Summary
Arbuscular mycorrhizal (AM) fungi form diverse communities and are known to influence above‐ground community dynamics and biodiversity. However, the multiscale patterns and drivers of AM fungal composition and diversity are still poorly understood.
We sequenced DNA markers from roots and root‐associated soil from Plantago lanceolata plants collected across multiple spatial scales to allow comparison of AM fungal communities among neighbouring plants, plant subpopulations, nearby plant populations, and regions. We also measured soil nutrients, temperature, humidity, and community composition of neighbouring plants and nonAM root‐associated fungi.
AM fungal communities were already highly dissimilar among neighbouring plants (c. 30 cm apart), albeit with a high variation in the degree of similarity at this small spatial scale. AM fungal communities were increasingly, and more consistently, dissimilar at larger spatial scales. Spatial structure and environmental drivers explained a similar percentage of the variation, from 7% to 25%. A large fraction of the variation remained unexplained, which may be a result of unmeasured environmental variables, species interactions and stochastic processes.
We conclude that AM fungal communities are highly variable among nearby plants. AM fungi may therefore play a major role in maintaining small‐scale variation in community dynamics and biodiversity.
Keywords: arbuscular mycorrhizal (AM) fungi, community composition, fungal community, Moran's eigenvector maps, plant community, soil microbial community, spatial structure
Short abstract
See also the Commentary on this article by https://doi.org/10.1111/nph.15212.
Introduction
Questions of how organisms are spatially distributed and what drives spatial patterns are central to community ecology (Nemergut et al., 2013). While most focus in the past has been on describing patterns above ground, we are now seeing a rise in studies investigating the spatial distribution of soil biota and the mechanisms driving their distribution (Nemergut et al., 2013; Tedersoo et al., 2014; Davison et al., 2015). How organisms are structured spatially will depend on their dispersal ability, interspecies interactions and the abiotic and biotic environments, and each of these potential drivers may regulate community composition at different spatial scales (Levin, 1992). While much is known about the spatial structure and drivers of above‐ground plant‐based communities, we still lack a comprehensive understanding of the multiscale patterns and drivers of plant‐associated communities below ground.
Arbuscular mycorrhizal (AM) fungi are root symbionts forming associations with the majority of plants (Smith & Read, 2008). These ubiquitous organisms are important as they facilitate plant uptake of growth‐limiting nutrients, in particular phosphorus, which can affect plant performance (Smith & Smith, 2011). In addition, increases in AM fungal diversity have been linked to increasing plant diversity, growth (van der Heijden et al., 1998; Hartnett & Wilson, 1999), and resistance to pathogens and drought (Newsham et al., 1995; Augé, 2001). There is also increasing evidence that the impact of AM fungi on plant defence and fitness depends on the composition of the AM fungal communities (Klironomos, 2000; Bever, 2002; Klironomos et al., 2004).
A global assessment of AM fungal communities showed low endemism, with the majority of AM taxa found on several continents (Davison et al., 2015; but see Bruns & Taylor, 2016; and Bruns et al., 2018). While this indicates that dispersal at a global scale does occur, it is generally thought that AM fungal communities are structured by local environmental filtering. However the key drivers may vary across spatial scales. AM fungi are dispersed by hyphal spread over smaller areas and by passive dispersal of spores by wind and animals, in some cases up to 2 km (Warner et al., 1987; Mangan & Adler, 2000). Previous work has found large variation in fungal community structure at the submetre scale (Anderson et al., 1983; Wolfe et al., 2007; Mummey & Rillig, 2008), while clustering and spatial autocorrelation were typically found up to the 1 m scale (Anderson et al., 1983). However, many of the studies on the spatial structure of AM fungi have focused on either small (1–2 m; Wolfe et al., 2007; Mummey & Rillig, 2008) or global scales (Davison et al., 2015), with few studies focusing at the mesoscale (cm to km). In addition, many studies have investigated the AM fungi in roots and soil associated with a mix of plant species (Anderson et al., 1983; Whitcomb & Stutz, 2007; Mummey & Rillig, 2008), and variation in AM fungal communities across spatial scales may thus be attributable to variation in plant species identity, rather than the impact of spatial scale per se. To the best of our knowledge, only a handful of studies have investigated the spatial structure of AM fungi associated with specific host plant species (Table 1). One study found that spatial autocorrelation accounted for 24% of the variation in AM fungal community composition associated with Zea mays in agricultural fields in Zimbabwe (Table 1; Lekberg et al., 2007). In another study, which investigated multiscale patterns of AM fungal communities, Chaudhary et al. (2014) found that AM fungi associated with two Artemisia species were similar at the smallest spatial scale (1 m apart) but varied significantly at larger scales, with the strongest differences between regions (c. 50–100 km apart; Table 1). To bridge the gap between small‐ and large‐scale studies, and disentangle the effect of spatial scale from that of plant species identity, in this study we sampled AM fungal communities associated with a single plant species across multiple spatial scales, ranging from tens of centimetres to tens of kilometres.
Table 1.
Overview of studies on the spatial structure of arbuscular mycorrhizal (AM) fungi associated with a specific host plant
| Study | Plant species | Vegetation type | AM fungi measured | Spatial scale measured | Spatial structure | Abiotic factors | |
|---|---|---|---|---|---|---|---|
| Sample area | Distance between samples | ||||||
| Chaudhary et al. (2014) | Artemisia filifolia and Artemisia tridentata | Semiarid shrubland (UT, USA) | Spore identification |
Samples taken in nested design within: (i) four regions (5000 ha); (ii) three sites within regions (1 ha); (iii) two microsites within sites (1 m2) |
(i) Regions were c. 50–100 km apart; (ii) sites were a few km apart; (iii) microsites were up to 1 m apart Nine random samples were taken within each 1 m microsite |
Positive spatial autocorrelation. Within‐group heterogeneity of AM fungal community composition higher for regions | P, mean annual temperature and precipitation, elevation and latitude influenced AM fungal community composition. pH, P, electric conductivity, and mean annual temperature influenced AM fungal spore richness. P influenced AM fungal spore evenness |
| Friese & Koske (1991) | Ammophila breviligulata | Sand dune (RI, USA) | Spore identification |
Samples taken within: (i) three plots of 40 × 40 × 40 cm |
(i) Distance between plots not listed. Sixteen samples were taken from each plot. Samples were 10 × 10 cm taken in 5 cm vertical increments |
No spatial autocorrelation, but spores tended to aggregate | No correlation to physical factors found |
| Hazard et al. (2013) | Trifolium repens and Lolium perenne | Pastures, arable fields, peatlands, forests (Republic of Ireland) | Molecular methods |
Samples taken within: (i) 40 sites of 30 m × 30 m |
(i) Sites were 7–392 km apart. Five randomly collected root samples (pool of five plants) were taken within each site |
No spatial autocorrelation | pH, rainfall and soil type influenced AM fungal community composition |
| Horn et al. (2017) | Festuca brevipila | Nature protection area of steppes and coastal habitats (Germany) | Molecular methods |
Samples taken in nested design within: (i) three macroplots (15 × 15 m) (ii) four plots within macroplots (3 × 3 m) |
(i) Macroplots were 20–500 m apart; (ii) plots were 9–15 m apart. Five samples were taken within plots 30 cm–3 m apart | The spatial structure explained up to 31% of the variation in the AM fungal community, with further variation explained by spatial‐phylogenetic effects | Environmental factors (pH, water content, C, N, C : N ratio, P, dehydrogenase activity) explained up to 10% of the variation in AM fungal community composition |
| Lekberg et al. (2007) | Maize (Zea mays) | Semi‐arid agricultural field (Zimbabwe) | Molecular methods |
Samples taken within: (i) 10 fields (0.5 ha each) |
(i) Fields were 25 km apart Twenty randomly collected root samples were taken within each field |
Positive spatial autocorrelation; 23.5% of variation in community composition explained by spatial autocorrelation | Soil variables (texture, moisture, organic C, pH, P, N) explained 38.6% of the variation in AM fungal community composition. |
| Sylvia (1986) | Uniola paniculata | Coastal foredunes (FL, USA) | Spore identification |
Samples taken in nested design within: (i) two sites (size not listed) (ii) three plots within sites (1 × 2 m) |
(i) Sites were c. 100 km apart; (ii) plots were 25–100 m apart. Thirty samples were taken along six diagonal transects within each plot. Each transect was 33 cm apart and each sample was 10 cm apart |
AM fungal species differed between sites and plots, but no results are reported for within‐plot differences. | – |
Arbuscular mycorrhizal fungal species differ in their responses to variation in soil properties, such as moisture, temperature and nutrient availability (Chaudhary et al., 2008), and this may in turn explain spatial variation in diversity and community composition (Wolfe et al., 2007; Hawkes et al., 2011; Kivlin et al., 2011). Lekberg et al. (2007) found that 38% of variation in AM fungal community composition in Zea mays was explained by the abiotic soil environment (Table 1). However, the biotic environment, including neighbouring plants and nonAM fungi in roots, may also be linked to the diversity and composition of AM fungal communities. While AM fungi are considered to colonize a wide range of host plants (Allen et al., 1995; Rosendahl, 2008; Helgason & Fitter, 2009), AM fungi in natural communities associate to a different degree with different host plant species, with some evidence of specialization (Helgason et al., 1998; Klironomos, 2000; Bever, 2002). Rodríguez‐Echeverría et al. (2017) found that vegetation type influenced AM fungal community composition in tropical African environments, and studies linking AM fungal and plant richness have found positive (Vogelsang et al., 2006; Hiiesalu et al., 2012), negative (Antoninka et al., 2011) or no relationships (Öpik et al., 2008, 2010; Lekberg et al., 2013). Another structuring factor may be the presence of root‐associated nonAM fungi. For example, Larimer et al. (2012) studied the interactions between AM fungi and endophytes in the grass Elymus hystrix and found that endophytes influenced AM fungal colonization while AM fungi affected fungal endophyte fitness. Other studies have shown similar reciprocal interactions between AM fungi and fungal pathogens (Newsham et al., 1995; Borowicz, 2001). It may thus be important to determine the relationship between different fungal groups in order to understand the community composition of AM fungi.
