Abstract
Cryptosporidium is a protozoan, apicomplexan, parasite that poses significant risk to humans and animals, as a common cause of potentially fatal diarrhea in immunodeficient hosts. The parasites have evolved a number of unique biological features that allow them to thrive in a highly specialized parasitic lifestyle. For example, the genome of Cryptosporidium parvum is highly reduced, encoding only 3,805 proteins, which is also reflected in its reduced cellular and organellar content and functions. As such, its remnant mitochondrion, dubbed a mitosome, is one of the smallest mitochondria yet found. While numerous studies have attempted to discover the function(s) of the C. parvum mitosome, most of them have been focused on in silico predictions. Here, we have localized components of a biochemical pathway in the C. parvum mitosome, in our investigations into the functions of this peculiar mitochondrial organelle. We have shown that three proteins involved in the mitochondrial iron‐sulfur cluster biosynthetic pathway are localized in the organelle, and one of them can functionally replace its yeast homolog. Thus, it seems that the C. parvum mitosome is involved in iron‐sulfur cluster biosynthesis, supporting the organellar and cytosolic apoproteins. These results spearhead further research on elucidating the functions of the mitosome and broaden our understanding in the minimalistic adaptations of these organelles.
Keywords: Apicomplexans, immunofluorescence assay, iron–sulfur clusters, mitochondria, mitosomes
IRON (Fe) and sulfur (S) are vital elements for every living cell. While iron‐sulfur clusters can be spontaneously formed in nature, both elements, when unbound, are toxic to the living cell, and thus specialized machineries have developed for their assembly. Current studies have identified four different biosynthetic pathways responsible for the assembly of iron‐sulfur clusters. In eukaryotes, the most common biosynthetic pathways are the Iron Sulfur Cluster (ISC) machinery that is present in most mitochondria investigated so far (Lill et al. 2012) in addition to the Cytosolic Iron‐sulfur cluster Assembly (CIA) machinery, components of which are found in the cytosol of all studied eukaryotes (Tsaousis et al. 2014). Furthermore, the SUlFur mobilization (SUF) machinery is localized in the plastids of plastid bearing organisms (including those apicomplexans, which still maintain their apicoplast), but it has recently been found to be present in various protists as well, including Blastocystis (cytosolic localization; Tsaousis et al. 2012), Pygsuia (cytosolic/mitochondrial localization; Stairs et al. 2014) and Stygiella (Leger et al. 2016). Even in the only currently confirmed amitochondriate organism, Monocercomonoides, a diverse homolog of the SUF machinery has been identified (Karnkowska et al. 2016). Lastly, components of the nitrogen fixation (NIF) machinery are encoded by the genomes of Entamoeba (Ali et al. 2004; van der Giezen et al. 2004) and its free‐living relative Mastigamoeba (Nyvltova et al. 2013).
The common feature among these machineries is the existence of three major components: a sulfur donor, an iron donor, and a scaffold protein for the assembly of the clusters. In the ISC pathway, these roles have been undertaken by the IscS, Frataxin, and IscU proteins, respectively. In eukaryotes, these proteins have been thoroughly characterized in humans (for review, see Rouault and Maio 2017), yeast (for review, see Braymer and Lill 2017), and trypanosomes (Basu et al. 2016). Recent reports have functionally characterized these proteins in other protists, including Blastocystis (Tsaousis et al. 2012) and microsporidia (Freibert et al. 2017; Goldberg et al. 2008).
Among apicomplexans, three of the different types of Fe‐S cluster assembly machineries can be found (Ali and Nozaki 2013): the mitochondrial ISC machinery, the apicoplast SUF machinery, and the cytosolic CIA machinery. To the best of our knowledge, the only studies on Fe‐S cluster assembly machineries in apicomplexans have been focused on the apicoplast‐localized SUF machinery in Plasmodium species (Charan et al. 2017). These studies have demonstrated the importance of this pathway in the survival of the parasite: the SUF machinery supports the apicoplast‐specific apoproteins (Gisselberg et al. 2013), some of which are vital for the sustainability of the parasites (Charan et al. 2014; Haussig et al. 2014). Importantly, however, with the exception of specific prediction analyses (Dellibovi‐Ragheb et al. 2013; Seeber 2002), functional investigations of the mitochondrial ISC machinery of apicomplexan parasites are completely absent.
Cryptosporidium is an intracellular, but extracytoplasmic apicomplexan parasite recently classified as a gregarine (Ryan et al. 2016). The parasite harbors a remnant mitochondrion, the mitosome, whose function(s) are still under debate (Mogi and Kita 2010). Cryptosporidium parvum and C. hominis mitosomes lack mitochondrial DNA, TCA cycle enzymes (with the exception of malate dehydrogenase), pyruvate dehydrogenase, complex 3 and 4 of the electron transport chain, and ATP synthase subunits with the exception of α and β (Liu et al. 2016; Mogi and Kita 2010). With the exclusion of two chaperone proteins (Cpn60 and Hsp70) that have been localized to the mitosome of the C. parvum sporozoite stage (Riordan et al. 2003; Slapeta and Keithly 2004), studies showing localization or biochemical characterization of Cryptosporidium mitochondria are lacking. With regard to the Fe‐S cluster assembly machinery, the only published study demonstrated heterologous localization of the C. parvum sulfur donor protein IscS and the scaffold protein IscU in Saccharomyces cerevisiae mitochondria (LaGier et al. 2003). Thus, direct localization studies of this machinery in Cryptosporidium are nonexistent.
Here, we employed various tools to characterize the ISC machinery of Cryptosporidium parvum. First, we generated specific antibodies against C. parvum IscS, IscU, and Frataxin. We then used an in vitro culture that can propagate the parasite in long term (Miller et al. 2018) to localize the proteins using western blotting and indirect immunofluorescence microscopy. This study represents the first localization of ISC biosynthetic pathway components in a Cryptosporidium mitosome, using not only the more frequently studied sporozoite stage but also other life stages of the parasite. As a result, this work leads the way to further studies on characterizing the peculiar mitochondrion of Cryptosporidium.
Materials and Methods
Bioinformatics analyses
To identify candidate ISC pathway proteins, we used CryptoDB (Heiges et al. 2006). BLASTP (Boratyn et al. 2012) were employed to search the database and identify full‐length sequences of the protein coding genes involved in the ISC pathway. The Cryptosporidium amino acid sequences were then aligned with homologous sequences from Homo sapiens, Saccharomyces cerevisiae, and Escherichia coli using MUSCLE (Edgar 2004). The secondary structures of the proteins were predicted using a combination of PSIPRED (McGuffin et al. 2000) and HMMTOP (Tusnady and Simon 2001). For the in silico prediction of potential mitochondrion‐targeted proteins, we employed MitoFates (Fukasawa et al. 2015), MitoProt (Claros 1995), Predotar (Small et al. 2004), and TargetP (Emanuelsson et al. 2007).
