Abstract
Lycopene attracts increasing interests in the pharmaceutical, food, and cosmetic industries due to its anti-oxidative and anti-cancer properties. Compared with other lycopene production methods, such as chemical synthesis or direct extraction from plants, the biosynthesis approach using microbes is more economical and sustainable. In this work, we engineered Haloferax mediterranei, a halophilic archaeon, as a new lycopene producer. H. mediterranei has the de novo synthetic pathway for lycopene but cannot accumulate this compound. To address this issue, we reinforced the lycopene synthesis pathway, blocked its flux to other carotenoids and disrupted its competitive pathways. The reaction from geranylgeranyl-PP to phytoene catalyzed by phytoene synthase (CrtB) was identified as the rate-limiting step in H. mediterranei. Insertion of a strong promoter PphaR immediately upstream of the crtB gene, or overexpression of the heterologous CrtB and phytoene desaturase (CrtI) led to a higher yield of lycopene. In addition, blocking bacterioruberin biosynthesis increased the purity and yield of lycopene. Knock-out of the key genes, responsible for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) biosynthesis, diverted more carbon flux into lycopene synthesis, and thus further enhanced lycopene production. The metabolic engineered H. mediterranei strain produced lycopene at 119.25 ± 0.55 mg per gram of dry cell weight in shake flask fermentation. The obtained yield was superior compared to the lycopene production observed in most of the engineered Escherichia coli or yeast even when they were cultivated in pilot scale bioreactors. Collectively, this work offers insights into the mechanism involved in carotenoid biosynthesis in haloarchaea and demonstrates the potential of using haloarchaea for the production of lycopene or other carotenoids.
Keywords: lycopene, biosynthesis, Haloferax mediterranei, rate-limiting steps, phytoene synthase, phytoene desaturase, bacterioruberin, PHBV
Introduction
Lycopene is a C40 isoprenoid compound in the carotenoid family. Due to its anti-oxidative and anti-cancer activities (Sies and Stahl, 1998; Gajowik and Dobrzynska, 2014), lycopene has been widely used for nutritional supplements, pharmaceutical and cosmetic products (Wei et al., 2017). The conventional methods for lycopene production include direct extraction from plants,chemical synthesis and microbial fermentation. Among these methods, microbial production of lycopene is more economical and sustainable (Chen et al., 2016). Recently, with the development of metabolic engineering techniques and synthetic biology, lycopene overproduction has been realized in Escherichia coli (Zhu et al., 2015; Wei et al., 2017; Wu et al., 2018; Xu et al., 2018), yeast (Xie et al., 2015a; Schwartz et al., 2017), Blakeslea trispora (Liu et al., 2012; Wang et al., 2016, 2017), and Rhodobacter sphaeroides (Su et al., 2018). However, the field is seeking a better platform for large-scale production of lycopene or other carotenoids. Halophilic archaea (haloarchaea) belong to the domain Archaea and are unique microorganisms that survive under the high salt condition (Singh and Singh, 2017). Many haloarchaeal species are capable of producing the compounds of the carotenoid family (Rodrigo-Banos et al., 2015), such as phytoene, β-carotene, lycopene, as well as the derivatives of bacterioruberin and salinixanthin (de Lourdes Moreno et al., 2012). Particularly, they hold several advantages for carotenoid production: the high-salt tolerance enables haloarchaea cultivation under non-sterile condition and thus reduces the energy cost (Singh and Singh, 2017). Additionally, the process of carotenoid extraction from haloarchaea is relatively simple, as the cell lysis undergoes in low sodium chloride (NaCl) condition. Consequently, haloarchaea are considered as an alternative producer for carotenoids (Naziri et al., 2014).
Haloferax mediterranei can use probably the largest range of single carbon sources and grows faster than other known members of the Halobacteriaceae (Oren and Hallsworth, 2014). Its complete genome information is available (Han et al., 2012), and the pyrF-based gene knockout system for genome-wide manipulation has also been well-established in this strain (Liu et al., 2011). With these merits, H. mediterranei has been one of the most common model strains for the study of physiology and metabolism in archaea. For example, it has been used to investigate poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) biosynthesis and its metabolism regulation processes (Han et al., 2013, 2017; Zhao et al., 2013; Bhattacharyya et al., 2014; Cai et al., 2015). However, only a few studies on its carotenoid production has been reported till recently. Fang et al. (2010) improved the C50 carotenoid production to 0.604 A494 nm/mL broth via a two-stage cultivation approach. Chen et al. (2015) used extruded rice bran and starch under optimal conductivity of brined medium for a high red pigment production of 556 mg/L. However, there is no work investigating either the production of other carotenoids (e.g., lycopene), or the pathway engineering to improve carotenoid production in H. mediterranei.
In this study, we explored the possibility to use H. mediterranei as a potential cell factory for lycopene production by multiple strategies (Figure 1). First, we attempted to identify and eliminate the rate-limiting steps involved in lycopene biosynthesis. Then we disrupted bacterioruberin synthesis to increase lycopene accumulation and purity. Heterologous phytoene synthase (CrtB) and phytoene desaturase (CrtI) encoding genes from other haloarchaea were employed for further enhancing lycopene production. Subsequently, we blocked PHBV synthesis to divert more acetyl-CoA flux to lycopene synthesis and also complemented the pyrF gene in the engineered strain for its future application in industrial scale. We finally obtained a metabolic engineered H. mediterranei strain with relatively high purity and production of lycopene.
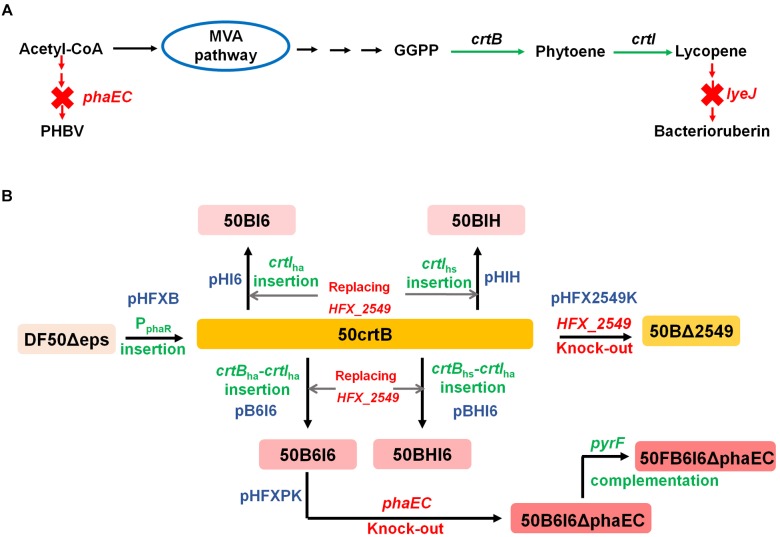
FIGURE 1.
Schematic illustration of engineered H. mediterranei for lycopene production. (A) Enhancement of lycopene synthesis by enhancing lycopene biosynthetic pathway and blocking its flux to bacterioruberin and its competitive pathway for PHBV biosynthesis. Green arrows represent enhanced steps and red arrows and crosses represent blocked steps. (B) Flowchart of engineering H. mediterranei strain for increasing lycopene synthesis. Green terms represent gene insertion and red ones represent gene deletion. The blue terms show the plasmids used for constructing the mutants of H. mediterranei.
Materials and Methods
Strains, Medium, and Culture Conditions
All the strains used in this study are listed in Supplementary Table S1. E. coli JM109 (Sambrook, 1989) was used for plasmids construction and E. coli JM110 was used to eliminate the methylated plasmid in vivo (Palmer and Marinus, 1994). Luria Broth (LB) medium was used for E. coli culture at 37°C. When needed, 100 μg/mL of ampicillin was added to LB medium. H. mediterranei was cultivated at 37°C in nutrient-rich AS-168 medium (per liter, 5 g casamino acids, 5 g yeast extract, 1 g sodium glutamate, 3 g trisodium citrate, 2 g KCl, 20 g MgSO4 ⋅ 7H2O, 200 g NaCl, 5 mg FeSO4 ⋅ 7H2O, and 0.036 mg MnCl2 ⋅ 4H2O [pH 7.0]). AS-168SY medium was similar to AS-168 medium, except that yeast extract was excluded. Plasmids were transformed into H. mediterranei with the polyethylene glycol-mediated transformation method (Cline et al., 1989). When required, AS-168 medium was supplemented with 50 mg/mL uracil (Sangon, China) and 250 mg/mL 5-Fluoroorotic acid (Sangon, China) for counter-selection of the recombinants without pyrF marker. For lycopene production, a 1% (V/V) seed culture of H. mediterranei or its mutants was inoculated into a shake flask containing 50 mL of MG medium (per liter, 110 g NaCl, 20.51 g MgCl2, 29.52 g MgSO4, 5 g KCl, 1 g CaCl2, 2 g NH4Cl, 0.0375 g KH2PO4, 10 g glucose, 15 g PIPES, Fe(III) citrate, and 1 mL trace element solution SL-6 [pH 7.2]) (Antón et al., 1988) and cultured at 37°C and 200 rpm for 7 days.