The community composition of AM fungi may differ between roots and the root‐associated soil. In general, communities of AM fungi found in soil have been considered to represent a common species pool for the entire plant community, with only a subset able to colonize the roots of a given plant species (Johnson et al., 2003; Davison et al., 2011). However, species diversity in the roots may also be higher than in soil if some species invest more in intraradical colonization with limited extraradical growth (Jakobsen et al., 1992; Munkvold et al., 2004), thereby limiting species detection in soil. Studies have found both higher (Hempel et al., 2007), similar (Busby et al., 2013), or lower (Saks et al., 2014; Varela‐Cervero et al., 2015) richness in soil as compared with roots.
We explored the multiscale patterns and drivers of root and root‐associated soil AM fungal communities associated with Plantago lanceolata in naturally fragmented plant populations across the Åland Islands (Finland). We assessed the spatial distribution of root colonization intensity, diversity indices and AM fungal community composition, in both roots and root‐associated soil, on scales ranging from neighbouring plants to regions. We further determined how much of the variation in AM fungal root colonization intensity, diversity indices and community composition was determined by spatial structure and the abiotic and biotic environments. More specifically we asked:
How does root colonization, diversity indices and composition change with distance across spatial scales? Is most variation found among neighbouring plants (c. 30 cm), among plant subpopulations (c. 10 m), among plant populations (c. 5 km), or among regions (c. 30 km)?
How do the abiotic (soil nutrients and climate) and biotic (i.e. neighbouring plants and nonAM root‐associated fungi) environments correlate with AM fungal root colonization, diversity indices and community composition, and how does this variation interact with spatial structure?
Do root and soil communities differ?
Materials and Methods
Study system and hierarchical sampling design
To investigate how communities of AM fungi vary across spatial scales, we focused on the perennial herb Plantago lanceolata L. in the Åland Islands, southwestern Finland (Fig. 1). This monoecious, rosette‐forming perennial herb has a cosmopolitan distribution and reproduces by outcrossing (Ross, 1973) and frequent clonal propagation through side rosettes (Mook et al., 1992). The plant has limited gene flow between populations (Bos et al., 1986), and seeds generally disperse within a metre from the mother plant (Bos, 1992). In the Åland Islands, P. lanceolata is typically found in small dry meadows (most of them < 1 ha) located within an agricultural landscape (Ojanen et al., 2013).
Figure 1.
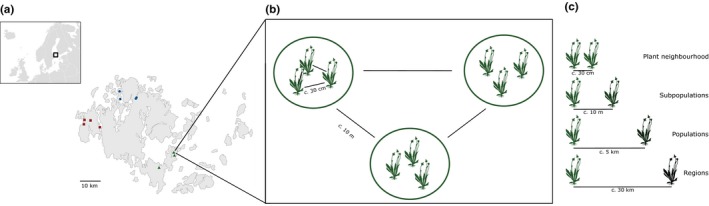
Location of sampled Plantago lanceolata individuals in the Åland Islands in the Baltic Sea, southwestern Finland. The top‐left inset shows the location of the Åland Islands within northern Europe. (a) Within the Åland Islands, three to four plant populations were sampled in three regions (with colours and symbols representing plant populations in different regions). (b) The inset illustrates that, within each plant population, we sampled three nearby plant individuals (separated by c. 30 cm) in each of three subpopulations within the plant population. (c) Schematic representation illustrating that individual plants at the neighbourhood scale were separated by c. 30 cm, plant subpopulations were separated by c. 10 m, plant populations within a region were separated by c. 5 km, and regions were separated by c. 30 km.
Root and soil samples of individual plants were collected in a nested design at multiple spatial scales (Fig. 1): the regional scale (three regions, c. 30 km apart), the population scale (three to four populations within each region, c. 5 km apart), the subpopulation scale (distinct sections within plant populations, c. 10 m apart), and the neighbourhood scale (individual plants within each subpopulation, c. 30 cm apart). In each subpopulation, three P. lanceolata plants and the surrounding soil (c. 200–500 ml; 25 mm radius around the plant; c. 100–150 mm deep) were collected in order to determine root colonization and community composition of root‐associated fungi within both roots and soil. Samples were collected from 7 to 10 July 2015. In total, we sampled 108 plants from 36 subpopulations, 11 populations, and three regions.
Roots were carefully separated from the soil, washed, cut into 2 cm pieces, and mixed thoroughly. Half of the roots from each sample were frozen at −20°C and subsequently freeze‐dried for later sequencing of DNA markers targeting both AM fungi and nonAM root‐associated fungi. The other half of the roots were stored in ethanol for later staining to determine colonization percentage. Roots were stained using trypan blue (Phillips & Hayman, 1970; Koske & Gemma, 1989) and scored using the gridline intersect method (McGonigle et al., 1990). The soil was thoroughly mixed and a representative subsample was frozen at −20°C and later freeze‐dried before determination of AM fungal community composition. To determine the community composition of root‐associated fungi in roots and the surrounding soil, DNA was extracted from 10–25 mg freeze‐dried root material (in a few samples below 10 mg) or 250–280 mg freeze‐dried soil using NucleoSpin Plant and Soil kits (Macherey‐Nagel, Düren, Germany).
Environmental drivers of AM fungal communities
In order to determine the effect of the environment on AM fungi, several abiotic and biotic measurements were conducted. A soil sample (300 ml) from each subpopulation was assessed for pH, plant available P, NH4, NO3, and total N (by Eurofins, Sweden). To determine the impact of climate, data loggers were placed in a majority of the subpopulations, both above ground (at a height of c. 5 cm) and below ground (c. 2 cm below the litter layer). Temperature and relative humidity were recorded above ground using Lascar loggers (EL‐USB‐2; Lascar Electronics, Wiltshire, UK), while below‐ground temperature was recorded using iButtons (DS1922L; Maxim Integrated, San Jose, CA, USA). The data loggers were set to record every 2 h for 1 yr, starting in June 2014. Mean below‐ground and above‐ground temperatures and relative humidities were calculated separately for the growing season (April–October) and nongrowing season (November–March). Owing to problems with recovering loggers, we were only able to retrieve data from half of the above‐ground loggers and three‐quarters of the below‐ground loggers. Soil moisture was measured (HH2, SM300; Delta‐T, Cambridge, UK) at three locations around each individual plant in June 2015 and the average value for each plant was calculated.
To investigate the effect of the surrounding vegetation on the community composition of AM fungi, we recorded plant species and their abundance in a 35 × 35 cm square around each sampled plant, and calculated plant species richness, Shannon's diversity index (Shannon, 1948) and Pielou's evenness index (Pielou, 1966). Plant species richness at each sample location was 5.3 ± 2.1 (mean ± SD), with the most common neighbouring plant species being Achillea millefolium, Agrostis capillaris and Galium verum. To investigate the link between AM and nonAM root‐associated fungi, a subset of root samples (n = 82) from eight different populations (Supporting Information Fig. S1) were selected.
Molecular methods
To assess the AM fungal community composition within roots and root‐associated soil, we used the primers NS31 and AML2 targeting a c. 560 bp central fragment of the small subunit rRNA gene in the Glomeromycota (Simon et al., 1992; Lee et al., 2008). These primers have often been used to assess the community composition of AM fungi (Öpik et al., 2010; Davison et al., 2015). Samples were sequenced at SciLifeLab/NGI (Solna, Sweden) on the MiSeq platform (Illumina Inc., San Diego, CA, USA), and sequences were clustered based on 99% similarity, a cutoff previously used in studies of AM fungal biogeographical patterns (Kivlin et al., 2011; but see Bruns & Taylor, 2016; and Bruns et al., 2018).To assess nonAM root‐associated fungi we used primers targeting the internal transcribed spacer (ITS) region using the primers fITS7 (Ihrmark et al., 2012) and ITS4 (White et al., 1990), which target a 250–450 bp fragment encompassing the entire ITS2 with flanking sequences in the 5.8 and large subunit genes. We followed the protocol of Clemmensen et al. (2016) and samples were sequenced at SciLifeLab/NGI (Uppsala, Sweden) on a PacBio RS II system (Pacific Biosciences, Menlo Park, CA, USA). Obtained sequences were analysed in the bioinformatics pipeline Scata (http://scata.mykopat.slu.se; Ihrmark et al., 2012), whereby they were clustered into operational taxonomic units (OTUs) using single linkage clustering with 98.5% sequence similarity. This resulted in a total of 104 149 sequences, clustering into 1247 OTUs. Sequences were obtained from 77 samples with an average of 1370 reads per sample. For further analyses we used a set of the 179 most common OTUs, making up 90% of the total number of reads (after removing plant and AM fungal OTUs). These OTUs were tentatively identified to the species level by comparing representative sequences with species hypotheses in the UNITE database (Kõljalg et al., 2005; Abarenkov et al., 2010).