Cryptosporidium parvum culturing and infection
The human cell line COLO‐680N was cultivated in RPMI‐1640 medium supplemented with 10% FBS, 100 U/ml of penicillin, 100 μg/ml of streptomycin. Cryptosporidium parvum oocysts (Iowa strain) were obtained from Bunch Grass Farm (Deary, Idaho, USA). For a typical infection, 1 × 105 oocysts were used to infect 25‐cm2 cell culture flasks at between 70% and 80% confluency (1.7 to 2 × 106 cells) giving a multiplicity of infection (MOI) of approximately 0.2. When smaller cell surfaces were used, the number of COLO‐680N and oocysts were altered proportionally. Excystation was done according to Miller et al. (2018).
Generation of antibodies used in this study
The predicted Cryptosporidium parvum frataxin, IscS, and IscU genes were amplified from genomic DNA (reference accession number XM_625594, AY029212, XM_627477, respectively) and cloned into an E. coli pET14b expression vector as a NdeI/BamHI fragment allowing His N‐terminal tagging. The recombinant DNA constructs were transformed chemically in the E. coli BL21(DE3) pLysS expression strain (Novagen, Millipore (UK) Limited, Watford, UK). Protein expression was done in 2 L of LB, seeded with an overnight inoculum and left at 37 °C for 16 h or until an OD600 reached 0.4–0.6, at which point IPTG was added to a final concentration of 0.5 mM. The cultures were grown for a further 4 h before centrifuging at 3,000 g for 30 min. The supernatant was then discarded and the pellet washed twice in PBS. The pellet was then weighed, before resuspending it in Novagen BugBuster reagent (Novagen) at 5 ml/g of pellet and 1 μl/ml of Benzonase, which was added to the mixture to inhibit proteolysis. The mixture was left to shake at a low setting for 20 min at room temperature and subsequently centrifuged at 16,000 g for 20 min at 4 °C. The His‐tagged proteins (Frataxin, IscU and IscS) were purified on Nickel affinity column. Eluted proteins were run onto a 15% SDS‐PAGE gels and the bands corresponding to the proteins of interest were then excised from the gels and used to inoculate a rabbit (for IscS) or rats (for IscU or Frataxin), which subsequently produced polyclonal antibodies to the target proteins.
For generation of antibodies against C. parvum 60S ribosomal protein L23A, peptide 1 (Aa‐29–43: nh2‐C+VKNSKRSSTTKIRTR‐conh2) and peptide 2 (58–73: ac‐PKYERKSIKTQKSLDC‐conh2) were synthesized. For peptide 1, a cysteine was added to target the coupling site at the carrier protein KHL (Keyhole Limpet Hemocyanin). BLASTP search revealed that neither peptide was present in the human protein homolog. Notably, peptide 1 was 100% identical with C. ubiquitum and C. hominis, but only 48% with C. muris; while peptide 2 shared 100% identity with C. hominis and 88% with C. ubiquitum. The conjugated peptides were raised in laying hens and isolated from the yolk of their eggs (Eurogentec, Southampton, UK).
Protein extraction and western blot analysis
Cryptosporidium parvum oocysts (5× 107) and COLO‐68ON cells (4–5 × 105) were lysed in 160 μl buffer L [20 mM Tris–Cl (pH 7.5), 10 mM EDTA, 10 mM EGTA, 150 mM NaCl and 1% (w/v) Triton and one tablet of protease inhibitor cocktail (Roche Products Limited, Welwyn Garden City, UK)] and sonicated at 14 watts/cm2, 3 × 30 s to break down the oocyst envelope, and shear the DNA. Subsequently, 40 μl of 5× SDS loading buffer (5% β‐mercaptoethanol, 0.02% bromophenol blue, 30% glycerol, 10% sodium dodecyl sulfate, 250 mM, pH 6.8) were added. Ten microliters of total proteins were analyzed on 15% SDS‐PAGE gels and stained with Coomassie Blue or transferred onto PVDF membrane (Roche Products Limited, Welwyn Garden City, UK) and probed with primary antibodies at 1:3,000 dilution. Anti‐chicken HPR‐conjugated antibodies (Sigma‐Aldrich Company Ltd, Gillingham, UK) were used as a secondary antibody (1:50,000) and the peroxidase reaction was catalyzed using ECL prime reagent (GE Healthcare, Amersham, UK).
Immunofluorescence microscopy
COLO‐680N infection was carried out in Nunc Lab‐Tek chamber slides (Permanox, 4.2 cm2), Nalgene Nunc International, Rochester, NY, USA. At harvesting points, cultures were washed with 1× PBS and then fixed in methanol for 10 min at room temperature. Next, the methanol was removed and the cells were permeabilized with 0.002% Triton‐X100 in 1× PBS at room temperature for 30 min. Cells were then washed three times prior to incubation for 1 h with Cryptosporidium‐specific commercial antibodies Crypt‐a‐glo (WaterborneTM; New Orleans, LA, USA, specific to Cryptosporidium oocysts) in dilution 1:10, and Sporo‐glo (WaterborneTM; New Orleans, specific to Cryptosporidium intracellular life cycle stages) in dilution 1:10 (Miller et al. 2018). Cells were subsequently washed three times with 1 × PBS, prior to incubation overnight with Cryptosporidium‐specific antibodies (anti‐Frataxin, dilution 1:200; anti‐IscS, dilution 1:100; anti‐IscU, dilution 1: 50). Cells were washed three times with 1xPBS prior to incubation with secondary antibodies Alexa 488 (Green; Molecular Probes, Thermo Fisher Scientific, Waltham, MA, USA) or Alexa 594 (Red; Molecular Probes, Thermo Fisher Scientific) at 1:200, for one hour. Cells were washed a further three times with 1× PBS and mounted with ProLong Gold antifade reagent (Thermo‐Fischer Scientific), using high‐performance coverslips (Carl Zeiss Microscopy, Carl Zeiss AG, Oberkochen, Germany). Slides were visualized by fluorescence microscopy using a Zeiss Elyra P1 confocal microscope (Carl Zeiss AG).
Yeast complementation studies of IscS
Yeast strains and growth conditions
The S. cerevisiae strain used in this study was the heterozygous diploid YCL017C (MATa/MATalpha his3delta1/his3delta1 leu2delta0/leu2delta0 lys2delta0/+ met15delta0/+ ura3delta0/ura3delta0 deltaNFS1:: KAN R ), the knockout diploid strain was purchased from Dharmacon, Horizon Discovery, Cambridge, UK, from the Yeast HetDip Knock Out collection, Clone ID: 23424. Typically, the YCL017C strain was grown at 30 °C in YEPD (1% Yeast Extract, 2% Peptone, 2% dextrose or glucose) in the presence of 200 μg/ml geneticin (G418).
