Abstract
Background: While data regarding expression of limb element and tissue markers during normal mouse limb development exist, few studies show expression patterns in upper and lower limbs throughout key limb development stages. A comparison to normal developmental events is essential when analyzing development of the limb in mutant mice models. Results: Expression patterns of the joint marker Gdf5, tendon and ligament marker Scleraxis, early muscle marker MyoD1, and blood vessel marker Cadherin5 (Cdh5) are presented during the most active phases of embryonic mouse limb patterning. Anti‐neurofilament staining of developing nerves in the fore‐ and hindlimbs and cartilage formation and progression also are described. Conclusions: This study demonstrates and describes a range of key morphological markers and methods that together can be used to assess normal and abnormal limb development. Developmental Dynamics 247:1217–1226, 2018. © 2018 The Authors. Developmental Dynamics published by Wiley Periodicals, Inc. on behalf of American Association of Anatomists
Keywords: Gene expression, limb development, forelimb, hindlimb, vascular, cartilage, nerve, muscle, tendon, joint, Alcian Blue/Alizarin Red
Key Findings
Expression patterns of molecular markers throughout both fore‐ and hindlimb development ‐ which can be used to assess normal and abnormal development.
Detailled description of innervation during fore‐ and hindlimb development confirming innervation first seen after limb patterning events have begun.
Description of cartilage development and progression indicates alizarin red staining not seen until E15.5 in both fore‐ and hindlimbs.
Hindlimb lags behind forelimb molecularly and morphologically until E14.5.
Detailled description of methods used to study fore‐ and hindlimb development.
Introduction
Although significant progress has been made in understanding the molecular and morphological mechanisms controlling limb development, just how neuronal innervation, cartilage formation and progression, vascular patterning, tendon development, and joint development are so intricately controlled and regulated is still unclear. Analysis of genetic mouse mutants is frequently used to identify factors essential for normal limb development and their mechanism of action, and requires a comparison to be made to normal development of limb elements.
C57BL/6 mouse embryos develop in 18.5 days, and the forelimb bud is apparent from embryonic day (E) 9.5. The hindlimb consistently lags about a half‐day behind forelimb development (Martin 1990). The limb develops in a proximodistal manner, with the proximal stylopod developing before the zeugopod and distal autopod (Muneoka et al., 1989; Martin 1990). Muscle precursors migrate in to the forelimb around E10.5, and expression of myogenic factors is detected in the limb at E11, with clear muscle blocks seen at E12.5 (Sassoon et al., 1989; Martin 1990). Nerve fascicles, originating from the brachial plexus and the spinal lumbosacral plexus, are present in the proximal mouse forelimb and hindlimb, respectively, at E11.5 (Martin 1990). Tendon progenitor cells in the limb condense and differentiate into tenocytes, with pre‐tendon mesenchymal masses detected at E13.5 (Perez et al., 2003; Birch et al., 2013). Tenocytes then produce extracellular matrix molecules, including type I and III collagen, and organize them into tendon fibrils (Birch et al., 2013; Liu et al., 2015). Fibrils become further organized into tendon fibers, and by E14.5 tendons are visible in the digits (Perez et al., 2003). By E14.5, distal muscles are also visible, the nerves have formed dense plexuses, and the epidermis is thickening. Skeletal patterning of the limbs begins with the formation of cartilage condensations, from around E11.5 (Martin, 1990), which are the precursors of bones of the limbs (Martin, 1990; Nowlan et al., 2010). These elements form in a proximal to distal manner: For example, the humerus/femur forms before the radius and ulna/tibia and fibula, which form before the digits (Martin, 1990; Vargesson and Hootnick, 2017). Joint development begins with the formation of the interzone, a region of flat, condensed mesenchymal cells. The formation of the interzone is followed by joint cavitation. Cavitation of mouse proximal limb joints is visible at E15.5 (Mitrovic 1977; Wang et al., 2001). Limb development is typically complete around E18.5 (Martin 1990).
We have performed a detailed spatiotemporal analysis of markers of joint (Gdf5), tendon (Scleraxis), muscle (MyoD1), vascular (Cdh5), neuronal (anti‐neurofilament), and cartilage (Alcian Blue/Alizarin Red staining) development. Although aspects of the expression patterns of these genes and tissue markers are published, single studies showing expression patterns of all of these markers in both upper and lower limbs are lacking, and there is limited analysis of the temporal changes in expression throughout limb development. Moreover, for several of these markers, distinct aspects of the expression patterns are reported in multiple publications, making determination of the spatiotemporal changes in expression difficult to ascertain.
Distinct aspects of Gdf5 expression have been reported in several publications that combined have shown Gdf5 expression between E11.5 and E14.5 in the presumptive sites of joint development and the forming joint interzone (Chen et al., 2016; Degenkolbe et al., 2013; Hellman et al., 2012; Houweling et al., 2001; Huang et al., 2016; Meech et al., 2005; Ota et al., 2007; Perez et al., 2010; Seemann et al., 2005; Sohaskey et al., 2008). Uniquely, we have shown side‐by‐side Gdf5 expression across a range of key stages of development in both the forelimb and hindlimb. MyoD1 expression, a myogenic helix‐loop‐helix protein family restricted to differentiating skeletal muscle (reviewed in Tapscott, 2005), has been examined in the context of comparison to mutant models but infrequently as a marker of muscle development alone (Grifone et al., 2005; Kablar et al., 1997; Laclef et al., 2003; L'honoré et al., 2010; Mayeuf‐Louchart et al., 2016), highlighting the requirement for a summative analysis of expression during mouse limb development. Scleraxis, a basic helix‐loop‐helix factor expressed in connective tissues that form muscle attachments (Schweitzer et al., 2001), is reexamined here alongside other limb element development markers to allow a comparison to the development of the other tissues (Murchison et al., 2007; Schweitzer et al., 2001). Cdh5 expression, an endothelial‐specific cadherin expressed during early vasculogenesis with expression maintained throughout adult life (Neuhaus et al., 2014), has not, as far as we are aware, been studied throughout mouse limb development, and indeed whole‐mount in situ analysis of blood vessel markers is rare, with the majority of studies describing expression at one or two time points as a comparison to mutant models (Eshkar‐Oren et al., 2009; Eshkar‐Oren et al., 2015; Ota et al., 2007; Vieira et al., 2007). Nerve patterns are usually assessed at specific time points to see the effect of a mutation or morphological interaction. Here, we show nerve innervation patterns throughout limb outgrowth. Similarly, cartilage and bone analysis is usually assessed in mouse limb studies at specific time points to study specific gene function and action (for example, Amarilio et al., 2007; Collinson et al., 2018; Firulli et al., 2005; Nowlan et al., 2010; Ray et al., 2015; Tavella et al., 2004). Here, we show in detail the progression of cartilage and bone development throughout fore‐ and hindlimb patterning. Taken together, our description of the spatiotemporal expression of these markers can be used as a toolbox to assess the formation and development of key tissues in normal and abnormal limb development.
