Summary
The WhiB‐like (Wbl) family of proteins are exclusively found in Actinobacteria. Wbls have been shown to play key roles in virulence and antibiotic resistance in Mycobacteria and Corynebacteria, reflecting their importance during infection by the human pathogens Mycobacterium tuberculosis, Mycobacterium leprae and Corynebacterium diphtheriae. In the antibiotic‐producing Streptomyces, several Wbls have important roles in the regulation of morphological differentiation, including WhiB, a protein that controls the initiation of sporulation septation and the founding member of the Wbl family. In recent years, genome sequencing has revealed the prevalence of Wbl paralogues in species throughout the Actinobacteria. Wbl proteins are small (generally ~80–140 residues) and each contains four invariant cysteine residues that bind an O2‐ and NO‐sensitive [4Fe–4S] cluster, raising the question as to how they can maintain distinct cellular functions within a given species. Despite their discovery over 25 years ago, the Wbl protein family has largely remained enigmatic. Here I summarise recent research in Mycobacteria, Corynebacteria and Streptomyces that sheds light on the biochemical function of Wbls as transcription factors and as potential sensors of O2 and NO. I suggest that Wbl evolution has created diversity in protein–protein interactions, [4Fe–4S] cluster‐sensitivity and the ability to bind DNA.
Introduction
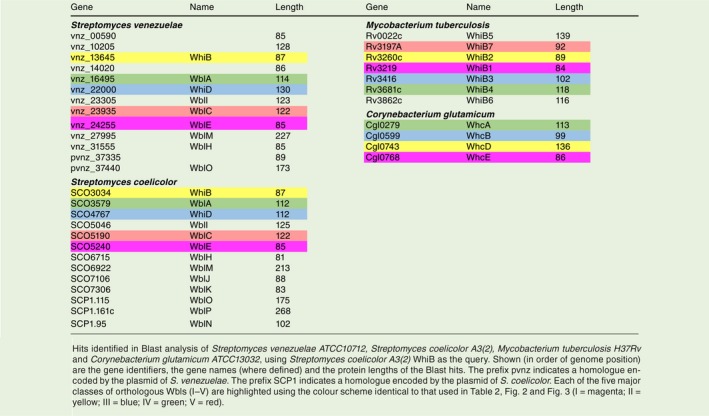
The WhiB‐like (Wbl) family of proteins, of which WhiB is the founding member, are exclusively found in Actinobacteria. Following the initial characterisation of WhiB in Streptomyces coelicolor (Sc) (Chater, 1972; Davis and Chater, 1992), multiple paralogues were identified first in Mycobacteria (Cole et al., 1998) and then in other species of Streptomyces (Soliveri et al., 2000). Since then, genome sequencing has revealed the prevalence of Wbl paralogues throughout the phylum (Table 1). In S. coelicolor, there are 14 Wbl proteins with 11 encoded on the chromosome and three encoded on the large linear plasmid, SCP1 (Bentley et al., 2002; 2004). In Mycobacteriun tuberculosis (Mtb) there are seven paralogues and in Corynebacterium glutamicum (Cg) there are four.
Table 1.
WhiB homologs in Actinobacteria.
Streptomyces are prolific producers of antibiotics and their production is both temporally and genetically linked to a complex process of differentiation (Fig. 1). Like other members of the Actinobacteria, Streptomyces employ a polar mode of growth (Flärdh et al., 2012), which, following germination and coupled to branching, establishes a vegetative mycelium. Upon nutrient depletion, Streptomyces initiate a series of complex developmental transitions to generate stress‐resistant spores (Flärdh and Buttner, 2009; Bush et al., 2015). Non‐branching aerial hyphae grow upwards away from the colony surface before multiple division septa divide the multi‐genomic compartments into uni‐genomic spores. Several wbl mutants show defects in either aerial hyphae formation or sporulation, reflecting the importance of Wbls in development (Fig. 1). Recently, a distinct ‘exploratory’ growth mode has been identified in which non‐branching vegetative hyphae of Streptomyces rapidly extend across surfaces, in response to glucose starvation and the presence of a volatile compound, trimethylamine, that raises pH (Jones et al., 2017; Jones and Elliot, 2018). However, S. venezuelae mutants lacking the Wbls WhiB or WhiD are still competent to explore, showing that they do not regulate this process (Jones et al., 2017).
Figure 1.
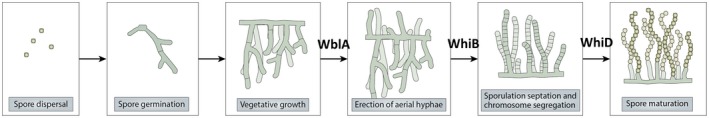
Regulation of Streptomyces differentiation by WhiB‐like proteins. 3 Wbl proteins have been shown to regulate separate stages in the developmental life‐cycle. WblA is involved in the early stages of aerial hyphae formation, WhiB controls the initiation of sporulation septation and WhiD is involved in the later stages of spore formation.
Like Streptomyces, Mycobacteria and Corynebacteria grow via the insertion of peptidoglycan at the poles. Cell division occurs as in other rod‐shaped bacteria, with the cell division machinery assembling at mid‐cell before a final mechanically driven ‘snapping’ event that separates the daughter cells (Zhou et al., 2016). The cell wall in Mycobacteria and Corynebacteria is particularly complex with an outer membrane consisting of mycolic acids that are covalently linked to the peptidoglycan (Hett and Rubin, 2008; Donovan and Bramkamp, 2014). This complexity contributes significantly to the resistance of both genera to antibiotics and other stresses, underlying the difficulty in the treatment of infections by the human pathogens M. tuberculosis, Mycobacterium leprae and C. diphtheriae. Significantly, Wbls have been shown to play key roles in virulence and antibiotic resistance, especially in Mycobacteria, further highlighting the value of research into this protein family (Morris et al., 2005; Singh et al., 2009; Burian et al., 2012; Chawla et al., 2012; Mehta et al., 2016).
Despite their discovery over 25 years ago, the Wbl protein family has largely remained enigmatic. Wbl proteins are relatively small, typically ranging from 81–139 residues in length (apart from the larger WblM, WblO and WblP proteins; Table 1). A unifying feature of all Wbls is the occurrence of four invariant cysteine residues that bind a [4Fe–4S] cluster (Fig. 2). The only other universally conserved sequence is a five residue (G[V/I]WGG) motif (Fig. 2), located at one end of a predicted loop that follows the last conserved cysteine. Based on predictions of conserved structural regions, this loop has previously been suggested to be a ‘prime candidate’ for interaction with another cellular component (Soliveri et al., 2000). More recently, research using model species in Streptomyces, Mycobacteria and Corynebacteria has revealed the central role of Wbls in the biology of Actinobacteria and shed light on their biochemical function as transcription factors and as potential sensors of O2 or nitic oxide (NO). Here I outline the findings of research into the five major classes of orthologous Wbls (Table 2 and Fig. 3) and summarise their contribution to our understanding of this important protein family.
Figure 2.
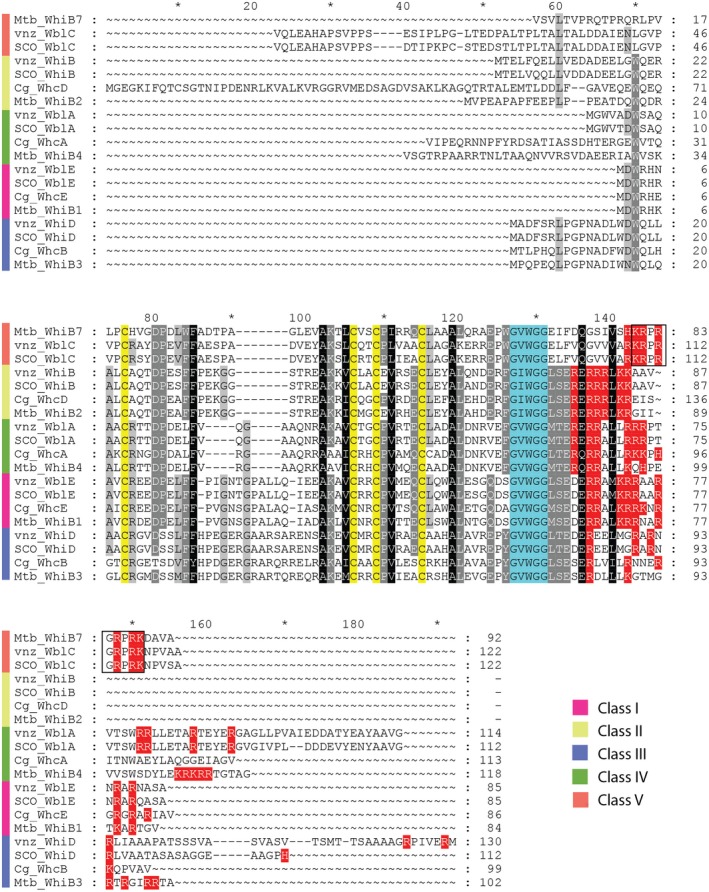
Protein sequence alignment of Wbl‐family members. Shown is an amino acid sequence alignments of Wbl proteins from Streptomyces venezuelae ATCC10712 (vnz), Streptomyces coelicolor A3(2) (SCO), Mycobacterium tuberculosis H37Rv (Mtb) and Corynebacterium glutamicum ATCC13032 (Cg). The alignment is restricted to members of the five classes of orthologous proteins (I–V) that have been the subject of significant study in Actinobacteria. As shown by the key, these classes are highlighted with the colour scheme identical to that used in Table 1, Table 2 and Fig. 3. The four invariant cysteines (C) are highlighted in yellow and the G(V/I)WGG motif is highlighted in turquoise. Positively charged residues that may facilitate DNA binding or interaction with DNA are highlighted in red. The ‘KRPRGRPRK’ AT‐hook motif, present in the WblC/WhiB7 family, is boxed.
