Abstract
Background
The mitochondrial DNA (mtDNA) seems to influence in a large number of diseases, including HIV infection. Moreover, there is a substantial inter-individual variability in the CD4+ recovery in HIV-infected patients on combination antiretroviral therapy (cART). Our study aimed to analyze the association between mtDNA haplogroups and CD4+ recovery in HIV-infected patients on cART.
Methods
This is a retrospective study of 324 naïve cART patients with CD4+ < 200 cells/mm3, who were followed-up during 24 months after initiating cART. All patients had undetectable HIV viral load during the follow-up. Besides, we included 141 healthy controls. MtDNA genotyping was performed by using Sequenom’s MassARRAY platform. The primary outcome variable was the slope of CD4+ recovery. Patients were stratified into two groups by the median slope value of CD4+ (9.65 CD4+ cells/mm3/month). Logistic regression analyses were performed to calculate the odds of CD4+ recovery according to mtDNA haplogroups.
Results
Our study included European HIV-infected patients within the N macro-cluster. The baseline values of CD4+ T-cells were similar between groups of patients stratified by the P50th of the slope of CD4+ T-cells recovery. Patients in the low CD4+ T-cells recovery group were older (p = 0.001), but this variable was included in the multivariate models. When we analyzed the frequencies of mtDNA haplogroups, no significant differences between HIV-infected individuals and healthy controls were found. We did not find any significant association between mtDNA haplogroups and the slope of CD4+ T-cells recovery by linear regression analysis. However, Patients carrying haplogroup H had a higher odds of having a better CD4+ recovery (> 9.65 CD4+ cells/mm3/month) than patients without haplogroup H (p = 0.032). The adjusted logistic regression showed that patients carrying haplogroup H had a higher likelihood of achieving a CD4+ recovery > 9.65 CD4+ cells/mm3/month [adjusted odds ratio (aOR) = 1.75 (95% CI = 1.04; 2.95); p = 0.035].
Conclusions
European mitochondrial haplogroup H was associated with the improved CD4+ recovery in HIV-infected patients starting cART with CD4+ < 200 cells/mm3.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1717-y) contains supplementary material, which is available to authorized users.
Keywords: HIV, Mitochondria, Haplogroups, mtDNA, Immune reconstitution, cART
Background
The human immunodeficiency virus (HIV) infects and destroys the CD4+ T cell, promoting a continuous loss of CD4+ T cells that leads to immunodeficiency, opportunistic diseases, and death [1, 2]. The cART reduces the plasma HIV-RNA to undetectable levels and restores immunologic function, decreasing clinical progression and prolonging life [3]. However, despite suppression of HIV replication, a fraction of cART-treated patients fail to reconstitute CD4+ T-cell numbers sufficiently [4].
Mitochondrial dysfunction is related to acquired immune deficiency syndrome (AIDS) progression, in which there is mitochondrial DNA (mtDNA) depletion, increased reactive oxygen species (ROS) formation, antioxidant enzyme deficiency, and increased oxidative damage in patients with accelerated AIDS disease [5]. Additionally, mitochondrial toxicity due to nucleoside reverse transcriptase inhibitors may contribute to severe side effects observed in HIV-infected individuals on combination antiretroviral therapy (cART) [5].
Variations in mtDNA sequence are associated with several disorders [6], including HIV infection [5]. The combinations of mtDNA polymorphisms define mitochondrial haplogroups, which have a well-defined phylogeny among human populations [7]. In European Caucasians (N macro-cluster), people stratify in 4 cluster or major-haplogroups (HV, U, JT, and IWX) and several main haplogroups (H, V, pre-V, J, T, U, W, X, I) [7]. In European HIV-infected patients, mtDNA haplogroup has been associated with viroimmunological parameters, metabolic alterations, and AIDS progression [5, 8–10]. To our knowledge, only a previous study of our group has reported data about European mtDNA haplogroups and immune recovery in a cohort of HIV-infected patients starting cART with CD4+ count < 350 cells/mm3 in a tertiary hospital in Madrid (Spain) [11]. In the current multicentric study, we analyzed the association between European haplogroups and CD4+ T-cell recovery in naïve HIV-infected patients starting cART with CD4+ count < 200 cells/mm3 collected throughout the Spanish national territory.
Methods
Study population
We performed a retrospective study in 324 European HIV-infected patients receiving cART and 141 healthy controls of age and sex similar to patients. The Institutional Ethics Committee approved the study in concordance with the Declaration of Helsinki. All subjects provided informed consent to participate in the study.