Results
We have analyzed the dynamic expression in mouse fore‐ and hindlimbs of Gdf5, MyoD1, and Cdh5 from E13.5 to E15.5; Scleraxis from E12.5 to E15.5; anti‐neurofilament from E10.5 to E14.5; and cartilage/bone development and progression from E11.5 to E16.5. Although mouse limb development begins around E9.0, the limb bud at this stage consists of a mass of undifferentiated cells, and the main limb structures—including tendons, skeleton, muscles, and vessels—are organized between E12.5 and E15.5 (Bénazet and Zeller 2009; Marcon et al., 2011; Martin, 1990; Tickle 2006).
Expression Pattern of the Joint Marker Gdf5
Cartilage elements develop in a proximodistal fashion, with the humerus/femur forming before the digits. Digits first develop as “rays” (digit and metacarpal/metatarsal as a continuous unit with webbing between the digits proper), and at E13.5, Gdf5 was expressed along rays in presumptive joint regions that develop to separate rays into proximal, intermediate, and distal phalanges (Fig. 1A yellow arrowhead, Fig. 1A–B`). By E14.5, there was no longer continuous Gdf5 expression along cartilage condensations, and thick bands of Gdf5 expression were visible in carpometacarpal/tarsometatarsal joints, metacarpophalangeal/metatarsophalangeal joints, and the developing distal interphalangeal joint in joint interzones (Fig. 1C white arrowhead, Fig. 1C`–E). At E15.5, joint patterning became more apparent and Gdf5 was expressed as thinner bands in the more defined carpometacarpal/tarsometatarsal joints, metacarpophalangeal/metatarsophalangeal joints, and proximal and distal interphalangeal joints. (Fig. 1F–J`). In sections through the joints, strong expression of Gdf5 was detected in joint interzones (Fig. 1E black arrow, Fig. 1H).
Figure 1.
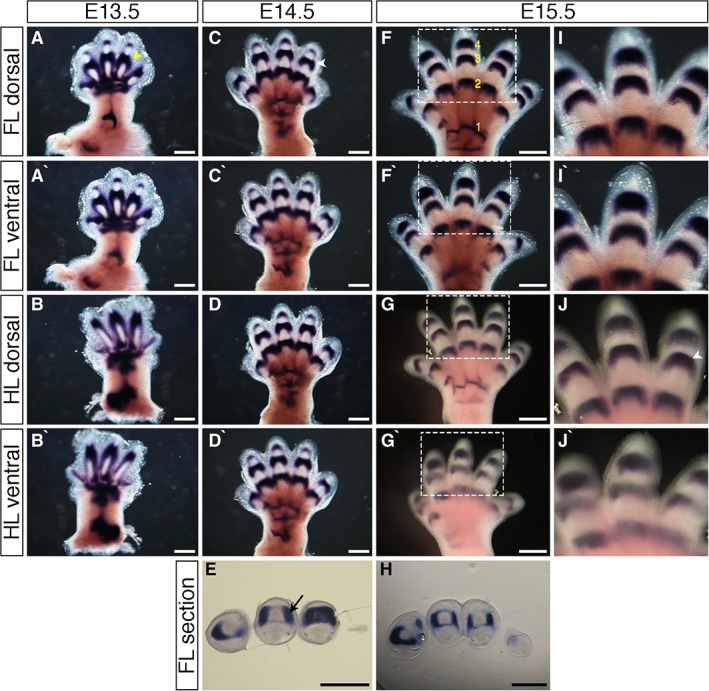
Expression patterns of Gdf5 during mouse limb development. A–D`,F–G`,I–J`: Whole‐mount in situ hybridization for Gdf5 in E13.5 (A–B`), E14.5 (C–D`), and E15.5 (F–G`) dorsal forelimbs (A,C,F), ventral forelimbs (A`,C`,F`), dorsal hindlimbs (B,D,G), and ventral hindlimbs (B`,D`,G`). I`,J`: Higher‐magnification images of regions highlighted in white dashed boxes in F–G`, respectively. E,H: Sections through distal interphalangeal joints of E14.5 (E) and E15.5 (H) forelimbs stained by in situ hybridization for Gdf5; dorsal at top, anterior to right. Gdf5 is expressed in prechondrogenic condensations at E13.5 (A yellow arrowhead), and in interphalangeal, metacarpophalangeal, and carpometacarpal joints at E14.5 and E15.5 (C,J white arrowhead), and in joint interzones (E black arrow). Developing joints are numbered (F yellow numbers) 1: carpometacarpal joint; 2: metacarpophalangeal joint; 3: proximal interphalangeal joint; 4: distal interphalangeal joint. Scale bars = 500 µm.
Expression Pattern of the Tendon and Ligament Marker Scleraxis
Expression patterns of Scleraxis were determined between E12.5 and E15.5, since the vast majority of connective tissue patterning in the limbs occurs between these stages. At E12.5, Scleraxis was expressed broadly throughout the early autopod (hand‐/footplate) (Fig. 2A white bracket), with weak expression in the primitive digits (Fig. 2A–B`). At E13.5, Scleraxis was expressed in the hand‐ and footplate (Fig. 2C white bracket) and in the digit rays, with thicker regions of expression at presumptive joint sites (compare to Fig. 1A–B`). Scleraxis expression domains were wider at presumptive joint sites of the forelimb compared to the hindlimb (Fig. 2C–D`), comparable to more defined Gdf5 expression at presumptive joint regions of the forelimb (Fig. 1A,A`). At E14.5, distinct bands of Scleraxis expression were visible through the autopod and extending to the digit tips. Expression was strongest at regions of joint development (Fig. 2F yellow arrow), where attachment sites between connective tissue and the skeleton form (Fig. 2E–F`). In E15.5 embryos, Scleraxis was expressed in distinct bands that extended along the digits, with strong areas of expression persisting around developing joint regions (Fig. 2H–I`,K–L`). Expression of Scleraxis corresponding to flexor tendons could be seen at the ventral side of section images (Fig. 2G,J, blue lines).
Figure 2.