Table 2.
The five classes of orthologous proteins in Mycobacteria, Streptomyces and Corynebacteria.
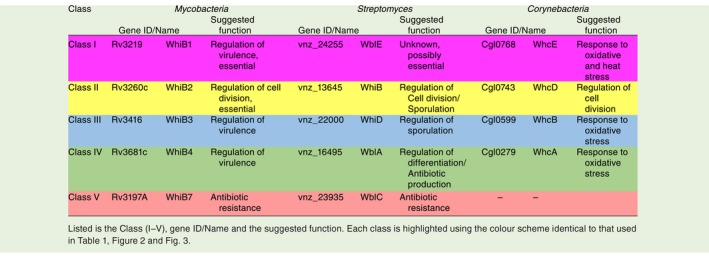
Figure 3.
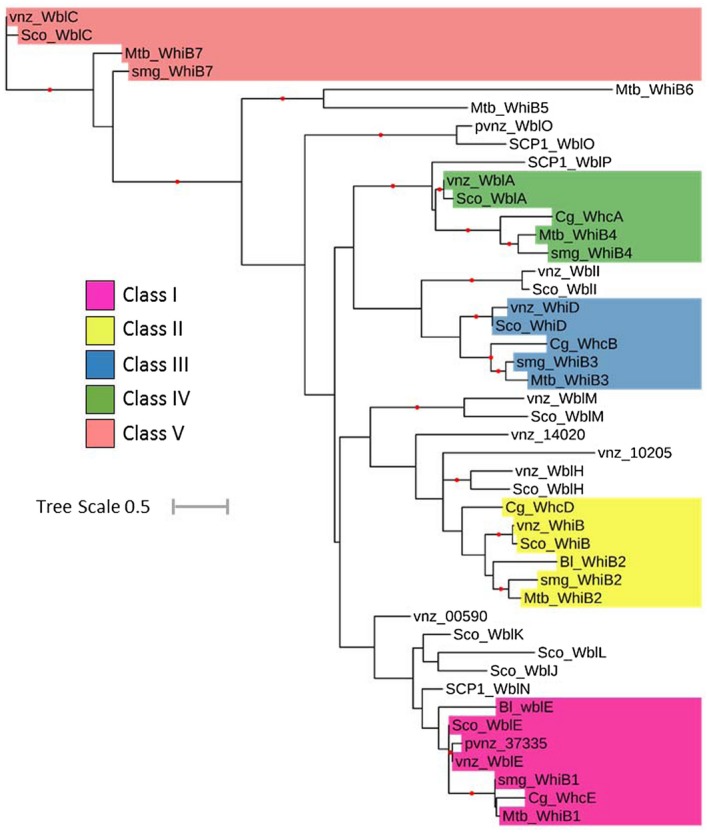
Phylogenetic tree of selected WhiB‐like proteins. Homologues were identified by Blast analysis using S. coelicolor A3(2) WhiB as the query. Searches were made against Streptomyces venezuelae ATCC10712 (vnz), Streptomyces coelicolor A3(2) (Sco), Mycobacterium tuberculosis H37Rv (Mtb), Mycobacterium smegmatis str. MC2 155 (smg), Bifidobacterium longum NCC2705 (Bl) and Corynebacterium glutamicum ATCC13032 (Cg). Amino acid sequences were aligned using CLUSTAL (Higgins & Sharp, 1988), and the resulting multiple sequence alignment was manually edited to ensure the alignment of four cysteine residues that are known to be completely conserved among Wbls. The resulting multiple sequence alignment was used to build a maximum likelihood phylogeny with 100 bootstrap replicates by PhyML 3.0 (Guindon et al., 2010) using the online server available at https://www.atgc-montpellier.fr/phyml/. WAG+G+I+F was selected as the best‐fit model based on a Smart Model Selection (Lefort et al., 2017) analysis of the multiple sequence alignment. iTOL software (https://itol.embl.de/) was used to generate the tree as displayed. Bootstrap values > 50% are indicated at their respective nodes by red dots (based on 100 replicates). The prefix pvnz indicates a homologue encoded by the plasmid of S. venezuelae. The prefix SCP1 indicates a homologue encoded by the plasmid of S. coelicolor. Tree Scale shown is 0.5 substitutions per site. As shown by the key, highlighted are members of the five major classes of orthologous Wbls (I–V) with the colour scheme identical to that used in Table 1, Table 2 and Fig. 2.
Class I (WhiB1/WblE/WhcE)
In M. tuberculosis, the whiB1 gene is essential (Smith et al., 2010). In Streptomyces, there have been two contrasting studies, one suggesting that the wblE gene may similarly be essential (Fowler‐Goldsworthy et al., 2011), and a second reporting that a ΔwblE mutant can be constructed and has no obvious phenotype (Homerová et al., 2003). Further study is therefore required to determine the importance of WblE. In C. glutamicum, deletion of the orthologous gene, whcE leads to an increased sensitivity to heat and oxidative stress (Kim et al., 2005). Oxidised WhcECg interacts with the efflux pump SpiE (Stress Protein Interacting with WhcE) and expression of the genes encoding both proteins increases under heat and oxidative stress, suggesting that WhcECg and SpiE collectively contribute to mediating the cellular response under these conditions (Park et al., 2016).
The [4Fe–4S] cluster of M. tuberculosis WhiB1 appears particularly O2‐stable (Smith et al., 2010; Crack et al., 2011), facilitating the application of NMR to generate the first structure of a WhiB‐like protein (Kudhair et al., 2017). This model indicates the [4Fe–4S] cluster holds three α‐helices in place, generating a compact structure (Fig. 4). The four invariant cysteines coordinate the cluster, with the loop between helix 3 and 4 (that includes the terminal two glycine residues of the conserved ‘middle domain’ G[V/I]WGG) running along one face of the cluster. A fourth, C‐terminal α‐helix includes positively charged residues, conserved to varying levels in other Wbl homologues (Fig. 2), which have been suggested to facilitate DNA‐binding. These include two arginine residues (R73 and R74) that are required for DNA‐binding by apo‐WhiB1Mtb in vitro (Smith et al., 2012).
Figure 4.
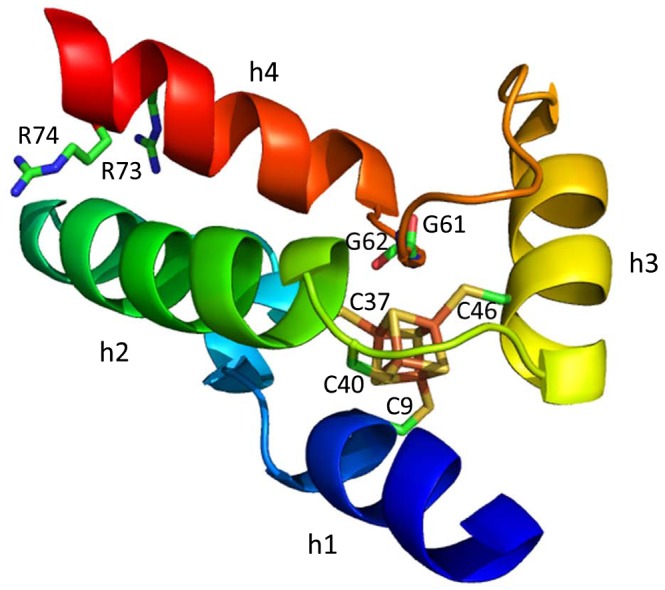
The Structure of WhiB1 from Mycobacterium tuberculosis (Kudhair et al., 2017). Labelled are the four conserved Cysteine residues (C9, C37, C40 and C46), two Glycines (G61 and G62) that are part of the conserved G(V/I)WGG turn and the C‐terminal Arginine residues (R73 and R74) that are proposed to interact with DNA in M. tuberculosis. Helices are labelled (h1‐h4). PDB ID:5oay.