Plasmid construction
The predicted Cryptosporidium parvum IscS gene was amplified from genomic DNA (reference accession number AY029212) and cloned into E. coli pET14b expression vector. For yeast complementation experiment, the fragment was subcloned into the bi‐directional yeast expression vector pBEVY‐L (Miller et al. 1998) as a SmaI/ SacI fragment. pBEVY‐L carries the LEU2 selection marker and can be propagated in synthetic complete media lacking leucine.
Yeast transformation and selection
Saccharomyces cerevisiae cells were transformed from overnight culture using the lithium acetate/ single stranded (ss) carrier DNA/ polyethylene glycol (PEG) method adapted from Gietz and Woods (Gietz and Woods 2002). Briefly, for each transformation, 1 ml of overnight culture was harvested at 13,000 g for 30 s and the pellet was washed once in sterile water. Then, reagents were added in the following order: 240 μl PEG 4000, 36 μl 1 M LiAc, 10 μl of 10 mg/ml ss DNA (recently boiled and sonicated), 2.5 μl β‐ mercaptoethanol, 2 μl plasmid (100–500 ng/μl), 70 μl sterile water. The mixture was left for 20 min at room temperature prior to heat shock at 42 °C for 20 min, then briefly spun down at 13,000 g to pellet the cells. Cells were resuspended in 200 μl of sterile water and 50 and 100 μl were plated out. Transformants were selected on synthetic dropout plates lacking leucine.
Sporulation (Adapted from Curran and Bugeja 2014)
Yeast transformants (one colony) were grown overnight in synthetic complete medium lacking leucine (Synthetic Complete (SC), ‐leu), with 2% glucose. Cells were then diluted 1/50 and 1/100 in 1 ml in SC, ‐leu medium enriched with 5% glucose and grown at 30 °C for approximately 8 h or until optical density reached OD600 = 0.1 to 0.3. Cells were spun at 3,000 g for 5 min and washed twice in sterile water. Cells were then resuspended in 1 ml of sterile 1% potassium acetate and left with vigorous shaking at 25 °C for 4–5 d. After 4 d, cells were examined under the microscope to evaluate the formation of tetrad cells or asci (in excess of 30%).
Isolation of spores (Adapted from Curran and Bugeja 2014)
To remove spores from asci, tetrad cells and nontetrad cells were harvested and resuspended in sterile water. Fifty microliters of lyticase (from a 100,000 units/ml stock) were added to digest the ascal wall, and the reaction was incubated at 37 °C for 10–15 min. One volume of sterile mineral oil was added and the mixture was vortexed for 2 min to mechanically disrupt asci and release spores. The oil/water mixture was spun at 3,000 g for 1 min in order to separate both phases. The mineral oil layer was enriched with hydrophobic spores, while the majority of vegetative cells and tetrad cells remained in the water. Spores were carefully removed from the mineral oil phase, mixed with sterile water and plated on appropriate media at dilutions ranging from 1/10 to 1/50. Spores were first plated out randomly on YEPD plates. Colonies were then individually picked up and transferred onto SC, ‐leu plates or YEPD, 200 μg/ml G418 plates. Cells (haploid or diploid) able to grow on both types of plates have a disrupted NFS1 gene (replaced by kanamycin marker) and carry the C. parvum putative homolog IscS (on pBEVY‐L).
Discrimination between haploid and diploid yeast transformants
This method was adapted from Huxley et al. (1990) and used here to ensure that cells growing on SC‐leu and YEPD, 200 μg/ml G418 were haploids and not diploids, thus demonstrating that C. parvum IscS can rescue delta NSF1 lethality in S. cerevisiae. For this analysis, the following primers were used: (1) 5′‐AGTCACATCAAGATCGTTTATGG‐3′, (2) 5′‐GCACGGAATATGGGACTACTTCG‐3′, (3) 5′‐ACTCCACTTCAAGTAAGAGTTTG‐3′. PCR mix: 1/5 colony, 0.2 μl each primer (100 pmole/μl), 2.0 μl dNTPs (10 mM), 0.25 μl GoTaq DNA polymerase, water to 20 μl. Hot start at 92 °C for 5 min, cycles 30 s at 92 °C, 30 s at 55 °C, 1 min at 72 °C. Expected product sizes were 404 bp for a Mat alpha haploid and 544 bp for a MAT a haploid. The presence of both bands indicated the presence of a diploid, which was not removed during the isolation of spores.
Results
Cryptosporidium ISC proteins
Due to the minimalistic properties of Cryptosporidium parvum mitosome, we sought to investigate the assembly of proteins constituting the ISC pathway of the parasite. Using BLASTP approach against various databases (nr and CryptoDB), we have identified a wide range of the components of the mitochondrial ISC machinery encoded by the C. parvum genome (Table S1). Interestingly, comparison with the C. andersoni and the C. ubiquitum genomes demonstrated that these species have a more complete range of components. To investigate the presence of compartment‐specific targeting signals, all the predicted Cryptosporidium spp. ISC proteins were analyzed using four different prediction programs (Table S2). These analyses revealed that all algorithms were abled to predict localization of IscS and IscU proteins in the mitochondria of all Cryptosporidium species, with the exception of MitoProt, and Frataxin was not predicted to be mitochondrial.
To predict the properties of these three proteins, the full‐length amino acid sequences of Cryptosporidium spp. IscS, IscU, and Frataxin proteins were aligned with yeast, human, and E. coli ISC proteins. Sequence alignment of all three proteins revealed conserved features associated with canonical ISC functions (Figs S1–S3). Interestingly, while all IscS homologs were predicted to harbor an N‐terminal mitochondrial targeting signal (MTS) (Fig. S1), in the case of the IscU, only C. ubiquitum was predicted to have one (Fig. S2). In the case of Frataxin, only C. ubiquitum and C. andersoni along with the yeast homolog were predicted to have an N‐terminal MTS (Fig. S3).
Cryptosporidium ISC machinery is expressed
To examine the expression and localization of these proteins in Cryptosporidium, we generated specific antibodies against the complete C. parvum IscS, IscU, and Frataxin proteins (named CpIscS, CpIscU, and CpFrataxin, respectively). To investigate the specificity of the antisera using western blots, we used proteins that were extracted from C. parvum Iowa strain oocysts. Western blotting experiments using the previously published anti‐CpCpn60 (Riordan et al. 2003) and newly generated anti‐CpL23A (control), anti‐CpIscS, anti‐CpIscU, and anti‐CpFrataxin antisera showed specific bands in molecular weights of 17, 29, 17, 16 kDa, respectively, in the protein extracts from the C. parvum oocysts, which were not present in the protein extracts from the noninfected COLO‐680N cells (Fig. 1).
Figure 1.