For full details on the molecular methods and bioinformatics, see Methods S1. Sequencing data for each sample in this study have been deposited at NCBI under accession numbers SRP132598 and SRP132591 for root and soil samples, respectively.
Statistical methods
All analyses were conducted in R v.3.4.2 (R Core Team, 2017). In all multivariate analyses, the community data were Hellinger pretransformed. This transformation allowed us to compare communities using response data in the same format, which facilitates the analyses and homogenizes the interpretation of the results (Legendre & Gallagher, 2001).
Spatial structure of AM fungal communities
To investigate how AM fungal root colonization, richness, diversity and evenness varied across multiple spatial scales for both root and soil samples, we modelled each response variable as a function of the fixed effects ‘region’, ‘population’ (nested within ‘region’), and ‘subpopulation’ (nested within ‘population’). To compare OTU richness (observed OTU count), Shannon's diversity, and Pielou's evenness for root and soil, we accounted for differences in sequencing depth by using the residual values of linear models, in which the response variables were expressed as a function of the square root of the total sequence number per sample (Tedersoo et al., 2014; Bálint et al., 2015). Sampling efficacy was further assessed using the function rarecurve in the R‐package vegan (Oksanen et al., 2015; Fig. S2).
To test how AM fungal communities varied across multiple spatial scales we used both presence‐absence and absolute count data. Communities were analysed by permutation‐based ANOVA using the function adonis in the R‐package vegan with the Euclidian dissimilarity index (Oksanen et al., 2015). To investigate if differences in sequencing depth influenced the results we added the square root of total sequences per sample to the models. Spatial variation of the AM fungal response variables (root colonization, species richness, diversity, evenness and community composition) was partitioned between the four hierarchical scales (neighbourhood, subpopulation, population and region) by dividing the sums of squares of each factor individually by the total sums of squares (Quinn & Keough, 2002). See Tables S1 and S2 for full ANOVA and permutation‐based ANOVA tables.
Environmental drivers of AM fungal communities
We started with assessing – akin to the analyses for the AM fungal community – the variation in the abiotic and biotic factors ascribed to spatial structure at each hierarchical scale by modelling each response variable as a function of the fixed effects ‘region’, ‘population’ (nested within ‘region’) and ‘subpopulation’ (nested within ‘population’). As soil nutrients and climate were assessed at the subpopulation scale, we did not include the factor ‘subpopulation’ in these models, and for those response variables, the residuals thus represent the pooled within‐population variation. For the nonAM fungal root‐associated diversity indices, we accounted for differences in sequencing depth by using the residuals of linear models with the square root of the total sequence number per sample as the explaining variable (Tedersoo et al., 2014).
To test how individual abiotic and biotic environmental factors influenced AM fungal richness, diversity, and evenness, we used multiple regression models. In order to avoid collinearity, we excluded a few of the predictor variables, and thereby modelled each of the response variables as a function of soil chemical characteristics (pH, P, NH4 and NO3), bioclimatic variables (mean below‐ and above‐ground temperatures in the growing and nongrowing seasons, mean annual relative humidity, and soil moisture), neighbouring plant richness, and nonAM fungal root‐associated richness. To determine how the environment influenced the composition of soil and root‐associated AM fungal communities, we used partial canonical redundancy analysis (partial RDA; Legendre & Legendre, 2012, section 11.1) using all environmental variables measured. We controlled for spatial structure using Moran's eigenvector maps (MEMs; see next section). The significance of each RDA axis was tested using the function anova.cca in the R‐package vegan.
In order to test if AM fungal community composition was related to the composition of the neighbouring plant community and to the composition of the nonAM root‐associated fungi we used Procrustes analysis and its associated test (Jackson, 1995) available in the R‐package vegan through the protest function.
Relative importance of spatial structure and environment in shaping AM fungal communities
We determined the importance of spatial structure and environmental factors, as well as their overlapping effect, in explaining variation in AM fungal colonization, richness, diversity, evenness, and community composition. To assess the spatial structure of the AM fungal community, we used MEMs, which were constructed through the diagonalization of a connection matrix weighted by the inverse of the Euclidean distance among samples (Fig. S3; Dray et al., 2006), resulting in both positive and negative MEMs (Borcard & Legendre, 2002; Dray et al., 2006). Following this, we constructed a separate set of MEMs for the root (n = 104) and soil (n = 96) AM fungal communities. The significance of the autocorrelation measured by each MEM was tested using the function moran.randtest in the R‐package adespatial (Dray et al., 2017), allowing identification of the MEMs that described the spatial pattern significantly (P < 0.05) better than randomly expected.
To choose the MEMs that were important in structuring the fungal community, we performed forward selection independently on the root and soil data, using the approach proposed by Blanchet et al. (2008). Only positively autocorrelated MEMs were selected and used for all other analyses.
For certain samples, missing values for specific environmental variables were interpolated by multiple regression with the other environmental variables as predictors. All interpolated values were well within the range of measured values.
Variation partitioning was carried out using the varpart function in the R‐package vegan, using the selected MEMs and the environmental variables. Through partial RDA we used isolated independent fractions of the variation partitioning analysis and assessed their significance with permutation‐based ANOVA, using the anova.cca function in the R‐package vegan. We used adjusted R 2 to evaluate the contribution of each fraction, as the adjusted R 2 has been recommended by Peres‐Neto et al. (2006) in variation partitioning using RDA models with Hellinger‐transformed data, and as it allowed for a more direct comparison between root and soil AM fungal communities, for which models were built on different numbers of explanatory variables. We note that a negative adjusted R 2 can be interpreted as an adjusted R 2 of 0 (Peres‐Neto et al., 2006).
Variation between AM fungal communities in roots and soil
To investigate how AM fungal richness, diversity, and evenness differed between root and soil communities, each response variable was modelled as a function of ‘sample type’ as a fixed effect and ‘plant individual’ as a random effect. In order to validate that differences in OTU richness between the roots and root‐associated soil were not an artefact of the differences in sequence number between root and root‐associated soil samples, rarefied OTU richness, based on resampling to the mean number of soil reads per sample (n = 4305; Fig. S2), was calculated using the function rarefy in the package vegan (Oksanen et al., 2015), which is based on Hurlbert (1971) and Heck et al. (1975). To test for differences in community composition between root and root‐associated soil we used linear discriminant analysis with the function lda in the R‐package mass (Venables & Ripley, 2002). We tested for significant differences between the two fungal communities using a chi‐squared test.
Results
We identified 1077 AM fungal OTUs from a total of 2 727 662 sequences. From these, 2 314 361 sequences were assigned to 1048 OTUs in the roots of P. lanceolata (n = 104 root samples) and 413 301 sequences were assigned to 812 OTUs in the surrounding soil (n = 96 soil samples). In total, 98.5% of all AM fungal OTUs were identified to genus level (Fig. 2). A majority (90%) of the OTUs were found in < 25% of all samples, while six OTUs were found in > 75% of all samples. These six very common OTUs all belonged to the genus Glomus.
Figure 2.
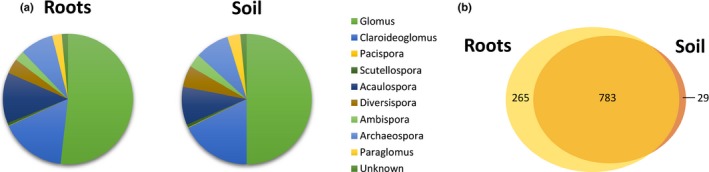
An overview of the fungal community in the roots and root‐associated soil of Plantago lanceolata. (a) The proportion of arbuscular mycorrhizal (AM) fungal operational taxonomic units (OTUs) within each genus in the roots and the root‐associated soil. (b) Venn diagram illustrating the number of OTUs found exclusively in roots, found in both roots and root‐associated soil, and found exclusively in the root‐associated soil.
Spatial structure of AM fungal communities
Arbuscular mycorrhizal fungal communities differed significantly between subpopulations and populations for most community descriptors (Table 2). Community composition in both roots and soil was significantly structured at all spatial scales, with small but significant variation also at the regional level (Table 2; Fig. 3). For all community descriptors, the majority of variation was found at the smallest spatial scale (i.e. between neighbouring plants), with variation decreasing at increasing spatial scales, a pattern consistent for both root and soil samples (Table 2). Differences in sequencing depth had a significant but small effect, explaining 1–2% of the variation in community composition (Table S2).
Table 2.