Experiments with protein expressed and purified from E. coli before anaerobic reconstitution to re‐assemble the cluster in vitro (Smith et al., 2010) and experiments with native cluster‐containing protein expressed and purified from M. smegmatis (Kudhair et al., 2017) revealed that the [4Fe–4S] cluster of WhiB1Mtb reacts with eight molecules of NO. Identical observations have been made for the Wbl paralogue WhiD from S. coelicolor (see below; Crack et al., 2011), suggesting that this may be a conserved property of WhiB‐like proteins. The apparent NO‐sensitivity of Wbls, including WhiB1Mtb, may be particularly significant in Mycobacteria where NO produced by lung macrophages is a major determinant of infection by M. tuberculosis. The significance of the NO‐sensing capability of Wbls (including WblE) in Streptomyces has yet to be investigated. In vitro, holo‐WhiB1Mtb cannot bind to DNA but the apo‐form is able to bind and repress transcription from its own promoter (Smith et al., 2010) as well as the promoters of groEL2 (encoding an essential chaperonin; Stapleton et al., 2012) and the espA operon that encodes for protein components required for ESX‐1 secretion, a major virulence factor of M. tuberculosis (Kudhair et al., 2017). Since holo‐WhiB1 is unable to bind to DNA in vitro, the structural model would suggest that cluster disassembly (e.g. via nitrosylation) is required to make the C‐terminal helix available for interaction with DNA. In support of this, NMR of apo‐WhiB1Mtb, prepared via nitrosylation, suggests a substantial conformational change upon addition of NO (Kudhair et al., 2017).
As has been shown for other Wbl paralogues in M. tuberculosis (Steyn et al., 2002; Burian et al., 2013), WhiB1Mtb interacts with the primary sigma factor, SigA (equivalent to σ70 in E. coli), via region 4 of the sigma (Kudhair et al., 2017). The conserved G[V/I]WGG motif has been speculated to be part of loop in Wbls that facilitates protein‐protein interaction (Soliveri et al., 2000), and therefore, the conformation of the loop between helix 3 and helix 4 that runs across the face of [4Fe–4S] cluster (Fig. 4) may be important for this interaction. In line with this, the WhiB1Mtb‐SigA interaction is dependent upon the presence of the [4Fe–4S] cluster and is disrupted under conditions of iron limitation or nitrosative stress (Kudhair et al., 2017). Since only apo‐WhiB1 has been shown to bind DNA, the significance of the interaction between holo‐WhiB1 and SigA requires further study. One possibility is that holo‐WhiB1Mtb can activate SigA‐dependent transcription in a manner that is distinct from apo‐WhiB1Mtb‐mediated gene repression (Kudhair et al., 2017).
Class II (WhiB2/WhiB/WhcD)
Streptomyces WhiB was the first identified Wbl family member (Chater, 1972). WhiB has since been shown to co‐control the cessation of aerial growth and the initiation of sporulation with an unrelated transcription factor, WhiA. whiA and whiB mutants of Streptomyces fail to halt aerial growth and do not initiate sporulation septation, giving long, extended aerial hyphae and no spores (Aínsa et al., 2000; Soliveri et al., 2000; Bush et al., 2013; Bush et al., 2016). These identical phenotypes led to the suggestion that WhiA and WhiB might function together to control a distinct pathway in the regulatory network underlying differentiation. Subsequently, in vivo ChIP‐seq in S. venezuelae (Sv) coupled with transcriptional profiling of whiA and whiB mutants revealed that WhiA and WhiB have identical regulons, binding to the promoter regions of the same ~250 transcriptional units, including those involved in cell division, e.g. ftsZ, ftsW (Bush et al., 2013; Bush et al., 2016). These WhiB ChIP‐seq results strongly suggests that WhiB, and therefore, probably all Wbls, function to regulate transcription in vivo. WhiA promoter binding in vivo was not observed in a whiB mutant, or vice versa, suggesting that the binding of WhiA and WhiB to DNA is co‐dependent. Furthermore, substitution of the invariant cysteine residues of WhiB prevents DNA binding in vivo by WhiB and, in‐turn, WhiA, demonstrating the importance of the [4Fe–4S] cluster in WhiB function (Bush et al., 2016).
The structure of WhiA consists of two distinct domains. The large N‐terminal domain is distantly related to the LAGLIDADG eukaryotic family of homing endonucleases but is non‐catalytic (it lacks key conserved residues) and cannot bind to DNA, meaning its function in WhiA is unknown (Kaiser et al., 2009; Kaiser and Stoddard, 2011). The ability of WhiA to bind to DNA is mediated by an additional C‐terminal helix‐turn‐helix domain that binds to DNA sequences containing the consensus motif GACAC (Kaiser and Stoddard, 2011; Bush et al., 2013). In vitro, WhiA is competent to bind to DNA containing such sequences in the absence of WhiB, but not in vivo (Bush et al., 2013). Given that there are ~15,000 GACAC sequences in the S. venezuelae genome, but only ~250 are bound by WhiA in vivo, it seems likely that the function of WhiB is to tailor the specificity of WhiA, directing its binding to specific promoters in vivo. WhiA is constitutively expressed throughout the Streptomyces lifecycle, so initiation of sporulation septation is controlled, at least in part, through the developmentally induced expression of its partner protein WhiB (Bush et al., 2017).
In Corynebacterium, the orthologue of WhiB, WhcDCg, similarly regulates the transcription of genes required for cell division and directly interacts with the orthologue of WhiA in vitro (Lee et al., 2017; 2018). WhcDCg cannot bind to promoter DNA by itself but stimulates the binding of WhiA to the ftsZ promoter, suggesting that WhcD enhances the DNA‐binding activity of WhiA through protein–protein interaction (Lee et al., 2018).
In Mycobacteria, WhiB2, the orthologue of WhiB, has been shown to be essential in both M. smegmatis and M. tuberculosis. Depletion of WhiB2 leads to irreversible filamentous growth and less frequent, aberrant septation (Gomez and Bishai, 2000). Interestingly, WhiB2 is highly similar to the WhiB‐like protein WhiBTM4 from the mycobacteriophage TM4. Apo‐WhiBTM4 competes with apo‐WhiB2 for binding to the whiB2 promoter, leading to the downregulation of whiB2 expression. Overexpression of WhiBTM4 in M. smegmatis gives the same phenotype as the whiB2 conditional mutant (Rybniker et al., 2010). In contrast, overexpression of WhiB2 leads to hyperseptation (Gomez and Bishai, 2000). Similar observations have been made in Streptomyces, in which overexpression of WhiB leads to hypersporulation (Bush et al., 2017). Collectively, these findings suggest that the WhiB‐orthologues in Mycobacteria also function to regulate cell division. Like whiB2, the gene encoding the Mycobacterial WhiA orthologue also appears to be essential (DeJesus et al., 2017). It therefore seems likely that WhiA and WhiB function cooperatively in Mycobacteria, as well as in Streptomyces and Corynebacteria.
Given that Wbl proteins are confined to the Actinomycetes, and that WhiA and WhiB seem to function together in these bacteria, it is perhaps surprising to find that WhiA orthologues are prevalent outside the Actinomycetes, being widely distributed in Gram‐positive bacteria. In Bacillus, the WhiA orthologue binds to the nucleoid but does not function as a transcription factor, instead regulating cell division and chromosome segregation by an as‐yet‐unknown mechanism (Surdova et al., 2013; Bohorquez et al., 2018). Indeed, the studies in Streptomyces and Corynebacteria suggest that the ability of WhiA to bind to specific promoters and function as a transcription factor depends upon WhiB, orthologues of which are restricted to the Actinomycete family, and therefore, absent from Bacillus sp.
Class III (WhiB3/WhiD/WhcB)
Like other members of the Wbl family, WhiB3Mtb has been shown to carry an O2 and NO‐sensitive [4Fe–4S] cluster (Singh et al., 2007). Expression of whiB3 is increased in the presence of NO and under O2‐deficient conditions, suggesting that M. tuberculosis might regulate whiB3 in response to environmental stimuli associated with infection (Steyn et al., 2002; Banaiee et al., 2006; Geiman et al., 2006; Larsson et al., 2012).
