Abstract
Interactions of multiple myeloma (MM) cells with endothelial cells (ECs) enhance angiogenesis and MM progression. Here, we investigated the role of Notch signaling in the cross talk between ECs and MM cells enabling angiogenesis. MMECs showed higher expression of Jagged1/2 ligands, of activated Notch1/2 receptors, and of Hes1/Hey1 Notch target genes than ECs from monoclonal gammopathy of undetermined significance patients, suggesting that homotypic activation of Notch pathway occurs in MM. MM cells co-cultured with MMECs triggered Notch activation in these cells through a cell-to-cell contact-dependent way via Jagged1/2, resulting in Hes1/Hey1 overexpression. The angiogenic effect of Notch pathway was analyzed through Notch1/2·siRNAs and the γ-secretase inhibitor MK-0752 by in vitro (adhesion, migration, chemotaxis, angiogenesis) and in vivo (Vk12598/C57B/6 J mouse model) studies. Activated Notch1/2 pathway was associated with the overangiogenic MMEC phenotype: Notch1/2 knockdown or MK-0752 treatment reduced Hes1/Hey1 expression, impairing in vitro angiogenesis of both MMECs alone and co-cultured with MM cells. MM cells were unable to restore angiogenic abilities of treated MMECs, proving that MMEC angiogenic activities closely rely on Notch pathway. Furthermore, Notch1/2 knockdown affected VEGF/VEGFR2 axis, indicating that the Notch pathway interferes with VEGF-mediated control on angiogenesis. MK-0752 reduced secretion of proangiogenic/proinflammatory cytokines in conditioned media, thus inhibiting blood vessel formation in the CAM assay. In the Vk12598/C57B/6 J mouse, MK-0752 treatment restrained angiogenesis by reducing microvessel density. Overall, homotypic and heterotypic Jagged1/2-mediated Notch activation enhances MMECs angiogenesis. Notch axis inhibition blocked angiogenesis in vitro and in vivo, suggesting that the Notch pathway may represent a novel therapeutic target in MM.
Introduction
In multiple myeloma (MM), malignant plasma cells (MM cells) interplay with bone marrow stromal cells (BMSCs), extracellular matrix, and soluble factors that result in the formation of a protumor niche favoring MM cell growth and survival, bone disease, and angiogenesis [1]. MM expansion depends on angiogenesis that accompanies progression from premalignant stage, i.e., monoclonal gammopathy of undetermined significance (MGUS) [2], to overt MM [3]. MM endothelial cells (MMECs) differ from the corresponding cells of MGUS patients (MGECs) because they are highly proliferative and display enhanced proangiogenic activity in vitro [4].
Notch is a transmembrane receptor (Notch1, 2, 3, and 4) that interacts with two different families of ligands: the Serrate-like ligands (Jagged1, 2) and the Delta-like ligands (DLL1, 3, 4) [5]. The receptor/ligand interaction determines the cleavage of Notch receptor by the γ-secretase complex, with the release of the Notch intracellular domains (ICDs) that translocate to the nucleus and modulate gene expression. The best described Notch target genes are the human hairy and enhancer of split (Hes) and hairy/enhancer of split related with the YRPW motif (Hey) transcription factors—namely, Hes1, Hes5, and Hey1 [6]—that regulate cell differentiation, cell cycle progression, survival, apoptosis, and angiogenesis [7], [8].
The role of Notch pathway in the coordination of physiologic angiogenesis through DLL4 and Jagged1 ligands has been studied [8], [9], [10], [11]. DLL4 expression is induced by the vascular endothelial growth factor (VEGF) in “tip” (highly proliferating) ECs, which drives vessel sprouts and filopodia protrusions. DLL4 activates the Notch pathway in neighboring ECs and reduces the expression of VEGF receptor 2 (VEGFR2), maintaining the quiescent “stalk” phenotype [5], [10], [11]. DLL4 is antagonized by Jagged1 which downregulates DLL4/Notch signaling and induces VEGFR2 expression [5], [12]. Indeed, Jagged1 is a positive regulator of “tip” ECs, hence increasing ECs proliferation, vessel sprouting, and branching [5], [12], [13].
Deregulation of Notch signaling occurs in tumor angiogenesis: overexpression of Jagged1 [14], DLL4 [15], [16], or Jagged2 [17] ligands has been described in several solid tumors. However, the involvement of Notch signaling in MM angiogenesis is still unclear. MM cells express Notch1/2 receptors and ligands [18], [19], [20], resulting in the homotypic activation of Notch pathway and in the heterotypic activation of surrounding stromal cells that contribute to MM cell proliferation [21], survival, and migration through the release of interleukin (IL)-6, insulin-like growth factor (IGF)-1, and VEGF [22].
Here, we investigated the role of the Notch axis in regulating angiogenesis through the cross talk between MMECs-MMECs and MMECs-MM cells. Overall, our results indicate a Jagged1/Jagged2-mediated homotypic and heterotypic activation of Notch signaling in MMECs. Blockade of the Notch pathway through Notch1/2·siRNAs or the γ-secretase inhibitor (GSI) MK-0752 restrains angiogenesis both in vitro and in vivo, suggesting that Notch may be a promising novel therapeutic target in MM.
Materials and Methods
Patients
The present study included 35 newly diagnosed MM patients and 17 MGUS patients who fulfilled the International Myeloma Working Group diagnostic criteria [23]. The study was approved by the Ethics Committee of the University of Bari Medical School (I.D. no. 5143/2016). All patients provided their informed consent in accordance with the Declaration of Helsinki.
Cell Cultures and Co-Cultures
Primary BM MGECs, MMECs, and MM cells were obtained by Ficoll gradient centrifugation of heparinized BM aspirates followed by incubation with magnetic microbeads coated with anti-CD31 or anti-CD138, respectively (Miltenyi Biotec, Bergisch Gladbach, Germany). MGECs or MMECs were cultured in Dulbecco's modified Eagle medium (DMEM) with 20% fetal bovine serum (FBS) (Sigma-Aldrich, St Louis, MN) and 1% antibiotic/antimycotic (Euroclone, Milan, Italy). Primary CD138+ MM cells were cultured in RPMI-1640 with 10% FBS and 1% antibiotic/antimycotic. RPMI-8226 MM cell line was purchased from ATCC (Manassas, VA) and cultured in RPMI-1640 medium with 10% FBS, sodium pyruvate 1 mM, high glucose, and HEPES 1 mM (all from Sigma-Aldrich).
In co-culture experiments, MMECs were cultured with RPMI-8226 cells or primary CD138+ MM cells at a 1:5 cell ratio in the presence/absence of transwell (0.4-μm pore size, Costar, Cambridge, MA). After co-culture experiments without transwell, MM cells were immunomagnetically depleted with anti-CD138 microbeads (Miltenyi Biotec); hence, experiments were carried out with purified MMECs.
