Abstract
The emerging phenomenon of cellular heterogeneity in tissue requires single-cell resolution studies. A specific challenge for suspension-based single-cell analysis is the preservation of intact cell states when single cells are isolated from tissue contexts, in order to enable downstream analyses to extract accurate, native information. We have developed DISSECT (Disaggregation for Intracellular Signaling in Single Epithelial Cells from Tissue) coupled to mass cytometry (CyTOF: Cytometry by Time-of-Flight), an experimental approach for profiling intact signaling states of single cells from epithelial tissue specimens. We have previously applied DISSECT-CyTOF to fresh mouse intestinal samples and to Formalin-Fixed, Paraffin-Embedded (FFPE) human colorectal cancer specimens. Here, we present detailed protocols for each of these procedures, as well as a new method for applying DISSECT to cryopreserved tissue slices. We present example data for using DISSECT on a cryopreserved specimen of the human colon to profile its immune and epithelial composition. These techniques can be used for high-resolution studies for monitoring disease-related alternations in different cellular compartments using specimens stored in cryopreserved or FFPE tissue banks.
Keywords: Single-cell, CyTOF, FFPE, cryopreserved, mass cytometry, epithelial cells, dissociation, disaggregation, signaling
1. Introduction
Analytic techniques have rapidly progressed to single-cell resolution, enabling assays that address cellular diversity within tissues. These approaches can characterize the increasingly appreciated heterogeneities that manifest in complex diseases. For instance, many cases of cancer are distinguished by intratumoral infiltration of immune, endothelial, mesenchymal, and endothelial cells. These microenvironmental influences, coupled to genetic alterations, present a landscape of diverse cancer cell states. Specifically, rare cell populations, such as cancer stem cells, possess specialized deleterious functions. Cancer stem cells frequently resist standard of care or targeted therapies, repopulate tumors during relapse, and contribute to distal metastases that eventually kill patients [1–4]. Addressing rare cancer cell populations, as well as microenvironmental infiltrates that modify prognosis, has become a necessity for better understanding cancer cell dynamics and devising more effective therapeutic strategies.
Until recently, results from single-cell analysis of epithelial cells have been skewed by the process of isolating individual cells from the native tissue context, which involves separating cells from the basement membrane and breaking cell-cell contacts [5]. Cellular junction disruption perturbs the characteristic polarity of epithelial cells and subsequently alters native morphology and signaling, which results in substantial artifacts during downstream analysis [6–8]. Our lab developed a technique, called DISSECT, to prevent these alterations by applying fixative to whole tissue to preserve cells in their native contexts prior to staining and dissociation. Single-cell suspensions from these preparations can be analyzed by mass cytometry (CyTOF), an advanced flow cytometry approach where heavy metal-tagged antibodies are utilized in place of fluorochrome-labeled ones to enable highly multiplexed measurements of protein expression [9]. We have previously demonstrated the application of DISSECT-CyTOF on fresh samples and FFPE specimens [8, 10]. FFPE has been a standard practice in clinical pathological analysis of tissues for almost a century [11]. Because its use in the long-term preservation of tissue morphologies has been widely demonstrated, large repositories of patient samples exist today. However, cryopreservation is now widely regarded as a superior method for tissue storage due to the preservation of sensitive antigens and nucleic acids that do not survive the process of formalin fixation, dehydration, and paraffinization. Here, we present detailed protocols for the application of DISSECT to fresh, FFPE, as well as cryopreserved tissue specimens. These procedures will allow single-cell signaling analysis of epithelial tissue and enable new insights for epithelial-derived diseases and solid cancers.
2. Materials
2.1. Common reagents to all DISSECT procedures
1× phosphate-buffered saline (1× PBS), without Ca+ and Mg+.
Deionized water (DI H2O).
15 mL and 50 mL conical tubes.
1 L containers.
1.5 mL Eppendorf tubes.
Cytometry tubes with 35-μm cell strainer caps.
Water bath set at 37 °C.
Rocker to apply gentle motion to samples.
Vortex.
Desktop centrifuge.
Minifuge.
Flow Block-2.5% donkey serum in 1× PBS: Add 1.25 mL of normal donkey serum [Jackson ImmunoResearch] into a 50 mL conical tube. Add 1× PBS to a final volume of 50 mL.
1% Triton X-100 in 1× PBS: Add 500 μL of Triton-X 100 [Sigma; Cat: T8787] into a 50 mL conical tube. Add 1× PBS to a volume of 50 mL.