Plasmid Construction for Gene Overexpression
The native candidate genes involved in lycopene synthesis were amplified with primers listed in Supplementary Table S2 from the H. mediterranei genomic DNA. The heterologous crt genes HAH_2563 (crtBha), HAH_1058 (crtIha) and OE_3093R (crtBhs), OE_3381R (crtIhs) were obtained via PCR from the genomic DNA of Haloarcula hispanica and Halobacterium salinarum, respectively. Amplified fragments were inserted into pWLR [derived from pWL502 by insertion of a strong promoter PphaR (Cai et al., 2012)] digested with BamHI and XbaI, by using One Step Cloning Kit (Yeasen, Co., Ltd., China), to generate plasmids for gene overexpression under the control of promoter PphaR (Supplementary Table S1).
Plasmid Construction for Gene Integration in Chromosome
All plasmids for gene knock-in or knock-out were constructed based on a suicide plasmid pHFX (Liu et al., 2011). A 583-bp DNA fragment located immediately upstream of crtBhm was amplified with primer pair crtB-in-1/crtB-in-2 from H. mediterranei genomic DNA. Another 580-bp fragment containing promoter PphaR and the partial 5′ region of crtB was amplified with primer pair crtB-in-3/crtB-in-4 from plasmid pW2547 used for crtBhm overexpression (Supplementary Table S1). Then, the two PCR products were inserted into the plasmid pHFX to construct the integration plasmid of pHFXB, which was used to replace the native promoter of HFX_2547 (crtBhm) in chromosome. For the heterologous crt gene integration, a 524-bp fragment up-stream of HFX_2549 and a 529-bp fragment down-stream of HFX_2549 were amplified by relevant primer pairs (Supplementary Table S2). Different crt gene fragments containing PphaR were amplified by using crtBha, crtIha, crtBhs, and crtIhs overexpression plasmids pWHA2563, pWHA1058, pWOE3093, and pWOE3381 (Supplementary Table S1) as PCR template, respectively. The corresponding homologous arm fragments and different crt genes, containing promoter PphaR, were assembled into pHFX to construct plasmids containing different crt genes used for their integration in chromosome (Supplementary Figure S1 and Supplementary Table S1). Similar to the plasmid construction method as described above, primer pairs HFX_2549-K1/HFX_2549-K2 and HFX_2549-K3/HFX_2549-K4 were used to construct the plasmid pHFX2549K for HFX_2549 knock-out. Primer pairs phaEC-K1/phaEC-K1 and phaEC-K3/phaEC-K4 were used for the construction of pHFXPK for phaEC knock-out (Supplementary Table S1).
Analysis of Carotenoids by Thin Layer Chromatography (TLC)
Carotenoids in acetone extract obtained from different cultures were analyzed by thin layer chromatography (TLC), following the protocol described by Strand et al. (1997) with slight modifications. Briefly, after cultivation in MG medium for 7 days, the cells (1 mL) were harvested by centrifugation (12,000 × g for 5 min, at 4°C), and resuspended in acetone (1 mL) under a reduced light condition to prevent photo-bleaching and degradation (Alper and Stephanopoulos, 2008). The acetone supernatant containing carotenoids was collected and transferred to a new tube. This process was repeated until the pellets were totally white. Acetone extracted carotenoids (10 μL) were analyzed by TLC on a silica plate (GF254, Qingdao Haiyang Chemical, Co., Ltd., China) with acetone and n-heptane (1:1, v/v) as the development liquid in fume hood at room temperature. In addition, the visualized spot on the resulting TLC plate was scraped off, and extracted with 200 μL of acetone. The obtained supernatant was then scanned under 350–550 nm.
Lycopene Quantification
The lycopene content in the extract was determined by using a HPLC system (Agilent, 1260, United States) equipped with a ZORBAX Eclipse XDB-C18 column (4.6 mm × 150 mm, 5 μm) and a UV/VIS detector. The absorption was detected at 450 nm. The mobile phase consisted of methanol-isopropanol (65:35 V/V) with a flow rate of 1 mL/min at 30°C. Injection volume of sample was 20 μL. The lycopene concentration was calculated based on the calibration curve of lycopene (Macklin Biochemical, Co., Ltd., China).
PHA Content Analysis
The cells were collected by centrifugation at 10,000 × g, 4°C, 15 min and lyophilized. The lyophilized cells were treated with a mixture of chloroform and methanol containing 3% (v/v) sulfuric acid at 95°C for 4 h. The resulting hydroxyacyl methylesters were then analyzed by GC-6820 instrument (Agilent, United States) as described by Han et al. (2007).
RNA Extraction and Quantitative Reverse Transcription-PCR (qRT-PCR)
The cells were cultured in AS-168 medium at 37°C for 12 h and subsequently harvested by centrifugation (12,000 × g, 4°C). The total RNA was extracted using TRIzol reagent (Invitrogen, United States) as previously described (Lu et al., 2008a). TURBO DNA-freeTM Kit (Thermo Fisher Scientific, United States) was used for removing DNA contamination. The cDNA was synthesized by reverse transcription with random hexamer primers from 1 μg of DNA-free total RNA using the Moloney Murine Leukemia Virus Reverse Transcriptase (M MLV-RT) (Promega, United States). The relative fold of gene expression was analyzed by ViiATM 7 Real-Time PCR System (Applied Biosystems, Inc., United States), using 7S RNA as an endogenous control to normalize the data of each sample. The primers used are listed in Supplementary Table S2.
Sequences Analysis and Databases
The DNA sequences were obtained from National Center for Biotechnology Information (NCBI) Genome Database. The information about the most identified enzymes involved in MVA and lycopene synthesis pathway (supported by evidence at protein level), was accessed from UniProt Database1. Sequence homology was assessed by BLASTN or BLASTP in NCBI (Altschul et al., 1990). Predictions of transmembrane helices in the proteins were performed by using the TMHMM Server v2.02 (Krogh et al., 2001).
The genome accession numbers deposited in GenBank are as following, H. mediterranei (CP001868.2), H. hispanica (NC_015948.1), and H. salinarum (AM774415.1).
Statistical Analysis
Experiments were performed in triplicate and data was analyzed by the GraphPad Prism 7 software and represented as mean ± standard deviation. Statistical analysis was done using a two-tailed t-test. Statistical significance was defined as ∗p < 0.05.
Results
Identifying the Rate-Limiting Steps Involved in Native Lycopene Biosynthesis
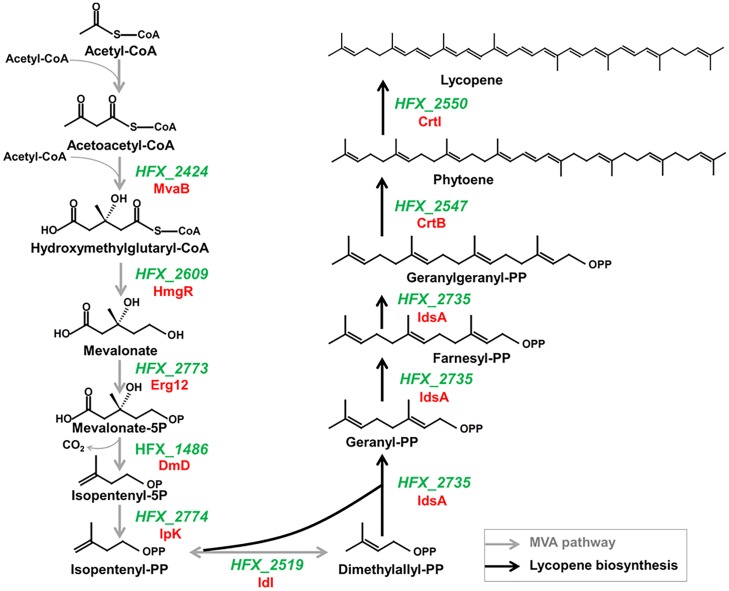
In silico metabolic pathway analysis reveals that H. mediterranei has a complete lycopene biosynthetic pathway, referring to the steps from isopentenyl-PP (IPP) and dimethylallyl-PP (DMAPP) to lycopene (Figure 2). Mevalonate (MVA) pathway provides the two important precursors, IPP and DMAPP, for lycopene synthesis. In MVA pathway, two molecules of acetyl-CoA are condensed to form acetoacetyl-CoA and a third acetyl-CoA molecule is then added to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) by Hydroxymethylglutaryl-CoA synthase (MvaB, HFX_2424). The next step involves the conversion of HMG-CoA to MVA by Hydroxymethylglutaryl-CoA reductase (HmgR, HFX_2609). MVA is then phosphorylated by Mevalonate kinase (Erg12, HFX_2773) to generate mevalonate phosphate (MVAP). Different from the classical MVA pathway, an alternative pathway is proposed for the IPP generation from MVAP, which is catalyzed by Diphosphomevalonate decarboxylase (DmD, HFX_1486) and Isopentenyl phosphate kinase (IpK, HFX_2774). IPP can be then isomerized to DMAPP by Isopentenyl-diphosphate delta-isomerase (IdI, HFX_2519). DMAPP and IPP are condensed to geranyl-PP (GPP) and then to farnesyl-PP (FPP) and finally to geranylgeranyl-PP (GGPP) by trifunctional prenyl diphosphate synthase (IdsA, HFX_2735). Two molecules of GGPPs are condensed by CrtB (HFX_2547) to form phytoene and then it undergoes four consecutive desaturation reactions catalyzed by CrtI (HFX_2550) to produce lycopene.
