Summary
MicroRNAs (miRNAs) are part of a complex regulatory network that modulates cellular lipid metabolism. Here, we identify miR-124 as a regulator of triglyceride (TG) metabolism. This study advances our knowledge of the role of miR-124 in human hepatoma cells. Transcriptional profiling of Huh7.5 cells overexpressing miR-124 reveals enrichment for host factors involved in fatty acid oxidation among repressed miRNA targets. In addition, miR-124 down-regulates arylacetamide deacetylase (AADAC) and adipose triglyceride lipase, lipases proposed to mediate breakdown of hepatic TG stores for lipoprotein assembly and mitochondrial β-oxidation. Consistent with the inhibition of TG and fatty acid catabolism, miR-124 expression promotes cellular TG accumulation. Interestingly, miR-124 inhibits the production of hepatitis C virus, a virus that hijacks lipid pathways during its life cycle. Antiviral activity of miR-124 is consistent with repression of AADAC, a pro-viral host factor. Overall, our data highlight miR-124 as a novel regulator of TG metabolism in human hepatoma cells.
Subject Areas: Biological Sciences, Biochemistry, Biochemical Mechanism
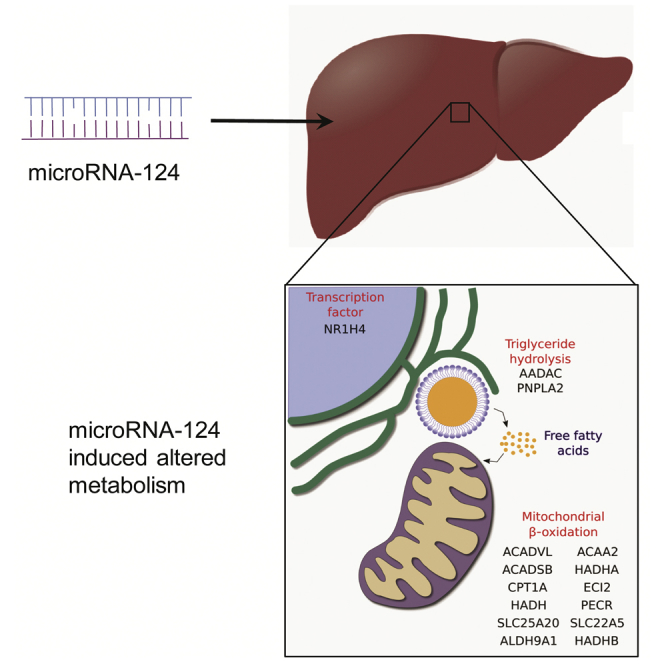
Graphical Abstract
Highlights
-
•
miR-124 regulates triglyceride and fatty acid metabolism
-
•
miR-124 represses genes associated with fatty acid and triglyceride breakdown
-
•
miR-124 promotes triglyceride accumulation in hepatoma cells
Biological Sciences; Biochemistry; Biochemical Mechanism
Introduction
Fatty acids serve as an important energy source for mammals in the fasted state (Houten and Wanders, 2010). The liver is the central organ in fatty acid metabolism, responsible for fatty acid uptake from serum, de novo lipid synthesis, and fatty acid oxidation or secretion in the form of lipoproteins (Mashek, 2013). Fatty acid oxidation serves to produce acetyl CoA, nicotinamide adenine dinucleotide (NADH), and flavin adenine dinucleotide (FADH2). NADH and FADH2 serve as electron carriers to feed into the electron transport chain to generate ATP. Both saturated and unsaturated fatty acids are broken down through reactions catalyzed in both mitochondria and peroxisomes. Although the key enzymes catalyzing fatty acid breakdown have been elucidated, the regulatory mechanisms governing fatty acid oxidation are not fully understood.
Under pathological conditions, hepatic fatty acid trafficking can be disrupted, resulting in increased fatty acid storage in the form of triglycerides (TGs). This manifests clinically as hepatic steatosis, as observed in hepatitis C virus (HCV) infection and fatty liver disease (Ress and Kaser, 2016). In the case of viral infection, the pathogen may manipulate fatty acid flux to create specific lipid-rich microenvironments to facilitate its life cycle (Chukkapalli et al., 2012). Proper regulatory controls on metabolic gene networks are integral to maintaining proper energy homeostasis and preventing hepatic metabolic disorders.
Recent work has demonstrated that microRNAs (miRNAs) are crucial to proper regulation of hepatic TG homeostasis (Moore et al., 2011). These small RNAs, ranging in size from 21 to 24 nucleotides, modulate gene expression through partial pairing with mRNAs, generally in the 3′ untranslated region (3′ UTR) (Pasquinelli, 2012). Canonical miRNA targeting of mRNAs results in a combination of transcript destabilization and translation repression (Pasquinelli, 2012). These small RNAs simultaneously regulate multiple targets within the same pathway (Ben-Hamo and Efroni, 2015). The critical role of miRNAs in the maintenance of hepatic lipid homeostasis has been established, with several miRNAs regulating aspects of fatty acid oxidation, lipid biosynthesis, and lipid excretion (Li et al., 2017, Moore et al., 2011, Singaravelu et al., 2014, Singaravelu et al., 2015a, Singaravelu et al., 2018). For example, miR-122 has been shown to extensively modulate hepatic lipid microenvironments (Esau et al., 2006). Given the abundance of the miRNA in the liver, it has been found to be directly involved in modulating cholesterol and hepatic fatty acid metabolism (Elmén et al., 2008, Esau et al., 2006, Krützfeldt et al., 2005). In addition, modulation of HCV replication by miR-122 has been well established (Jopling et al., 2005). A thorough understanding of the functional role of miRNAs in hepatic fatty acid metabolism is critical to properly define the etiology of metabolic disorders.
