Abstract
Increasing evidence indicates that long non-coding RNAs (lncRNAs) regulate gene or protein expression; however, their function in the progression of hepatic fibrosis remains unclear. Hepatic fibrosis is a continuous wound-healing process caused by numerous chronic hepatic diseases, and the activation of hepatic stellate cells (HSCs) is generally considered to be a pivotal step in hepatic fibrosis. In the process of hepatic fibrosis, some lncRNAs regulates diverse cellular processes. Here are several examples: the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and liver fibrosis-associated lncRNA1 (lnc-LFAR1) promote HSC activation in the progression of hepatic fibrosis via the transforming growth factor-β signaling pathway; the lncRNA HIF 1 alpha-antisense RNA 1 (HIF1A-AS1) and Maternally expressed gene 3 reduce HSC activation which are associated with DNA methylation; the lncRNA plasmacytoma variant translocation 1, Homeobox (HOX) transcript antisense RNA and MALAT1 promote HSC activation as competing endogenous RNAs (ceRNAs); the long intergenic non-coding RNA-p21 (lncRNA-p21) and Growth arrest-specific transcript 5 reduce HSC activation as ceRNAs. As we get to know more about the function of lncRNAs in hepatic fibrosis, more and more ideas for the molecular targeted therapy in hepatic fibrosis will be put forward.
Keywords: Long non-coding RNAs, Hepatic fibrosis, Hepatic stellate cell, TGF-β signaling pathway, DNA methylation, ceRNA
Background
Hepatic fibrosis is a continuous wound-healing process that results in the dysregulation of extracellular matrix (ECM) proteins and the distortion of normal liver architecture [1]. Many chronic hepatic diseases, such as viral hepatitis, alcohol toxicity, drug abuse, metabolic syndrome, hereditary disorders of metabolism, autoimmune hepatitis and Clonorchis sinensis infection, lead to hepatic fibrosis and even cirrhosis [2], which is the primary stage of hepatic carcinoma, leading to one of the major causes of mortality in cancer worldwide.
Although extensive studies on hepatic fibrosis have been reported, their regulatory mechanisms are still partially understood. The activation of hepatic stellate cells (HSCs), the resident perisinusoidal cell type, is generally considered to be a pivotal step in hepatic fibrosis [3]. In normal hepatic tissue, HSCs with abundant vitamin A stores are quiescent. Following with hepatic injury of any etiology, the quiescent HSCs lose their stored vitamin A and trans-differentiate into fibrogenic myofibroblast-like cells. The activated HSCs are identified as proliferative cells that express ECM, and secrete profibrogenic mediators, thereby contributing to the fibrosis [4]. Therefore, the suppression of the HSC activation is regarded to be a potential therapeutic target for hepatic fibrosis.
Genome tiling arrays and cap analysis gene expression showed that non-protein coding RNAs (ncRNAs), which were considered to be ‘‘evolutionary junk” in the past, have more functions in transcription [5, 6]. Recently, a large number of ncRNA molecules have been identified by RNA microarrays and next-generation sequencing of transcriptomes [7]. NcRNAs are classified into two types based on their relative sizes. Those less than 200 nucleotides (nt) are called small or short non-coding RNAs, while those longer than 200 nt are called long non-coding RNAs (lncRNAs) [8]. LncRNAs are considered to play roles in physiological conditions as well as in several human diseases, including cancer, metabolic diseases, cardiovascular diseases and so on [9]. Increasing evidence has suggested that lncRNAs regulate gene or protein expression by coordinating epigenetic, transcriptional, or post-transcriptional processes [10]. But the function of lncRNAs in hepatic fibrosis remains elusive.
The goal of this review is to summarize the roles of lncRNAs in hepatic fibrosis, including the regulation on transforming growth factor (TGF)-β signaling pathway, DNA methylation and competing endogenous RNAs (ceRNAs) (Fig. 1) according to current knowledge. It will illustrate some important information for the treatment hepatic fibrosis and present novel guidance in future researches.
Fig. 1.