FIGURE 2.
Proposed main steps in the mevalonate and lycopene biosynthesis pathways of H. mediterranei based on the KEGG pathway database. For simplicity, cofactors and ATP consumption are not shown. Gray arrows show the MVA pathway and black ones show the lycopene biosynthetic pathway descripted in this study.
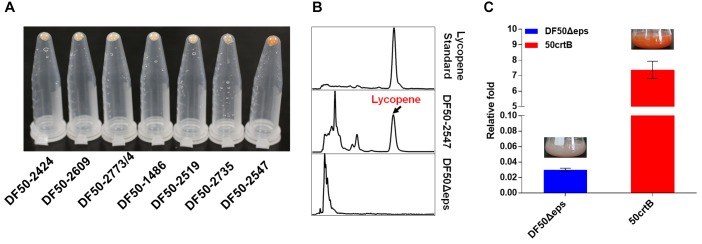
Although H. mediterranei possessed the complete lycopene synthetic pathway, we could not detect lycopene accumulation in this strain (Figure 3B). The extremely low lycopene production might be due to the rate-limiting steps in its lycopene biosynthesis. According to the previous studies about rate-limiting steps in MVA and lycopene synthesis pathway (Goldstein and Brown, 1990; Kang et al., 2005; Steussy et al., 2005; Anthony et al., 2009; Lombard and Moreira, 2011; Berthelot et al., 2012), we selected all the predicted genes as our candidates to be overexpressed in H. mediterranei (Figure 2 and Table 1). To rapidly identify the rate-limiting steps, we used a plasmid-based expression system for candidate genes overexpression under the control of a strong constitutive promoter PphaR. The expression plasmid containing each gene was transformed into the strain DF50Δeps (Zhao et al., 2013) individually and correct transformants were confirmed by PCR and Sanger sequencing. Functional overexpression of the genes encoding rate-limiting enzymes made transformants orange or even red, so it was easy to identify the genes encoding rate-limiting enzymes. Obviously, only the overexpression of gene crtBhm resulted in an orange colored phenotype (Figure 3A). Meanwhile the lycopene accumulation in this crtBhm-overexpressed strain DF50-2547 was further confirmed by HPLC (Figure 3B), whereas no lycopene accumulation was detected in other gene overexpressed strains (Supplementary Figure S2). These results indicated that the step from GGPP to phytoene was the rate-limiting step in lycopene synthesis in H. mediterranei.
FIGURE 3.
Identification of the rate-limiting steps involved in lycopene biosynthesis in H. mediterranei. (A) Collected H. mediterranei cells (1 mL) with a single gene overexpressed in eppendorf tubes. HFX_2773 and HFX_2774 are overlapped by four nucleotides and thus they are co-expressed. (B) HPLC analysis of the lycopene produced by DF50-2547 and DF50Δeps. (C) qRT-PCR analysis of crtBhm transcriptional level in DF50Δeps and 50crtB. The relative fold of crtBhm gene expression level is calculated by normalization to the expression of inner control 7S RNA. Insets are the photographs of 7-day shake flask cultures.
Table 1.
Identity analysis between candidate enzymes involved in lycopene biosynthesis in H. mediterranei and idendified enzymes.
| Enzyme in H. mediterranei | Identified enzymes | Protein identity | ||
|---|---|---|---|---|
| Enzyme | Gene | Gene | Reference strain | |
| MvaB | HFX_2424 | HVO_2419 | H. vocanii (VanNice et al., 2013) | 95% |
| HmgR | HFX_2609 | HVO_2583 | H. vocanii (Bischoff and Rodwell, 1996) | 89% |
| Erg12 | HFX_2773 | HVO_2761 | H. vocanii (Azami et al., 2014) | 92% |
| DmD | HFX_1486 | HVO_1412 | H. vocanii (VanNice et al., 2014) | 90% |
| IpK | HFX_2774 | HVO_2762 | H. vocanii (VanNice et al., 2014) | 89% |
| IdI | HFX_2519 | idi | E. coli (Hemmi et al., 1998) | 32% |
| IdsA | HFX_2735 | ispA | E. coli (Hosfield et al., 2004) | 31% |
| crtE | E. vulneris | 30% | ||
| CrtB | HFX_2547 | crtB | Synechococcus elongatus (Chamovitz et al., 1992) | 32% |
| CrtI | HFX_2550 | crtI | Erwinia uredovora (Fraser et al., 1992) | 31% |
Reinforcing the Rate-Limiting Step in Lycopene Synthesis by Insertion of a Strong Promoter
The plasmid-based overexpression of crtBhm could reinforce the rate-limiting step and thus enhanced the production of lycopene in H. mediterranei. However, this plasmid-based system is not genetically stable and probably brings a metabolic burden. To address this issue, we therefore constructed the plasmid pHFXB (Supplementary Table S1) and used a two-step homologous recombination method to insert promoter PphaR into the chromosome immediately up-stream of crtBhm in DF50Δeps. The engineered strain, termed 50crtB, was easy to be distinguished visually, because of its orange color, which was different from the light pink color of its parental strain DF50Δeps (Figure 3C).
Next, we analyzed the transcription level of crtBhm in strain 50crtB and DF50Δeps by qRT-PCR. The result showed that the insertion of promoter PphaR dramatically increased the transcription level of crtBhm by 245 times, when compared with the DF50Δeps strain (Figure 3C). As expected, the high CrtBhm expression level significantly promoted the conversion from GGPP to phytoene and subsequently improved lycopene biosynthesis. After cultivation in shake flasks containing 50 mL MG medium for 7 days, a lycopene production of 6.05 ± 0.18 mg/g dry cell weight (DCW) (35.15 ± 0.43 mg/L) was achieved (Figure 4C).
FIGURE 4.
Analysis of lycopene production of 50crtB and 50BΔ2549. (A) TLC analysis of the carotenoids extracted from 50crtB and 50BΔ2549. Spot 1 contains mainly lycopene, Spot 2–Spot 4 consists of bacterioruberin and its derivates. (B) UV-Vis absorption spectra of the carotenoids extracted from the pigment spot of 50BΔ2549 in TLC plate. The wavelength range is set as 350 ∼ 550 nm. (C) Lycopene titer and production of 50crtB and 50BΔ2549. Strains are cultured for 7 days in 50 mL MG medium at 37°C in shake flasks. ∗p < 0.05, lycopene production and titer of the group 50BΔ2549 are compared with the 50crtB. Data is expressed as Mean ± SD of triplicate determinations.
Disrupting Bacterioruberin Biosynthesis to Improve the Accumulation and Purity of Lycopene
Lycopene is the last shared intermediate in bacterioruberin and retinal biosynthesis in some haloarchaea (Peck et al., 2017). However, H. mediterranei lacks the genes, crtY (encoding lycopene cyclase) (Peck et al., 2002), brp and blh (encoding β-carotene dioxygenase) (Peck et al., 2001), involved in the retinal biosynthesis pathway and therefore, lycopene can only flux into bacterioruberin biosynthesis (Figure 1A). Thus, we next blocked bacterioruberin biosynthesis to enhance lycopene accumulation and its purity. Bioinformatic analysis revealed the presence of two genes (HFX_1501 and HFX_2549) potentially involved in bacterioruberin synthesis in H. mediterranei. Both of them are annotated as putative prenyltransferases, which can transfer 5-carbon prenyl groups to various substrates. HFX_1501 encodes a protein of 284 amino acid residues, which showed 30 and 28% identity to the LyeJ of H. salinarum and H. japonica, respectively (Supplementary Figure S3A). In the case of HFX_2549, it encodes a 292-amino acid protein exhibiting 64 and 61% identity to the LyeJ of H. salinarum and H. japonica, respectively (Supplementary Figure S3B). Moreover, it is located within the carotenoid biosynthetic gene cluster. Furthermore, a membrane topology analysis using TMHMM revealed that HFX_2549 encoded an integral membrane protein containing seven transmembrane domains (Supplementary Figure S3B), consistent with the LyeJ of H. salinarum or H. japonica. This suggested that HFX_2549 is likely to encode LyeJ in H. mediterranei.
We knocked out the gene, HFX_2549, and obtained the strain 50BΔ2549. The carotenoid components of the 50crtB and 50BΔ2549 strains were analyzed qualitatively by TLC. The sample from 50crtB displayed multiple spots on the silica plate (Figure 4A, Spots 1–4), which represented lycopene, bacterioruberin and its derivates. In contrast, the sample from 50BΔ2549 only contained a single spot 1 (Figure 4A, Spot 1). Subsequently, spot 1 on the silica plate from the 50BΔ2549 strain was recovered and extracted by using acetone. The UV-Vis absorbance spectrum of the extracted sample had a typical three-finger shape of lycopene at 442, 470 and 501 nm, similar to the absorption spectrum of lycopene standard (Figure 4B). These results indicated that 50BΔ2549 could not synthesize bacterioruberin and its derivates, and the reaction was terminated at the lycopene step. Consequently, HFX_2549 was the key enzyme for bacterioruberin synthesis in H. mediterranei. Lycopene production of 50BΔ2549 was further quantified by HPLC. The production significantly increased to 45.54 ± 1.23 mg/g DCW, which was about 6.5 times higher than that of 50crtB (Figure 4C).
