiotherapy
To identify potential miRNA signatures as a prognostic tool for radiotherapy patients, the miRNA profiles of plasma screened by high-throughput real-time miRNA PCR array were first reviewed. The plasma from patients was collected in prior to radiotherapy and after completion of radiotherapy (Additional file 1: Fig. S1). We monitored each patient’s condition at 6 months after radiotherapy, and the patients were characterized as poor responsive or responsive according to RECIST criteria by medical doctors. The screening set includes eight plasma samples collected prior to radiotherapy and seven plasma samples collected after radiotherapy (Table 1). The miRNA expression profiles in the plasma from poor responsive and responsive patients were compared. Total 22 candidate miRNAs were selected from the screening results (Additional file 1: Table S1).
Table 1.
Distribution of the clinical status of patients in this study
| Before radiation | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Screening set (n = 8) | Training set (n = 38) | |||||||||||||
| Poor response | Response | P | Poor response | Response | P | |||||||||
| N | Mean | SD | N | Mean | SD | N | Mean | SD | N | Mean | SD | |||
| Typeb | ||||||||||||||
| H&N | 2 | 2 | 1 | 8 | 17 | 0.69 | ||||||||
| Colorectal | 2 | 2 | 5 | 8 | ||||||||||
| Agea | 4 | 69.75 | 16.24 | 4 | 62 | 16.81 | 0.52 | 13 | 61.54 | 15.03 | 25 | 61.08 | 15.21 | 0.93 |
| Sexb | ||||||||||||||
| Female | 1 | 1 | 1 | 6 | 13 | 0.732 | ||||||||
| Male | 3 | 3 | 7 | 12 | ||||||||||
| Stageb | ||||||||||||||
| I | 1 | 1 | 0.23 | 1 | 4 | 0.111 | ||||||||
| II | 0 | 2 | 0 | 6 | ||||||||||
| III | 3 | 1 | 4 | 8 | ||||||||||
| IV | 0 | 0 | 8 | 7 | ||||||||||
| Total dosagea (Gy) | 3 | 63.87 | 11.68 | 4 | 60.25 | 11.37 | 0.69 | 13 | 66.18 | 7.58 | 24 | 63.61 | 9.29 | 0.398 |
| Chemotherapyb | 4 | 4 | 1 | 13 | 25 | 0.728 | ||||||||
| Before radiation | |||||||
|---|---|---|---|---|---|---|---|
| Testing (n = 24) | |||||||
| Poor response | Response | P | |||||
| N | Mean | SD | N | Mean | SD | ||
| Typeb | |||||||
| H&N | 1 | 10 | 0.902 | ||||
| Colorectal | 1 | 12 | |||||
| Agea | 2 | 69.5 | 9.19 | 22 | 54.45 | 11.99 | 0.1 |
| Sexb | |||||||
| Female | 2 | 15 | 0.343 | ||||
| Male | 0 | 7 | |||||
| Stageb | |||||||
| I | 0 | 3 | 0.081 | ||||
| II | 0 | 6 | |||||
| III | 0 | 8 | |||||
| IV | 2 | 5 | |||||
| Total dosagea (Gy) | 2 | 60.2 | 13.86 | 22 | 58.03 | 14.7 | 0.843 |
| Chemotherapyb | 2 | 22 | 0.577 | ||||
| After radiation | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Screening set (n = 7) | Training set (n = 31) | |||||||||||||
| Poor response | Response | P | Poor response | Response | P | |||||||||
| N | Mean | SD | N | Mean | SD | N | Mean | SD | N | Mean | SD | |||
| Typeb | ||||||||||||||