The Spanish HIV BioBank of the CoRIS and the AIDS Research Institute IrsiCaixa-HIVACAT, Institut de Recerca en Ciències de la Salut Germans Trias i Pujol (Barcelona, Spain) provided patients. Epidemiological and clinical data were collected from medical records. The inclusion criteria were: (1) naïve for cART at inclusion in the cohort; (2) plasma HIV-RNA > 200 copies/mL; (3) starting cART with CD4+ counts < 200 cells/µL; (4) complete viral suppression (plasma HIV-RNA < 50 copies/mL) for two years after starting cART; (5) regular follow up of CD4+ counts and plasma HIV-RNA for two years after starting cART; (6) Individuals who were within the European N macro-haplogroup.
DNA genotyping
The Spanish National Genotyping Center (CeGen; http://www.cegen.org/) genotyped DNA samples by using the iPLEX® Gold technology and Agena Bioscience’s MassARRAY platform (San Diego, CA, USA). We selected 14 mtDNA polymorphisms within N macro-cluster [7, 11], to stratify patients by major-haplogroups or cluster (HV, IWX, U, and JT) and haplogroups (H, V, pre-V, J, T, I, W and X) (see Additional file 1).
Outcome variables
The primary outcome variable was the slope of CD4+ T-cells (summary measure) during 2 years of follow-up. Patients were stratified in two groups by the median value of CD4+ T-cells slope in the whole population of patients (9.65 CD4+ cells/mm3/month): high CD4+ recovery (≥ 9.65 CD4+ cells/mm3/month) and low CD4+ recovery (< 9.65 CD4+ cells/mm3/month).
Statistical analysis
Statistical analysis was performed with SPSS 22.0 software (SPSS INC, Chicago, IL, USA). All tests were two-tailed with p-values ≤ 0.05 considered significant. Categorical data and proportions were analyzed by using Chi squared test or Fisher´s exact test. Mann–Whitney U test was used to compare data between independent groups when the variables were continuous.
Generalized Linear Models (GLM) with a gamma distribution (log-link) were used to evaluate the differences in the slope of CD4+ T-cells recovery (continuous variable). Logistic regression models were used to calculate the odds of higher or lower CD4+ recovery according to mtDNA haplogroups, one for each haplogroup evaluated. The multivariate regression tests were adjusted by the main clinical characteristics at baseline: gender, age, length of HIV-infection, baseline CD4+ cells/mm3, HIV transmission route, hepatitis C and hepatitis B coinfection, and type of cART regimen.
Results
Characteristics of the study population
Our study included European HIV-infected patients within the N macro-cluster [7]. From the total Spanish AIDS Research Network (CoRIS) and AIDS Research Institute IrsiCaixa-HIVACAT Cohorts (n = 6160 HIV-patients) we only selected the HIV-patients meeting the inclusion criteria described in Methods section (n = 418). Lastly, 94 individuals were excluded from the analysis either because genotyping was not valid (n = 13) or because the haplogroup did not belong to the N-cluster or to a European haplogroup (n = 81). Thus, the final analysis included 324 patients (see Additional file 2). The baseline characteristics of patients stratified by the P50th of the slope of CD4+ T-cells recovery are shown in Table 1. Patients in the low CD4+ T-cells recovery group were older (p = 0.001), but this variable was included in the multivariate models. Moreover, the baseline values of CD4+ T-cells were similar between groups, but after 2 years of successful cART, the values CD4+ T-cells were higher in patients with high CD4+ recovery than low CD4+ recovery [513 (418.4; 636.5) vs. 274 (176.7; 365.6); p < 0.001].
Table 1.