Lycopene Production Improvement by Importing Phytoene Desaturase From Other Haloarchaea
To improve the yield of the target products, it is often necessary to enhance the availability of essential precursors (Xie et al., 2015a). In our study, overexpression of crtBhm supplied more precursor, phytoene, for lycopene synthesis, and enhancement of the flux from phytoene to lycopene is another strategy for lycopene production improvement. Here, we strengthened the process by importing heterologous gene crtIha or crtIhs from carotenogenic haloarchaea, H. hispanica or H. salinarum. First, we confirmed the function of crtIha and crtIhs by transforming their expression plasmid pWHA1058 or pWOE3381 (Supplementary Table S1) into the 50BΔ2549 strain. The positive transformants with the functional expression of heterologous crtI showed enhanced color intensity (Figure 5A). Afterward, we constructed two plasmids, pHI6 and pHIH (Supplementary Table S1), for the integration of crtIha and crtIhs into the chromosome of H. mediterranei (Supplementary Figure S1A). To avoid unpredictable effects brought by the insertion site, we used 50crtB as the host and integrated these two genes separately in the chromosome by replacing HFX_2549 (Figure 1B). The two resultant strains were named 50BI6 and 50BIH (Supplementary Table S1).
FIGURE 5.
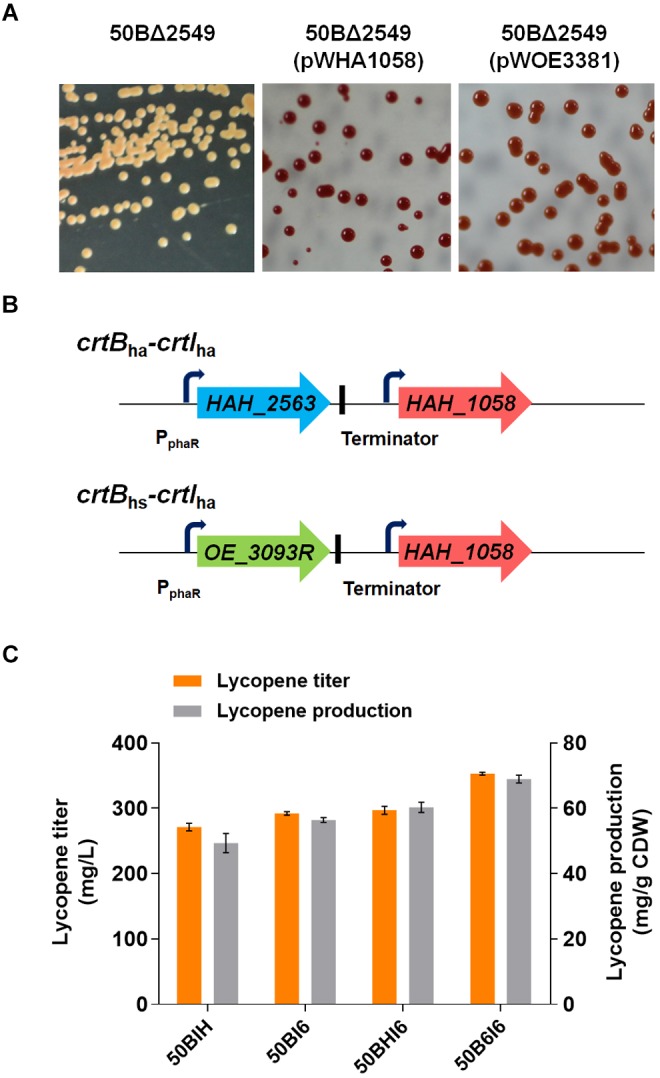
Importation of heterologous crt genes to improve lycopene production. (A) Colony color of H. mediterranei 50BΔ2549 cultured in AS-168 medium and the strains after introducing the expression plasmid containing crtI from Haloarcula hispanica or Halobacterium salinarum cultured in AS-168Y medium. 50BΔ2549 with pWHA1058 means crtIha expressed strain and pWOE3381 means crtIhs expressed strain. (B) crtB-crtI expression cassettes. Each gene was under the control of PphaR promoter and a terminator was in the downstream of crtBha or crtBhs. (C) Lycopene titer and production of engineered H. mediterranei by insertion of heterologous crt genes in the chromosome of 50crtB by replacing HFX_2549. Strains are cultured for 7 days in 50 mL MG medium at 37°C in shake flasks.
Lycopene production by the engineered strains were then determined by HPLC. Lycopene yields of 56.46 ± 0.74 mg/g DCW (292.21 ± 2.88 mg/L) and 49.37 ± 2.95 mg/g DCW (271.43 ± 6.07 mg/L) were obtained in the engineered strains, 50BI6 and 50BIH, respectively. This result showed that the heterologous expression of crtIha in H. mediterranei was more effective for enhancing lycopene synthesis compared to crtIhs. Finally, 50BI6 got a 24.0% increase in lycopene production and 10.2% increase in lycopene titer compared to 50BΔ2549 (Figure 5C).
Heterologous crtB Overexpression for Further Optimizing Lycopene Production
The high lycopene production strain 50BI6 contained two copies of crtI (crtIhm and crtIha). To investigate whether CrtB was still the rate-limiting enzyme in 50BI6, another copy of crtB from H. hispanica or H. salinarum was introduced into 50BI6. We constructed two types of crtB-crtI expression cassettes, crtBha-crtIha and crtBhs-crtIha (Figure 5B) and inserted these expression cassettes in the chromosome of 50crtB by replacing HFX_2549 to obtain new strains, 50B6I6 and 50BHI6 (Figure 1B and Supplementary Figure S1B). We analyzed the transcriptional status of crtBha, crtBhs, and crtIha in these two strains and found that all the expected genes were successfully transcribed base on the RT-PCR analysis (Supplementary Figure S4). The lycopene yields of 50B6I6 and 50BHI6 were further increased to 68.95 ± 1.19 mg/g DCW (353.06 ± 2.39 mg/L) and 60.33 ± 1.56 mg/g DCW (297.05 ± 6.50 mg/L), respectively (Figure 5C). This suggested that the strains with co-introduced crtBha and crtIha could produce more lycopene. Lycopene titer displayed a 20.8% increase in 50B6I6 compared to 50BI6.
Disruption of PHBV Biosynthesis to Divert More Acetyl-CoA Flux to Lycopene
H. mediterranei can accumulate a large amount of PHBV when cultured in MG medium (Zhao et al., 2013) and acetyl-CoA is an important precursor for its biosynthesis (Don et al., 2006). Blocking acetyl-CoA flux to PHBV biosynthesis may be able to further maximize the lycopene production (Figure 1B). To prove this, we knocked out the PHBV synthase encoding genes, phaE and phaC (Lu et al., 2008b), in 50B6I6 and obtained a new strain, named 50B6I6ΔphaEC (Supplementary Table S1). It could not produce PHBV as determined by gas chromatography (Table 2) and this result suggested that the deletion of phaEC blocked PHBV synthesis. Moreover, HPLC analysis showed that 50B6I6ΔphaEC synthesized lycopene with a production level of 119.25 ± 0.55 mg/g DCW, which was 73.0% higher than that of 50B6I6. Meanwhile, lycopene titer of 50B6I6ΔphaEC increased to 429.41 ± 5.81 mg/L, which was 21% enhancement compared to 50B6I6. This result indicated that the disruption of PHBV biosynthesis could enhance the acetyl-CoA flux to lycopene biosynthesis.
Table 2.
PHBV content and lycopene production in 50B6I6 and 50B6I6ΔphaEC.
| Strains | PHBV % | Lycopene |
|
|---|---|---|---|
| Titer (mg/L) | Production (mg/g) | ||
| 50B6I6 | 62.84 ± 1.11 | 353.06 ± 2.39 | 68.95 ± 1.19 |
| 50B6I6ΔphaEC | 0 | 429.41 ± 5.81 | 119.25 ± 0.55 |
Effect of Auxotrophy on Lycopene Production
In this work, all the genetic manipulation was based on the pyrF-deleted strain DF50Δeps (uracil auxotrophic mutant). Uracil addition was required to culture these engineered strains in MG medium. However, this approach was not suitable for high-cell density fermentation and increased the production cost. To address this issue, we restored the functional expression of pyrF in 50B6I6ΔphaEC. A linear DNA fragment containing pyrF and homologous arms, 500 bp in up-stream or down-stream of pyrF, was amplified by PCR using the genomic DNA of H. mediterranei as a template and transferred into the 50B6I6ΔphaEC strain. The screening process was carried out using AS-168SY medium, in which the negative colonies could not grow. We obtained the correct pyrF complementary strain 50FB6I6ΔphaEC and it gave a lycopene yield of 107.37 ± 2.37 mg/g DCW (396.70 ± 13.39 mg/L), while there was no difference in biomass between 50B6I6ΔphaEC and 50FB6I6ΔphaEC (Figure 6). Although the complementation of pyrF did not increase the biomass and led to a little decrease in lycopene content, 50FB6I6ΔphaEC could be cultured in MG medium without uracil, which was more feasible for industrial application.