Percentage of variation explained at each spatial scale for root colonization and community descriptors in both root and root‐associated soil of Plantago lanceolata using either ANOVA (root colonization, richness, diversity and evenness) or PERMANOVA (community composition for both the presence‐absence and abundance of operational taxonomic units (OTUs))
| Root colonization | Richness | Diversity | Evenness | Community composition presence‐absence | Community composition abundance | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Root | Soil | Root | Soil | Root | Soil | Root | Soil | Root | Soil | ||
| Region | 1.0 | 3.9 | 0.1 | 1.2 | 0.5 | 1.5 | 0.1 | 4.0 | 3.5 | 4.4 | 4.4 |
| Population | 14.7 | 11.2 | 28.8 | 22.7 | 29.4 | 18.6 | 29.8 | 14.6 | 14.1 | 20.7 | 18.5 |
| Subpopulation | 37.9 | 22.7 | 16.2 | 27.7 | 27.5 | 30.5 | 23.1 | 29.0 | 28.3 | 35.1 | 31.2 |
| Plant neighbourhooda | 46.4 | 62.1 | 54.8 | 48.4 | 42.5 | 49.4 | 47.1 | 52.5 | 54.1 | 39.9 | 46.0 |
Significant estimates (P < 0.05) are shown in bold.
No significance levels were calculated for plant neighbourhood as these estimates are based on the residual variation.
Figure 3.
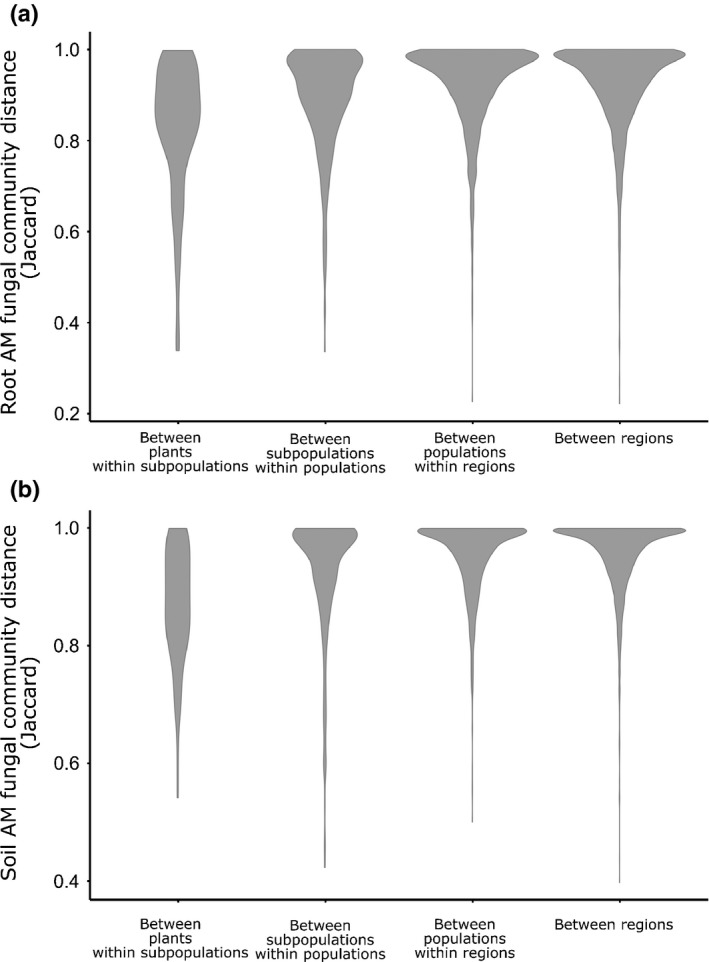
Violin plots showing the pairwise Jaccard distance between arbuscular mycorrhizal (AM) fungal communities in root (a) and soil (b) samples of Plantago lanceolata.
Environmental drivers of AM fungal communities
The soil chemical characteristics and climatic variables differed in their variation across the three measured spatial scales (subpopulation, population and region): NO3 and total N varied mostly among regions; NH4, below‐ground and above‐ground temperatures and relative humidities in the growing season varied mostly among populations; and pH and above‐ground temperatures in the nongrowing season varied mostly within populations (Table S3; Fig. S4). Most variation in AM fungal community descriptors was found at the smallest spatial scale measured (among neighbouring plants). Similar to this, vegetation and nonAM root‐associated fungi varied mostly at the smaller spatial scales measured (at the plant neighbourhood and subpopulation scales) with decreasing variation at the population and regional scales (Table S3).
Descriptors of the AM fungal community were differently related to the abiotic and biotic variables (Table S4). Colonization of roots by AM fungi increased whereas AM fungal evenness decreased with increasing soil moisture. Strikingly, AM fungal richness was not affected by any of the environmental variables. AM fungal diversity and evenness in the roots were negatively affected by increasing temperatures below ground in the nongrowing season and above ground in the growing season. AM fungal diversity in the root‐associated soil and AM fungal evenness in both root and root‐associated soil increased under less acidic conditions. Accordingly, AM fungal community composition in both roots and soil appeared to be strongly influenced by pH and NH4, with opposing effects (Fig. 4). Furthermore, soil moisture, temperature, and plant and nonAM fungal diversity indices were related to the composition of AM fungal communities in roots (Fig. 4a), whereas humidity, temperature and plant diversity indices correlated with AM fungal community composition in the root‐associated soil (Fig. 4b).
Figure 4.
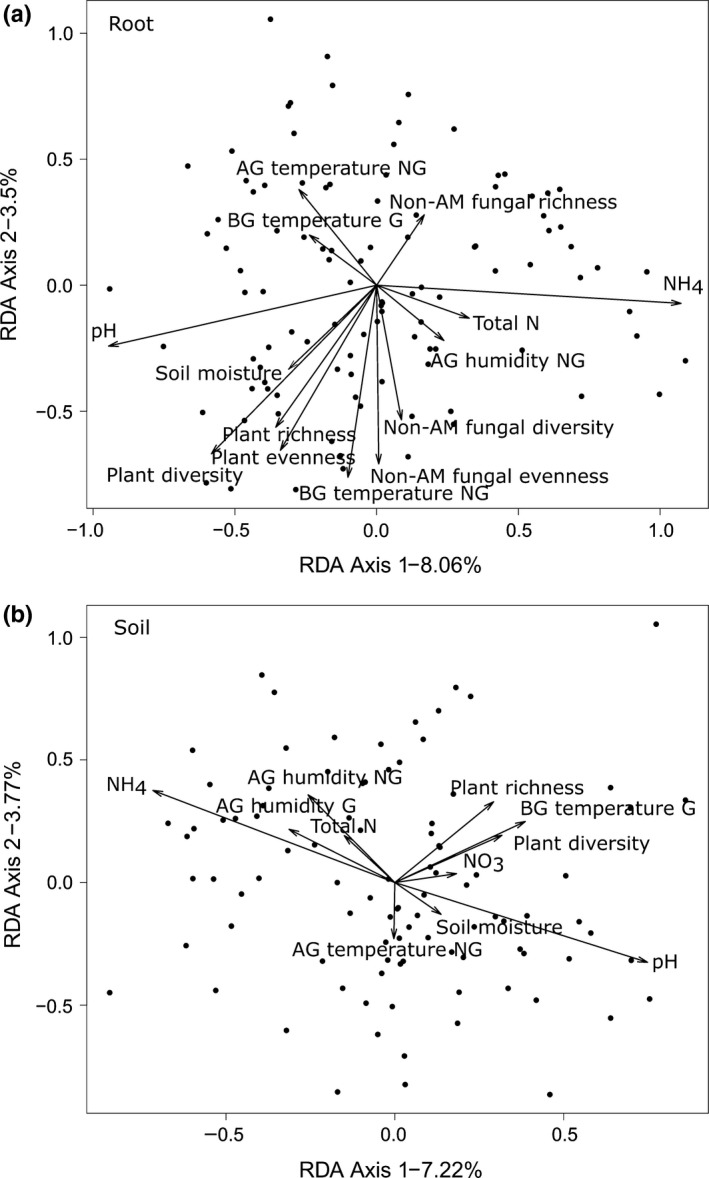
Partial canonical redundancy analysis (partial RDA) ordination plots for arbuscular mycorrhizal (AM) fungal communities in root (a) and root‐associated soil (b) of Plantago lanceolata where the main environmental variables are shown. All environmental variables with arrows very close to the centre were removed for visual clarity and because no interpretation could be gained from them. For each analysis, the Moran's eigenvector maps were used to control for spatial structure. Each point represents an AM fungal community found within a single sample, while vectors show the main environmental drivers. A correlation scaling was used to draw each ordination plot so that angles between variables could be interpreted directly. AG, above ground; BG, below ground; G, growing season; NG, nongrowing season.
Arbuscular mycorrhizal fungal communities in both roots and root‐associated soil were correlated to the neighbouring plant communities (r = 0.53, n = 104, P = 0.001; and r = 0.60, n = 96, P = 0.001, respectively) and to community composition of the nonAM root‐associated fungi (r = 0.73, n = 104, P = 0.001; r = 0.80, n = 96, P = 0.001, respectively).