Recent studies have demonstrated the involvement of miR-124-3p (miR-124) in hepatobiliary pathologies (Liu et al., 2016, Ning et al., 2014). Expression of miR-124 has been shown to be directly regulated by the liver-enriched transcription factor, hepatocyte nuclear factor 4α (Ning et al., 2014). Recent work demonstrated that hepatic delivery of the miRNA suppresses tumorigenesis in mice (Ning et al., 2014). To date, miR-124 has mainly been examined in the context of processes in the central nervous system, where it is highly expressed (Sun et al., 2015), whereas its physiological function in the liver remains poorly studied.
Herein, we examine the role of miR-124 in hepatic lipid homeostasis. Our data suggest a novel role for this miRNA in the metabolic stress response. Genome-wide expression profiling reveals that miR-124 concertedly represses multiple genes involved in fatty acid oxidation and TG hydrolysis. Through repression of these catabolic pathways, miR-124 promotes hepatocellular TG storage. Interestingly, we also demonstrate that miR-124 impairs the infectivity of HCV, a hepatotropic virus with a strong dependence on hepatic lipid pathways for its propagation. Overall, our work demonstrates that miR-124 is a novel regulator of fatty acid homeostasis and HCV infection.
Results
miR-124 Expression Is Modulated during HCV Infection and Serum Depletion
The antiviral oxysterol 25-hydroxycholesterol (25-HC) plays an important role in regulating the immune response to viral infection. The oxysterol has multifaceted effects through modulation of lipid metabolism and inflammatory and stress response pathways (Singaravelu et al., 2015b). Recent work by our group identified numerous miRNAs that are differentially expressed following treatment with 25-HC (Singaravelu et al., 2015a). One of these 25-HC-regulated miRNAs, hsa-miR-124-3p (miR-124), has been shown to play a role in liver disease (Liu et al., 2016, Ning et al., 2014).
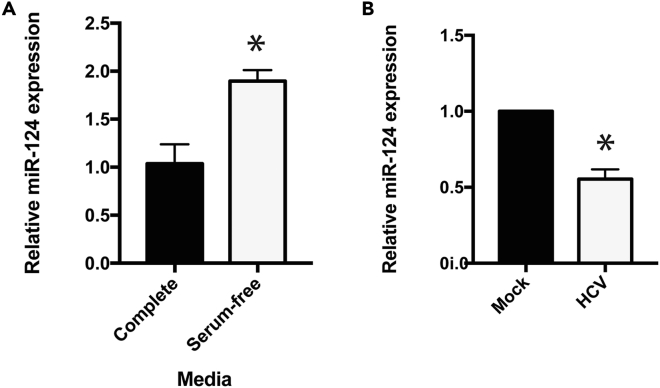
We sought to examine whether miR-124 plays a functional role in the liver. Based on its regulation by 25-HC, we hypothesized that miR-124 is dysregulated during metabolic stresses in hepatocytes. We first measured the expression of the miRNA during serum depletion. We found that cultivation of Huh7.5 hepatoma cells for 48 hr in serum-free media increased miR-124 expression approximately 2-fold compared with normal culturing conditions (Figure 1A). We also considered viral infection as another source of metabolic stress, which could influence miR-124 expression. HCV is a hepatotropic virus that alters liver metabolism to facilitate its viral life cycle. Our previous work (Singaravelu et al., 2015a), along with others' (Vallianou et al., 2016), demonstrated that HCV infection decreases miR-124 expression. We reproduced this finding using a cell-culture-adapted high-titer strain of HCV (Russell et al., 2008). Forty-eight hours post-infection, we observed a 2-fold decrease in miR-124 expression levels (Figure 1B). Taken together, these results demonstrate that miR-124 levels are modulated during metabolic stresses (i.e., serum depletion and HCV infection) and point toward a metabolic regulatory role for the miRNA in liver cells.
Figure 1.
Serum Starvation and HCV Infection Modulate miR-124 Expression
(A) Huh7.5 cells were cultured in complete or serum-free media for 48 hr. qRT-PCR was performed to measure relative miR-124 expression (n = 3).
(B) Huh7.5 cells were infected with HCV, and miR-124 expression levels relative to mock were measured 48 hr post-infection (n = 3).
Data are represented as the mean ± SEM (*p < 0.05).
