Summary
The carrier state is an alternative bacteriophage life cycle by which virulent bacteriophage can persist in association with host bacteria. Campylobacter jejuni carrier state strains exhibit growth phase dependent motility due to a truncated flagella phenotype. Genome sequencing identified a T368A substitution in the G3 domain of the SRP‐like GTPase FlhF from C. jejuni PT14CP30A carrier state strains, which we hypothesized to be the cause of the complex motility phenotype. We have analyzed the role of this mutation in C. jejuni PT14 and demonstrated that flhF(T368A) leads to a large proportion of cells unable to synthesize flagella, while the remaining cells form a single flagellum at one pole leading to significantly reduced motility. The flhF(T368A) mutation causes a reduction in the phage adsorption constant, which leads to a decrease in infection efficiency. Down‐regulation of σ28 and σ54 dependent flagellar genes were observed as responses to the flhF(T368A) mutation. FlhF(T368A) protein is impaired in GTPase activity and exhibits reduced stability. C. jejuni carrying flhF(T368A) are less sensitive to bacteriophage infection and formation of the carrier state. The acquisition of flhF(T368A) in carrier state strains acts to prevent super‐infection and maintain association with the bacteriophage that provoked the interaction.
Introduction
Campylobacter jejuni is a bacterial pathogen commonly responsible for foodborne gastroenteritis across the world. Infection can arise from a variety of food or water‐borne sources but is often associated with the consumption of contaminated poultry meat (Newell et al., 2011). Campylobacter are Gram‐negative spiral shaped bacteria that are motile by means of a single unsheathed flagellum at one or both poles of the cell (Nachamkin et al., 1993). During infection Campylobacter are frequently located within the thick mucus layer lining the intestine (Berry et al., 1988). Motility is clearly a significant phenotype for bacteria migrating to, and moving within the mucus to reach desired microenvironments that can facilitate growth (Lertsethtakarn et al., 2011). Intact flagella that support motility are essential for colonization by Campylobacter, and non‐motile Campylobacter either do not colonize the intestines of animal hosts (Mertins et al., 2013), or in some cases, only lead to short‐term colonization that cannot persist more than seven days postinfection (Hendrixson and DiRita, 2004).
Flagella biosynthesis is a complex process that requires the coordinate expression of over fifty genes to produce a functional flagella (Chevance and Hughes, 2008). Flagellar export in C. jejuni is controlled by a two‐component regulator FlgSR, where the FlgS histidine kinase autophosphorylates to activate the response regulator FlgR. Phosphorylated FlgR in conjunction with the σ54–RNA polymerase complex activates the transcription of genes encoding flagellar components, including the flagellar hook and hook‐associated proteins, and the minor flagellin FlaB. FlgSR and σ54 are also required for the full expression of σ28–dependent genes involved in flagellar biosynthesis, including the expression of flaA encoding the major flagellin protein (Gilbreath et al., 2011; Lertsethtakarn et al., 2011). At the front of the transcriptional cascade there are several genes transcribed from general σ70 promoters that are required for σ54 transcription, these include flhA, flhB, fliP, fliR as well as flhF (Hendrixson and Dirita, 2003). FlhF is reported to impart spatial and/or numerical control of flagella biosynthesis in C. jejuni (Balaban and Hendrixson, 2011), and to function at an early stage of flagella biosynthesis (Green et al., 2009).
Various strategies have been developed and evaluated in order to reduce the contamination of poultry by Campylobacter (Newell et al., 2011). Using bacteriophages, which are natural predators of bacteria and ubiquitous in environment, as a therapy against Campylobacter has demonstrated promise over the past decade (Loc Carrillo et al., 2005; Connerton et al., 2011; Kittler et al., 2013; Hammerl et al., 2014). Phage infection is initiated by phage attaching to a specific receptor followed by injection of its genome into the host cell (Choi et al., 2013). Bacteriophage life cycles are classed as lytic or lysogenic depending on whether the injected phage nucleic acid commits to replication and release of new phage particles upon host cell lysis or can integrate into the host genome and replicate together with the host cell (Boyd and Brussow, 2002). However, alternative bacteriophage life cycles have been described, which includes the carrier state life cycle (Abedon, 2009). The carrier state life cycle describes a situation where the host bacteria and bacteriophage persist in an equilibrium, where some of the bacteria are resistant to phage infection while others are sensitive and support phage replication, with the result that bacteria and phage maintain similar population levels in serial culture (Siringan et al., 2014).
Carrier state C. jejuni PT14CP30ACS show impaired motility and the development of a growth phase dependent sub‐population in broth culture that are associated with phage resistance (Siringan et al., 2014). Transmission electron microscope (TEM) images show the non‐motile bacteria to have truncated flagella (Siringan et al., 2014). However, the reason for this defect in flagellar biosynthesis remains unknown. This study identifies a point mutation within the flhF gene of C. jejuni carrier state cultures harboring bacteriophage CP30A and subsequently examines the mechanisms by which the flhF(T368A) allele modulates flagella function, bacteriophage infection and affects persistence of the carrier state of C. jejuni PT14.
Results
The FlhF(T368A) mutation is a feature of carrier state PT14CP30ACS
Whole genome sequencing of three independent C. jejuni bacteriophage carrier state isolates exhibiting typical impaired motility (PT14CP30ACS‐1, PT14CP30ACS‐2 and PT14CP30ACS‐3) identified a shared adenine to guanine substitution at nucleotide 1102 within the flhF gene. Mapping the carrier state sequence data to the wild type C. jejuni PT14 genome (Brathwaite et al., 2013) with reference to the incumbent error profiles of the Illumina sequencing platform (Schirmer et al., 2016), revealed four further genes to feature G or A indels at varying frequencies within homopolymer regions that signify phase variation (Table 1). The genome sequences otherwise retained the phase variable gene profiles of the motile progenitor. The flhF mutation creates a T368A substitution in the FlhF protein when compared to the wild type sequence of C. jejuni PT14. The NCBI database currently contains 604 protein sequences of FlhF from Campylobacter and Helicobacter species, which are well conserved, and all of which feature a threonine residue at the equivalent sequence position. The substitution is therefore unique and has the potential to modify the function of FlhF (Fig. 1).
Table 1.
Nucleotide changes present in the genome sequences of bacteriophage carrier state cultures.
| Accession number | Gene product | Changes in coding region | Frequency | Reading frame change | Homopolymer change | Occurence in CSLC cultures |
|---|---|---|---|---|---|---|
| A911_00305 | FlhF GTPase | A to G substitution pos.1102 | 100% | T368A | – | 1,2,3 |
| A911_05520 | 1,3‐galactosyltransferase | G deletion pos.341 | 28–33% | G114 fs | 11G→10G | 1,2,3 |
| A911_06880 | KpsC capsular polysaccharide exporter | A deletion pos.439 | 48–56% | R147 fs | 6A→5A | 1,2,3 |
| A911_06906 | SAM dependent methyltransferase | G deletion pos.402 | 84–88% | Y135 fs | 9G→8G | 1,2,3 |
| A911_07000 | alpha‐2,3‐sialyltransferase | A insertion pos.544 | 16–18% | I182 fs | 8A→9A | 1,3 |
| A911_08080 | Lipoprotein | G deletion pos.506 | 61–69% | G169 fs | 10G→9G | 1,2,3 |
PT14CP30ACS‐1, PT14CP30ACS‐2 and PT14CP30ACS‐3 relative to the reference sequence of C. jejuni PT14 (CP003871). fs indicates a frameshift (phase‐off).
Figure 1.
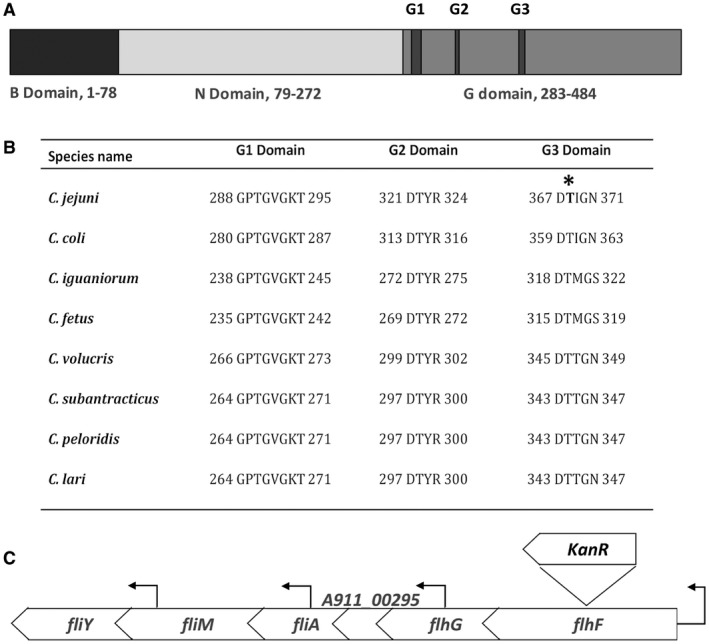
FlhF domain organisation, sequence alignment between Campylobacter species and flhF gene locus. Panel A. Domain structure of C. jejuni PT14 FlhF protein with reference to the amino acid residue locations: Basic domain located in the N‐terminus; N domain located in the central region; and the G domain located at the C‐terminus. The G domain functions as a GTPase and contains multiple conserved subdomains as indicated by G1, G2 and G3. Panel B. Clustal Omega alignment of the G1, G2 and G3 subdomains of the FlhF protein orthologues from C. jejuni subsp. jejuni PT14 (GenBank accession NC_018709.4), C. coli RM4661 (GenBank accession CP007181.1), C. iguaniorum 1485E (GenBank accession CP009043.1), Campylobacter fetus subsp. fetus 04/554 (GenBank accession CP008808.1), C. volucris LMG24379 (GenBank accession CP007774.1), C. subantarcticus LMG24374 (GenBank accession CP007772.1), C. peloridis LMG23910 (GenBank accession CP007766.1), C. lari RM2100 (GenBank accession CP000932.1). Panel C. The structure of the flhF locus of C. jejuni PT14 with the insertion site of the kanamycin resistant gene positioned in the same orientation of flhF, and the location of the transcription start sites marked by directional arrows above the bar (Hooton and Connerton, 2015).