Relative importance of spatial structure and environment in shaping AM fungal communities
Spatial structure and environmental variables explained a similar amount of variation in the majority of community descriptors (7.0–22.5% and 7.2–25.1%, respectively; Figs 4, S5). However, spatial structure had a significant influence on most aspects of the AM fungal communities, whereas environmental variables were only significant in relation to community composition, colonization and diversity in soil (Table S4). Combined spatial and environmental variation explained 0–13.4% of the variation in AM fungal community variables. Despite the use of a large set of spatial and environmental variables in the analyses, a major fraction of the variation in the AM fungal community remained unexplained (Figs 5, S5).
Figure 5.
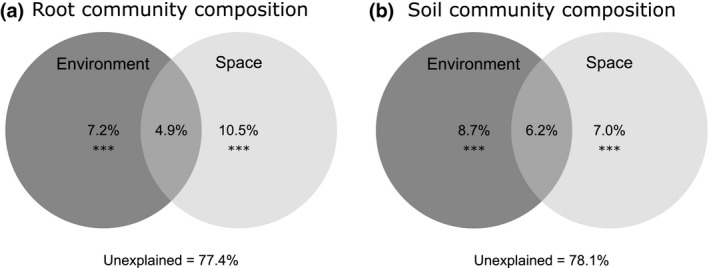
The relative importance of space and the environment in structuring the arbuscular mycorrhizal (AM) fungal community composition in the roots (a) and the root‐associated soil (b) of Plantago lanceolata. All values are presented using the adjusted coefficient of determination (R 2 adjusted). The variation is partitioned into four fractions: purely environmental variation, purely spatial variation, both environmental and spatial variation, and unexplained variation (residuals). Asterisks indicate that a significant amount of variation is explained by the given fraction (***, P < 0.001).
Variation between AM fungal communities in roots and soil
Roots hosted higher overall richness of AM fungal OTUs than the root‐associated soil (mean ± SD per sample: 120 ± 70 and 65 ± 45, respectively), as confirmed by a significantly higher richness in roots when using both residual and rarefied richness (χ 2 = 12.46, d.f. = 1, P < 0.001 and χ 2 = 34.45, d.f. = 1, P < 0.001, respectively). Evenness was lower in roots than in root‐associated soil (χ 2 = 27.56, d.f. = 1, P < 0.001). Most OTUs were found in both roots and the root‐associated soil, although some OTUs were found exclusively in either roots (25%) or root‐associated soil (3%; Fig. 2b). While there was a large overlap in OTUs and a fairly equal representation of species within genera (Fig. 2), AM fungal communities differed significantly between roots and root‐associated soil (χ 2 = 153.41, d.f. = 1, P < 0.001). AM fungal communities in the roots and root‐associated soil were more similar but more variable at small spatial scales, whereas communities were increasingly dissimilar at larger spatial scales (Fig. S6).
Discussion
In this study we investigated the spatial variation and drivers of AM fungal communities associated with a wild plant species in a fragmented landscape. We found that AM fungal communities were already highly dissimilar among neighbouring plants, even though there was considerable variation in similarity at this small spatial scale. AM fungal communities were increasingly, and more consistently, dissimilar at larger spatial scales. This pattern matched the spatial variation in some of the key abiotic (pH) and biotic (plant neighbourhood and nonAM fungal community) predictors of AM fungal community structure, which also varied mostly within populations. Spatial structure and environmental parameters were equally important predictors of AM fungal communities, and largely explained different parts of the variation. Finally, even though there was a large overlap in AM fungal OTUs between root and root‐associated soil communities, community composition in root and soil differed, and we found a higher species richness in the roots.
Spatial structure of AM fungal communities
Communities of AM fungi in both the roots and root‐associated soil were already found to vary strongly among neighbouring P. lanceolata plants, with substantial additional variation among plant subpopulations. There was less, but still significant, spatial structuring in the AM fungal communities associated with the focal plant species at scales ranging from a few to tens of kilometres (i.e. among populations or regions). Chaudhary et al. (2014) also observed spatial structure in AM fungal abundance and diversity at different scales in a semiarid region of southern Utah (Table 1). Similarly, we found that AM fungal communities furthest apart were consistently highly dissimilar, while similarity between closely located samples was higher and more variable. However, in contrast to our findings, Chaudhary et al. (2014) found most variation at the larger spatial scales (between regions c. 50–100 km apart). This discrepancy may be a result of differences in methodology between the two studies, such as sampling of different habitats (small dry meadows in an agricultural landscape vs a semiarid shrub land) or the investigation of the community in roots and root‐associated soil from single plants in the present study, as compared with abundance and diversity measures per microsite (1 m2) in Chaudhary et al. (2014).
Most other studies on the spatial distribution of AM fungi have focused on either very small or very large scales. In agreement with our findings of large variation in the AM fungal community at small spatial scales, several studies have found large variation at the submetre and metre scales (Whitcomb & Stutz, 2007; Mummey & Rillig, 2008). However, it is important to note that these studies sampled AM fungal communities associated with mixed vegetation. Hence, the small‐scale spatial variation in the AM fungal community structure may have been a result of specificity in plant–fungal associations rather than spatial variation of the AM fungal community associated with a single plant species. At the larger regional scale (25 km), communities of AM fungi in cultivated maize fields were found to become increasingly dissimilar with increasing distance (Lekberg et al., 2007). Studies at the global scale have similarly found that AM fungal communities become more dissimilar with increasing geographic distance (Kivlin et al., 2011; Davison et al., 2015), which is in general agreement with our results.
Drivers of AM fungal communities
The abiotic and biotic variables pH, neighbouring vegetation and nonAM root‐associated fungi were spatially structured in a similar way as the AM fungi, being highly variable at the smallest spatial scales at which they were measured (within populations), and much less variable across larger spatial scales (among populations and among regions). In comparison, other measured abiotic factors (such as NO3, total N, temperature, and relative humidity) varied primarily among populations and regions. Overall, spatial structure and environmental factors explained c. 9% and 11%, respectively, of the variation in AM fungal community structure.
Our exploratory analysis of the environmental drivers of AM fungal communities pinpoints several important abiotic and biotic factors. Like several other studies, we found that pH and nitrogen availability strongly influenced AM fungal community composition (Jumpponen et al., 2005; Lekberg et al., 2007, 2011; Dumbrell et al., 2010; van Diepen et al., 2011). We also found that higher temperatures led to a decrease in diversity and evenness of AM fungi and influenced the AM fungal community composition. Given projected increases in temperatures (European Environment Agency, 2017) and the observation that warmer temperatures seem to favour potential dominants in the AM fungal community, these findings suggest that we may expect less diverse AM fungal communities in the future.
Our findings show a strong relationship between AM fungal communities and plant and nonAM fungal communities. Evidence for a connection between communities of plant and AM fungi have been found both in experimental studies (Stampe & Daehler, 2003; Johnson et al., 2004) and in natural systems (Öpik et al., 2006; Kivlin et al., 2011). Such a link may indicate both specificity between AM fungi and plants (Kivlin et al., 2011; Velázquez et al., 2013; Rodríguez‐Echeverría et al., 2017) and a similar response of plants and AM fungi to the abiotic environment. While the relationship between AM fungal and plant communities has been studied frequently, the relationship between the AM and nonAM fungal communities has been studied to a much lesser extent. In this study, we found a strong relationship between AM and nonAM fungal root‐associated communities. The relationship between these distinct fungal groups may have two nonmutually exclusive explanations: AM and nonAM root‐associated fungi respond to the same abiotic drivers; or positive and negative interactions between AM and nonAM fungi result in predictable co‐occurrence patterns (Larimer et al., 2012).
The high variation at the small spatial scale reported here may be a result of both the expected low dispersal abilities of AM fungi over short distances, in addition to competitive interactions between AM fungal species and priority effects (Fukami et al., 2010). Lekberg et al. (2012) found that mainly stochastic processes were shaping AM fungal communities at the local scale. Similarly, reanalysing the data of Davison et al. (2015), Powell & Bennett (2016) found that AM fungal community composition was highly unpredictable within similar environments. In the current study we found that c. 80% of the variation in AM fungal community composition could not be explained by spatial structure or the environment. This seems to match other studies on plant‐associated fungal communities, which generally report a relatively large fraction (c. 35–80%) of unexplained variation (e.g. Lekberg et al., 2007; Sterkenburg et al., 2015; Horn et al., 2017). Such unexplained variation is probably a result of unmeasured environmental variables, species interactions and stochastic processes.
Variation between AM fungal communities in roots and soil
We found that the composition of AM fungal communities was largely similar between roots and the root‐associated soil, with roots and soil hosting similar proportions of AM fungal genera and sharing most of the OTUs. However, roots hosted an overall higher richness than the root‐associated soil. This is in contrast to some studies which have suggested that the soil contains a species pool of AM fungi from which the plants can be colonized (Davison et al., 2011). While many studies have found higher AM fungal richness in soil than in roots (Bainard et al., 2011; Martínez‐García et al., 2011; Varela‐Cervero et al., 2015), others have found equal or lower richness in the soil (Verbruggen et al., 2012; Saks et al., 2014). While these findings may reflect real ecological patterns, the patterns could also be ascribed to methodological differences. Methodological differences in sequencing depth between root and soil samples may be a result of biological characteristics of AM fungi: AM fungi generally contain lower densities of biomass in soil than in roots (Olsson et al., 2010), and the distribution of nuclei in the extraradical mycelium can be highly variable (Gamper et al., 2008). Moreover, divergent composition of AM fungal communities in roots and root‐associated soil may be caused by differences among taxa in their relative abundance of intra‐ or extraradical structures (Hart et al., 2001) and seasonality of spore production (Bever et al., 2001; Jansa et al., 2002).