Western Blotting
Protein lysates from MGECs and MMECs were obtained with a lysis buffer that preserves transmembrane proteins. Thirty-five micrograms of protein lysates was separated on 4%-12% NuPAGE gels (Invitrogen Corp.), electrotransferred to a polyvinylidene difluoride membrane (PerkinElmer Life Science Inc., Boston, MA), immunoblotted overnight with primary antibodies (Supplementary Table 1), and incubated with horseradish peroxidase–labeled secondary antibodies for 1 hour (Bio-Rad, Hercules, CA). Immunoreactive bands were visualized by enhanced chemiluminescence (Bio-Rad) with the Gel Logic 1,500 Imaging System (Eastman Kodak Co., Rochester, NY), and quantified as optical density units with Kodak Molecular Imaging Software.
Immunofluorescence
MGECs or MMECs (5 × 103) were seeded on chamber slides (Lab Tek II Chamber Slides, Thermo Scientific Fisher Scientific Inc.), fixed, permeabilized, and incubated overnight with anti-Notch1 and anti-Notch2 (Cell Signaling Technology Inc., Danvers, MA) primary antibodies. The next day, ECs were incubated with secondary FITC-conjugated anti-rabbit antibody (Sigma-Aldrich). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen Corp.).
Real-Time Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated by the RNeasy Micro Kit (Qiagen Venlo, Netherlands) and reverse transcribed with the iScript cDNA Synthesis Kit (Bio-Rad). Real-time PCRs were carried out with the “StepOne Real-Time RT-PCR System” (Applied Biosystems) and performed with TaqMan assays (Supplementary Table 2). The gene expression (fold change) was analyzed by the 2−ΔΔCt formula.
Flow Cytometry
The expression of Notch ligands on BM CD45+CD38+CD138+ MM cells, RPMI-8226 cells, MMECs, and MGECs and of VEGFR2/pVEGFR2 on MMECs was detected using the monoclonal antibodies listed in Supplementary Table 3. Samples were acquired to flow cytometry (FACScanto II, BD) and analyzed using FACS Diva software (BD).
Jagged1, Jagged2, Notch1, and Notch2 Small Interfering RNA Transfection (siRNA) and MK-0752 Treatment
MMECs were transiently transfected with control siRNAs, Jagged1∙siRNA 25 nM, Jagged2∙siRNA 25 nM, Notch1·siRNA 25 nM, Notch2·siRNA 50 nM (SMART-pool; Dharmacon RNA Technologies, Lafayette, CO), or the transfection reagent alone (Lipofectamine, RNAiMAX siRNA transfection reagent, Invitrogen Corp.) for 72 hours. For co-culture experiments, MM cells were added the day after the treatment. MM cells were transiently transfected with control siRNAs, Jagged1∙siRNA 25 nM, Jagged2∙siRNA 25 nM, or the transfection reagent alone for 48 hours.
MMECs cultured alone or co-cultured with RPMI-8226 cells with/without transwell were treated with vehicle or MK-0752 (Selleckchem, Houston, TX) 5 nM for 48 hours.
Apoptosis Assay
Apoptosis was assessed by Annexin-V-PE/7-AAD (Becton Dickinson-BD, San Jose, CA) staining and flow cytometry analysis according to manufacturer's instructions.
Matrigel Angiogenesis
MMECs treated with Notch1·siRNA or Notch2·siRNA or with MK-0752 were seeded (3.5 × 104) on 48-well plates coated with growth factors–reduced Matrigel (BD Biosciences) in serum-free medium (SFM). After 16 hours, the skeletonization on Matrigel was followed by measurement of mesh areas, branching points, and vessel lengths in three randomly chosen fields with the EVOS inverted microscope (Euroclone) at ×10.
Adhesion Assay
MMECs treated with Notch1·siRNA or Notch2·siRNA or with MK-0752 were stained with Calcein AM (Invitrogen, ThermoFisher Scientific) for 1 hour and then plated (1 × 103 cells/well) in quadruplicate in fibronectin-coated 96-well plates. After 45 minutes, nonadherent cells were washed away, and the rate of adherent cells was established reading fluorescence at 495 nm at VICTOR X3 Multilabel Plate Reader (PerkinElmer Inc., Waltham, MA).
Chemotaxis Assay
MMECs (5 × 104) treated with Notch1·siRNA or Notch2·siRNA or with MK-0752 were tested in a Boyden chamber assay to assess their migration toward SFM (negative control) or DMEM added with 1.5% FBS, VEGF, and fibroblast growth factor-2 (FGF-2) (both 10 ng/ml; Miltenyi Biotec) as chemoattractants (positive control). After 16 hours at 37°C, the migrated cells were fixed, stained, and counted by the EVOS microscope at ×400.
Wound-Healing Assay
MMECs (4 × 104)/24-well plates were treated with Notch1·siRNA or Notch2·siRNA or with MK-0752. Sixteen hours before the end of the treatment, a wound was made by scraping the cell monolayer with a sterile pipette tip. After 16 hours, MMECs were fixed and stained with Crystal Violet (0.1% in 20% methanol). Cell migration was determined by counting the MMECs that moved into the “wound” and indicated as migrated cells/field.
Detection of Cytokines
Conditioned media from MMECs treated with Notch1·siRNA or Notch2·siRNA or with MK-0752 were tested for 55 cytokines involved in angiogenesis using the Human Angiogenesis Array (R&D System) according to the manufacturer's instructions. Secreted levels of cytokines were quantified with Kodak Molecular Imaging Software. Released VEGF was measured by using human VEFG-A Bio-Plex platform (Bioclarma, Turin, Italy). Total protein content of the conditioned media was evaluated by protein detergent compatible assay (Bio-Rad).
In Vivo Chorioallantoic Membrane (CAM) Assay
Fertilized chicken eggs were incubated at 37°C and constant humidity. On day 8, sterilized gelatin sponges adsorbed with conditioned media from MMECs, treated or not with MK-0752 5 nM for 48 hours, were implanted on the top of the CAM. CAMs were examined daily until day 12 and photographed in ovo with a stereomicroscope. Blood vessels entering the sponges within the focal plane of the CAMs were counted at ×50 magnification [24].
The MM Vk*MYC Mouse for In Vivo Inhibition of the Notch Pathway and Challenge with Tumor Cells
Three days before intravenous tumor cell challenge (1 × 106 Vk12598 cells derived from one MM Vk*MYC mouse [25]), MK-0752 (5 mg/kg) was injected intraperitoneally (i.p.) in 6-week-old C57BL/6 J recipients. Treatment was performed every 3 days throughout the duration of the experiment. As control, mice were injected with PBS 10% DMSO i.p. Mice were sacrificed within 5 weeks. Periodical retro-orbital sampling of blood served to perform serum protein electrophoresis. Undiluted sera were loaded on agarose gel (Hydragel, Sebia electrophoresis, Norcross, GA). Electrophoresis was performed by the semiautomated multiparameter Hydrasys system Sebia, and gels were analyzed by densitomer/scanner Gelscan Sebia and Phoresis software for the flat-bed scanner. For immunohistochemistry, femurs of treated and untreated C57BL/6J mice were formalin-fixed, paraffin-embedded, and decalcified with Ion-Exchange Decal Unit (Biocare Medical, Pacheco, CA). Three-micrometer sections were stained with CD31 (R&D system). Images were acquired using ImageScope, and analysis was performed using Aperio Image Scope Software.