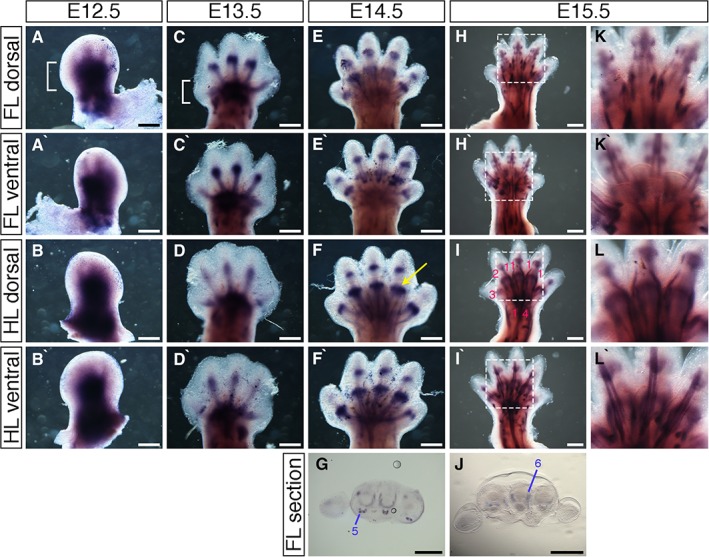
Scleraxis expression pattern during mouse limb development. A–F`,H–I`,K–L`: Whole‐mount in situ hybridization for Scleraxis at E12.5 (A–B`), E13.5 (C–D`), E14.5 (E–F`), and E15.5 (H–I`) in dorsal forelimbs (A,C,E,H), ventral forelimbs (A`,C`,E`,H`), dorsal hindlimbs (B,D,F,I), and ventral hindlimbs (B`,D`,F`,I`). K–L`: Higher magnification of regions highlighted in white dashed boxes in H–I`, respectively. G,J: Section images through proximal metacarpal joints at E14.5 (G) and E15.5 (J) of forelimbs stained by in situ hybridization for Scleraxis; dorsal at top, anterior to left. Blue lines indicate position of flexor tendons. Wrist tendon progenitors (A white bracket) at E12.5 give rise to zeugopod tendons (C white bracket). Phalangeal insert sites for connective tissues can be distinguished (F yellow arrow). Tendons are numbered (I pink numbers; G–J blue numbers) 1: extensor digitorium communis, 2: extensor carpi radialis longus, 3: extensor pollicis, 4: extensor carpi ulnaris, 5: flexor digitorum profundus, 6: lumbrical tendon. Scale bars = 500 µm.
Expression Pattern of the Muscle Marker MyoD1
At E13.5, whole‐mount in situ hybridization analysis of MyoD1 revealed developing muscle patterning in the fore‐ and hindlimb stylopod and zeugopod, with stronger staining in regions of developing muscle blocks (Fig. 3B yellow arrow). Faint expression was visible in the hand‐ and footplate (Fig. 3A–B`). At E14.5, MyoD1 showed strong expression in the proximal zeugopod (forearm/leg), with stronger expression where bands of muscle are developing (Fig. 3 red arrow, Fig. 3C`), with stronger expression in the forelimb zeugopod compared to the hindlimb zeugopod. MyoD1 was also expressed in the hand‐ and footplate, showing muscle bands from the center radiating to digit bases. A thin band of expression through the middle of the wrist connected expression regions of the zeugopod to the autopod in the forelimb, but no such expression band from the zeugopod to the autopod is seen in the hindlimb (Fig. 3C–D`). Expression at E15.5 was similar to that at E14.5 in the zeugopod and autopod. The band of expression connecting the two regions was stronger in the forelimb in E15.5 compared to E14.5 (Fig. 3E blue arrow, Fig.E`–H`). MyoD1 expression in the limbs, when viewed at higher magnification, demonstrates the definition and organization of fibers into defined muscle sets by E15.5 (Fig. 3G–H`).
Figure 3.
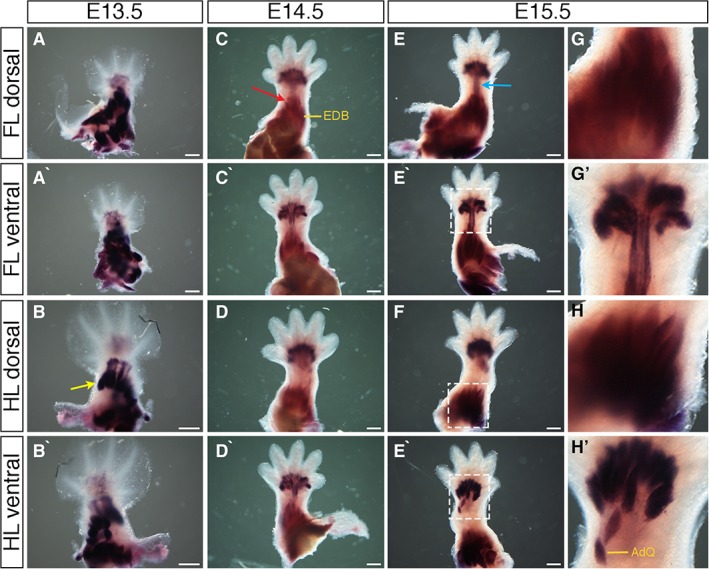
Expression of MyoD1 during mouse limb development. A–H`: Whole‐mount in situ hybridization for MyoD1 at E13.5 (A–B`), E14.5 (C–D`), and E15.5 (E–F`) in dorsal forelimbs (A,C,E), ventral forelimbs (A`,C`,E`), dorsal hindlimbs (B,D,F), and ventral hindlimbs (B`,D`,F`). G–H`: Higher magnification of regions highlighted in white dashed boxes in E–F`, respectively. Expression is seen in developing muscle blocks (B yellow arrow; C red arrow) and in a band connecting zeugopod and autopod muscle blocks in the forelimb of E15.5 embryos (E blue arrow). Some distinct muscles are highlighted (C, EDB: extensor digitorum brevis; H`, AdQ: abductor quinti muscle). Scale bars = 500 µm.
Expression Pattern of the Endothelial Cadherin Cdh5
Cdh5 expression was visible through the zeugopod and in the hand‐ and footplate at E13.5 and along the digit rays, but little expression was apparent interdigitally (Fig. 4A–B`). Stronger regions of expression existed in between digits at the digit base (Fig. 4A` white arrow). Larger vessels and more complex networks developed from these relatively simple early networks by E14.5. Thick bands of expression radiated from the hand‐ and footplate and along the digit borders. Expression was also visible at presumptive joint sites along the digits (Fig. 4C–D, Fig. 4D` blue arrow). At E15.5, strong bands of Cdh5 expression existed along the wrist/ankle, into the hand‐ and footplate, and along digit borders. Bands of expression along the presumptive joint sites were no longer present (Fig. 4E–F`), confirmed when viewing the limbs at higher magnification (Fig. 4G–H`). At all ages, no expression was observed in the distal digit tips (Fig. 4E–F, Fig. 4F` red arrow).
Figure 4.