| H&N | 2 | 2 | 0.66 | 7 | 15 | 0.94 | ||||||||
| Colorectal | 1 | 2 | 3 | 6 | ||||||||||
| Agea | 3 | 62.67 | 9.71 | 4 | 62 | 16.81 | 0.95 | 10 | 58.6 | 13.23 | 21 | 60.29 | 14.73 | 0.75 |
| Sexb | ||||||||||||||
| Female | 1 | 1 | 0.81 | 5 | 11 | 0.9 | ||||||||
| Male | 2 | 3 | 5 | 10 | ||||||||||
| Stageb | ||||||||||||||
| I | 0 | 1 | 0.14 | 0 | 3 | 0.042* | ||||||||
| II | 0 | 2 | 0 | 5 | ||||||||||
| III | 3 | 1 | 2 | 7 | ||||||||||
| IV | 0 | 0 | 8 | 6 | ||||||||||
| Total dosagea (Gy) | 2 | 70.6 | 0.85 | 4 | 60.25 | 11.37 | 0.29 | 10 | 66.88 | 6.63 | 20 | 64.28 | 9.01 | 0.43 |
| Chemotherapyb | 3 | 4 | 0.81 | 10 | 21 | 0.36 | ||||||||
| After radiation | |||||||
|---|---|---|---|---|---|---|---|
| Testing (n = 24) | |||||||
| Poor response | Response | P | |||||
| N | Mean | SD | N | Mean | SD | ||
| Typeb | |||||||
| H&N | 1 | 10 | 0.902 | ||||
| Colorectal | 1 | 12 | |||||
| Agea | 2 | 69.5 | 9.19 | 22 | 54.45 | 11.99 | 0.1 |
| Sexb | |||||||
| Female | 2 | 15 | 0.343 | ||||
| Male | 0 | 7 | |||||
| Stageb | |||||||
| I | 0 | 3 | 0.081 | ||||
| II | 0 | 6 | |||||
| III | 0 | 8 | |||||
| IV | 2 | 5 | |||||
| Total dosagea (Gy) | 2 | 60.2 | 13.86 | 22 | 58.03 | 14.7 | 0.843 |
| Chemotherapyb | 2 | 22 | 0.577 | ||||
Each groups were well matched for age, gender. Mean, average of each samples
SD standard deviation, M male, F female
*P value < 0.05
aIndependent samples test
bPearson Chi Square
Changes in the miRNA expression levels after radiotherapy
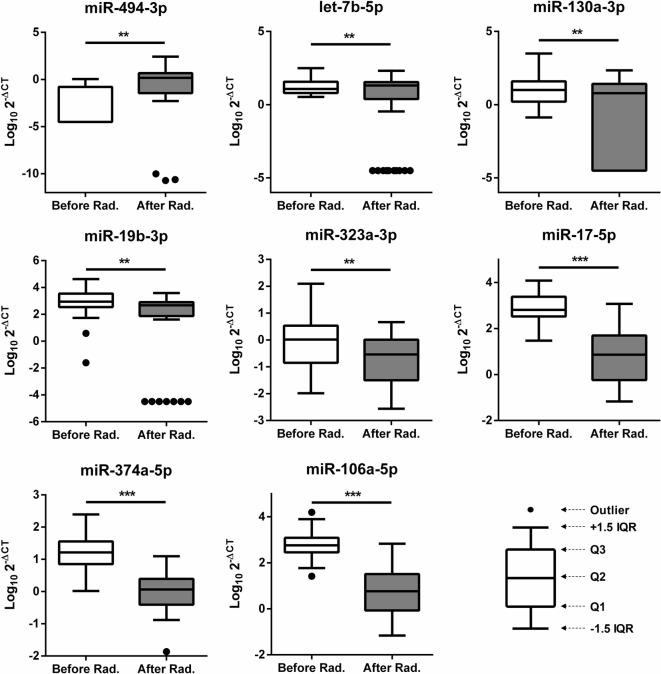
Investigations on the possible influence of radiotherapy on miRNA expression patterns were of primary interest. To validate the data from our screen, we checked the expression of 22 candidate miRNAs from the training set by single qRT-PCR. The training set included 38 different plasma samples collected from patients prior to radiotherapy and 31 different plasma samples collected from patients after radiotherapy (Table 1). First, we examined whether radiotherapy exerted any changes in these candidate miRNA expressions. Our results showed that eight miRNAs had significantly different expression levels after radiotherapy. miR-494-3p and let-7b-5p expression was increased, but the other six miRNAs—miR-130a-3p, miR-19b-3p, miR-323a-3p, miR-17-5p, miR-374a-5p, and miR-106a-5p—had significantly decreased expression (Fig. 1). We also assessed miRNA changes among the same patients before and after radiotherapy by the paired t-test. Nine miRNAs expressions including miR-299-5p and eight miRNAs above showed significant difference before and after radiotherapy (Additional file 1: Fig. S2). Interestingly, radiation-triggered deregulation of miR-494-3p, let-7b-5p, and miR-106a-5p has also been reported in previous studies [5, 13, 24, 25, 29–32].
Fig. 1.