miR-124 Targets Genes with Functional Roles in Fatty Acid Catabolism
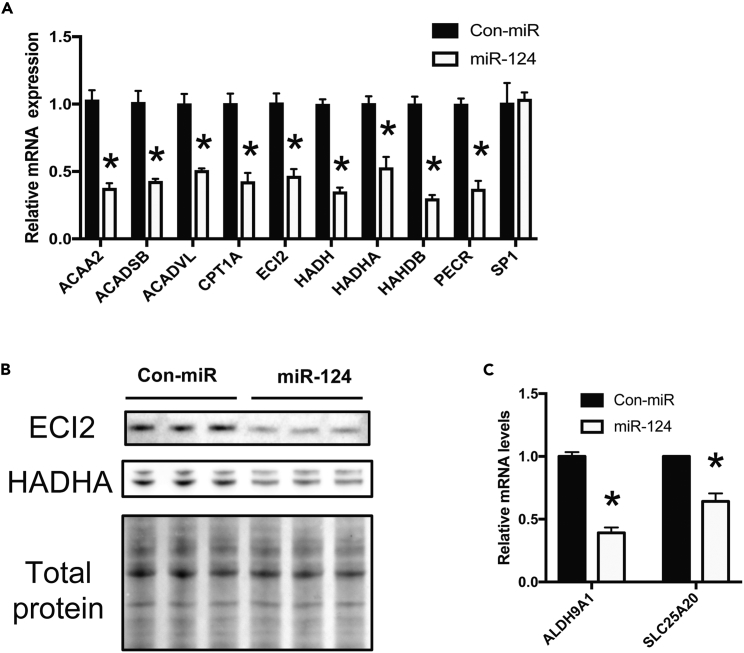
To investigate the hepatic pathways regulated by miR-124, we performed gene expression profiling in Huh7.5 cells transfected with control or miR-124 mimic. A summary of the top 10 up- and down-regulated genes is given in Table S1. To identify the direct targets of miR-124 in the hepatic cells, we examined the overlap between predicted targets of miR-124 (4,450 genes as per TargetScan; Agarwal et al., 2015) and genes repressed by miR-124 mimic transfection by over 1.5-fold (1,693 genes). For our analysis, a cutoff of 1.5-fold was utilized given that direct targeting of a specific gene by a single miRNA may induce modest overall changes in the gene's expression. However, cooperative targeting of multiple genes, involved in the same pathway, by a single miRNA may induce more pronounced phenotypic changes (Bonci et al., 2008, Hashimoto et al., 2013). The overlap produced a list of 425 genes. Bioinformatics analysis via the ToppGene Suite (Chen et al., 2009), was performed to identify associated pathways enriched in this list of repressed miR-124 targets. Interestingly, fatty acid degradation and metabolism were the only two significantly enriched pathways (Table S2; p < 0.05). We then used qRT-PCR to validate miR-124-mediated repression of genes in the fatty acid β-oxidation pathway bearing putative miR-124-binding sites in their 3′ UTR (Figure 2A). We analyzed the expression of HADH, HADHA, HADHB, PECR, ACADVL, ECI2, CPT1A, ACAA2, and ACADSB. Consistent with the microarray results, each of these genes' expression was found to be down-regulated by qRT-PCR (Figures 2A, S1, and S2). Notably, miR-124's repression of CPT1A expression is consistent with previous work demonstrating that this gene is a direct miR-124 target in prostate cancer cells (Valentino et al., 2017). Immunoblot analyses further confirmed similar reduction of HADHA and ECI2 protein levels (Figure 2B). Interestingly, serum starvation of Huh7.5 cells, which promotes miR-124 expression, results in a significant decrease in ACADSB expression, consistent with physiologically relevant miR-124 repression of this gene during metabolic stress (Figure S3A). This is further supported by rescue of ACADSB expression during serum starvation by miR-124 inhibition (Figure S3B). The sum of miR-124's repressive effects on these individual genes should result in a greater overall effect of miR-124 on fatty acid oxidation.
Figure 2.
miR-124 Down-Regulates Fatty Acid Oxidation
(A) Relative mRNA expression levels of miR-124 predicted targets with roles in fatty acid degradation in Huh7.5 cells transfected with control (Con-miR) or miR-124 mimic, as measured by qRT-PCR (n > 5).
(B) Immunoblot analysis of protein expression levels of ECI2 and HADHA (lower band) in miR-124- or control mimic- (Con-miR) transfected Huh7.5 cells. Three independent biological replicates are shown, and total protein detection serves as the loading control. The blot is cropped to emphasize relevant bands.
(C) Relative mRNA expression levels of miR-124 predicted targets with roles in carnitine biosynthesis (n > 6).
Data are represented as the mean ± SEM (*p < 0.05). See also Tables S1 and S2.
Carnitine is a key cofactor that enables shuttling of acyl-CoAs into the mitochondria. Long-chain fatty acids are otherwise impermeable to the mitochondria. Closer examination of the microarray results revealed a down-regulation in the expression of SLC25A20, a gene encoding carnitine-acylcarnitine translocase, which mediates transport of acyl carnitines across the inner mitochondrial membrane; ALDH9A1, a gene encoding 4-trimethylaminobutyraldehyde dehydrogenase, which is proposed to be involved in the hepatic synthesis of carnitine, a key cofactor for the transfer of long-chain fatty acids to mitochondria for oxidation; and SLC22A5, a transporter for carnitine uptake from serum (Figures S1 and S2) (Strijbis et al., 2010). qRT-PCR also validated the down-regulation of these three miR-124 targets in Huh7.5 cells transfected with miR-124 mimics (Figure 2C). Collectively, these data demonstrate that miR-124 is involved in the repression of carnitine homeostasis and fatty acid oxidation.
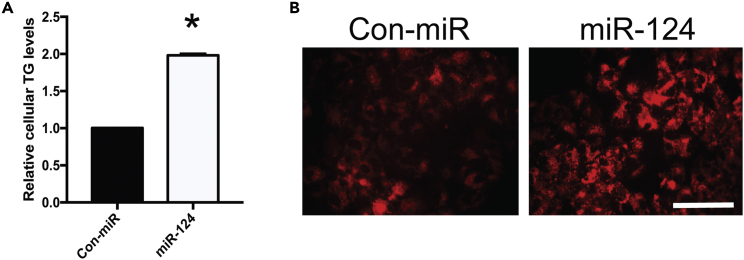
miR-124 Promotes Cellular TG Accumulation
Inhibition of fatty acid oxidation results in an accumulation of free fatty acids, which can be incorporated into TGs for storage. Therefore, we hypothesized that miR-124 overexpression would influence cellular TG levels. We assayed TG levels in control and miR-124-mimic-transfected Huh7.5 cells and observed a significant increase in cellular TG levels (Figure 3A), in line with miR-124's repression of fatty acid oxidation (Figure 2). Similarly, oil red O staining revealed an increase in cellular lipid levels during miR-124 mimic transfection (Figure 3B). Collectively, our data demonstrate that miR-124 promotes hepatocellular TG accumulation.