Baseline clinical and epidemiological characteristics of HIV infected patients
| Characteristics | All patients | HIV groups | ||
|---|---|---|---|---|
| Low CD4+ recovery | High CD4+ recovery | p-value | ||
| No. | 324 | 162 | 162 | |
| Male | 264 (81.5%) | 138 (85.2%) | 126 (77.8%) | 0.086 |
| Age (years) | 41.0 (34.7; 49.2) | 43 (36.4; 51.3) | 39.9 (33.8; 46.5) | 0.001 |
| Time since HIV diagnosis (years) | 1 (1.0; 1.0) | 1 (1.0; 2.0) | 1 (1.0; 1.0) | 0.117 |
| CD4+ cell count at baseline (cells/μL) | 105 (41; 159) | 93.3 (37.5;147) | 114.5 (41.5;165) | 0.230 |
| Hepatitis C infection | 29 (8.9%) | 16 (9.9%) | 13 (8.0%) | 0.559 |
| Hepatitis B infection | 14 (4.3%) | 8 (4.9%) | 6 (3.7%) | 0.585 |
| cART regimen | ||||
| PI-based | 103 (31.8%) | 45 (28.0%) | 58 (35.8%) | 0.469 |
| NNRTI-based | 165 (51.1%) | 88 (54.7%) | 77 (47.5%) | |
| PI+ NNRTI-based | 38 (11.8%) | 20 (12.4%) | 18 (11.1%) | |
| Others | 17 (5.3%) | 8 (5.0%) | 9 (5.6%) | |
| HIV transmission route | ||||
| IDU | 51 (17.1%) | 30 (20.8%) | 21 (13.4%) | 0.219 |
| Homosexual transmission | 152 (50.8%) | 68 (47.2%) | 84 (54.2%) | |
| Heterosexual transmission | 96 (32.1%) | 46 (31.9%) | 50 (32.3%) | |
IDU intravenous drug users, HIV human immunodeficiency virus, cART combination antiretroviral therapy, PI HIV protease inhibitor, NNRTI non-nucleoside analogue HIV reverse transcriptase inhibitor
Statistical: Values were expressed as absolute number (percentage) and median (percentile 25; percentile 75). Significant differences are shown in bold. p-values were calculated by Chi square and Mann–Whitney tests
Characteristics of mtDNA haplogroups
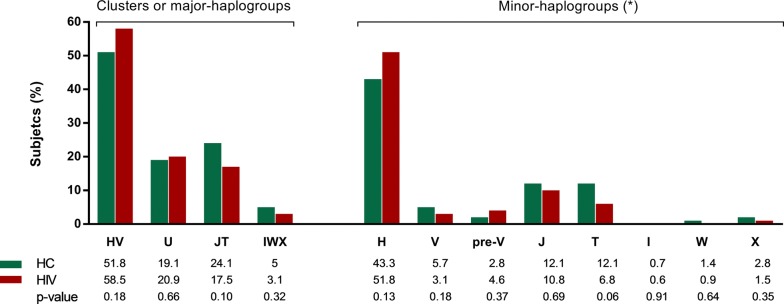
When we analyzed the frequencies of mtDNA haplogroups, no significant differences between HIV-infected individuals and healthy controls were found (Fig. 1). Note that healthy controls were similar to HIV-infected patients regarding gender and age. Moreover, the cluster IWX and minor haplogroups V, pre-V, I, X, and W were discarded for the genetic association study because these mtDNA haplogroups had low frequencies (< 5%) in HIV infected patients (Fig. 1). Thus, the genetic association tests were performed on the clusters HV, U, and JT; and on the haplogroups H, J, and T.
Fig. 1.
Frequencies of mtDNA haplogroups in HIV infected patients and healthy controls. Statistical: p-values were calculated by Chi square test. *The percentages of mtDNA haplogroups, for both HIV and HC groups, do not add to 100% because the cluster U was not stratified in minor-haplogroups. HIV human immunodeficiency virus, HC healthy controls
mtDNA haplogroups and CD4+ T-cell recovery
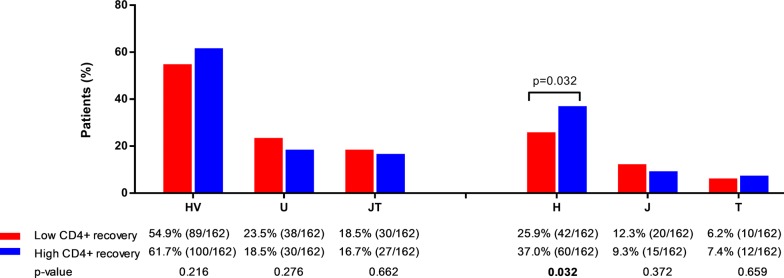
We did not find any significant association between mtDNA haplogroups and the slope of CD4+ T-cells recovery by linear regression analysis (Table 2). However, we found significant values when patients were stratified by the P50th of the slope of CD4+ T-cells recovery (Fig. 2). We found higher frequency of H haplogroup in patients with high CD4+ recovery (≥ 9.65 CD4+ cells/mm3/month) (p = 0.032). On the other hand, in multivariate analysis adjusted by the main clinical characteristics at baseline (see statistical analysis section), patients with haplogroup H had higher odds of having a better CD4+ T-cell recovery than patients without haplogroup H [adjusted odds ratio (aOR) = 1.75 (95% CI = 1.04; 2.95); p = 0.035]. No significant associations were found for the others haplogroups (Fig. 3; full description in Additional file 3).