Significant changes in the miRNAs expression levels in plasma after radiotherapy. miRNA levels from the plasma of patients detected by qRT-PCR using RNU6 as a control. The Y axis presents the expression level (Log10 2−ΔCT). Rad radiation. Student’s t-test: *P value < 0.05; **P value < 0.01; ***P value < 0.001
miRNAs expression levels linked to radiotherapy responses
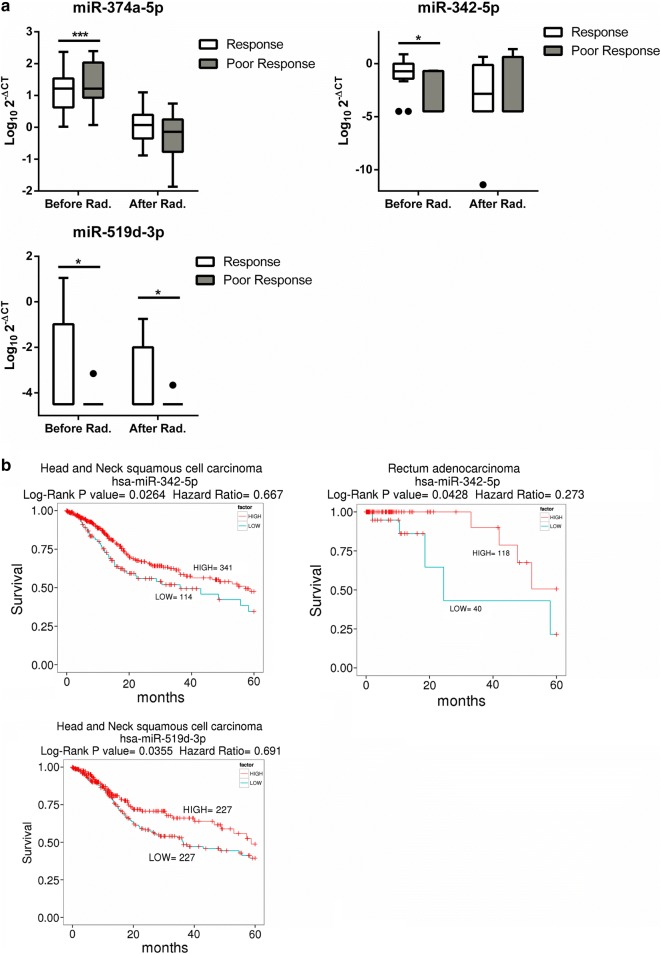
We further analyzed whether the plasma miRNA expression levels were associated with prognostic responses 6 months after radiotherapy. We investigated which candidate miRNA expression levels were different between the poor responsive and responsive groups (Fig. 2a). High expression levels of miR-374a-5p and low expression levels of miR-342-5p and miR-519d-3p prior to radiotherapy were observed in the poor responsive group (p < 0.0001, p = 0.044 and p = 0.014, respectively). In addition, low expression levels of miR-519d-3p after radiation were also shown in the poor responsive group (p = 0.0251). These results suggest that higher levels of miR-374a-5p and lower levels of miR-342-5p or miR-519d-3p in plasma could be linked to worse prognosis. Interestingly, the previous study reported that miR-374b-5p expression is linked to the radiation resistance in HNSCC [13]. Further, we utilized a public website of smRNA-seq analysis of the clinical cancer specimens [28]. Kaplan–Meier plot was analyzed to check for an association between miR-374a-5p, miR-342-5p or miR-519d-3p expression and 5-year survival. (Fig. 2b and Additional file 1: Fig. S3). Interestingly, both of head and neck squamous cell carcinoma and rectum adenocarcinoma patients with low miR-342-5p expression had significantly shorter survival than those in higher expression group (p = 0.0264 and 0.0428, respectively). Lower miR-519d-3p expression also had significantly shorter 5-year survival (p = 0.0355).
Fig. 2.
The difference in the miRNA expression levels between the poor responsive and responsive groups. a Difference in the miRNA expression in the poor responsive vs responsive groups before or after radiation. miRNA levels from the plasma of patients detected by qRT-PCR using RNU6 as a control. b The Kaplan–Meier survival curve of head and neck patients: low miRNA expression versus high miRNA expression. The statistical significance of the difference between the two groups was showed. The Y axis presents the expression level (Log10 2−ΔCT). Rad radiation. Student’s t-test: *P value < 0.05; **P value < 0.01; ***P value < 0.001
To develop a miRNA signature-based predicative model for patients with unknown radiation responses, we carried out a ROC analysis for the all candidate miRNAs (Additional file 1: Table S2). The AUC values of let-7b-5p and miR-342-5p were 0.722 and 0. 762, respectively, in the pre-radiotherapy samples (Table 2A). These values suggest that plasma let-7b-5p and miR-342-5p levels are good potential candidates for radiotherapy biomarkers.
Table 2.
