Summary
Global warming is resulting in increased frequency of weather extremes. Root‐associated fungi play important roles in terrestrial biogeochemical cycling processes, but the way in which they are affected by extreme weather is unclear. Here, we performed long‐term field monitoring of the root‐associated fungus community of a short rotation coppice willow plantation, and compared community dynamics before and after a once in 100 yr rainfall event that occurred in the UK in 2012.
Monitoring of the root‐associated fungi was performed over a 3‐yr period by metabarcoding the fungal internal transcribed spacer (ITS) region. Repeated soil testing and continuous climatic monitoring supplemented community data, and the relative effects of environmental and temporal variation were determined on the root‐associated fungal community.
Soil saturation and surface water were recorded throughout the early growing season of 2012, following extreme rainfall. This was associated with a crash in the richness and relative abundance of ectomycorrhizal fungi, with each declining by over 50%. Richness and relative abundance of saprophytes and pathogens increased.
We conclude that extreme rainfall events may be important yet overlooked determinants of root‐associated fungal community assembly. Given the integral role of ectomycorrhizal fungi in biogeochemical cycles, these events may have considerable impacts upon the functioning of terrestrial ecosystems.
Keywords: extreme weather, mycorrhizal fungi, root‐associated fungi, soil fungi, temporal variation in microbial communities
Short abstract
See also the Commentary on this article by https://doi.org/10.1111/nph.15086.
Introduction
There is mounting evidence that global warming is directly increasing the frequency of weather extremes, including heavy rainfall events, droughts and high temperatures across Europe and North America (Mallakpour & Villarini, 2015; Shepherd, 2015), which may be further increased during El Nino events (Cai et al., 2014). Terrestrial ecosystems play a key role in determining global climate–ecosystem feedbacks, due to their role in the exchange of greenhouse gases (GHG) including CO2, methane and nitrous oxide, and storage of carbon (C) in soil and vegetation. Extreme weather may have major effects on climate–terrestrial ecosystem interactions, with links to reductions of C stocks through a range of feedbacks operating on plant productivity and soil processes (Reichstein et al., 2013). However, studies of the impacts of extreme weather events on soil systems are limited due to their rarity and our inability to capture such events in space and time. As a result, very little is known of the ways in which extreme weather, particularly extreme rainfall events, affect soil communities and associated processes, and the consequences for climate–ecosystem feedbacks (Heimann & Reichstein, 2008).
Plants live in close association with distinct microbial communities in the rhizosphere, the radial gradient spanning from the plant root into the soil (Hartmann et al., 2008) into which microorganisms are selectively recruited (Berendsen et al., 2012). These root‐associated communities are diverse, and although most attention has focused on their bacterial and fungal communities (Smit et al., 1999; Smalla et al., 2001), they include a plethora of other components including protists and nematodes (Bonkowski, 2004). These communities likely play key roles in terrestrial ecosystems, connecting above‐ and belowground diversity (Bardgett & van der Putten, 2014).
Mycorrhizal fungi, particularly ectomycorrhizal (ECM) and arbuscular mycorrhizal (AM) types are some of the best‐characterized fungal inhabitants of the rhizosphere. These fungi form mutualistic symbioses with their host plants (Smith & Read, 2010), conferring a range of benefits, particularly by increasing host uptake of nutrients (Garcia et al., 2014) and providing disease resistance in exchange for C (Chakravarty & Unestam, 1987; Liu et al., 2007). These root‐associated fungi can be major components of terrestrial ecosystems through their role in driving biogeochemical cycles (Högberg et al., 2010). For example within temperate forest soils, they may contribute a third of ecosystem C and nitrogen (N) mineralization (Finzi et al., 2015). ECM dominate the root zone in temperate and boreal forest systems, supporting plant nutrition and productivity while acting as major conduits by which C flows from the plant to the soil on a global scale (Leake et al., 2004). Nonmycorrhizal fungi also inhabit the rhizosphere, but their diversity and functional significance are typically less well understood (Kubartová et al., 2008; Dean et al., 2012). Some such fungi occur as endophytes within the root and can also confer growth benefits to the host plant, whilst saprophytes promote mineralization processes, altering nutrient availability and indirectly influencing plant growth (Smith & Read, 2010; Bardgett & van der Putten, 2014). Pathogens, meanwhile, can directly reduce plant growth and development. Even within each of these differing ‘life strategies’, fungi can also show considerable functional variability (Kiers et al., 2011; Dean et al., 2012), and changes in the composition of root‐associated fungal communities may have profound ecosystem‐level effects on soil processes such as C, N and phosphorus (P) cycles within natural and agricultural systems.
Assembly of root‐associated fungi has been shown to be regulated by a wide range of spatial and temporal variables. Soil pH has a near ubiquitous effect on soil microbial community assembly (Coughlan et al., 2000; Gosling et al., 2013; Tedersoo et al., 2014), whilst available P and N have been linked to changes in the composition of root‐associated fungal communities, with increases of both linked to reduced mycorrhizal abundance (Gosling et al., 2013). Climatic factors such as rainfall and temperature also have been indicated to drive fungal richness on a global level (Swaty et al., 1998), whilst increasing geographical distance between soil fungal communities has been linked with increasing dissimilarity, independent to that of changing environmental factors (Lilleskov et al., 2004; Barnes et al., 2016b). Root‐associated fungal communities also change over time, with strong evidence for seasonality of community structure (Bohrer et al., 2004; Dumbrell et al., 2011), and inter‐annual shifts which can take place across multiple years (Last et al., 1984; Visser, 1995; Daniell et al., 2001; Husband et al., 2002), adding further complexity to understanding the factors that regulate community assembly.
Given that soil biodiversity and functioning remains largely ‘a black box’, it is unsurprising that to date little is known of the way in which extreme weather affects the assembly of root‐associated fungi, and the consequences for climate–ecosystem feedbacks (Bardgett et al., 2008). The data that are currently available on extreme weather–soil interactions are limited and derived from experimental manipulations (Barnard et al., 2013; Amend et al., 2016); however, drought and rewetting of soil cores has revealed a high resilience of the fungal community to desiccation. Soil saturation following intense rainfall can have profound effects on plant productivity, and exposure for long periods can even cause plant death (Rivest et al., 2013). Furthermore, soil saturation leads to hypoxic or anoxic conditions, promoting microbial reduction reactions, and in turn influencing the abundance and composition of soil microbiota (Coutts & Nicoll, 1990; Unger et al., 2009; Wilson et al., 2011; Wagner et al., 2015). Fungi can have varied responses to waterlogging within terrestrial ecosystems, with ‘high moisture’ specialist AMF and saprophytic species that are seemingly unaffected or even thrive in waterlogged and anoxic conditions (Camy et al., 2003; Fougnies et al., 2007; García et al., 2008; Yang et al., 2016). ECM fungal species are known to have variable hydrophobicity (Lilleskov et al., 2011), which may be an adaptation to moisture amounts within habitats and across seasons (Unestam & Sun, 1995). Some ECM fungi inhabit environments which are permanently waterlogged (Baar et al., 2002). However, increasing saturation may reduce ECM fungal abundance across ecosystems (Tedersoo et al., 2009), and even induce death of ECM fungal mycelium within an ecosystem as soil becomes waterlogged (Coutts & Nicoll, 1990). However, the way in which change in weather, and in particular weather extremes, affects the composition and function of the rhizosphere fungal community remains unknown, representing a considerable obstacle in understanding future climate‐ecosystem feedbacks.
Here, we quantify the relative effects of spatial (geographical distance between sampling locations), environmental (soil and climatic properties) and temporal variation (seasonal and interannual) in determining the root‐associated fungal community of willow over a 3‐yr period. Soil cores were taken across line transects in October 2010, July 2011, October 2011, July 2012 and October 2012. Fine roots were extracted from soil cores and their associated fungal communities were profiled by high‐throughput sequencing. Significantly, sampling in 2012 coincided with record levels of rainfall across the UK, and locally at our field site, rainfall was at its highest annual levels since records began in 1947. This provided an unique opportunity to elucidate the magnitude to which extreme rainfall and associated soil saturation influences assembly of communities in situ, including the responses of fungi with distinct nutritional modes (ectomycorrhiza, pathogens, endophytes and saprophytes), relative to spatial and seasonal variation in soil and climatic factors.