FlhF together with the signal sequence‐binding protein Ffh and signal recognition particle (SRP) receptor FtsY, form a unique subfamily within the SIMIBI‐type nucleoside triphosphate‐binding protein class referred to as SRP GTPase proteins, which are universally conserved (Leipe et al., 2002; Zanen et al., 2004). Unlike Ffh and FtsY that function as a SRP‐SRP Receptor (SRP‐SR) heterodimer in the presence of GTP, FlhF proteins form a stable homodimer structure through interaction of their GTP binding domains (Bange et al., 2007). The GTP binding domain (G domain) is located toward the C‐terminus of FlhF, which shares conserved amino acid residues with the G domains of Ffh and FtsY. FlhF also contains conserved B and N domains (Fig. 1A). The B domain functions to regulate dimerization in the presence of GTP (Bange et al., 2007), while the N domain is thought to stabilize the GTP bound state by rearranging the SRP conformation upon ribosome binding, and further prime it for the formation of subsequent homodimer or heterodimer complexes (Halic et al., 2006). The G domain defines the GTP binding site, within which are five conserved nucleotide‐binding elements (G1‐G5) (Eichler and Moll, 2001). The functions of the SRP GTPases include translation, protein translocation, signal transduction, regulation of cell polarity and possibly cell division (Bulyha et al., 2011). The observed T368A mutation is identified in the third GTP binding element of the G domain within FlhF (G3, Fig. 1B).
FlhF is required for motility of C. jejuni PT14 but FlhF(T368A) has a negative impact on motility
In order to determine the effect of flhF(T368A) on the motility of C. jejuni independent of phage replication or phase variable changes in the carrier state, three flhF constructs were prepared in C. jejuni PT14. C. jejuni PT14flhF::kan was created by inserting a kanamycin resistance gene in the same orientation to inactivate flhF (Fig. 1C). Complements of the flhF::kan mutant with either flhF wild type or flhF(T368A) alleles were prepared by directed insertion into the A911_00230 pseudogene. As the flhF gene sequence is identical between C. jejuni PT14 and C. jejuni NCTC11168, similar constructs were prepared in C. jejuni NCTC11168 in parallel experiments.
The swarming motility of wild type C. jejuni PT14 and its flhF mutants were assessed on 0.4% w/v agar plates. As expected, the wild type C. jejuni strain was fully motile (Fig. 2A) with the cells exhibiting typical bipolar flagella when observed using TEM (Fig. 2F). Inactivation of the flhF gene resulted in a non‐motile phenotype (PT14flhF::kan) where flagella structures were absent from all the cells imaged by TEM (Fig. 2B and G). The flhF mutant cells also appeared curved rather than the spiral form of the wild type that typifies campylobacters. Morphometric measurements indicate the reduced curvature is associated with a significant increase in the helical pitch of the cells compared to wild type (Table 2). Complementation of the flhF mutation in C. jejuni PT14 (flhF::kan 00230::flhF‐cat) restored 90% motility compared to wild type C. jejuni PT14 with 88% of cells exhibiting polar flagella structures by TEM (Fig. 2C and H). These cells had also notably regained spiral morphology. Alternatively, complementation with flhF(T368A) produced strains (flhF::kan 00230::flhF(T368A)‐cat) with approximately 50% of the swarming motility of the wild type with TEM showing 24% of the cells to have single polar flagella structures (Fig. 2D and I) or none (Supplementary File 1D). The mean contour lengths of the flagella of these strains were also significantly reduced compared to the wild type and the wild type complement strain, as determined from TEM images (Table 2). Motility in C. jejuni shows temperature dependence, with growth temperature of 42°C favoring motility with longer flagella compared to lower growth temperatures (Wösten et al., 2010). The swarming motility of the wild type, flhF mutant and complement strains were assessed at 30, 37 and 42°C. Wild type and wild type complement strains showed the greatest motility at 42°C, reduced motility at 37°C and no motility at 30°C. Whereas the flhF(T368A) complement stain was only measurably motile at 42°C (Table 2).
Figure 2.
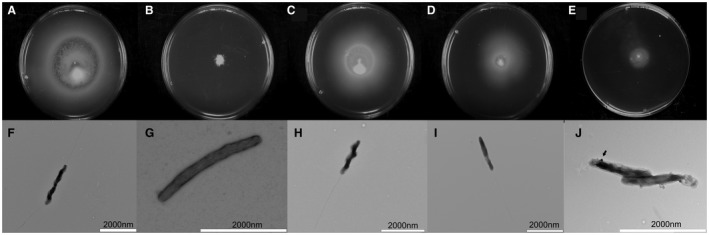
Swarming motility assays and TEM images. Panels A to E show swarming growth after 48 h from center inoculums of 0.4% of agar (w/v) MH plates, where the diameters of the bacterial halos formed are recorded in the panel key below. Panels F to J show TEMs of uranyl acetate stained bacteria and the flagellated bacteria population were determined (n = 100). Panels A and F are wild type C. jejuni PT14 (d = 77 mm); B and G C. jejuni PT14flhF::kan (d = 6 mm); C and H C. jejuni PT14flhF::kan 00230::flhF‐cat (d = 68 mm); D and I C. jejuni PT14flhF::kan 00230::flhF(T368A)‐cat (d = 39 mm); E and J C. jejuni PT14CP30ACS (d = 17 mm). The arrowhead in panel J indicates a bacteriophage binding to the surface of a C. jejuni PT14CP30ACS cell.
Table 2.
Summary of motility characteristics of flhF mutant, complement and carrier state strains.
| C. jejuni strain | Cell shape | Cell pitch (µm) | Flagellated cells (%) | Swarming motility (mm)a | Flagellar length (µm) | ||
|---|---|---|---|---|---|---|---|
| 42oC | 37oC | 30oC | |||||
| Wild type PT14 | Spiral | 0.91 ± 0.16 | 94 ± 3 | 75 ±7 | 23 ± 3 | ≤3 | 4.74 0.73 |
| flhF::kan | Curved | 2.36 ± 0.33b | 0 | ≤3 | ≤3 | ≤3 | 0 |
| flhF::kan,00230::flhF‐cat | Spiral | 0.91 ± 0.12 | 88 ± 4 | 68 ± 4 | 17 ± 2 | ≤3 | 4.51 ± 1.3 |
| flhF::kan,00230::flhF(T368A)‐cat | Curved | 1.90 ± 0.37b | 24 ± 3 | 38 ± 6 | ≤3 | ≤3 | 3.16 ± 0.91c |
| PT14CP30ACS‐1 | Curved | 1.67 ± 0.47b | 5 ± 3 | 14 ± 2 | ≤3 | ≤3 | 0.69 ± 0.14b , c |
Means ± SD;
Growth diameters on 48 h MH motility plates
indicates p < 0.05 relative to wild type
measurements from the minority of cells with flagella.
By comparison, the carrier state strain C. jejuni PT14CP30ACS was non‐motile under these conditions (Fig. 2E). TEM revealed 95% of the cells to have no flagella (Fig. 2J), with the remaining cells exhibiting either short or aberrant flagella positioned at non‐polar locations (Supplementary File 1A and B). Phage CP30A was also often observed to be adsorbed to the surface of the cells as indicated by the arrow in Fig. 2J and occasionally encapsulated (Supplementary File 1C).
The swarming motility of C. jejuni NCTC11168 and its flhF mutants were also determined. Similar to C. jejuni PT14, wild type C. jejuni NCTC11168 was motile while the flhF::kan derivative was non‐motile. Complementation the flhF::kan mutant with either flhF or flhF(T368A) successfully restored its motility but as observed for C. jejuni PT14, wild type flhF complementation produced a greater recovery in motility than flhF(T368A) complementation (data not shown).
FlhF is required for full expression of flagellar associated σ28 and σ54 dependent genes but FlhF(T368A) is impaired
Insertion of the kanamycin gene within flhF resulted in 2‐fold lower expression from the native promoter of flhF compared to wild type C. jejuni PT14 as measured by qRT‐PCR (p < 0.01; Fig. 3). Ectopic complementation of flhF (flhF::kan 00230::flhF‐cat) restored flhF transcription to produce levels 2‐fold greater than wild type C. jejuni PT14 (p < 0.01). The second gene of the flhF operon is flhG, which is reported to encode an ATPase that represents a member of the ParA superfamily of proteins that regulate cell division, and in the case of C. jejuni is reported to mediate the site and numerical control of flagellar synthesis (Gulbronson et al., 2016). Inactivation of flhF similarly reduced the transcription of the flhG gene relative to wild type (p < 0.01), and transformant cells carrying ectopic copies of flhF also restored flhG transcription to wild type levels. The increase in transcription of flhG as a consequence of second site flhF expression would mitigate against any effect of the kanamycin gene insertion. Ectopic complementation with flhF(T368A) (flhF::kan 00230::flhF(T368A)‐cat) also rescued transcription of flhF and flhG but the levels were significantly lower than for the wild type complement (p < 0.01).
Figure 3.
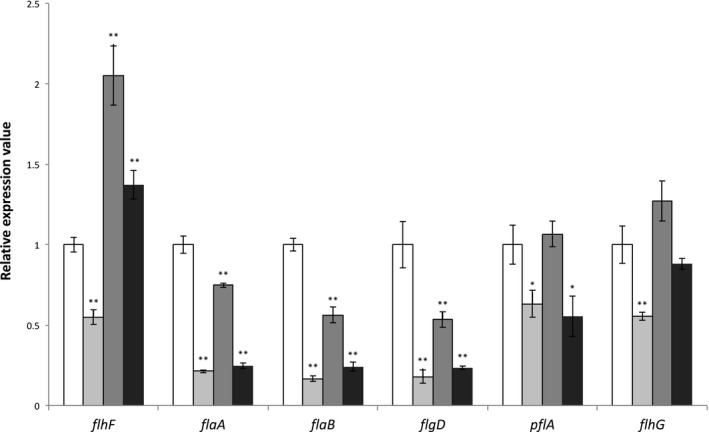
Transcript levels of flagellar associated genes. Relative transcription levels of flhF, flaA, flaB, flgD pflA and flhG were determined by qPCR. Values represent the means of RNA populations extracted from triplicate cultures. The housekeeping gene encoding phosphoglycerokinase (pgk) was used to normalize the data. C. jejuni PT14 wild type (□); C. jejuni PT14flhF::kan (■); C. jejuni PT14 flhF::kan 00230::flhF‐cat (■); C. jejuni PT14flhF::kan 00230::flhF(T368A)‐cat (■). * indicates gene expression value of flhF derivative is significantly different to that of wild type p < 0.05; ** indicates p < 0.01.
We next determined if flhF was essential for the expression of σ28 and σ54 –dependent flagellar associated genes, and whether the expression of flhF(T368A) affected the regulation of these genes (Fig. 3). The flaA gene encoding the major flagellin FlaA was down‐regulated 4.7‐fold in the flhF mutant (p < 0.01). Ectopic expression of flhF restored flaA expression to 75% of wild type, whereas ectopic expression of flhF(T368A) did not produce a significant increase in flaA expression over the flhF mutant. In contrast to flaA that has a σ28‐dependent promoter, the minor flagellin encoding gene, flaB, has a σ54‐dependent promoter (Guerry et al., 1991; Wassenaar et al., 1993). The flaB gene was down‐regulated 6‐fold in the flhF mutant (p < 0.01), and ectopic expression of flhF also increased flaB expression over the flhF mutant but this increase was not evident upon ectopic expression of the flhF(T368A) allele (p > 0.05). Similarly, flgD (encoding a hook assembly protein) uses a σ54‐dependent promoter (Balaban et al., 2009) that is reduced in the flhF mutant and increases upon ectopic expression of wild type flhF but not flhF(T368A). Expression of the pflA gene encoding the paralyzed flagella protein was also significantly reduced in the flhF mutant (p < 0.05) and recovered upon expression of wild type flhF but not flhF(T368A). In the absence of pflA expression cells produce intact flagella structures but are non‐motile (Gao et al., 2014). In summary, ectopic complementation of inactivated flhF mutants of C. jejuni PT14 (flhF::kan 00230::flhF‐cat) resulted in significant increases in the expression of σ28 and σ54 regulated genes (p < 0.05), but these increases were not evident upon expression of flhF(T368A).