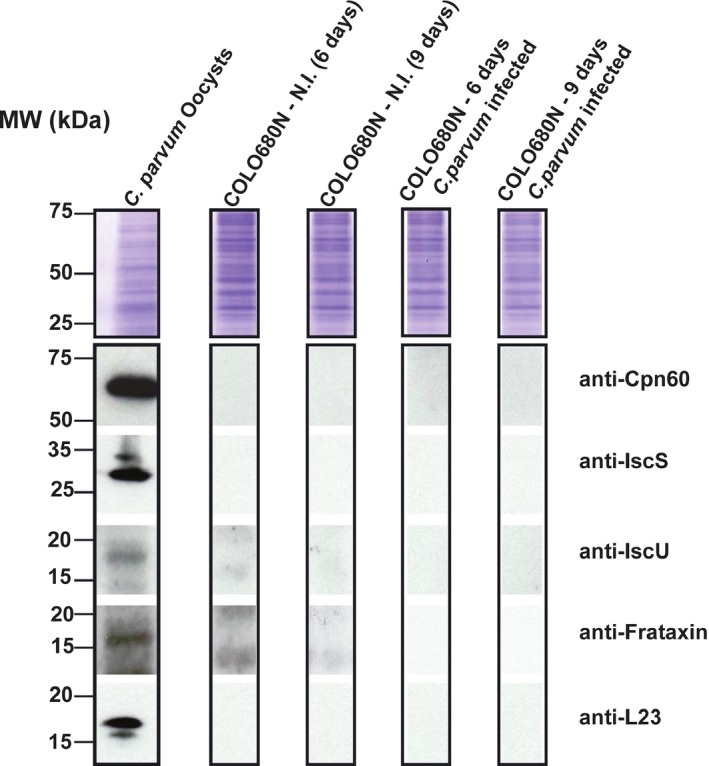
Resolution of total proteins by SDS‐PAGE and downstream analysis: Top panel, the protein gel was stained with Coomassie Blue, as a loading control reference. Other panels, Western blot analysis was done to probe for individual proteins, using corresponding antibodies against: Cpn60, IscS, IscU, frataxin, and L23A. Cryptosporidium parvum oocysts were isolated from calves infected with the C. parvum Iowa strain. COLO680N‐N.I corresponds to host control cells. COLO680N infected corresponds to host cells infected with C. parvum. Host cells noninfected and infected were cultured in parallel and harvested at day 6 and day 9 postinfection.
The ISC machinery is localized in Cryptosporidium mitosomes
To determine the cellular location of the C. parvum ISC proteins, indirect immunofluorescence analyses were carried out on 10‐day infected COLO‐680N cells and/or on isolated oocysts. Co‐localization of the various proteins within the oocyst of C. parvum was achieved by co‐staining with Crypt‐a‐glo, a FITC conjugated, monoclonal antibody with high specificity for a Cryptosporidium oocyst wall protein (Miller et al. 2018). Localization within other stages of C. parvum, sporozoites and intracellular stages, for example, was achieved by co‐staining with Sporo‐glo, a sensitive, FITC‐conjugated polyclonal antibody raised against C. parvum sporozoites.
CpIscS
Incubation of the infected cultures with the anti‐CpIscS revealed a life cycle‐dependent localization of CpIscS in immunofluorescent assays (Fig. 2A; Fig. S4). During the extracellular stages (e.g. sporozoites and merozoites), the signal from IscS was punctate (Fig. S4) and restricted to regions typically between 500 and 800 nm in diameter. DAPI‐stained oocysts of C. parvum showed successful permeation as the four nuclei of the contained sporozoites could reliably be observed in an oocyst. However, the absence of detectable fluorescence, within these sporozoites, suggested absence of IscS signal. It may, therefore, be assumed that IscS expression levels are either negligible so as not to be detectable or IscS is not being expressed at all, while the sporozoite remains within a matured (thick) oocyst. Immunofluorescent assays of the other intracellular stages revealed that while IscS remained expressed at detectable levels, its localization did not remain consistent. In detectable extracellular life cycle stages, determined as any detectable Sporo‐Glo not within/closely associated with a host cell and <4 μm in diameter, IscS was found concentrated in a region similar to that observed in the sporozoites. This indicates the presence of an organelle matching the physical descriptions of the mitosome, corroborating the hypothesis that the ISC pathway localizes to the mitosome of C. parvum. However, in occasions, immunofluorescent assays of intracellular life cycle stages, determined by positive Sporo‐Glo detection, proximity to host nucleus, amorphous/round shape, and an approximate parasite size exceeding 4 μm showed that IscS was more diffuse (Fig. S4). Indeed, it was hard to discern any regularly observable structural association with IscS expression in intracellular stages. Furthermore, in areas where later developmental stages of C. parvum were present, IscS staining appeared to decrease inversely proportional to the diffusion of genetic material, indicating that IscS expression declined as the sporozoites were formed.
Figure 2.
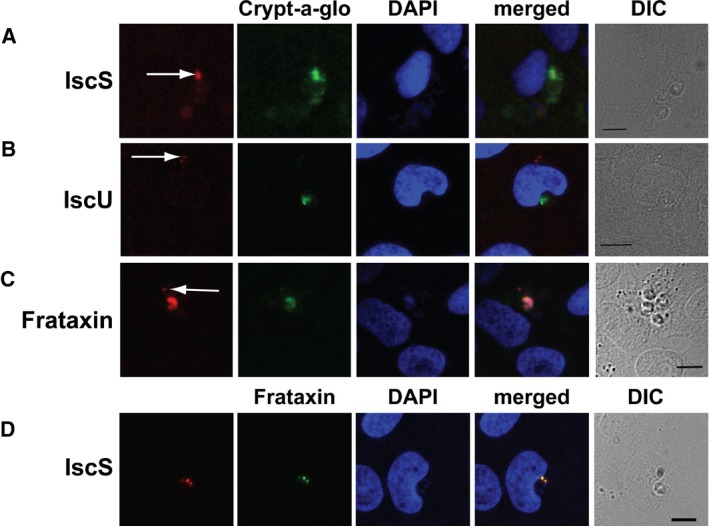
Cellular localization of IscS, IscU, and Frataxin‐like proteins in Cryptosporidium parvum by immunofluorescence microscopy. The nuclei of COLO‐680N host cells (large nuclei) and parasites (small nuclei) were labeled with DAPI (blue). (A) Rabbit antisera to CpIscS detects discreet structures (red) on C. parvum, whereas the Crypt‐a‐glo detects (green) the parasites. (B) Rat antisera to CpIscU detect discreet structures (red) on C. parvum, whereas the Crypt‐a‐glo detects (green) the parasites. (C) Rat antisera to CpFrataxin detect discreet structures (red) on C. parvum, whereas the Crypt‐a‐glo detects (green) the parasites. Differential interference contrast (DIC) images of the cells used for immunofluorescence. (D) Rabbit antisera to CpIscS detects discreet structures (red) on C. parvum that colocalize with the same discreet structures detected by CpFrataxin rat antisera (green). Scale bar 10 μm.