Materials and Methods
Study site and experimental design
The study was performed in a short rotation coppice willow plantation near Lincoln, UK (53.3163°N, 0.5777°W), which was subject to minimal land management practices. The site had a 30‐yr mean air temperature of 9.9°C and a fine loam over clay soil (15% clay, 49% sand and 36% silt). In 2000, the willow was planted at a density of 15 000 stools ha−1, covering an area of c. 9.44 ha. Salix cuttings were planted in paired rows 0.75 m apart, with 1.5 m spaces between the rows. Six closely related Salix viminalis L. genotypes were planted in order to limit disease spread, with Tora (60%) being the most abundant (the others being Bjorn (10%), Bowles Hybrid (10%), Jorr (10%) and Jorunn (10%)). Willow genotypic variation was assessed along the line of the trees where line transects were performed, and no pattern was observed in planting (data not shown). Given the density of trees found within the field site, and that roots can spread over 9 m from individual S. viminalis trees (Phillips et al., 2014), roots were considered an homogenous mixture of multiple genotypes within the upper soil profile. Coppicing began in 2001, with further events in 2004, 2007 and 2010, providing an average yield of 6.72 t ha−1. In February 2010, 660 kg ha−1 PK fertilizer (Fibrophos, Melton, UK), 20 t ha−1 of lime and 20 t ha−1 of locally sourced green waste compost were added to the field site; no exogenous nutrient inputs were added during the duration of the experiment. After planting, soil remained untilled throughout the lifetime of the crop. Therefore, soil microbial communities developed over this period without major disruption of the soil matrix, in a system with limited aboveground diversity.
Sampling was performed along single line transects (as per Barnes et al., 2016b) at each sampling time. Starting from the southernmost edge of the field heading due north, single line transects started 25 m into the field in order to avoid edge effects, sampling eight locations every 20 m along the line. At each location, four subsamples were taken 1 m apart from north, south, east and west directions from the central location, using a 4.5‐cm diameter soil auger (Van Walt Equipment, Haslemere, UK) to a depth of 15 cm, which was wiped clean with ethanol between samples to limit contamination. Transects were taken in October 2010, July 2011, October 2011, July 2012 and October 2012, shifting a further 3 m north each time in order to avoid sampling of previously disturbed areas.
Soil nutrient analysis
Soil cores were homogenized evenly by gloved hand, before 100 g was removed and loosely covered to air‐dry, and sieved to < 2 mm particle size. Soil pH and nutrient analysis were performed as described previously in Barnes et al. (2016a), with pH, NO3, Mg, available K and available P determined for each individual subsample (32 per transect) and averaged at each of the eight sampling locations. This was repeated for each line transect performed at the different times. These soil properties were analysed as they have repeatedly been shown to influence root‐associated fungal community assembly (Geisseler & Scow, 2014).
Root‐associated fungal DNA extraction
Following removal of c. 100 g (from a total of between 200 and 300 g) of soil for nutrient analysis, the remaining soil from each subsample was soaked in deionized water at room temperature for 1 h before all roots were hand extracted using forceps (between 2 and 5 g of roots per subsample). Nonsenescent fine roots (< 2 mm in diameter), which were identified by their colour, texture, turgor and branching structures, were washed over a 6‐mm sieve to remove adherent soil. By selecting these nonsenescent fine roots, sampling of contaminating roots from the herbaceous understory species was also avoided. Entire pools of selected roots were cut into 1‐cm lengths, mixed, and a 0.5 g randomly selected subsample was used for DNA extraction. Roots were initially exposed to mechanical lysis of two periods of 30 s at 30 hz using a TissueLyser (Qiagen) before extraction using the PowerSoil DNA extraction kit (MP Biomedicals, Cambridge, UK) as per the manufacturer's instructions. After extraction, the four subsamples at each sampling location were equilibrated to 25 ng μl−1 using a nanodrop ND1000 (Fischer Scientific, Loughborough, UK) and 10 μl of each was pooled to make the DNA template for each of the eight sampling locations for each sampling time, with 40 samples in total used for sequencing (eight sampling locations, five time points).
Weather data collection
Meteorological data were provided by a weather station located at the field site. These consisted of a cup anemometer, wind vane, air and wet bulb temperatures (Didcot Instruments AWS, Didcot, UK) and a rain gauge (Rimco, Malton, UK). Data were logged as 30‐min averages with the exception of rainfall, which was logged as the 30 min total. Temperature and rainfall data were measured continuously from April 2010 to December 2012. Measurements of volumetric soil moisture content (SWC; m3 m−3) were only recorded over the main growth period (June–November). SWC was recorded by two CS616 time domain reflectometer (TDR) probes (Cambell Scientific Inc., Logan, UT, USA), which measure the upper 0.3 m of the soil profile. SWC sensors scanned every 10 s and were logged as 30‐min averages. All meteorological data were recorded using CR10 data loggers (Campbell Scientific, UT, USA).
In order to test the soil saturation point, six plastic 66‐mm diameter and 30‐cm depth tubes were hammered into the soil to remove intact cores. Cores were bagged and returned to the lab, before being cut into 0–15 cm and 15–30 cm sections. Volumetric moisture content (VMC) was determined as per Laird et al. (2010), with cores saturated within a solution of 0.001 M CaCl2 and weighed, before being incubated in an oven until completely dry and reweighed. VMC was subsequently determined as a percentage of bulk density of soil.
High‐throughput amplicon sequencing and processing of sequencing data
DNA extracts underwent sequencing and subsequent bioinformatics as described previously in Barnes et al. (2016b) using ITS1F and ITS4 primers (Gardes & Bruns, 1993; Yang et al., 2012), before sequencing on a Roche 454 GS Junior pyrosequencer (454 Life Sciences/Roche Applied Biosystems, Nurley, NJ, USA) at Micropathology Ltd (Coventry, UK). Raw sequences were deposited at the NCBI sequence read archive under the accession numbers SRP056724 for October 2010, and SRP062101 for July 2011, October 2011, July 2012 and October 2012.
Operational taxonomic units (OTUs) were picked de novo from sequences using the Uclust algorithm at a 97% similarity level, and chimera checked using the Uchime algorithm in de novo mode (Edgar, 2010; Edgar et al., 2011). Taxonomic assignments were initially made using the UNITE database (27.08.2013 release from the UNITE project) using Blast (with an e‐value > 0.001), before OTUs of >0.1% relative abundance underwent a further Blast search against the continuously updated Unite database and taxonomy reassigned if e‐value > 0.001. OTUs with ≤3 reads assigned were also removed to limit the effects of sequencing errors and artefacts (Huse et al., 2007).
After processing, 76.6% (92 595) of total reads remained, averaging 2341 reads per sample, across 931 OTUs (mean length = 496.1, standard deviation = 22.5). Chao1 estimates were calculated to estimate the theoretical maximum number of OTUs per sampling location (Chao, 1987), revealing that 63.8% of OTUs were detected after rarefaction and many rare OTUs were undetected. Given the issues McMurdie & Holmes, 2014 raise in performing community analyses with variable library sizes, the rarefied OTU table underwent normalization using the DeSeq2 package as part of Qiime (Anders & Huber, 2010). Negative values were considered zeros before data were finally converted to relative abundances (as percentage of normalized read numbers) as per Fernandez et al. (2017). The negative binomial model performed within DeSeq2 can simultaneously account for variation in library sizes and biological variability better than simply subsampling to a uniform sequencing depth alone.
Some members of the Ascomycota, Basidiomycota and Zygomycota can form ECM associations or exist as root endophytes, whereas others act as saprophytes or pathogens. Therefore, a likely life strategy (i.e. nutritional mode) was determined for OTUs in a two‐step process. Initially an automated approach was taken through parsing the OTU table through the FUNGuild database (v.1.0) (Tedersoo et al., 2014; Nguyen et al., 2016), before a secondary round of assignments were made on a manual basis using previous literature findings (Supporting Information Table S1). These were performed on OTUs with family level or greater specificity of taxonomic assignments and consensus of life strategy within the taxonomic unit. Groups considered were ECM fungi, endophytes, pathogens and saprotrophs, whilst other fungi that were very low in OTU richness such as lichen symbionts and yeasts, or those that were not taxonomically assigned at genus level, were not included within the life strategy analyses.
In order to further understand the ECM fungal response to extreme rainfall, ECM fungal OTUs also were divided by exploration type (Agerer, 2001) and by their mycelial hydrophobicity (Lilleskov et al., 2011), with extramatrical exploration type divided into contact‐short, contact‐medium and medium‐long groupings as performed by Fernandez et al. (2017).
Statistical analyses
Repeated measures one‐way ANOVAs were performed on climatic data (soil moisture, precipitation and temperature) to test for significant differences between years, whilst two‐way ANOVAs with sampling time (repeated measure) and distance across transects were performed for assessing significant spatio‐temporal variation in soil properties (Oksanen et al., 2007).
Repeated measures one‐way ANOVAs were performed on the OTU data for phylum, family, genus, species, life strategy (known ectomycorrhizal, pathogen, saprophyte and endophyte taxa), hydrophobicity and extramatrical exploration type in order to analyse variation between sampling locations (df = 39) and sampling times (df = 4), and post‐hoc (Tukey's honestly significant difference) tests were performed when significant differences were observed. Due to the high number of analyses performed on individual OTU data, P‐values from individual OTU analyses were adjusted using Benjami and Hochberg's false discovery rate procedure in order to limit false‐positives, using the p.adjust function within the stats package of R (Benjamini & Hochberg, 1995). Finally, general linear modelling (GLM) was used to ascertain which temporal (time as weeks after sampling) and environmental parameters (soil pH, P, K, Mg, NO3 and SWC) significantly explained variance in the relative abundances of individual OTUs that were found to significantly change between time points using repeated measure ANOVAs (Grafen and Hails, 2002). GLM was performed in Xlstat (Addinsoft, Paris, France) with the aim of disentangling the effects of sampling time from that of SWC.