Conclusion
Arbuscular mycorrhizal fungal communities differed strongly among neighbouring plants and among plant subpopulations, but there was also strong variation in dissimilarity at these small spatial scales. At the larger spatial scales, AM fungal communities were more consistently dissimilar. Spatial structure and environmental predictors explained equal parts of variation in AM fungal communities. For example, pH, vegetation and nonAM root‐associated fungi, which were just like the AM fungal community most variable at small spatial scales, were correlated with the structure of the AM fungal community. While pH may affect the AM fungal community composition directly, the question of ‘who drives who’ is more difficult to answer for the biotic factors: while we focused on predictors of the AM fungal community, it may well be that the AM fungi themselves contribute to shaping the nonAM fungal and plant communities. A promising avenue for future research would be to conduct experimental studies to disentangle the causal drivers and reciprocal interactions among AM fungi, nonAM fungi, and the neighbouring plant community. Given the small scale at which the AM fungal communities vary, a promising avenue for future research may be to measure the abiotic and biotic environments at a finer scale than in the current study, and link this fine‐scale variation in the environment to the AM fungal community structure. Such studies may also target within‐plant variation in the AM fungal community. Overall, our findings contribute fundamental knowledge by highlighting the large variation in AM fungal community structure at small spatial scales, which may in turn add to the local diversity of both plants and plant‐associated organisms. From an applied perspective, understanding the distribution and drivers of the AM fungi may be important for conservation and agroecological management.
Author contributions
P.U.R., A.J.M.T., A.F.A. and L.W.H. conceived and designed the experiment. P.U.R. conducted the empirical work. P.U.R. and L.W.H. conducted the molecular work, and P.U.R., L.W.H. and B.D.L. performed the bioinformatic analyses. P.U.R., A.J.M.T. and F.G.B. analysed the data. P.U.R. wrote the first draft, and all authors contributed to the final manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 The proportion of nonAM root‐associated fungal classes within the roots of Plantago lanceolata.
Fig. S2 Rarefaction curves for AM fungi in roots and root‐associated soil.
Fig. S3 Connection diagram of sampled Plantago lanceolata individuals in the Åland Islands in the Baltic Sea, southwestern Finland.
Fig. S4 Variation of abiotic and biotic factors for each population.
Fig. S5 Variation partitioning explaining AM fungal community descriptors.
Fig. S6 Violin plot showing the distance between AM fungal communities in root and root‐associated soil for each hierarchical spatial scale.
Table S1 ANOVA table for partitioning of variance for each hierarchical spatial scale
Table S2 PERMANOVA table for partitioning of variance for each hierarchical spatial scale
Table S3 Percentage of variation explained at each spatial scale for soil nutrients, bioclimatic variables, vegetation and nonAM root‐associated fungi at the regional, population, and subpopulation levels.
Table S4 P‐values for the impact of environmental variables on the arbuscular mycorrhizal fungal root colonization and diversity indices in both root and root‐associated soil
Methods S1 Full description of molecular methods and bioinformatics.
Acknowledgements
The authors acknowledge Science for Life Laboratory, the National Genomics Infrastructure, the Swedish National Infrastructure for Computing (SNIC 2017/7‐300), and Uppmax (b2016102) for assistance in sequencing and computational infrastructure; Giulia Zacchello for assistance in the laboratory; and the Maj and Tor Nessling foundation (2014211 to A.J.M.T.), the Academy of Finland (265761 to A.J.M.T.) and the Swedish Research Council Vetenskapsrådet (2015‐03993 to A.J.M.T.) for funding.
See also the Commentary on this article by https://doi.org/10.1111/nph.15212.
References
- Abarenkov K, Henrik Nilsson R, Larsson K‐H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T et al 2010. The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytologist 186: 281–285. [DOI] [PubMed] [Google Scholar]
- Allen EB, Allen MF, Helm DJ, Trappe JM, Molina R, Rincon E. 1995. Patterns and regulation of mycorrhizal plant and fungal diversity. Plant and Soil 170: 47–62. [Google Scholar]
- Anderson RC, Liberta AE, Dickman LA, Katz AJ. 1983. Spatial variation in vesicular‐arbuscular mycorrhiza spore density. Bulletin of the Torrey Botanical Club 110: 519–525. [Google Scholar]
- Antoninka A, Reich PB, Johnson NC. 2011. Seven years of carbon dioxide enrichment, nitrogen fertilization and plant diversity influence arbuscular mycorrhizal fungi in a grassland ecosystem. New Phytologist 192: 200–214. [DOI] [PubMed] [Google Scholar]
- Augé RM. 2001. Water relations, drought and vesicular‐arbuscular mycorrhizal symbiosis. Mycorrhiza 11: 3–42. [Google Scholar]
- Bainard LD, Koch AM, Gordon AM, Newmaster SG, Thevathasan NV, Klironomos JN. 2011. Influence of trees on the spatial structure of arbuscular mycorrhizal communities in a temperate tree‐based intercropping system. Agriculture, Ecosystems & Environment 144: 13–20. [Google Scholar]
- Bálint M, Bartha L, O'Hara RB, Olson MS, Otte J, Pfenninger M, Robertson AL, Tiffin P, Schmitt I. 2015. Relocation, high‐latitude warming and host genetic identity shape the foliar fungal microbiome of poplars. Molecular Ecology 24: 235–248. [DOI] [PubMed] [Google Scholar]
- Bever JD. 2002. Host‐specificity of AM fungal population growth rates can generate feedback on plant growth. Plant and Soil 244: 281–290. [Google Scholar]
- Bever JD, Schultz PA, Pringle A, Morton JB. 2001. Arbuscular mycorrhizal fungi: more diverse than meets the eye, and the ecological tale of why. BioScience 51: 923–932. [Google Scholar]
- Blanchet FG, Legendre P, Borcard D. 2008. Forward selection of explanatory variables. Ecology 89: 2623–2632. [DOI] [PubMed] [Google Scholar]
- Borcard D, Legendre P. 2002. All‐scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling 153: 51–68. [Google Scholar]
- Borowicz VA. 2001. Do arbuscular mycorrhizal fungi alter plant–pathogen relations? Ecology 82: 3057–3068. [Google Scholar]
- Bos M. 1992. Gene flow characters and population structure in Plantago lanceolata In: Kuiper PJC, Bos M, eds. Plantago: a multidisciplinary study. Berlin, Heidelberg, Germany: Springer‐Verlag, 222–231. [Google Scholar]
- Bos M, Harmens H, Vrieling K. 1986. Gene flow in Plantago I. Gene flow and neighbourhood size in P. lanceolata . Heredity 56: 43–54. [Google Scholar]
- Bruns TD, Corradi N, Redecker D, Taylor JW, Öpik M. 2018. Glomeromycotina: what is a species and why should we care? New Phytologist 220: 963–967. [DOI] [PubMed] [Google Scholar]
- Bruns TD, Taylor JW. 2016. Comment on ‘Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism’. Science 351: 826. [DOI] [PubMed] [Google Scholar]
- Busby RR, Stromberger ME, Rodriguez G, Gebhart DL, Paschke MW. 2013. Arbuscular mycorrhizal fungal community differs between a coexisting native shrub and introduced annual grass. Mycorrhiza 23: 129–141. [DOI] [PubMed] [Google Scholar]
- Chaudhary VB, Lau MK, Johnson NC. 2008. Macroecology of microbes – biogeography of the Glomeromycota In: Varma A, ed. Mycorrhiza. Berlin, Heidelberg, Germany: Springer‐Verlag, 529–563. [Google Scholar]
- Chaudhary VB, O'Dell TE, Rillig MC, Johnson NC. 2014. Multiscale patterns of arbuscular mycorrhizal fungal abundance and diversity in semiarid shrublands. Fungal Ecology 12: 32–43. [Google Scholar]
- Clemmensen KE, Ihrmark K, Durling MB, Lindahl BD. 2016. Sample preparation for fungal community analysis by high‐throughput sequencing of barcode amplicons In: Martin F, Uroz S, eds. Methods in molecular biology, vol 1399. Microbial Environmental Genomics (MEG). New York, NY, USA: Humana Press, 61–88. [DOI] [PubMed] [Google Scholar]
- Davison J, Moora M, Öpik M, Adholeya A, Ainsaar L, Bâ A, Burla S, Diedhiou AG, Hiiesalu I, Jairus T et al 2015. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349: 970–973. [DOI] [PubMed] [Google Scholar]
- Davison J, Öpik M, Daniell TJ, Moora M, Zobel M. 2011. Arbuscular mycorrhizal fungal communities in plant roots are not random assemblages: selectivity in AMF‐plant associations. FEMS Microbiology Ecology 78: 103–115. [DOI] [PubMed] [Google Scholar]
- van Diepen LTA, Lilleskov EA, Pregitzer KS. 2011. Simulated nitrogen deposition affects community structure of arbuscular mycorrhizal fungi in northern hardwood forests. Molecular Ecology 20: 799–811. [DOI] [PubMed] [Google Scholar]
- Dray S, Blanchet G, Borcard D, Clappe S, Guenard G, Jombart T, Larocque G, Legendre P, Madi N, Wagner HW. 2017. adespatial: Multivariate multiscale spatial analysis. R package version 0.0‐9. [WWW document] URL https://CRAN.R-project.org/package=adespatial [accessed 27 November 2017].