Statistical Analysis
Analyses were performed using GraphPadPrism5 software. Results were analyzed using the Wilcoxon signed-rank test. P < .05 was considered statistically significant.
Results
Homotypic Activation of the Notch Pathway in MMECs
To investigate Notch1/2 pathway in MGECs and MMECs, we analyzed the cleavage of full-length (FL) Notch1/2 into their ICDs and the expression of Hes1 and Hey1 Notch target genes [6] as sign of Notch activation. Western blotting analysis demonstrated a higher expression of the ICDs of both Notch1 and Notch2 in MMECs compared to MGECs (+43.6% and+ 61.9%, respectively) (Figure 1A), suggesting that the Notch pathway is activated in MMECs. Immunofluorescence analysis confirmed western blotting results, showing a brighter intracellular signal of Notch1 and Notch2 ICDs in MMECs (Figure 1B). Accordingly, real-time RT-PCR of Notch1/2 target genes indicated a significant increase of Hes1 and Hey1 mRNA levels in MMECs, thus proving the activation of Notch signaling (Figure 1C).
Figure 1.

Notch pathway homotypic activation in MMECs. (A) Western blotting analysis of Notch1 and Notch2 expression in MGECs (n = 12) and MMECs (n = 17) (β-actin as loading control). Representative pictures from the same experiments are shown. Data expressed as relative intensity of FL Notch1 and the ICD (left panel) and Notch2 FL and Notch2 ICD (right panel) in MMECs vs. MGECs show the high expression of Notch1/2 ICDs in MMECs. (B) Representative images of three independent immunofluorescence experiments of Notch1/2 ICDs (green) expression by MGECs and MMECs. Nuclei were counterstained with DAPI. Original magnification: ×400. Note the intense expression of Notch1/2 ICDs in MMECs. (C) Hes1 and Hey1 mRNA expression by MGECs and MMECs was analyzed by real-time RT-PCR and normalized to endogenous GAPDH. Gene expression analysis reveals the activation of Notch signaling in MMECs. (D) Flow cytometry analysis of Jagged1, Jagged2, DLL1, DLL3, and DLL4 expression by MGECs (n = 8) and MMECs (n = 11). Data are expressed as mean ± S.D. Note the strong expression of Jagged1/2 in MMECs. (E) MMECs were treated with Jagged1·siRNA and Jagged2·siRNA 25 nM for 72 hours. Hes1 and Hey1 mRNA expression by siRNA-treated MMECs was analyzed by real-time RT-PCR and normalized to endogenous GAPDH. Data are expressed as mean ± S.D. Note that Jagged1/2 knockdown inhibits Notch signaling activation.
* P < .05, ** P < .001 MMECs vs. MGECs.
Since Notch pathway activation is mediated through the receptors/ligands interaction [6], we next evaluated the expression of Jagged1, Jagged2, DLL1, DLL3, and DLL4. Flow cytometry analysis showed that the percentages of Jagged1/2-positive cells were higher in MMECs than in MGECs (Figure 1D), whereas DLL1, DLL3, and DLL4 expression was irrelevant. This suggests a conceivable involvement of Jagged1/2 ligands in the activation of Notch pathway in MMECs. Similar results were obtained by analyzing Notch ligand expression in real-time RT-PCR (data not shown). To evaluate the involvement of Jagged1/2 ligands, we silenced Jagged1 and Jagged2 through siRNA transfection in MMECs. Treatment of MMECs with Jagged1∙siRNA 25 nM and Jagged2∙siRNA 25 nM for 72 hours significantly reduced both mRNA and protein levels (data not shown). Analysis of Notch pathway activation in Jagged1/2 knockdown MMECs showed a significant reduction of Hes1 and Hey1 mRNA levels (Figure 1E).
Overall, these data indicate that the Notch pathway is activated in MMECs through homotypic interactions among nearby ECs mediated by Jagged1 and Jagged2 ligands.
Heterotypic activation of Notch pathway in MMECs
Because Notch pathway is deregulated in MM [22], we wondered whether the cross talk between MMECs and MM cells triggers Notch signaling activation. According to literature data [19], [20], RPMI-8226 cells exhibited a high expression of Jagged1 (94% ± 4%) and Jagged2 (65% ± 7%) and a marginal expression of DLL1, DLL3, and DLL4 (Supplementary Figure 1A). Likewise, flow cytometry analysis of Notch ligands on primary MM cells from 10 newly diagnosed patients showed that CD38+CD138+ cells were positive for Jagged1 (91.7% ± 10%), Jagged2 (63.3% ± 36%), and DLL3 (24% ± 10%), while no expression of DLL1 and DLL4 was observed (Supplementary Figure 1, B and C).
In order to investigate the role of MM cells in MMECs Notch activation, we set up co-culture experiments using MMECs and RPMI-8226 cells at a 1:5 cell ratio, with/without transwell. As illustrated in Figure 2A, RPMI-8226 cells significantly raised Notch1 and Notch2 ICDs in MMECs, suggesting that an increased Notch1/2 cleavage occurs in MMEC:MM cell co-cultures without transwell. Accordingly, real-time RT-PCR revealed higher levels of Notch target genes Hey1 and Hes1 in MMECs co-cultured with MM cells (Figure 2B). Similar results were obtained by co-culturing paired MMECs and primary CD138+ cells from 6 MM patients at a 1:5 cell ratio, with/without transwell. Indeed, analysis of Notch target gene expression by real-time RT-PCR showed that the CD138+ cells increased both Hes1 and Hey1 gene expression in MMECs (Figure 2C), implying that both CD138+ MM cells and RPMI-8226 cells activate the Notch pathway in MMECs. Next, to verify the role of Jagged1/2 in the heterotypic activation of Notch pathway, we set up co-culture experiments using MMECs and RPMI-8226 transfected with Jagged1·siRNA and Jagged2·siRNA without transwell. Analysis of Hes1 and Hey1 expression revealed that Jagged1/2 knockdown MM cells did not activate Notch pathway in MMECs (Figure 2D).
Figure 2.
MM cells activate Notch signaling in MMECs. MMECs (n = 5) were cultured alone or co-cultured with MM cells at 1:5 cell ratio, with/without transwell, to prevent cell-to-cell contact for 48 hours. (A) Western blotting analysis of Notch1 and Notch2 in MMECs (β-actin as loading control). Representative images from the same experiments are shown. Data are expressed as relative intensity of FL Notch1 and its ICD (left panel) and Notch2 FL and Notch2 ICD (right panel) in MMECs co-cultured with MM cells vs. MMECs cultured alone. (B) Hes1 and Hey1 mRNA expression by MMECs was analyzed by real-time RT-PCR and normalized to endogenous GAPDH. Data are expressed as mean ± S.D. Note that MM cells stimulate Notch signaling. (C) MM cells were treated with Jagged1∙siRNA and Jagged2∙siRNA 25 nM for 48 hours and co-cultured with MMECs. Hes1 and Hey1 mRNA expression in MMECs was analyzed by real-time RT-PCR and normalized to endogenous GAPDH. Data are expressed as mean ± S.D. Note that Jagged1/2 knockdown inhibits Notch signaling activation.
*P < .05, **P < .001 MMECs cultured alone vs. co-cultured cells.