Table 2.
Summary of the slopes of CD4+ T-cells recovery (cells/mm3/month) for each mtDNA haplogroup and the relationship between them in HIV-infected patients who started combined antiretroviral therapy
| Mt DNA haplogroups | Unadjusted analysis* | Adjusted analysis** | |||
|---|---|---|---|---|---|
| No | Yes | p-value | aAMR (95% CI) | p-value | |
| Clusters or major haplogroups | |||||
| HV | 9.1 (5.5; 14) | 10 (6.4; 14.7) | 0.288 | 0.98 (0.86; 1.13) | 0.839 |
| U | 9.8 (6.2; 14.1) | 9.1 (5.1; 13.9) | 0.321 | 0.91 (0.77; 1.08) | 0.300 |
| JT | 9.8 (6.2; 14) | 9.2 (5.6; 13.6) | 0.765 | 1.12 (0.94; 1.33) | 0.205 |
| Haplogroups | |||||
| H | 9.1 (5.6; 13.7) | 10.2 (6.9; 14.1) | 0.075 | 1.04 (0.91; 1.21) | 0.544 |
| J | 9.8 (6.2; 14) | 8.8 (5.2; 12.2) | 0.522 | 1.07 (0.86; 1.32) | 0.537 |
| T | 9.6 (6.1; 14) | 9.7 (6.8; 14.2) | 0.735 | 1.17 (0.90; 1.51) | 0.225 |
HIV human immunodeficiency virus, aAMR adjusted arithmetic mean ratio, CI confidence interval
Statistical: * Values were expressed as median (cells/mm3/month) (percentile 25; percentile 75). p-values were calculated by Mann–Whitney tests. ** Values were expressed as adjusted arithmetic mean ratio (aAMR) and 95% of confidence interval (95% CI) of the slopes of CD4+ T-cells recovery (cells/mm3/month). p-values were calculated by Generalized Linear Models test with a normal distribution (log-link). These regressions were adjusted by the most important clinical and epidemiological characteristics (gender, age, length of HIV-infection, baseline CD4+ cells/mm3, HIV transmission route related to IDU, hepatitis C and hepatitis B coinfection, and type of cART regimen)
Fig. 2.
Percentage of HIV-infected patients according to mtDNA haplogroups and CD4+ T cell recovery during the follow-up. Statistical: p-values were calculated by Chi squared test or Fisher’s exact test
Fig. 3.
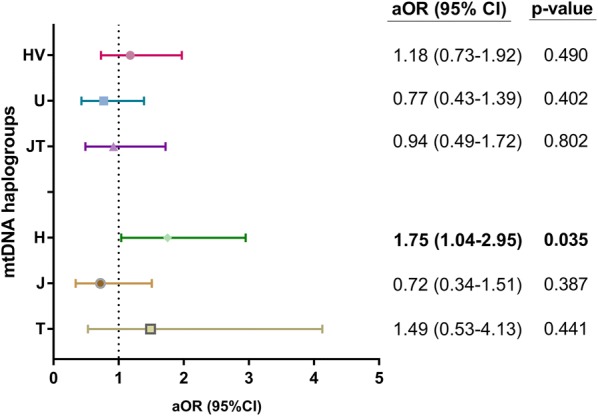
Summary of the association between mtDNA haplogroups and CD4+ T cell recovery in HIV-infected patients who started combination antiretroviral therapy. Statistical: Logistic regression models were used to calculate the odds of higher or lower CD4+ recovery according to mtDNA haplogroups, one for each haplogroup evaluated. These regressions were adjusted by the most important clinical and epidemiological characteristics (gender, age, length of HIV-infection, baseline CD4+ cells/mm3, HIV transmission route, hepatitis C and hepatitis B coinfection, and cART regimen). aOR adjusted odds ratio, 95% CI 95% of confidence interval
Discussion
In this study, haplogroup H was related to better CD4+ T-cells counts recovery in HIV-infected patients starting cART with low CD4 counts and followed during the first 24 months after initiating cART. This association was found both in the univariate and in the multivariate analysis adjusted by the most important baseline characteristics. However, we did not find any association when the continuous outcome measure (slope of CD4+ T-cells recovery) was used, possibly because there is no linear relationship between the analyzed variables.