- Dray S, Legendre P, Peres‐Neto PR. 2006. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecological Modelling 196: 483–493. [Google Scholar]
- Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. 2010. Relative roles of niche and neutral processes in structuring a soil microbial community. The ISME Journal 4: 337–345. [DOI] [PubMed] [Google Scholar]
- European Environment Agency . 2017. Climate change. Impacts and vulnerability in Europe 2016: an indicator‐based report. Copenhagen, Denmark: European Environment Agency. [Google Scholar]
- Friese CF, Koske RE. 1991. The spatial dispersion of spores of vesicular‐arbuscular mycorrhizal fungi in a sand dune: microscale patterns associated with the root architecture of American beachgrass. Mycological Research 95: 952–957. [Google Scholar]
- Fukami T, Dickie IA, Paula Wilkie J, Paulus BC, Park D, Roberts A, Buchanan PK, Allen RB. 2010. Assembly history dictates ecosystem functioning: evidence from wood decomposer communities: carbon dynamics and fungal community assembly. Ecology Letters 13: 675–684. [DOI] [PubMed] [Google Scholar]
- Gamper HA, Young JPW, Jones DL, Hodge A. 2008. Real‐time PCR and microscopy: are the two methods measuring the same unit of arbuscular mycorrhizal fungal abundance? Fungal Genetics and Biology 45: 581–596. [DOI] [PubMed] [Google Scholar]
- Hart MM, Reader RJ, Klironomos JN. 2001. Life‐history strategies of arbuscular mycorrhizal fungi in relation to their successional dynamics. Mycologia 93: 1186–1194. [Google Scholar]
- Hartnett DC, Wilson GWT. 1999. Mycorrhizae influence plant community structure and diversity in tallgrass prairie. Ecology 80: 1187–1195. [Google Scholar]
- Hawkes CV, Kivlin SN, Rocca JD, Huguet V, Thomsen MA, Suttle KB. 2011. Fungal community responses to precipitation. Global Change Biology 17: 1637–1645. [Google Scholar]
- Hazard C, Gosling P, Van Der Gast CJ, Mitchell DT, Doohan FM, Bending GD. 2013. The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. The ISME Journal 7: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck KL, van Belle G, Simberloff D. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56: 1459–1461. [Google Scholar]
- van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf‐Engel R, Boller T, Wiemken A, Sanders IR. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72. [Google Scholar]
- Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW. 1998. Ploughing up the wood‐wide web? Nature 394: 431. [DOI] [PubMed] [Google Scholar]
- Helgason T, Fitter AH. 2009. Natural selection and the evolutionary ecology of the arbuscular mycorrhizal fungi (Phylum Glomeromycota). Journal of Experimental Botany 60: 2465–2480. [DOI] [PubMed] [Google Scholar]
- Hempel S, Renker C, Buscot F. 2007. Differences in the species composition of arbuscular mycorrhizal fungi in spore, root and soil communities in a grassland ecosystem. Environmental Microbiology 9: 1930–1938. [DOI] [PubMed] [Google Scholar]
- Hiiesalu I, Öpik M, Metsis M, Lilje L, Davison J, Vasar M, Moora M, Zobel M, Wilson SD, Pärtel M. 2012. Plant species richness belowground: higher richness and new patterns revealed by next‐generation sequencing. Molecular Ecology 21: 2004–2016. [DOI] [PubMed] [Google Scholar]
- Horn S, Hempel S, Verbruggen E, Rillig MC, Caruso T. 2017. Linking the community structure of arbuscular mycorrhizal fungi and plants: a story of interdependence? The ISME Journal 11: 1400–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert SH. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52: 577–586. [DOI] [PubMed] [Google Scholar]
- Ihrmark K, Bödeker ITM, Cruz‐Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandström‐Durling M, Clemmensen KE et al 2012. New primers to amplify the fungal ITS2 region – evaluation by 454‐sequencing of artificial and natural communities. FEMS Microbiology Ecology 82: 666–677. [DOI] [PubMed] [Google Scholar]
- Jackson DA. 1995. PROTEST: a PROcrustean randomization TEST of community environment concordance. Écoscience 2: 297–303. [Google Scholar]
- Jakobsen I, Abbott LK, Robson AD. 1992. External hyphae of vesicular‐arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. New Phytologist 120: 371–380. [Google Scholar]
- Jansa J, Mozafar A, Anken T, Ruh R, Sanders I, Frossard E. 2002. Diversity and structure of AMF communities as affected by tillage in a temperate soil. Mycorrhiza 12: 225–234. [DOI] [PubMed] [Google Scholar]
- Johnson D, Vandenkoornhuyse PJ, Leake JR, Gilbert L, Booth RE, Grime JP, Young JPW, Read DJ. 2004. Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosms. New Phytologist 161: 503–515. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Rowland DL, Corkidi L, Egerton‐Warburton LM, Allen EB. 2003. Nitrogen enrichment alters mycorrhizal allocation at five mesic to semiarid grasslands. Ecology 84: 1895–1908. [Google Scholar]
- Jumpponen A, Trowbridge J, Mandyam K, Johnson L. 2005. Nitrogen enrichment causes minimal changes in arbuscular mycorrhizal colonization but shifts community composition ‐ evidence from rDNA data. Biology and Fertility of Soils 41: 217–224. [Google Scholar]
- Kivlin SN, Hawkes CV, Treseder KK. 2011. Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biology and Biochemistry 43: 2294–2303. [Google Scholar]
- Klironomos JN. 2000. Host‐specificity and functional diversity among arbuscular mycorrhizal fungi In: Bell CR, Brylinsky M, Johnson‐Green P, eds. Proceedings of the Eighth International Symposium on Microbial Ecology. Microbial Biosystems: New Frontiers. Halifax, Canada: Atlantic Canada Society for Microbial Ecology, 845–851. [Google Scholar]
- Klironomos JN, McCune J, Moutoglis P. 2004. Species of arbuscular mycorrhizal fungi affect mycorrhizal responses to simulated herbivory. Applied Soil Ecology 26: 133–141. [Google Scholar]
- Kõljalg U, Larsson K‐H, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E et al 2005. UNITE: a database providing web‐based methods for the molecular identification of ectomycorrhizal fungi. New Phytologist 166: 1063–1068. [DOI] [PubMed] [Google Scholar]
- Koske RE, Gemma JN. 1989. A modified procedure for staining roots to detect VA mycorrhizas. Mycological Research 92: 486–505. [Google Scholar]
- Larimer AL, Bever JD, Clay K. 2012. Consequences of simultaneous interactions of fungal endophytes and arbuscular mycorrhizal fungi with a shared host grass. Oikos 121: 2090–2096. [Google Scholar]
- Lee J, Lee S, Young JPW. 2008. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiology Ecology 65: 339–349. [DOI] [PubMed] [Google Scholar]
- Legendre P, Gallagher ED. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129: 271–280. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre L. 2012. Numerical ecology. Amsterdam, the Netherlands: Elsevier. [Google Scholar]
- Lekberg Y, Gibbons SM, Rosendahl S, Ramsey PW. 2013. Severe plant invasions can increase mycorrhizal fungal abundance and diversity. The ISME Journal 7: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekberg Y, Koide RT, Rohr JR, Aldrich‐Wolfe L, Morton JB. 2007. Role of niche restrictions and dispersal in the composition of arbuscular mycorrhizal fungal communities. Journal of Ecology 95: 95–105. [Google Scholar]
- Lekberg Y, Meadow J, Rohr JR, Redecker D, Zabinski CA. 2011. Importance of dispersal and thermal environment for mycorrhizal communities: lessons from Yellowstone National Park. Ecology 92: 1292–1302. [DOI] [PubMed] [Google Scholar]
- Lekberg Y, Schnoor T, Kjøller R, Gibbons SM, Hansen LH, Al‐Soud WA, Sørensen SJ, Rosendahl S. 2012. 454‐sequencing reveals stochastic local reassembly and high disturbance tolerance within arbuscular mycorrhizal fungal communities. Journal of Ecology 100: 151–160. [Google Scholar]
- Levin SA. 1992. The problem of pattern and scale in ecology. Ecology 73: 1943–1967. [Google Scholar]
- Mangan SA, Adler GH. 2000. Consumption of arbuscular mycorrhizal fungi by terrestrial and arboreal small mammals in a Panamanian cloud forest. Journal of Mammalogy 81: 563–570. [Google Scholar]
- Martínez‐García LB, Armas C, Miranda J de D, Padilla FM, Pugnaire FI. 2011. Shrubs influence arbuscular mycorrhizal fungi communities in a semi‐arid environment. Soil Biology and Biochemistry 43: 682–689. [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. 1990. A new method which gives an objective measure of colonization of roots by vesicular‐arbuscular mycorrhizal fungi. New Phytologist 115: 495–501. [DOI] [PubMed] [Google Scholar]
- Mook JH, Haeck J, van der Toorn J, van Tienderen PH. 1992. The demographic structure of populations In: Kuiper PJC, Bos M, eds. Plantago: a multidisciplinary study. Berlin/Heidelberg, Germany: Springer, 69–87. [Google Scholar]
- Mummey DL, Rillig MC. 2008. Spatial characterization of arbuscular mycorrhizal fungal molecular diversity at the submetre scale in a temperate grassland: arbuscular mycorrhizal fungal molecular diversity. FEMS Microbiology Ecology 64: 260–270. [DOI] [PubMed] [Google Scholar]
- Munkvold L, Kjøller R, Vestberg M, Rosendahl S, Jakobsen I. 2004. High functional diversity within species of arbuscular mycorrhizal fungi. New Phytologist 164: 357–364. [DOI] [PubMed] [Google Scholar]
- Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P et al 2013. Patterns and processes of microbial community assembly. Microbiology and Molecular Biology Reviews 77: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsham KK, Fitter AH, Watkinson AR. 1995. Arbuscular mycorrhiza protect an annual grass from root pathogenic fungi in the field. The Journal of Ecology 83: 991–1000. [Google Scholar]
- Ojanen SP, Nieminen M, Meyke E, Pöyry J, Hanski I. 2013. Long‐term metapopulation study of the Glanville fritillary butterfly (Melitaea cinxia): survey methods, data management, and long‐term population trends. Ecology and Evolution 3: 3713–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2015. vegan: community ecology package. R package version 2.2‐1. [WWW document] URL http://CRAN.R-project.org/package=vegan [accessed 16 August 2017].