These results indicate that Jagged1/2-expressing MM cells trigger a cell-to-cell contact-dependent activation of Notch signaling in MMECs.
Involvement of the Notch Pathway in MM Angiogenesis
To evaluate the involvement of Notch pathway in MM angiogenesis, we silenced Notch1 and Notch2 expression in MMECs through siRNA transfection and analyzed their ability to produce capillary-like structures on Matrigel. Treatment of MMECs with Notch1·siRNA 25 nM and Notch2·siRNA 50 nM for 72 hours significantly reduced Notch1 and Notch2 mRNA levels (Supplementary Figure 2, A and B), switching off Notch signaling, as demonstrated by the reduction of Hes1 and Hey1 mRNA (Supplementary Figure 2, C and D). No effect on cell viability was observed (Supplementary Figure 2E).
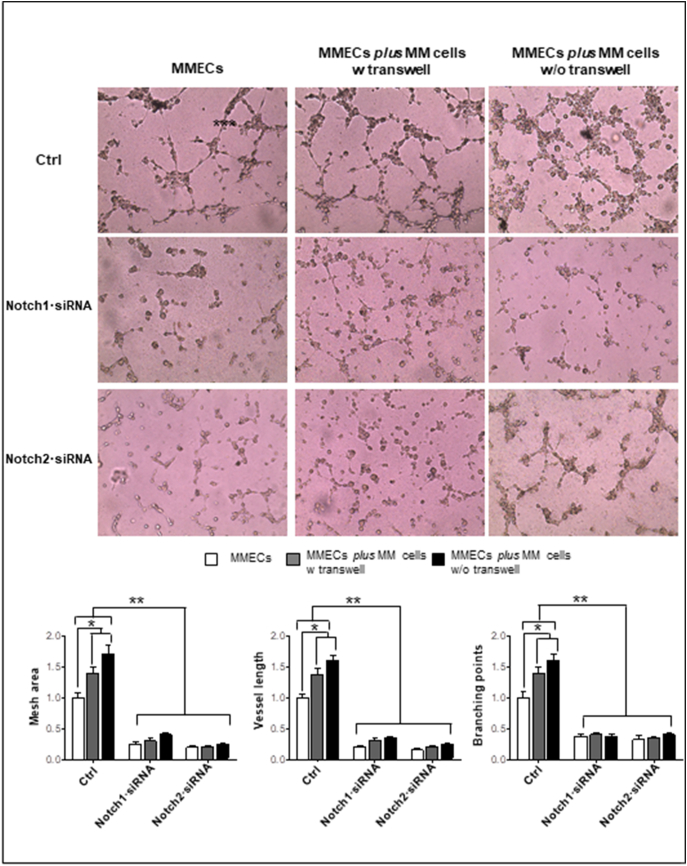
Functionally, Notch1/2·siRNAs MMECs did not exhibit angiogenic ability (Figure 3). Control MMECs formed a complex network by developing many junctions. By contrast, MMECs knocked down for Notch1/2 lost this ability: very few disorganized tubes appeared on Matrigel, as demonstrated by the significant reduction of mesh area, branching points, and vessel length. RPMI-8226 cells co-cultured with control MMECs improved the angiogenic network due to their proangiogenic activity. On the contrary, RPMI-8226 cells co-cultured with Notch1/2·siRNAs MMECs failed to recover MMECs angiogenesis.
Figure 3.
Notch1·siRNA and Notch2·siRNA affect MMEC in vitro angiogenesis. MMECs were treated with Notch1·siRNA 25 nM and Notch2·siRNA 50 nM for 72 hours and cultured alone or co-cultured with MM cells at 1:5 cell ratio with/ without transwell. Representative images of five independent in vitro angiogenesis assays of MMECs seeded on Matrigel-coated 48-well plates are shown. Original magnification: ×200. Bar graphs represent relative mesh area, vessel length, and branching points in MMECs co-cultured with MM cells vs. MMECs cultured alone, analyzed by EVOS software. Data are expressed as mean ± S.D. Note the reduction of angiogenic sprouting and vessel branching, and the failure of MM cells to restore angiogenesis in Notch1·siRNA and Notch2·siRNA MMECs.
*P < .05, **P < .001 vs. other groups.
As angiogenesis is a complex process involving several MMECs functions, we next evaluated the effect of Notch knockdown on each single angiogenesis-related function. As illustrated in Supplementary Figure 3, silencing Notch1 and Notch2 affected MMECs’ spontaneous migration and chemotaxis. Notch2·siRNA, but not Notch1·siRNA, inhibited MMECs’ adhesion. RPMI-8226 cells co-cultured with Notch1/2·siRNA MMECs did not restore MMECs’ angiogenic properties.
These data suggest a direct involvement of Notch1/2 in the regulation of angiogenesis in MM.
Notch1/2 and VEGF/VEGFR2 pathways interference in MMECs
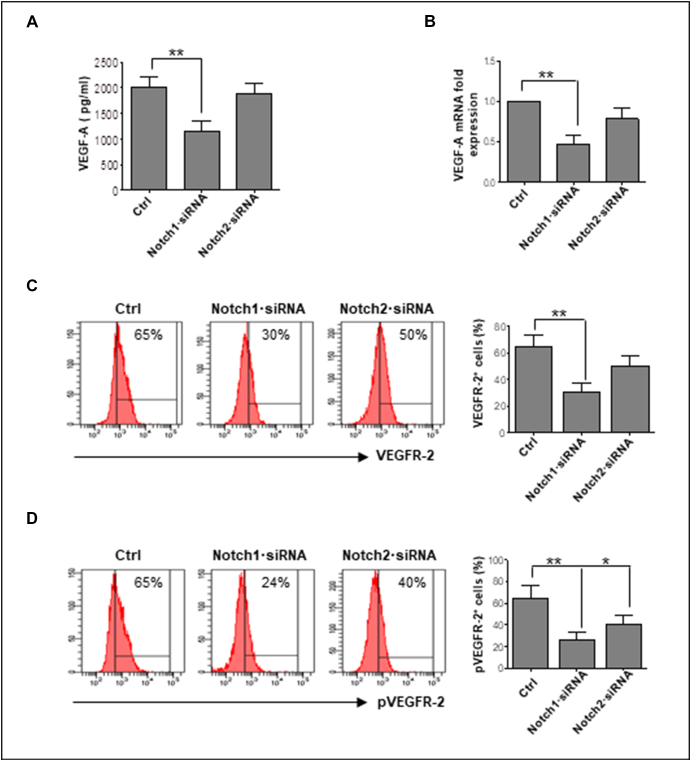
Since VEGF/VEGFR2 axis is the most important autocrine and paracrine loop for ECs’ angiogenic activities [26], [27] and Notch1/2 inhibition strongly reduced angiogenesis, we wondered whether Notch signaling may modulate VEGF pathway in MMECs. As shown in Figure 4A, Notch1·siRNA reduced the VEGF release by MMECs. Overlapping results were obtained by normalization of VEGF-A levels to total protein content of conditioned media (Supplementary Figure 4). Real time RT-PCR confirmed ELISA results, showing lower VEGF mRNA levels in Notch1 knockdown MMECs (Figure 4B). As a consequence, flow cytometry analysis of VEGFR2/pVEGFR2 expression demonstrated that Notch1/2 knockdown significantly decreased the percentages of VEGFR2- and pVEGFR2-positive MMECs (Figure 4, B and C).