Figure 3.
miR-124 Promotes Cellular Triglyceride Accumulation
(A) Relative cellular TG content in control and miR-124-mimic-transfected Huh7.5 cells as assessed by TG assays (n = 3). Data are representative of the mean ± SEM (*p < 0.05).
(B) Oil red O staining of cellular lipid droplet content in control and miR-124-mimic-transfected Huh7.5 cells. Cells were visualized using fluorescence microscopy. Scale bar, 100 μm.
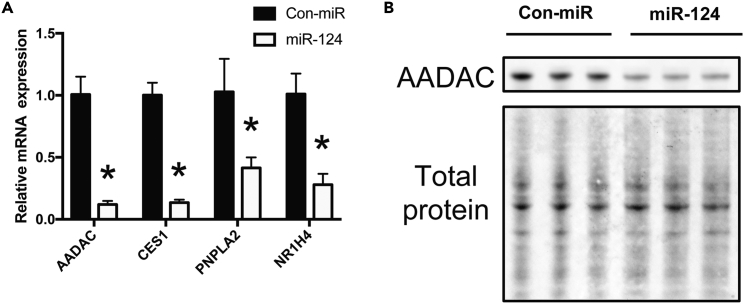
miR-124 Inhibits Expression of Triglyceride Hydrolases
We sought to identify additional targets of miR-124, which could contribute to miR-124's induction of TG accumulation. Hydrolysis of TG stores mediates mobilization of fatty acids for very-low-density lipoprotein synthesis or as substrates for fatty acid oxidation. Therefore, repressed expression of TG hydrolases (TGHs) could also contribute to miR-124-induced increase in cellular TG levels. We noted that arylacetamide deacetylase (AADAC), an endoplasmic reticulum-localized carboxylester hydrolase that hydrolyzes TGs (Lo et al., 2010), was among the most highly miR-124-repressed genes in our microarray data (Table S1). Down-regulation at the transcriptional level was verified by qRT-PCR (Figure 4A), and immunoblot analysis confirmed that protein levels were also decreased in the miR-124-mimic-treated cells (Figure 4B). The microarray data also revealed decreased carboxylesterase 1 (CES1) and NR1H4 expression. CES1 is another hydrolase responsible for TG breakdown, whereas NR1H4 encodes for the farnesoid X receptor (FXR). Neither gene possesses miR-124-binding sites in their 3′ UTR. FXR is a nuclear hormone receptor, which regulates the clearance of hepatic TGs, in part through induced expression of CES1 (Phulukdaree et al., 2015). qRT-PCR analysis validated an over 7.6-fold decrease in CES1 and an over 3.5-fold decrease in NR1H4 expression levels (Figure 4A). Surprisingly, we did not observe a down-regulation of CES1 protein expression levels; however, a recent study suggests that the half-life of the CES1 protein is approximately 96 hr (Ross et al., 2012). This suggests that the timescale of our miR-124 overexpression experiments (72 hr) may not have been sufficient to enable observation of miR-124's repressive effect on CES1 expression. Lastly, we observed miR-124-mediated repression of PNPLA2 expression in the microarray data. This miR-124 target gene encodes adipose triglyceride lipase (ATGL), a hepatic lipase responsible for mediating hepatic TG hydrolysis and channeling hydrolyzed fatty acids to β-oxidation (Ong et al., 2011). This is consistent with our qRT-PCR data (Figure 4A), and previous reports demonstrating miR-124-mediated repression of PNPLA2 expression in adipocytes (Das et al., 2015). Collectively, concordant repression of TG hydrolysis and fatty acid oxidation should contribute to TG accumulation mediated by miR-124 overexpression in hepatic cells.
Figure 4.
miR-124 Down-Regulates Genes associated with Triglyceride Hydrolysis
(A) Relative mRNA expression levels of PNPLA2, CES1, NR1H4, and AADAC in control and miR-124-mimic-transfected cells as assessed by qRT-PCR analysis (n > 5). Data are represented as the mean ± SEM (*p < 0.05).
(B) Immunoblot analysis of protein expression levels of AADAC in miR-124- or control mimic- (Con-miR) transfected Huh7.5 cells. Three independent biological replicates are shown, and total protein detection serves as loading control. Blot is cropped to emphasize relevant bands.
miR-124 Inhibits HCV Infection
We and others have identified several miRNAs that are differentially expressed during infection with HCV, promoting either pro- or antiviral environments via modulation of genes involved in lipid pathways (Li et al., 2017, Shirasaki et al., 2013, Singaravelu et al., 2014, Singaravelu et al., 2015a). We postulated, given HCV's strong reliance on hepatic lipid pathways to facilitate its life cycle, that miR-124, through regulation of TG homeostasis, could influence viral infectivity. miR-124 overexpression in Huh7.5 cells decreased viral infectivity and repressed the expression of miR-124 targets (Figure S4). This is consistent with a previous study demonstrating that decreased expression of AADAC impairs production of HCV (Nourbakhsh et al., 2013). Our work suggests that miR-124 inhibits HCV infection through inhibition of AADAC expression.