- Olsson PA, Rahm J, Aliasgharzad N. 2010. Carbon dynamics in mycorrhizal symbioses is linked to carbon costs and phosphorus benefits. FEMS Microbiology Ecology 72: 125–131. [DOI] [PubMed] [Google Scholar]
- Öpik M, Moora M, Liira J, Zobel M. 2006. Composition of root‐colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe: arbuscular mycorrhizal fungal communities around the globe. Journal of Ecology 94: 778–790. [Google Scholar]
- Öpik M, Moora M, Zobel M, Saks Ü, Wheatley R, Wright F, Daniell T. 2008. High diversity of arbuscular mycorrhizal fungi in a boreal herb‐rich coniferous forest. New Phytologist 179: 867–876. [DOI] [PubMed] [Google Scholar]
- Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier Ü, Zobel M. 2010. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytologist 188: 223–241. [DOI] [PubMed] [Google Scholar]
- Peres‐Neto PR, Legendre P, Dray S, Borcard D. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87: 2614–2625. [DOI] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS. 1970. Improved procedures for clearing roots and staining parasitic and vesicular‐arbuscular mycorrhizal fungi for rapid assessment of infection. Transactions of the British Mycological Society 55: 158–160. [Google Scholar]
- Pielou EC. 1966. The measurement of diversity in different types of biological collections. Journal of Theoretical Biology 13: 131–144. [Google Scholar]
- Powell JR, Bennett AE. 2016. Unpredictable assembly of arbuscular mycorrhizal fungal communities. Pedobiologia 59: 11–15. [Google Scholar]
- Quinn GP, Keough MJ. 2002. Experimental design and data analysis for biologists. Cambridge, UK: Cambridge University Press. [Google Scholar]
- R Core Team . 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rodríguez‐Echeverría S, Teixeira H, Correia M, Timóteo S, Heleno R, Öpik M, Moora M. 2017. Arbuscular mycorrhizal fungi communities from tropical Africa reveal strong ecological structure. New Phytologist 213: 380–390. [DOI] [PubMed] [Google Scholar]
- Rosendahl S. 2008. Communities, populations and individuals of arbuscular mycorrhizal fungi. New Phytologist 178: 253–266. [DOI] [PubMed] [Google Scholar]
- Ross MD. 1973. Inheritance of self‐incompatability in Plantago lanceolata . Heredity 30: 169–176. [Google Scholar]
- Saks Ü, Davison J, Öpik M, Vasar M, Moora M, Zobel M. 2014. Root‐colonizing and soil‐borne communities of arbuscular mycorrhizal fungi in a temperate forest understorey. Botany‐Botanique 92: 277–285. [Google Scholar]
- Shannon CE. 1948. A mathematical theory of communication. Bell System Technical Journal 27: 379–423. [Google Scholar]
- Simon L, Lalonde M, Bruns TD. 1992. Specific amplification of 18S fungal ribosomal genes from vesicular‐arbuscular endomycorrhizal fungi colonizing roots. Applied and Environmental Microbiology 58: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. 2008. Mycorrhizal symbiosis. New York, NY, USA: Academic Press. [Google Scholar]
- Smith SE, Smith FA. 2011. Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annual Review of Plant Biology 62: 227–250. [DOI] [PubMed] [Google Scholar]
- Stampe ED, Daehler CC. 2003. Mycorrhizal species identity affects plant community structure and invasion: a microcosm study. Oikos 100: 362–372. [Google Scholar]
- Sterkenburg E, Bahr A, Brandström Durling M, Clemmensen KE, Lindahl BD. 2015. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytologist 207: 1145–1158. [DOI] [PubMed] [Google Scholar]
- Sylvia DM. 1986. Spatial and temporal distribution of vesicular‐arbuscular mycorrhizal fungi associated with Uniola paniculata in Florida foredunes. Mycologia 78: 728–734. [Google Scholar]
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco‐Palacios AM, Thu PQ, Suija A et al 2014. Global diversity and geography of soil fungi. Science 346: 1256688. [DOI] [PubMed] [Google Scholar]
- Varela‐Cervero S, Vasar M, Davison J, Barea JM, Öpik M, Azcón‐Aguilar C. 2015. The composition of arbuscular mycorrhizal fungal communities differs among the roots, spores and extraradical mycelia associated with five Mediterranean plant species. Environmental Microbiology 17: 2882–2895. [DOI] [PubMed] [Google Scholar]
- Velázquez MS, Cabello MN, Barrera M. 2013. Composition and structure of arbuscular‐mycorrhizal communities in El Palmar National Park, Argentina. Mycologia 105: 509–520. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. 2002. Modern applied statistics with S. New York, NY, USA: Springer. [Google Scholar]
- Verbruggen E, van der Heijden MGA, Weedon JT, Kowalchuk GA, Röling WFM. 2012. Community assembly, species richness and nestedness of arbuscular mycorrhizal fungi in agricultural soils. Molecular Ecology 21: 2341–2353. [DOI] [PubMed] [Google Scholar]
- Vogelsang KM, Reynolds HL, Bever JD. 2006. Mycorrhizal fungal identity and richness determine the diversity and productivity of a tallgrass prairie system. New Phytologist 172: 554–562. [DOI] [PubMed] [Google Scholar]
- Warner NJ, Allen MF, MacMahon JA. 1987. Dispersal agents of vesicular‐arbuscular mycorrhizal fungi in a disturbed arid ecosystem. Mycologia 79: 721–730. [Google Scholar]
- Whitcomb S, Stutz JC. 2007. Assessing diversity of arbuscular mycorrhizal fungi in a local community: role of sampling effort and spatial heterogeneity. Mycorrhiza 17: 429–437. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. [Google Scholar]
- Wolfe BE, Mummey DL, Rillig MC, Klironomos JN. 2007. Small‐scale spatial heterogeneity of arbuscular mycorrhizal fungal abundance and community composition in a wetland plant community. Mycorrhiza 17: 175–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 The proportion of nonAM root‐associated fungal classes within the roots of Plantago lanceolata.
Fig. S2 Rarefaction curves for AM fungi in roots and root‐associated soil.
Fig. S3 Connection diagram of sampled Plantago lanceolata individuals in the Åland Islands in the Baltic Sea, southwestern Finland.
Fig. S4 Variation of abiotic and biotic factors for each population.
Fig. S5 Variation partitioning explaining AM fungal community descriptors.
Fig. S6 Violin plot showing the distance between AM fungal communities in root and root‐associated soil for each hierarchical spatial scale.
Table S1 ANOVA table for partitioning of variance for each hierarchical spatial scale
Table S2 PERMANOVA table for partitioning of variance for each hierarchical spatial scale
Table S3 Percentage of variation explained at each spatial scale for soil nutrients, bioclimatic variables, vegetation and nonAM root‐associated fungi at the regional, population, and subpopulation levels.
Table S4 P‐values for the impact of environmental variables on the arbuscular mycorrhizal fungal root colonization and diversity indices in both root and root‐associated soil
Methods S1 Full description of molecular methods and bioinformatics.