Figure 4.
Notch pathway interferes with the VEGF/VEGFR-2 axis. MMECs (n = 10) were treated with Notch1·siRNA 25 nM and Notch2·siRNA 50 nM for 72 hours. (A) ELISA assay for VEGF detection in conditioned media from Notch1·siRNA and Notch2·siRNA MMECs vs. control MMECs. (B) VEGF mRNA expression by MMECs was analyzed by real-time RT-PCR and normalized to endogenous GAPDH. Data are expressed as mean ± S.D. Note the reduction of VEGF expression and release in MMECs treated with Notch1·siRNA. Flow cytometry analysis of (C) VEGFR2 and (D) pVEGFR2 expression. Histogram analysis of a representative MMECs is shown. Bar graphs show the reduction of VEGFR2 in MMECs treated with Notch1/2·siRNAs and of pVEGFR2 in MMECs treated with Notch1·siRNA vs. control MMECs. Data are expressed as mean ± S.D.
*P < .05, **P < .001 vs. others group.
Overall, these results indicate that Notch1/2 inhibition affects the VEGF/VEGFR2 loop in MMECs.
MK-0752 inhibition of MMEC angiogenesis
The Notch pathway has been recently considered a suitable therapeutic target for cancer treatment [28], [29]. Therefore, we wondered whether the selective GSI MK-0752 could affect MMECs angiogenesis. Preliminary dose-finding experiments revealed a significant inhibition of the Notch pathway in MMECs treated with MK-0752 5 nM for 48 hours, as demonstrated by the reduction of Notch1/2 ICDs and of Hes1 and Hey1 mRNA levels (Supplementary Figure 5, A and B). The effect of MK-0752 on angiogenesis was analyzed by treating MMECs, cultured alone or co-cultured with RPMI-8226 cells, with/without transwell. MK-0752 treatment significantly reduced MMECs’ spontaneous and chemotactic migration (Figure 5, A and B), adhesion (Figure 5C), and Matrigel angiogenesis (Figure 5D). The antiangiogenic effect of MK-0752 was also observed in MMECs:RPMI-8226 co-cultures, implying that MM cells were unable to switch on again Notch signaling via ligand binding and/or via soluble factors (Figure 5, A-D). No effect on MMECs and MM cells viability was observed (Supplementary Figure 5, C and D).
Figure 5.
MK-0752 reduces angiogenic functions of MMECs. MMECs (n = 12) were treated with MK0752 5 nM for 48 hours, cultured alone or co-cultured with RPMI-8226 MM cells at a 1:5 cell ratio, with/without transwell, and tested for angiogenic assays. (A) Spontaneous migration in the “wound-healing assay.” Representative images of wound closure 16 hours after the scratch. Original magnification: ×200. Bar graphs represent migrated cells/field expressed as mean ± S.D. (B) Chemotaxis toward chemoattractive medium (FGF-2, VEGF, and FBS) of treated vs. untreated MMECs. Data are expressed as mean ± S.D. (C) Adhesion on fibronectin-coated 96-well plates of MMECs stained with Calcein AM. Data are expressed as mean ± S.D. (D) Representative images of in vitro angiogenesis assay of MMECs seeded on Matrigel-coated 48-well plates. Original magnification: ×200. Bar graphs represent relative mesh area, vessel length, and branching points in treated vs. untreated MMECs analyzed by EVOS software. Data are expressed as mean ± S.D. The decrease of angiogenic sprouting and vessel branching formation and the inability of MM cells to trigger angiogenesis are evident. (E) CAM assay shows a reduction of vessel formation toward the sponge induced by conditioned media from treated vs. untreated cell cultures. Representative pictures on a stereomicroscope. Original magnification: ×50. Data are expressed as the mean ± S.D. of five independent experiments.
*P < .05, **P < .001.
In ovo analysis of CAMs implanted with a gelatin sponge soaked with the conditioned media of untreated MMECs showed many newly formed capillaries spreading radially toward the sponge that were significantly enhanced after the addition of conditioned media of MMECs:RPMI-8226 co-cultures (Figure 5E). By contrast, conditioned media of MK-0752-treated cell cultures induced only poor angiogenesis, suggesting that MK-0752 affects the secretion of angiogenesis-related cytokines. Analysis of cytokine content in the conditioned media revealed that MK-0752 inhibited the release of several proangiogenic and proinflammatory cytokines or growth factors: VEGF, hepatocyte growth factor (HGF), angiopoietin-2 (Ang-2) [30], endothelin-1 (ET-1) [31], insulin-like growth factor-binding protein (IGFBP-1/3) [32], monocyte chemoattractant protein-1 (MCP-1) [33], urokinase plasminogen activator (uPA) [2], IL-1β [34], and IL-8 [30]. MK-0752 also reduced the levels of the antiangiogenic pentraxin-3 [35] and thrombospondin-1 [36] (Supplementary Figure 6).
Antiangiogenic Effect of MK-0752 in Mice
C57BL/6J mice engrafted with Vk12598 cells derived from one MM Vk*MYC were used to assess whether MK-0752 had antiangiogenic effects in vivo. This murine model represents a suitable preclinical system for the study of MM: it exhibits similar clinical and biological features to human MM, i.e., rearrangements of c-myc associated with the spontaneous progression from MGUS to MM, the strong dependence of Vk12598 MM cell on BM microenvironment, and the secretion of a serum monoclonal Ig that represents a tumor burden marker [25]. The antiangiogenic effect was analyzed by evaluating the microvessel density on femur sections from untreated and MK-0752–treated mice. As illustrated in Figure 6A, sections from untreated mice showed a strong vascularization with arborized and tortuous vessels. Drug treatment significantly reduced the size of the vessels that appeared small and round. Analysis of CD31-positive cells by AperioScope software demonstrated a significant decrease of the ECs number/area. Furthermore, the antiangiogenic effect of MK-0752 was related to a reduction of the tumor burden, as demonstrated by the lower levels of M-protein in treated mice (Figure 6B).
Figure 6.

In vivo antiangiogenic effect of MK-0752. C57BL/6J mice engrafted with Vk12598 cells were treated every 3 days with MK-0752 5 mg/kg or with vehicle as control for 5 weeks. (A) Immunohistochemistry of CD31+ cells (brown) in mice femur sections, as the microvessel density index, shows a significant reduction of ECs on the surface area in treated vs. untreated mice. Bar graph indicates the number of CD31+ ECs/area ratio analyzed by Aperio Scope software. (B) Serum protein electrophoresis of M-protein levels in MK-0752-treated vs. vehicle-treated mice at 3 weeks and 5 weeks after the injection of tumor cells.
*P < .05, **P < .001 treated vs. untreated mice.