Hellinger transformations downweight rare species and perform well with high‐throughput sequencing datasets in which there are many zero values, and thus were performed on the community data before Bray–Curtis similarity matrices were generated (Bray & Curtis, 1957). Nonmetric multidimensional scaling analysis (nMDS) was performed from this matrix to visualize trends in community similarity. Variation within the overall community and each life strategy group of the root‐associated fungi separately was quantified via PERMANOVA (using the adonis function within R) against temporal (sampling year and season), environmental properties (pH, NO3, Mg, K and P) and geographical distance (between sampling locations) using sampling time as the strata (as October 2010, July 2011, October 2011, July 2012 or October 2012). As the order of explanatory variables affects the outcome of PERMANOVA analyses, variables were placed in order of the largest proportion of variation explained before running the final analysis. The nMDS analyses and PERMANOVA were performed using the vegan package of R (v.2.4‐2; Oksanen et al., 2007), whilst nMDS and volcano plots were created using the package ggplots2 (v.2.2.1; Wickham, 2016).
Results
Edaphic properties
Biogeochemical values were calculated as the mean of the four subsamples at each sampling location, before undergoing two‐way repeated measures ANOVA that used distance across transect as a factor, and sampling time (as October 2010, July 2011, October 2011, July 2012 and October 2012) as a repeated measure. There were strong significant gradients across the transects for pH (df = 7, F = 14.45, P < 0.001), available P (df = 7, F = 3.51, P < 0.001), available K (df = 7, F = 8.59, P < 0.001) and Mg (df = 7, F = 2.74, P = 0.024), all of which were significantly greater in sampling locations 1–3 relative to sampling locations 5–8 at all time points (data not shown). Between sampling times, pH (df = 4, F = 21.61, P < 0.001) was significantly raised in 2011 and 2012, available P decreased significantly after 2010 (df = 4, F = 16.07, P < 0.001), whilst NO3 showed significant seasonal variation (df = 4, F = 13.95, P < 0.001), with substantially higher quantities in soil in July, relative to October transects. Available K also varied over time, peaking in October 2010 and July 2012 time points (df = 4, F = 4.14, P = 0.030) (Fig. S1a–e).
Climatic data
The average daily temperature at the field site varied throughout the years (df = 10, F = 14.1, P < 0.001), from c. –1°C in December 2010 to 17°C in July 2012 (Fig. S2a); however, temperature did not vary between years (df = 2, F = 0.325, P = 0.725) despite 2011 having a warmer spring and autumn compared to the other years. Precipitation did not consistently differ between months (Fig. S2b), yet it differed strongly between years (df = 2, F = 3.86, P = 0.036), with 2012 being substantially wetter in April (140 mm per month) and June (130 mm per month), which was over double that of the previous year's averages. SWC was only monitored between June and November, but reflected rainfall data, with 2012 significantly higher than 2010 and 2011 in all months (df = 2, F = 5.47, P = 0.020), but with no apparent consistent monthly differences between years (df = 4, F = 0.669, P = 0.628; Fig. S2c). The average monthly SWC across the entire monitoring period was 0.313 m3 m−3; however, for 2012 this figure was 0.406 m3 m−3 and reached as high as 0.484 m3 m−3 in both June and July 2012. Saturation tests within the laboratory suggested that complete saturation can occur from 0.510 m3 m−3, and thus in June and July 2012 soil was 94.9% of minimum saturation (Fig. S3). Furthermore, observations in the field showed a considerable presence of standing surface water throughout this period (Fig. S4).
Investigating spatial, temporal and environmental factors regulating root‐associated fungal assemblage
In order to understand the spatio‐temporal variation of the root‐associated fungal community composition over the sampling period, an initial Bray–Curtis similarity matrix was created from the sequencing data and visualized using multidimensional scaling (limited to two dimensions, stress = 0.145, 999 iterations). Samples taken in 2010 and 2011 grouped together, and were distinct from those taken in 2012 (Fig. 1). There was no evidence for seasonal changes in community composition for either the 2011 or 2012 time points, as there was no grouping of October or July time points within the nMDS ordinations. PERMANOVA was subsequently performed against temporal data (sampling year and season as factors), environmental properties (pH, NO3, Mg, K and P) and geographical distance (as location on transect), whilst sampling time point was used as the strata (Table 1). In this analysis, sampling year (2010, 2011 or 2012) had the greatest effect on the community composition, explaining 21.1% of variation, whilst pH explained a further 10.8% of variation (Fig. S5), with a total of 31.9% total community variation explained by measured parameters. There was no significant effect of seasonality (i.e. sampling in either July or October) on community structure over the 3‐yr sampling period. Neither geographical distance between sampling locations nor the remaining soil properties affected community assembly.
Figure 1.
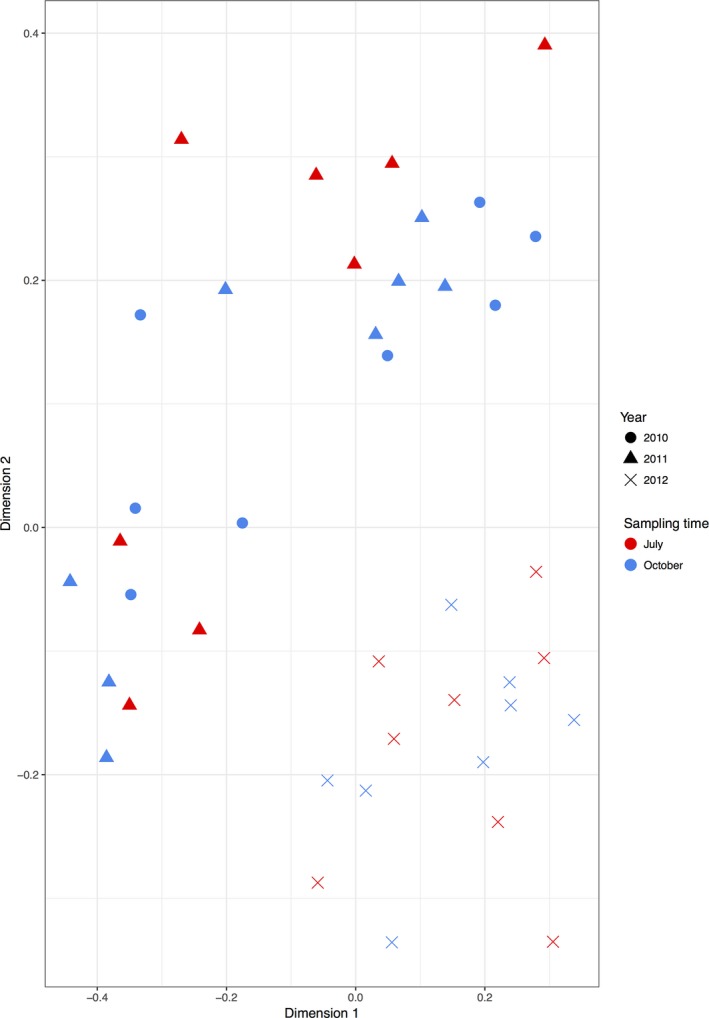
Nonmetric dimensional scaling showing clustering based on similarity of the root‐associated fungal communities between sampling time points.
Table 1.
The relative importance of time, geographical distance (between samples) and soil properties for the root‐associated fungal community of willow as revealed by PERMANOVA
| Parameter | Degrees of freedom | F‐value | R 2 | P‐value |
|---|---|---|---|---|
| Season | 1 | 1.338 | 0.026 | 0.155 |
| Year | 3 | 3.640 | 0.211 | 0.001 |
| pH | 1 | 5.586 | 0.108 | 0.001 |
| Distance | 1 | 0.772 | 0.015 | 0.736 |
| Nitrate (NO3) | 1 | 0.859 | 0.017 | 0.612 |
| Magnesium (Mg) | 1 | 0.759 | 0.015 | 0.771 |
| Potassium (K) | 1 | 1.045 | 0.020 | 0.350 |
| Phosphorus (P) | 1 | 1.356 | 0.026 | 0.153 |
| Residuals | 29 | 0.562 | ||
| Total | 39 | 1.000 |
Bold indicates significance (P < 0.05).