The discriminatory ability of the miRNA expression profile for the poor responsive and responsive groups
| A | ||
|---|---|---|
| miRNA | AUC | |
| Before radiation | After radiation | |
| miR-494-3p | 0.552 | 0.636 |
| let-7b-5p | 0.722 | 0.552 |
| miR-323a-3p | 0.562 | 0.512 |
| miR-19b-3p | 0.603 | 0.547 |
| miR-342-5p | 0.762 | 0.527 |
| miR-374a-5p | 0.568 | 0.625 |
| miR-519d | 0.658 | 0.647 |
| B | |
|---|---|
| miRNA ratio | AUC |
| Before radiation | |
| 130a-3p/let-7b-5p | 0.788 |
| 130a-3p/19b-3p | 0.763 |
| 130a-3p/374a-5p | 0.763 |
| 130a-3p/17-5p | 0.745 |
| 106a-5p/130a-3p | 0.732 |
| After radiation | |
| 130a-3p/let-7b-5p | 0.752 |
| 628-5p/let-7b-5p | 0.714 |
| 130a-3p/148a-3p | 0.686 |
| 148a-3p/494-3p | 0.676 |
| 148a-3p/628-5p | 0.671 |
The expression of 22 candidate miRNAs were changed to the ratio form to eliminate normalization issue in plasma. The top five miRNA ratios were statistically calculated their AUC values by ROC analysis form (A) before radiation group and (B) after radiation group
Set up two classifiers to predict radiotherapy responses
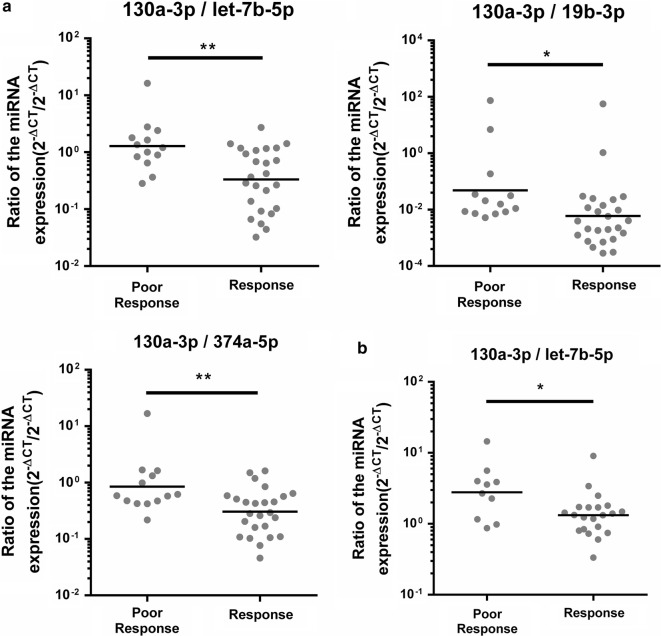
There is no strong evidence to indicate which miRNA or non-coding RNA is the appropriate internal control to normalize miRNA expression levels in plasma so far. Therefore, we utilized the ratio method, which divided two miRNAs expression levels from the same sample to eliminate the normalization issue. We calculated all miRNAs combination ratios and selected the miRNAs combination ratios with top five values of AUC (Table 2B). In the pre-radiation samples, the ratio levels of miR-130a-3p/let-7b-5p, miR-130a-3p/miR-19b-3p, and miR-130a-3p/miR-374a-5p were significantly different between poor responsive and responsive patients (p = 0.00122, 0.0419, and 0.0087, respectively) (Fig. 3a), and their AUC values were 0.788, 0.763, and 0.763, respectively. Interestingly, in the post-radiotherapy samples, the ratio levels of miR-130a-3p/let-7b-5p was also significantly different (p = 0.03147) (Fig. 3b), and its AUC value to discriminate poor responsive from responsive patients was 0.752.
Fig. 3.