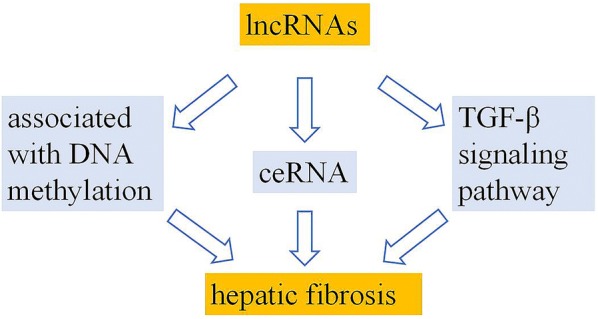
It is the summary of the mechanism of lncRNAs that regulate hepatic fibrosis through TGF-β signaling pathway, DNA methylation and ceRNA in this review
The interaction of lncRNAs and the transforming growth factor (TGF)-β signaling pathway in hepatic fibrosis
TGF-β is a key regulator of liver physiology and pathology during the process of initial liver injury-inflammation-fibrosis [11]. TGF-β1, a potent fibrogenic cytokine from the autocrine or paracrine pathway, is a crucial signal that promotes HSC activation [12]. Some studies have shown that lncRNAs interact with the TGF-β signaling pathway to promote HSC activation and then induce hepatic fibrosis.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) which is located in human chromosome 11q13.1 (mouse chromosome 19qA), also known as nuclear-enriched abundant transcript 2 (NEAT2), is a widely expressed lncRNA, and was firstly identified through subtractive hybridization in stage I of non-small cell lung cancer [13, 14]. A growing number of evidence indicated that MALAT1 was closely related to various pathological processes, including diabetes complications and hepatic carcinoma [15], and could influence the progression of hepatic fibrosis by repressing the expression and function of silent information regulator 1 [SIRT1, a Nicotinamide adenine dinucleotide (NAD)-dependent class III protein deacetylase] [16]. As one of the best characterized deacetylase enzymes, SIRT1 can protect cultured cells against metabolic, geneotoxic, hypoxic, and heat stress by deacetylating a number of key transcription factors [17], while it can induce the deacetylation of Smad3 (a downstream mediator of TGF-β signaling pathway) and weaken the ability of Smad3 binding to the promoter of fibrogenic genes, such as collagen type I gene promoters, which means that the activation of SIRT1 attenuates TGF-β signaling and then reduces TGF-β-stimulated collagen expression [18, 19]. In summary, MALAT1 can promote the HSC activation through blocking the SIRT1 mediated inhibition of TGF-β signaling pathway in the progression of hepatic fibrosis (Fig. 2a). What’s more, MALAT1 is also reported that it acts as a competing endogenous RNA for miR-101b to regulate RAS-related C3 botulinum substrate 1 (Rac1) and contributes to hepatic fibrosis [20] (Fig. 4).
Fig. 2.
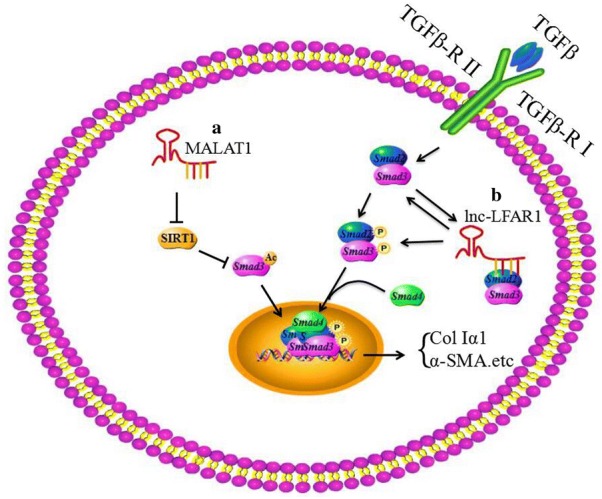
lncRNAs regulate hepatic fibrosis via TGF-β signaling pathway. a MALAT1 represses SIRT1 to inhibit the deacetylation of Smad3. Then the deacetylation of Smad3 binds to fibrogenic genes, such as colIα1, to induce the expression of colIα1. b lnc-LFAR1 is induced by Smad2/3 and in turn to promote the phosphorylation of Smad2/3, that provides a positive feedback loop to enhance Smad2/3 binding to the target gene, therefore causing the high expression of colIα1 and α-SMA. The deposition of ECM and activation of HSCs contribute to the hepatic fibrosis
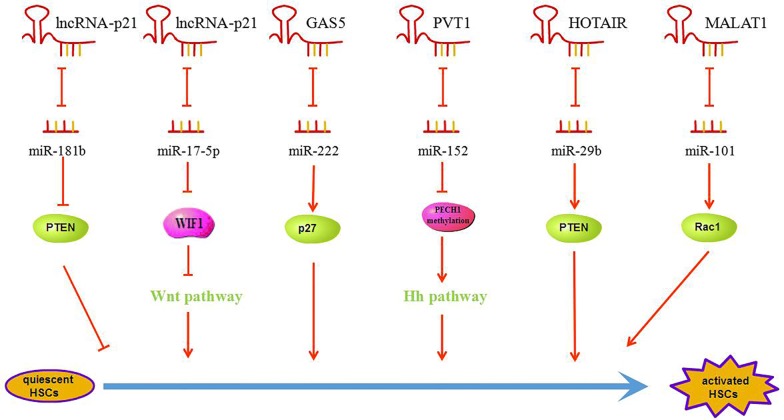
Fig. 4.