1% Triton-X 100 in H2O: Add 500 μL of Triton-X 100 into a 50 mL conical tube. Add DI H2O to a final volume of 50 mL.
10% Sodium Dodecyl Sulfate (SDS) in H2O: Add 25 mL of 20% SDS to a 50 mL conical tube. Add DI H2O to a volume of 50 mL.
XX Detergent-1% saponin, 0.05% Triton-X 100, and 0.01% SDS in 1× PBS: Weigh and add 0.5 g of saponin [Sigma] to a 50 mL conical tube. Add 2.5 mL of 1% Triton X-100 in 1× PBS and 50 μL of 10% SDS to the 50 mL conical tube. Add 1× PBS to a final volume of 50 mL.
27.5 gage needles.
1 mL syringes.
0.5% Bovine Serum Albumin (BSA) in 1× PBS: Weigh and add 0.25 g of BSA into a 50 mL conical. Add 1× PBS to a final volume of 50 mL. Dissolve.
4% Paraformaldehyde (PFA) stored at 4 °C.
0.01% Triton-X 100 in 1× PBS: Add 10 mL of 1% Triton X-100 in 1× PBS into a 1 L container and add 1× PBS to a final volume of 1000 mL.
0.003% Triton-X 100 in 1× PBS: Add 150 μL of 1% Triton X-100 in 1× PBS solution into a 50 mL conical tube and add 1× PBS to a volume of 50 mL.
0.003% Triton-X 100 in DI H2O: Add 10 mL of the 1% Triton X-100 in DI H2O into a 1L container and add DI H2O to a volume of 1000 mL.
Dissociation solution-1 mg/mL of collagenase and dispase in 1× PBS: Collagenase [CalBio; Activity: 1260 U/mg] and dispase [Gibco; Activity: 1.73 U/mg] are each dissolved in DI H2O at 100 mg/mL and frozen in 50 μL aliquots at −20 °C. (See Note 1) The day of the procedure, prepare the working solution by adding 2 μL of both collagenase and dispase to the same 1.5 mL Eppendorf tube followed by 200 μL of 1× PBS. 200 μL of dissociation solution is necessary per sample therefore scale accordingly.
Hemocytometer.
Ir Intercalator: 1000× (125 μM) intercalator [Fluidigm; Cat: 201192A] is aliquoted at 25 μL each in 1.5mL Eppendorf tubes and stored at −20 °C. To prepare a 50× stock solution, add 500 μL of 1× PBS to a 25 μL aliquot. (See Note 2). The day of procedure, prepare a 1× working solution by adding 2 μL of the 50× stock solution to 100 μL of 1× PBS in a 1.5 mL Eppendorf tube. 100 μL of intercalator is necessary per sample, therefore, scale accordingly.
Normalization Beads: The day of the procedure, prepare a 1× bead solution by adding 1000 μL of a 10× stock solution of normalization bead [DVS; Cat: 201078] to a 15 mL conical tube. Add DI H2O until a volume of 10 mL. This volume is used for 5–10 samples; therefore, adjust accordingly.
Antibodies: Refer to Table 1.
Mass cytometer (Fluidigm).
Data in this manuscript were analyzed using Cytobank (http://www.cytobank.org).
Table 1.
Analytes measured in a human cryopreserved colon specimen for this study.
| Tag | Target | Cat# | Clone |
|---|---|---|---|
| 154Sm | CD45 | 3154001B | H130 |
| 156Gd | PDGFRb | 3156018A | 18A2 |
| 158Gd | CDH1 (E-CAD) | 3158018B | DECMA-1 |
| 162Dy | Pan-Keratin | 3162027A | C11 |
| 165Ho | ITGb3 | 3165010B | VI-PL2 |
| 167Er | CD11b | 3167011B | ICRF44 |
| 168Er | Ki67 | 3168007B | B56 |
| 170Er | CK20 | CS13063 | D9Z1Z |
| 174Yb | CK8/18 | 3174014A | C51 |
2.2. FFPE-DISSECT
Heat block at 65 °C.
Pressure cooker that has a warm and heat setting along with a timer [Cuisinart; Model: EPC-1200PC].
Histoclear [National Diagnostics; Cat:5989-27-5].
100% ethanol (Histology grade).
70% ethanol: Add 700 mL of 100% ethanol into a 1L glass jar and add DI H2O to a final volume of 1000 mL.