WhiB3Mtb expression is also upregulated under acidic conditions, both in vitro and inside macrophages (Geiman et al., 2006; Rohde et al., 2007), implicating WhiB3Mtb in the response to low pH within phagosomes. This pH‐dependent increase in whiB3 expression is mediated by the response regulator PhoP (Feng et al., 2018). The ability of M. tuberculosis to survive conditions of acidic stress within host macrophages is a key feature of persistence during infection. In line with this, deletion of whiB3 leads to decreased intraphagosomal survival of M. tuberculosis at low pH (Mehta et al., 2016). WhiB3Mtb has been shown to mediate redox homeostasis in the phagosome by upregulating the biosynthesis of surface‐associated lipids (Singh et al., 2009), by downregulating genes involved in the innate immune response, and by blocking phagosomal maturation (Mehta et al., 2016).
In common with most other Wbls, WhiB3Mtb lacks a recognisable DNA‐binding motif and, like many of its paralogues, it also interacts with the primary sigma factor, SigA (Steyn et al., 2002). In vitro, holo‐WhiB3Mtb binds DNA weakly in both oxidised and reduced states but the oxidised (disulphide form) of apo‐WhiB3 binds DNA very strongly. Thus, the activity of WhiB3Mtb as a transcription factor may be controlled by a thiol‐based redox switch (Singh et al., 2009). Under oxidising conditions associated with active infection, apo‐WhiB3Mtb is likely to be transcriptionally active, whereas under reducing conditions associated with latency, holo‐WhiB3Mtb is likely to be transcriptionally inactive (Green et al., 2014).
The WhiB3 orthologue in Streptomyces, WhiD, was the first Wbl protein shown to carry an [4Fe–4S] cluster (Jakimowicz et al., 2005; Crack et al., 2009). WhiD is one of three Wbl proteins in Streptomyces that play a role in the transition from vegetative to reproductive growth that ultimately leads to the formation of spores (Fig. 1; Flärdh and Buttner, 2009; Bush et al., 2015). A whiD mutant forms irregularly sized spores that are more sensitive to heat, prone to lysis and show variability in cell wall deposition (Molle et al., 2000). This suggests that WhiD, either directly or indirectly, regulates the expression of genes involved in the later stages of sporulation in Streptomyces. Furthermore, the [4Fe–4S] cluster is required for the activity of WhiDSc – substitution of any one of the conserved cysteine residues gives a null mutant phenotype (Jakimowicz et al., 2005). Like WhiB1 of M. tuberculosis (see above), the [4Fe–4S] cluster of WhiDSc reacts with eight molecules of NO. This nitrosylation is extremely rapid, with the reaction occurring 104 times faster than that with O2 (Crack et al., 2011). However, as is the case for its mycobacterial orthologue, the physiological significance of the NO‐sensitivity of WhiD has yet to be established.
In Corynebacterium, the WhiB3/WhiD orthologue, WhcB, has not been extensively studied. WhcBCg is expressed during stationary phase and has been suggested to have a role in controlling the expression of gene(s) in the oxidative stress response pathway (Lee et al., 2012).
Class IV (WhiB4/WblA/WhcA)
In M. tuberculosis, WhiB4 has been shown to regulate the response to oxidative stress to modulate virulence. Transcriptional analysis identified WhiB4Mtb‐repressed genes that are differentially expressed under various stress conditions. Deletion of whiB4 leads to hyper‐induction of antioxidants, increased resistance to oxidative stress in vitro and enhanced survival in macrophages, reflecting the importance of WhiB4Mtb in maintaining redox homeostasis (Chawla et al., 2012). As has been shown for other Wbl‐family members, WhiB4 contains an O2‐ and NO‐sensitive [4Fe–4S] cluster (Alam et al., 2007; Chawla et al., 2012). However, the [4Fe–4S] cluster of WhiB4Mtb appears to be more sensitive to O2 than reported for other Wbls, such as WhiB3/WhiD (Singh et al., 2007; Crack et al., 2009) and WhiB1(Kudhair et al., 2017), suggesting a specific role for WhiB4Mtb in redox‐sensing (Chawla et al., 2012). In line with this, holo‐WhiB4Mtb is not competent to bind to DNA but the loss of the [4Fe–4S] cluster and oxidation of the coordinating cysteines strongly stimulates DNA binding. In vitro and in isolation, oxidised apo‐WhiB4 does not appear to bind to DNA in a sequence‐specific manner but rather preferentially binds to GC‐rich sequences in the minor‐groove. Oligomerisation of WhiB4Mtb, driven by disulfide bond formation, has been observed both in vitro and in vivo in M. smegmatis, suggesting a possible mechanism for the control of WhiB4Mtb activity (Chawla et al., 2012).
The WhiB4 orthologue, WblA, is one of three Wbl‐family members that regulate differentiation in Streptomyces. Whereas WhiB and WhiD regulate the initiation of sporulation septation and spore maturation, respectively, WblA appears to play an earlier role in development, during the formation of aerial hyphae (Fig. 1). In S. coeilcolor, a wblA mutant exhibits a defect in sporulation, with some aerial hyphae failing to sporulate and appearing thinner compared to wild‐type (Fowler‐Goldsworthy et al., 2011). Microarray analysis indicates that WblA influences the expression of genes involved in antibiotic production, morphological differentiation and oxidative stress (Kang et al., 2007; Kim et al., 2012a; Yu et al., 2014). However, WblA binding to the promoters of target genes has not yet been demonstrated, either in vivo (e.g. by ChIP‐seq) or in vitro (e.g. by EMSA).
Like M. tuberculosis WhiB4 and S. coelicolor WblA, the Corynebacterium orthologue, WhcA, appears to negatively regulate the expression of genes involved in the response to oxidative stress (Choi et al., 2009). WhcA directly interacts with SpiA (Stress protein interacting with WhcA, annotated as a dioxygenase/oxido‐reductase and suggested to be involved in signal perception) and this interaction can be disrupted in the presence of the oxidant diamide (Park et al., 2011; 2012). This mechanism appears to be conserved in Streptomyces, in which a SpiA orthologue has been identified and similarly demonstrated to be a negative regulator of the WblA‐dependent oxidative stress response (Kim et al., 2013). However, in Mycobacteria, there is no clear SpiA‐orthologue nor have other protein partners of WhiB4 been identified.
Class V (WhiB7/WblC)
WhiB7 and its orthologue, WblC, control innate multi‐drug resistance in Mycobacteria and Streptomyces respectively (Morris et al., 2005; Ramon‐Garcia et al., 2013). There is no obvious orthologue in Corynebacteria (Table 2 and Fig. 3). In M. tuberculosis, expression of whiB7 is induced by exposure to sub‐inhibitory concentrations of translation‐inhibiting antibiotics, such as erythromycin, streptomycin and tetracycline (Morris et al., 2005). As well as controlling its own expression, WhiB7Mtb has been shown to control the expression of a plethora of antibiotic resistance genes (Morris et al., 2005; Burian et al., 2013). In this way, induction of WhiB7Mtb by a specific antibiotic can give rise to a broad spectrum of resistance. Overexpression of whiB7 promotes multi‐drug resistance in M. tuberculosis, making WhiB7Mtb an attractive potential target for new therapeutics (Morris et al., 2005). In addition to antibiotics, whiB7 is also upregulated in response to fatty acids that may be accumulated internally or encountered within eukaryotic hosts during infection by M. tuberculosis (Morris et al., 2005).
In Streptomyces, WblC protein levels are induced in the presence of translation‐inhibiting antibiotics. How this induction occurs is not known, but it may well involve the extremely long 5′ UTR of the wblC mRNA, which appears to contain an ORF encoding a small protein (Yoo et al., 2016). Recently, the first direct target of WblCSc was characterised (Yoo et al., 2016). Unexpectedly, this target was the sigR‐rsrA operon, encoding the sigma factor SigR and its redox‐sensitive antisigma factor, RsrA, which together control the principal oxidative stress response in Streptomyces (Paget et al., 1998; Kang et al., 1999). Under oxidative stress conditions, SigR is liberated from RsrA and activates transcription of a large regulon of genes that help the bacterium survive oxidative stress and re‐establish normal redox poise (Paget et al., 1998; 2001; Kim et al., 2012b). This rapid and transient response depends upon the autoregulatory function of SigR that induces expression of a longer (but unstable) isoform of SigR (SigR′) from a SigR‐target promoter (sigRp2) (Kim et al., 2009). In contrast, in the presence of antibiotics, WblCSc mediates a longer‐lasting response by binding to a more downstream (and SigR‐independent) promoter, sigRp1, thereby increasing the expression of the shorter (but stable) isoform of SigR (Yoo et al., 2016).