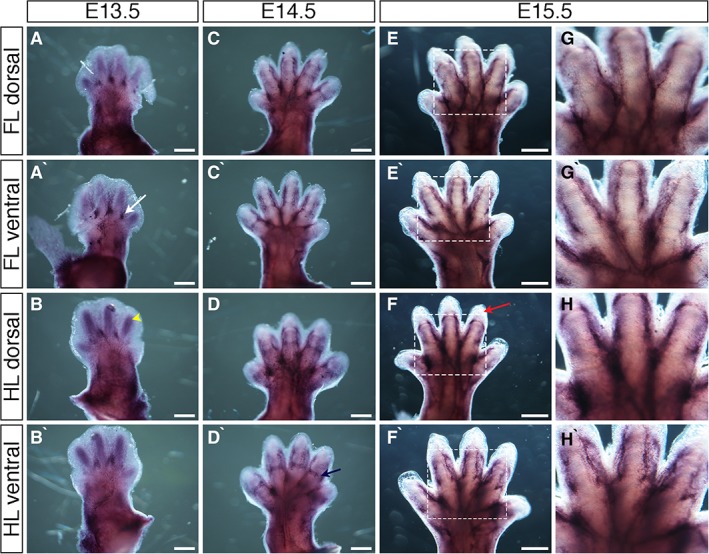
Expression of Cadherin5 (Cdh5) during mouse limb development. A–H`: Whole‐mount in situ hybridization for Cdh5 at E13.5 (A–B`), E14.5 (C–D`), and E15.5 (E–F`) in dorsal forelimbs (A, C, E), ventral forelimbs (A`,C`,E`), dorsal hindlimbs (B,D,F), and ventral hindlimbs (B`,D`,F`). Strong regions of expression are seen at the digit base, between digits (A` white arrow). Blood vessels are visible in regions of developing cartilage at E13.5 (B yellow arrowhead). Bands of expression across presumptive joint sites are visible at E14.5 (D navy arrow). Avascular distal mesoderm is indicated (F` red arrow). G–H`: Higher magnification of regions highlighted in white dashed boxes in E–F`, respectively. Scale bars = 500 µm.
Neuronal Patterning in the Developing Mouse Limb
Innervation of neuronal development in the mouse embryonic limb was examined using an anti‐neurofilament antibody between E10.5 and E14.5 by whole‐mount immunofluorescence (Fig. 5A–G`) and immunoperoxidase staining (Fig. 5H–O`). Both methods gave similar results in terms of neuronal innervation patterns. Forelimbs obtain innervation from spinal nerves via the brachial plexus, and hindlimbs from spinal nerves via the lumbar plexus. At E10.5, no nerves were present in the early fore‐ or hindlimb buds (data not shown). The first signs of limb innervation from spinal nerves were evident in the E11.5 forelimb (Fig. 5A,H arrowheads). In the hindlimb, innervation was more variable, where it was evident in some limbs (Fig. 5I arrowheads) but not others (Fig. 5B). This likely reflects differences in the precise age of the embryos, which can vary up to 0.5 day between embryos of a single litter. However, if present, nerves in the E11.5 hindlimb always extended for a shorter distance into the limb compared to the forelimb (Fig. 5H,I). Innervation of forelimbs and hindlimbs then continues at a rapid pace; however, innervation in the hindlimb lags behind that in the forelimb until E14.5 (Fig. 5A–0`). Main nerve trunks were present at E12.5, including branches that are targeted to proximal tissues in both forelimbs and hindlimbs (Fig. 5C,J,K). By E13.5, more distal nerve branches could be seen, proximal patterning of nerves had become more intricate, and nerves had reached the hand‐ and footplate (Fig. 5D,E,L,M). By E14.5, nerves had reached digit tips in both forelimbs and hindlimbs, thicker nerve branches were visible, and patterning was substantially more complex (Fig. 5F,G,N,O), further confirmed when looking at the hand‐ and footplates in higher magnification (Fig. 5F`,G`,N`,O`).
Figure 5.
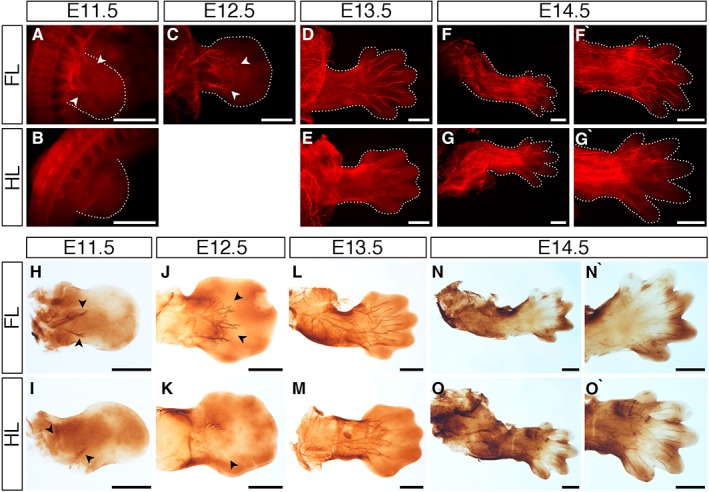
Development of limb innervation I. A–O`: Staining with an anti‐neurofilament antibody using a fluorescent secondary antibody (A–G’) or peroxidase conjugated secondary antibody (H–O`) to visualize limb innervation patterns in E11.5 (A,B,H,I), E12.5 (C,J,K), E13.5 (D,E,L,M), and E14.5 (F–G`,N–O`) forelimbs (A,C,D,F,F`,H,J,L,N,N`) and hindlimbs (B,E,G,G’,I,K,M,O,O`). Nerves enter the limb bud at E11.5 (A,B,H,I arrowheads) and handplate at E12.5 (C,J,K arrowheads). Innervation is more advanced in the forelimbs up to E13.5, where nerves have reached the digits in the forelimb (D,L) and remain at the base of the footplate in the hindlimb (E,M). Nerve innervation is throughout the limbs by E14.5 (F,G,N,O). High‐magnification images of the E14.5 forelimb handplate (F`,N`) and hindlimb footplate (G`,O`) show nerves have reached the tips of the condensing digits. Limbs are attached to the embryo body in A–E, and detached in F–O`. Dotted lines indicate the outline of the limbs in the fluorescent images. Scale bars = 500 µm.
Cartilage Formation and Progression in the Developing Mouse Limb
Formation of cartilage condensations and differentiation into bone was assessed in the mouse embryonic limb between E12.5 and E16.5 by Alcian Blue and Alizarin Red staining (Fig. 6). Alcian Blue stains cartilage and Alizarin Red stains differentiating bone. Alcian Blue staining was first observed in some forelimb elements at E12.5, namely the humerus, radius, ulna, and digits 2, 3, and 4 (Fig. 6A); the hindlimb had no obvious Alcian Blue staining (Fig. 6B). By E13.5, all cartilage elements in both forelimbs and hindlimbs (long bones and digits) were stained by Alcian Blue (Fig. 6C,D). Alizarin Red staining was not detected until E15.5, when it stained a region in the medial part of the humerus (Fig. 6G asterisk) and a very small domain in the medial part of the femur (Fig. 6H black arrow). By E16.5, all the long bones in both forelimb (humerus, radius, and ulna) and hindlimb (femur, tibia, and fibula) displayed strong region of Alizarin Red staining in the medial parts of the long bones (Fig. 6I,J black asterisks).