50% ethanol: Add 500 mL of 100% ethanol into a 1L glass jar and add DI H2O to a final volume of 1000 mL.
1× antigen retrieval buffer-1× DAKO solution: Prepare the day of the procedure, by adding 1mL of a 10× DAKO target retrieval solution (pH 6) [Agilent; Cat: S169984–2] into a 15 mL conical. Add DI H2O to a final volume of 10 mL. 1.5 mL solution per sample is necessary therefore adjust accordingly.
0.3% Triton X-100 in 1× PBS: Add 15 mL of the 1% Triton X-100 in 1× PBS solution into a 50 mL conical tube and add 1× PBS to a volume of 50 mL.
2.3. Cryo-DISSECT and DISSECT on fresh tissue
Acetone stored at −20 °C in a 50 mL conical.
3. Methods
For FFPE applications, standard procedures for FFPE embedding were assumed to be followed. In summary, resected tissue is immediately fixed in formalin overnight (16–24 h), followed by dehydration, clearing, and embedding in paraffin. For cryopreservation, resected tissue is immediately embedded in Optimal Cutting Temperature compound (OCT) and stored at −80 °C in mold cassettes. For fresh tissue, follow the DISSECT protocol to 3.1.3, then switch to the Cryo-DISSECT protocol starting at 3.3.5. For FFPE specimens, follow the FFPE-DISSECT protocol to 3.2.20, then switch to the Cryo-DISSECT protocol starting at 3.3.17. For cryopreserved specimens, follow the Cryo-DISSECT protocol. An overview of the protocols is shown in Fig. 1A. Be sure to process and analyze the samples in due time (see Note 3).
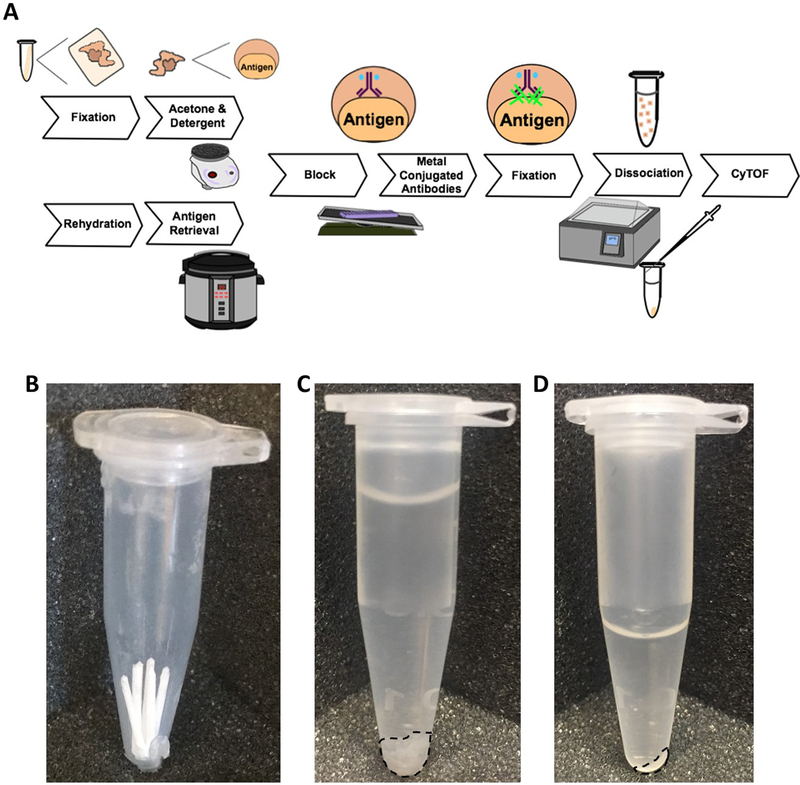
Fig. 1. Overview of methodologies.
(A) Cryopreserved or fresh tissue undergoes fixation followed by acetone and detergent incubation, while FFPE tissue initially undergoes rehydration and antigen retrieval. Both protocols merge for block, staining, signal fixation and dissociation. Under each step is the equipment used for performing the techniques, while tissue states are diagramed above the steps. Samples are normalized by: (B) number of curls before fixation, (C) tissue volume before dissociation, (D) and number of cells after dissociation prior to cytometry analysis.
3.1. Starting DISSECT from fresh tissue
Isolate tissue and divide it into small (<1mm) portions.