LncRNA represses miRNA to regulate the process of liver fibrosis. LncRNA-p21 inhibits the activation of HSCs through miR-181b and miR-17-5p; GAS5 inhibits the activation of HSCs through miR-222. PVT1, HOTAIR and MALAT1 promote the activation of HSCs through miR-152, miR-29b and miR-101 respectively
The liver fibrosis-associated lncRNA1 (lnc-LFAR1) is a 734 nt transcript and it was originally identified as a liver-enriched lncRNA in fibrotic liver of mice. The lnc-LFAR1 of mice is located in chromosome 4q25, and it adjacents to the CYP2U1 and HADH genes which are the same as human [21]. ALGGEN-PROMO and JASPAR software analysis have shown that there are three potential Smad2/3 binding sites (SBE) in the promoter of lnc-LFAR1, which means that Smad2/3 can bind to the promoter of lnc-LFAR1 to increase its expression. Furthermore, the lnc-LFAR1 in turn up regulates the expression of Smad2/3 and promotes Smad2/3 phosphorylation in liver fibrogenesis [21]. The phosphorylation of Smad2/3 promotes its nuclear translocation and the ability binding to the target promoters, such as collagen type I gene [22]. All in all, lnc-LFAR1 induces the activation of HSCs to promote hepatic fibrosis by interacting with TGF-β signaling pathway (Fig. 2b).
lncRNA associates with DNA methylation to inhibit the activation of HSCs in hepatic fibrosis
DNA methylation is a type of epigenetic modification in mammals and involves in numerous biological processes, including transposable element silencing, genomic imprinting and X chromosome inactivation [23]. The process of DNA methylation is regulated by methyltransferases, for examples, DNA methyltransferases (DNMT1, DNMT3a, DNMT3b) induce de novo methylation and ten–eleven translocation methylcytosine dioxygenase (TET) family member enzymes (TET1, 2 and 3) induce DNA demethylation re-activated or re-expressed silenced genes. DNMTs and TETs are critical in the cycle of DNA methylation and demethylation [24]. Advancing studies indicate that DNMTs and TETs play significant roles to change 5-methylcytosine and 5-hydroxymethylcytosine during HSC transdifferentiation to myofibroblast-like cells [25]. Increasing evidence shows that several lncRNAs are associated with DNA methylation to inhibit the activation of HSCs in hepatic fibrosis.
Maternally expressed gene 3 [MEG3, which is also known as gene trap locus 2 (GTL2)], is a lncRNA with the length of 1.6 kb nucleotides. It is a part of the DLK1–MEG3 imprinting locus and located at human chromosome 14q32 and at mouse distal chromosome 12 [26]. It is expressed in virous human tissues and acts as a tumor suppressor [27]. Recently, the loss of MEG3 expression has been gradually proved in various types of human cancers, such as hepatic cancer, gastric cancer, lung cancer, glioma, cervical cancer, bladder cancer [28–34]. On the one hand, MEG3 can selectively regulate p53 target gene expression resulting in the accumulation of p53 protein, and leading to cell growth inhibition [35]; on the other hand, MEG3 activates p53, and then intervenes in the p53-dependent mitochondrial apoptosis pathway to increase mitochondrial cytochrome c release and to culminate in direct caspase activation [35]. The expression of MEG3 was negatively correlated with the differentially methylated regions (DMRs) hypermethylation level, suggesting that DNA methylation plays an important role in silencing the MGE3 gene [28]. DNA methyltransferase 1 (DNMT1) can maintain methylation pattern on the daughter strand after DNA methylation and contribute to hypermethylation of MEG3 gene promoter and decrease the expression of MEG3 [30]. MEG3 could activate p53 to cause caspase-3-dependent apoptosis and reduce the expression of alpha-1 type I collagen (colIα1) and α-smooth muscle actin (α-SMA) in activeted HSCs induced by TGF-β1 [36]. It is indicated that MEG3 plays a critical role in HSC activation and hepatic fibrogenesis (Fig. 3). Therefore, it reveals that high-expression of MEG3 are potentially regarded as a novel therapeutic target for treating liver fibrosis.
Fig. 3.