The data found in this study confirm the positive influence of European haplogroup H on CD4+ T-cells count recovery in cART-treated patients, which was previously described by our group in a different cohort [11]. In addition to the influence of haplogroup H, in this previous article of Guzmán-Fulgencio et al. [11], we also found a worse CD4+ recovery in patients with haplogroup J and T. However, we did not observe any significant association for haplogroups J and T in the current study. The small sample size could explain this discrepancy in these haplogroups, which may have impaired the ability to detect less robust associations. However, we must also consider other factors. On the one hand, the article of Guzmán-Fulgencio et al. [11] studied a cohort of HIV-infected patients provided by a tertiary referral hospital center (Spain), with baseline CD4+ values < 350 cells/mm3, and followed during at least 24 months. On the other hand, we now analyzed a cohort of HIV-infected patients provided by a large number of hospitals spread throughout Spain (a sample more representative of Spanish population), with baseline CD4+ values < 200 cells/mm3 (a more restricted criteria), and followed during the first 24 months after starting cART (the same follow-up in all patients). Moreover, the type of statistical analysis applied was different in our previous study. In the article of Guzmán-Fulgencio et al. [11], a survival analysis was performed with CD4+ > 500 cells/mm3 as the primary outcome, whereas in the present study, we calculated the slope of CD4+ T-cells count for each patient and compared groups by logistic regression analysis.
Moreover, mtDNA haplogroups in other cohort have also been related to CD4+ T-cells recovery in HIV-infected patients who started cART. Grady et al. found that African L2 haplogroup was associated with decreased odds of CD4+ T-cells recovery after cART (CD4+ count change of ≥ 100 cells/mm3 and median CD4+ T-cell increases at 48 weeks of follow-up) in the AIDS Clinical Trials Group study 384 [12]. However, they did not find any significant association with European mitochondrial haplogroups in non-Hispanic white participants.
In the present study, the age of HIV-infected patients with worse CD4+ T-cells recovery was higher than patients with better CD4+ T-cells recovery, which could have had a negative impact since a poorer CD4+ T-cell counts recovery has been found in older HIV-infected patients starting cART [13]. However, we think that only 3 years of difference between groups is not relevant in adults of 35–45 years. In fact, we included the age in the multivariate analysis, and the significant association between haplogroup H and CD4+ T-cells recovery was maintained.
Variants in mtDNA which are not silent may have an essential role in adaptation to environmental conditions, by modulating specific mitochondrial functions [14]. Thus, the differences found in mtDNA haplogroups distribution among the different groups of patients in our study may be because haplogroup H is related to higher activity in the electron transport chain, producing higher levels of ATP and ROS than other haplogroups [15–17], increasing the immune response against HIV infection [9]. Furthermore, ROS production may lead to an up-regulation of antioxidant defenses without causing severe immune damage [18], contributing to proper immune function, ensuring control of HIV replication and, in turn, decreasing oxidative stress and apoptosis [19, 20]. Additionally, a higher degree of energetic efficiency could have a substantial impact on HIV infection since the efficiency of the metabolism regulate T-cell function and susceptibility to infection [21]. The functional role of mtDNA haplogroups is controversial, but it is possible that the effects of mitochondrial haplogroups may emerge under special conditions such as CD4+ T-cells recovery in HIV-infected patients starting cART. However, we could not perform functional experiments to determinate the energetic efficiency in isolated mitochondria of T-cells from patients.
Nowadays or in the future, our results may have an impact on clinical practice in those cases where cART starts with very low CD4+ T-cells. cART should begin in all HIV-infected patients, regardless of the CD4+ T-cells count in order to decrease the risk of HIV transmission and prevent AIDS-related illness [22]. However, late presentation for HIV care is a significant and persistent issue throughout the world [23–25], including developed countries with health systems with good access to health services [24]. Late presenters have delayed initiation of cART, CD4 values below 350 cells/mm3 and below 200 cells/mm3 in many cases [26], and higher risk of AIDS progression and death [27]. The initiation of cART with very low CD4+ T-cell counts is consistently associated with poorer outcomes of cART [28]. Thus, our data could provide information to improve the management of HIV-infected patients with poorer prognosis of CD4+ T-cells recovery.
Study limitations
Our study has some limitations that we should take into account to make a correct interpretation of the results. Firstly, this is a retrospective study and, therefore, the case record is selected a priori from patients surviving long enough to yield sufficient follow-up (at least 24 months after cART). Secondly, the sample size is limited, particularly in some haplogroups, which may have impaired the ability to detect less robust associations. Thirdly, although our results suggest that some variants in mtDNA may influence CD4+ T-cell recovery, it would be necessary to perform functional ROS and ATP measurement in the patients to provide additional confirmatory data. Fourthly, this study was carried out on Caucasian patients, and our conclusions are only truly applicable to this population.