FIGURE 6.
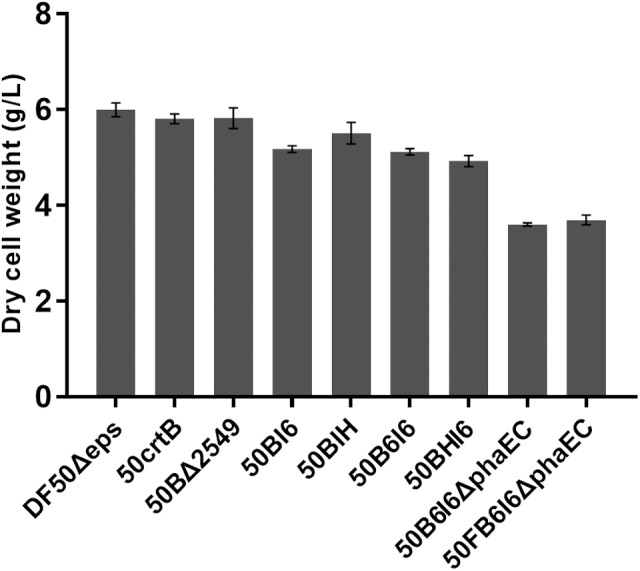
Dry cell weights of engineered H. mediterranei strains.
Discussion
Regarding to food safety issues, lycopene from natural source, such as watermelon, gac fruit, tomato, and so on (Perkins-Veazie and Davis, 2004; Viuda-Martos et al., 2014; Lv et al., 2015; Papaioannou et al., 2016; Wimalasiri et al., 2017), is superior to that from chemical synthesis. Among these fruits or vegetables, tomato is a major source of lycopene, but its total lycopene content is too low to meet the market requirements. Multiple strategies of engineering the carotenoid synthesis pathway in tomato fruit to improve its lycopene content were adopted in several studies. Fraser et al. (2002) introduced an additional crtB from Erwinia uredovora into tomato in a fruit-specific manner and obtained a 1.8-fold increase of lycopene content. Enfissi et al. (2005) got a 1.6-fold increase of lycopene content by overexpression of a bacterial 1-deoxy-D-xylulose-5-phosphate synthase (DXS) encoding gene. Namitha and Negi (2018) introduced a bacterial crtY gene from Pantoea agglomerans into tomato fruit to enhance lycopene production by 2.1-fold. In addition, other strategies were also used to alter carotenoid content in tomato fruit. Overexpression of blue light photoreceptor, cryptochrome 2, resulted in a 1.7-fold increase of lycopene content in tomato fruit (Giliberto et al., 2005). Importing the pepper fibrillin gene into tomato fruit led to a 118% increase in lycopene level (Simkin et al., 2007). Additionally, Mehta et al. (2002) demonstrated that higher level of polyamines in tomato fruit by fruit-specific overexpression of a yeast S-adenosylmethionine decarboxylase gene (ySAMdc) enhanced the lycopene content by 2 ∼ 3 folds in tomato fruit. In the same way, Neily et al. (2011) overexpressed the spermidine synthase gene in tomato and also got an unexpected increase of 1.3 ∼ 2.2 folds in lycopene prodcution. The increased polyamines were revealed to affect multiple cell pathways and broad gene expression levels, thereby enhancing lycopene accumulation (Mattoo et al., 2006, 2007; Kolotilin, 2008; Kolotilin et al., 2011; Neily et al., 2011; Guo et al., 2018). However, these transgenic plants are still far from large-scale industrial application for lycopene production (Table 3). Lycopene production by microbial fermentation is an attractive alternative to use of plants. Moreover, the strains also need to be engineered for improving its production to make the fermentation process more cost competitive. In engineered E. coli, the highest lycopene production of 448 mg/g DCW was obtained by employing a new combinatorial multi-gene pathway assembly scheme (Coussement et al., 2017). In yeast, through engineering host and pathway, the highest lycopene yield, 55.56 mg/g DCW was achieved in 5-L bioreactors (Chen et al., 2016). In B. trispora, a lycopene production of 103.58 mg/g DCW was realized by the modification of the bifunctional gene, carRA, combined with addition of tripropylamine (Wang et al., 2017). In this study, a carotenogenic haloarchaea, H. mediterranei was chosen as a novel chassis cell for lycopene overproduction, due to its several superior features, such as high salt tolerance capability, easy lysis, etc. These can contribute to reduce the energy cost brought by strict sterilization and to simplify the process for lycopene extraction. We adopted multiple strategies to engineer H. mediterranei for lycopene production enhancement and finally our best strain gave a lycopene yield of 119.25 mg/g DCW, which was even higher than the yields of most well-studied strains (Table 3). Although engineered E. coli produced the highest level of lycopene, it is controversial to use it for lycopene synthesis, since this strain would release endotoxin (Ray and Raetz, 1987). In contrast, haloarchaea has an extremely low endotoxin level because of its special structure of cell envelope (Xue et al., 2018). Thus, H. mediterranei is a promising microbial host for lycopene biosynthesis.
Table 3.
Summary of lycopene production in transgenic plant and engineered microorganisms.
| Organism | Strategies | Culture condition | Lycopene production | Reference | |
|---|---|---|---|---|---|
| Plant | |||||
| Tomato | Engineering carotenoids synthesis pathway | Grown in the glasshouse | 5.22 mg/g DCW | Fraser et al., 2002 | |
| Tomato | Grown in the glasshouse | 6.7 mg/g DCW | Enfissi et al., 2005 | ||
| Tomato | Grown in green house | ∼0.11 mg/g DCW | Namitha and Negi, 2018 | ||
| Tomato | Manipulation of the blue light photoreceptor cryptochrome 2 | Grown in green house | 1.35 mg/g DCW | Giliberto et al., 2005 | |
| Tomato | Importing the pepper fibrillin gene in tomato | Grown in green house | ∼0.48 mg/g FW∗ | Simkin et al., 2007 | |
| Tomato | Enhancement of polyamine accumulation | Grown in green house | ∼0.11 mg/g FW∗ | Mehta et al., 2002 | |
| Tomato | Grown in green house | 1.72 mg/g DCW | Neily et al., 2011 | ||
| Microbe | |||||
| 50B6I6ΔphaEC | Pathway engineering | Shake flask fermentation | 119.25 mg/g DCW | This study | |
| E. coli | Microtiter plate fermentation | 448 mg/g DCW | Coussement et al., 2017 | ||
| E. coli | Chromosomal evolution | Shake flask fermentation | 33.4 mg/g DCW | Chen et al., 2013 | |
| E. coli | Pathway balancing | Fed-batch fermentation | 43.7 mg/g DCW | Zhu et al., 2015 | |
| E. coli | Pathway engineering combined with NADPH and ATP balancing | Fed-batch fermentation | 50.6 mg/g DCW | Tao et al., 2014) | |
| E. coli | Plasmid based overexpression of carotenoids synthesis genes | Shake flask fermentation | 67 mg/g DCW | Xu et al., 2018 | |
| S. cerevisiae | Directed evolution and metabolic engineering | Fed-batch fermentation | 24.41 mg/g DCW | Xie et al., 2015a | |
| S. cerevisiae | Pathway engineering | Fed-batch fermentation | 55.56 mg/g DCW | Chen et al., 2016 | |
| Yarrowia lipolytica | Fed-batch fermentation | 21.1 mg/g DCW | Schwartz et al., 2017 | ||
| B. trispora | Genetically manipulated the bifunctional protein gene, carRA | Shake flask fermentation | 103.58 mg/g DCW | Wang et al., 2017 | |
∗FW, fresh weight.
However, in optimal culture conditions for growth, H. mediterranei, is less pigmented and no lycopene can be detected (Figure 3B). This might be due to the presence of a rate-limiting step involved in lycopene synthesis. Then we investigated the overexpression of the possible rate-limiting enzyme encoding genes in the MVA and lycopene synthesis pathway and found that only crtB overexpression resulted in significant lycopene accumulation. This result suggested that the MVA pathway in H. mediterranei is efficient to produce essential precursors of IPP and DMAPP for lycopene synthesis. Thus, in the next step, the first strategy we adopted to improve lycopene production was reinforcing lycopene synthesis pathway. First, we eliminated the rate-limiting step by inserting a strong promoter in the chromosome ahead of crtB and realized a lycopene production of 6.05 mg/g DCW. Then to avoid the limitation of final target products, brought by insufficient ability of downstream pathway (Leonard et al., 2010), we imported heterologous crtI and enhanced the lycopene production to 56.46 mg/g DCW. Next, we integrated heterologous haloarchaeal crtB-crtI cassettes into the chromosome and got a lycopene yield of 68.95 mg/g DCW. Similarly, Xie et al. (2015a) adjusted the copy number of crt genes to get more than 80% increase of lycopene production in Saccharomyces cerevisiae. They reported that multiple copies of crt genes led to an about 13% decrease in biomass (Xie et al., 2015a). Similar results were also obtained in the present study (Figure 6). The dry cell weight of the engineered strains 50BI6, 50BIH, 50B6I6, and 50BHI6 showed a decrease of about 10%. This might be due to the metabolic burden brought by the overexpression of crt genes. However, the significant increase of lycopene titer overweighed the slight biomass decrease in these engineered strains.