C. jejuni PT14 flhF mutants are less sensitive to phage infection
Campylobacter‐specific bacteriophages of the Myoviridae subfamily Eucampyvirinae (Javed et al. 2014) were applied to bacterial lawns formed from either C. jejuni PT14, or PT14flhF::kan or PT14flhF::kan 00230::flhF‐cat or PT14flhF::kan 00230::flhF(T368A)‐cat at a test dilution of log10 7 pfu ml–1 in order to investigate the impact of the flhF related genotypes on their ability to act as a host for bacteriophage infection. Data are presented as the efficiency of plating (EOP) for each phage relative to the titer on wild type C. jejuni PT14 for each of the host strains in Table 3. Phages CP220, ɸ3 and ɸ15 are classified as group II (Cp220likevirus) based on their genome sizes (180 and 190 kb) and head diameters (Sails et al., 1998), and the remaining phages are group III (Cp8unalikevirus) with genome sizes of approximately 140 kb (Connerton et al., 2004; Siringan et al., 2011; Firlieyanti et al., 2016). Group II phages are flagellotropic, which require a functional flagellar to infect (Coward et al., 2006; Scott et al., 2007a; Baldvinsson et al., 2014; Lis and Connerton, 2016), whereas group III phages can be affected by loss of motility they show dependence on capsular polysaccharide structures (Sørenson et al., 2011; Lis and Connerton, 2016). The flhF inactive strain showed reduced EOP for all group III phages compared to wild type (p < 0.05), and no replication of the group II phages (Table 3). Complementation of the flhF mutant with wild type flhF restored sensitivity to all the phages tested, however, the EOPs of CP220, Ø3 and CLP6 remained significantly reduced compared to wild type C. jejuni PT14. The flhF(T368A) complement strain exhibited significantly reduced EOPs for the group III phages similar to that observed for the knock‐out flhF mutant, and similarly no plaques were formed for the group II phages on flhF(T368A) (Table 3). Group III phage retained the ability to replicate on the C. jejuni PT14 flhF mutant and have been demonstrated to enter the carrier state (Siringan et al., 2014). Group III phage CP30A were therefore selected to investigate the replication parameters of the flhF derivatives. The adsorption constants (k) for phage CP30A binding to C. jejuni PT14 flhF mutant or the flhF(T368A) complement show a 1.7‐fold decrease compared to wild type (p < 0.05; Table 4), whereas the wild type complement of flhF showed no significant difference (p > 0.05). The burst size revealed no difference between C. jejuni PT14 (1.95 ± 0.54 pfu cell–1) and its flhF mutant derivatives. The latent period for all the C. jejuni strains tested was approximately 60 min.
Table 3.
Efficiency of plating of bacteriophages replicating on flhF mutant and derivatives compared to C. jejuni PT14.
| C. jejuni strains | Group II phage | Group III phage | ||||||
|---|---|---|---|---|---|---|---|---|
| CP220 | Ø3 | Ø15 | CP30A | CP8 | CPX | CLP6 | CLP47 | |
| PT14 | 1 ± 0.10 | 1 ± 0.13 | 1 ± 0.33 | 1 ± 0.12 | 1 ± 0.13 | 1 ± 0.07 | 1 ± 0.13 | 1 ± 0.06 |
| flhF::kan | ND | ND | ND | 0.57 ± 0.03a | 0.50 ± 0.22a | 0.40 ± 0.18a | 0.26 ± 0.09a | 0.17 ± 0.04a |
| flhF::kan,00230::flhF‐cat | 0.06 ± 0.02a | 0.33 ± 0.07a | 0.72 ± 0.12 | 0.74 ± 0.13 | 0.88 ± 0.22 | 0.76 ± 0.25 | 0.63 ± 0.05a | 0.88 ± 0.17 |
| flhF::kan,00230::flhF(T368A)‐cat | ND | ND | ND | 0.51 ± 0.06a | 0.63 ± 0.13a | 0.44 ± 0.07a | 0.57 ± 0.09a | 0.35 ± 0.10a |
ND indicates none detected;
indicates EOP in the test group is significantly different to the EOP of the control wild type C. jejuni PT14 group (p < 0.05).
Table 4.
Replication parameters for phage CP30A on different host strains.
| C. jejuni host strain | Adsorption constant (k) × 10–10 (ml min–1) | Burst size (pfu cell–1) | Latent period (min) |
|---|---|---|---|
| PT14 | 1.13 ± 0.42 | 1.95 ± 0.54 | 60 |
| PT14flhF::kan | 0.65 ± 0.02a | 1.91 ± 0.69 | 60 |
| PT14flhF::kan 00230::flhF‐cat | 1.19 ± 0.24 | 1.91 ± 0.38 | 60 |
| PT14flhF::kan 00230::flhF(T368A)‐cat | 0.63 ± 0.24a | 1.91 ± 0.87 | 60 |
indicates p < 0.05 relative to wild type.
We further investigated the growth of host cultures in response to phage replication. Phage CP30A was used to infect C. jejuni PT14, PT14flhF::kan, PT14flhF::kan 00230::flhF‐cat and PT14flhF::kan 00230::flhF(T368A)‐cat in broth cultures. Increases in phage titer were observed in all cases when the density of the bacteria was > 7 log10 CFU ml–1 – the phage proliferation threshold (Cairns et al., 2009). In the wild type C. jejuni PT14 cultures, an increase in phage titer was accompanied by a 2.2 log10 CFU ml–1 fall in the bacterial count at 8 h, whereafter the counts remained static for 6 h before recommencing growth after 16 h (Fig. 4A). Phage replication was impaired in the flhF knock‐out mutant and the flhF(T368A) complement as indicated by delays of 2–4 h in the bacterial population crash and concomitant rise in phage titer. As observed previously for laboratory cultures, bacterial growth recommenced after the crash with the development of phage resistance (Scott et al., 2007b). However, the static period following the phage‐induced population crash of the flhF mutant was reduced to 4 h followed by a comparatively reduced rate of recovery (Fig. 4B). The static period was reinstated in the complement strain PT14flhF::kan 00230::flhF‐cat and to a lesser extent for PT14flhF::kan 00230::flhF(T368A)‐cat (Fig. 4C and D), suggesting more efficient reinfection of the C. jejuni with flagellar structures.
Figure 4.
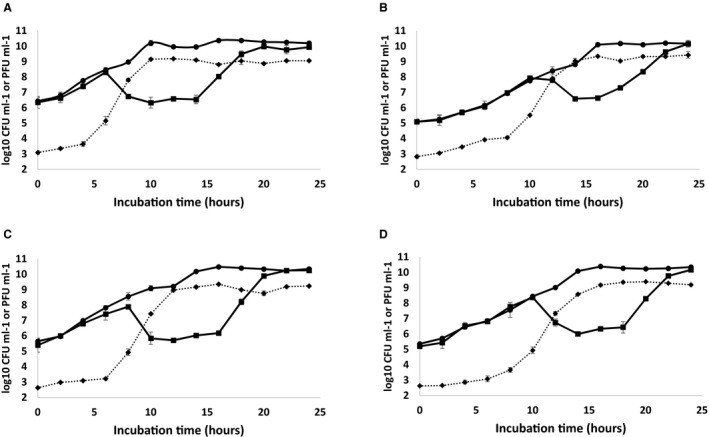
Growth characteristics of C. jejuni PT14 wild type and its flhF mutants upon infection by bacteriophage CP30A. Broth cultures containing 4 log10 CFU C. jejuni were pre‐incubated for 2 h before infection with 2 log10 PFU bacteriophage CP30A (time zero). Sample aliquots were removed every 2 h to determine the bacterial counts and phage titers. Panels A. C. jejuni PT14, B. C. jejuni PT14flhF::kan, C. C. jejuni PT14flhF::kan 00230::flhF‐cat, D. C. jejuni PT14flhF::kan 00230:: flhF(T368A)‐cat. The panels show the viable counts for a parallel uninfected control culture (●); viable counts for the bacteriophage CP30A infected C. jejuni cultures (■); bacteriophage CP30A titers from the infected cultures (◆).
Individual colonies were recovered from the enumeration plates of the cultures post static phase (n = 100) and their sensitivity to infection by group II (CP220, Φ3, Φ15) and III (CP30A, CPX, CLP6 and CLP47) phages compared to non‐infected cultures. Of these, 10% of the isolates recovered post phage CP30A infection of C. jejuni PT14 wild type and 7% for the flhF complement (PT14flhF::kan 00230::flhF‐cat) had entered carrier state life cycle with continued CP30A phage association and impaired motility. The remaining isolates retained their susceptibility to phage CP220, Φ3, Φ15, CP30A and CPX. However, 53% of the colonies recovered post CP30A infection of C. jejuni PT14 wild type had developed resistance to phages CLP6 and CLP47, which distinguished them from other group III phage. Wild type complement (PT14flhF::kan 00230::flhF‐cat) isolates did not develop resistance to phage CLP6 and CLP47. No carrier state isolates emerged from the flhF mutant (PT14flhF::kan) or the flhF(T368A) complement (PT14flhF::kan 00230::flhF(T368A)‐cat). The frequency of infection of these strains is reduced, which is also likely to effect the formation of the carrier state. However, isolates from these cultures had acquired resistance to phages CLP6 and CLP47, again marking the phages CLP6 and CLP47 as different to the group III phages CP30A and CPX.
To assess if the carrier state strains emerging from CP30A infection of wild type cultures had acquired the flhF(T368) mutation, the flhF gene was PCR amplified from genomic DNAs prepared from plate swabs of carrier state cultures and the amplicons sequenced with reference to the presence of the base calls for the wild type and flhF(T368) alleles. The flhF(T368A) allele appeared as the dominant flhF genotype in these cultures but the wild type allele was also evident at frequencies ≤ 10–3. The presence of a wild type subpopulation would provide a mechanism by which phage titers are maintained through bacteriophage replication in a subpopulation of cells, and an explanation for the emergence of motile types observed in synchronous broth cultures (Siringan et al., 2014).