Given the community shift that occurred between 2011 and 2012, community data were partitioned into pre‐ (October 2010, July 2011 and October 2011) and post‐extreme rainfall (July 2012 and October 2012), and reanalysed for seasonal, environmental and edaphic effects that may have been masked by the weather event (Fig. S6; Table S2). The two separate PERMANOVAs revealed that the pre‐extreme rainfall communities were regulated only by pH (21.6%), whilst seasonality (as July or October) and geographical distance (as location on the line transect) had no effect. The post‐extreme rainfall community remained entirely unexplained by all measured parameters.
Determining the taxonomic shifts associated with the change in community composition in 2012
Total OTU richness varied from 80 to 299 OTUs between samples, and there was no significant difference between sampling times (F = 0.98, P = 0.431). However, the Basidiomycota, which represented the largest phylum by OTU richness, differed significantly between sampling times (F = 4.24, P = 0.008), with between 116.4 and 98.6 OTUs per sampling location recorded across the 2010 and 2011 communities (known as the pre‐extreme rainfall samples hereafter) and only 68.0 and 69.2 in the 2012 communities (known as the post‐extreme rainfall samples hereafter; Fig. 2). The relative abundance of Basidiomycota declined significantly from 65.4% in the pre‐extreme to 37.7% in the post‐extreme rainfall samples (F = 12.27, P < 0.001).
Figure 2.
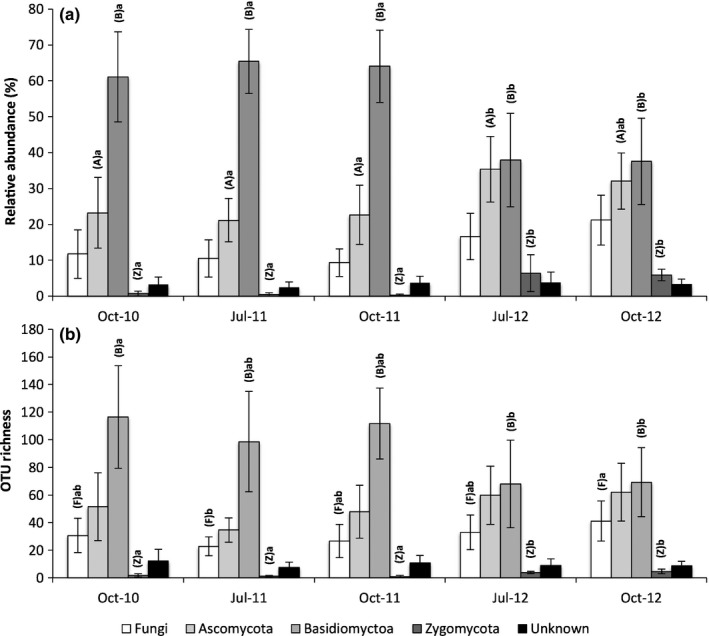
Average (a) relative abundance and (b) operational taxonomic unit (OTU) richness of the willow root‐associated fungal community separated at the phylum level over a 3‐yr sampling period. October 2010, July 2011 and October 2011 were pre‐extreme rainfall at the site, whilst July 2012 and October 2012 were post‐extreme rainfall. Error bars represent ± 1 SD of the mean. Capital letters in parentheses represent the taxonomic groups that differ between sampling times and different lower case letters indicate the associated significant differences, by Tukey's test (α = 0.05).
The Ascomycota were the second most OTU‐rich and abundant phylum. Whilst OTU richness ranged from 34.6 to 62.0, there was no significant change over time (F = 2.46, P = 0.068). However, the Ascomycota varied significantly in abundance between time points (F = 5.04, P = 0.004), from 22.3% in the pre‐extreme rainfall samples to 33.7% of the post‐extreme rainfall samples. The Zygomycota were the least abundant and diverse phylum, but OTU richness increased (F = 16.45, P < 0.001) from an average of 1.4 per sampling location in the pre‐extreme to 4.2 per sampling location in the post‐extreme rainfall samples. Furthermore, relative abundance of Zygomycota reads increased significantly (F = 13.06, P < 0.001) from just 0.4% in the pre‐extreme to 6.2% in the post‐extreme rainfall samples.
There were a number of OTU that could only be assigned to the fungal kingdom, or had no taxonomic information at all. They accounted for between 9.3% and 21.2% of reads (F = 5.48, P = 0.002, respectively) and were lowest in July 2012 and highest in October 2012. These averaged 30.8 OTUs per sample and remained stable over time in OTU richness (F = 2.68, P = 0.052). There also was an additional 3.3% average abundance of reads that could not be assigned even at the kingdom level (varying between 2.5% and 3.8% between sampling times), accounting for an average of 9.6 OTUs per sample (ranging from 7.5 and 12.1) which were retained in measures of richness and compositional analyses.
Determining the functional shifts associated with the change in community composition in 2012
Relative abundance of ECM fungi remained stable throughout the pre‐extreme rainfall samples, varying from 52.0% to 56.1%. However, there was a significant decline to 23.5% in the samples after the extreme rainfall event (F = 7.71, P < 0.001) (Fig. 3a). There was also a significant decline in ECM fungal OTU richness in the post‐extreme rainfall samples relative to pre‐extreme rainfall samples (F = 20.14, P < 0.001), falling from 79–95 to 43–45 OTUs (Fig. 3b).
Figure 3.
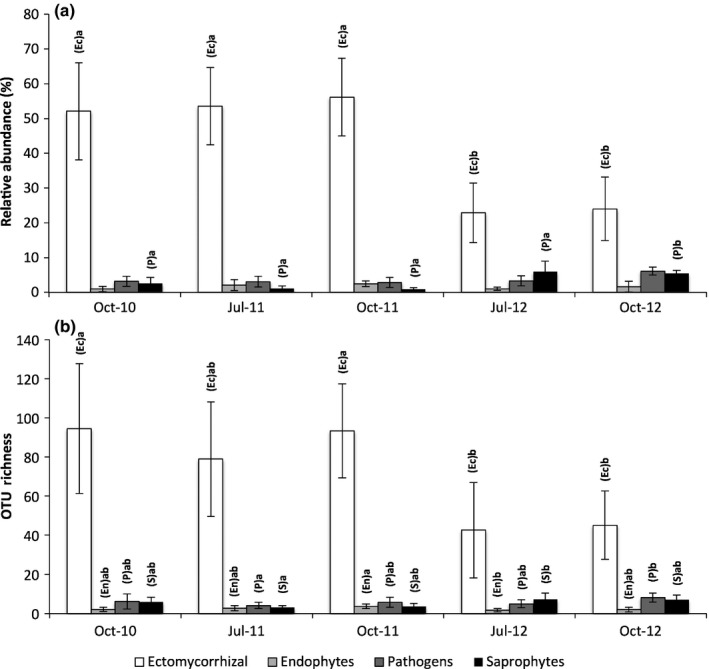
Average (a) relative abundance and (b) operational taxonomic unit (OTU) richness of differing life strategies of root‐associated fungi extracted from willow roots over a 3‐yr sampling period. October 2010, July 2011 and October 2011 were pre‐extreme rainfall at the site, whilst July 2012 and October 2012 were post‐extreme rainfall. Error bars represent ± 1 SD of the mean. Capital letters in parentheses represent the taxonomic groups that differ between sampling times and different lower case letters indicate the associated significant differences, by Tukey's test (α = 0.05).
Pathogens were the second most abundant and OTU‐rich life strategy, accounting for an average of 3.7% of relative abundance. Pathogen relative abundance differed over time (F = 6.93, P < 0.001), from 3.1% pre‐extreme to 4.7% post‐extreme rainfall. The number of pathogen OTUs within the community varied significantly over time (F = 3.38, P = 0.022), with 5.3 OTUs pre‐extreme and 6.6 OTUs post‐extreme rainfall.
Saprophytes accounted for a further 3.2% of relative abundance and also varied significantly over time (F = 2.94, P = 0.038), ranging from 2.7% and 5.7% in the pre‐extreme and post‐extreme rainfall communities, respectively. Saprophyte OTU richness increased significantly (F = 5.84, P = 0.013) from 5.0% pre‐extreme rainfall to 7.1% afterwards.
Endophytes were consistently found in low abundance and low OTU richness. Endophyte OTU richness varied significantly between sampling points, ranging from an average of 2.5 OTUs pre‐extreme rainfall to 1.9 OTUs post‐extreme rainfall (F = 3.08, P = 0.032), but did not vary in relative abundance, with an average of just 0.56% of total abundance (F = 1.59, P = 0.205).
Closer inspection of ECM fungal families also revealed declines in both OTU richness and abundance in nearly every family. The overall trend of decline in ECM fungal abundance post‐extreme rainfall was driven primarily by changes in the Cortinariaceae (Table S3) whose abundance fell from 32.3% to 9.8% of total relative abundance, whilst OTU richness dropped from 65.5 to 27.5.