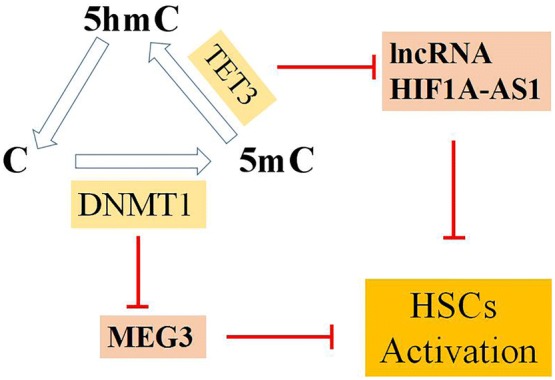
The transform among Cytosine (C), 5-methylcytosine (5-mC) and 5-hydroxymethylcytosine (5-hmC) forms the basis of the cycle of DNA methylation and demethylation. As the critical regulation enzymes in the cycle of DNA methylation and demethylation, DNMT1 and TET3 contribute to hepatic fibrosis through repressing MEG3 and lncRNA HIF1A-AS1
The lncRNA HIF 1 alpha-antisense RNA 1 (HIF1A-AS1) was initially reported in human kidney cancers and it is located on chromosome 14, with 2100 nt [37]. It has been demonstrated that the HIF1A-AS1 involves in the proliferation and apoptosis of vascular smooth muscle cells and the vascular endothelial cells [38, 39], and it is also related with the process of the non-small cell lung cancer and the colorectal carcinoma [40, 41]. HIF1A-AS1 acts as an inhibitor or an activator of cell proliferation and apoptosis depending on its binding partners and cell types. As one of the ten to eleven translocation (TET) family members, TET3 can catalyze 5-methylcytosine (5-mC) demethylate to 5-hydroxymethylcytosines (5-hmCs), which leads to cancer suppression [42]. In hepatic fibrosis, TET3 promotes the activation of HSCs through suppressing the expression of 1ncRNA1A-AS1 [43]. Thus, lncRNA HIF1A-AS1 interacts with the partner TET3 associated with DNA methylation to inhibit the activation of HSCs. These indicate that the up-regulation of lncRNA HIF1A-AS1 may be a potential therapy pathway for hepatic fibrosis (Fig. 3).
lncRNAs act as ceRNAs in hepatic fibrosis
MicroRNAs (miRNAs) pair with miRNA response elements (MREs) on target RNA transcripts resulting in degradation or translational repression of the target transcripts [44]. A competing endogenous RNA (ceRNA) is a endogenous origin transcript targeted by a miRNA that sequesters the activity of the bound miRNA, effectively de-repressing other targets of that miRNA [45]. LncRNAs have been gained substantial attention as ceRNAs to sponge miRNAs to consequently modulate the derepression of miRNA targets, thereby protecting their target mRNAs [46]. Recently, several studies have shown that lncRNAs act as ceRNAs which play an important regulatory role in the process of hepatic fibrosis.
The long intergenic non-coding RNA-p21 (lncRNA-p21), which resides 15 Kb upstream of the gene encoding the critical cell cycle regulator Cdkn1a (also known as p21), contains two exons comprising 3.1 Kb [47]. LncRNA-p21 functions as a downstream transcriptional repressor in the p53 pathway via activating p53 to promote apoptosis [48]. It has been reported that lncRNA-p21 has been deregulated in various human diseases, such as skin tumors, prostate cancer and hepatocellular carcinoma [49–51]. It also acts as a tumor suppressor in cancers, but the mechanism of the process remains unclear. LncRNA-p21 directly binds target mRNA to regulate the translation as a post-transcriptional inhibitor [52], while it also acts as a locus-restricted coactivator for p53-mediated p21 expression in regulating the G1/S checkpoint [53]. Furthermore, it has been proposed that the lncRNA-p21 is also able to regulate gene expression by directing the chromatin localization of protein binding partners [54]. As a tumor suppressor, the phosphatase and tensin homologue deleted on chromosome 10 (PTEN) is often deregulated in various cancers, and it is a direct target of miR-181b that has been reported in hepatic fibrosis [55]. Yu et al. demonstrated that the lncRNA-p21 enhanced PTEN expression through competitively binding miR-181b as a ceRNA and inhibited the activation of HSCs via PTEN/Akt pathway in hepatic fibrosis [56]. It is also reported that the lncRNA-p21 sponges miR-17-5p to inhibit WIF1 through Wnt/β-catenin pathway resulting in suppression the HSC activation [57]. According to the research results above, we conclude that the lncRNA-p21 acts as a ceRNA to prevent the HSC activation in hepatic fibrosis.
Growth arrest-specific transcript 5 (GAS5) was initially discovered in a screen for potential tumor suppressor genes which expressed at high levels during growth arrest and it was originally isolated from mouse embryo NIH 3T3 cells using subtraction hybridization [58]. GAS5 has been reported as a tumor suppressor in some kinds of cancers, and it has been shown the GAS5 is involved in proliferation, apoptosis, and migration of tumor cells in breast cancer, gastric cancer and prostate cancer [59–61]. The GAS5 directly binds miR-21 to down-regulate its expression at exon 4 of GAS5 and negatively regulate the expression of miR-21 in hepatocellular carcinoma [62]. Moreover, it was also demonstrated that the GAS5 acts as ceRNA to control cardiac fibroblast activation and cardiac fibrosis by targeting miR-21 through PTEN/MMP-2 signaling pathway [63]. In hepatic fibrosis, the GAS5 through interacting with miR-222 to promote the expression of p27 protein, thereby inhibiting the activation and proliferation of HSCs [64].