Fix tissue in 4% PFA for 20 min at room temperature (RT).
Follow the Cryo-DISSECT protocol from 3.3.5.
3.2. Starting DISSECT from FFPE tissue (FFPE-DISSECT)
Cut 2–5 50 μm curls using a microtome and put them into a 1.5 mL Eppendorf tube (will appear similar to Fig. 1B) (see Note 4).
Incubate tubes of FFPE curls on heat block at 65 °C for 25 min.
Add 1 mL of histoclear to each pre-warmed tube. Vortex and check the tubes to determine if paraffin wax is melted (see Note 5). If paraffin wax is not melted, incubate tubes containing histoclear and curls for 8 min on the heat block at 65 °C. If paraffin wax is melted, incubate tubes containing histoclear and curls for 8 min at RT (see Note 6).
Pellet tissue at 2000 xg for 2 min and remove supernatant. Repeat twice at RT (see Note 7 prior to rehydration and antigen retrieval).
Suspend tissue in 1mL of 100% ethanol, vortex and incubate at RT for 8 min. Pellet tissue at 2000 xg for 2 min or allow it to settle and carefully remove supernatant. Repeat once.
Suspend tissue in 1 mL of 70% ethanol (see 2.2.5), vortex and incubate at RT for 8 min. Pellet tissue at 2000 xg for 2 min or allow it to settle and carefully remove supernatant. Repeat once.
Suspend tissue in 1 mL of 50% ethanol (see 2.2.6), vortex and incubate at RT for 8 min. Pellet tissue at 2000 Xg for 2 min or allow it to settle and carefully remove supernatant. Repeat once.
Suspend tissue in 1 mL 1× PBS.
Pellet tissue at 2000 xg for 2 min and remove supernatant. Repeat twice. If sample is greater than 300 μL when pelleted, split up sample into individual tubes.
While washing samples, make a 1× DAKO antigen retrieval buffer (see 2.2.7) and heat the solution with a loosened lid in pressure cooker on the “keep warm” setting.
Suspend tissue in 0.3% Triton X-100 (see 2.2.8). Incubate at RT for 10 min.
Pellet tissue at 2000 xg for 2 min and remove supernatant.
Suspend tissue in 1 mL 1× PBS.
Pellet tissue at 2000 xg for 2 min and remove supernatant.
Suspend tissue in 1mL of hot 1× antigen retrieval buffer. Place tubes with lids open in rack in pressure cooker, lock lid, and set for 4 min at high pressure. Leave tubes in pressure cooker for 20 min total.
Remove tubes from pressure cooker and allow 20 min to cool on the bench at room temperature. After this step, tissue should assume new appearance and be much easier to pellet because any agar will have melted away.
Pellet tissue at 2000 xg for 2 min and remove supernatant.
Suspend tissue in 1 mL 1× PBS and transfer to a new pre-labeled tube.
Pellet tissue at 2000 xg for 2 min and remove supernatant. Repeat twice. Tissue can be stored at 4 °C on 1× PBS until use.
Follow the Cryo-DISSECT protocol from 3.3.17.
3.3. Starting DISSECT from frozen tissues (Cryo-DISSECT)
Store tissues at −80 °C until processing.
Cut 2–5 50 μm sections using a cryostat at −20 °C (Fig. 1B) (see Note 4).
Place sections into a 1.5 mL Eppendorf tube and keep at −20 or −80 °C until needed.
If necessary, thaw samples into 4% PFA for 20 min at RT (see Note 8).
Pellet tissue at 2000 xg for 2 min and remove supernatant (for general advice, see Notes 9 and 10).
Suspend tissue in 1 mL 1× PBS; incubate for 5 min.
Pellet tissue at 2000 xg for 2 min and remove supernatant. Repeat twice. Wash more times for longer periods if OCT remains in the sample (see Note 6).
If sample is greater than 300 μL when pelleted, split up sample.
Pellet tissue at 2000 xg for 2 min and remove supernatant (see Notes 7 and 11 prior to acetone).
Vortex sample vigorously to ensure that the tissue does not remain pelleted.
Add 1 mL of acetone that was kept at −20 °C, vortex briefly, and immediately place the tube into a minifuge.
Spin for approximately 10–20 seconds at 2000 xg to pellet tissue.
Pour off acetone and let dry for approximately 30 seconds.
Suspend tissue in 1 mL of XX detergent solution (see 2.1.16).