Discussion
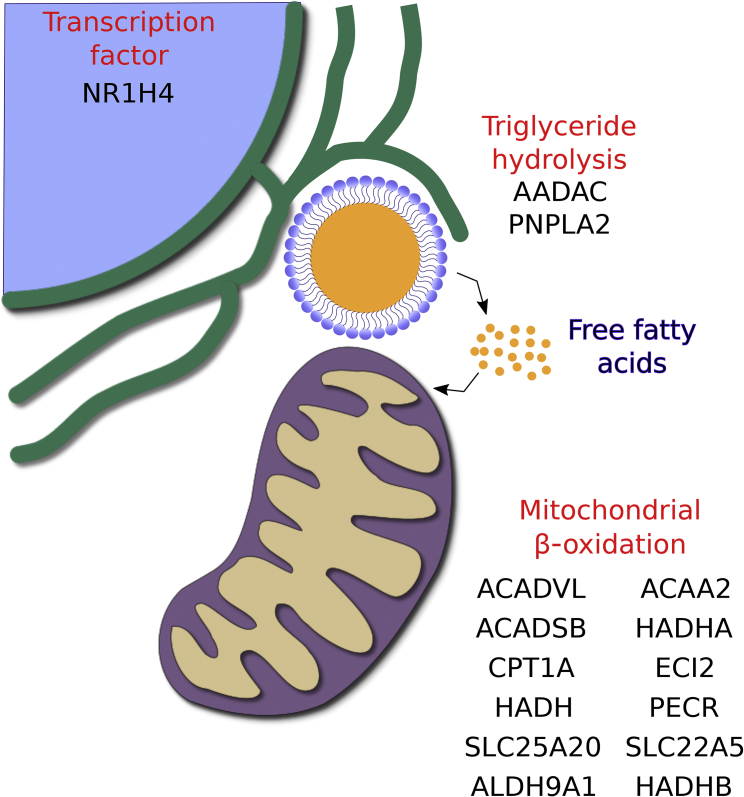
miRNAs have emerged as a key regulatory layer in the maintenance of hepatic lipid homeostasis (Moore et al., 2011). However, there are limited examples of miRNAs playing key roles in the regulation of TG catabolism. Hepatic miR-33 has been shown to regulate several key enzymes in the fatty acid oxidation pathway (Davalos et al., 2011). Herein, we identify a novel function for miR-124 in the regulation of TG catabolism via multiple key enzymes involved in fatty acid oxidation and TG hydrolysis. Through concerted targeting of multiple genes, miR-124 produces a cooperative inhibitory effect on lipid catabolism, resulting in hepatocellular TG accumulation (Figure 5). This highlights an emerging theme in miRNA regulation whereby miRNAs have evolved to target multiple genes in the same pathway to produce a greater overall regulatory effect.
Figure 5.
miR-124 Down-Regulates Fatty Acid and Triglyceride Catabolism
Schematic depicting concerted regulation by miR-124 of genes involved in triglyceride hydrolysis and fatty acid catabolism.
Our work demonstrates that miR-124 suppresses the expression of several key enzymes in the mitochondrial β-oxidation pathway (ACADVL, ACADSB, HADHA, HADHB, HADH, and ACAA2). ACADVL encodes the mitochondrial membrane-bound long-chain acyl-CoA dehydrogenase (VLCAD), which is responsible for the first step of the mitochondrial fatty acid oxidation cycle for saturated long-chain fatty acids (14–20 carbons). VLCAD catalyzes the dehydrogenation of acyl-CoAs to trans-Δ2-enoyl-CoA. ACADSB performs an analogous dehydrogenation reaction for short/branched fatty acids. HADHA and HADHB form a hetero-octamer complex, called the mitochondrial trifunctional protein, responsible for catalyzing the last three steps of fatty acid oxidation for long-chain fatty acids. Specifically, HADHA is responsible for catalyzing trans-Δ2-enoyl CoA hydration to form L-3-hydroxyacyl-CoA, and the subsequent dehydrogenation to 3-ketoacyl CoA; then HADHB sequentially catalyzes long-chain keto-acyl-CoA thiolase activity producing an acetyl-CoA and an acyl-CoA. For short- and medium-chain fatty acids, HADH and ACAA2 catalyze the last two steps of the oxidation. For unsaturated fatty acids, cis-enoyl-CoA esters have to be transformed to trans configurations to enable their catabolism. ECI2 contributes to the catabolism of unsaturated fatty acids by catalyzing an isomerization of cis-Δ3 and trans-Δ3-enoyl CoA esters into trans-Δ2-enoyl-CoA esters. Fatty acid oxidation can also be catalyzed in the peroxisome, with the participation of a different set of enzymes, including PECR, which encodes a peroxisomal trans-Δ2-enoyl-CoA reductase. Collectively, our data highlight miR-124 as a regulator of multiple key nodes in both peroxisomal and mitochondrial β-oxidation.
Our data demonstrate that miR-124 down-regulates proteins responsible for carnitine uptake from plasma (SLC22A5), carnitine synthesis (ALDH9A1), and acyl-CoA conjugation of carnitine (CPT1A), as well as translocation of acyl-CoA-carnitine through the inner mitochondrial membrane (SLC25A20). As the CPT1A-catalyzed acyl transfer to carnitine represents one of the rate-limiting steps of fatty acid oxidation, this suggests that targeting of CPT1A by miR-124 should enable the miRNA to exert greater regulatory control over fatty acid oxidation.