Plasmacytoma variant translocation 1 (PVT1) in size of > 300 nt is transcribed from a locus adjacent to the MYC locus on human chromosome 8q24 (mouse chromosome 15) [65]. Recently, the PVT1 is found to be up-regulated in a series of human tumors, such as hepatocellular carcinoma, ovarian cancer, malignant pleural mesothelioma, non-small lung cancer and renal cancer [66–70]. PVT1 was deemed as a mediator of ECM in the diabetic kidney [71], which suggested that the PVT1 might involve in fibrosis. Epithelial–mesenchymal transition (EMT) process is considered as a key event in the activation of HSCs and hepatic fibrosis via activating Hedgehog (Hh) signaling pathway [72]. Patched1 (PTCH1), a member of Hh family, is also a negative regulator of Hh pathway. PVT1 can indirectly enhance PTCH1 methylation and down-regulate the expression of PTCH1 via competitively binding miR-152. Therefore, the PVT1 may serve as a ceRNA for miR-152 through Hh pathway to regulate the activation of HSC in hepatic fibrosis [65].
Homeobox (HOX) transcript antisense RNA (HOTAIR) is a 2158 nt lncRNA that locates to a boundary of the HOXC locus, one of the four chromosomal loci (HOX A to D) containing the clustered HOX genes [73]. Accumulating studies have indicated that HOTAIR is up-regulated in multiple cancers, including breast cancer, lung adenocarcinoma, renal cell carcinoma, pancreatic cancer, hepatocellular carcinoma [74–78]. MiR-29b can up-regulate the expression of PTEN via DNMT3b to suppress liver fibrosis [79]. The HOTAIR acts as a ceRNA to sponge miR-29b and then attenuates DNMT3b, leading to enhancement of PTEN methylation that contributes to liver fibrosis [80].
Whether ceRNA is an inhibitor or an activator to HSC activation depends on the spongy of miRNAs (Fig. 4). The model of how lncRNA works as ceRNA to sponge miRNAs may be widely accepted in (Fig. 5). The binding sites between lncRNA and miRNA shown in this review are displayed in the table (Table 1).
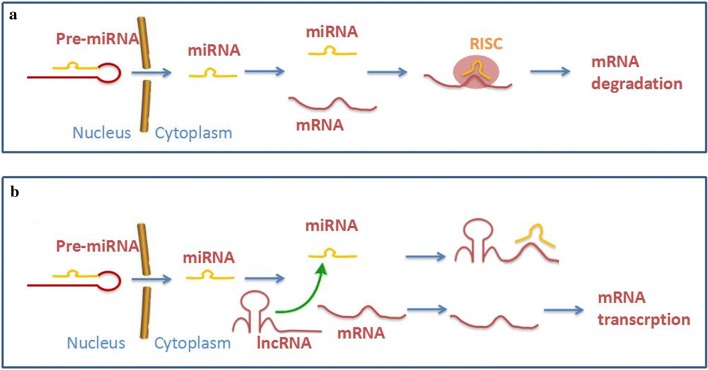
Fig. 5.
a The pre-miRNA gets out from the nucleus to be the mature miRNA. The mature miRNA incorporates into the RISC and the target mRNA to the target mRNA. b The activity of the miRNAs is inhibited by the presence of lncRNAs, which act as ceRNAs by sharing common MREs. Low levels of available miRNAs for the target mRNA translations
Table 1.
lncRNA as ceRNA in hepatic fibrosis
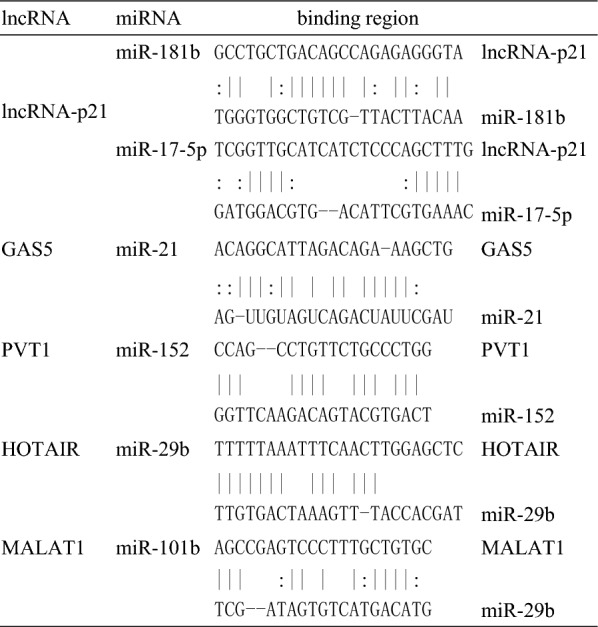
Only these lncRNAs that are described in this review are shown
Conclusion
Attention has been paid to lncRNA structure, function and evolution. Although there is a great interest in new lncRNAs, it is a scientific topic in the future. To explore new therapies of hepatic fibrosis, more lncRNAs have been found to be involved in the activation of HSCs.