Discussion
This study demonstrates the direct involvement of Notch pathway in MM angiogenesis. To date, dysregulation of Notch signaling in MM has been related to MM cell growth, survival, self-renewal, and bone disease [22]. MM cells establish a Notch1/2-Jagged1/2–mediated cross talk with BMSCs that leads to a reciprocal activation of the Notch pathway and to the release by BMSCs of several cytokines/growth factors—i.e., IGF1, IL-6, stromal cell-derived factor1 alpha, and VEGF—that modulate MM cell proliferation, drug resistance, and migration [20], [21], [22]. Berenstein et al. have shown that co-cultures of MM cells with BM mesenchymal stromal cells decreased the expression of miR-223 in a contact-dependent manner via Notch/Jagged2 activation that was correlated with an overexpression of the tumor-supportive cytokines VEGF and IL-6. Inhibition of the Notch signaling strongly affected this supportive mechanism, suggesting that the Notch/Jagged2/miR-223 axis plays a crucial role in MM [37]. Although several factors may activate the Notch pathway in BM microenvironment [6], [7], [8], here we proved a canonical activation of the Notch signaling in MMECs via Jagged1/2 ligands and Notch1/2 receptors.
Higher expression of Notch1/2 and Jagged1/2 was observed on MMECs compared to MGECs, suggesting that their expression parallels the MGUS to MM transition. In addition, the increase of Notch1/2 ICDs and their intracellular localization in MMECs as well as the rise of Hey1 mRNA levels imply a homotypic activation of Notch signaling in MMECs through the MMEC-MMEC interactions via Jagged1/2 ligands. Activation of Notch1/2 pathway is closely involved in the MMEC overangiogenic phenotype. Indeed, Notch1/2 knockdown in MMECs reduces mRNA levels of Hes1 and Hey1 and affects MMECs adhesion, migration, and angiogenesis in vitro.
BM angiogenesis is a constant hallmark of MM progression and is enhanced by the autocrine and paracrine VEGF loop in MMECs [26], [27]. Due to the high secretion of VEGF by MM cells and MMECs, VEGFR2 is constitutively phosphorylated in MMECs and associated with the activation of ERK1/2 that induces MMECs proliferation and chemotaxis [26], [27]. Increasing evidence points to a cross talk between the VEGF and Notch pathways during physiological and tumor angiogenesis [38], [39]. VEGF stimulation increases the expression of DLL4, which activates Notch receptors in adjacent ECs and represses the VEGFR2 transcription [5], [10], [11]. Thus, the DLL4/Notch pathway functions as a negative regulator of angiogenesis through the downmodulation of VEGF-induced responses [40], [41], [42]. MMECs secrete large amounts of VEGF, express high levels of Jagged1/2, but do not express DLL4. Exposure of MMECs to exogenous VEGF further increases the expression of the “proangiogenic” Jagged1/2 but has no effect on DLL4 expression (data not shown). Jagged1 is a positive regulator of angiogenesis based on its ability to antagonize DLL4/Notch signaling and to induce VEGFR2 expression [12], [13], [43].
Here, we show that Notch1/2 knockdown in MMECs reduces VEGF mRNA and release, and VEGFR2/pVEGFR2 expression, thus interfering with the autocrine VEGF/VEGFR2 loop. Accordingly homotypic Notch1/2 activation contributes to the overangiogenic phenotype of MMECs via the low expression of “antiangiogenic” DLL4 and the high expression of “proangiogenic” Jagged1/2 that modulate VEGFR2/pVEGFR2 expression.
Otherwise, Notch heterotypic activation occurs in BM microenvironment [22]. RPMI-8226 and primary CD138+ MM cells showed a strong expression of Jagged1 and Jagged2 ligands [18], [19], [20], [21], [22]. Here, primary CD138+ MM cells and RPMI-8226 cells increase the expression of Hes1 and Hey1 Notch target genes and, thus, activate Notch pathway in paired co-cultured MMECs. Knockdown experiments with Jagged1/2∙siRNAs demonstrated that activation of Notch pathway by MM cells occurs via Jagged1/2-mediated heterotypic cell-to-cell interactions.
MM cells activate several proangiogenic pathways in MMECs through the release of growth factors such as VEGF and HGF and via cell-to-cell contacts that prompt EC migration, chemotaxis, adhesion to extracellular matrix, spreading, and formation of an angiogenic network [2], [3], [44]. Our results indicate that MM cells trigger the angiogenic ability of MMECs through the activation of Jagged1/2-Notch1/2 axis. Indeed, Notch1/2·siRNAs strongly reduced the in vitro Matrigel angiogenesis assay of MMECs co-cultured with MM cells. Co-cultured RPMI-8226 cells are not able to reactivate Notch signaling and to restore the angiogenic activity in Notch1/2·siRNAs MMECs. Thus, the proangiogenic activity of MM cells and the angiogenic properties of MMECs closely rely on Notch pathway activation in MMECs.
Targeting the Notch pathway is currently offering new opportunities for drug development [28]. Preclinical and ongoing clinical studies using Notch inhibitors are under consideration in cancer treatment, including MM [29]. Two alternative therapeutic approaches have been developed to block Notch signaling, i.e., GSIs and specific mAbs targeting DLL4, Jagged1, and Notch receptors [29]. In this study, we investigated the pharmacological effect of the GSI MK-0752 on Notch-induced angiogenesis through in vitro and in vivo studies. MK-0752 is a new GSI currently employed in phase I clinical studies for the treatment of breast, head and neck squamous cell, and pancreatic carcinomas [45]. It is the analogue of the MRK003 that exerts proapoptotic and antiproliferative effects on non-Hodgkin lymphoma and MM cells and is able to overcome the protective effect of BMSCs [46].
In vitro inhibition of the Notch pathway through MK-0752 reduced the proangiogenic activities of MMECs, cultured alone or co-cultured with RPMI-8226 cells: MK-0752 significantly affected MMEC adhesion, migration, chemotaxis, and angiogenesis.
Notch signaling interacts with several pathways, i.e., NF-κB, VEGF, Wnt, and HIF-1α, thus modulating cell proliferation, apoptosis, migration, survival, and cytokines/growth factors release. Maniati et al. described a reciprocal cross talk between NF-κB and Notch signaling that modulates the expression of proinflammatory cytokines/chemokines through downregulation of the peroxisome proliferator-activated receptor, a repressor of inflammatory genes, induced by Hes1 [47]. Furthermore, Wnt/β-catenin activation contributes to Hes1 overexpression [48] and stimulates the release of proangiogenic cytokines and cell cycle progression [49]. A mutual interplay between Notch and HIF-1α is induced by hypoxia: HIF-1α binds Notch ICD and increases the expression of Notch downstream genes; conversely, Notch ICD sequesters the factor inhibiting HIF-1α (FIH-1), resulting in HIF-1α stabilization and the expression of HIF-1α–responsive genes [50]. In our study, MK-0752 modulates the secretion of angiogenic and inflammatory cytokines, i.e., VEGF, HGF, Ang-2 [30], ET-1 [31], IGFBP-1/3 [32], MCP-1 [33], uPA [2], IL-1β [34], and IL-8 [30], by MMECs and MM cells, thus reducing the formation of blood vessels in the CAM assay.