Gently vortex or agitate for 30 min at RT.
Pellet tissue at 2000 xg for 2 min and remove supernatant. Add 1mL of 1× PBS. Repeat once. Tissue can be stored at this step at 4 °C in 1× PBS for a few days if needed.
Suspend tissue in 300 μL of Flow Block (see 2.1.12) (Fig. 1C).
Incubate tubes upright for 30 min with gentle rocking.
Add 1mL of 1× PBS.
Pellet tissue at 2000 xg for 2 min and remove supernatant.
Suspend tissue in 100 μL of primary antibodies in Flow Block.
Incubate overnight, upright, covered (or in a dark room), with gentle rocking. Be sure to confirm that stain has penetrated by microscopy (Fig. 2A) (see Note 12).
Add 1 mL of 1× PBS.
Pellet tissue at 2000 xg for 2 min and remove supernatant. Repeat twice.
If needed, suspend tissue in secondary antibody in 100–200 μL of Flow Block.
Incubate for 1 h covered with gentle rocking.
Add 1 mL of 1× PBS.
Pellet tissue at 2000 xg for 2 min and remove supernatant. Repeat twice.
To fix signals, suspend tissue in 4% PFA.
Incubate for 30 min with gentle rocking.
Add 1 mL of 1× PBS.
Pellet tissue at 2000 xg for 2 min and remove supernatant. Repeat twice.
Enzymatic dissociation: Suspend tissue in 200 μL dissociation solution (see 2.1.24) (see Note 13).
Incubate for 1 h in the 37 °C water bath. For FFPE tissue, incubate for 20 min.
Add 800 μL of 0.003% Triton X-100 in 1× PBS (see 2.1.22).
Mechanical dissociation: passage tissue through a 27.5 gage needle connected to a 1mL syringe 5–10 times until solution is cloudy and most the tissue is dissociated.
To remove large debris and undigested material, filter cells through a 35 μm-nylon mesh filter. This can be done with a tube cap filter (designed for a 5 mL flow cytometry tube) directly into a clean 1.5 mL Eppendorf tube (Fig. 1D).
Pellet cells at 5000 xg for 5 min and remove supernatant.
Suspend cells in 100 μL of 0.5% BSA (see 2.1.29) in PBS overnight at 4 °C if not running immediately. If running immediately, go to next step.
Add 1 mL 0.003% Triton X-100 in 1× PBS. Single cells can be imaged via fluorescently labelled secondary antibodies to confirm staining (Fig. 2B) (see Note 12).
Pellet cells at 5000 xg for 5 min and remove supernatant.
Suspend cells in 100 μL of intercalator (see 2.1.26).
Incubate for at least 20 min and at most 48 hh.
Add 1 mL of 0.003% Triton X-100 in 1× PBS.
Pellet cells at 5000 xg for 5 min and remove supernatant.
Add 1 mL of 0.003% Triton X-100 in DI H2O (see 2.1.23).
Determine the number of cells in each sample by loading 10 μL onto a hemocytometer.
Pellet cells at 5000 xg for 5 min and remove supernatant. Leave samples on ice in the dark until they are ready to load.
Re-suspend cells in a volume of normalization bead solution (2.1.27) to achieve a concentration of 2.5–5 × 105 cells/mL, or, if cell density is insufficient, into the minimum sample volume for the instrument (usually 350–450 μL).
Transfer to a 5 mL filter top tube immediately before loading on the CyTOF. (Alternatively, fluorescent flow cytometry can be used with replacing the intercalator with a DNA dye such as Hoechst).
After samples are finished running on the CyTOF instrument, FSC data files can be uploaded to http://www.cytobank.org for analysis. Optionally, these files can be normalized using elemental beads [12].