FlhF(T368A) shows reduced thermal stability and GTPase activity
FlhF protein is reported to have GTPase activity that is required for the correct biosynthesis of functional flagella (Balaban et al., 2009). Wild type FlhF and FlhF(T368A) proteins were expressed as N‐terminal His6‐tagged fusions in Escherichia coli using a pET28b vector. For wild type FlhF, E. coli BL21DE3 Star cells were used for protein expression, but for FlhF(T368A) it was necessary to use Origami 2 DE3 pLysS cells to overcome inherent protein instability and prevent inclusion body formation. The proteins were purified by a combination of Ni‐affinity and Superose 12 size‐exclusion chromatography. GTPase activities were measured by detecting the release of free phosphate (P i) as a product of GTP hydrolysis. The temperature optimum of FlhF GTPase was 50°C with a specific activity of 45 nmol min–1 mg–1 compared to a temperature of optimum of 42°C and a specific activity of 17.5 nmol min–1 mg–1 for FlhF(T368A) (Fig. 5A and B respectively). The specific GTPase specific activity of wild type FlhF was 2‐fold greater than that of FlhF(T368A) at 42°C that represents the growth temperature that C. jejuni exhibits the greatest motility. Consistent with the relative thermal sensitivity of the FlhF proteins, the wild type protein produced a 7 ± 0.03% increase in GTPase activity upon pre‐incubation at 42°C for 5 min that was not observed with FlhF(T368A). However, pre‐incubation with 0.2 mM GTP at 42°C for 5 min with correction for substrate conversion at time zero, produced a 12 ± 0.1% increase in the GTPase activity for wild type FlhF compared to a 14 ± 0.5% increase for FlhF(T368A). The enzymes show substrate stabilization but FlhF(T368A) shows a significantly greater response.
Figure 5.
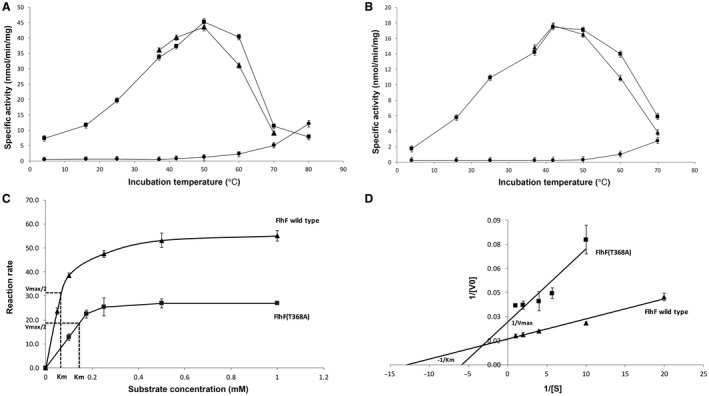
GTPase activities of purified FlhF and FlhF(T368A) proteins. Panels A and B show temperature profiles of the GTPase activities for FlhF proteins: Panel A. FlhF wild type protein; Panel B. FlhF(T368A) protein, where (■) denotes the FlhF GTPase activity temperature profile; (▲) denotes FlhF protein preheated at the recorded temperature for 5 min before commencing the reaction; (●) represents the GTP self‐hydrolysis control. Panels C (Michaelis‐Menten plots) and D (linear regression Lineweaver‐Burk plots) show kinetic plots for GTPase activities of FlhF (▲) and FlhF(T368A) (■) proteins.
Kinetic parameters at the optimum growth temperature of C. jejuni of 42°C were determined for both FlhF and FlhF(T368A) (Fig. 5C and D). The K M for FlhF was 0.075 mM and the V max 62.5 nmol min–1 mg–1, and by comparison the GTPase activity of FlhF(T368A) was impaired with a K M of 0.1736 mM and V max 37.74 nmol min–1 mg–1. The k cat for FlhF and FlhF(T368A) were calculated as 0.0115 s–1 and 0.007 s–1 respectively.
Discussion
The complex motility phenotypes of bacteriophage carrier state C. jejuni prompted this study (Siringan et al., 2014). Motility is essential for C. jejuni to colonize animal hosts and cause disease (Lertsethtakarn et al., 2011), and accordingly carrier state strains show reduced adhesion and invasion of human colonic epithelial cells (Brathwaite et al., 2015) and fail to colonize chickens (Siringan et al., 2014). C. jejuni possess a characteristic high‐torque flagellar motor that enables swimming at viscosities that effectively immobilize other enteric bacteria, and has been proposed to represent a disproportional leap in the evolution of the flagellar motor structure (Chaban et al., 2018). The flagellar patternation of C. jejuni is characterized as amphitrichous with a single flagellum located at each of the cell poles. Campylobacter flagella biosynthesis is complex but control of the number of flagellar and their polar location are dependent upon the proper expression of the flhFflhG operon (Balaban and Hendrixson, 2011). We have identified a point mutation in the flhF gene of carrier state C. jejuni strains in association with bacteriophage CP30A that we hypothesized was responsible for the observed impaired motility. The mutation represents a unique substitution of T368A within the third GTP binding element of the conserved G domains of FlhF (Bange et al., 2007).
As reported previously inactivation of flhF produces a non‐motile phenotype in C. jejuni (Balaban et al., 2009). Ectopic complementation of the flhF mutation at the 00230 pseudogene locus of C. jejuni PT14 restored motility. However, complementation with flhF(T368A) led to reduced motility compared to wild type and the wild type complement, indicating that the flhF(T368A) mutation present in CP30A associated carrier state strains will confer an impaired motility phenotype independent of the phage association/replication or the presence of phase variable reading frames. Transmission electron microscopy of C. jejuni PT14 and the flhF mutant derivatives confirmed C. jejuni PT14 to have a typical bi‐polar flagella and that the flhF::kan inactivated mutant was devoid of flagella structures. Wild type complement cells (PT14flhF::kan 00230::flhF‐cat) regained a polar flagella, whereas the cells expressing the flhF(T368A) allele (PT14flhF::kan00230::flhF(T368A)‐cat) showed single flagella structures in 24% of the cells examined and the rest none. These data confirm the critical importance of the GTPase component of FlhF in C. jejuni (Balaban et al., 2009).
Insertional inactivation of flhF significantly reduced the expression of the flhF and the downstream gene flhG (Fig. 1C). The transcript levels of flhF and flhG could be restored to wild type or greater levels by ectopic expression of flhF under the native σ70 promoter. The wild type flhF complement achieves double the relative expression of wild type implying the two flhF promoters present are functioning similarly to additively increase the transcript level but this does not extend to flhG, which remains dependent on the resident flhF promoter. Ectopic expression of the flhF(T368A) allele also increased flhF and flhG transcription levels over the flhF inactivated mutant but were significantly less than the wild type flhF complement. These data support the contention that the flhF gene product exerts a positive feedback on flhF and flhG expression. This could be accomplished either by direct or indirect activation of the flhF flhG promoter, or alternatively via specific stabilization of the flhF flhG mRNA.
To gain a greater understanding of the role of flhF(T368A) on flagellar biosynthesis we examined the expression of σ28 (the flagellar sigma factor regulating the major flagellin encoding gene flaA) and σ54 (sigma factor regulating genes encoding flagellar basal body, hook, the minor flagellin flaB and anti‐sigma factor flgM) genes in the mutant and wild type cultures. In the absence of flhF, the expression levels of flaA (σ28 transcription‐dependent), flaB and flgD (σ54 transcription‐dependent) were all significantly reduced, and less severe but still significant downregulation was observed for pflA. Hendrixson and DiRita (2004) hypothesized that flhF exerted it’s influence at the start of the σ54 transcriptional cascade, and therefore deletion of flhF would result in improper transcription of the downstream genes and reduced levels of σ54 dependent gene expression, which would account for the observations in this study that flaB and flgD transcription becomes downregulated. However, Correa et al. (2005) reported that in Vibrio cholerae, flhF is also essential for the expression of several σ28 dependent flagellar genes including the major flagellin homologue. In contrast Hendrixson and DiRita (2003) found that in C. jejuni 81–176, σ28 dependent flaA was largely unaffected by deletion of flhF. Subsequently Balaban et al., (2009) reported that a flhF deletion mutant of C. jejuni 81–176 expressed 50% less flaA but point mutations located in the G1 and G2 domains of flhF increased flaA expression. More recently Ren et al. (2018) have reported 5.3‐fold down regulation of the flaA gene in a flhF insertional inactivation mutant constructed in C. jejuni NCTC11168 (as we also observed). In this study, disruption of flhF led to a significant reduction in the expression of flaA. Ectopic expression of wild type flhF in the flhF mutant led to increases in the expression of σ28 and σ54 dependent genes that was not observed upon expression of the flhF(T368A) allele. Instead, the flaA, flaB and flgD transcript levels were not significantly different to those recorded for the flhF::kan mutant. The mechanism by which FlhF enables the transcription of the flagellar associated genes appears dependent on assembling the FlhF GTPase domain since the flaA, flaB and flgD transcript levels fail to recover upon expression of FlhF(T368A) exhibiting impaired GTPase. We have noted instability in the FlhF(T368A) protein that may hinder recruitment of components required to affect the expression of downstream flagellar components, which may also provide an explanation as to the differential effects of mutants affected in FlhF GTPase activity (Balaban et al., 2009), if the mutants also alter FlhF structural dependencies. The expression of the σ28 and σ54 dependent genes determined in this study for the flhF(T368A) complement relative to wild type C. jejuni PT14 are consistent with the transcriptome (RNA‐seq) study reported for a C. jejuni PT14CP30A carrier state culture (Brathwaite et al., 2015). The exception to this is the expression of flaB, which is down regulated in the flhF(T368A) complement strain relative to wild type C. jejuni PT14, as compared to a 3‐fold up regulation reported for the C. jejuni PT14CP30A carrier state strain. Phage transcription indicative of the progression toward replication was evident in the carrier state cultures (0.2% of the total reads), and where the abundance of the major capsid protein transcripts represented > 95% of all the normalized read counts (Brathwaite et al., 2015). We therefore posit that the up regulation of flaB in the carrier state C. jejuni is a consequence phage transcription/replication. In this context FlaB has recently been reported to confer defensive properties against phage infection, which affects regrowth of the host following lysis of laboratory cultures (Lis and Connerton, 2016).
Bange et al. (2007) reported that FlhF of B. subtilis forms a stable homodimer structure in the presence of GTP, and that the G domains form a composite active site harboring two GTP molecules. Differences in the predicted protein structures of FlhF and FlhF(T368A) are presented, respectively, in Fig. 6A and B, which are based on models of the C. jejuni FlhF protein calculated from the template flagellar biosynthesis protein FlhF‐ 2px0.1 of B. subtilis (Bange et al., 2007). Substitution of the threonine residue with alanine prevents the formation of the hydrogen bond between L287 and T368 that tethers two adjacent β‐sheets that flank a returning loop that constitute the G3 domain. The effect of this is likely to destabilize GTP binding, and as a consequence the formation of the functional homodimer. Figure 6C shows the location of T368 at the monomer interface. The N domains remain free of the interface to enable interaction with other cellular components, and are separated by a distance of 12 Å in the dimer structure. The dependence of the G domains to stabilize the dimer is consistent with the reduced thermal stability and greater substrate dependence observed for the GTPase activity of FlhF(T368A). Impaired GTP binding would not only reduce the catalytic activity but the stability of any complex incorporating FlhF. Indeed wild type FlhF protein had a 2.3‐fold lower K M for the GTP substrate than FlhF(T368A). The k cat of 0.0115 s–1 for the FlhF protein was in excess of the k cat of 0.007 s–1 for FlhF(T368A).