In vitro data were confirmed in the in vivo Vk*MYC MM mouse model. MK-0752 decreased angiogenesis by reducing microvessel density and CD31-positive ECs: tumor vessels appeared round and smaller compared to the arborized and tortuous vessels observed in untreated mice. Based on our in vitro results and on Notch pathway role in the cross talk between MM cells and BM microenvironment [20], [21], [22], the in vivo antiangiogenic activity of MK-0752 may be related to several factors, including the inhibition of proangiogenic/proinflammatory cytokines in MMECs and MM cells as well as the apoptotic and antiproliferative effect on tumor cells [21], [46].
In conclusion, homotypic and heterotypic activation of the Notch1/2 pathway is actively involved in MM angiogenesis. The overexpression of the proangiogenic Jagged1/2 ligands on MMECs and MM cells and the interference of Notch1/2 with the VEGF/VEGFR2 loop indicate that Notch1/2-mediated angiogenesis may represent a mechanism of MMECs to escape from the VEGF/VEGFR2 axis control, providing the rationale for a Notch-targeted approach as a new antiangiogenic therapy.
Author Contributions
I. S., M. A. F. and R. R. conceived and designed the study. R. R., V. R., F. D., and A. V. contributed to study design. A. L., A. B., P. L., V. D, L. D. M., M. B., D. D., D. R., R. C., M. T. P., A. N., M. A. M., and R. F. conducted part of the experiments and were involved in the collection, analysis, and interpretation of the data. I. S. and M. A. F. wrote the manuscript. R. R., V. R., F. D., and A. V. revised the manuscript. All authors reviewed the report, approved the draft submission, and agreed to be accountable for all aspects of this study.
Acknowledgments
Acknowledgements
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) Investigator Grant (no. 20441) to V. R., Milan, Italy; the Special Program Molecular Clinical Oncology 5 per 1,000 (grant no. 9965) to A. V., Milan, Italy; Associazione Italiana Ricerca sul Cancro Investigator Grant (no. 20614) to R.C.; and University of Milan-Line 2B-Project 2017 to R. C.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.10.011.
Appendix A. Supplementary data
Supplementary figures
Supplementary tables
References
- 1.Ribatti D, Nico B, Vacca A. Importance of the bone marrow microenvironment in inducing the angiogenic response in multiple myeloma. Oncogene. 2006;25:4257–4266. doi: 10.1038/sj.onc.1209456. [DOI] [PubMed] [Google Scholar]
- 2.Kyle RA, Rajkumar SV. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma. Curr Hematol Malig Rep. 2010;5(2):62–69. doi: 10.1007/s11899-010-0047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vacca A, Ribatti D, Roncali L, Ranieri G, Serio G, Silvestris F, Dammacco F. Bone marrow angiogenesis and progression in multiple myeloma. Br J Haematol. 1994;87:503–508. doi: 10.1111/j.1365-2141.1994.tb08304.x. [DOI] [PubMed] [Google Scholar]
- 4.Vacca A., Ria R., Semeraro F., Merchionne F., Coluccia M., Boccarelli A., Scavelli C., Nico B., Gernone A., Battelli F. Endothelial cells in the bone marrow of patients with multiple myeloma. Blood. 2003;102:3340–3348. doi: 10.1182/blood-2003-04-1338. [DOI] [PubMed] [Google Scholar]
- 5.D'Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275:652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- 7.Dufraine J, Funahashi Y, Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene. 2008;27:5132–5137. doi: 10.1038/onc.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 9.Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Jakobsson L., Franco C.A., Bentley K., Collins R.T., Ponsioen B., Aspalter I.M., Rosewell I., Busse M., Thurston G., Medvinsky A. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol. 2010;12:943–953. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]
- 11.Sainson RC, Harris AL. Regulation of angiogenesis by homotypic and heterotypic notch signalling in endothelial cells and pericytes: from basic research to potential therapies. Angiogenesis. 2008;11:41–51. doi: 10.1007/s10456-008-9098-0. [DOI] [PubMed] [Google Scholar]
- 12.Pedrosa A.R., Trindade A., Carvalho C., Graça J., Carvalho S., Peleteiro M.C., Adams R.H., Duarte A. Endothelial Jagged1 promotes solid tumor growth through both pro-angiogenic and angiocrine functions. Oncotarget. 2015;6:24404–24423. doi: 10.18632/oncotarget.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benedito R, Roca C, Sörensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Q., Li S., Chepeha D.B., Giordano T.J., Li J., Zhang H., Polverini P.J., Nor J., Kitajewski J., Wang C.Y. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell. 2005;8:13–23. doi: 10.1016/j.ccr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Patel NS, Li JL, Generali D, Poulsom R, Cranston DW, Harris AL. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res. 2005;65:8690–8697. doi: 10.1158/0008-5472.CAN-05-1208. [DOI] [PubMed] [Google Scholar]
- 16.Mailhos C, Modlich U, Lewis J, Harris A, Bicknell R, Ish-Horowicz D. Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation. 2001;69:135–144. doi: 10.1046/j.1432-0436.2001.690207.x. [DOI] [PubMed] [Google Scholar]
- 17.Pietras A, von Stedingk K, Lindgren D, Påhlman S, Axelson H. JAG2 induction in hypoxic tumor cells alters Notch signaling and enhances endothelial cell tube formation. Mol Cancer Res. 2011;9:626–636. doi: 10.1158/1541-7786.MCR-10-0508. [DOI] [PubMed] [Google Scholar]
- 18.Skrtić A, Korać P, Krišto DR, Ajduković Stojisavljević R, Ivanković D, Dominis M. Immunohistochemical analysis of NOTCH1 and JAGGED1 expression in multiple myeloma and monoclonal gammopathy of undetermined significance. Hum Pathol. 2010;41:1702–1710. doi: 10.1016/j.humpath.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Houde C., Li Y., Song L., Barton K., Zhang Q., Godwin J., Nand S., Toor A., Alkan S., Smadja N.V. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood. 2004;104:3697–3704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- 20.Jundt F., Pröbsting K.S., Anagnostopoulos I., Muehlinghaus G., Chatterjee M., Mathas S., Bargou R.C., Manz R., Stein H., Dörken B. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- 21.Xu D., Hu J., Xu S., De Bruyne E., Menu E., Van Camp B., Vanderkerken K., Van Valckenborgh E. Dll1/Notch activation accelerates multiple myeloma disease development by promoting CD138+ MM-cell proliferation. Leukemia. 2012;26:1402–1405. doi: 10.1038/leu.2011.332. [DOI] [PubMed] [Google Scholar]
- 22.Colombo M., Mirandola L., Platonova N., Apicella L., Basile A., Figueroa A.J., Cobos E., Chiriva-Internati M., Chiaramonte R. Notch-directed microenvironment reprogramming in myeloma: a single path to multiple outcomes. Leukemia. 2013;27:1009–1018. doi: 10.1038/leu.2013.6. [DOI] [PubMed] [Google Scholar]