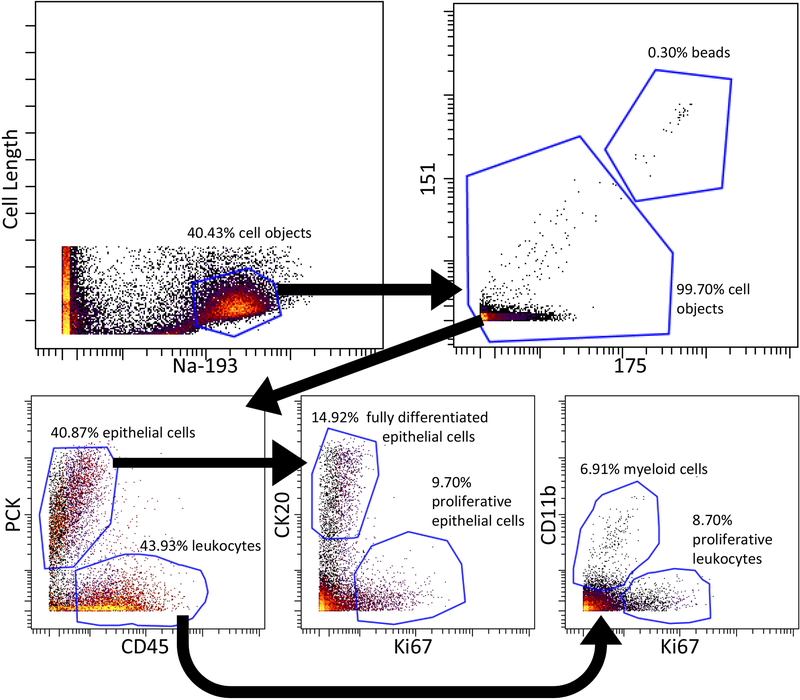
The intact cell population is determined through a bivariate plot comparing cell length and Ir intercalator (channel 193 or 191). Once the cell population is identified, elemental beads can be gated out via metals 151 and 175 (and others). The cell population can then be analyzed by bivariate plots for marker expression and further sub-population classification (Fig. 3).
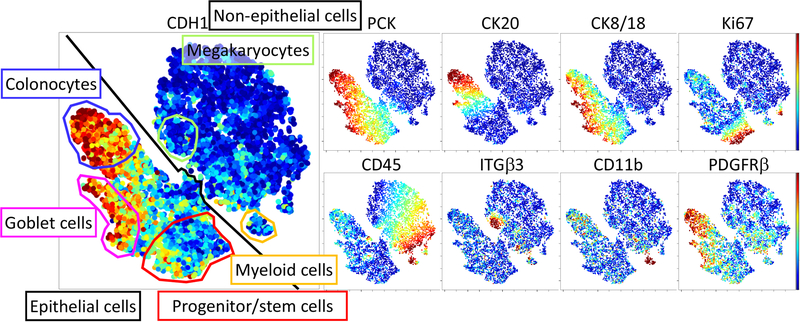
Different cell populations can also be identified by t-distributed stochastic neighbor embedding (t-SNE) analysis, allowing the visualization of multidimensional data in 2-D space. Overlaying different markers on a t-SNE map allows for the identification of cell clusters by their properties (Fig. 4). (see Note 14).
Fig. 2. Immunofluorescence validation at different stages of processing.
(A) Whole mount tissue staining before dissociation. (B) Single cell suspension after dissociation. Columnar epithelial morphologies are observed.
Fig. 3. Bivariate plot strategies for analyzing cell populations from Cryo-DISSECT mass cytometry data of the human colon.
Cells were gated as objects with positive staining of Ir intercalator and exceeding a certain ion cloud size. Elemental normalization beads were further excluded in the 155 and 175 channels from cell objects. Within the cell object population, epithelial cells were PCK-positive, while PCK-negative cells represented non-epithelial cells, a population of which was CD45+ leukocytes. Amongst epithelial cells, differentiated cells (on the luminal surface) were identified by CK20 expression while proliferative cells were marked by Ki67 expression. Amongst leukocytes, myeloid (monocyte and others) cells were marked by CD11b, while proliferative leukocytes were marked by Ki67.
Fig. 4. t-SNE analysis of cell populations from Cryo-DISSECT mass cytometry data of the human colon.
Heat overlays (low to high) represent the Arcsinh-transformed expression level of indicated proteins determined by mass cytometry. Cell populations (color-coded gates) were manually identified on the t-SNE plot based on marker expression, as detailed in Fig. 3.
Acknowledgements
This work was supported by NIH grants R01DK103831 and P50CA095103 (K.S.L., C.R.S.), U01CA215798 (A.J.S.), and T32AI007281 (C.R.S.).
4. Notes
Once thawed, collagenase and dispase aliquots are good for at least a month stored at 4 °C.
The 50× Ir Intercalator stock solution is good for at least 6 months stored in the dark at 4 ˚C between uses.