ECM fungi were divided by hydrophobicity and exploration type, and dynamics analysed over time. The hydrophobic ECM fungi were present in considerably greater relative abundance and OTU richness compared to hydrophilic types (Fig 4). Hydrophobic ECM fungi declined significantly in both relative abundance (F = 6.49, P < 0.001) and OTU richness (F = 6.20, P < 0.001) over time, from averages of 15.7% and 39.1 OTUs pre‐extreme rainfall to just 4.1% and 13.4 OTUs, respectively, post‐extreme rainfall. The relative abundance and OTU richness of hydrophilic ECM fungi did not change significantly over time (F = 1.67, P = 0.180 and F = 1.74, P = 0.163, respectively).
Figure 4.
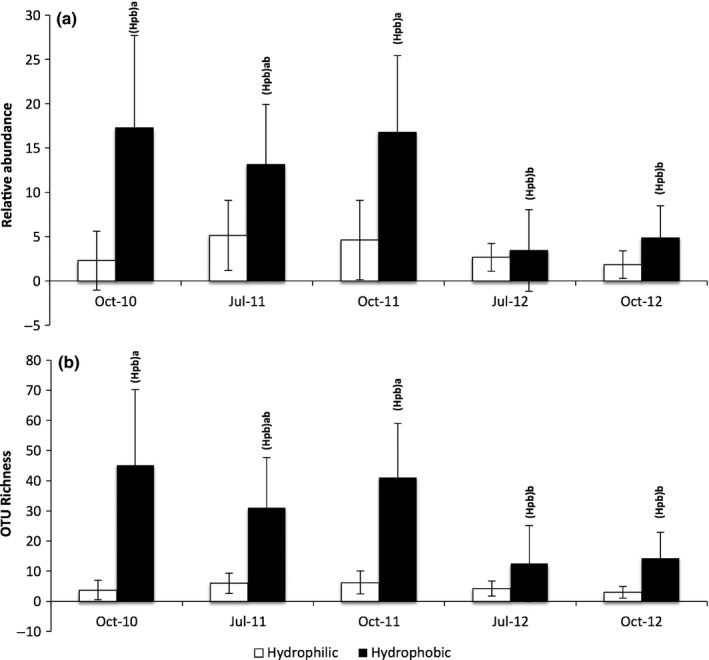
Average (a) relative abundances and (b) operational taxonomic unit (OTU) richness of the ectomycorrhizal (ECM) fungi according to hydrophobicity. ECM fungi were extracted from willow roots over a 3‐yr sampling period. October 2010, July 2011 and October 2011 were pre‐extreme rainfall at the site, whilst July 2012 and October 2012 were post‐extreme rainfall. Error bars represent ± 1 SD of the mean. Capital letters in parentheses represent the taxonomic groups that differ between sampling times and different lower case letters indicate the associated significant differences, by Tukey's test (α = 0.05).
There also were significant differences in the contact type of ECM fungi over the sampling period (Fig. S7). The contact‐medium type ECM fungi were the most abundant, and declined significantly in relative abundance (F = 3.26, P = 0.023) but not in OTU richness (F = 6.49, P = 0.131), from averages of 31.6% and 64.1 OTUs pre‐extreme rainfall to 8.4% and 25.5 OTUs, respectively, post‐extreme rainfall. The contact‐short ECM fungi also declined in relative abundance (F = 9.35, P < 0.001), from an average of 5.1% pre‐extreme rainfall to 2.8% post‐extreme rainfall, whilst OTU richness also declined significantly over this period (F = 6.27, P < 0.001), from averages of 6.4 to 4.6 OTU. The long‐range ECM fungi were present in too low abundance and OTU richness to be meaningfully analysed, with an average richness of just 0.1 OTU and relative abundance of 0.1%.
Investigating the key fungal OTUs within the pre‐ and post‐extreme rainfall root‐associated communities
We further analysed shifts after the extreme rainfall event by performing repeated measure ANOVAs for individual OTUs over the sampling period and visualized this variation using volcano plots (Fig. S8; Newbold et al., 2017). In total, 42 OTUs were shown to vary significantly between time points (q < 0.001), with 41 increasing in abundance and only one decreasing in the post‐extreme rainfall samples (Table S4). Despite the overall significant decline in ECM fungal relative abundance and OTU richness, no single ECM fungal OTU declined significantly post‐extreme rainfall at the q < 0.001 level, although nearly all showed a numerical decline in 2012. Meanwhile four pathogens (two Truncatella angustata OTUs, a Plectosphaerella OTU and a Pilidium OTU) and five saprophytic OTU (four Mortierellaceae OTUs and a Sporormiaceae OTU) increased significantly in abundance. A further four soil yeasts (All Cryptococcus OTUs) and two lichenized fungi (Venturiaceae OTU and Verrucaria andesiatica OTU) also increased in relative abundance, in addition to 26 OTUs with an unknown life strategy. The only OTU to significantly decrease in relative abundance within the post‐extreme rainfall samples could not be assigned to a life strategy.
Finally, GLM was performed on the 42 OTUs that varied significantly between sampling times, with individual OTU abundances analysed against soil pH, P, K, Mg, NO3, SWC and time (weeks) (Table S4). As expected, time correlated with 23 of these 42 OTUs, and accounted for the largest percentage of variation of these OTUs, 20.3% on average. However, SWC correlated with more of these 42 OTUs than time, with a total of 33 OTUs correlating with SWC and an average of 16.7% of variation. Whilst every OTU correlated with either time or SWC, or both, soil pH, P, K and Mg correlated with relatively few OTU abundances, with just two, six, five, four and nine OTUs, respectively.
Discussion
This work demonstrates that a root‐associated fungal community structure was relatively consistent between October 2010 and October 2011, before a dramatic taxonomic and functional transition took place between October 2011 and July 2012, which coincided with an extreme rainfall event that left soils saturated and likely under anoxic soil conditions. As part of this transition in the root‐associated fungal communities, there was a substantial decline in the relative abundance and richness of the ectomycorrhizal (ECM) fungal community, and a concomitant increase in the relative abundance and richness of pathogens and saprophytes. These changes persisted until at least October 2012.
The driver for the community transition between 2011 and 2012 was not directly elucidated. A strong gradient in soil properties existed across transects, which affected composition of the root‐associated fungi (pH) and may have overshadowed any seasonal changes in composition, which have previously been observed in studies of root‐associated fungi (Bohrer et al., 2004; Dumbrell et al., 2011), and which were not detected here. Previous studies have demonstrated extensively that soil nutrients heavily regulate the composition of root‐associated fungal communities (Klamer et al., 2002; van der Gast et al., 2011). The field site underwent coppicing in the spring of 2010, alongside fertilization, compost application and liming, which was the most likely cause of the reduction in phosphorus (P) availability between 2010 and the later time points. However, major soil physico‐chemical properties did not differ significantly in 2012 compared to 2010 and 2011, and although a delayed response to the intense management practices of spring 2010, 2 yr earlier, cannot be ruled out, there were no farm management practices imposed during the sampling period, such as crop harvest, fungicide application or tillage that could have directly or indirectly altered root‐associated fungal biota in such a sudden manner.
However, importantly, climate records show that although temperature was comparable between 2010, 2011 and 2012, there were major differences in rainfall between the 2010/2011 and 2012 growing seasons. Heavy rainfall in early 2012 resulted in peak soil moisture content (SWC) field measurements throughout June and July, and a considerable amount of surface water was present at the field site throughout this period (Fig. S4). Nationally, 2012 was the wettest summer, and second wettest year in the UK since records began in 1910 (Parry et al., 2013), whilst locally it was the highest rainfall year since records began in 1947 (Waddington monitoring station, MetOffice, UK; c. 14 km away from study site). When general linear modelling (GLM) were used to partition variation of the temporally variable operational taxonomic units (OTUs), SWC was the best predictor of OTU abundance, correlating with more OTU than time and all other environmental variables. The taxonomic and functional transition in the root fungal community in 2012 was therefore associated with an extreme weather event, and the extreme rainfall clearly resulted in the prolonged soil saturation we observed during the early summer of 2012.
Soil saturation can result in hypoxic or anoxic soil conditions which can result in changes in soil microbial communities and death of biomass, dependent on the extent and longevity of O2 depletion (Wagner et al., 2015). Relative to the pre‐extreme rainfall samples, in the post‐extreme rainfall samples ECM fungal OTU richness halved and relative abundance declined by nearly two‐thirds, driven primarily by basidiomycete ECM fungi, and the Cortinariaceae in particular. Laboratory studies have shown that ECM fungus hyphal growth and ECM formation are reduced at high soil moisture contents (Theodorou, 1978; Lodge, 1989; Boucher & Malajczuk, 1990) and within experimental plots, extra‐matrical hyphae of ECM fungi have been shown to die under waterlogged conditions (Coutts & Nicoll, 1990). The mycelia produced by fungal species, particularly ECM fungi, range from hydrophobic to hydrophilic. This may be an adaptation to widely contrasting soil moisture availability between habitats and across seasons, with hydrophobicity an adaptation to retain water in dry soil and prevent inundation in wet soil. Fungal species with hydrophilic mycelium may be better able to resist, or show preference for, water saturated soil (Unestam & Sun, 1995). Importantly, we found that the extreme rainfall in 2012 was associated with losses in richness and relative abundance of hydrophobic but not hydrophilic ECM fungi, with losses from both the contact‐short and contact‐medium exploration types, providing further support that the transitions in community structure which we detected could be the result of soil water saturation in 2012.