MALAT1 and lncRNA-LFAR1 activate HSCs through the TGF-β signaling pathway. Some lncRNAs will be discovered to induce the activation of HSCs via the TGF-β signaling pathway in the future. Other signaling pathways, such as Wnt, NF-κB and Notch signaling pathway, are also related with the HSC activation. The lncRNA AC067945.2 down-regulates collagen expression in skin fibroblasts and it possibly correlates with the VEGF and Wnt signalling pathways [81]. Therefore, it is worthwhile to explore some new lncRNAs via other signaling pathways that take part in the activation of HSCs.
MEG3 and HIF1A-AS1 inhabit the HSC activation with DNA methylation. MEG3 and HIF1A-AS1 are repressed by DNMT1 and TET3 respectively. MEG3 causes the accumulation and activation of p53 that decreases proliferation and increases apoptosis of activated HSCs. The mechanism of HIF1A-AS1 inhabiting the HSC activation is still need to be explored. It is also possible that DNMT1 or TET3 regulates other lncRNAs in the process of hepatic fibrosis. Thus, the interaction between lncRNAs and other enzymes associated with methylation is also worth studying in hepatic fibrosis.
The last role of lncRNAs regulating the HSC activation is to be ceRNAs for miRNAs. PVT1, HOTAIR and MALAT1 promote the activation of HSCs. LncRNA-p21 and GAS5 reduce the activation of HSCs. The miRNAs are inhibited by the lncRNAs which act as ceRNAs via sharing common MREs. The interaction between lncRNAs and miRNAs is not a one-to-one relationship, for example, lncRNA-p21 represses miR-181b and miR-17-5p. In addition, lncRNA MIR100HG has been confirmed to encode miR-100, let-7a-2 and miR-125b-1. It is worth exploring whether any lncRNAs regulate miR-cluster in liver fibrosis or not.
The differences in the expression of lncRNA between normal and hepatic fibrotic tissues not only imply that lncRNAs may take part in the progression of the hepatic fibrosis, but also suggest that lncRNAs may be the biomarkers for the clinical diagnosis of hepatic fibrosis. Moreover, it is possible that not only lncRNAs itself, but also both the binding proteins and the target genes will be new therapeutic targets, which may lead to the development of new anti-fibrosis treatments.
Authors’ contributions
HP and LYW contributed equally to this work and wrote the manuscript. JJL and YQZ gave some helps for this work. WBA and JFW revised and approved the article prior to its being submitted for publication. All authors read and approved the final manuscript.
Acknowledgements
We are thankful for the financial support of the National Natural Science Foundation of China (Grant Number: 81670555) and the Science Research Innovation Foundation of Graduate Student of China Three Gorges University (No. 2018SSPY106). We also thank Shi-Zhen Zhao and Shan-Bing Yin for English language polishing.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Funding
The National Natural Science Foundation of China (Grant Numbers: 81670555) and the Science Research Innovation Foundation of Graduate Student of China Three Gorges University (No. 2018SSPY106) has given financial support.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- lncRNAs
long non-coding RNAs
- ECM
extracellular matrix
- HSCs
hepatic stellate cells
- ceRNAs
competing endogenous RNAs
- TGF-β
transforming growth factor-β
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- SIRT1
silent information regulator 1
- lnc-LFAR1
liver fibrosis-associated lncRNA1
- SBE
Smad2/3 binding sites
- DNMT
DNA methyltransferases
- TET
ten–eleven translocation methylcytosine dioxygenase
- MEG3
maternally expressed gene 3
- α-SMA
α-smooth muscle actin colIα1: alpha-1 type I collagen
- miRNAs
MicroRNAs
- HIF1A-AS1
lncRNA HIF 1 alpha-antisense RNA 1
- lncRNA-p21
long intergenic non-coding RNA-p21
- PTEN
the phosphatase and tensin homologue deleted on chromosome 10
- GAS5
growth arrest-specific transcript 5
- PVT1
plasmacytoma variant translocation 1
- HOTAIR
Homeobox (HOX) transcript antisense RNA
- MREs
miRNA response elements
Contributor Information
Hu Peng, Email: 315210671@qq.com.
Lin-Yan Wan, Email: wanlinyan0224@163.com.
Jia-Jie Liang, Email: 707817508@qq.com.
Yan-Qiong Zhang, Email: zhangyanqiong@ctgu.edu.cn.
Wen-Bing Ai, Phone: +86-781-639-6668, Email: 1043642574@qq.com.
Jiang-Feng Wu, Phone: +86-717-639-7198, Email: wujiangfeng@ctgu.edu.cn.