Samples should be processed to completion within a week of thawing or de-paraffinization. As with any mass cytometry experiment, to avoid the possibility of cross-labeling between heavy metals and antibodies, run samples within 24 h of staining. DISSECT samples can be more prone to cross-labeling artifacts due to the level of processing required.
Under some circumstances, cutting 50 μm sections from FFPE- or cryo-blocks can present problems with crumbling. Weigh paper (or something similar) can be used to collect tissue fragments which do not remain in the curl and fall during cutting. Cutting can be made easier by trimming down the amount of embedded material with a razor blade. Too many curls in 1 tube during de-paraffinization can result in incomplete wax removal and rehydration. The initial melted or thawed embedding material should be less than 250 μL in volume. Samples can be pooled after retrieval if necessary.
Tissues in histoclear may refuse to settle and even swirl around in the tube in response to electrostatic charges from gloved fingers in near contact. This can make removal of solution without disturbing the tissue pellet difficult. There are a number of ways this could be overcome. For instance, biopsy bags have been used with some success, but they tend to wick and carry solutions over between washes, and it can be difficult to recover the tissue from the mesh [13]. Larger volumes or different types of tubes may also help, though it’s important to choose materials with respect to their resistance to histoclear. Even polypropylene tubes are somewhat damaged by histoclear, so it is helpful to change tubes after the tissue is hydrated. Furthermore, it is recommended to avoid spinning tissues faster than 5000 xg to prevent tissue from pelleting too densely or being damaged. If tissue is still not settling, carefully remove the histoclear, rotating and tilting the tube to allow for liquid to move away from tissue as it sticks to the side of the tube. If tissues are still difficult to pellet and aspirate before antigen retrieval, it is suggested to prepare excess antigen retrieval solution, and buffer exchange it onto the tissue (add a volume, carefully remove as much as possible, then add another volume). It may be helpful to repeat (with hot buffer) after antigen retrieval as well, as it is possible to still find wax precipitating in the cooling solution.
The first washes of both procedures are intended to remove the embedding matrix (paraffin for FFPE-DISSECT and OCT for Cryo-DISSECT), and subsequent staining is dependent on its complete removal. Because of this, users should normalize samples such that each tube contains a comparable amount of matrix, and split any samples that may contain too much material. Washes can be lengthened or added to assist in removal of the embedding matrix. If possible, embedding material should be reduced prior to the cutting of sections by trimming the tissue block.
Before beginning antigen retrieval (FFPE-DISSECT) or acetone (Cryo-DISSECT) procedures, sample should be normalized such that each tube contains roughly the same pelleted volume of tissue. This can be done by splitting or pooling like samples, and ensures that all samples consistently experience the same concentration of reagents. Likewise, samples should be normalized by cell number (determined in 3.3.47) through dilution immediately before loading onto CyTOF for analysis.
For cryopreserved tissue, it is important to make a distinction between tissue which has or has not been fixed prior to freezing. It is recommended to fix tissue for no longer than 1 h if doing so prior to freezing. Curls cut from tissue that has never been fixed will need to be fixed gently in a large volume of 4% PFA while undergoing thawing for 20 min before further PBS washes. Users should follow cryopreservation guidelines concerning the use of sucrose and freezing speeds that are appropriate for their tissue of interest, and cut 5–10 μm sections for slides (in addition to thick sections for cytometry) to verify that morphology and cell integrity have been properly preserved.
Always check the lid and sides of tubes between washes for adherent tissue. If tissue is sticking to the side, adding Triton-X 100 or BSA to the 1× PBS may help to release it. It is advised to use Triton-X 100 below its critical micelle concentration in a wash to reduce its capacity to damage lipids [14]. Incubating tubes upright ensures the tissue remains submerged. Once hydrated in 1× PBS with Triton-X 100, tissues tend to pellet much easier.
If tissue is difficult to pellet, for any given incubation or wash, aspirate the most difficult samples first. This would leave a large residual volume for the first sample to avoid tissue loss. All other samples should be aspirated to that volume to normalize wash efficiency. Add an additional wash or two of the given buffer for all samples to compensate for the large residual volume at each step. This ensures that (a) samples are treated the same and (b) samples are given enough buffer exchanges to sufficiently wash the tissue.
The acetone and XX detergent wash sequence utilized for Cryo-DISSECT is somewhat time and temperature sensitive. To ensure that samples are handled consistently, prep them in batches of 4–6 samples maximum. It is crucial not to begin with too much tissue: a residual 1× PBS volume as little as 100 μL can dilute the acetone and prevent cells from pelleting properly, resulting in tissue loss or difficulty decanting.