This work showed that activation of the SigR regulon not only increased resistance to oxidative stress, but also resistance to translation‐inhibiting antibiotics, revealing a perhaps unexpected overlap between the types of cellular damage caused by oxidative stress and by the inhibition of translation (Yoo et al., 2016). It seems likely that the transient expression of the regulon mediated by the unstable isoform SigR′ is sufficient to cope with oxidative stress, but the more sustained expression of the regulon mediated by the stable SigR isoform is required to deal with the consequences of the inhibition of ribosomes (Yoo et al., 2016). In Mycobacteria, the SigR homologs (SigEMtb and SigHMtb) are also likely to be targets of WhiB7Mtb since expression of both also increases upon addition of translation‐inhibiting antibiotics (Yoo et al., 2016). In both Streptomyces and Mycobacteria, this SigR‐dependent shift to reducing conditions would be expected to play a role in maintaining the [4Fe–4S] cluster of WhiB7/WblC, meaning that WhiB7/WblC would function in an autoregulatory loop. In line with this, treatment of M. smegmatis with erythromycin creates a highly reducing environment in the cytoplasm and this shift is dependent upon whiB7 (Burian et al., 2012; 2013).
Of all the Wbls, the way in which WhiB7/WblC binds DNA is most clear. This is because WhiB7/WblC is the only member of the Wbl family that carries a recognised DNA‐binding motif, the AT‐hook (Fig. 2). AT‐hooks do not bind to specific consensus sites but rather bind to the minor groove of AT‐rich DNA. They are often found in proteins that carry additional DNA‐binding domains, suggesting that their function is to fine‐tune the specificity or affinity of DNA binding (Aravind and Landsman, 1998). In common with many other Wbls, holo‐WhiB7Mtb interacts with the primary sigma factor in M. tuberculosis, SigA. WhiB7Mtb ‐mediated multi‐drug resistance is dependent both on the holo‐WhiB7‐SigA interaction and on the AT‐rich binding site of WhiB7Mtb, which is found just upstream of the ‐35 element bound by SigA in M. tuberculosis. Therefore, it is likely that in the presence of antibiotics, the function of WhiB7/WblC is to recruit the primary sigma factor to a specific subset of promoters to induce the expression of resistance genes (Burian et al., 2013).
Discussion
Despite the fact that Wbl proteins regulate key processes across the Actinobacteria, including virulence, antibiotic resistance and morphogenesis, for many years their biochemical function has remained elusive. In the past, it has been suggested that Wbls can function as disulphide reductases (Alam et al., 2007; Garg et al., 2007) or chaperones (Konar et al., 2012). However, given the many studies summarised in this review, it seems certain that WhiB‐like proteins function primarily to regulate transcription. In vitro studies, mostly in Mycobacteria, reveal the ability of Wbls to bind to DNA and in vivo ChIP‐seq in Streptomyces confirms that WhiB, and therefore, likely all Wbls, function to regulate transcription (Bush et al., 2016). Recently, induced expression of transcription factors followed by ChIP‐seq has been coupled to transcriptional profiling in M. tuberculosis to identify genome‐wide DNA‐binding events (Rustad et al., 2014; Minch et al., 2015). Careful analysis of these data and similar experimental approaches are needed to further reveal the regulatory roles of Wbl proteins in vivo. Much of the original uncertainty regarding the biochemical function of Wbls arose from the absence of a recognisable DNA‐binding domain. WhiB7/WblC is the only exception, containing an AT‐hook, a motif that typically binds AT‐rich sequences in the minor groove (Aravind and Landsman, 1998). The other Wbls contain a series of positively charged amino acids at varying frequencies towards the C‐terminus (Fig. 2). Given this, it is hard to understand how such proteins in isolation could bind specific target promoters in vivo. In line with this, several studies suggest that binding to DNA in vitro is non‐specific (Chawla et al., 2012), or else occurs with a low‐degree of sequence discrimination (Singh et al., 2009) or to a large promoter region with no core motif (Smith et al., 2010). An emerging theme, therefore, is that the ability of Wbls to function as site‐specific transcription factors seems to depend on partner or accessory proteins.
In Mycobacteria, WhiB1, WhiB3 and WhiB7 have all been shown to interact directly with the primary sigma factor SigA. In the case of WhiB7Mtb, the AT‐hook mediates binding to an AT‐rich sequence upstream of the SigA ‐35 element, likely increasing the specificity of the sigma factor at a subset of promoters (Burian et al., 2013). The interactions between SigA and WhiB1/WhiB3/WhiB7 have all been shown to occur via region 4 of the sigma factor (Steyn et al., 2002; Burian et al., 2013; Kudhair et al., 2017), the domain responsible for contacting the ‐35 element and a well‐established target of Class II activators (Browning and Busby, 2016). For SigA‐WhiB7, the interaction requires a triplet of residues (EPW), adjacent to the conserved GVWGG‐motif of WhiB7Mtb (Fig. 2) and part of the loop region previously predicted to facilitate protein‐protein interaction (Soliveri et al., 2000; Burian et al., 2013). WhiB3Mtb contains a similar (EPY) motif, in line with the observed SigA‐WhiB3 interaction (Steyn et al., 2002). For SigA‐WhiB3 and SigA‐WhiB7, the interactions can be eliminated by a specific SigA substitution (R515H) in region 4 (Steyn et al., 2002; Burian et al., 2013). In contrast, in WhiB1Mtb, the triplet motif of the ‘middle‐domain’ is not conserved (Fig. 2). Furthermore, the R515H SigA variant is viable in Mycobacteria, suggesting that WhiB1, which is essential for viability, interacts with SigA via different residues of the Wbl ‘middle‐domain’ and SigA region 4. Similarly, WhiB2M tbis essential (DeJesus et al., 2017) and carries a different (ERF) triplet of residues adjacent to the GVWGG‐motif (Fig. 2). Therefore, if WhiB2Mtb also interacts with SigA, it is likely to do so via different amino acid contacts, or else it interacts with a different sigma factor. Indeed, another member of the Wbl family, M. tuberculosis WhiB5, does not seem to interact with SigA (Casonato et al., 2012), suggesting that it might interact with an alternative sigma factor. Considering this, it is interesting to note that three of the genes encoding Wbls in S. coelicolor (whiD, wblE and wblJ) are located close to sigma factor genes and one wbl gene, located on the SCP1 plasmid, encodes a protein (WblP) consisting of a Wbl domain fused to an ECF sigma factor (Bentley et al., 2004). Wbl‐sigma interactions have not been studied in Streptomyces and Corynebacteria, although it seems likely that the Wbl‐sigma interactions observed in Mycobacterium would be conserved between the clear orthologues in Streptomyces and Corynebacteria discussed in this review (Table 2 and Fig. 3). In this context, it is interesting to note that the triplet of residues adjacent to the G[V/I]WGG‐motif is conserved among orthologues in each of the five classes of Wbl proteins (Fig. 2).
Individual actinobacterial species typically contain multiple WhiB‐like paralogues (Table 1 and Fig. 3), all carrying an [4Fe–4S] cluster (Fig. 2). This raises the question as to how Wbls can maintain distinct regulatory functions in the same cell. In other paralogous families of transcription factors, innate operator DNA sequence discrimination plays a central role. The limited ability of Wbls to function as site‐specific transcription factors (except for WhiB7/WblC) means that the ability to direct a specific cellular response based on innate DNA sequence recognition is at least limited. Clearly, the regulation of wbl gene expression plays an important role. In Mycobacteria and Corynebacteria, the expression of different wbl genes has been shown to be activated by various stress conditions (e.g. low nutrient availability, heat, oxidative stress) (Geiman et al., 2006; Lee et al., 2013) and, in both Mycobacteria and Streptomyces, transcription of whiB7/wblC has been shown to be activated by translation‐inhibiting antibiotics (Morris et al., 2005; Yoo et al., 2016). In Streptomyces, the expression of developmental genes, including whiB, whiD and wblA, is controlled as part of a complex regulatory network (Flärdh and Buttner, 2009; Bush et al., 2015; 2017). The overall effect of such control is that few Wbl paralogues are likely to be present at the same time in the cell. This partly ensures that the correct Wbl‐mediated transcriptional response is mounted on exposure to a given signal or stress.