Reduced ECM fungal abundance under flooding could have negative implications for plant vitality for a number of reasons: (1) Flooding has been linked to increased nutrient availability to plants (Mendoza et al., 2005), and under these conditions, the ECM fungal continuum suggests that ECM fungi could become slightly parasitic to their hosts (Karst et al., 2008). (2) ECM fungi show functional diversity, particularly in soil resource uptake and contributions to host nutrition, and thus major community transitions effecting a wide phylogenetic range of ECM fungi, such as that observed here, could impact upon host nutrient uptake (Leake et al., 2004). (3) Increased diversity and relative abundance of root pathogens were associated with the rainfall event. Mycorrhizal fungi may play a vital role in protecting plants from attack by pathogens, and increased colonization by pathogens could be a consequence of reductions in the diversity and composition of ECM fungi (Nagy & Fossdal, 2013).
Changes in the abundance and composition of ECM fungi of the magnitude observed in the post‐extreme rainfall samples could represent shifts in the pathway by which carbon (C) passes from the plant to the soil, given that ECM fungi are large sinks of C within forest ecosystems (Durall et al., 1994; Högberg et al., 2010). Additionally, the increased diversity and relative abundance of saprophytes could also indicate the increased importance of assimilate reaching soil decomposer pathways following the rainfall event, through increased decomposition of root hairs, roots or other root‐associated fungi in response to flooding (Sauter, 2013; Jaiphong et al., 2016).
It should be noted that the shift in the ECM fungal community persisted for the duration of the growing season of 2012. Clearly the timescales and extent of community recovery will be of fundamental importance in determining the ecosystem‐level implications and legacy of the extreme weather. This study was conducted within an homogenous willow plantation. There is evidence that ECM fungal richness increases with plant richness (Johnson et al., 2005; Tedersoo et al., 2014), and the extent to which the resistance and resilience responses of willow‐associated fungal communities reflects that of more diverse natural ecosystems is unclear.
Extensive research into the biotic and abiotic factors regulating root‐associated fungi has been performed (Tedersoo et al., 2014), yet the weather‐induced shift in taxonomic and functional composition has, to the best of the authors' knowledge, not been observed before within environmental samples. Studies of the effects of disturbances on soil fungal community composition generally have been limited to persistent changes, such as fertilization (Brearley et al., 2007; Gosling et al., 2013) and pollution (van der Gast et al., 2011; Thion et al., 2012). However, precipitation has been linked to changes in AM fungal community composition (Hazard et al., 2013) and sporulation (Lovelock et al., 2003), and ECM fungal richness (Tedersoo et al., 2012), in the field. Studies of the effects of manipulation of soil moisture content on fungal communities are limited to just a handful of studies. Whilst soil fungal communities had considerable resilience to droughts (Evans & Wallenstein, 2012; Barnard et al., 2013; Amend et al., 2016), increasing soil water content has had mixed results, reducing fungal diversity in a study by Hawkes et al. (2011) but having a nominal effect in a study by Fry et al. (2016). These results together with those presented in our study indicate that ECM fungal communities are sensitive to precipitation, with pronounced effects at very high soil water contents.
Extreme weather may therefore be an often overlooked regulator in microbial community development over annual time frames, potentially overshadowing the more frequently observed seasonal community variations (Bell et al., 2009; Dumbrell et al., 2011; Gilbert et al., 2012). With projections suggesting increases in the frequency of extreme weather patterns associated with climate change (Hansen et al., 2012), and extreme weather already known to impact C cycling, leading to losses in terrestrial C stocks (Reichstein et al., 2013), there is a pressing need to understand the impacts of extreme weather events on soil fungal processes and their environmental significance. The decreasing costs of performing large‐scale metagenomic and metatranscriptomic studies should allow the comprehensive long‐term environmental monitoring studies urgently required to understand climate‐rhizosphere feedbacks to be performed.
Author contributions
G.D.B., C.J.v.d.G. and N.P.M. conceived the study; G.B., C.J.B., N.P.M. and C.J.v.d.G. designed the sampling strategy; C.J.B. performed the sampling and analysis; C.J.v.d.G. advised on spatial statistical methods; R.R. and N.P.M. performed soil saturation tests and provided assistance in the field; C.J.B and G.D.B. wrote the manuscript; and all authors contributed to revisions.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Boxplots of nutrient content for each sampling time points pH, (b) P, K, Mg and NO3 kg−1.
Fig. S2 Monthly average temperature, monthly precipitation and monthly soil water content from the willow site over the 3‐yr sampling period.
Fig. S3 Boxplot showing distribution of soil VMC which corresponded with full saturation.
Fig. S4 Evidence of surface water found within July 2012 that was not present in any of the other sampling events.
Fig. S5 Nonmetric dimensional scaling showing clustering based on similarity of the root‐associated fungal communities and pH shown by colour.
Fig. S6 Nonmetric dimensional scaling plots with clustering based on similarity of the root‐associated fungal communities, partitioned into before and after extreme rainfall.
Fig. S7 Average relative abundances and OTU richness of the ECM fungi divided by exploration type, sampled from willow roots over the 3‐yr period.
Fig. S8 Volcano plots of change in relative abundance of each OTU between seasons plotted against ‐log10(q) values generated from repeated measure ANOVAs (using data from all sampling times).
Table S2 Relative importance of season, geographical distance (between samples) and soil properties on the root‐associated fungal communities of willow as revealed by PERMANOVA, with the pre‐shift communities (October 2010, July 2011 and October 2012) and post‐shift communities (July 2012 and October 2012) also analysed again in separate analyses
Table S3 Average relative abundance and OTU richness of ECM families within SRC willow root‐associated fungal communities sampled over the 3‐yr period
Table S1 List of taxonomic assignments and references, in addition to P‐values (and subsequent q‐values) produced via repeated measure ANOVAs for each individual OTU
Table S4 Table of OTUs which significantly varied in relative abundance over time (as found with repeated measure ANOVAs), whilst GLM models were also performed for each of these OTU to correlate their abundance against time and environmental properties
Acknowledgements
This project was part of the Carbo‐Biocrop project funded by the Natural Environment Research Council (NERC). We acknowledge NERC Centre for Ecology and Hydrology National Capability funding through project NEC03487, Ross Morrison for providing meterological data and the landowner, Jonathan Wright for access to the commercial plantation.
See also the Commentary on this article by https://doi.org/10.1111/nph.15086.