Figure 6.
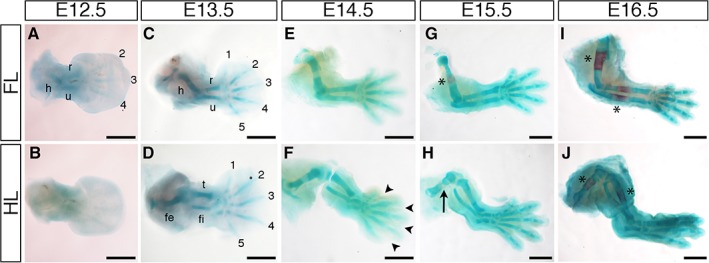
Cartilage and bone analysis. A–J: Staining with Alcian Blue and Alizarin Red to visualize onset and development of cartilage and bone in E12.5 (A,B), E13.5 (C,D), E14.5 (E,F), E15.5 (G,H), and E16.5 (I,J) forelimbs (A,C,E,G,I) and hindlimbs (B,D,F,H,J). Alcian Blue–stained cartilage condensations are observed in proximal forelimb tissue and in some distal digits from E12.5 (A). Like the nerve innervation patterns, the hindlimb lags behind the forelimb (B). All cartilage elements are seen in both forelimb and hindlimb by E13.5 (C,D), and elements in both limbs have lengthened considerably and look almost identical by E14.5, even though digit separation is yet to complete in the hindlimb (F arrowheads). Alizarin Red is first seen in the middle of the humerus (forelimb) and femur (hindlimb) at E15.5; staining is stronger and broader in the humerus (G asterisk) and only just detectable in a small part of the femur (H black arrow). By E16.5, Alizarin Red staining can be seen in the middle of all the long bones in both the forelimb and hindlimb (I,J, asterisk). h: humerus; r: radius; u: ulna; fe: femur; t: tibia; fi: fibula; 1: digit 1; 2: digit 2; 3: digit 3; 4: digit 4; 5: digit 5. Scale bars = 500 µm.
Discussion
Previous research has focused on comparison of wild‐type expression patterns with mutant mouse models rather than on wild‐type expression alone through development. Analysis of limb development in mutant mice models requires comparison to a model of “normal” development, and this article provides a useful, informative resource for the research community to refer to when studying limb development of mutant mouse models. We have described expression of markers of a range of important tissues essential for limb development: Gdf5, MyoD1, and Cdh5 from E13.5 to E15.5; Scleraxis from E12.5 to E15.5; and anti‐neurofilament marker from E10.5 to E14.5. Although previous expression patterns are published in part, we show the dynamic expression of these markers and the related tissue development throughout limb development. Expression patterns in fore‐ and hindlimbs are shown, along with section images and images of limb innervation through development.
Expression patterns analyzed are largely supported by previous publications. Meech et al., (2005) reported Gdf5 expression in digit rays at E11.5 and in developing cartilage condensations by E13.5; however, we do not see Gdf5 expression exclusively across developing joint sites until E14.5 (Meech et al., 2005; Ota et al., 2007; Seemann et al., 2005). This difference may be due to strain differences in embryonic development, which can exhibit differences in gestation periods (Degenkolbe et al., 2013; Huang et al., 2016). Gdf5 is expressed across developing joint sites, including interphalangeal joints, from E14.5 (Chen et al., 2016; Houweling et al., 2001; Huang et al., 2016; Sohaskey et al., 2009). Our findings for Scleraxis expression are in agreement with previously published analyses of Scleraxis, indicating expression in the autopod and in developing digit rays and cartilage condensations at E12.5, as well as defined expression in developing tendons at E13.5 (Huang et al., 2015; McGlinn et al., 2005; Schweitzer et al., 2001) and along developing digits, with strongest expression across sites of developing interzones at E14.5 (Murchison et al., 2007; Schweitzer et al., 2001). MyoD1 is expressed in muscle progenitors in the zeugopod, not the autopod, at E12.5. By E13.5, muscle blocks are visible in the zeugopod, with early expression visible in the autopod, supporting patterns observed in this study (Behrens et al., 2003; Hasson et al., 2010; L'honoré et al., 2010; Wood et al., 2013). Expression patterns of other blood vessel markers support the Cdh5 expression patterns finding we have observed. Platelet endothelial cell adhesion molecule (PECAM; also known as CD‐31) is expressed strongly at the base of interdigital regions at E12.5 (Vieira et al., 2007), and anti‐CD31 staining is also seen along the periphery of developing digits, with expression strongest at the base of interdigital regions and at regions of developing interzones at E14.5 (Ben et al., 2012; Eshkar‐Oren et al., 2015). Nerve patterning of the limbs in our study is consistent with previous work, indicating innervation in the limbs from around E11.5 (Martin, 1990; Vieira et al., 2007), and nerve branching is seen in the autopod in both forelimb and hindlimb by E13.5, reaching to the base of prechondrogenic condensations (Vieira et al., 2007). Our work also shows that innervation occurs in the forelimb before the hindlimb, and the hindlimb lags behind in innervation extent until around E14.5 (Fig. 5).
Comparing the expression patterns between fore‐ and hindlimbs, patterning of Gdf5 and Scleraxis is well established in the hand‐ and footplate by E13.5, and MyoD1 expression is seen later in the hand‐ and footplate from E14.5. Gdf5 expression is seen more distally than that of Scleraxis during limb development. At E14.5, Gdf5 expression can be seen in the distal interphalangeal joints, but Scleraxis is seen only as distal as the proximal interphalangeal joints in expression domains that overlap with those of Gdf5. Cdh5 expression is seen in the regions of developing digit cartilage from E13.5, and from E14.5 some stronger bands of expression can be seen across digits in regions that may also express Gdf5 and Scleraxis.
Far later than the expression of Cdh5, Gdf5, and Scleraxis, the neural plexus enters the digit tips at E14.5 (Martin, 1990) (Fig. 5F–G`,N–O`). Nerves enter the limbs at E11.5, and progression is delayed behind the migration of myocytes into the limb (Martin, 1990) (Fig. 5). The delayed migration of nerves into the limb bud, behind that of migrating myocytes, is also reported in the developing chicken limb (Mahony et al., 2018). This suggests a conserved mechanism whereby nerves enter the limb after initial tissue outgrowth from the flank, possibly allowing for myocyte and cartilage differentiation in the aneural environment before rapid innervation proceeds. In agreement with our finding that limb patterning has already begun before nerves enter the limb, in chickens, the presence of nerves is not required for limb patterning (Harsum et al., 2001; Mahony et al., 2018; Strecker and Stephens, 1983). We used two assays to observe neuronal innervation into the mouse fore‐ and hindlimbs: immunofluorescence and immunoperoxidase (Fig. 5). Both assays clearly show nerve patterning in detail and largely complement each other, though it is also clear that there is a marked variability in staging of early limbs between litters: Some E11.5 limbs show some proximal innervation of hindlimbs, while embryos from other litters do not. This is likely due to differences of embryonic age within and between litters.