As highlighted in this review, another key contributory factor may be the need for Wbls to interact with partner proteins. Several Wbl paralogues have been shown to interact with the primary sigma factor, SigA. Other protein partners for Wbls have also been identified; SpiE interacts with WhcE in Corynebacterium (Park et al., 2016) and SpiA interacts with WhcA/WblA in Corynebacterium and Streptomyces (Park et al., 2012; Kim et al., 2013). In Streptomyces, WhiB has been shown to co‐control the expression of genes required for sporulation with an unrelated protein, WhiA. One of the consequences of Class II activation is that the α‐CTDs of the RNAP holoenzyme are unable to bind in their preferred positions, upstream of the ‐35 promoter element. This means that additional transcription factors, so‐called Class I activators, that bind further upstream and interact with the α‐CTDs, can also influence transcription, creating synergy in bacterial regulatory networks (Browning & Busby, 2016). It is tempting to speculate that WhiB functions as a Class II activator to bind and recruit a sigma factor, perhaps SigA (called σHrdB in Streptomyces), while WhiA binds further upstream akin to a Class I activator. It may therefore be a WhiA‐WhiB‐SigA tripartite complex that regulates gene expression to initiate sporulation septation in Streptomyces. Further research is required to establish whether other members of the Wbl family function in concert with partner proteins and what the functional consequences of such interactions are.
Interestingly, the presence and status of the [4Fe–4S] cluster seems to influence DNA‐binding in different ways across the Wbl family. The cluster appears to be required for S. venezuelae WhiB and M. tuberculosis WhiB7 to bind DNA (Burian et al., 2013; Bush et al., 2016), whereas the mycobacterial apo‐WhiB1, apo‐WhiB2, apo‐WhiB3 and apo‐WhiB4 all bind DNA, at least in vitro (Singh et al., 2009; Rybniker et al., 2010; Smith et al., 2010; Chawla et al., 2012; Stapleton et al., 2012; Kudhair et al., 2017). In the case of apo‐WhiB3Mtb and apo‐WhiB4Mtb, the oxidation state of the coordinating cysteine residues appears important – the oxidised, disulphide forms have increased affinity for DNA (Singh et al., 2009; Chawla et al., 2012). Despite these findings, it cannot be ruled out that individual Wbls have different regulatory roles depending upon the state of the [4Fe–4S] cluster. For example, WhiB6 performs such a dual function in Mycobacteria, differentially regulating the expression of two sets of genes (Chen et al., 2016). Further in vivo identification of the genes under the direct control of Wbls is required to grasp the full scope of Wbl‐mediated regulation of gene expression.
The function of Wbl proteins may also be controlled in part by their relative sensitives to O2 and NO. Although Wbls all contain a [4Fe–4S] cluster, some are relatively tolerant of aerobic conditions, such as WhiB1Mtb (Kudhair et al., 2017), whereas others are more sensitive to O2, such as WhiB4Mtb (Chawla et al., 2012). In the case of WhiB1Mtb, the aerobic stability of the protein further strengthens the hypothesis that NO, and not O2 controls its activity. In Streptomyces, the [4Fe–4S] cluster of WhiD reacts several orders of magnitude more rapidly with NO than with O2 (Crack et al., 2011). Although the reaction with NO in vitro may be a conserved feature of Wbls, it seems unlikely that all Wbl proteins respond to NO in vivo, given their diverse biological roles. The ability of some Wbls to respond to NO makes sense in pathogenic Mycobacteria and Corynebacteria, which encounter NO as part of the host defence response. Streptomyces encounter endogenously produced NO (Johnson et al., 2008; Sasaki et al., 2016) and, like many other bacteria, use a dedicated NO sensor (NsrR) that controls the expression of detoxifying enzymes (Crack et al., 2015). However, further research is required to understand the in vivo significance of NO sensing in the Wbl family. Indeed, the [4Fe–4S] cluster of Wbl proteins is likely to be sensitive to other reactive oxygen species (Crack et al., 2009) and reactive nitrogen species, and major future challenges are to decipher not only the network of protein–protein interactions through which Wbl proteins effect their functions, but also the precise nature of the cellular signal(s) to which they respond.
Note
Whilst this review was under consideration, Chawla et al., reported that WhiB4Mtb dynamically couples genome condensation to the oxidative stress response in Mtb (Chawla, M., Mishra, S., Anand, K., Parikh, P., Mehta, M., Vij, M., Verma, T., Singh, P., Jakkala, K., Verma, H.N., AjitKumar, P., Ganguli, M., Narain Seshasayee, A.S., Singh, A. 2018. 19: 116‐133). Extending their work reported in this review, the authors demonstrate, in vitro, that under conditions of oxidative stress, disulphide‐linked oligomerisation of WhiB4Mtb stimulates the protein’s ability to condense DNA. In line with these findings, in vivo ChIP‐seq coupled to microarray transcriptional profiling revealed non‐specific binding of WhiB4Mtb to GC‐rich genomic regions leading to both direct and indirect effects on transcription. These findings suggest that WhiB4Mtb links the oxidative stress response of Mtb to DNA organisation and gene expression.
Acknowledgements
I would like to thank Kelley Gallagher and Govind Chandra for assistance with bioinformatic and phylogenetic analysis, Susan Schlimpert for figure construction, and Jeff Green, Nick Le Brun and Mark Buttner for helpful comments on the manuscript. I would also like to acknowledge the discussions that inspired this review, generated during the France–UK seed meeting on iron‐sulfur cluster proteins, funded by the Higher Education, Research and Innovation Department of the French Embassy. MJB is supported by BBSRC Institute Strategic Programme Grant BB/J004561/1 to the John Innes Centre.
References
- Aínsa, J.A. , Ryding, N.J. , Hartley, N. , Findlay, K.C. , Bruton, C.J. and Chater, K.F. (2000) WhiA, a protein of unknown function conserved among gram‐positive bacteria, is essential for sporulation in Streptomyces coelico lor A3(2). Journal of Bacteriology, 182, 5470–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam, M.S. , Garg, S.K. and Agrawal, P. (2007) Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe‐4S] cluster co‐ordinating protein disulphide reductase. Molecular Microbiology, 63, 1414–1431. [DOI] [PubMed] [Google Scholar]
- Aravind, L. and Landsman, D. (1998) AT‐hook motifs identified in a wide variety of DNA‐binding proteins. Nucleic Acids Research, 26, 4413–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaiee, N. , Jacobs, W.R. Jr. and Ernst, J.D. (2006) Regulation of Mycobacterium tuberculosis whiB3 in the mouse lung and macrophages. Infection and Immunity, 74, 6449–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, S.D. , Brown, S. , Murphy, L.D. , Harris, D.E. , Quail, M.A. , Parkhill, J. , et al. (2004) SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelico lor A3(2). Molecular Microbiology, 51, 1615–1628. [DOI] [PubMed] [Google Scholar]
- Bentley, S.D. , Chater, K.F. , Cerdeno‐Tarraga, A.M. , Challis, G.L. , Thomson, N.R. , et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelico lor A3(2). Nature, 417, 141–147. [DOI] [PubMed] [Google Scholar]
- Bohorquez, L.C. , Surdova, K. , Jonker, M.J. and Hamoen, L.W. (2018) The conserved DNA binding protein WhiA influences chromosome segregation in Bacillus subtilis . Journal of Bacteriology, 200, e00633–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, D.F. and Busby, S.J. (2016) Local and global regulation of transcription initiation in bacteria. Nature Reviews Microbiology, 14, 638–650. [DOI] [PubMed] [Google Scholar]
- Burian, J. , Ramon‐Garcia, S. , Sweet, G. , Gomez‐Velasco, A. , Av‐Gay, Y. and Thompson, C.J. (2012) The mycobacterial transcriptional regulator whiB7 gene links redox homeostasis and intrinsic antibiotic resistance. Journal of Biological Chemistry, 287, 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian, J. , Yim, G. , Hsing, M. , Axerio‐Cilies, P. , Cherkasov, A. , Spiegelman, G.B. and Thompson, C.J. (2013) The mycobacterial antibiotic resistance determinant WhiB7 acts as a transcriptional activator by binding the primary sigma factor SigA (RpoV). Nucleic Acids Research, 41, 10062–10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M.J. , Bibb, M.J. , Chandra, G. , Findlay, K.C. and Buttner, M.J. (2013) Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae . MBio, 4, e00684–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M.J. , Chandra, G. , Bibb, M.J. , Findlay, K.C. and Buttner, M.J. (2016) Genome‐wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces . MBio, 7, e00523–00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M.J. , Chandra, G. , Findlay, K.C. and Buttner, M.J. (2017) Multi‐layered inhibition of Streptomyces development: BldO is a dedicated repressor of whiB. Molecular Microbiology, 104, 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M.J. , Tschowri, N. , Schlimpert, S. , Flärdh, K. and Buttner, M.J. (2015) c‐di‐GMP signalling and the regulation of developmental transitions in streptomycetes. Nature Reviews Microbiology, 13, 749–760. [DOI] [PubMed] [Google Scholar]