References
- 1.Wang J, et al. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget. 2015;6:7325–7338. doi: 10.18632/oncotarget.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, et al. Proteomic identification of potential Clonorchis sinensis excretory/secretory products capable of binding and activating human hepatic stellate cells. Parasitol Res. 2014;113:3063–3071. doi: 10.1007/s00436-014-3972-z. [DOI] [PubMed] [Google Scholar]
- 3.Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henderson NC, Iredale JP. Liver fibrosis: cellular mechanisms of progression and resolution. Clin Sci (Lond) 2007;112:265–280. doi: 10.1042/cs20060242. [DOI] [PubMed] [Google Scholar]
- 5.Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2006;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 7.Koh W, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad of Sci USA. 2014;111:7361–7366. doi: 10.1073/pnas.1405528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beermann J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 9.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi: 10.1042/bst20120020. [DOI] [PubMed] [Google Scholar]
- 10.Signal B, Gloss BS, Dinger ME. Computational approaches for functional prediction and characterisation of long noncoding RNAs. Trends Genet. 2016;32:620–637. doi: 10.1016/j.tig.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Dooley S, Dijke PT. TGF-β in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang JW, Hien TT, Lim SC, Jun DW, Choi HS, Yoon JH, Cho IJ, Kang KW. Pin1 induction in the fibrotic liver and its roles in TGF-β1 expression and Smad2/3 phosphorylation. J Hepatol. 2014;60:1235–1241. doi: 10.1016/j.jhep.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;39:1471–2164. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 15.Liu JY, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, et al. Silent information regulator 1 (SIRT1) ameliorates liver fibrosis via promoting activated stellate cell apoptosis and reversion. Toxicol Appl Pharmacol. 2015;289:163–176. doi: 10.1016/j.taap.2015.09.028. [DOI] [PubMed] [Google Scholar]
- 17.Guarente L. Hypoxic Hookup. Science. 2009;324:1281–1282. doi: 10.1126/science.1175679. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, et al. Transcriptional repression of SIRT1 by protein inhibitor of activated STAT 4 (PIAS4) in hepatic stellate cells contributes to liver fibrosis. Sci Rep. 2016;6:28432. doi: 10.1038/srep28432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei J, et al. The histone deacetylase sirtuin 1 is reduced in systemic sclerosis and abrogates fibrotic responses by targeting transforming growth factor β signaling. Arthritis Rheumatol. 2015;67:1323–1334. doi: 10.1002/art.39061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu F, et al. MALAT1 functions as a competing endogenous RNA to mediate Rac1 expression by sequestering miR-101b in liver fibrosis. Cell Cycle. 2015;24:3885–3896. doi: 10.1080/15384101.2015.1120917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, et al. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch pathways. Nat Commun. 2017;8:1–16. doi: 10.1038/s41467-017-00204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng XM, et al. Smad2 protects against TGF-β/Smad3-mediated renal fibrosis. J Am Soc Nephrol. 2010;21:1477–1487. doi: 10.1681/asn.2009121244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, et al. The real culprit in systemic lupus erythematosus: abnormal epigenetic regulation. Int J Mol Sci. 2015;16:11013–11033. doi: 10.3390/ijms160511013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page A, et al. Hepatic stellate cell transdifferentiation involves genome-wide remodeling of the DNA methylation landscape. J Hepatol. 2016;64:661–673. doi: 10.1016/j.jhep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–R53. doi: 10.1530/jme-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U. Loss of Imprinting and Allelic Switching at the DLK1-MEG3Locus in Human Hepatocellular Carcinoma. PLoS ONE. 2012;7:e49462. doi: 10.1371/journal.pone.0049462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun M, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tomor Biol. 2014;35:1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 30.Yan J, et al. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. 2014;31:879. doi: 10.1007/s12032-014-0879-6. [DOI] [PubMed] [Google Scholar]
- 31.Lu KH, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 33.Qin R, et al. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013;60:486–492. doi: 10.4149/neo_2013_063. [DOI] [PubMed] [Google Scholar]
- 34.Ying L, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol BioSyst. 2013;9:407. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.m702029200. [DOI] [PubMed] [Google Scholar]
- 36.He Y, et al. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta. 2014;1842:2204–2215. doi: 10.1016/j.bbadis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Bertozzi D, et al. Characterization of novel antisense HIF-1α transcripts in human cancers. Cell Cycle. 2011;10:3189–3197. doi: 10.4161/cc.10.18.17183. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, et al. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur J Cardiothorac Surq. 2015;47:439–446. doi: 10.1093/ejcts/ezu215. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, et al. Clopidogrel reduces apoptosis and promotes proliferation of human vascular endothelial cells induced by palmitic acid via suppression of the long non-coding RNA HIF1A-AS1 in vitro. Mol Cell Biochem. 2015;404:203–210. doi: 10.1007/s11010-015-2379-1. [DOI] [PubMed] [Google Scholar]