Skeletal patterning progresses in a proximal‐to‐distal manner, and once all the cartilage condensations (detected by Alcian Blue) have been patterned, they are gradually replaced by bone (detected by Alizarin Red) as limb development proceeds, again in a proximal‐to‐distal manner. The forelimb has cartilage condensations before the hindlimb in the humerus, radius, ulna, and middle digits (Fig. 6A,B). However, the hindlimb rapidly catches up and both limbs are difficult to differentiate by E12.5 (Fig. 6C,D). Alizarin Red staining is first seen in the proximal long bones (humerus and femur, respectively) before the medial long bones, again underlining that the patterning and differentiation of limb elements is in a proximal‐to‐distal manner (Fig. 6G–J).
In this article, we have described the expression patterns and developmental progression of markers of limb elements through the most active stages of mouse limb development. This array of expression patterns can be used to further understand normal mouse limb development and outgrowth, as well as to understand tissue changes and gene regulation following functional misexpression experiments.
Experimental Procedures
Mice
All experimental procedures and conditions were in accordance with institutional Animal Welfare and Ethical Review Bodies (AWERB) and UK Home Office guidelines. Wild‐type C57BL/6J mice were maintained in in‐house breeding colonies. Mice were mated and the morning of vaginal plug formation counted as E0.5. Pregnant mothers were killed by cervical dislocation and embryos fixed overnight in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS). Experimental protocols were each repeated at least twice, and the number of embryos used was a minimum of three on each occasion, and in most cases totaled between five and eight embryos per protocol.
Whole‐mount In Situ Hybridization
To generate templates for the MyoD1, Cdh5, and Gdf5 riboprobes, RNA was extracted from E14.5 and E15.5 mouse limbs using the Qiagen RNeasy Mini Kit and cDNA synthesized using SuperScript III (Thermo Fisher Scientific) according to the manufacturer's instructions. DNA fragments were isolated by Polymerase Chain Reaction (PCR) (94˚C for 5 min, 35 cycles of 94˚C for 40 sec, 55˚C for 1 min, 72˚C for 90 sec, followed by a final extension at 72˚C for 5 min) and cloned into either pGEM‐T Easy (Promega; Gdf5) or pBluescript KS + (MyoD1 and Cdh5). The following primers were used: Gdf5 forward, AAAGGGCAAGATGACCGAGG; reverse, ACGTTGTTGGCAGAGTCGAT; MyoD1 forward, TTTTTGAGGACCTGGACCCG; reverse, TGCCATCAGAGCAGTTGGAG; Cdh5 forward, TTGCCCTGAAGAACGAGGAC; reverse, GTCGGAGGAATTGGTGCTCA. The riboprobe template for Scleraxis was a gift from Dr. Ronen Schweitzer, Portland, Oregon. Antisense RNA probes were then prepared by taking plasmid DNA and using PCR with M13 forward and M13 reverse primers to generate the probe template. in vitro transcription with relevant RNA polymerase was then carried out to generate the riboprobe labeled with digoxigenin (we used a Boehringer Mannheim kit), followed by removal of unincorporated nucleotides by placing solution through a G‐50 spin column. Whole‐mount in situ hybridization was performed as follows: Embryos were dissected and fixed overnight in 4% PFA, dehydrated in 50% methanol in PBT (PBS with 0.1% tween), and then stored in 100% methanol after three times at ‐20C˚. Dehydrated embryos were bleached in 6% hydrogen peroxide in 100% methanol for 1 hr, then rehydrated via washes in 50% methanol:PBT and 25% methanol:PBT to PBT. Embryos (up to E12.5) were treated with 10 ug/ml of proteinase K for 20 min, E13.5 and E14.5 embryos were treated with proteinase K at 20 ug/ml for 15 min, and E15.5 embryos were treated with proteinase K at 40 ug/ml for 15 min. Embryos were rinsed in PBT and post‐fixed in 4% PFA for 20 min. Embryos were then incubated in prehybridization solution (50% formamide, 5X SSC pH4.5, 50 ug/ml yeast RNA, 1% SDS, 50 ug/ml heparin) at 70C˚ for 1 hr and then incubated in fresh prehybridization solution containing the relevant riboprobe that had been preheated to 85C˚ for 5 min prior to addition to the prehybridization mix (we use 10 ul of an in vitro transcription reaction per 1 ml of prehybridization mix). Hybridization was carried out overnight at 65˚C. Following hybridization, the probe can be removed and stored at ‐20C˚ for another in situ hybridization and reused up to five times before being remade. Embryos are then placed in solutions with varying stringency at 65C˚ to remove nondescript probe binding. First into solution 1 (50% formamide, 5X SSC, 1% SDS, pH4.5) for three washes of 30 min each, then into solution 3 (50% formamide, 5X SSC, pH4.5) for 3 washes of 30 min each, before being placed in TBST (tris‐buffered saline with 1% tween). Embryos are then blocked in a solution of 10% inactivated goat serum in TBST for 1 hr and then placed in the antibody solution of 1% inactivated goat serum in TBST containing anti‐DIG antibody at 1:5000 overnight at 4C˚. The following day, embryos are washed in TBST at least 10 times throughout the day. The next day, embryos are washed in NTMT (100 mM NaCl, 100 mM Tris‐HCl pH9.5; 50 mM MgCl2, 1% tween) three times (10 min each) and then placed in the color solution (1 ml NTMT with 4.5 ul BCIP [5’Bromo‐4‐chloro‐3‐indoly phosphate] (50 mg/ml) and 2.5 ul NBT [Nitroblue tetrazolium]; (5 mg/ml) at room temperature in the dark. Embryos were checked every 20 min, and the reaction was terminated when complete by washing in PBT, then post‐fixed in 4% PFA and stored in PBT.
Immunofluorescence
Immunofluorescence staining was carried out based on previous protocols with some modifications (Kardon, 1998) using an anti‐neurofilament antibody (Millipore AB1987) and IgG goat anti‐rabbit Cy3 secondary antibody (Jackson ImmunoResearch). No antigen retrieval was carried out. Following overnight fixation in 4% PFA at 4˚C, embryos were washed in PBS and then placed in Dent's bleach (1 part hydrogen peroxide, 2 parts Dent's fix) overnight at 4˚C. Embryos were washed in 100% methanol and fixed in Dent's fixative (1 part Dimethyl sulfoxide, 4 parts methanol) overnight at 4˚C. After being rinsed multiple times in PBS, embryos were placed in the primary antibody in blocking solution overnight (18 hr minimum) at 4˚C. Following washes in PBS (over 6 hr), embryos were placed in the secondary antibody overnight at 4˚C. Embryos were then rinsed in PBS multiple times over a 6‐hr period, then placed in a 50% methanol:PBS solution, followed by 100% methanol washes, before being placed in BABB (33.3% benzyl alcohol, 66.6% benzyl benzoate) to clear and allow imaging under a fluorescence microscope.