A subset of the genes repressed by miR-124 do not possess canonical seed sites, suggesting a non-canonical or indirect mode of regulation. A previous study, which utilized Argonaute 2 (Ago2) co-immunoprecipitation followed by mRNA microarray analysis to identify miR-124 targets in HEK293 cells, suggested that miR-124-loaded Ago2 directly interacted with the 3′ UTR of HADHB mRNA (Karginov et al., 2007). A subsequent study confirmed this interaction in mice brains using high-throughput sequencing of RNAs isolated by Ago cross-linking immunoprecipitation (Chi et al., 2009). Close examination of the 3′ UTR reveals a potential miR-124 “G-bulge” site (Figure S5), which is a recently described non-canonical miRNA-binding motif where a G nucleotide bulge is allowed in the mRNA (in the corresponding nucleotide position 5–6 of the miRNA) (Chi et al., 2012). Another potently repressed gene, AADAC, possesses no mRNA-binding sites in the 3′ UTR to the best of our knowledge; however, examination of the open reading frame (ORF) reveals the presence of one canonical site. Typically, ORF-binding sites are less prone to miRNA regulation; however, if the sites are preceded by a stretch of rare codons, the recognition site may be functional (Gu et al., 2009). Examination of the three codons upstream of the binding site reveal three rare codons (usage <12%; Figure S6), suggesting that miR-124 may regulate AADAC through a functional ORF-binding site. This implies that miR-124 regulates a subset of its metabolic gene targets using non-canonical mode of target recognition.
Several of the miR-124 targets have been established as targets of the transcription factor peroxisome proliferator-activated receptor α (PPAR-α), including HADHA, HADHB, ACAA2, ACADVL, SLC25A20, SLC22A5, and CPT1A (Rakhshandehroo et al., 2010). This nuclear hormone receptor acts as a master transcriptional regulator of mitochondrial and peroxisomal fatty acid oxidation and is activated as part of the fasting response (Kersten et al., 1999). Although several of our described targets have been validated as direct targets of miR-124 in other systems, we postulate that decreased PPAR-α signaling is a major contributor to the observed reduction in fatty acid oxidation gene expression during miR-124 overexpression. This is in line with decreased expression of the TGHs ATGL and CES1, which has previously been correlated with decreased PPAR-α activity (Xu et al., 2014). Also, AADAC overexpression has been shown to promote fatty acid oxidation in rat hepatoma cells (Lo et al., 2010). Therefore miR-124-mediated repression of these TGHs should result in an indirect repression of fatty acid oxidation.
Our miRNA expression analysis revealed that serum depletion activates miR-124 expression (Figure 1A). Interestingly, a recent study demonstrated that statin treatment of hepatoma cells induces miR-124 expression (Phulukdaree et al., 2015). Together, these data suggest that metabolic stresses regulate miR-124 expression. In statin-treated mice, increased mitochondrial and peroxisomal fatty acid oxidation has been observed in the liver (Park et al., 2016), suggesting that statin-induced miR-124 expression may serve to buffer this increase in fatty acid oxidation.
Previous work suggested that miR-124 promotes hepatic TG synthesis through regulation of TRIB3 (Liu et al., 2016) and enhanced hepatic lipogenesis. We did not, however, observe increased expression of lipogenic genes (SREBPs and FASN; Figures S1 and S5) or repression of TRIB3 (Figure S1). This could be a result of a variation in models used (Huh7.5 vs. mice), but our data strongly suggest that the observed miR-124-induced TG accumulation in Huh7.5 cells is due to decreased TG and fatty acid breakdown, as opposed to increased lipogenesis.
Also, miR-124 was previously identified as a broadly antiviral miRNA against murine cytomegalovirus, murine herpesvirus 68, and herpes simplex virus 1 during a genome-wide functional miRNA screen (Santhakumar et al., 2010). The same group later confirmed that the miRNA's antiviral activity extended to influenza A virus strains and respiratory syncytial virus (McCaskill et al., 2017). In the latter study, the authors ascribed this antiviral activity, in part, to the regulation of p38 mitogen-activated protein kinase (MAPK) signaling. Although we cannot exclude the contribution of the p38 MAPK signaling pathway to the observed anti-HCV effects of miR-124, given the strong dependence of HCV on hepatic lipid pathways, miR-124's regulation of hepatocellular metabolism likely contributes to the its influence on HCV. It is important to note that our previous work, using an HCV replicon model, suggests that miR-124 promotes HCV replication (Singaravelu et al., 2015a). In contrast, this work employing a full-length virus (JFH-1) suggests that, in a model fully recapitulating the entire viral life cycle, miR-124 inhibits HCV infectivity. This is consistent with miR-124 regulation of AADAC, the only positive regulator of virion production identified among our miR-124 targets (Nourbakhsh et al., 2013).
Overall, our data highlight a novel functional role for miR-124 in hepatic TG catabolism. This metabolic-stress-regulated miRNA concordantly regulates multiple nodes within the TG hydrolysis and fatty acid oxidation pathways, yielding a greater overall effect on TG homeostasis. It is interesting to note that there are several miRNAs that target metabolic pathways in the liver at different nodes. These miRNAs cooperate, often in a redundant fashion, to fine-tune metabolic networks in the liver and manage the homeostatic response to multiple inputs (e.g., from the muscle and fat). Future work should examine the function of miR-124 in the context of systemic energy homeostasis.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Limitations of Study
We demonstrated that miR-124 inhibits the expression of genes associated with fatty acid and TG metabolism in the Huh7.5 hepatoma model. Further analysis is required to elucidate the role of this miRNA regulatory axis in vivo.