Users with immunohistochemistry or immunofluorescent staining on FFPE and cryopreserved tissues are well-suited to perform these procedures, as many quality control steps involve imaging. It may be helpful for users to modify the buffers, antigen retrieval, and staining conditions to those where the staining can be visualized in imaging (either FFPE- or Cryo-DISSECT). Generally, antibodies that work in the respective imaging applications have a better chance of working in FFPE- or Cryo-DISSECT coupled to cytometry. Validation of antibodies should occur before any experiment. Additionally, it is important and advantageous to validate the presence of signals at every step by checking a portion of the sample suspension on the microscope with a fluorescently-labeled secondary antibody (Fig. 2).
The enzymatic and mechanical dissociation procedure may require optimization to accommodate different tissues, fixations, and antigen retrieval techniques. While a variety of tissues have been successfully processed using the dissociation solution described above, the duration has varied significantly. FFPE intestinal tissue, presumably due to the high level of processing, dissociates much faster (20 min dissociation) than the same tissue that has only been lightly fixed (1h dissociation time). FFPE tissue that has been hydrated, but not antigen retrieved, however, will not dissociate. To optimize dissociation, multiple digests of the same tissue should be set up and mechanical dissociated at selected time points to identify the best conditions. Tissues should mostly pass through the syringe after some light trituration (moving the plunger a short distance in and out 10–20 times quickly). For lightly fixed mouse intestinal tissue, the muscle normally does not dissociate and is filtered out. Samples should become cloudy with cells after 5–10 full-volume passages with a syringe, and mostly single cells bearing intact morphology should be observed under the microscope (Fig. 2B). For some tissues, pipetting alone may be enough to separate cells.
The human colon tissue analyzed here has two major cell populations: epithelial and non-epithelial cells. Within epithelial cells, there are distinct absorptive, secretory, and progenitor cell populations, while in the non-epithelial cells myeloid and megakaryocyte cell populations can be observed.
5. References
- 1.Shukla S, Meeran SM (2014) Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim Biophys Acta 1840:3494–3502 [DOI] [PubMed] [Google Scholar]
- 2.Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M et al. (2017) Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545:187–192 [DOI] [PubMed] [Google Scholar]
- 3.de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. (2017) A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 543:676–680 [DOI] [PubMed] [Google Scholar]
- 4.Singh A, Settleman J (2010) EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29:4741–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simmons AJ, Lau KS (2017) Deciphering tumor heterogeneity from FFPE tissues: Its promise and challenges. Mol Cell Oncol 4:e1260191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrahamsen I, Lorens JB (2013) Evaluating extracellular matrix influence on adherent cell signaling by cold trypsin phosphorylation-specific flow cytometry. BMC Cell Biol 14:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng J, Mohammadreza A, Gao W, Merza S, Smith D, Kelbauskas L, et al. (2014) A minimally invasive method for retrieving single adherent cells of different types from cultures. Sci Rep 4:5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons AJ, Banerjee A, McKinley ET, Scurrah CR, Herring CA, Gewin LS et al. (2015) Cytometry-based single-cell analysis of intact epithelial signaling reveals divergent MAPK activation during TNF-α-induced apoptosis. Mol Syst Biol 11:835–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendall SC, Davis KL, Amir el-AD, Tadmor MD, Simonds EF, Chen TJ et al. (2014) Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157:714–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons AJ, Scurrah CR, McKinley ET, Herring CA, Irish JM, Washington MK et al. (2016) Impaired coordination between signaling pathways is revealed in human colorectal cancer using single-cell mass cytometry of archival tissue blocks. Sci Signal 9:rs11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW (2013) Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank 11:101–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, et al. (2013) Normalization of mass cytometry data with bead standards. Cytometry A 83:483–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corver WE, ter Haar NT (2011) High-resolution multiparameter DNA flow cytometry for the detection and sorting of tumor and stromal subpopulations from paraffin-embedded tissues. Curr Protoc Cytom Chapter 7:Unit 7.37 [DOI] [PubMed] [Google Scholar]
- 14.Koley D, Bard AJ (2010) Triton X-100 concentration effects on membrane permeability of a single HeLa cell by scanning electrochemical microscopy (SECM). Proc Natl Acad Sci U S A 107:16783–7 [DOI] [PMC free article] [PubMed] [Google Scholar]