The second strategy we used is disruption of the lycopene flux to other carotenoids or deletion of the competing pathways sharing common precursors with the lycopene synthetic pathway. Wang et al. (2016) inhibited the activity of lycopene cyclase, the enzyme responsible for conversion of lycopene to β-carotene, and increased the lycopene content by 90.1%. In E. coli, the knockout of gdhA, accE, and fdhF gave a 37% increase in lycopene content (Alper et al., 2005). In this work, we knocked out the gene lyeJ to block the bacterioruberin biosynthesis and thus improved the lycopene purity and got a 6.5-fold increase of lycopene production. MVA pathway commences with acetyl-CoA, which is also the important precursor for PHBV synthesis in H. mediterranei. Removing the competing pathways for lycopene synthesis can theoretically facilitate lycopene accumulation. So we disrupted the PHBV synthesis in H. mediterranei, by deleting the key genes phaE and phaC. As expected, the engineered strain 50B6I6ΔphaEC did not synthesis PHBV and showed an increase of lycopene titer as expected. The loss of PHBV caused a decrease in dry cell weight by 42% (Figure 6). On the other hand, the lycopene production was sharply enhanced by 73%. This indicated that more acetyl-CoA could flux to lycopene synthesis via MVA pathway with the disruption of PHBV synthesis.
High biomass is necessary to achieve a high yield of lycopene. However, the presence of auxotrophies can cause an organism to grow more slowly than the equivalent prototroph (Pronk, 2002). Furthermore, it is not feasible for high-cell density fermentation and practical application because of the requirement of additional uracil. In Y. lipolytica, the alleviation of both leu2 and ura3 auxotrophies gave a 1.9-fold enhancement in lycopene titer (Schwartz et al., 2017). In this work, we complemented the pyrF auxotrophy in strain 50B6I6ΔphaEC. The engineered strain 50FB6I6ΔphaEC showed no difference in dry cell weight compared with 50B6I6ΔphaEC (Figure 6), but gave a little decrease in lycopene titer and production. The low lycopene accumulation after the complementation of pyrF gene might be ascribed to the fact that the utilization efficiency of the uracil synthesized in vivo was lower than that of the uracil added for cell growth. However, no need for uracil overweighed the slight decrease of lycopene production as for developing a cost-effective industrial strain.
This work reveals that H. mediterranei possesses a great potential for lycopene biosynthesis and much more efforts are needed to further increase its lycopene production. The reduction of FPP flux to the squalene biosynthetic pathway (a competing pathway for carotenoid synthesis) is expected to further increase lycopene yield. This strategy has been used to improve lycopene production in yeast (Xie et al., 2015b). Besides, modulation of the NADPH and ATP levels is another alternative approach for further enhancing lycopene synthesis in H. mediterranei. Additionally, optimization of culture conditions and fed-batch fermentation might also be adopted for maximizing lycopene production in H. mediterranei.
Conclusion
In this work, we engineered a haloarchaeon, H. mediterranei, as a novel host for lycopene overproduction by adopting multiple strategies. Introducing a constitutive promoter enhanced the expression level of the rate-limiting enzyme encoding gene, crtB, and disrupting the bacterioruberin synthesis significantly increased the lycopene production and purity. Importing different heterologous crt expression cassettes were also an effective method for improving lycopene production. Further blocking PHBV synthesis to direct more acetyl-CoA flux into carotenoid synthesis showed a dramatic increase in lycopene production, up to 119.25 ± 0.55 mg/g DCW, in shake flask fermentation. Complementation of pyrF in the engineered strain 50B6I6ΔphaEC had no increase in both dry cell weight and lycopene production, but it could grow without addition of uracil and thus is more suitable for industrial application. The engineering pathway that we developed in this study shows the potential for high-level production of lycopene and offers biological insights into carotenoid production in haloarchaea.
Author Contributions
Z-QZ, JH, and HX conceived the project, analyzed the data, and drafted the manuscript. Z-QZ and JZ performed the study. QX, D-HZ, and JH critically revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yeh-Hsing Lao at Columbia University, Sumit Kumar at Indian Institute of Technology and Ruchira Mitra for assistance in manuscript writing and Tong Sun for his technical support.
Funding. This work was financially supported by the National Natural Science Foundation of China (Grant Nos. NSFC-31330001 and NSFC-91751201), National Science and Technology Foundation Project of China (Grant No. 2015FY110100), and the Youth Innovation Promotion Association of CAS (Grant No. 2015070).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02893/full#supplementary-material
References
- Alper H., Jin Y.-S., Moxley J. F., Stephanopoulos G. (2005). Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 7 155–164. 10.1016/j.ymben.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Alper H., Stephanopoulos G. (2008). Uncovering the gene knockout landscape for improved lycopene production in E. coli. Appl. Microbiol. Biotechnol. 78 801–810. 10.1007/s00253-008-1373-x [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Anthony J. R., Anthony L. C., Nowroozi F., Kwon G., Newman J. D., Keasling J. D. (2009). Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab. Eng. 11 13–19. 10.1016/j.ymben.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Antón J., Meseguer I., Rodríguezvalera F. (1988). Production of an extracellular polysaccharide by Haloferax mediterranei. Appl. Environ. Microbiol. 54 2381–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azami Y., Hattori A., Nishimura H., Kawaide H., Yoshimura T., Hemmi H. (2014). (R)-mevalonate 3-phosphate is an intermediate of the mevalonate pathway in Thermoplasma acidophilum. J. Biol. Chem. 289 15957–15967. 10.1074/jbc.M114.562686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot K., Estevez Y., Deffieux A., Peruch F. (2012). Isopentenyl diphosphate isomerase: a checkpoint to isoprenoid biosynthesis. Biochimie 94 1621–1634. 10.1016/j.biochi.2012.03.021 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Saha J., Haldar S., Bhowmic A., Mukhopadhyay U. K., Mukherjee J. (2014). Production of poly-3-(hydroxybutyrate-co-hydroxyvalerate) by Haloferax mediterranei using rice-based ethanol stillage with simultaneous recovery and re-use of medium salts. Extremophiles 18 463–470. 10.1007/s00792-013-0622-9 [DOI] [PubMed] [Google Scholar]
- Bischoff K. M., Rodwell V. W. (1996). 3-Hydroxy-3-methylglutaryl-coenzyme a reductase from Haloferax volcanii: purification, characterization, and expression in Escherichia coli. J. Bacteriol. 178 19–23. 10.1128/jb.178.1.19-23.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Cai L., Liu H., Liu X., Han J., Zhou J., et al. (2012). Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl. Environ. Microbiol. 78 1946–1952. 10.1128/AEM.07114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. F., Cai L., Zhao D. H., Liu G. M., Han J., Zhou J., et al. (2015). A novel DNA-binding protein, PhaR, plays a central role in the regulation of polyhydroxyalkanoate accumulation and granule formation in the haloarchaeon Haloferax mediterranei. Appl. Environ. Microbiol. 81 373–385. 10.1128/aem.02878-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz D., Misawa N., Sandmann G., Hirschberg J. (1992). Molecular cloning and expression in Escherichia coli of a cyanobacterial gene coding for phytoene synthase, a carotenoid biosynthesis enzyme. FEBS Lett. 296 305–310. 10.1016/0014-5793(92)80310-D [DOI] [PubMed] [Google Scholar]
- Chen C. W., Hsu S. H., Lin M. T., Hsu Y. H. (2015). Mass production of C-50 carotenoids by Haloferax mediterranei in using extruded rice bran and starch under optimal conductivity of brined medium. Bioprocess. Biosyst. Eng. 38 2361–2367. 10.1007/s00449-015-1471-y [DOI] [PubMed] [Google Scholar]