IscU
Anti‐CpIscU localization studies produced similar patterns to that of CpIscS when observed via immunofluorescent assays (Fig. 2B; Fig. S5). Life‐cycle stage‐dependent expression continued to be present; with Crypt‐a‐glo stained oocysts presenting DAPI‐stained sporozoite nuclei, but no detectable CpIscU signal. Conversely, several extracellular life cycle stages could be detected by residual Crypt‐a‐glo labeling or punctate DAPI stain, which did display detectable levels of CpIscU. However, in occasions, CpIscU signal appeared to be more diffused than that of CpIscS throughout the life cycle of the parasite (Fig. S5). This could indicate that CpIscU may not only be localized to a specific structure within the parasite, or at least not exclusively during the development of C. parvum.
Frataxin
Anti‐CpFrataxin localization studies showed similar patterns to those of CpIscS and CpIscU. CpFrataxin antibody labeling of infected cultures showed, again, a lack of CpFrataxin in mature oocysts, but abundance in other life cycle stages (Fig. 2C; Fig. S6). Most notably, CpFrataxin signal remained intense even in late stage zygotes, denoted by the intense Crypt‐a‐glo signal that lacked the characteristic “ring” of a mature oocyst. This is in contrast to both CpIscS and CpIscU that showed decreasing levels of expression leading up to zygote maturation. CpFrataxin signal also remained relatively diffuse throughout the life cycle, although dramatically more so in the intracellular stages, similarly to CpIscU. Despite this, fluorescence microscopy using both anti‐CpFrataxin the anti‐CpIscS antibodies demonstrated clear co‐localization of both proteins in distinct, punctuated structures within C. parvum intracellular stages (Fig. 2D). In addition, using confocal microscopy, the CpFrataxin labeling in sporozoites appeared considerably more localized than any other life cycle stage, appearing to congregate in a defined, approximately 500‐nm oval near the apical end of the parasite (Fig. 3; Video S1).
Figure 3.
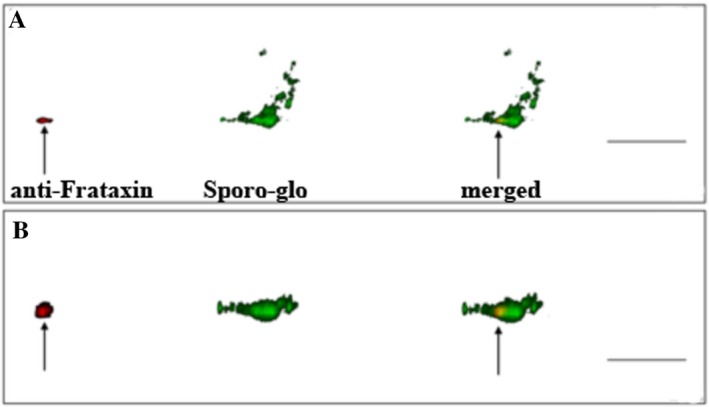
Confocal microscopy of an infected COLO‐680N culture, labeled with anti‐CpFxn (red) and SporoGlo (green). (A) A 3D, high‐resolution confocal image of a Cryptosporidium parvum extracellular life cycle stage in an infected COLO‐680N culture. Clear labeling of CpFrataxin (red) highlights an approximately 700‐nm wide oval (arrow) at the apical end of the parasite, a red outline has been drawn to illustrate the shape of the parasite and to highlight the location of the mitosome. (B) The same image rotated through 90° on the x‐axis. Scale bar: 5 μm.
Cryptosporidium IscS can functionally replace the yeast homolog
The CpIscS recombinant construct was transformed into a heterozygous ScNfs1p (ScIscS) knockout, strain YCL017C, where a homozygous knockout is lethal. In the knockout strain of YCL017C, the Nfs1 gene was replaced by a G418 resistance marker on chromosome III. Colonies of transformants were grown on SC‐leucine plates to select for S. cerevisiae containing the plasmid pBEVY‐L/IscS. Sporulation to isolate haploid, which might rescue Nfs1 lethality, in the presence of C. parvum IscS, was carried out in a nitrogen‐deprived liquid culture. Spores were removed from the ascus by enzymatic digestion of the cell wall, separated from nondigested tetrads and plated out on YEPD plates, as described in Materials and Methods. Spores were first plated out on YEPD before being individually picked onto SC‐leu and YEPD/G418 and tested for mating type via colony PCR (Fig. S7). We observed that CpIscS recovered the fatal ScNfs1 knockout, as determined from the matching colonies present on both leucine deficient media and G418 resistance plates (Fig. 4).
Figure 4.
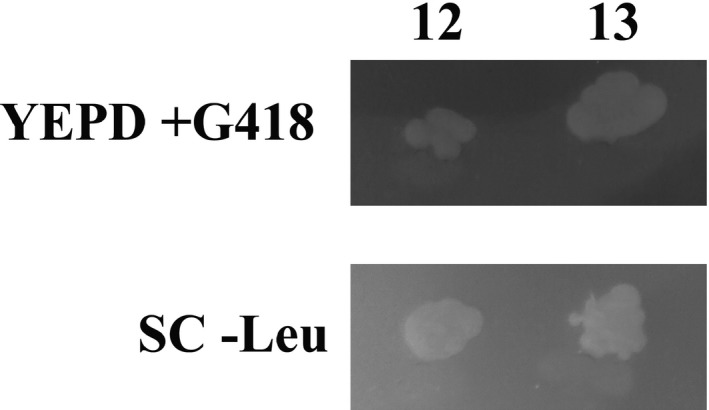
Complementation of Saccharomyces cerevisiae nfs1:: KANMX4 knockout haploids with Cryptosporidium parvum IscS and random spore analysis. Top panel, spores‐derived colonies were streaked onto YEPD, 200 μg/ml G418 plates to select for clones that lack the NFS1 yeast gene. Bottom panel, spores‐derived were streaked onto SC‐Leu plates, in parallel to select for strains that had taken in the plasmid pBEVY‐L/C. parvum IscS. All colonies were checked for haploidy (see Fig. S7).
Discussion
The recently established system for maintaining C. parvum in culture allows investigating further the cell biology of this parasite, within and outside the host (Miller et al. 2018). During invasion, Cryptosporidium becomes intracellular, but extracytoplasmic, and it has a constant dependence on the host. Due to this reliance, the parasite has reduced metabolic capabilities, including mitochondrial functions, as shown by the absence of most canonical pathways. For example, C. parvum and C. hominis remnant mitochondria (mitosomes) have lost the protein coding genes involved in the TCA cycle, thus ATP synthesis depends solely on glycolysis (Abrahamsen et al. 2004; Xu et al. 2004). The only complete biosynthetic pathways predicted to be present in these mitosomes are those of ubiquinone and Fe‐S cluster biosynthesis. A more extreme case is the C. ubiquitum mitosome, which also lacks the capacity of ubiquinone biosynthesis (Liu et al. 2016).