- Casonato, S. , Cervantes Sanchez, A. , Haruki, H. , Rengifo Gonzalez, M. , Provvedi, R. , Dainese, E. , et al. (2012) WhiB5, a transcriptional regulator that contributes to Mycobacterium tuberculosis virulence and reactivation. Infection and Immunity, 80, 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater, K.F. (1972) A morphological and genetic mapping study of white colony mutants of Streptomyces coelico lor . Journal of General Microbiology, 72, 9–28. [DOI] [PubMed] [Google Scholar]
- Chawla, M. , Parikh, P. , Saxena, A. , Munshi, M. , Mehta, M. , Mai, D. , et al. (2012) Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo . Molecular Microbiology, 85, 1148–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Hu, Y. , Cumming, B.M. , Lu, P. , Feng, L. , Deng, J. , et al. (2016) Mycobacterial WhiB6 differentially regulates ESX‐1 and the Dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Reports, 16, 2512–2524. [DOI] [PubMed] [Google Scholar]
- Choi, W.W. , Park, S.D. , Lee, S.M. , Kim, H.B. , Kim, Y. and Lee, H.S. (2009) The whcA gene plays a negative role in oxidative stress response of Corynebacterium glutamicum . FEMS Microbiology Letters, 290, 32–38. [DOI] [PubMed] [Google Scholar]
- Cole, S.T. , Brosch, R. , Parkhill, J. , Garnier, T. , Churcher, C. , Harris, D. , et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Crack, J.C. , den Hengst, C.D. , Jakimowicz, P. , Subramanian, S. , Johnson, M.K. , Buttner, M.J. , et al. (2009) Characterization of [4Fe‐4S]‐containing and cluster‐free forms of Streptomyces WhiD. Biochemistry, 48, 12252–12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crack, J.C. , Munnoch, J. , Dodd, E.L. , Knowles, F. , Al Bassam, M.M. , Kamali, S. , et al. (2015) NsrR from Streptomyces coelico lor is a nitric oxide‐sensing [4Fe‐4S] cluster protein with a specialized regulatory function. Journal of Biological Chemistry, 290, 12689–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crack, J.C. , Smith, L.J. , Stapleton, M.R. , Peck, J. , Watmough, N.J. , Buttner, M.J. , et al. (2011) Mechanistic insight into the nitrosylation of the [4Fe‐4S] cluster of WhiB‐like proteins. Journal of the American Chemical Society, 133, 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, N.K. and Chater, K.F. (1992) The Streptomyces coelico lor whiB gene encodes a small transcription factor‐like protein dispensable for growth but essential for sporulation. Molecular & General Genetics, 232, 351–358. [DOI] [PubMed] [Google Scholar]
- DeJesus, M.A. , Gerrick, E.R. , Xu, W. , Park, S.W. , Long, J.E. , Boutte, C.C. , et al. (2017) Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. MBio, 8, e02133–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan, C. and Bramkamp, M. (2014) Cell division in Corynebacterineae. Frontiers in Microbiology, 5, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, L. , Chen, S. and Hu, Y. (2018) PhoPR positively regulates whiB3 expression in response to low pH in pathogenic mycobacteria. Journal of Bacteriology, 200, e00766–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh, K. and Buttner, M.J. (2009) Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nature Reviews Microbiology, 7, 36–49. [DOI] [PubMed] [Google Scholar]
- Flärdh, K. , Richards, D.M. , Hempel, A.M. , Howard, M. and Buttner, M.J. (2012) Regulation of apical growth and hyphal branching in Streptomyces . Current Opinion in Microbiology, 15, 737–743. [DOI] [PubMed] [Google Scholar]
- Fowler‐Goldsworthy, K. , Gust, B. , Mouz, S. , Chandra, G. , Findlay, K.C. and Chater, K.F. (2011) The actinobacteria‐specific gene wblA controls major developmental transitions in Streptomyces coelico lor A3(2). Microbiology, 157, 1312–1328. [DOI] [PubMed] [Google Scholar]
- Garg, S.K. , Suhail Alam, M. , Soni, V. , Radha Kishan, K.V. and Agrawal, P. (2007) Characterization of Mycobacterium tuberculosis WhiB1/Rv3219 as a protein disulfide reductase. Protein Expression and Purification, 52, 422–432. [DOI] [PubMed] [Google Scholar]
- Geiman, D.E. , Raghunand, T.R. , Agarwal, N. and Bishai, W.R. (2006) Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB‐like genes. Antimicrobial Agents and Chemotherapy, 50, 2836–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, J.E. and Bishai, W.R. (2000) whmD is an essential mycobacterial gene required for proper septation and cell division. Proceedings of the National Academy of Sciences USA, 97, 8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, J. , Rolfe, M.D. and Smith, L.J. (2014) Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide. Virulence, 5, 794–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. , Dufayard, J.F. , Lefort, V. , Anisimova, M. , Hordijk, W. and Gascuel, O. (2010) New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Hett, E. C. , and Rubin, E. J. (2008) Bacterial growth and cell division: a mycobacterial perspective. Microbiology and Molecular Biology Reviews, 72, 126–156, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D.G. and Sharp, P.M. (1988) CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene, 73, 237–244. [DOI] [PubMed] [Google Scholar]
- Homerová, D. , Sevcikova, J. and Kormanec, J. (2003) Characterization of the Streptomyces coelico lor A3(2) wblE gene, encoding a homologue of the sporulation transcription factor. Folia Microbiologica (Praha), 48, 489–495. [DOI] [PubMed] [Google Scholar]
- Jakimowicz, P. , Cheesman, M.R. , Bishai, W.R. , Chater, K.F. , Thomson, A.J. and Buttner, M.J. (2005) Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe‐4S] cluster. Journal of Biological Chemistry, 280, 8309–8315. [DOI] [PubMed] [Google Scholar]
- Johnson, E.G. , Sparks, J.P. , Dzikovski, B. , Crane, B.R. , Gibson, D.M. and Loria, R. (2008) Plant‐pathogenic Streptomyces species produce nitric oxide synthase‐derived nitric oxide in response to host signals. Chemistry & Biology, 15, 43–50. [DOI] [PubMed] [Google Scholar]
- Jones, S.E. and Elliot, M.A. (2018) ‘Exploring’ the regulation of Streptomyces growth and development. Current Opinion in Microbiology, 42, 25–30. [DOI] [PubMed] [Google Scholar]
- Jones, S.E. , Ho, L. , Rees, C.A. , Hill, J.E. , Nodwell, J.R. and Elliot, M.A. (2017) Streptomyces exploration is triggered by fungal interactions and volatile signals. eLife, 6, e21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, B.K. , Clifton, M.C. , Shen, B.W. and Stoddard, B.L. (2009) The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease. Structure, 17, 1368–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, B.K. and Stoddard, B.L. (2011) DNA recognition and transcriptional regulation by the WhiA sporulation factor. Scientific Reports, 1, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.G. , Paget, M.S. , Seok, Y.J. , Hahn, M.Y. , Bae, J.B. , Hahn, J.S. , et al. (1999) RsrA, an anti‐sigma factor regulated by redox change. The EMBO Journal, 18, 4292–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, S.H. , Huang, J. , Lee, H.N. , Hur, Y.A. , Cohen, S.N. and Kim, E.S. (2007) Interspecies DNA microarray analysis identifies WblA as a pleiotropic down‐regulator of antibiotic biosynthesis in Streptomyces . Journal of Bacteriology, 189, 4315–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.S. , Lee, H.N. , Kim, P. , Lee, H.S. and Kim, E.S. (2012a) Negative role of wblA in response to oxidative stress in Streptomyces coelico lor . Journal of Microbiology and Biotechnology, 22, 736–741. [DOI] [PubMed] [Google Scholar]
- Kim, J.S. , Lee, H.N. , Lee, H.S. , Kim, P. and Kim, E.S. (2013) A WblA‐binding protein, SpiA, involved in Streptomyces oxidative stress response. Journal of Microbiology and Biotechnology, 23, 1365–1371. [DOI] [PubMed] [Google Scholar]
- Kim, M.S. , Dufour, Y.S. , Yoo, J.S. , Cho, Y.B. , Park, J.H. , Nam, G.B. , et al. (2012b) Conservation of thiol‐oxidative stress responses regulated by SigR orthologues in actinomycetes. Molecular Microbiology, 85, 326–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.S. , Hahn, M.Y. , Cho, Y. , Cho, S.N. and Roe, J.H. (2009) Positive and negative feedback regulatory loops of thiol‐oxidative stress response mediated by an unstable isoform of sigmaR in actinomycetes. Molecular Microbiology, 73, 815–825. [DOI] [PubMed] [Google Scholar]