- Chen Y., Xiao W. H., Wang Y., Liu H., Li X., Yuan Y. J. (2016). Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb. Cell Fact. 15:113. 10.1186/s12934-016-0509-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Y., Shen H. J., Cui Y. Y., Chen S. G., Weng Z. M., Zhao M., et al. (2013). Chromosomal evolution of Escherichia coli for the efficient production of lycopene. BMC Biotechnol. 13:6. 10.1186/1472-6750-13-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S. W., Lam W. L., Charlebois R. L., Schalkwyk L. C., Doolittle W. F. (1989). Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35 148–152. 10.1139/m89-022 [DOI] [PubMed] [Google Scholar]
- Coussement P., Bauwens D., Maertens J., De Mey M. (2017). Direct combinatorial pathway optimization. ACS Synth. Biol. 6 224–232. 10.1021/acssynbio.6b00122 [DOI] [PubMed] [Google Scholar]
- de Lourdes Moreno M., Sanchez-Porro C., Garcia M. T., Mellado E. (2012). Carotenoids’ production from halophilic bacteria. Methods Mol. Biol. 892 207–217. 10.1007/978-1-61779-879-5-12 [DOI] [PubMed] [Google Scholar]
- Don T. M., Chen C. W., Chan T. H. (2006). Preparation and characterization of poly(hydroxyalkanoate) from the fermentation of Haloferax mediterranei. Sci. Polym. Ed. 17 1425–1438. 10.1163/156856206778937208 [DOI] [PubMed] [Google Scholar]
- Enfissi E. M. A., Fraser P. D., Lois L.-M., Boronat A., Schuch W., Bramley P. M. (2005). Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 3 17–27. 10.1111/j.1467-7652.2004.00091.x [DOI] [PubMed] [Google Scholar]
- Fang C. J., Ku K. L., Lee M. H., Su N. W. (2010). Influence of nutritive factors on C-50 carotenoids production by Haloferax mediterranei ATCC 33500 with two-stage cultivation. Bioresour. Technol. 101 6487–6493. 10.1016/j.biortech.2010.03.044 [DOI] [PubMed] [Google Scholar]
- Fraser P. D., Misawa N., Linden H., Yamano S., Kobayashi K., Sandmann G. (1992). Expression in Escherichia coli, purification, and reactivation of the recombinant Erwinia uredovora phytoene desaturase. J. Biol. Chem. 267 19891–19895. [PubMed] [Google Scholar]
- Fraser P. D., Romer S., Shipton C. A., Mills P. B., Kiano J. W., Misawa N., et al. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. U.S.A. 99 1092–1097. 10.1073/pnas.241374598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajowik A., Dobrzynska M. M. (2014). Lycopene - antioxidant with radioprotective and anticancer properties. A review. Rocz. Panstw. Zakl. Hig. 65 263–271. [PubMed] [Google Scholar]
- Giliberto L., Perrotta G., Pallara P., Weller J. L., Fraser P. D., Bramley P. M., et al. (2005). Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 137 199–208. 10.1104/pp.104.051987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. (1990). Regulation of the mevalonate pathway. Nature 343 425–430. 10.1038/343425a0 [DOI] [PubMed] [Google Scholar]
- Guo J., Wang S., Yu X., Dong R., Li Y., Mei X., et al. (2018). Polyamines regulate strawberry fruit ripening by abscisic acid, auxin, and ethylene. Plant Physiol. 177 339–351. 10.1104/pp.18.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Hou J., Zhang F., Ai G., Li M., Cai S., et al. (2013). Multiple propionyl coenzyme A-supplying pathways for production of the bioplastic poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in Haloferax mediterranei. Appl. Environ. Microbiol. 79 2922–2931. 10.1128/aem.03915-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lu Q., Zhou L., Zhou J., Xiang H. (2007). Molecular characterization of the phaEC(Hm) genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui. Appl. Environ. Microbiol. 73 6058–6065. 10.1128/AEM.00953-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Wu L. P., Liu X. B., Hou J., Zhao L. L., Chen J. Y., et al. (2017). Biodegradation and biocompatibility of haloarchaea-produced poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymers. Biomaterials 139 172–186. 10.1016/j.biomaterials.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Han J., Zhang F., Hou J., Liu X., Li M., Liu H., et al. (2012). Complete genome sequence of the metabolically versatile halophilic archaeon Haloferax mediterranei, a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) producer. J. Bacteriol. 194 4463–4464. 10.1128/JB.00880-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H., Ohnuma S., Nagaoka K., Nishino T. (1998). Identification of genes affecting lycopene formation in Escherichia coli transformed with carotenoid biosynthetic genes: candidates for early genes in isoprenoid biosynthesis. J. Biochem. 123 1088–1096. 10.1093/oxfordjournals.jbchem.a022047 [DOI] [PubMed] [Google Scholar]
- Hosfield D. J., Zhang Y., Dougan D. R., Broun A., Tari L. W., Swanson R. V., et al. (2004). Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J. Biol. Chem. 279 8526–8529. 10.1074/jbc.C300511200 [DOI] [PubMed] [Google Scholar]
- Kang M. J., Lee Y. M., Yoon S. H., Kim J. H., Ock S. W., Jung K. H., et al. (2005). Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol. Bioeng. 91 636–642. 10.1002/bit.20539 [DOI] [PubMed] [Google Scholar]
- Kolotilin I. (2008). Assessment of Transgenic Approaches to Increase Lycopene Content in Ripe Tomato Fruits. Ph.D. thesis. Hebrew University, Jerusalem. [Google Scholar]
- Kolotilin I., Koltai H., Bar-Or C., Chen L., Nahon S., Shlomo H., et al. (2011). Expressing yeast SAMdc gene confers broad changes in gene expression and alters fatty acid composition in tomato fruit. Physiol. Plant 142 211–223. 10.1111/j.1399-3054.2011.01458.x [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Leonard E., Ajikumar P. K., Thayer K., Xiao W.-H., Mo J. D., Tidor B., et al. (2010). Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc. Natl. Acad. Sci. U.S.A. 107 13654–13659. 10.1073/pnas.1006138107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. L., Han J., Liu X. Q., Zhou J., Xiang H. (2011). Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J. Genet. Genomics 38 261–269. 10.1016/j.jgg.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Liu X.-J., Liu R.-S., Li H.-M., Tang Y.-J. (2012). Lycopene production from synthetic medium by Blakeslea trispora NRRL 2895 (+) and 2896 (-) in a stirred-tank fermenter. Bioprocess. Biosyst. Eng. 35 739–749. 10.1007/s00449-011-0654-4 [DOI] [PubMed] [Google Scholar]
- Lombard J., Moreira D. (2011). Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 28 87–99. 10.1093/molbev/msq177 [DOI] [PubMed] [Google Scholar]
- Lu Q., Han J., Zhou L., Coker J. A., DasSarma P., DasSarma S., et al. (2008a). Dissection of the regulatory mechanism of a heat-shock responsive promoter in haloarchaea: a new paradigm for general transcription factor directed archaeal gene regulation. Nucleic Acids Res. 36 3031–3042. 10.1093/nar/gkn152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Han J., Zhou L., Zhou J., Xiang H. (2008b). Genetic and biochemical characterization of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthase in Haloferax mediterranei. J. Bacteriol. 190 4173–4180. 10.1128/jb.00134-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv P., Li N., Liu H., Gu H., Zhao W.-E. (2015). Changes in carotenoid profiles and in the expression pattern of the genes in carotenoid metabolisms during fruit development and ripening in four watermelon cultivars. Food Chem. 174 52–59. 10.1016/j.foodchem.2014.11.022 [DOI] [PubMed] [Google Scholar]
- Mattoo A. K., Chung S. H., Goyal R. K., Fatima T., Solomos T., Srivastava A., et al. (2007). Overaccumulation of higher polyamines in ripening transgenic tomato fruit revives metabolic memory, upregulates anabolism-related genes, and positively impacts nutritional quality. J. AOAC Int. 901456–1464. [PubMed] [Google Scholar]
- Mattoo A. K., Sobolev A. P., Neelam A., Goyal R. K., Handa A. K., Segre A. L. (2006). Nuclear magnetic resonance spectroscopy-based metabolite profiling of transgenic tomato fruit engineered to accumulate spermidine and spermine reveals enhanced anabolic and nitrogen-carbon interactions. Plant Physiol. 142 1759–1770. 10.1104/pp.106.084400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R. A., Cassol T., Li N., Ali N., Handa A. K., Mattoo A. K. (2002). Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality, and vine life. Nat. Biotechnol. 20 613–618. 10.1038/nbt0602-613 [DOI] [PubMed] [Google Scholar]
- Namitha K. K., Negi P. S. (2018). Transformation of tomato cv. arka ahuti (Solanum lycopersicum L.) with phytoene desaturase (CrtI) and lycopene β-cyclase (CrtY) genes increases carotenoid content and antioxidant potential. J. Plant Biochem. Biotechnol. 27 68–77. 10.1007/s13562-017-0417-7 [DOI] [Google Scholar]