References
- Agerer R. 2001. Exploration types of ectomycorrhizae. Mycorrhiza 11: 107–114. [Google Scholar]
- Amend AS, Martiny AC, Allison SD, Berlemont R, Goulden ML, Lu Y, Treseder KK, Weihe C, Martiny JBH. 2016. Microbial response to simulated global change is phylogenetically conserved and linked with functional potential. ISME Journal 10: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar J, Bastiaans T, van de Coevering M, Roelofs J. 2002. Ectomycorrhizal root development in wet Alder carr forests in response to desiccation and eutrophication. Mycorrhiza 12: 147–151. [DOI] [PubMed] [Google Scholar]
- Bardgett RD, Freeman C, Ostle NJ. 2008. Microbial contributions to climate change through carbon cycle feedbacks. ISME Journal 2: 805–814. [DOI] [PubMed] [Google Scholar]
- Bardgett RD, van der Putten WH. 2014. Belowground biodiversity and ecosystem functioning. Nature 515: 505–511. [DOI] [PubMed] [Google Scholar]
- Barnard RL, Osborne CA, Firestone MK. 2013. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME Journal 7: 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CJ, Burns CA, van der Gast CJ, McNamara NP, Bending GD. 2016a. Spatio‐temporal variation of core and satellite arbuscular mycorrhizal fungus communities in Miscanthus giganteus . Microbial Symbioses 7: 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CJ, van der Gast CJ, Burns CA, McNamara NP, Bending GD. 2016b. Temporally variable geographical distance effects contribute to the assembly of root‐associated fungal communities. Microbial Symbioses 7: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CW, Acosta‐Martinez V, McIntyre NE, Cox S, Tissue DT, Zak JC. 2009. Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a chihuahuan desert grassland. Microbial Ecology 58: 827–842. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B (Methodological) 57: 289–300. [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends in Plant Science 17: 478–486. [DOI] [PubMed] [Google Scholar]
- Bohrer KE, Friese CF, Amon JP. 2004. Seasonal dynamics of arbuscular mycorrhizal fungi in differing wetland habitats. Mycorrhiza 14: 329–337. [DOI] [PubMed] [Google Scholar]
- Bonkowski M. 2004. Protozoa and plant growth: the microbial loop in soil revisited. New Phytologist 162: 617–631. [DOI] [PubMed] [Google Scholar]
- Boucher NL, Malajczuk N. 1990. Effects of high soil moisture on formation of ectomycorrhizas and growth of karri (Eucalyptus diversicolor) seedlings inoculated with Descolea maculata, Pisolithus tinctorius and Laccaria laccata . New Phytologist 114: 87–91. [DOI] [PubMed] [Google Scholar]
- Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecological Monographs 27: 325–349. [Google Scholar]
- Brearley FQ, Scholes JD, Press MC, Palfner G. 2007. How does light and phosphorus fertilisation affect the growth and ectomycorrhizal community of two contrasting dipterocarp species? Plant Ecology 192: 237–249. [Google Scholar]
- Cai W, Borlace S, Lengaigne M, van Rensch P, Collins M, Vecchi G, Timmermann A, Santoso A, McPhaden MJ, Wu L et al 2014. Increasing frequency of extreme El Nino events due to greenhouse warming. Nature Climate Change 4: 111–116. [Google Scholar]
- Camy C, Dreyer E, Delatour C, Marçais B. 2003. Responses of the root rot fungus Collybia fusipes to soil waterlogging and oxygen availability. Mycological Research 107: 1103–1109. [DOI] [PubMed] [Google Scholar]
- Chakravarty P, Unestam T. 1987. Differential influence of ectomycorrhizae on plant growth and disease resistance in Pinus sylvestris seedlings. Journal of Phytopathology 120: 104–120. [Google Scholar]
- Chao A. 1987. Estimating the population size for capture‐recapture data with unequal catchability. Biometrics 43: 783–791. [PubMed] [Google Scholar]
- Coughlan AP, Dalpé Y, Lapointe L, Piché Y. 2000. Soil pH‐induced changes in root colonization, diversity, and reproduction of symbiotic arbuscular mycorrhizal fungi from healthy and declining maple forests. Canadian Journal of Forest Research 30: 1543–1554. [Google Scholar]
- Coutts MP, Nicoll BC. 1990. Waterlogging tolerance of roots of Sitka spruce clones and of strands from Thelephoraterrestris mycorrhizas. Canadian Journal of Forest Research 20: 1894–1899. [Google Scholar]
- Daniell TJ, Husband R, Fitter AH, Young JPW. 2001. Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microbiology Ecology 36: 203–209. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JaL, Pretorius ZA, Hammond‐Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J et al 2012. The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbrell AJ, Ashton PD, Aziz N, Feng G, Nelson M, Dytham C, Fitter AH, Helgason T. 2011. Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytologist 190: 794–804. [DOI] [PubMed] [Google Scholar]
- Durall DM, Jones MD, Tinker PB. 1994. Allocation of 14C‐carbon in ectomycorrhizal willow. New Phytologist 128: 109–114. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SE, Wallenstein MD. 2012. Soil microbial community response to drying and rewetting stress: does historical precipitation regime matter? Biogeochemistry 109: 101–116. [Google Scholar]
- Fernandez CW, Nguyen NH, Stefanski A, Han Y, Hobbie SE, Montgomery RA, Reich PB, Kennedy PG. 2017. Ectomycorrhizal fungal response to warming is linked to poor host performance at the boreal‐temperate ecotone. Global Change Biology 23: 1598–1609. [DOI] [PubMed] [Google Scholar]
- Finzi AC, Abramoff RZ, Spiller KS, Brzostek ER, Darby BA, Kramer MA, Phillips RP. 2015. Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Global Change Biology 21: 2082–2094. [DOI] [PubMed] [Google Scholar]
- Fougnies L, Renciot S, Muller F, Plenchette C, Prin Y, de Faria SM, Bouvet JM, Sylla SN, Dreyfus B, Bâ AM. 2007. Arbuscular mycorrhizal colonization and nodulation improve flooding tolerance in Pterocarpus officinalis Jacq. seedlings. Mycorrhiza 17: 159–166. [DOI] [PubMed] [Google Scholar]
- Fry EL, Manning P, Macdonald C, Hasegawa S, De Palma A, Power SA, Singh BK. 2016. Shifts in microbial communities do not explain the response of grassland ecosystem function to plant functional composition and rainfall change. Soil Biology and Biochemistry 92: 199–210. [Google Scholar]
- Garcia K, Delteil A, Conéjéro G, Becquer A, Plassard C, Sentenac H, Zimmermann S. 2014. Potassium nutrition of ectomycorrhizal Pinus pinaster: overexpression of the Hebeloma cylindrosporum HcTrk1 transporter affects the translocation of both K+ and phosphorus in the host plant. New Phytologist 201: 951–960. [DOI] [PubMed] [Google Scholar]
- García I, Mendoza R, Pomar MC. 2008. Deficit and excess of soil water impact on plant growth of Lotus tenuis by affecting nutrient uptake and arbuscular mycorrhizal symbiosis. Plant and Soil 304: 117–131. [Google Scholar]
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- van der Gast CJ, Gosling P, Tiwari B, Bending GD. 2011. Spatial scaling of arbuscular mycorrhizal fungal diversity is affected by farming practice. Environmental Microbiology 13: 241–249. [DOI] [PubMed] [Google Scholar]
- Geisseler D, Scow KM. 2014. Long‐term effects of mineral fertilizers on soil microorganisms – a review. Soil Biology and Biochemistry 75: 54–63. [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I et al 2012. Defining seasonal marine microbial community dynamics. ISME Journal 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosling P, Mead A, Proctor M, Hammond JP, Bending GD. 2013. Contrasting arbuscular mycorrhizal communities colonizing different host plants show a similar response to a soil phosphorus concentration gradient. New Phytologist 198: 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafen A, Hails R. 2002. Modern statistics for the life sciences. Oxford, UK: Oxford University Press. [Google Scholar]
- Hansen J, Sato M, Ruedy R. 2012. Perception of climate change. Proceedings of the National Academy of Sciences, USA 109: E2415–E2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Rothballer M, Schmid M. 2008. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant and Soil 312: 7–14. [Google Scholar]
- Hawkes CV, Kivlin SN, Rocca JD, Huguet V, Thomsen MA, Suttle KB. 2011. Fungal community responses to precipitation. Global Change Biology 17: 1637–1645. [Google Scholar]
- Hazard C, Gosling P, van der Gast CJ, Mitchell DT, Doohan FM, Bending GD. 2013. The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. ISME Journal 7: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann M, Reichstein M. 2008. Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 451: 289–292. [DOI] [PubMed] [Google Scholar]
- Högberg MN, Briones MJI, Keel SG, Metcalfe DB, Campbell C, Midwood AJ, Thornton B, Hurry V, Linder S, Näsholm T et al 2010. Quantification of effects of season and nitrogen supply on tree below‐ground carbon transfer to ectomycorrhizal fungi and other soil organisms in a boreal pine forest. New Phytologist 187: 485–493. [DOI] [PubMed] [Google Scholar]
- Husband R, Herre EA, Turner SL, Gallery R, Young JPW. 2002. Molecular diversity of arbuscular mycorrhizal fungi and patterns of host association over time and space in a tropical forest. Molecular Ecology 11: 2669–2678. [DOI] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology 8: R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiphong T, Tominaga J, Watanabe K, Nakabaru M, Takaragawa H, Suwa R, Ueno M, Kawamitsu Y. 2016. Effects of duration and combination of drought and flood conditions on leaf photosynthesis, growth and sugar content in sugarcane. Plant Production Science 19: 427–437. [Google Scholar]
- Johnson D, Ijdo M, Genney DR, Anderson IC, Alexander IJ. 2005. How do plants regulate the function, community structure, and diversity of mycorrhizal fungi? Journal of Experimental Botany 56: 1751–1760. [DOI] [PubMed] [Google Scholar]