Here, we sought to investigate the localization and function of proteins involved in the mitochondrial Fe‐S cluster (ISC) biosynthesis in C. parvum. We amplified, cloned, and expressed the genes encoding for CpIscS, CpIscU, and CpFrataxin and generated specific antibodies against these proteins. Subsequently, using western blotting and indirect immunofluorescence assays, we demonstrated that during the extracellular life cycle stages (sporozoites/merozoites), these proteins are strongly expressed (western blots; Fig. 1), while there is much less (immunofluorescent assays; Fig. 2, Figs S3–S6) or no expression (western blots; Fig. 1) in the intracellular stages of invasion. This could be due to the fact that there are relatively more host proteins in the extracts than the C. parvum samples. Even the previously published CpCpn60 antibody was not able to detect any protein in host cells infected with C. parvum. The lack of detection in western blots was not comparable to the immunofluorescent assays experiments, where all the ISC proteins were localized in most life cycle stages. This could be due to the metabolic capacity of the parasite during the different stages of its life cycle. Cryptosporidium is typically more metabolically active during its intracellular/epicellular stage, where there is constant exchange of nutrients with the host. This is consistent with previous transcriptomic studies, which have demonstrated that enzymes involved in the ubiquinone biosynthesis are more highly expressed at later stages of the in vitro infection [36–72 h] (Mauzy et al. 2012). The higher expression of these enzymes, which require Fe‐S clusters, could explain the need for the ISC machinery and its presence during these life cycle stages.
Using higher resolution confocal microscopy, we determined the localization of these ISC proteins in the mitosome of the sporozoites as well. Previous studies speculated that ISC machinery might be the only biosynthetic pathway present in all mitochondria (Lillig and Lill 2009). In the case of Cryptosporidium, indirect evidence has suggested that CpIscS and CpIscU are localized in their mitosomes (LaGier et al. 2003). All three generated antibodies have shown punctuate localizations of CpIscS, CpIscU, and CpFrataxin in a structure that is no larger than 500 nm in diameter, consistent with the identified size of the mitosome in C. parvum sporozoite (Slapeta and Keithly 2004). This is the first direct localization of components of a biosynthetic pathway in the mitosome of C. parvum. It is also the first documentation of the presence of a mitosome in other life cycle stages than the sporozoite in Cryptosporidium. Using electron microscopy on C. parvum‐infected COLO‐680N cells, we have shown the presence of mitosomes in some of these intracellular/extracytoplasmic life cycle stages (Fig. S8). Interestingly, the immunofluorescent assays experiments were inconsistent in the localization pattern of the ISC proteins during the various life cycle stages in C. parvum infection (either punctuate or cytosolic). There are two potential explanations for this phenomenon: either the ISC machinery is translocated to the cytosol in various life cycle stages of Cryptosporidium or there is a morphological change of Cryptosporidium mitosomes in later‐stage parasites. The latter scenario might be more plausible, since it is not unusual for apicomplexan parasites. For example, the morphology of Toxoplasma mitochondria changes radically during the transition of the parasite from the host cell to the extracellular matrix (Ovciarikova et al. 2017). The mitochondria of Plasmodium falciparum grow from single, small, discrete organelles into highly branched structures in the later stages of its life cycle (van Dooren et al. 2005). Our current knowledge on the morphology of Cryptosporidium remnant mitochondria during the life cycle of the parasite remains limited and is worthy of further investigations. For example, the recently developed CRISPR/Cas9 system for C. parvum (Vinayak et al. 2015) could enable us to monitor fluorescent‐tagged single‐cells to investigate further the cellular geography of the parasite through its life cycle. Here, we have developed Cryptosporidium‐specific mitochondrial markers for broaden exploration of the mitosomal function and morphology.
Although the primary sequences of CpIscS, CpIscU, and CpFrataxin proteins have conserved sequence features associated with canonical ISC functions, we also investigated whether these functions are preserved, compared with other organisms. We have examined whether CpIscS could functionally replace the S. cerevisiae homolog in a knockout strain. Nfs1 knockout is lethal for the organism (Li et al. 1999), and thus, we have demonstrated that the C. parvum homolog can functionally replace the yeast counterpart, suggesting that the proteins have conserved functions.
In summary, our results have exhibited the localization of the ISC biosynthetic pathway in C. parvum mitosome, the first of its kind for this parasite, which will initiate new investigations on the characterization of the minimalistic organelle in Cryptosporidium species and could be potentially used as a model for exploring the cell biology of mitochondria of other organisms within gregarines. The ISC machinery is present in C. parvum mitochondria to support apoproteins that are potentially part of the ubiquinone biosynthesis. In the case of C. andersoni, the ISC machinery could also support the apoproteins that are present in the respiratory complex, which has been predicted to be present as well (Liu et al. 2016). The presence of both Isa1/Isa2 proteins in the predicted proteome of C. andersoni (Table S1), which are responsible for the delivery of [4Fe‐4S] clusters to the respiratory complex, provides further support for the presence of this complex in their mitochondria. On the other hand, it is unclear why C. ubiquitum has to contain the ISC machinery in its mitochondria, since none of the apoproteins that require Fe‐S clusters (including ubiquinone biosynthesis) are encoded by its genome (Liu et al. 2016). To our knowledge, there are no electron micrographs revealing the C. ubiquitum organelle, which would be worthy of further investigations, in our quest to explore the minimal functions of mitochondrial organelles.
Supporting information
Figure S1. Conservation of functionally important residues in Cryptosporidium cysteine desulphurase (IscS) sequences.
Figure S2. Conservation of functionally important residues in Cryptosporidium scaffold protein (IscU) sequences.
Figure S3. Identification of candidate functionally important residues in Cryptosporidium Frataxin sequences.
Figure S4. Detection of CpIscS via indirect immunofluorescence.
Figure S5. Detection of CpIscU via indirect immunofluorescence.
Figure S6. Detection of CpFrataxin via indirect immunofluorescence.
Figure S7. Work flow for the yeast‐based functional complementation assay.
Figure S8. Electron Microscope Images of Cryptosporidium parvum life cycle stages in an infected COLO‐680N culture.
Table S1. Identified proteins potentially involved in mitochondrial Fe‐S cluster assembly in various Cryptosporidium species as extracted from NCBI and CryptoDB using BLASTP.
Table S2. In silico mitochondrial prediction of proteins involved in Fe‐S cluster assembly in Cryptosporidium species.
Video S1. Animation of cellular staining of Cryptosporidium parvum cell using confocal microscopy.
Acknowledgments
This research was supported by BBSRC research grant (BB/M009971/1) to ADT. CNM was funded by an award from the School of Biosciences at the University of Kent. LJ is supported by a Bill and Melinda Gates Foundation grant to ADT. We thank Eleni Gentekaki for proof‐reading the manuscript and the reviewers for their constructive comments.