Conclusions
In conclusion, European haplogroup H was associated with the improved CD4+ recovery in HIV-infected patients starting cART with CD4+ < 200 cells/mm3. This finding could be useful to understand the host genetic factors involved in the immune recovery of cART-treated patients. Additionally, it also may help to improve the management of patients with a poorer prognosis of immune recovery.
Additional files
Additional file 1. Summary of European mitochondrial DNA (mtDNA) haplogroups with their defining polymorphisms.
Additional file 2. Flow chart showing the sequential steps to select the HIV-patients included in the study.
Additional file 3. Summary of full multivariate model results for the regression between mitochondrial DNA (mtDNA) haplogroups and CD4+ T cell recovery in HIV-infected patients who started combination antiretroviral therapy.
Authors’ contributions
LMM and MGR: Investigation, methodology, writing—original draft. JB, MG, YMP, MM, JAI, EB and OJM: Data curation, methodology. MAJS: Data curation, methodology, review and editing. JMB, NR and SR: Conceptualization, formal analysis, writing—original draft, visualization, supervision. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank the Spanish National Genotyping Center (CEGEN-PRB2-ISCIII) for providing SNP genotyping services (http://www.cegen.org). CEGEN is supported by grant PT13/0001, ISCIII-SGEFI/FEDER. We also acknowledge the patients in this study for their participation and the Centro de Transfusión of Comunidad de Madrid for the healthy donor blood samples provided.
We acknowledge the Spanish HIV-1 BioBank integrated into the Spanish AIDS Research Network (RIS) and collaborating centers for the clinical samples provided. The HIV BioBank, integrated in the Spanish AIDS Research Network, is supported by ISCIII, Spanish Health Ministry (Grant nº RD06/0006/0035 and RD12/0017/0037) as part of the State Plan for Scientific and Technical Research and Innovation and co-financed by ISCIII–Sub-Directorate General for Research Assessment and Promotion and European Regional Development Fund (ERDF) and Foundation for Research and Prevention of AIDS in Spain (FIPSE). The RIS Cohort (CoRIS) is funded by the ISCIII through the Spanish AIDS Research Network (RISC03/173 and RD12/0017/0018) as part of the State Plan for Scientific and Technical Research and Innovation and co-financed by ISCIII–Sub-Directorate General for Research Assessment and Promotion and European Regional Development Fund (ERDF).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets analyzed during the current study available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Research Ethic Committee of the Instituto de Salud Carlos III and was conducted in accordance with the Declaration of Helsinki. All patients gave their written informed consent.
Funding
This work has been (partially) funded by Grants RD12/0017/0024 and RD16CIII/0002/0002 to SR, and RD12/0017/0031 and RD16/0025/0013 to JMB as part of the Health Research and Development Strategy, State Plan for Scientific and Technical Research and Innovation (2008–2011; 2013–2016) and co-financed by Institute of Health Carlos III, ISCIII–Sub-Directorate General for Research Assessment and Promotion and European Regional Development Fund (ERDF).
Luz Mª Medrano is supported by Spanish Carlos III Institute of Health (ISCIII) Madrid, Spain [Grant Number CD14/00002]. N Rallón is a Miguel Servet investigator from the ISCIII [Grant Number CP14/00198]; M. García is a predoctoral student co-funded by CP14/00198 Grant and Intramural Research Scholarship from IIS-FJD.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- HIV
human immunodeficiency virus
- AIDS
acquired immune deficiency syndrome
- mtDNA
mitochondrial DNA
- cART
combination antiretroviral therapy
- ROS
reactive oxygen species
Contributor Information
Luz M. Medrano, Email: luzmedranodios@gmail.com
Mónica Gutiérrez-Rivas, Email: mgutierrez19@hotmail.com.
Julià Blanco, Email: jblanco@irsicaixa.es.
Marcial García, Email: marcial.garcia@hospitalreyjuancarlos.es.
María A. Jiménez-Sousa, Email: majimenezsousa@yahoo.es
Yolanda M. Pacheco, Email: yolandam.pacheco.exts@juntadeandalucia.es
Marta Montero, Email: martamontero72@gmail.com.
José Antonio Iribarren, Email: joseantonio.iribarrenloyarte@osakidetza.eus.
Enrique Bernal, Email: ebm.hgurs@gmail.com.