Figure 6.
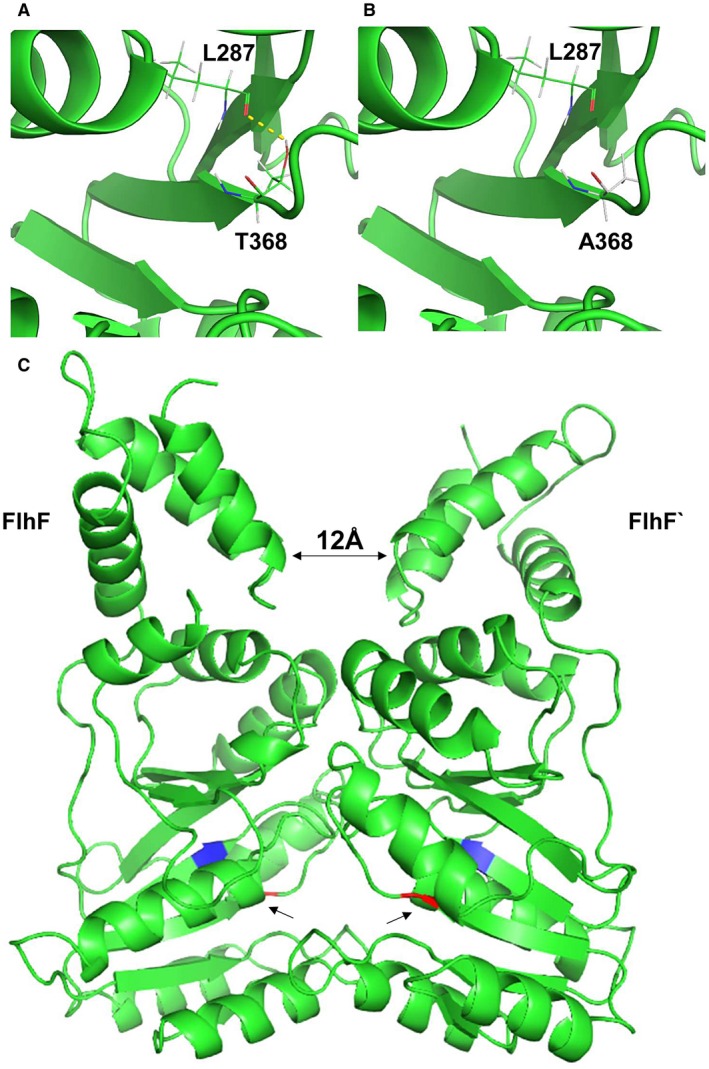
Predicted structures of FlhF and FlhF(T368A). Predicted location of the T368A mutation within a C. jejuni FlhF model derived from FlhF‐2px0.1.A using Swiss‐Model (Waterhouse et al., 2018) and rendered in PYMOL (DeLano, 2002). Panel A. Location of T368 in the FlhF wild type structure with the hydrogen bond marked as a yellow dotted line to the main chain oxygen of L287 (red). Panel B. Location of A368 in the FlhF(T368A) mutant protein that does not permit the formation of the hydrogen bond. Panel C. Predicted FlhF protein homodimer structure (FlhF and FlhF`) with the N domains orientated toward the top of the figure and the G domains at the bottom. The arrow heads in G domains indicate the locations of T368A substitution colored red in juxtaposition to L287 colored blue, to which the threonine residue is hydrogen bonded.
The mechanism by which FlhF and FlhG regulate the polar flagella number and localization is not fully understood. In V. cholera FlhF was reported as solely required for the recruitment and correct localization of the flagellar MS ring inner membrane protein FliF, which is presumed one of the earliest flagellar structural components (Green et al., 2009). C. jejuni FlhF has also been proposed to operate at an early stage in flagellar biosynthesis on the basis that mutational changes that alter GTPase activity can lead to flagellar mislocalization or increased numbers of flagella (Balaban et al., 2009). The likely first step of flagellar biosynthesis is the formation of a multiple inner membrane protein complex, known as the flagellar export apparatus, which is able to secrete most of the flagellar proteins (Hendrixson, 2008), and it is to this complex that the GTP‐bound FlhF homodimer may be recruited to initiate flagellar biosynthesis. Once triggered, the flagellar export apparatus is proposed to release GDP‐bound FlhF, which can no longer act to stimulate flagellar formation (Schuhmacher et al., 2015). FlhF GTPase activity is stimulated by membrane located FlhG that itself is subject to ATP dependent homodimerization to affect flagellar assembly (Gulbronson et al., 2016). FlhF may also actively participate in the export assembly as a recent report demonstrates flhF deficient mutants unable to localize components of flagella export apparatus to the cell pole (Ren et al., 2018).
Surface structures are candidates for adhesion targets and receptors necessary for bacteriophage infection. In C. jejuni, flagella and capsular polysaccharide have been proposed to constitute essential attachment sites for the respective infection of group II and group III Campylobacter phages (Rakhuba et al., 2010; Sørensen et al., 2015). Group II phages CP220, Φ3 and Φ15 failed to infect non‐flagellated strains that included the largely non‐flagellated flhF(T368A) expressing strain. We further observed that group III phages showed a greater efficiency of plaque formation on wild type C. jejuni PT14 than the flhF inactivated mutant derivatives, suggesting that although flagella may not be essential, the presence of a functional flagella increases infection efficiency. Group III phages have been observed to enter the carrier state where the flhF(T368A) mutation would not strictly prevent phage infection but would certainly reduce the efficiency (Table 3).
Comparing the growth dynamics of group III phage infected wild type and flhF mutant C. jejuni PT14 in broth cultures demonstrated the mutant bacteria supported phage replication but did not exhibit reductions in the population count to the same degree as wild type. Complemetation of the flhF mutation with the flhF and flhF(T368A) allele restored wild type behavior with phage‐induced bacterial population crashes of 3.5 and 2.3 log10 CFU ml–1 respectively. The static period post lysis represents the growth stage when the probability of a bacterial cell division is similar to the chance of it being infected and lysed by phage particles. This period is reduced in the flhF mutant (2 h) compared to wild type (6 h), where the rate of infection is reduced. The static period was reinstated upon flhF complementation (6 h) but not so for flhF(T368A) complementation (4 h). The infection rate of the strain carrying flhF(T368A) is reduced. Calculation of the phage adsorption constants revealed a significant difference between the wild type and flhF mutant. The wild type complement of flhF regained the adsorption rate of the wild type, whereas the complement carrying flhF(T368A) remained similar to the inactivated flhF mutant. The root cause of these differences is the absence of the flagellar structure leading to inefficient interaction between the phage and the host bacteria.
Phage resistance and the formation of carrier state life cycle emerge under these conditions. The phase variable genes identified in the carrier state cultures (Table 1) are consistent with the emergence of phage resistance; kpsC mutants are impaired in capsular polysaccharide biosynthesis and have previously been reported to give rise to phage resistance (Coward et al., 2006), and the additional four phase variable genes identified have previously been reported to show shifts to phase‐off in bacteriophage escape mutants (Lis and Connerton, 2016). We have analyzed the frequency of carrier state formation in C. jejuni PT14 cells recovering post bacteriophage CP30A infection (the static phase). For wild type C. jejuni PT14 and the wild type complement PT14flhF::kan 00230::flhF‐cat, 7–10% of the recovered cells had entered carrier state life cycle on the basis of the continued association with phage CP30A. These carrier state cells showed resistance against super‐infecting phage CP220, Φ3, Φ15, CP30A, CPX, CLP6, and CLP47 having lost motility. However, no carrier state cells were recovered from the flhF mutant or the strain expressing flhF(T368A). These strains exhibit inefficient phage adsorption, which results in fewer infected cells compared with cells with intact flagella. These data suggest that the flhF(T368A) mutation does not provoke carrier state formation but is rather a consequence that enables limited motility in sub‐populations of cells that may be phage infected, while maintaining a reservoir of insensitive cells that will continue to divide in the presence of the phage. The acquisition of the flhF(T368A) mutation will also tend to maintain the specific phage association in the carrier state cultures as they appear to resist super‐infection. Phase variable genes that alter phage sensitivity will also act to prevent super‐infection and stabilize bacterial populations in the presence of high phage titers.
This study has demonstrated the independent selection of the point mutation flhF(T368A) in C. jejuni PT14CP30A carrier state isolates. FlhF(T368A) protein shows catalytically compromised GTPase activity with reduced stability that affects the ability of FlhF to produce a functional flagella. Non‐flagellated bacterial populations are resistant to group II bacteriophage and show diminished sensitivity to group III bacteriophage due to inefficient adsorption. However, C. jejuni carrying flhF(T368A) produce a subpopulation of bacteria with flagellar structures that can support phage adsorption and replication. Finely balanced structural/catalytic instability of the FlhF(T368A) GTPase represents a pivotal point in the cellular commitment to flagellar assembly that confers motility and phage sensitivity. The balance in carrier state cultures is tipped against motility in favor of phage insensitivity in the presence of high phage titers. Indeed CP30A carrier state bacteria are profoundly impaired in motility, and as a consequence exhibit poor cellular adhesion and invasion, and generally fail to colonize the intestinal tract of chickens (Hooton et al., 2016). However, at lower phage titers, as evident upon dilution in early exponential phase carrier state broth cultures, reversion to wild type motility may arise until the phage density increases and these motile bacteria become infected and lysed (Siringan et al., 2014). Under these circumstances reversion to the wild type allele will be subject to counter selection and the flhF(T368A) allele maintained. Consistent with this view we have used targeted amplicon sequencing to establish the dominance of the flhF(T368A) allele in carrier state isolates, and that the wild type allele can arise or persist but at low frequencies (≤ 10–3). The flhF(T368A) mutation is maintained because it produces adaptive phenotypes that allow bacteria and phage to coexist, and accord the abilities to escape phage super‐infection and survive extra‐intestinal environments (Siringan et al., 2014). We have demonstrated the complex mechanism by which carrier state cultures can arise and persist in C. jejuni due to the acquisition of a subtle mutation in a gene essential for the control of a key physiological attribute to this species; we expect that as other examples of bacteria/phage carrier state associations are established the nature of the mutational changes may vary but there will be parallels in the mechanisms that permit the host bacteria to express adaptive phenotypes to enable the bacteria and bacteriophage to coexist and survive.