The scatter plots of miRNAs expression ratio. a The scatter plots of miR-130a-3p/let-7b-5p, miR-130a-3p/miR-19b-3p and miR-130a-3p/miR-374a-5p were shown to distinguish responsive or poor responsive in the pre-radiation samples. b The scatter plots of miR-130a-3p/let-7b-5p was shown to distinguish responsive or poor responsive in the post-radiation samples. The Y axis was presents the ratio (2−ΔCT/2−ΔCT). Student’s t-test: *P value < 0.05; **P value < 0.01; ***P value < 0.001
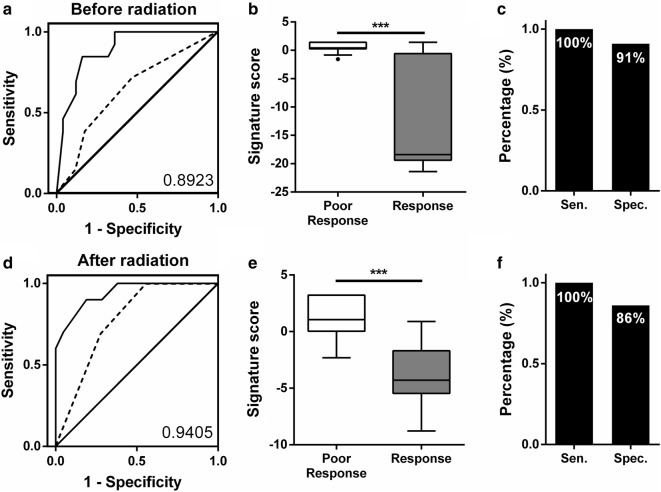
To establish a proper model to further estimate the radiotherapy responses, the different ratios of the miRNA data from the training set were combined to calculate the formula using multiple logistic regression. Therefore, we established two classifiers that could significantly distinguish the poor responsive from responsive patients, and these two classifiers could predict the radiation responses 6 months after radiotherapy (Fig. 4). For the pre-radiotherapy samples (n = 38), the classifier including three miRNA ratios—miR-130a-3p/let-7b-5p, miR-130a-3p/miR-19b-3p and miR-130a-3p/miR-374a-5p—with tumor stage data, and the AUC values was 0.8923 (95% CI 0.7910 to 0.9936) (Fig. 4a). For post-radiotherapy samples (n = 31), the AUC of the classifier, which included two miRNA ratios—miR-130a-3p/let-7b-5p and miR-130a-3p/miR-148a-3p—with tumor stage data, reached 0.9405 (95% CI 0.8591 to 1.022) (Fig. 4d). We further analyzed the distribution of the two signatures (Fig. 4b, e). Moreover, we validated these two signatures by testing another sample set (n = 24). In the pre-radiation samples, the classifier could detect poor responsive from responsive patients at a cut-off point of 0.2145 with 100% sensitivity and 91% specificity (Fig. 4c). In the post-radiation samples, the classifier could detect poor responsive from responsive patients at a cut-off point of 2.865 with 100% sensitivity and 86% specificity (Fig. 4f). Poor responsive patients were identified by both classifiers.
Fig. 4.
The ROC analysis of miRNA combinations. a–c The ROC analysis for miR-130a-3p/let-7b-5p, miR-130a-3p/miR-19b-3p, miR-130a-3p/miR-374a-5p and tumor stage were shown to distinguish responsive or poor response patients before radiation treatment. d–f The ROC analysis for the miR-130a-3p/let-7b-5p, miR-130a-3p/miR-148a-3p and tumor stage were shown to distinguish responsive or poor responsive patients after radiation treatment. Dashed line presents the ROC of tumor stage. b, e The box plots show the classifier distribution that combines the miRNA ratios with clinical data from training set. c, f The histogram shows the best accuracy of the sensitivity and specificity from the testing set. Res responsive patients, Poor poor responsive patients, Sen sensitivity, Spec specificity, AUC area under the curve
Discussion
In this study, we aimed to establish plasma miRNAs as ancillary predictive biomarkers for radiotherapy. Furthermore, we compared miRNA expression before and after treatment, and revealed that the expression levels of eight miRNAs had significant changes after radiotherapy. Interestingly, in the pre-radiation samples, we revealed that the expression levels of miRNA-374-5p, miR-342-5p and miR-519d-3p were significantly different between the responsive and poor responsive groups. These data suggested that the expression levels of three miRNAs may influence radiation sensitivity.
The let-7 family of miRNAs is a group of well-known tumor suppressor miRNAs, and many studies showed its levels are affected by radiation in vitro and in vivo [20]. Among them, let-7b is transcriptionally repressed by p53, and this mechanism depends on functional p53 and radiation-activated ATM signaling [33]. In mice with functional p53, a decrease in let-7b levels was observed in the more radiosensitive tissues upon radiation. These results are consistent with our finding that the let-7b-5p levels significantly decreased only in the plasma of the radiotherapy responsive group. Previous studies showed that the levels of miR-494-3p increased upon radiation in glioma cells [30]. Moreover, miR-494-3p could induce the radiosensitivity of oral squamous cell carcinoma by downregulating Bmi1 [25]. We similarly observed that the levels of miR-494-3p are increased after radiotherapy, and higher levels of miR-494-3p were expressed in the responsive group. Furthermore, it has also been reported that levels of miR-19b and miR-17 decreased in lymphocytes after radiation [20, 31]. However, changes in the miR-106 levels were observed in lung, thyroid MCF-7 and blood cells after radiation [20, 21, 34, 35]. In addition, the decrease or increase in these miRNA levels may not be consistent between cells and plasma, which may be due to tissue-specific or functional differences between cells and extracellular conditions.