- 23.Rajkumar S.V., Dimopoulos M.A., Palumbo A., Blade J., Merlini G., Mateos M.V., Kumar S., Hillengass J., Kastritis E., Richardson P. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15(12):e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 24.Ribatti D. The chick embryo chorioallantoic membrane (CAM) assay. Reprod Toxicol. 2017;70:97–101. doi: 10.1016/j.reprotox.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Chesi M., Robbiani D.F., Sebag M., Chng W.J., Affer M., Tiedemann R., Valdez R., Palmer S.E., Haas S.S., Stewart A.K. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ria R., Vacca A., Russo F., Cirulli T., Massaia M., Tosi P., Cavo M., Guidolin D., Ribatti D., Dammacco F. A VEGF-dependent autocrine loop mediates proliferation and capillarogenesis in bone marrow endothelial cells of patients with multiple myeloma. Thromb Haemost. 2004;92:1438–1445. doi: 10.1160/TH04-06-0334. [DOI] [PubMed] [Google Scholar]
- 27.Vacca A, Ria R, Ribatti D, Semeraro F, Djonov V, Di Raimondo F, Dammacco F. A paracrine loop in the vascular endothelial growth factor pathway triggers tumor angiogenesis and growth in multiple myeloma. Haematologica. 2003;88:176–185. [PubMed] [Google Scholar]
- 28.Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med. 2011;17(11):1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 29.Colombo M., Galletti S., Garavelli S., Platonova N., Paoli A., Basile A., Taiana E., Neri A., Chiaramonte R. Notch signaling deregulation in multiple myeloma: A rational molecular target. Oncotarget. 2015;6(29):26826–26840. doi: 10.18632/oncotarget.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giuliani N, Storti P, Bolzoni M, Palma BD, Bonomini S. Angiogenesis and multiple myeloma. Cancer Microenviron. 2011;4(3):325–337. doi: 10.1007/s12307-011-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salani D, Taraboletti T, Rosanò L, Di Castro V, Borsotti P, Giavazzi R, Bagnato A. Endothelin-1 induces an angiogenic phenotype in cultured endothelial cells and stimulates neovascularization in vivo. Am J Pathol. 2000;157(5):1703–1711. doi: 10.1016/S0002-9440(10)64807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granata R., Trovato L., Lupia E., Sala G., Settanni F., Camussi G., Ghidoni R., Ghigo E. Insulin-like growth factor binding protein-3 induces angiogenesis through IGF-I– and SphK1-dependent mechanisms. J Thromb Haemost. 2007;5(4):835–845. doi: 10.1111/j.1538-7836.2007.02431.x. [DOI] [PubMed] [Google Scholar]
- 33.Hong KH, Ryu J, Han KH. Monocyte chemoattractant protein-1–induced angiogenesis is mediated by vascular endothelial growth factor-A. Blood. 2005;105(4):1405–1407. doi: 10.1182/blood-2004-08-3178. [DOI] [PubMed] [Google Scholar]
- 34.Voronov E, Carmi Y, Apte RN. Role of IL-1–mediated inflammation in tumor angiogenesis. Adv Exp Med Biol. 2007;601:265–270. doi: 10.1007/978-0-387-72005-0_28. [DOI] [PubMed] [Google Scholar]
- 35.Basile A., Moschetta M., Ditonno P., Ria R., Marech I., De Luisi A., Berardi S., Frassanito M.A., Angelucci E., Derudas D. Pentraxin 3 (PTX3) inhibits plasma cell/stromal cell cross-talk in the bone marrow of multiple myeloma patients. J Pathol. 2013;229(1):87–98. doi: 10.1002/path.4081. [DOI] [PubMed] [Google Scholar]
- 36.Lawler PR, Lawler J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb Perspect Med. 2012;2(5):a006627. doi: 10.1101/cshperspect.a006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berenstein R., Nogai A., Waechter M., Blau O., Kuehnel A., Schmidt-Hieber M., Kunitz A., Pezzutto A., Dörken B., Blau I.W. Multiple myeloma cells modify VEGF/IL-6 levels and osteogenic potential of bone marrow stromal cells via Notch/miR-223. Mol Carcinog. 2016;55(12):1927–1939. doi: 10.1002/mc.22440. [DOI] [PubMed] [Google Scholar]
- 38.Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, Wiegand SJ. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci U S A. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridgway J., Zhang G., Wu Y., Stawicki S., Liang W.C., Chanthery Y., Kowalski J., Watts R.J., Callahan C., Kasman I. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 40.Thurston G, Kitajewski J. VEGF and Delta-Notch: interacting signalling pathways in tumour angiogenesis. Br J Cancer. 2008;99:1204–1209. doi: 10.1038/sj.bjc.6604484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noguera-Troise I., Daly C., Papadopoulos N.J., Coetzee S., Boland P., Gale N.W., Lin H.C., Yancopoulos G.D., Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 42.Kuhnert F, Kirshner JR, Thurston G. Dll4-Notch signaling as a therapeutic target in tumor angiogenesis. Vasc Cell. 2011;3:20. doi: 10.1186/2045-824X-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Funahashi Y., Hernandez S.L., Das I., Ahn A., Huang J., Vorontchikhina M., Sharma A., Kanamaru E., Borisenko V., Desilva D.M. A notch1 ectodomain construct inhibits endothelial notch signaling, tumor growth, and angiogenesis. Cancer Res. 2008;68:4727–4735. doi: 10.1158/0008-5472.CAN-07-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moschetta M., Basile A., Ferrucci A., Frassanito M.A., Rao L., Ria R., Solimando A.G., Giuliani N., Boccarelli A., Fumarola F. Novel targeting of phospho-cMET overcomes drug resistance and induces antitumor activity in multiple myeloma. Clin Cancer Res. 2013;19(16):4371–4382. doi: 10.1158/1078-0432.CCR-13-0039. [DOI] [PubMed] [Google Scholar]
- 45.Piha-Paul S.A., Munster P.N., Hollebecque A., Argilés G., Dajani O., Cheng J.D., Wang R., Swift A., Tosolini A., Gupta S. Results of a phase 1 trial combining ridaforolimus and MK-0752 in patients with advanced solid tumours. Eur J Cancer. 2015;51(14):1865–1873. doi: 10.1016/j.ejca.2015.06.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramakrishnan V., Ansell S., Haug J., Grote D., Kimlinger T., Stenson M., Timm M., Wellik L., Halling T., Rajkumar S.V. MRK003, a γ-secretase inhibitor exhibits promising in vitro pre-clinical activity in multiple myeloma and non-Hodgkin's lymphoma. Leukemia. 2012;26(2):340–348. doi: 10.1038/leu.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maniati E., Bossard M., Cook N., Candido J.B., Emami-Shahri N., Nedospasov S.A., Balkwill F.R., Tuveson D.A., Hagemann T. Crosstalk between the canonical NF-κB and Notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice. J Clin Invest. 2011;121(12):4685–4699. doi: 10.1172/JCI45797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, Taga T. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol. 2008;28:7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qu B, Liu BR, Du YJ, Chen J, Cheng YQ, Xu W, Wang XH. Wnt/β-catenin signaling pathway may regulate the expression of angiogenic growth factors in hepatocellular carcinoma. Oncol Lett. 2014;7(4):1175–1178. doi: 10.3892/ol.2014.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng X., Linke S., Dias J.M., Zheng X., Gradin K., Wallis T.P., Hamilton B.R., Gustafsson M., Ruas J.L., Wilkins S. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci U S A. 2008;105:3368–3373. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary tables