Experimental procedures
Bacterial strains, growth conditions and antibiotic concentrations
C. jejuni PT14 and NCTC11168 were routinely grown on blood agar base No.2 plates (Oxoid, Basingstoke, UK) containing 5% (v/v) defibrinated horse blood (BA) at 42°C under microaerobic conditions (5% v/v oxygen, 2% v/v hydrogen, 88% v/v nitrogen, 5% v/v carbon dioxide) for 18 h. Escherichia coli TOP10, BL21DE3 Star and Origami 2 DE3 pLysS were cultured in Luria‐Bertani (LB) broth with appropriate antibiotic at 37°C with 150 r.p.m. shaking for 18 h. Kanamycin (25 μg ml–1) and chloramphenicol (12.5 μg ml–1) were used for selection of C. jejuni. For selection of E. coli, kanamycin (50 μg ml–1), chloramphenicol (25 μg ml–1) and ampicillin (100 μg ml–1) were applied as required.
Whole genome and amplicon sequencing
Genomic DNAs from C. jejuni carrier state isolates PT14CP30ACS‐1, PT14CP30ACS‐2, and PT14CP30ACS‐3 recovered from biofilms and subsequently colony purified (Siringan et al., 2014), were prepared using a GenElute Bacterial Genomic DNA Kit (Sigma‐Aldrich, Gillingham, UK) from BA plate swabs. DNA sequencing was performed using the Illumina MiSeq platform. The data consisted of 2.2–2.8 million paired‐end sequence reads of 250 bp in length. Initial processing of the raw data, mapping of the sequence reads to C. jejuni PT14 (GenBank accession CP003871) and variant detection were performed using CLC Genomics Workbench version 8.0 (Qiagen, Aarhus, Denmark). The flhF gene sequence containing the flhF(T368A) allele was PCR amplified from each carrier state culture using flhF(T368A) primers (flhF(T368A) F, 5′‐CATGTTTGGTGTTTGCAG‐3′, and flhF(T368A) R, 5′‐AGAATCGGTGCAGTTGAG‐3′) in three independent reactions, which were pooled in equal volumes and purified using the Wizard SV Gel and PCR Clean‐Up System (Promega, Madison, USA). Sequence libraries were generated for paired‐end sequencing.
Construction of C. jejuni PT14 flhF mutants
Wild type flhF and flhF(T368A) genes were PCR amplified from wild type C. jejuni PT14 and C. jejuni PT14CP30ACS respectively using flhF forward and reverse primers containing BsmBI and NgoMIV sites (flhF F, 5′‐CGTCTCACATGGTGATAAGTGGTGTGAGGTG‐3′, and flhF R, 5′‐GCCGGCTCATCTGGCACTTCTTGTCC‐3′). PCR products were cloned into pCR 2.1TOPO vector supplied from Invitrogen to create construct pCR2.1::flhF and pCR2.1::flhF(T368A). Point mutation of flhF(T368A) was confirmed by PCR sequencing using flhF, flhF(T368A) and M13 universal primers, and aligning with the wild type flhF sequence.
To construct a flhF inactive mutant, a flhF cassette was first obtained by digesting pCR2.1::flhF with EcoRI. This cassette was then cloned as an EcoRI‐EcoRI fragment into an EcoRI‐digested pUC4K backbone to replace the kanamycin resistance gene to create pUC4KΔkan::flhF. A kanamycin resistance cassette without a promoter was amplified from the pUC4k vector by PCR using primers containing XbaI sites (Kan F, 5′‐TCTAGAGCAAGGAACAGTGAATTGGAG‐3′, and Kan R, 5′‐TCTAGAGTGCGTAAGAACATAGAAAGG‐3′). The resulting kanamycin cassette was firstly cloned into pCR2.1TOPO, followed by digestion with XbaI to obtain the kanamycin resistance gene as a XbaI‐XbaI fragment and ligating the kanamycin resistance cassette into XbaI digested pUC4KΔkan::flhF to construct pUC4KΔkan::flhF::kan. This plasmid was naturally transformed into C. jejuni PT14 with the aim of targeting cross‐over of the flhF sequences flanking the kanamycin cassette to form C. jejuni PT14flhF::kan.
To create the flhF complementation constructs, BsmBI‐ NgoMIV fragments containing either flhF or flhF(T368A) were obtained by digesting pCR2.1::flhF or pCR2.1::flhF(T368A) with BsmBI and NgoMIV. The flhF and flhF(T368A) BsmBI‐ NgoMIV fragments were then inserted into BsmBI‐ NgoMIV digested pCfdxA::PerR plasmid (Gaskin et al., 2007) to form pCfdxA::flhF and pCfdxA::flhF(T368A) constructs. The constructs were naturally transformed into C. jejuni PT14flhF::kan and targeted to the pseudogene Cj0046 (A911_00230) using flanking homologous sequences within the vector.
Natural transformation
Overnight grown Campylobacter was harvested into 10 ml Müller‐Hinton (MH) broth using sterile cotton swab and an OD600 reading of the suspension was measured. The bacterial concentration was then estimated based on optical density using an empirical equation previously stated by Scott (2006).
Approximately 7.5 log10 CFU Campylobacter cells were dispensed in to the center of a MH blood agar plate and incubated at 42°C under microaerobic conditions for 6 h. A 1 μg aliquot of DNA were dispensed onto the surface of the bacteria halo and allowed to air dry. The plate was then incubated for another 18 h at 42°C under microaerobic conditions. After incubation for three to five days, the transformants were selected using MH blood agar with appropriate antibiotics, and further confirmed by polymerase chain reaction and DNA sequencing.
Motility assay
Motility assay was carried out by stabbing an inoculum of overnight grown C. jejuni into the center of a semi‐solid motility agar plate (0.4% w/v MH agar) using a sterilized pipette tip. The plates were incubated at 42°C under microaerobic conditions for 48 h before the diameter of the motility halo was measured and recorded.
Transmission electron microscopy
C. jejuni was firstly fixed with 3% (v/v) glutaraldehyde in 0.1 M cacodylate buffer, negative stained by uranyl acetate and then examined by TEM. To fix bacteria cells, fixative solution was prepared by adding 2.5 ml of 0.2 M cacodylate buffer, 1.9 ml of distilled water into 600 μl 25% (v/v) EM glutaraldehyde. Overnight grown C. jejuni cells were harvested into 600 μl of fixative solution using a 10 μl of inoculating loop. The C. jejuni pellet was fixed in the fixative solution at room temperature for 1 h and then centrifuged at 10,000 × g for one minute. The supernatant was discarded and 1 ml of 0.1 M cacodylate buffer was added to gently wash the pellet. The pellet was left in 0.1 M cacodylate buffer for 10 min and then centrifuged at 10,000 × g for one minute. The supernatant was removed and the pellet was then re‐suspended gently into 600 μl of 0.1 M cacodylate buffer. A 14 μl aliquot of the fixed C. jejuni suspension was transferred onto formvar/ carbon film on copper 200 mesh grid and left for 30 s. The suspension was removed by lens paper and 14 μl of 0.5% (w/v) uranyl acetate was added onto the grids to negative stain the C. jejuni cells for one minute. After staining, uranyl acetate was removed by lens paper and the grid was ready to be examined by TEM.
Real‐time PCR
C. jejuni PT14, PT14flhF::kan, PT14flhF::kan 00230::flhF‐cat and PT14flhF::kan 00230::flhF(T368A)‐cat were grown on blood agar plates containing appropriate antibiotics at 42°C under microaerobic conditions for 18 h. Bacteria were then harvested into MH broth using a sterile cotton swab and the total RNA was isolated with Trizol reagent. First strand cDNA was subsequently synthesized by protocol modified from Untergasser (2008). Firstly, 20 μl of enzyme mix containing 8 μl of 5x First Strand Buffer, 4 μl of dithiothreitol (DTT), 2 μl of dNTPs (10 mM each), 1 μl of SUPERase inhibitor, 1 μl of SuperScript II and 4 μl of water was prepared and stored at room temperature. Annealing mix was prepared in 20 μl of volume containing 1 μg of total RNA, 25 ng of μl–1 random hexamers and the primers were annealed in a thermocycler at 70°C for 10 min followed by 25°C for 10 min. To which 20 μl of enzyme mix was then added to the reaction and the cDNA was synthesized in a thermocycler at 25°C for 10 min, 37°C for 45 min, 42°C for 45 min and 70°C for 15 min. The cDNA samples were diluted one in five prior to use in real‐time PCR.
To prepare samples for real‐time PCR, PowerUp SYBR Green Master Mix from ThermoFisher Scientific was used according to manufacturer’s protocol. Briefly, samples were prepared in a 96 well micro‐titer plate by mixing 10 μl of PowerUp SYBR Green Master Mix with 10 pmol of gene specific forward, reverse primer, and 2 μl of cDNA template in a total volume of 20 μl. Then real‐time PCR was carried out using a Light Cycler 480 instrument from Roche with settings of pre‐incubation at 95°C for 6 min; followed by 40 cycles of amplification including incubation at 95°C for 30 s, 58°C for 30 s and 72°C for one minute; and then one cycle of melting curve including incubation at 45°C for 5 s and 65°C for one minute; and the run was finished by cooling at 40°C for 30 s. The primer pairs used in real‐time PCR analysis were flhF F, 5′‐CCGTTGAAGATACAGAACAAAT‐3′, and flhF R, 5′‐GGCTACCATAACCTCATAAAG‐3′; flaA F, 5′‐CAGCTGAGTCACAAATCCGT‐3′, and flaA R, 5′‐CCATGGCATAAGAGCCACTT‐3′; flaB F, 5′‐GTTAAAGCAGCAGAATCAACCA‐3′, and flaB R, 5′‐ACTCATAGCATAAGAACCTGACTG‐3′; flgD F, 5′‐AATGGCTGGACAAGAAGTTCC‐3′, and flgD R, 5′‐CTCCATCGCTTGAACCACCA‐3′; pflA F, 5′‐TGCCTTATGTTGGAGCTTTGG‐3′, and pflA R, 5′‐TGTGCATCAATCACCACTTGA‐3′; and flhG F, 5′‐AGCGCGAATCTAGCCAATGT‐3′, flhG R, 5′‐AAGGAGCATTCTCCGCGTAA‐3′, pgk F, 5′‐TAGACGCATAAGATCAGCTATTCC‐3′ and pgk R, 5′‐ AAGTCTAGCAAGACGCTTAGC‐3′.
Efficiency of plating
Bacteria were overnight grown on BA plates and subsequently harvested into 10 ml of 10 mM MgSO4 solution using a sterile cotton swab. From this suspension, 500 μl of aliquots were mixed with 5 ml of aliquots of NZCYM top agar tempered at 55°C and then poured onto a pre‐dried NZCYM plate. The plate was allowed to dry at room temperature and then incubated at 42°C for 30 min. After incubation, 10 μl of decimally diluted bacteriophage samples were spotted onto the bacterial lawn in triplicate and the spots were allowed to dry at room temperature. The plate was further incubated at 42°C under microaerobic conditions for 18 h. The plaques for each spot were measured and the plaque forming units per ml was then calculated.