Acknowledgments
J.P.P. is supported by grant funding from the Canadian Institutes of Health Research (CIHR), Canada (220886) and Natural Sciences and Engineering Research Council of Canada (NSERC), Canada (210719). R.S. received graduate student funding from the Canadian Network on hepatitis C and is the recipient of post-doctoral funding from the Life Science Research Foundation. M.H.P. received post-doctoral funding from the Canadian Network on Hepatitis C. D.Ö. is the recipient of post-doctoral funding from the CIHR. N.A. is the recipient of doctoral postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada. mRNA microarray profiling was performed by The Center for Applied Genomics (TCAG), The Hospital for Sick Children, Toronto, ON, Canada. Gene expression profiling data from miR-124 and control mimic-transfected Huh7.5 cells have been deposited to NCBI Gene Expression Omnibus under the following accession number: GSE122134.
Author Contributions
Conceptualization, J.P.P., R.S., and M.H.P.; Methodology, R.S., M.H.P, and T.A.S.; Formal Analysis, R.S., M.H.P., T.A.S., D.Ö., and J.P.P.; Investigation, M.H.P., T.S., J.N., N.A., and D.Ö.; Writing – Original Draft, R.S., T.A.S., M.H.P., N.A., and J.P.P.; Review & Editing, all authors; Funding Acquisition, R.S. and J.P.P.; Supervision, R.S., M.H.P., and J.P.P.
Declaration of Interests
The authors declare no competing interests.
Published: December 21, 2018
Footnotes
Supplemental Information includes Transparent Methods, six figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.isci.2018.11.028.
Supplemental Information
References
- Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hamo R., Efroni S. MicroRNA regulation of molecular pathways as a generic mechanism and as a core disease phenotype. Oncotarget. 2015;6:1594–1604. doi: 10.18632/oncotarget.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D., Coppola V., Musumeci M., Addario A., Giuffrida R., Memeo L., D’Urso L., Pagliuca A., Biffoni M., Labbaye C. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W., Hannon G.J., Darnell R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V., Heaton N.S., Randall G. Lipids at the interface of virus-host interactions. Curr. Opin. Microbiol. 2012;15:512–518. doi: 10.1016/j.mib.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.K., Stadelmeyer E., Schauer S., Schwarz A., Strohmaier H., Claudel T., Zechner R., Hoefler G., Vesely P.W. MicroRNA-124a regulates lipolysis via adipose triglyceride lipase and comparative gene identification 58. Int. J. Mol. Sci. 2015;16:8555–8568. doi: 10.3390/ijms16048555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos A., Goedeke L., Smibert P., Ramirez C.M., Warrier N.P., Andreo U., Cirera-Salinas D., Rayner K., Suresh U., Pastor-Pareja J.C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjärn M., Hansen J.B., Hansen H.F., Straarup E.M. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., Watts L., Booten S.L., Graham M., McKay R. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Gu S., Jin L., Zhang F., Sarnow P., Kay M.A. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Akiyama Y., Yuasa Y. Multiple-to-multiple relationships between MicroRNAs and target genes in gastric cancer. PLoS One. 2013;8:e62589. doi: 10.1371/journal.pone.0062589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten S.M., Wanders R.J. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J. Inherit. Metab. Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;1:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Karginov F.V., Conaco C., Xuan Z., Schmidt B.H., Parker J.S., Mandel G., Hannon G.J. A biochemical approach to identifying microRNA targets. Proc. Natl. Acad. Sci. U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S., Seydoux J., Peters J.M., Gonzalez F.J., Desvergne B., Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Li Q., Lowey B., Sodroski C., Krishnamurthy S., Alao H., Cha H., Chiu S., El-Diwany R., Ghany M.G., Liang T.J. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat. Commun. 2017;8:1789. doi: 10.1038/s41467-017-01954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhao J., Liu Q., Xiong X., Zhang Z., Jiao Y., Li X., Liu B., Li Y., Lu Y. MicroRNA-124 promotes hepatic triglyceride accumulation through targeting tribbles homolog 3. Sci. Rep. 2016;6:37170. doi: 10.1038/srep37170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo V., Erickson B., Thomason-Hughes M., Ko K.W., Dolinsky V.W., Nelson R., Lehner R. Arylacetamide deacetylase attenuates fatty-acid-induced triacylglycerol accumulation in rat hepatoma cells. J. Lipid Res. 2010;51:368–377. doi: 10.1194/jlr.M000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashek D.G. Hepatic fatty acid trafficking: multiple forks in the road. Adv. Nutr. 2013;4:697–710. doi: 10.3945/an.113.004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill J.L., Ressel S., Alber A., Redford J., Power U.F., Schwarze J., Dutia B.M., Buck A.H. Broad-spectrum inhibition of respiratory virus infection by microRNA mimics targeting p38 MAPK signaling. Mol. Ther. Nucleic Acids. 2017;7:256–266. doi: 10.1016/j.omtn.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.J., Rayner K.J., Suárez Y., Fernández-Hernando C. The role of microRNAs in cholesterol efflux and hepatic lipid metabolism. Ann. Rev. Nutr. 2011;31:49–63. doi: 10.1146/annurev-nutr-081810-160756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning B.F., Ding J., Liu J., Yin C., Xu W.-P., Cong W.-M., Zhang Q., Chen F., Han T., Deng X. Hepatocyte nuclear factor 4α-nuclear factor-κB feedback circuit modulates liver cancer progression. Hepatology. 2014;60:1607–1619. doi: 10.1002/hep.27177. [DOI] [PubMed] [Google Scholar]