Immunoperoxidase Staining of Nerves
Immunoperoxidase staining was based on previously described protocols with modifications (Kardon, 1998) using an anti‐neurofilament antibody (Millipore AB1987) and a peroxidase conjugated secondary antibody (Sigma). Following overnight fixation in 4% PFA at 4˚C, embryos were washed in PBS and then placed in Dent's bleach overnight at 4˚C. Embryos were washed in methanol and fixed in Dent's fixative overnight at 4˚C. After being rinsed in PBS, embryos were blocked and then placed in the primary antibody for 2 days at 4˚C. Following multiple washes in PBS, embryos were placed in the secondary antibody overnight at 4˚C. Following PBS washes, embryos were incubated in fresh 3,3‐diaminobenzidine (DAB; Sigma) until reaction was complete, rinsed in methanol, and cleared and stored in BABB.
Vibratome Sectioning
Embryos were embedded in 4% agarose in water and sectioned transversely using a Vibratome 1000 Plus at 100 µm. The sections were collected in 48 well plates containing PBS and mounted on Superfrost slides (VWR) with Vectashield mounting medium (Vector Labs).
Alcian Blue and Alizarin Red Staining of Mouse Embryos
To observe cartilage and appearance of bone, we stained embryos with Alcian Blue and Alizarin Red using a protocol we modified from Rigueur and Lyons (2014).
Embryos were fixed in 70% ethanol overnight, placed into 95% ethanol for 1 hr, and incubated in acetone overnight at room temperature. Embryos were stained with 0.03% Alcian Blue for 4 hr, then with 0.005% Alizarin Red for 4 hr before being cleared in fresh 1% KOH for 1 hr (E11.5–E13.5) or 2 hr (E14.5–E16.5), then stored in glycerol.
Imaging
Whole‐mount and section images were captured using a Nikon SMZ1500 microscope with Nikon DS‐L1 camera. Whole‐mount antibody stains were imaged using a Nikon SMZ1500 and a Nikon DXM1200 camera. Figures were prepared using Adobe Photoshop.
Acknowledgements
The authors thank Yolanda Ramirez Casas and J. Martin Collinson for technical advice, and past and present lab staff for helpful discussions.
References
- Amarilio R, Viukov SV, Sharir A, Eshkar‐Oren I, Johnson RS, Zelzer E. 2007. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechrondrogenic cells during early skeletogenesis. Development 134:3917–3928. [DOI] [PubMed] [Google Scholar]
- Behrens A, Haigh J, Mechta‐Grigoriou F, Nagy A, Yaniv M. Wagner EF. 2003. Impaired intervertebral disc formation in the absence of Jun. Development 130:108–109. [DOI] [PubMed] [Google Scholar]
- Ben AS, Malkinson G, Krief S, Shwartz Y, Ely Y, Ferrara N, Yaniv K. Zelzer E. 2012. S1P1 inhibits sprouting angiogenesis during vascular development. Development 139:3859–3869. [DOI] [PubMed] [Google Scholar]
- Bénazet JD, Zeller R. 2009. Vertebrate limb development: moving from classical morphogen gradients to an integrated 4‐dimensional patterning system. Cold Spring Harb Perspect Biol 1:a001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch HL, Thorpe CT, Rumian AP. 2013. Specialisation of extracellular matrix for function in tendons and ligaments. Muscles Ligaments Tendons J 3:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Capellini TD, Schoor M, Mortlock DP, Reddi AH, Kingsley DM. 2016. Heads, Shoulders, Elbows, Knees, and Toes: Modular Gdf5 Enhancers Control Different Joints in the Vertebrate Skeleton. PLOS Genet 12:e1006454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson JM, Lindstrom NO, Neves C, Wallace K, Mehang C, Charles RH, Ross ZK, Fraser AM, Mbogo I, Oras K, Nakamoto M, Barker S, Duce S, Miedzybrodzka Z, Vargesson, N . 2018. The developmental and genetic basis of ‘clubfoot’ in the peroneal muscular atrophy mutant mouse. Development 145:pii160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenkolbe E, König J, Zimmer J, Walther M, Reißner C, Nickel J, Plöger F, Raspopovic J, Sharpe J, Dathe K, Hecht JT, Mundlos S, Doelken SC, Seemann P. 2013. A GDF5 point mutation strikes twice—causing BDA1 and SYNS2. PLoS Genet 9:e1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshkar‐Oren I, Krief S, Ferrara N, Elliott AM, Zelzer E. 2015. Vascular patterning regulates interdigital cell death by a ROS‐mediated mechanism. Development 142:672–680. [DOI] [PubMed] [Google Scholar]
- Eshkar‐Oren I, Viukov SV, Salameh S, Krief S, Oh C, Akiyama H, Gerber H‐P, Ferrara N, Zelzer E. 2009. The forming limb skeleton serves as a signaling center for limb vasculature patterning via regulation of Vegf. Development 136:1263–1272. [DOI] [PubMed] [Google Scholar]
- Firulli, BA , Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. 2005. Altered Twist1 and Hand2 dimerization is associated with Sathre–Chotzen syndrome and limb abnormalities. Nat Genet 37:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. 2005. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development 132:2235–2249. [DOI] [PubMed] [Google Scholar]
- Harsum S, Clarke JD, Martin P. 2001. A reciprocal relationship between cutaneous nerves and repairing skin wounds in the developing chick embryo. Dev Biol 238:27–39. [DOI] [PubMed] [Google Scholar]
- Hasson P, DeLaurier A, Bennett M, Grigorieva E, Naiche LA, Papaioannou VE, Mohun MJ, Logan MP. 2010. Tbx4 and Tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev Cell 18:148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman TV, Nickel J, Mueller TD. 2012. Missense Mutations in GDF‐5 Signaling: Molecular Mechanisms Behind Skeletal Malformation In: Cooper D, Chen J‐M, editors. Mutations in Human Genetic Disease. London: IntechOpen Limited; p 11–54. [Google Scholar]
- Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AF, Rüther U, Christoffels VM. 2001. Gene and cluster‐specific expression of the Iroquois family members during mouse development. Mech Dev 107:169–174. [DOI] [PubMed] [Google Scholar]
- Huang AH, Riordan TJ, Pryce B, Weibel JL, Watson SS, Long F, Lefebvre W, Harfe BD, Stadlet S, Akiyama H, Tufa SF, Keene DR. Schweitzer R. 2015. Musculoskeletal integration at the wrist underlies the modular development of limb tendons. Development 142:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B‐L, Trofka A, Furusawa A, Norrie JL, Rabinowitz AH, Vokes SA, Taketo M, Zakany J, Mackem S. 2016. An interdigit signalling centre instructs coordinate phalanx‐joint formation governed by 5’Hoxd‐Gli3 antagonism. Nat Commun 7:12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. 1997. MyoD and Myf‐5 differentially regulate the development of limb versus trunk skeletal muscle. Development 124:4729–4738. [DOI] [PubMed] [Google Scholar]