Our results showed that three initial miRNAs in plasma—miR-374a-5p, miR-342-5p and miR-519d-3p—are involved in the prognosis of radiation responses as shown in Fig. 2. Summerer et al. demonstrated that high expression of miR-374b-5p in the plasma of individuals with HNSCC correlated with worse prognosis. Interestingly, miR-374a-5p and miR-374b-5p are present in the same seed region, so both of them may regulate the same radiation response-related genes. However, the mechanisms of miR-374a-5p and the other two miRNAs, miR-342-5p and miR-519d-3p, involved in radiotherapy responses were unclear until now, and our results show that three miRNAs have low AUC values for predicting radiotherapy outcomes. In addition, previous studies showed the expression of miR-296-5p and miR-16 have changed after radiotherapy and proposed that their expressions were related to the patients’ survival [10, 36]. However, small sample size or lack of sufficient predictive model to assess prognosis of radiotherapy in these studies limited the application in clinical use.
We applied each candidate miRNAs expression level to the combination of the ratio of miRNAs expression and tumor stage data, which produced two classifiers to predict radiotherapy outcomes 6 months after radiotherapy. The combination of the expression ratios levels of miR-130a-3p/let-7b-5p, miR-130a-3p/miR-19b-3p, and miR-130a-3p/miR-374a-5p and the tumor stage were up-regulated in poor responsive patients’ pre-radiotherapy samples. Moreover, the combination of the expression ratios of miR-130a-3p/let-7b-5p and miR-130a-3p/miR-148a-3p were up-regulated in poor responsive patients’ post-radiotherapy samples. It is noted that both classifiers contained miR-130 expression. High miR-130 expression has been found in radiation-resistant lung and prostate cells [5, 37]. We observed that miR-130 expression levels was significantly decreased in plasma but no significant differences were observed between the poor responsive and responsive groups after radiation. Therefore, we established two miRNA bio-signature models that could act as ancillary prognostic tools for radiotherapy patients, to predict responses 6 months after radiotherapy, which revealed 100% sensitivity in the testing set. If poor responsive can be identified before or just after initial radiotherapy, the patient may receive an alternative radiation process or other active therapy. However, any bio-signature requires multiple cohorts to validate its reproducibility, and then it can be applied as a clinical biomarker. The two classifiers in this study to predict radiotherapy outcomes require more validation in different cohorts and different types of cancer.
Conclusions
To date, no clinical tools could predict the therapeutic effects of radiation therapy. This study applied the miRNAs expression in plasma as ancillary predictive biomarkers for prognosis of radiotherapy. The expressions of miR-374a-5p, miR-342-5p and miR-519d-3p were observed the significant difference between the radiotherapy outcomes in prior of radiotherapy. Patients with lower miR-342-5p or miR-519d-3p expression had significantly shorter 5-year survival. Two classifiers were established from pre- and post-radiotherapy samples to predict radiotherapy outcome 6 months after radiotherapy with area under the curve (AUC) values of 0.8923 and 0.9405.
Additional file
Additional file 1. Additional figures and tables.
Authors’ contributions
NHM, TSC, CLC and CCC designed and supervised this study. ALL, YNC, YRC and CHL implemented the experiment and established predictive model. SCL, ALL and CHL analyzed clinical data. NHM and ALL wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Sung-Nine Wang and Yuan-Fu Chan of National Central University for assisting experiments. The authors thank the technical supports provided by Core Facilities for High Throughput Experimental Analysis of Institute of Systems Biology and Bioinformatics, National Central University.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All research methods and analytical data are available from the authors.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All subjects were informed and agreed to participate in this program. The study was approved by the Institutional Review Board (IRB) of Taiwan Landseed Hospital.
Funding
This work was supported by the following programs: Ministry of Science and Technology, Taiwan (MOST106-2320-B-008-005-MY3), the National Central University-Landseed Hospital United Research Center (NCU-LSH-101-A-021, NCU-LSH-106-A-004), Landseed Hospital (2016-02), the Delta Research Center (NCU-DEL-105-A-004), Academia Sinica, Taiwan (BM10701010023) and NCU-Landseed Chronic Disease Research Center.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- miRNA
microRNA
- AUC
area under the curve
- CT
computed tomography
- MRI
magnetic resonance imaging
- PET
positron emission tomography
- RECIST
response evaluation criteria in solid tumors
- ROC
receiver operating characteristic
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
Contributor Information
An-Lun Li, Email: t982020@gmail.com.
Tao-Sang Chung, Email: samchungs@gmail.com.