Literature Cited
- Abrahamsen, M. S. , Templeton, T. J. , Enomoto, S. , Abrahante, J. E. , Zhu, G. , Lancto, C. A. , Deng, M. , Liu, C. , Widmer, G. , Tzipori, S. , Buck, G. A. , Xu, P. , Bankier, A. T. , Dear, P. H. , Konfortov, B. A. , Spriggs, H. F. , Iyer, L. , Anantharaman, V. , Aravind, L. & Kapur, V. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum . Science, 304:441–445. [DOI] [PubMed] [Google Scholar]
- Ali, V. & Nozaki, T. 2013. Iron‐sulphur clusters, their biosynthesis, and biological functions in protozoan parasites. Adv. Parasitol., 83:1–92. [DOI] [PubMed] [Google Scholar]
- Ali, V. , Shigeta, Y. , Tokumoto, U. , Takahashi, Y. & Nozaki, T. 2004. An intestinal parasitic protist, Entamoeba histolytica, possesses a non‐redundant nitrogen fixation‐like system for iron‐sulfur cluster assembly under anaerobic conditions. J. Biol. Chem., 279:16863–16874. [DOI] [PubMed] [Google Scholar]
- Basu, S. , Horakova, E. & Lukes, J. 2016. Iron‐associated biology of Trypanosoma brucei . Biochem. Biophys. Acta., 1860:363–370. [DOI] [PubMed] [Google Scholar]
- Boratyn, G. M. , Schaffer, A. A. , Agarwala, R. , Altschul, S. F. , Lipman, D. J. & Madden, T. L. 2012. Domain enhanced lookup time accelerated BLAST. Biol. Direct, 7:12‐6150‐7‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braymer, J. J. & Lill, R. 2017. Iron‐sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem., 292:12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charan, M. , Choudhary, H. H. , Singh, N. , Sadik, M. , Siddiqi, M. I. , Mishra, S. & Habib, S. 2017. Fe‐S cluster assembly in the apicoplast and its indispensability in mosquito stages of the malaria parasite. FEBS J., 284:2629–2648. [DOI] [PubMed] [Google Scholar]
- Charan, M. , Singh, N. , Kumar, B. , Srivastava, K. , Siddiqi, M. I. & Habib, S. 2014. Sulfur mobilization for Fe‐S cluster assembly by the essential SUF pathway in the Plasmodium falciparum apicoplast and its inhibition. Antimicrob. Agents Chemother., 58:3389–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M. G. 1995. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci., 11:441–447. [DOI] [PubMed] [Google Scholar]
- Curran, B. P. & Bugeja, V. 2014. Basic investigations in Saccharomyces cerevisiae . Methods Mol. Biol., 1163:1–14. [DOI] [PubMed] [Google Scholar]
- Dellibovi‐Ragheb, T. A. , Gisselberg, J. E. & Prigge, S. T. 2013. Parasites FeS up: iron‐sulfur cluster biogenesis in eukaryotic pathogens. PLoS Pathog., 9:e1003227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren, G. G. , Marti, M. , Tonkin, C. J. , Stimmler, L. M. , Cowman, A. F. & McFadden, G. I. 2005. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum . Mol. Microbiol., 57:405–419. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5:113‐2105‐5‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O. , Brunak, S. , von Heijne, G. & Nielsen, H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc., 2:953–971. [DOI] [PubMed] [Google Scholar]
- Freibert, S. A. , Goldberg, A. V. , Hacker, C. , Molik, S. , Dean, P. , Williams, T. A. , Nakjang, S. , Long, S. , Sendra, K. , Bill, E. , Heinz, E. , Hirt, R. P. , Lucocq, J. M. , Embley, T. M. & Lill, R. 2017. Evolutionary conservation and in vitro reconstitution of microsporidian iron‐sulfur cluster biosynthesis. Nat. Commun., 8:13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa, Y. , Tsuji, J. , Fu, S. C. , Tomii, K. , Horton, P. & Imai, K. 2015. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics, 14:1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D. & Woods, R. A. 2002. Transformation of yeast by lithium acetate/single‐stranded carrier DNA/Polyethylene glycol method. Methods Enzymol., 350:87–96. [DOI] [PubMed] [Google Scholar]
- van der Giezen, M. , Cox, S. & Tovar, J. 2004. The Iron‐sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol. Biol., 4:7‐2148‐4‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselberg, J. E. , Dellibovi‐Ragheb, T. A. , Matthews, K. A. , Bosch, G. & Prigge, S. T. 2013. The suf iron‐sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites. PLoS Pathog., 9:e1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, A. V. , Molik, S. , Tsaousis, A. D. , Neumann, K. , Kuhnke, G. , Delbac, F. , Vivares, C. P. , Hirt, R. P. , Lill, R. & Embley, T. M. 2008. Localization and functionality of microsporidian iron‐sulphur cluster assembly proteins. Nature, 452:624–628. [DOI] [PubMed] [Google Scholar]
- Haussig, J. M. , Matuschewski, K. & Kooij, T. W. 2014. Identification of vital and dispensable sulfur utilization factors in the Plasmodium apicoplast. PLoS ONE, 9:e89718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiges, M. , Wang, H. , Robinson, E. , Aurrecoechea, C. , Gao, X. , Kaluskar, N. , Rhodes, P. , Wang, S. , He, C. Z. , Su, Y. , Miller, J. , Kraemer, E. & Kissinger, J. C. 2006. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res., 34:D419–D422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley, C. , Green, E. D. & Dunham, I. 1990. Rapid assessment of S. Cerevisiae mating type by PCR. Trends Genet., 6:236. [DOI] [PubMed] [Google Scholar]
- Karnkowska, A. , Vacek, V. , Zubacova, Z. , Treitli, S. C. , Petrzelkova, R. , Eme, L. , Novak, L. , Žárský, V. , Barlow, L. D. , Herman, E. K. , Soukal, P. , Hroudová, M. , Doležal, P. , Hroudová, M. , Stairs, C. W. , Roger, A. J. & Hampl, V. 2016. A eukaryote without a mitochondrial organelle. Curr. Biol., 26:1274–1284. [DOI] [PubMed] [Google Scholar]
- LaGier, M. J. , Tachezy, J. , Stejskal, F. , Kutisova, K. & Keithly, J. S. 2003. Mitochondrial‐type iron‐sulfur cluster biosynthesis genes (IscS and IscU) in the apicomplexan Cryptosporidium parvum . Microbiology, 149:3519–3530. [DOI] [PubMed] [Google Scholar]
- Leger, M. M. , Eme, L. , Hug, L. A. & Roger, A. J. 2016. Novel hydrogenosomes in the microaerophilic jakobid Stygiella incarcerata . Mol. Biol. Evol., 33:2318–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Kogan, M. , Knight, S. A. , Pain, D. & Dancis, A. 1999. Yeast mitochondrial protein, Nfs1p, coordinately regulates iron‐sulfur cluster proteins, cellular iron uptake, and iron distribution. J. Biol. Chem., 274:33025–33034. [DOI] [PubMed] [Google Scholar]