- Nourbakhsh M., Douglas D.N., Pu C.H., Lewis J.T., Kawahara T., Lisboa L.F., Wei E., Asthana S., Quiroga A.D., Law L.M. Arylacetamide deacetylase: a novel host factor with important roles in the lipolysis of cellular triacylglycerol stores, VLDL assembly and HCV production. J. Hepatol. 2013;59:336–343. doi: 10.1016/j.jhep.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Ong K.T., Mashek M.T., Bu S.Y., Greenberg A.S., Mashek D.G. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology. 2011;53:116–126. doi: 10.1002/hep.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.S., Jang J.E., Ko M.S., Woo S.H., Kim B.J., Kim H.S., Park H.S., Park I.S., Koh E.H., Lee K.U. Statins increase mitochondrial and peroxisomal fatty acid oxidation in the liver and prevent non-alcoholic steatohepatitis in mice. Diabetes Metab. J. 2016;40:376–385. doi: 10.4093/dmj.2016.40.5.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Phulukdaree A., Moodley D., Khan S., Chuturgoon A.A. Atorvastatin increases miR-124a expression: a mechanism of Gamt modulation in liver cells. J. Cell. Biochem. 2015;116:2620–2627. doi: 10.1002/jcb.25209. [DOI] [PubMed] [Google Scholar]
- Rakhshandehroo M., Knoch B., Muller M., Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010:612089. doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ress C., Kaser S. Mechanisms of intrahepatic triglyceride accumulation. World J. Gastroenterol. 2016;22:1664–1673. doi: 10.3748/wjg.v22.i4.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross M.K., Borazjani A., Wang R., Crow J.A., Xie S. Examination of the carboxylesterase phenotype in human liver. Arch. Biochem. Biophys. 2012;522:44–56. doi: 10.1016/j.abb.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R.S., Meunier J.C., Takikawa S., Faulk K., Engle R.E., Bukh J., Purcell R.H., Emerson S.U. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. U S A. 2008;105:4370–4375. doi: 10.1073/pnas.0800422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhakumar D., Forster T., Laqtom N.N., Fragkoudis R., Dickinson P., Abreu-Goodger C., Manakov S.A., Choudhury N.R., Griffiths S.J., Vermeulen A. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc. Natl. Acad. Sci. U S A. 2010;107:13830–13835. doi: 10.1073/pnas.1008861107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki T., Honda M., Shimakami T., Horii R., Yamashita T., Sakai Y., Sakai A., Okada H., Watanabe R., Murakami S. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J. Virol. 2013;87:5270–5286. doi: 10.1128/JVI.03022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu R., Chen R., Lyn R.K., Jones D.M., O'Hara S., Rouleau Y., Cheng J., Srinivasan P., Nasheri N., Russell R.S. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98–108. doi: 10.1002/hep.26634. [DOI] [PubMed] [Google Scholar]
- Singaravelu R., O'Hara S., Jones D.M., Chen R., Taylor N.G., Srinivasan P., Quan C., Roy D.G., Steenbergen R.H., Kumar A. MicroRNAs regulate the immunometabolic response to viral infection in the liver. Nat. Chem. Biol. 2015;11:988–993. doi: 10.1038/nchembio.1940. [DOI] [PubMed] [Google Scholar]
- Singaravelu R., Quan C., Powdrill M.H., Shaw T.A., Srinivasan P., Lyn R.K., Alonzi R.C., Jones D.M., Filip R., Russell R.S. MicroRNA-7 mediates cross-talk between metabolic signaling pathways in the liver. Sci. Rep. 2018;8:361. doi: 10.1038/s41598-017-18529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu R., Srinivasan P., Pezacki J.P. Armand-Frappier Outstanding Student Award — the emerging role of 25-hydroxycholesterol in innate immunity. Can. J. Microbiol. 2015;61:521–530. doi: 10.1139/cjm-2015-0292. [DOI] [PubMed] [Google Scholar]
- Strijbis K., Vaz F.M., Distel B. Enzymology of the carnitine biosynthesis pathway. IUBMB Life. 2010;62:357–362. doi: 10.1002/iub.323. [DOI] [PubMed] [Google Scholar]
- Sun Y., Luo Z.M., Guo X.M., Su D.F., Liu X. An updated role of microRNA-124 in central nervous system disorders: a review. Front. Cell. Neurosci. 2015;9:193. doi: 10.3389/fncel.2015.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentino A., Calarco A., Di Salle A., Finicelli M., Crispi S., Calogero R.A., Riccardo F., Sciarra A., Gentilucci A., Galderisi U. Deregulation of MicroRNAs mediated control of carnitine cycle in prostate cancer: molecular basis and pathophysiological consequences. Oncogene. 2017;36:6030–6040. doi: 10.1038/onc.2017.216. [DOI] [PubMed] [Google Scholar]
- Vallianou I., Dafou D., Vassilaki N., Mavromara P., Hadzopoulou-Cladaras M. Hepatitis C virus suppresses Hepatocyte Nuclear Factor 4 alpha, a key regulator of hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 2016;78:315–326. doi: 10.1016/j.biocel.2016.07.027. [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y., Chen W.D., Xu Y., Yin L., Ge X., Jadhav K., Adorini L., Zhang Y. Hepatic carboxylesterase 1 is essential for both normal and farnesoid X receptor-controlled lipid homeostasis. Hepatology. 2014;59:1761–1771. doi: 10.1002/hep.26714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.