- Kardon G. 1998. Muscle and tendon morphogenesis in the avian hind limb. Development 125:4019–4032. [DOI] [PubMed] [Google Scholar]
- L'honoré A, Ouimette J‐F, Lavertu‐Jolin M, Drouin J, Buckingham M, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y. 2010. Pitx2 defines alternate pathways acting through MyoD during limb and somitic myogenesis. Development 137:3847–3856. [DOI] [PubMed] [Google Scholar]
- Laclef C, Hamard G, Demignon J, Souil E, Houbron C, Maire P. 2003. Altered myogenesis in Six1‐deficient mice. Development 130:2239–2252. [DOI] [PubMed] [Google Scholar]
- Liu H, Xu J, Liu C‐F, Lan Y, Wylie C, Jiang R. 2015. Whole transcriptome expression profiling of mouse limb tendon development by using RNA‐seq. J Orthop Res 33:840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinn E, Lammerts van Bueren K, Fiorenza S, Mo R, Poh AM, Forrest A, Soares MB, de Fatima Bonaldo M, Grimmond S, Hui C‐C, Wainwright, B. Wicking C. 2005. Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech Dev 122:1218–1233. [DOI] [PubMed] [Google Scholar]
- Mahony C, McMenemy S, Rafipay AJ, Beedie SL, Fraga LR, Gutschow M, Figg WD, Erskine L, Vargesson N. 2018. CPS49‐induced neurotoxicity does not cause limb patterning anomalies in developing chicken embryos. J Anat 232:568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon L, Arqués C., Torres MS, Sharpe J. 2011. A Computational Clonal Analysis of the Developing Mouse Limb Bud. PLoS Comput Biol 7:e1001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P. 1990. Tissue patterning in the developing mouse limb. Int J Dev Biol 34:323–336. [PubMed] [Google Scholar]
- Mayeuf‐Louchart A, Montarras D, Bodin C, Kume T, Vincent SD, Buckingham M. 2016. Endothelial cell specification in the somite is compromised in Pax3‐positive progenitors of Foxc1/2 conditional mutants, with loss of forelimb myogenesis. Development 143:872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech R, Edelman DB, Jones FS, Makarenkova HP. 2005. The homeobox transcription factor Barx2 regulates chondrogenesis during limb development. Development 132:2135–2146. [DOI] [PubMed] [Google Scholar]
- Mitrovic DR. 1977. Development of the metatarsophalangeal joint of the chick embryo: morphological, ultrastructural and histochemical studies. Am J Anat 150:333–347. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Wanek N, Bryant SV. 1989. Mammalian limb bud development: in situ fate maps of early hindlimb buds. J Exp Zool 249:50–54. [DOI] [PubMed] [Google Scholar]
- Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R. 2007. Regulation of tendon differentiation by scleraxis distinguishes force‐transmitting tendons from muscle‐anchoring tendons. Development 134:2697–2708. [DOI] [PubMed] [Google Scholar]
- Neuhaus H, Metikala S, Hollemann T. 2014. Xenopus cadherin 5 is specifically expressed in endothelial cells of the developing vascular system. Int J Dev Biol 58:51–56. [DOI] [PubMed] [Google Scholar]
- Nowlan NC, Sharpe J, Roddy KA, Prendergast PJ, Murphy P. 2010. Mechanobiology of embryonic skeletal development: Insights from animal models. Birth Defects Res C Embryo Today 90:2013‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota S, Zhou Z‐Q, Keene DR, Knoepfler P, Hurlin PJ. 2007. Activities of N‐Myc in the developing limb link control of skeletal size with digit separation. Development 134:1583–1592. [DOI] [PubMed] [Google Scholar]
- Perez AV, Perrine M, Brainard N, Vogel KG. 2003. Scleraxis (Scx) directs lacZ expression in tendon of transgenic mice. Mech Dev 120:1153–1163. [DOI] [PubMed] [Google Scholar]
- Perez WD, Weller CR, Shou S, Stadler HS. 2010. Survival of Hoxa13 homozygous mutants reveals a novel role in digit patterning and appendicular skeletal development. Dev Dyn 239:446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Singh PN, Sohaskey ML, Harland RM, Bandyopadhyay A. 2015. Precise spatial regulation of BMP signaling is essential for articular cartilage differentiation. Development 142:1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigueur D, Lyons KM. 2014. Whole‐mount skeletal staining. Methods Mol Biol 1130:113‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M. 1989. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature 341:303–307. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Chyun, JH , Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. 2001. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128:3855–3866. [DOI] [PubMed] [Google Scholar]
- Seemann P, Schwappacher R, Kjaer KW, Krakow D, Lehmann K, Dawson K, Stricker S, Pohl J, Plöger F, Staub E, Nickel J, Sebald W, Knaus P, Mundlos S. 2005. Activating and deactivating mutations in the receptor interaction site of GDF5 cause symphalangism or brachydactyly type A2. J Clin Invest 115:2373–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey ML, Yu J, Diaz MA, Plaas AH, Harland RM. 2008. JAWS coordinates chondrogenesis and synovial joint positioning. Development 135:2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker TR, Stephens TD. 1983. Peripheral nerves do not play a trophic role in limb skeletal morphogenesis. Teratology 27:159–167. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132:2685–2695. [DOI] [PubMed] [Google Scholar]
- Tavella S, Biticchi R, Schito A, Minina E, Di Martino, D , Pagano A, Vortkamp V, Horton WA, Cancedda R, Garofalo S. 2004. Targeted expression of SHH affects chrondrocyte differentiation, growth plate organization and Sox9 expression. J Bone Miner Res 19:1678–1688. [DOI] [PubMed] [Google Scholar]
- Tickle C. 2006. Making digit patterns in the vertebrate limb. Nat Rev Mol Cell Biol 7:45–53. [DOI] [PubMed] [Google Scholar]
- Vargesson N, Hootnick D. 2017. Arterial dysgenesis and limb defects: Clinical and experimental examples. Reprod Toxicol 70:21–29. [DOI] [PubMed] [Google Scholar]
- Vieira JM, Schwarz Q, Ruhrberg C. 2007. Selective requirements for NRP1 ligands during neurovascular patterning. Development 134:1833–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Green RP, Zhao G. Ornitz DM. 2001. Differential regulation of endochondral bone growth and joint development by FGFR1 and FGFR3 tyrosine kinase domains. Development 128:3867–3876. [DOI] [PubMed] [Google Scholar]
- Wood WM, Etemad S, Yamamoto M, Goldhamer DJ. 2013. MyoD‐expressing progenitors are essential for skeletal myogenesis and satellite cell development. Dev Biol 384:114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]