- Lill, R. , Hoffmann, B. , Molik, S. , Pierik, A. J. , Rietzschel, N. , Stehling, O. , Uzarska, M. A. , Webert, H. , Wilbrecht, C. & Muhlenhoff, U. 2012. The role of mitochondria in cellular iron‐sulfur protein biogenesis and iron metabolism. Biochem. Biophys. Acta., 1823:1491–1508. [DOI] [PubMed] [Google Scholar]
- Lillig, C. H. & Lill, R. 2009. Lights on iron‐sulfur clusters. Chem. Biol., 16:1213–1214. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Roellig, D. M. , Guo, Y. , Li, N. , Frace, M. A. , Tang, K. , Zhang, L. , Feng, Y. & Xiao, L. 2016. Evolution of mitosome metabolism and invasion‐related proteins in Cryptosporidium . BMC Genom., 17:1006‐016‐3343‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauzy, M. J. , Enomoto, S. , Lancto, C. A. , Abrahamsen, M. S. & Rutherford, M. S. 2012. The Cryptosporidium parvum transcriptome during in vitro development. PLoS ONE, 7:e31715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin, L. J. , Bryson, K. & Jones, D. T. 2000. The PSIPRED protein structure prediction server. Bioinformatics, 16:404–405. [DOI] [PubMed] [Google Scholar]
- Miller, C. N. , Jossé, L. , Brown, I. , Blakeman, B. , Povey, J. , Yiangou, L. , Price, M. , Cinatl, J. , Xue, W. F. , Michaelis, M. & Tsaousis, A. D. 2018. A cell culture platform for Cryptosporidium that enables long‐term cultivation and new tools for the systematic investigation of its biology. Int. J. Parasitol., 48:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller 3rd, C. A. , Martinat, M. A. & Hyman, L. E. 1998. Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: expressionvectors with bi‐directional promoters for use in Saccharomyces cerevisiae . Nucleic Acids Res., 26:3577–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi, T. & Kita, K. 2010. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium . Parasitol. Int., 59:305–312. [DOI] [PubMed] [Google Scholar]
- Nyvltova, E. , Sutak, R. , Harant, K. , Sedinova, M. , Hrdy, I. , Paces, J. , Vlcek, C. & Tachezy, J. 2013. NIF‐type iron‐sulfur cluster assembly system is duplicated and distributed in the mitochondria and cytosol of Mastigamoeba balamuthi . Proc. Natl Acad. Sci. USA, 110:7371–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovciarikova, J. , Lemgruber, L. , Stilger, K. L. , Sullivan, W. J. & Sheiner, L. 2017. Mitochondrial behaviour throughout the lytic cycle of Toxoplasma gondii . Sci. Rep., 7:42746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan, C. E. , Ault, J. G. , Langreth, S. G. & Keithly, J. S. 2003. Cryptosporidium parvum Cpn60 targets a relict organelle. Curr. Genet., 44:138–147. [DOI] [PubMed] [Google Scholar]
- Rouault, T. A. & Maio, N. 2017. Biogenesis and functions of mammalian iron‐sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem., 292:12744–12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, U. , Paparini, A. , Monis, P. & Hijjawi, N. 2016. It's official – Cryptosporidium is a gregarine: what are the implications for the water industry? Water Res., 105:305–313. [DOI] [PubMed] [Google Scholar]
- Seeber, F. 2002. Biogenesis of iron‐sulphur clusters in amitochondriate and apicomplexan protists. Int. J. Parasitol., 32:1207–1217. [DOI] [PubMed] [Google Scholar]
- Slapeta, J. & Keithly, J. S. 2004. Cryptosporidium parvum mitochondrial‐type HSP70 targets homologous and heterologous mitochondria. Eukaryot. Cell, 3:483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I. , Peeters, N. , Legeai, F. & Lurin, C. 2004. Predotar: a tool for rapidly screening proteomes for N‐terminal targeting sequences. Proteomics, 4:1581–1590. [DOI] [PubMed] [Google Scholar]
- Stairs, C. W. , Eme, L. , Brown, M. W. , Mutsaers, C. , Susko, E. , Dellaire, G. , Soanes, D. M. , van der Giezen, M. & Roger, A. J. 2014. A SUF Fe‐S cluster biogenesis system in the mitochondrion‐related organelles of the anaerobic protist Pygsuia . Curr. Biol., 24:1176–1186. [DOI] [PubMed] [Google Scholar]
- Tsaousis, A. D. , Gentekaki, E. , Eme, L. , Gaston, D. & Roger, A. J. 2014. Evolution of the cytosolic iron‐sulfur cluster assembly machinery in Blastocystis species and other microbial eukaryotes. Eukaryot. Cell, 13:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaousis, A. D. , Ollagnier de Choudens, S. , Gentekaki, E. , Long, S. , Gaston, D. , Stechmann, A. , Vinella, D. , Py, B. , Fontecave, M. , Barras, F. , Lukeš, J. & Roger, A. J. 2012. Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis . Proc. Natl Acad. Sci. USA, 109:10426–10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady, G. E. & Simon, I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics, 17:849–850. [DOI] [PubMed] [Google Scholar]
- Vinayak, S. , Pawlowic, M. C. , Sateriale, A. , Brooks, C. F. , Studstill, C. J. , Bar‐Peled, Y. , Cipriano, M. J. & Striepen, B. 2015. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum . Nature, 523:477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Widmer, G. , Wang, Y. , Ozaki, L. S. , Alves, J. M. , Serrano, M. G. , Puiu, D. , Manque, P. , Akiyoshi, D. , Mackey, A. J. , Pearson, W. R. , Dear, P. H. , Bankier, A. T. , Peterson, D. L. , Abrahamsen, M. S. , Kapur, V. , Tzipori, S. & Buck, G. A. 2004. The genome of Cryptosporidium hominis . Nature, 431:1107–1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Conservation of functionally important residues in Cryptosporidium cysteine desulphurase (IscS) sequences.
Figure S2. Conservation of functionally important residues in Cryptosporidium scaffold protein (IscU) sequences.
Figure S3. Identification of candidate functionally important residues in Cryptosporidium Frataxin sequences.
Figure S4. Detection of CpIscS via indirect immunofluorescence.
Figure S5. Detection of CpIscU via indirect immunofluorescence.
Figure S6. Detection of CpFrataxin via indirect immunofluorescence.
Figure S7. Work flow for the yeast‐based functional complementation assay.
Figure S8. Electron Microscope Images of Cryptosporidium parvum life cycle stages in an infected COLO‐680N culture.
Table S1. Identified proteins potentially involved in mitochondrial Fe‐S cluster assembly in various Cryptosporidium species as extracted from NCBI and CryptoDB using BLASTP.
Table S2. In silico mitochondrial prediction of proteins involved in Fe‐S cluster assembly in Cryptosporidium species.
Video S1. Animation of cellular staining of Cryptosporidium parvum cell using confocal microscopy.