- Kim, T.H. , Park, J.S. , Kim, H.J. , Kim, Y. , Kim, P. and Lee, H.S. (2005) The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochemical and Biophysical Research Communications, 337, 757–764. [DOI] [PubMed] [Google Scholar]
- Konar, M. , Alam, M.S. , Arora, C. and Agrawal, P. (2012) WhiB2/Rv3260c, a cell division‐associated protein of Mycobacterium tuberculosis H37Rv, has properties of a chaperone. The FEBS Journal, 279, 2781–2792. [DOI] [PubMed] [Google Scholar]
- Kudhair, B.K. , Hounslow, A.M. , Rolfe, M.D. , Crack, J.C. , Hunt, D.M. , Buxton, R.S. , et al. (2017) Structure of a Wbl protein and implications for NO sensing by M. tuberculosis. Nature Communications, 8, 2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, C. , Luna, B. , Ammerman, N.C. , Maiga, M. , Agarwal, N. and Bishai, W.R. (2012) Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1‐7 in redox environments. PLoS One, 7, e37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.S. , Kim, P. , Kim, E.S. , Kim, Y. and Lee, H.S. (2018) Corynebacterium glutamicum WhcD interacts with WhiA to exert a regulatory effect on cell division genes. Antonie Van Leeuwenhoek, 111, 641–648. [DOI] [PubMed] [Google Scholar]
- Lee, D.S. , Kim, Y. and Lee, H.S. (2017) The whcD gene of Corynebacterium glutamicum plays roles in cell division and envelope formation. Microbiology, 163, 131–143. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y. , Kim, H.J. , Kim, E.S. , Kim, P. , Kim, Y. and Lee, H.S. (2013) Regulatory interaction of the Corynebacterium glutamicum whc genes in oxidative stress responses. Journal of Biotechnology, 168, 149–154. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y. , Park, J.S. , Kim, H.J. , Kim, Y. and Lee, H.S. (2012) Corynebacterium glutamicum whcB, a stationary phase‐specific regulatory gene. FEMS Microbiology Letters, 327, 103–109. [DOI] [PubMed] [Google Scholar]
- Lefort, V. , Longueville, J.E. and Gascuel, O. (2017) SMS: smart model selection in PhyML. Molecular Biology and Evolution, 34, 2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, M. , Rajmani, R.S. and Singh, A. (2016) Mycobacterium tuberculosis WhiB3 responds to vacuolar pH‐induced changes in mycothiol redox potential to modulate phagosomal maturation and virulence. Journal of Biological Chemistry, 291, 2888–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minch, K.J. , Rustad, T.R. , Peterson, E.J. , Winkler, J. , Reiss, D.J. , Ma, S. , et al. (2015) The DNA‐binding network of Mycobacterium tuberculosis . Nature Communications, 6, 5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle, V. , Palframan, W.J. , Findlay, K.C. and Buttner, M.J. (2000) WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelico lor A3(2). Journal of Bacteriology, 182, 1286–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R.P. , Nguyen, L. , Gatfield, J. , Visconti, K. , Nguyen, K. , Schnappinger, D. , et al. (2005) Ancestral antibiotic resistance in Mycobacterium tuberculosis . Proceedings of the National Academy of Sciences USA, 102, 12200–12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget, M.S. , Kang, J.G. , Roe, J.H. and Buttner, M.J. (1998) sigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelico lor A3(2). The EMBO Journal, 17, 5776–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget, M.S. , Molle, V. , Cohen, G. , Aharonowitz, Y. and Buttner, M.J. (2001) Defining the disulphide stress response in Streptomyces coelico lor A3(2): identification of the sigmaR regulon. Molecular Microbiology, 42, 1007–1020. [DOI] [PubMed] [Google Scholar]
- Park, J.C. , Park, J.S. , Kim, Y. , Kim, P. , Kim, E.S. and Lee, H.S. (2016) SpiE interacts with Corynebacterium glutamicum WhcE and is involved in heat and oxidative stress responses. Applied Microbiology and Biotechnology, 100, 4063–4072. [DOI] [PubMed] [Google Scholar]
- Park, J.S. , Lee, J.Y. , Kim, H.J. , Kim, E.S. , Kim, P. , Kim, Y. and Lee, H.S. (2012) The role of Corynebacterium glutamicum spiA gene in whcA‐mediated oxidative stress gene regulation. FEMS Microbiology Letters, 331, 63–69. [DOI] [PubMed] [Google Scholar]
- Park, J.S. , Shin, S. , Kim, E.S. , Kim, P. , Kim, Y. and Lee, H.S. (2011) Identification of SpiA that interacts with Corynebacterium glutamicum WhcA using a two‐hybrid system. FEMS Microbiology Letters, 322, 8–14. [DOI] [PubMed] [Google Scholar]
- Ramon‐Garcia, S. , Ng, C. , Jensen, P.R. , Dosanjh, M. , Burian, J. , Morris, R.P. , et al. (2013) WhiB7, an Fe‐S‐dependent transcription factor that activates species‐specific repertoires of drug resistance determinants in actinobacteria. Journal of Biological Chemistry, 288, 34514–34528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, K.H. , Abramovitch, R.B. and Russell, D.G. (2007) Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host & Microbe, 2, 352–364. [DOI] [PubMed] [Google Scholar]
- Rustad, T.R. , Minch, K.J. , Ma, S. , Winkler, J.K. , Hobbs, S. , Hickey, M. , et al (2014) Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression‐derived regulatory network. Genome Biology, 15, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybniker, J. , Nowag, A. , van Gumpel, E. , Nissen, N. , Robinson, N. , Plum, G. , et al (2010) Insights into the function of the WhiB‐like protein of mycobacteriophage TM4 – a transcriptional inhibitor of WhiB2. Molecular Microbiology, 77, 642–657. [DOI] [PubMed] [Google Scholar]
- Sasaki, Y. , Oguchi, H. , Kobayashi, T. , Kusama, S. , Sugiura, R. , Moriya, K. , et al. (2016) Nitrogen oxide cycle regulates nitric oxide levels and bacterial cell signaling. Scientific Reports, 6, 22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. , Crossman, D.K. , Mai, D. , Guidry, L. , Voskuil, M.I. , Renfrow, M.B. , et al. (2009) Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathogens, 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. , Guidry, L. , Narasimhulu, K.V. , Mai, D. , Trombley, J. , Redding, K.E. , et al. (2007) Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe‐4S] cluster and is essential for nutrient starvation survival. Proceedings of the National Academy of Sciences USA, 104, 11562–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.J. , Stapleton, M.R. , Buxton, R.S. and Green, J. (2012) Structure‐function relationships of the Mycobacterium tuberculosis transcription factor WhiB1. PLoS One, 7, e40407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.J. , Stapleton, M.R. , Fullstone, G.J. , Crack, J.C. , Thomson, A.J. , Le Brun, N.E. , et al. (2010) Mycobacterium tuberculosis WhiB1 is an essential DNA‐binding protein with a nitric oxide‐sensitive iron‐sulfur cluster. Biochemical Journal, 432, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliveri, J.A. , Gomez, J. , Bishai, W.R. and Chater, K.F. (2000) Multiple paralogous genes related to the Streptomyces coelico lor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology, 146(Pt 2), 333–343. [DOI] [PubMed] [Google Scholar]
- Stapleton, M.R. , Smith, L.J. , Hunt, D.M. , Buxton, R.S. and Green, J. (2012) Mycobacterium tuberculosis WhiB1 represses transcription of the essential chaperonin GroEL2. Tuberculosis (Edinb), 92, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn, A.J. , Collins, D.M. , Hondalus, M.K. , Jacobs, W.R. Jr. , Kawakami, R.P. and Bloom, B.R. (2002) Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proceedings of the National Academy of Sciences USA, 99, 3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surdova, K. , Gamba, P. , Claessen, D. , Siersma, T. , Jonker, M.J. , Errington, J. , et al. (2013) The conserved DNA‐binding protein WhiA is involved in cell division in Bacillus subtilis . Journal of Bacteriology, 195, 5450–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, J.S. , Oh, G.S. , Ryoo, S. and Roe, J.H. (2016) Induction of a stable sigma factor SigR by translation‐inhibiting antibiotics confers resistance to antibiotics. Scientific Reports, 6, 28628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, P. , Liu, S.P. , Bu, Q.T. , Zhou, Z.X. , Zhu, Z.H. , Huang, F.L. and Li, Y.Q. (2014) WblAch, a pivotal activator of natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis L10, is positively regulated by AdpAch. Applied and Environmental Microbiology, 80, 6879–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Halladin, D.K. and Theriot, J.A. (2016) Fast mechanically driven daughter cell separation is widespread in actinobacteria. MBio, 7, e00952–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