Yao-Ning Chan, Email: jimmy1010374@gmail.com.
Chien-Lung Chen, Email: chencl4922@gmail.com.
Shih-Chieh Lin, Email: jaylin@mail.ncku.edu.tw.
Yun-Ru Chiang, Email: wendy91000@gmail.com.
Chen-Huan Lin, Email: chlpig27@gmail.com.
Chi-Ching Chen, Email: chen3971@landseed.com.tw.
Nianhan Ma, Phone: +886-34227151, Email: nianhan.ma@g.ncu.edu.tw.
References
- 1.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. 2005;104:1129–1137. doi: 10.1002/cncr.21324. [DOI] [PubMed] [Google Scholar]
- 2.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8:545–554. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 3.Toustrup K, Sorensen BS, Metwally MA, Tramm T, Mortensen LS, Overgaard J, Alsner J. Validation of a 15-gene hypoxia classifier in head and neck cancer for prospective use in clinical trials. Acta Oncol. 2016;55:1091–1098. doi: 10.3109/0284186X.2016.1167959. [DOI] [PubMed] [Google Scholar]
- 4.Ishigami T, Uzawa K, Higo M, Nomura H, Saito K, Kato Y, Nakashima D, Shiiba M, Bukawa H, Yokoe H, et al. Genes and molecular pathways related to radioresistance of oral squamous cell carcinoma cells. Int J Cancer. 2007;120:2262–2270. doi: 10.1002/ijc.22561. [DOI] [PubMed] [Google Scholar]
- 5.Wang XC, Du LQ, Tian LL, Wu HL, Jiang XY, Zhang H, Li DG, Wang YY, Wu HY, She Y, et al. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer. 2011;72:92–99. doi: 10.1016/j.lungcan.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Salim H, Akbar NS, Zong D, Vaculova AH, Lewensohn R, Moshfegh A, Viktorsson K, Zhivotovsky B. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br J Cancer. 2012;107:1361–1373. doi: 10.1038/bjc.2012.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu JQ, Yi HM, Ye X, Zhu JF, Yi H, Li LN, Xiao T, Yuan L, Li JY, Wang YY, et al. MiRNA-203 reduces nasopharyngeal carcinoma radioresistance by targeting IL8/AKT Signaling. Mol Cancer Ther. 2015;14:2653–2664. doi: 10.1158/1535-7163.MCT-15-0461. [DOI] [PubMed] [Google Scholar]
- 8.Tommelein J, De Vlieghere E, Verset L, Melsens E, Leenders J, Descamps B, Debucquoy A, Vanhove C, Pauwels P, Gespach CP, et al. Radiotherapy-activated cancer-associated fibroblasts promote tumor progression through paracrine IGF1R activation. Cancer Res. 2018;78:659–670. doi: 10.1158/0008-5472.CAN-17-0524. [DOI] [PubMed] [Google Scholar]
- 9.Jacob NK, Cooley JV, Yee TN, Jacob J, Alder H, Wickramasinghe P, Maclean KH, Chakravarti A. Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS ONE. 2013;8:e57603. doi: 10.1371/journal.pone.0057603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Q, Li B, Li P, Shi Z, Vaughn A, Zhu L, Fu S. Plasma microRNAs to predict the response of radiotherapy in esophageal squamous cell carcinoma patients. Am J Transl Res. 2015;7:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 11.Moertl S, Mutschelknaus L, Heider T, Atkinson MJ. MicroRNAs as novel elements in personalized radiotherapy. Transl Cancer Res. 2016;5:S1262–S1269. doi: 10.21037/tcr.2016.11.37. [DOI] [Google Scholar]
- 12.Kerns SL, Dorling L, Fachal L, Bentzen S, Pharoah PD, Barnes DR, Gomez-Caamano A, Carballo AM, Dearnaley DP, Peleteiro P, et al. Meta-analysis of genome wide association studies identifies genetic markers of late toxicity following radiotherapy for prostate cancer. EBioMedicine. 2016;10:150–163. doi: 10.1016/j.ebiom.2016.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summerer I, Unger K, Braselmann H, Schuettrumpf L, Maihoefer C, Baumeister P, Kirchner T, Niyazi M, Sage E, Specht HM, et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer. 2015;113:76–82. doi: 10.1038/bjc.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Schena FP, Serino G, Sallustio F. MicroRNAs in kidney diseases: new promising biomarkers for diagnosis and monitoring. Nephrol Dial Transplant. 2014;29:755–763. doi: 10.1093/ndt/gft223. [DOI] [PubMed] [Google Scholar]