Onofre Juan Martínez, Email: onofrejmartinez@hotmail.com.
José M. Benito, Phone: +34 91 544 37 20, Email: jbenito1@hotmail.com, Email: jose.benito@fjd.es
Norma Rallón, Email: normaibon@yahoo.com.
Salvador Resino, Phone: +34 918 223 266, Email: sresino@isciii.es.
References
- 1.Saag MS, Holodniy M, Kuritzkes DR, O’Brien WA, Coombs R, Poscher ME, Jacobsen DM, Shaw GM, Richman DD, Volberding PA. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 2.Brien WA, Hartigan PM, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff MS, Hamilton JD. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet. 2013;382:1525–1533. doi: 10.1016/S0140-6736(13)61809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cenderello G, De Maria A. Discordant responses to cART in HIV-1 patients in the era of high potency antiretroviral drugs: clinical evaluation, classification, management prospects. Expert Rev Anti Infect Ther. 2016;14:29–40. doi: 10.1586/14787210.2016.1106937. [DOI] [PubMed] [Google Scholar]
- 5.Hart AB, Samuels DC, Hulgan T. The other genome: a systematic review of studies of mitochondrial DNA haplogroups and outcomes of HIV infection and antiretroviral therapy. AIDS Rev. 2013;15:213–220. [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hulgan T, Samuels DC, Bush W, Ellis RJ, Letendre SL, Heaton RK, Franklin DR, Straub P, Murdock DG, Clifford DB, et al. Mitochondrial DNA haplogroups and neurocognitive impairment during HIV infection. Clin Infect Dis. 2015;61:1476–1484. doi: 10.1093/cid/civ527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, Kingsley L, Goedert JJ, Vlahov D, Donfield S, et al. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman-Fulgencio M, Jimenez JL, Garcia-Alvarez M, Bellon JM, Fernandez-Rodriguez A, Campos Y, Rodriguez C, Gonzalez-Garcia J, Riera M, Viciana P, et al. Mitochondrial haplogroups are associated with clinical pattern of AIDS progression in HIV-infected patients. J Acquir Immune Defic Syndr. 2013;63:178–183. doi: 10.1097/QAI.0b013e3182893f74. [DOI] [PubMed] [Google Scholar]
- 11.Guzmán-Fulgencio M, Berenguer J, Micheloud D, Fernández-Rodríguez A, García-Álvarez M, Jiménez-Sousa MA, Bellón JM, Campos Y, Cosín J, Aldámiz-Echevarría T, et al. European mitochondrial haplogroups are associated with CD4+ T cell recovery in HIV-infected patients on combination antiretroviral therapy. J Antimicrob Chemother. 2013;68:2349–2357. doi: 10.1093/jac/dkt206. [DOI] [PubMed] [Google Scholar]
- 12.Grady BJ, Samuels DC, Robbins GK, Selph D, Canter JA, Pollard RB, Haas DW, Shafer R, Kalams SA, Murdock DG, et al. Mitochondrial genomics and CD4 T-cell count recovery after antiretroviral therapy initiation in AIDS clinical trials group study 384. J Acquir Immune Defic Syndr. 2011;58:363–370. doi: 10.1097/QAI.0b013e31822c688b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micheloud D, Berenguer J, Bellón JM, Miralles P, Cosin J, de Quiros JC, Conde MS, Muñoz-Fernández MA, Resino S. Negative influence of age on CD4+ cell recovery after highly active antiretroviral therapy in naive HIV-1-infected patients with severe immunodeficiency. J Infect. 2008;56:130–136. doi: 10.1016/j.jinf.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arning L, Haghikia A, Taherzadeh-Fard E, Saft C, Andrich J, Pula B, Höxtermann S, Wieczorek S, Akkad DA, Perrech M, et al. Mitochondrial haplogroup H correlates with ATP levels and age at onset in Huntington disease. J Mol Med (Berl) 2010;88:431–436. doi: 10.1007/s00109-010-0589-2. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Redondo D, Marcuello A, Casajús JA, Ara I, Dahmani Y, Montoya J, Ruiz-Pesini E, López-Pérez MJ, Díez-Sánchez C. Human mitochondrial haplogroup H: the highest VO2max consumer—is it a paradox? Mitochondrion. 2010;10:102–107. doi: 10.1016/j.mito.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Marcuello A, Martinez-Redondo D, Dahmani Y, Casajus JA, Ruiz-Pesini E, Montoya J, Lopez-Perez MJ, Diez-Sanchez C. Human mitochondrial variants influence on oxygen consumption. Mitochondrion. 2009;9:27–30. doi: 10.1016/j.mito.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Ballard JW, Katewa SD, Melvin RG, Chan G. Comparative analysis of mitochondrial genotype and aging. Ann N Y Acad Sci. 2007;1114:93–106. doi: 10.1196/annals.1396.011. [DOI] [PubMed] [Google Scholar]