- 40.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Patho. 2015;7:7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 41.Gong W, Tian M, Qiu H, Yang Z. Elevated serum level of lncRNA-HIF1A-AS1 as a novel diagnostic predictor for worse prognosis in colorectal carcinoma. Cancer Biomark. 2017;12:54. doi: 10.3233/cbm-170179. [DOI] [PubMed] [Google Scholar]
- 42.Hsu CH, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 43.Zhang QQ, et al. TET3 mediates the activation of human hepatic stellate cells via modulating the expression of long non-coding RNA HIF1A-AS1. Int J Clin Exp Patho. 2014;11:7744–7751. [PMC free article] [PubMed] [Google Scholar]
- 44.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 46.Shi X, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Riley T, et al. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 48.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang YJ, Bikle DD. LncRNA profiling reveals new mechanism for VDR protection against skin cancer formation. J Steroid Biochem Mol Biol. 2014;144:87–90. doi: 10.1016/j.jsbmb.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 50.Işın M, Uysaler E, Özgür E, Köseoğlu H, Şanlı Öner, Yücel Ömer B, Gezer Uğur, Dalay N. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:168. doi: 10.3389/fgene.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ning Y, Yong F, Haibin Z, Hui S, Nan Z, Guangshun Y. LincRNA-p21 activates endoplasmic reticulum stress and inhibits hepatocellular carcinoma. Oncotarget. 2015;6:28151–28163. doi: 10.18632/oncotarget.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon J, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 In cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2013;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 55.Zheng J, et al. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol Cell Biochem. 2015;398:1–9. doi: 10.1007/s11010-014-2199-8. [DOI] [PubMed] [Google Scholar]
- 56.Yu F, et al. Identification of a novel lincRNA-p21-miR-181b-PTEN signaling cascade in liver fibrosis. Mediators Inflamm. 2016;2016:1–10. doi: 10.1155/2016/9856538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu F, et al. LincRNA-p21 inhibits the Wnt/β-catenin pathway in activated hepatic stellate cells via sponging MicroRNA-17-5p. Cell Physiol Biochem. 2017;41:1970–1980. doi: 10.1159/000472410. [DOI] [PubMed] [Google Scholar]
- 58.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/S0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 59.Mourtada-Maarabouni M, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 60.Sun M, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75:693–705. doi: 10.1002/pros.22952. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao H, et al. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. 2017;386:11–18. doi: 10.1016/j.tox.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 64.Yu F, et al. Long non-coding RNA growth arrest-specific transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism of competing endogenous RNA. J Biol Chem. 2015;290:28286–28298. doi: 10.1074/jbc.m115.683813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barsotti AM, et al. p53-dependent Induction of PVT1 and miR-1204. J Biol Chem. 2012;287:2509–2519. doi: 10.1074/jbc.m111.322875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, Yang N, Zhou WP, Li WL, Li W, Sun SH. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 67.Liu E, Liu Z, Zhou Y. Carboplatin-docetaxel-induced activity against ovarian cancer is dependent on up-regulated lncRNA PVT1. Int J Clin Exp Pathol. 2015;4:3803–3810. [PMC free article] [PubMed] [Google Scholar]
- 68.Riquelme E, et al. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol. 2014;9:998–1007. doi: 10.1097/jto.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang YR, et al. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;10:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Q, Yang F, Yang Z, Fang Z, Fu W, Chen W, Liu X, Zhao J, Wang Q, Hu X, Li L. Long noncoding RNA PVT1 inhibits renal cancer cell apoptosis by up-regulating Mcl-1. Oncotarget. 2017;8:101865–101875. doi: 10.18632/oncotarget.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS ONE. 2011;6:e18671. doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi SS, et al. Leptin promotes the myofibroblastic phenotype in hepatic stellate cells by activating the hedgehog pathway. J Biol Chem. 2010;285:36551–36560. doi: 10.1074/jbc.m110.168542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu L, et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 75.Yuanshun LIU, et al. Lentivirus-mediated silencing of HOTAIR lncRNA restores gefitinib sensitivity by activating Bax/Caspase-3 and suppressing TGF-α/EGFR signaling in lung adenocarcinoma. Oncol Lett. 2018;15:2829–2838. doi: 10.3892/ol.2017.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dasgupta P, et al. MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther. 2018 doi: 10.1158/1535-7163.mct-17-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim K, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Z, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 79.Zheng J, et al. Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation—a novel mechanism suppressing liver fibrosis. FEBS J. 2014;281:88–103. doi: 10.1111/febs.12574. [DOI] [PubMed] [Google Scholar]
- 80.Yu F, et al. HOTAIR epigenetically modulates PTEN expression via MicroRNA-29b: a novel mechanism in regulation of liver fibrosis. Mol Ther. 2017;25:205. doi: 10.1016/j.ymthe.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen L, Li J, et al. Overexpression of LncRNA AC067945.2 down-regulates collagen expression in skin fibroblasts and possibly correlates with the VEGF and Wnt signalling pathways. Cell Physiol Biochem. 2018;45(2):761–771. doi: 10.1159/000487167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.