Enumeration of C. jejuni and bacteriophage
C. jejuni sample was ten‐fold serial diluted with MH broth and 10 μl of each dilution was spotted in triplicate onto a CCDA plate with 2% (w/v) agar to reduce swarming. After 48 h incubation at 42°C under microaerobic conditions, C. jejuni colonies were counted. To enumerate bacteriophage, the phage sample was ten‐fold serial diluted. A bacterial lawn of C. jejuni PT14 was prepared and 10 μl of each dilution phage sample was spotted onto the lawn in triplicate. Plates were then incubated at 42°C under microaerobic conditions for 48 h and plaques were counted.
Growth characteristics of C. jejuni PT14 and its flhF mutants against bacteriophage CP30A
Overnight cultured C. jejuni PT14, PT14flhF::kan, PT14flhF 00230::flhF‐cat and PT14flhF 00230::flhF(T368A)‐cat mutants were transferred into 100 ml of MH broth with appropriate antibiotics to give a final concentration of approximately 4 log10 CFU ml–1 and incubated at 42°C under microaerobic conditions with 150 r.p.m. shaking for 2 h. After incubation, bacteriophage CP30A was applied into each C. jejuni culture to give a final concentration of 102 PFU ml–1 and the cultures were carried on incubating for another 24 h. Sample aliquots were removed every 2 h for C. jejuni and bacteriophage enumeration.
One step growth curve for bacteriophage CP30A against C. jejuni PT14 and its flhF mutant derivatives
Overnight cultured C. jejuni PT14, PT14flhF::kan, PT14flhF 00230::flhF‐cat and PT14flhF 00230::flhF(T368A)‐cat mutants were transferred into 100 ml of MH broth with appropriate antibiotics to give a final concentration of approximately 7 log10 CFU ml–1 and incubated at 42°C under microaerobic conditions with 150 r.p.m. shaking for 2 h. The viable count was then measured after incubation as described above. For the experiment, bacteriophage CP30A was diluted and added to bacterial suspension at the titer of 106 PFU ml–1. The bacteria/ phage mix was further incubated at 42°C under microaerobic conditions with 150 r.p.m. shaking for 3 h and aliquoted samples were taken every 15 min. Aliquots were centrifuged at 13,000 × g for 5 min and the supernatant containing free phages were removed for enumeration. The adsorption constant was calculated using equation k = –ln (P t/P 0)/N t, where P t = phage titer at time point t (PFU ml–1), P 0 = initial phage titer (PFU ml–1), N = bacterial viable count (CFU ml–1) and t = time (min).
FlhF protein purification
Both flhF wild type and flhF(T368A) genes were PCR amplified including their promoter using flhF his6‐tag forward and reverse primer respectively containing in‐frame 5′ NdeI and BamHI site (flhF His‐tag F, 5′‐AACATATGGGACAACTTATACATACTT‐3′, and flhF His‐tag R, 5′‐AAGGATCCATTGCGAAGTTTATTTGCTTGG‐3′). The flhF PCR products were cloned into pCR 2.1 TOPO vector and the flhF(T368A) point mutation was confirmed by PCR sequencing. The flhF wild type and flhF(T368A) fragments were obtained by digesting the plasmids with NdeI‐BamHI and then ligated into a NdeI‐BamHI digested pET28b vector backbone to create N‐terminal His6‐tagged fusion. The pET28b construct containing wild type flhF was transformed into chemically competent E. coli BL21DE3 Star cells. The pET28b construct containing the His6‐tagged FlhF(T368A) gene was transformed into Origami2 DE3 pLysS cells to overcome inclusion body formation. To express the protein, a fresh colony harboring the pET28bflhF plasmid was cultured in 10 ml of LB broth at 37°C with 150 r.p.m. shaking for 18 h with the presence of kanamycin and chloramphenicol. After incubation, overnight grown culture was diluted 1:50 in one liter LB broth containing kanamycin, and chloramphenicol. The culture was incubated at 37°C with 150 r.p.m. shaking until OD600 reached 0.6 and then induced with isopropyl‐β‐D‐thiogalacto‐pyranoside (IPTG) at a final concentration of 0.4 mM at 37°C for 4 h. The bacteria cells were pelleted by centrifuging at 4,000 × g for 15 min at 4°C and then purified using a HisTrap HP column, supplied from GE Healthcare Life Sciences. Briefly, a pellet derived from one liter culture volume was resuspended in 10 ml of lysis buffer (50 mM Tris‐HCl, 150 mM NaCl, 20 mM imidazole, pH 7.2) containing 1 ml of lysozyme stock solution (10 mg ml–1) and 300 units of Benzonase Endonuclease supplied from Sigma‐Aldrich. The suspension was incubated on ice for 30 min and centrifuged at 12,000 × g for 30 min and 4°C. The clear lysate was collected and applied to HisTrap HP column, which was pre‐equilibrated with five column volumes of water and five column volumes of binding buffer (50 mM Tris‐HCl, 150 mM NaCl, 20 mM imidazole, pH 7.2). The column was then washed with 15 column volumes of binding buffer and eventually eluted with elution buffer (50 mM Tris‐HCl, 150 mM NaCl, 250 mM imidazole, pH 7.2) for five column volumes. After this, 500 μl of this protein was size fractionated into 24 fractions using Superose 12 chromatography (GE healthcare Life Sciences). Purified protein was analyzed by 12% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) against Precision Plus Protein Dual Color Standards (Bio‐rad) and protein concentration was measured by Bradford assay.
GTP hydrolysis assay
The GTP hydrolysis assay was carried out using a High Throughput Colorimetric GTPase Assay Kit from Innova Biosciencs. Prior to the GTP hydrolysis assay, a standard curve was prepared to convert the absorbance reading into released P i from hydrolysis reaction. A set of P i standards was prepared to give a final concentration of 1, 2, 5, 7.5, 10, 20, 50 and 75 μM. Triplicate samples were prepared for each P i concentration by mixing 200 μl of each standard with 50 μl of PiColorLock in a 96‐well microtiter plate. After two minutes, 20 μl of stabilizer was added and mixed thoroughly. After 30 min, the absorbance at wavelength of 612 nm was measured and the standard curve of absorbance against P i concentration constructed. For the GTP hydrolysis assay, the substrate buffer mix was first prepared in 100 μl of volume containing 100 mM Tris buffer, 5 mM MgCl2 and a fixed concentration of GTP. For the temperature assay, 0.25 mM of GTP was applied as a standard substrate concentration. The reactions were initiated by rapid mixing of 100 μl of enzyme with 100 μl of substrate buffer mix and incubated immediately at 4, 16, 25, 37, 42, 50, 60, 70 and 80°C for 30 min. A 50 μl of aliquot of PiColorLock solution containing 0.5 μl of Accelerator solution was added to stop the enzyme reaction. After two minutes, 20 μl of stabilizer was added and mixed thoroughly to prevent color development from background P i. The color reached a maximum signal after 30 min incubation at room temperature and the absorbance of the sample was then determined at a wavelength of 612 nm in a Tecan plate reader. Pre‐heated samples were measured to determine the thermal stability of the enzyme activities. In this experiment, protein samples were heated at different temperatures for 5 min prior to mixing with substrate buffer. For substrate pre‐incubation 0.2 mM GTP was added to the incubation mixes at 42°C, and either stopped as a pre‐reaction control or diluted into the GTPase assay containing 0.5 mM GTP. The contribution of GTP self‐hydrolysis was monitored by control incubations without protein and the corrected enzymatic activities recorded. For kinetic assays, 0.05, 0.1, 0.25, 0.5 and 1 mM of GTP was used as substrate concentrations, and after 0, 2, 4, 6, 8 and 10 min the enzyme reaction at 42°C were stopped before the absorbance was measured. Released P i was subsequently determined from OD612 readings against a standard curve. The elution buffer for the His6‐tag protein purification was applied as negative control in this enzymatic assay.
Author contributions
LL carried out the experiments. LL and IC designed the study, interpreted the data and wrote the manuscript.
Supporting information
Acknowledgement
This work was supported by the Biotechnology and Biological Sciences Research Council [grant numbers BB/I024682/1; BB/P02355X/1], UK.