- Karst J, Marczak L, Jones MD, Turkington R. 2008. The mutualism–parasitism continuum in ectomycorrhizas: a quantitative assessment using meta‐analysis. Ecology 89: 1032–1042. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A et al 2011. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882. [DOI] [PubMed] [Google Scholar]
- Klamer M, Roberts MS, Levine LH, Drake BG, Garland JL. 2002. Influence of elevated CO2 on the fungal community in a coastal scrub oak forest soil investigated with terminal‐restriction fragment length polymorphism analysis. Applied and Environmental Microbiology 68: 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubartová A, Ranger J, Berthelin J, Beguiristain T. 2008. Diversity and decomposing ability of saprophytic fungi from temperate forest litter. Microbial Ecology 58: 98–107. [DOI] [PubMed] [Google Scholar]
- Laird DA, Fleming P, Davis DD, Horton R, Wang B, Karlen DL. 2010. Impact of biochar amendments on the quality of a typical Midwestern agricultural soil. Geoderma 158: 443–449. [Google Scholar]
- Last FT, Mason PA, Ingleby K, Fleming LV. 1984. Succession of fruitbodies of sheathing mycorrhizal fungi associated with Betula pendula . Forest Ecology and Management 9: 229–234. [Google Scholar]
- Leake J, Johnson D, Donnelly D, Muckle G, Boddy L, Read D. 2004. Networks of power and influence: the role of mycorrhizal mycelium in controlling plant communities and agroecosystem functioning. Canadian Journal of Botany 82: 1016–1045. [Google Scholar]
- Lilleskov EA, Bruns TD, Horton TR, Taylor D, Grogan P. 2004. Detection of forest stand‐level spatial structure in ectomycorrhizal fungal communities. FEMS Microbiology Ecology 49: 319–332. [DOI] [PubMed] [Google Scholar]
- Lilleskov EA, Hobbie EA, Horton TR. 2011. Conservation of ectomycorrhizal fungi: exploring the linkages between functional and taxonomic responses to anthropogenic N deposition. Fungal Ecology 2: 174–183. [Google Scholar]
- Liu J, Maldonado‐Mendoza I, Lopez‐Meyer M, Cheung F, Town CD, Harrison MJ. 2007. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant Journal 50: 529–544. [DOI] [PubMed] [Google Scholar]
- Lodge DJ. 1989. The influence of soil moisture and flooding on formation of VA‐endo‐and ectomycorrhizae in Populus and Salix . Plant and Soil 117: 243–253. [Google Scholar]
- Lovelock CE, Andersen K, Morton JB. 2003. Arbuscular mycorrhizal communities in tropical forests are affected by host tree species and environment. Oecologia 135: 268–279. [DOI] [PubMed] [Google Scholar]
- Mallakpour I, Villarini G. 2015. The changing nature of flooding across the central United States. Nature Climate Change 5: 250–254. [Google Scholar]
- McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Computational Biology 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza R, Escudero V, García I. 2005. Plant growth, nutrient acquisition and mycorrhizal symbioses of a waterlogging tolerant legume (Lotus glaber Mill.) in a saline‐sodic soil. Plant and Soil 275: 305–315. [Google Scholar]
- Nagy NE, Fossdal CG. 2013. Host responses in Norway spruce roots induced to the pathogen Ceratocystis polonica are evaded or suppressed by the ectomycorrhizal fungus Laccaria bicolor . Plant Biology 15: 99–110. [DOI] [PubMed] [Google Scholar]
- Newbold LK, Burthe SJ, Oliver AE, Gweon HS, Barnes CJ, Daunt F, van der Gast CJ. 2017. Helminth burden and ecological factors associated with alterations in wild host gastrointestinal microbiota. ISME Journal 11: 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG. 2016. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology 20: 241–248. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2007. The vegan package. [WWW document] URL http://cran.r-project.org/.
- Parry S, Marsh T, Kendon M. 2013. 2012: from drought to floods in England and Wales. Weather 68: 268–274. [Google Scholar]
- Phillips CJ, Marden M, Suzanne LM. 2014. Observations of root growth of young poplar and willow planting types. New Zealand Journal of Forestry Science 44: 15. [Google Scholar]
- Reichstein M, Bahn M, Ciais P, Frank D, Mahecha MD, Seneviratne SI, Zscheischler J, Beer C, Buchmann N, Frank DC et al 2013. Climate extremes and the carbon cycle. Nature 500: 287–295. [DOI] [PubMed] [Google Scholar]
- Rivest D, Lorente M, Olivier A, Messier C. 2013. Soil biochemical properties and microbial resilience in agroforestry systems: effects on wheat growth under controlled drought and flooding conditions. Science of The Total Environment 463–464: 51–60. [DOI] [PubMed] [Google Scholar]
- Sauter M. 2013. Root responses to flooding. Current Opinion in Plant Biology 16: 282–286. [DOI] [PubMed] [Google Scholar]
- Shepherd TG. 2015. Climate science: the dynamics of temperature extremes. Nature 522: 425–427. [DOI] [PubMed] [Google Scholar]
- Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant‐dependent enrichment and seasonal shifts revealed. Applied and Environmental Microbiology 67: 4742–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit E, Leeflang P, Glandorf B, van Elsas JD, Wernars K. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR‐amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Applied and Environmental Microbiology 65: 2614–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read DJ. 2010. Mycorrhizal symbiosis. London, UK: Academic Press. [Google Scholar]
- Swaty RL, Gehring CA, Van Ert M, Theimer TC, Keim P, Whitham TG. 1998. Temporal variation in temperature and rainfall differentially affects ectomycorrhizal colonization at two contrasting sites. New Phytologist 139: 733–739. [Google Scholar]
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco‐Palacios AM, Thu PQ, Suija A et al 2014. Global diversity and geography of soil fungi. Science 346: 1256688. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Toots M, Diédhiou AG, Henkel TW, Kjøller R, Morris MH, Nara K, Nouhra E, Peay KG et al 2012. Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Molecular Ecology 21: 4160–4170. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Suvi T, Jairus T, Ostonen I, Põlme S. 2009. Revisiting ectomycorrhizal fungi of the genus Alnus: differential host specificity, diversity and determinants of the fungal community. New Phytologist 182: 727–735. [DOI] [PubMed] [Google Scholar]
- Theodorou C. 1978. Soil moisture and the mycorrhizal association of Pinus radiata D. don. Soil Biology and Biochemistry 10: 33–37. [Google Scholar]
- Thion C, Cébron A, Beguiristain T, Leyval C. 2012. Long‐term in situ dynamics of the fungal communities in a multi‐contaminated soil are mainly driven by plants. FEMS Microbiology Ecology 82: 169–181. [DOI] [PubMed] [Google Scholar]
- Unestam T, Sun Y‐P. 1995. Extramatrical structures of hydrophobic and hydrophilic ectomycorrhizal fungi. Mycorrhiza 5: 301–311. [Google Scholar]
- Unger IM, Kennedy AC, Muzika R‐M. 2009. Flooding effects on soil microbial communities. Applied Soil Ecology 42: 1–8. [Google Scholar]
- Visser S. 1995. Ectomycorrhizal fungal succession in jack pine stands following wildfire. New Phytologist 129: 389–401. [Google Scholar]
- Wagner D, Eisenhauer N, Cesarz S. 2015. Plant species richness does not attenuate responses of soil microbial and nematode communities to a flood event. Soil Biology and Biochemistry 89: 135–149. [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York, NY, USA: Springer. [Google Scholar]
- Wilson JS, Baldwin DS, Rees GN, Wilson BP. 2011. The effects of short‐term inundation on carbon dynamics, microbial community structure and microbial activity in floodplain soil. River Research and Applications 27: 213–225. [Google Scholar]
- Yang H, Koide RT, Zhang Q. 2016. Short‐term waterlogging increases arbuscular mycorrhizal fungal species richness and shifts community composition. Plant and Soil 404: 373–384. [Google Scholar]
- Yang H, Zang Y, Yuan Y, Tang J, Chen X. 2012. Selectivity by host plants affects the distribution of arbuscular mycorrhizal fungi: evidence from ITS rDNA sequence metadata. BMC Evolutionary Biology 12: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Boxplots of nutrient content for each sampling time points pH, (b) P, K, Mg and NO3 kg−1.
Fig. S2 Monthly average temperature, monthly precipitation and monthly soil water content from the willow site over the 3‐yr sampling period.
Fig. S3 Boxplot showing distribution of soil VMC which corresponded with full saturation.
Fig. S4 Evidence of surface water found within July 2012 that was not present in any of the other sampling events.
Fig. S5 Nonmetric dimensional scaling showing clustering based on similarity of the root‐associated fungal communities and pH shown by colour.
Fig. S6 Nonmetric dimensional scaling plots with clustering based on similarity of the root‐associated fungal communities, partitioned into before and after extreme rainfall.
Fig. S7 Average relative abundances and OTU richness of the ECM fungi divided by exploration type, sampled from willow roots over the 3‐yr period.
Fig. S8 Volcano plots of change in relative abundance of each OTU between seasons plotted against ‐log10(q) values generated from repeated measure ANOVAs (using data from all sampling times).
Table S2 Relative importance of season, geographical distance (between samples) and soil properties on the root‐associated fungal communities of willow as revealed by PERMANOVA, with the pre‐shift communities (October 2010, July 2011 and October 2012) and post‐shift communities (July 2012 and October 2012) also analysed again in separate analyses
Table S3 Average relative abundance and OTU richness of ECM families within SRC willow root‐associated fungal communities sampled over the 3‐yr period
Table S1 List of taxonomic assignments and references, in addition to P‐values (and subsequent q‐values) produced via repeated measure ANOVAs for each individual OTU
Table S4 Table of OTUs which significantly varied in relative abundance over time (as found with repeated measure ANOVAs), whilst GLM models were also performed for each of these OTU to correlate their abundance against time and environmental properties