- 19.Treitinger A, Spada C, Verdi JC, Miranda AF, Oliveira OV, Silveira MV, Moriel P, Abdalla DS. Decreased antioxidant defence in individuals infected by the human immunodeficiency virus. Eur J Clin Invest. 2000;30:454–459. doi: 10.1046/j.1365-2362.2000.00642.x. [DOI] [PubMed] [Google Scholar]
- 20.Moretti S, Marcellini S, Boschini A, Famularo G, Santini G, Alesse E, Steinberg SM, Cifone MG, Kroemer G, De Simone C. Apoptosis and apoptosis-associated perturbations of peripheral blood lymphocytes during HIV infection: comparison between AIDS patients and asymptomatic long-term non-progressors. Clin Exp Immunol. 2000;122:364–373. doi: 10.1046/j.1365-2249.2000.01375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craveiro M, Clerc I, Sitbon M, Taylor N. Metabolic pathways as regulators of HIV infection. Curr Opin HIV AIDS. 2013;8:182–189. doi: 10.1097/COH.0b013e32835fc53e. [DOI] [PubMed] [Google Scholar]
- 22.Saag MS, Benson CA, Gandhi RT, Hoy JF, Landovitz RJ, Mugavero MJ, Sax PE, Smith DM, Thompson MA, Buchbinder SP, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2018 recommendations of the international antiviral society—USA Panel. JAMA. 2018;320:379–396. doi: 10.1001/jama.2018.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kranzer K, Govindasamy D, Ford N, Johnston V, Lawn SD. Quantifying and addressing losses along the continuum of care for people living with HIV infection in sub-Saharan Africa: a systematic review. J Int AIDS Soc. 2012;15:17383. doi: 10.7448/IAS.15.2.17383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croxford S, Yin Z, Burns F, Copas A, Town K, Desai S, Skingsley A, Delpech V, Opt TP. Linkage to HIV care following diagnosis in the WHO European Region: a systematic review and meta-analysis, 2006–2017. PLoS ONE. 2018;13:e0192403. doi: 10.1371/journal.pone.0192403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suarez-Garcia I, Sobrino-Vegas P, Dalmau D, Rubio R, Iribarren JA, Blanco JR, Gutierrez F, Montero Alonso M, Bernal E, Vinuesa Garcia D, et al. Clinical outcomes of patients infected with HIV through use of injected drugs compared to patients infected through sexual transmission: late presentation, delayed anti-retroviral treatment and higher mortality. Addiction. 2016;111:1235–1245. doi: 10.1111/add.13348. [DOI] [PubMed] [Google Scholar]
- 26.Antinori A, Coenen T, Costagiola D, Dedes N, Ellefson M, Gatell J, Girardi E, Johnson M, Kirk O, Lundgren J, et al. Late presentation of HIV infection: a consensus definition. HIV Med. 2011;12:61–64. doi: 10.1111/j.1468-1293.2010.00857.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Pan X, Ma Q, Yang J, Xu Y, Zheng J, Wang H, Zhou X, Jiang T, Jiang J, et al. HIV cause-specific deaths, mortality, risk factors, and the combined influence of HAART and late diagnosis in Zhejiang, China, 2006–2013. Sci Rep. 2017;7:42366. doi: 10.1038/srep42366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Molina JA, Diaz-Menendez M, Plana MN, Zamora J, Lopez-Velez R, Moreno S. Very late initiation of HAART impairs treatment response at 48 and 96 weeks: results from a meta-analysis of randomized clinical trials. J Antimicrob Chemother. 2012;67:312–321. doi: 10.1093/jac/dkr478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Summary of European mitochondrial DNA (mtDNA) haplogroups with their defining polymorphisms.
Additional file 2. Flow chart showing the sequential steps to select the HIV-patients included in the study.
Additional file 3. Summary of full multivariate model results for the regression between mitochondrial DNA (mtDNA) haplogroups and CD4+ T cell recovery in HIV-infected patients who started combination antiretroviral therapy.
Data Availability Statement
The datasets analyzed during the current study available from the corresponding author on reasonable request.