- Naziri D., Hamidi M., Hassanzadeh S., Tarhriz V., Maleki Zanjani B., Nazemyieh H., et al. (2014). Analysis of carotenoid production by Halorubrum sp. TBZ126; an extremely halophilic archeon from Urmia Lake. Adv. Pharm. Bull. 4 61–67. 10.5681/apb.2014.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neily M. H., Matsukura C., Maucourt M., Bernillon S., Deborde C., Moing A., et al. (2011). Enhanced polyamine accumulation alters carotenoid metabolism at the transcriptional level in tomato fruit over-expressing spermidine synthase. J. Plant Physiol. 168 242–252. 10.1016/j.jplph.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Oren A., Hallsworth J. E. (2014). Microbial weeds in hypersaline habitats: the enigma of the weed-like Haloferax mediterranei. FEMS Microbiol. Lett. 359 134–142. 10.1111/1574-6968.12571 [DOI] [PubMed] [Google Scholar]
- Palmer B. R., Marinus M. G. (1994). The dam and dcm strains of Escherichia coli - a review. Gene 143 1–12. 10.1016/0378-1119(94)90597-5 [DOI] [PubMed] [Google Scholar]
- Papaioannou E. H., Liakopoulou-Kyriakides M., Karabelas A. J. (2016). Natural origin lycopene and its “Green” downstream processing. Crit. Rev. Food Sci. Nutr. 56 686–709. 10.1080/10408398.2013.817381 [DOI] [PubMed] [Google Scholar]
- Peck R. F., Echavarri-Erasun C., Johnson E. A., Ng W. V., Kennedy S. P., Hood L., et al. (2001). Brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J. Biol. Chem. 276 5739–5744. 10.1074/jbc.M009492200 [DOI] [PubMed] [Google Scholar]
- Peck R. F., Johnson E. A., Krebs M. P. (2002). Identification of a lycopene beta-cyclase required for bacteriorhodopsin biogenesis in the archaeon Halobacterium salinarum. J. Bacteriol. 184 2889–2897. 10.1128/jb.184.11.2889-2897.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck R. F., Plesa A. M., Graham S. M., Angelini D. R., Shaw E. L. (2017). Opsin-mediated inhibition of bacterioruberin synthesis in halophilic archaea. J. Bacteriol. 199:e00303-17. 10.1128/jb.00303-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins-Veazie P., Davis A. R. (2004). In search of high lycopene watermelon. Cucurbit Genet. Coop. Rep. 27 51–53. [Google Scholar]
- Pronk J. T. (2002). Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 68 2095–2100. 10.1128/AEM.68.5.2095-2100.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B. L., Raetz C. R. (1987). The biosynthesis of gram-negative endotoxin. A novel kinase in Escherichia coli membranes that incorporates the 4′-phosphate of lipid A. J. Biol. Chem. 262 1122–1128. [PubMed] [Google Scholar]
- Rodrigo-Banos M., Garbayo I., Vilchez C., Bonete M. J., Martinez-Espinosa R. M. (2015). Carotenoids from haloarchaea and their potential in biotechnology. Mar. Drugs 13 5508–5532. 10.3390/md13095508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook H. C. (1989). Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schwartz C., Frogue K., Misa J., Wheeldon I. (2017). Host and pathway engineering for enhanced lycopene biosynthesis in Yarrowia lipolytica. Front. Microbiol. 8:11. 10.3389/fmicb.2017.02233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H., Stahl W. (1998). Lycopene: antioxidant and biological effects and its bioavailability in the human. Proc. Soc. Exp. Biol. Med. 218 121–124. 10.3181/00379727-218-44285a [DOI] [PubMed] [Google Scholar]
- Simkin A. J., Gaffé J., Alcaraz J.-P., Carde J.-P., Bramley P. M., Fraser P. D., et al. (2007). Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 68 1545–1556. 10.1016/j.phytochem.2007.03.014 [DOI] [PubMed] [Google Scholar]
- Singh A., Singh A. K. (2017). Haloarchaea: worth exploring for their biotechnological potential. Biotechnol. Lett. 39 1793–1800. 10.1007/s10529-017-2434-y [DOI] [PubMed] [Google Scholar]
- Steussy C. N., Vartia A. A., Burgner J. W. II, Sutherlin A., Rodwell V. W., Stauffacher C. V. (2005). X-ray crystal structures of HMG-CoA synthase from Enterococcus faecalis and a complex with its second substrate/inhibitor acetoacetyl-CoA. Biochemistry 44 14256–14267. 10.1021/bi051487x [DOI] [PubMed] [Google Scholar]
- Strand A., Shivaji S., Liaaen-Jensen S. (1997). Bacterial carotenoids 55. C50-carotenoids 25.† revised structures of carotenoids associated with membranes in psychrotrophic Micrococcus roseus. Biochem. Syst. Ecol. 25 547–552. 10.1016/S0305-1978(97)00039-2 [DOI] [Google Scholar]
- Su A., Chi S., Li Y., Tan S., Qiang S., Chen Z., et al. (2018). Metabolic redesign of Rhodobacter sphaeroides for lycopene production. J. Agric. Food Chem. 66 5879–5885. 10.1021/acs.jafc.8b00855 [DOI] [PubMed] [Google Scholar]
- Tao S., Miao L., Li Q., Dai G., Lu F., Tao L., et al. (2014). Production of lycopene by metabolically-engineered Escherichia coli. Biotechnol. Lett. 36 1515–1522. 10.1007/s10529-014-1543-0 [DOI] [PubMed] [Google Scholar]
- VanNice J. C., Skaff D. A., Keightley A., Addo J. K., Wyckoff G. J., Miziorko H. M. (2014). Identification in Haloferax volcanii of phosphomevalonate decarboxylase and isopentenyl phosphate kinase as catalysts of the terminal enzyme reactions in an archaeal alternate mevalonate pathway. J. Bacteriol. 196 1055–1063. 10.1128/jb.01230-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanNice J. C., Skaff D. A., Wyckoff G. J., Miziorko H. M. (2013). Expression in Haloferax volcanii of 3-hydroxy-3-methylglutaryl coenzyme A synthase facilitates isolation and characterization of the active form of a key enzyme required for polyisoprenoid cell membrane biosynthesis in halophilic archaea. J. Bacteriol. 195 3854–3862. 10.1128/jb.00485-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viuda-Martos M., Sanchez-Zapata E., Sayas-Barbera E., Sendra E., Perez-Alvarez J. A., Fernandez-Lopez J. (2014). Tomato and tomato byproducts. Human health benefits of lycopene and its application to meat products: a review. Crit. Rev. Food Sci. Nutr. 54 1032–1049. 10.1080/10408398.2011.623799 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen X., Hong X., Du S., Liu C., Gong W., et al. (2016). Cyclase inhibitor tripropylamine significantly enhanced lycopene accumulation in Blakeslea trispora. J. Biosci. Bioeng. 122 570–576. 10.1016/j.jbiosc.2016.05.001 [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Pang J., Zheng Y. M., Jiang P. P., Gong W. F., Chen X. W., et al. (2017). Genetic manipulation of the bifunctional gene, carRA, to enhance lycopene content in Blakeslea trispora. Biochem. Eng. J. 119 27–33. 10.1016/j.bej.2016.12.011 [DOI] [Google Scholar]
- Wei Y., Mohsin A., Hong Q., Guo M., Fang H. (2017). Enhanced production of biosynthesized lycopene via heterogenous MVA pathway based on chromosomal multiple position integration strategy plus plasmid systems in Escherichia coli. Bioresour. Technol. 250 382–389. 10.1016/j.biortech.2017.11.035 [DOI] [PubMed] [Google Scholar]
- Wimalasiri D., Brkljaca R., Piva T. J., Urban S., Huynh T. (2017). Comparative analysis of carotenoid content in Momordica cochinchinensis (Cucurbitaceae) collected from Australia, Thailand and Vietnam. J. Food Sci. Technol. 54 2814–2824. 10.1007/s13197-017-2719-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Ye L., Zhao D., Li S., Li Q., Zhang B., et al. (2018). Engineering membrane morphology and manipulating synthesis for increased lycopene accumulation in Escherichia coli cell factories. 3 Biotech 8:269. 10.1007/s13205-018-1298-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Lv X., Ye L., Zhou P., Yu H. (2015a). Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering. Metab. Eng. 30 69–78. 10.1016/j.ymben.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Xie W., Ye L., Lv X., Xu H., Yu H. (2015b). Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab. Eng. 28 8–18. 10.1016/j.ymben.2014.11.007 [DOI] [PubMed] [Google Scholar]
- Xu J., Xu X., Xu Q., Zhang Z., Jiang L., Huang H. (2018). Efficient production of lycopene by engineered E. coli strains harboring different types of plasmids. Bioprocess. Biosyst. Eng. 41 489–499. 10.1007/s00449-017-1883-y [DOI] [PubMed] [Google Scholar]
- Xue Q., Liu X. B., Lao Y. H., Wu L. P., Wang D., Zuo Z. Q., et al. (2018). Anti-infective biomaterials with surface-decorated tachyplesin I. Biomaterials 178 351–362. 10.1016/j.biomaterials.2018.05.008 [DOI] [PubMed] [Google Scholar]
- Zhao D., Cai L., Wu J., Li M., Liu H., Han J., et al. (2013). Improving polyhydroxyalkanoate production by knocking out the genes involved in exopolysaccharide biosynthesis in Haloferax mediterranei. Appl. Microbiol. Biotechnol. 97 3027–3036. 10.1007/s00253-012-4415-3 [DOI] [PubMed] [Google Scholar]
- Zhu F., Lu L., Fu S., Zhong X., Hu M., Deng Z., et al. (2015). Targeted engineering and scale up of lycopene overproduction in Escherichia coli. Process Biochem. 50 341–346. 10.1016/j.procbio.2014.12.008 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.