- 16.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mall C, Rocke DM, Durbin-Johnson B, Weiss RH. Stability of miRNA in human urine supports its biomarker potential. Biomark Med. 2013;7:623–631. doi: 10.2217/bmm.13.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandellini P, Rancati T, Valdagni R, Zaffaroni N. miRNAs in tumor radiation response: bystanders or participants? Trends Mol Med. 2014;20:529–539. doi: 10.1016/j.molmed.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Templin T, Paul S, Amundson SA, Young EF, Barker CA, Wolden SL, Smilenov LB. Radiation-induced micro-RNA expression changes in peripheral blood cells of radiotherapy patients. Int J Radiat Oncol Biol Phys. 2011;80:549–557. doi: 10.1016/j.ijrobp.2010.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S, Wang J, Ding N, Hu W, Zhang X, Wang B, Hua J, Wei W, Zhu Q. Exosome-mediated microRNA transfer plays a role in radiation-induced bystander effect. RNA Biol. 2015;12:1355–1363. doi: 10.1080/15476286.2015.1100795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu SY, Wu AT, Liu SH. MicroRNA-17-5p regulated apoptosis-related protein expression and radiosensitivity in oral squamous cell carcinoma caused by betel nut chewing. Oncotarget. 2016;7:51482–51493. doi: 10.18632/oncotarget.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang T, Yin L, Wu J, Gu JJ, Wu JZ, Chen D, Yu HL, Ding K, Zhang N, Du MY, et al. MicroRNA-19b-3p regulates nasopharyngeal carcinoma radiosensitivity by targeting TNFAIP3/NF-kappaB axis. J Exp Clin Cancer Res. 2016;35:188. doi: 10.1186/s13046-016-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng JH, Yu CC, Lee YC, Lin CW, Chang WW, Kuo YL. miR-494-3p induces cellular senescence and enhances radiosensitivity in human oral squamous carcinoma cells. Int J Mol Sci. 2016;17:1092. doi: 10.3390/ijms17071092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Liu SM, Lu J, Lee HC, Chung FH, Ma N. miR-524-5p suppresses the growth of oncogenic BRAF melanoma by targeting BRAF and ERK2. Oncotarget. 2014;5:9444–9459. doi: 10.18632/oncotarget.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung IF, Chang SJ, Chen CY, Liu SH, Li CY, Chan CH, Shih CC, Cheng WC. YM500v3: a database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2017;45:D925–D931. doi: 10.1093/nar/gkw1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marta GN, Garicochea B, Carvalho AL, Real JM, Kowalski LP. MicroRNAs, cancer and ionizing radiation: where are we? Rev Assoc Med Bras. 1992;2015(61):275–281. doi: 10.1590/1806-9282.61.03.275. [DOI] [PubMed] [Google Scholar]
- 30.Kwak SY, Yang JS, Kim BY, Bae IH, Han YH. Ionizing radiation-inducible miR-494 promotes glioma cell invasion through EGFR stabilization by targeting p190B rhoGAP. Biochim Biophys Acta. 2014;1843:508–516. doi: 10.1016/j.bbamcr.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Girardi C, De Pitta C, Casara S, Sales G, Lanfranchi G, Celotti L, Mognato M. Analysis of miRNA and mRNA expression profiles highlights alterations in ionizing radiation response of human lymphocytes under modeled microgravity. PLoS ONE. 2012;7:e31293. doi: 10.1371/journal.pone.0031293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 33.Saleh AD, Savage JE, Cao L, Soule BP, Ly D, DeGraff W, Harris CC, Mitchell JB, Simone NL. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS ONE. 2011;6:e24429. doi: 10.1371/journal.pone.0024429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abou-El-Ardat K, Monsieurs P, Anastasov N, Atkinson M, Derradji H, De Meyer T, Bekaert S, Van Criekinge W, Baatout S. Low dose irradiation of thyroid cells reveals a unique transcriptomic and epigenetic signature in RET/PTC-positive cells. Mutat Res. 2012;731:27–40. doi: 10.1016/j.mrfmmm.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, Park IC, Jin YW, An S. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35:81–86. [PubMed] [Google Scholar]
- 36.Maia D, de Carvalho AC, Horst MA, Carvalho AL, Scapulatempo-Neto C, Vettore AL. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J Transl Med. 2015;13:262. doi: 10.1186/s12967-015-0621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDermott N, Meunier A, Wong S, Buchete V, Marignol L. Profiling of a panel of radioresistant prostate cancer cells identifies deregulation of key miRNAs. Clin Transl Radiat Oncol. 2017;2:63–68. doi: 10.1016/j.ctro.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional figures and tables.
Data Availability Statement
All research methods and analytical data are available from the authors.