References
- Abedon, S. (2009) Disambiguating bacteriophage pseudolysogeny: an historical analysis of lysogeny, pseudolysogeny and the phage carrier state In: Adams H.T. (Ed.) Contemporary Trends in Bacteriophage Research. New York: Nova Science Publishers, pp. 285–307. [Google Scholar]
- Balaban, M. and Hendrixson, D.R. (2011) Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni . PLoS Pathogens, 7, e1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban, M. , Joslin, S.N. and Hendrixson, D.R. (2009) FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni . Journal of Bacteriology, 191, 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldvinsson, S.B. , Sørensen, M.C. , Vegge, C.S. , Clokie, M.R. and Brøndsted, L. (2014) Campylobacter jejuni motility is required for infection of the flagellotropic bacteriophage F341. Applied and Environmental Microbiology, 80, 7096–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange, G. , Petzold, G. , Wild, K. , Parlitz, R.O. and Shinning, I. (2007) The crystal structure of the third signal‐recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proceedings of the National Academy of Sciences of the United States of America, 104, 13621–13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, J.T. , Hugdahl, M.B. and Doyle, M.P. (1988) Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni . Applied and Environmental Microbiology, 54, 2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, E.F. and Brussow, H. (2002) Common themes among bacteriophage‐encoded virulence factors and diversity among the bacteriophages involved. Trends in Microbiology, 10, 521–529. [DOI] [PubMed] [Google Scholar]
- Brathwaite, K.J. , Siringan, P. , Moreton, J. , Wilson, R. and Connerton, I. (2013) Complete genome sequence of universal bacteriophage host strain Campylobacter jejuni subsp jejuni PT14. Genome Announcements, 6, e00969–e00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brathwaite, K.J. , Siringan, P. , Connerton, P.L. and Connerton, I. (2015) Host adaption to the bacteriophage carrier state of Campylobacter jejuni . Research in Microbiology, 166, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyha, I. , Hot, E. , Huntley, S. and Sogaard‐Andersen, L. (2011) GTPases in bacterial cell polarity and signalling. Current Opinion in Microbiology, 14, 726–733. [DOI] [PubMed] [Google Scholar]
- Cairns, B.J. , Timms, A.R. , Jansen, V.A. , Connerton, I.F. and Payne, R.J. (2009) Quantitative models of in vitro bacteriophage‐host dynamics and their application to phage therapy. PLoS Pathogens, 5, e1000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban, B. , Coleman, I. and Beeby, M. (2018) Evolution of higher torque in Campylobacter‐type bacterial flagellar motors. Scientific Reports, 8, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance, F.F. and Hughes, K.T. (2008) Coordinating assembly of a bacterial macromolecular machine. Nature Reviews Microbiology, 6, 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y. , Shin, H. , Lee, J. and Ryu, S. (2013) Identification and characterization of a novel flagellum‐dependent Salmonella‐infecting bacteriophage, iEPS5. Applied and Environmental Microbiology, 79, 4829–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerton, P.L. , Loc Carrillo, C. , Swift, C. , Dillon, E. , Scott, A. , Rees, C.E. et al (2004) Longitudinal study of Campylobacter jejuni bacteriophages and their hosts from broiler chickens. Applied and Environmental Microbiology, 70, 3877–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerton, P.L. , Timms, A.R. and Connerton, I.F. (2011) Campylobacter bacteriophages and bacteriophage therapy. Journal of Applied Microbiology, 111, 255–265. [DOI] [PubMed] [Google Scholar]
- Correa, N.E. , Peng, F. and Klose, K.E. (2005) Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. Journal of Bacteriology, 187, 6324–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward, C. , Grant, A.J. , Swift, C. , Philp, J. , Towler, R. , Heydarian, M. et al (2006) Phase‐variable surface structures are required for infection of Campylobacter jejuni by bacteriophages. Applied and Environmental Microbiology, 72, 4638–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. (2002) Pymol: An open‐source molecular graphics tool. CCP4 Newsletter. On Protein Crystallography, 40, 82–92. [Google Scholar]
- Eichler, J. and Moll, R. (2001) The signal recognition particle of Archaea. Trends in Microbiology, 9, 130–136. [DOI] [PubMed] [Google Scholar]
- Firlieyanti, A.S. , Connerton, P.L. and Connerton, I. (2016) Campylobacters and their bacteriophages from chicken liver: the prospect for phage biocontrol. International Journal of Food Microbiology, 237, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, B. , Lara‐Tejero, M. , Lefebre, M. , Goodman, A.L. and Galán, J.E. (2014) Novel components of the flagellar system in epsilonproteobacteria. mBio, 5, e01349–01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin, D.J. , Van Vliet, A.H. and Pearson, B.M. (2007) The Campylobacter genetic toolbox: development of tractable and generally applicable genetic techniques for Campylobacter jejuni . Zoonoses and Public Health, 54, 101. [Google Scholar]
- Gilbreath, J.J. , Cody, W.L. , Merrell, D.S. and Hendrixson, D.R. (2011) Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter . Microbiology and Molecular Biology Reviews, 75, 84–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, J.C. , Kahramanoglou, C. , Rahman, A. , Pender, A.M. , Charbonnel, N. and Fraser, G.M. (2009) Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane‐associated signal recognition particle family GTP‐binding protein. Journal of Molecular Biology, 391, 679–690. [DOI] [PubMed] [Google Scholar]
- Guerry, P. , Alm, R.A. , Power, M.E. , Logan, S.M. and Trust, T.J. (1991) Role of two flagellin genes in Campylobacter motility. Journal of Bacteriology, 173, 4757–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbronson, C.J. , Ribardo, D.A. , Balaban, M. , Knauer, C. , Bange, G. and Hendrixson, D.R. (2016) FlhG employs diverse intrinsic domains and influences FlhF GTPase activity to numerically regulate polar flag. Molecular Microbiology, 99, 291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic, M. , Blau, M. , Becker, T. , Mielke, T. , Pool, M.R. , Wild, K. et al (2006) Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature, 444, 507–511. [DOI] [PubMed] [Google Scholar]
- Hammerl, J.A. , Jackel, C. , Alter, T. , Janzcyk, P. , Stingl, K. , Knuver, M.T. et al (2014) Reduction of Campylobacter jejuni in broiler chicken by successive application of group II and group III phages. PLoS One, 9, e114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson, D.R. (2008) Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni . Molecular Microbiology, 70, 519–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson, D.R. and Dirita, V.J. (2003) Transcription of σ54 ‐dependent but not σ28 ‐dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Molecular Microbiology, 50, 687–702. [DOI] [PubMed] [Google Scholar]
- Hendrixson, D.R. and DiRita, V.J. (2004) Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Molecular Microbiology, 52, 471–484. [DOI] [PubMed] [Google Scholar]
- Hooton, S.P.T. , Brathwaite, K.J. and Connerton, I.F. (2016) The bacteriophage carrier state of Campylobacter jejuni features changes in host non‐coding RNAs and the acquisition of new host‐derived CRISPR spacer sequences. Frontiers in Microbiology, 7, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton, S.P.T. and Connerton, I.F. (2015) Campylobacter jejuni acquire host‐derived CRISPR spacers when in changes in association with bacteriophages harboring CRISPR‐like Cas4 protein. Frontiers in Microbiology, 5, 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed, M.A. , Ackermann, H.W. , Azeredo, J. , Carvalho, C.M. , Connerton, I. , Evoy, S. , et al (2014) A suggested classification for two groups of Campylobacter myoviruses. Archives of Virology, 159, 181–190. [DOI] [PubMed] [Google Scholar]
- Kittler, S. , Fischer, S. , Abdulmawjood, A. , Glunder, G. and Klein, G. (2013) Effect of bacteriophage application on Campylobacter jejuni loads in commercial broiler flocks. Applied and Environmental Microbiology, 79, 7525–7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe, D.D. , Wolf, Y.I. , Koonin, E.V. and Aravind, L. (2002) Classification and evolution of P‐loop GTPases and related ATPases. Journal of Molecular Biology, 317, 41–72. [DOI] [PubMed] [Google Scholar]
- Lertsethtakarn, P. , Ottemann, K.M. and Hendrixson, D.R. (2011) Motility and chemotaxis in Campylobacter and Helicobacter . Annual Review of Microbiology, 65, 389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis, L. and Connerton, I.F. (2016) The minor flagellin of Campylobacter jejuni (FlaB) confers defensive properties against bacteriophage infection. Frontiers in Microbiology, 7, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc Carrillo, C. , Atterbury, R.J. , El‐Shibiny, A. , Connerton, P.L. , Dillon, E. , Scott, A. et al (2005) Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Applied and Environmental Microbiology, 71, 6554–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins, S. , Allan, B.J. , Townsend, H.G. , Koster, W. and Potter, A.A. (2013) Role of motAB in adherence and internalization in polarized Caco‐2 cells and in cecal colonization of Campylobacter jejuni . Avian Diseases, 57, 116–122. [DOI] [PubMed] [Google Scholar]
- Nachamkin, I. , Yang, X.H. and Stern, N.J. (1993) Role of Campylobacter jejuni flagella as colonization factors for three‐ day‐old chicks: Analysis with flagellar mutants. Applied and Environmental Microbiology, 59, 1269–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell, D.G. , Elvers, K.T. , Dopfer, D. , Hansson, I. , Jones, P. , James, S. et al (2011) Biosecurity‐based interventions and strategies to reduce Campylobacter spp. on poultry farms. Applied and Environmental Microbiology, 77, 8605–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhuba, D.V. , Kolomiets, E.I. , Dey, E.S. and Novik, G.I. (2010) Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Journal of Molecular Biology, 59, 145–155. [PubMed] [Google Scholar]
- Ren, F. , Lei, T. , Song, Z. , Yu, T. , Li, Q. , Huang, J. et al (2018) Could FlhF be a key element that controls Campylobacter jejuni flagella biosynthesis in the initial assembly stage? Microbiological Research, 207, 240–248. [DOI] [PubMed] [Google Scholar]
- Sails, A.D. , Wareing, D.R. , Bolton, F.J. , Fox, A.J. and Curry, A. (1998) Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. Journal of Medical Microbiology, 47, 123–128. [DOI] [PubMed] [Google Scholar]
- Schirmer, M. , D’Amore, R. , Ijaz, U.Z. , Hall, N. and Quince, C. (2016) Illumina error profiles: resolving fine‐scale variation in metagenomic sequencing data. BMC Bioinformatics, 17, 125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher, J.S. , Thormann, K.M. and Bange, G. (2015) How bacteria maintain location and number of flagella? FEMS Microbiology Reviews, 39, 812–822. [DOI] [PubMed] [Google Scholar]
- Scott, A. (2006) Genome changes associated with bacteriophage resistance in Campylobacter. PhD Thesis, University of Nottingham, Nottingham. [Google Scholar]
- Scott, A. , Timms, A.R. , Connerton, P.L. , Loc Carrillo, C. , Radzum, K.A. and Connerton, I.F. (2007a) Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLOS Pathogens, 3, 1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A. , Timms, A.R. , Connerton, P.L. , El‐Shibiny, A. and Connerton, I. (2007b) Bacteriophage influence Campylobacter jejuni types populating broiler chickens. Environmental Microbiology, 9, 2341–2353. [DOI] [PubMed] [Google Scholar]
- Siringan, P. , Connerton, P.L. , Cummings, N.J. and Connerton, I.F. (2014) Alternative bacteriophage life cycles: the carrier state of Campylobacter jejuni . Open Biology, 4, 130200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siringan, P. , Connerton, P.L. , Payne, R.J. and Connerton, I.F. (2011) Bacteriophage‐mediated dispersal of Campylobacter jejuni biofilms. Applied and Environmental Microbiology, 77, 3320–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, M.C.H. , Gencay, Y.E. , Birk, T. , Baldvinsson, S.B. , Jäckel, C. , Hammerl, J.A. et al (2015) Primary isolation strain determines both phage type and receptors recognised by Campylobacter jejuni bacteriophage. PLoS One, 10, e0116287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen, M.C. , van Alphen, L.B. , Harboe, A. , Li, J. , Christensen, B.B. , Szymanski, C.M. , et al (2011) Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. Journal of Bacteriology, 193, 6742–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser, A. (2008) cDNA synthesis using superscript II. Available at: https://www.untergasser.de/lab/protocols/cdna_synthesis_superscript_ii_v1_0.htm [Google Scholar]
- Wassenaar, T.M. , van der Zeijst, B.A. , Ayling, R. and Newell, D.G. (1993) Colonization of chicks by motility mutantas of Campylobacter jejuni demonstrates the importance of flagellin A expression. Journal of general microbiology, 139, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Waterhouse, A. , Bertoni, M. , Bienert, S. , Studer, G. , Tauriello, G. , Gumienny, R. , et al (2018) SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten, M.M. , vanDijk, L. , Veenendaal, A.K. , deZoete, M.R. , Bleumink‐Pluijm, N.M. , and vanPutten, J.P. (2010) Temperature‐dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Molecular Microbiology, 75, 1577–1591. [DOI] [PubMed] [Google Scholar]
- Zanen, G. , Antelmann, H. , Westers, H. , Hecker, M. , van Dijl, J.M. and Quax, W.J. (2004) FlhF, the third signal recognition particle‐GTPase of Bacillus subtilis, is dispensable for protein secretion. Journal of Bacteriology, 186, 5956–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


