Abstract
The origin of eukaryotes stands as a major open question in biology. Central to this question is the nature and timing of the origin of the mitochondrion, an ubiquitous eukaryotic organelle originated by the endosymbiosis of an alphaproteobacterial ancestor. Different hypotheses disagree, among other aspects, on whether mitochondria were acquired early or late during eukaryogenesis. Similarly, the nature and complexity of the receiving host is debated, with models ranging from a simple prokaryotic host to an already complex proto‐eukaryote. Here, I will discuss recent findings from phylogenomics analyses of extant genomes that are shedding light into the evolutionary origins of the eukaryotic ancestor, and which suggest a later acquisition of alpha‐proteobacterial derived proteins as compared to those with different bacterial ancestries. I argue that simple eukaryogenesis models that assume a binary symbiosis between an archaeon host and an alpha‐proteobacterial proto‐mitochondrion cannot explain the complex chimeric nature that is inferred for the eukaryotic ancestor. To reconcile existing hypotheses with the new data, I propose the “pre‐mitochondrial symbioses” hypothesis that provides a framework for eukaryogenesis scenarios involving alternative symbiotic interactions that predate the acquisition of mitochondria. © 2018 The Authors. IUBMB Life published by Wiley Periodicals, Inc. on behalf of International Union of Biochemistry and Molecular Biology, 70(12):1188–1196, 2018
Keywords: evolution, eukaryotic evolution, mitochondria
Abbreviations
- LECA
Last Eukaryotic Common Ancestor
- FECA
First Eukaryotic Common Ancestor
- HGT
Horizontal Gene Transfer
INTRODUCTION
In the second edition of their seminal textbook “The microbial world”, the famous microbiologist Roger Stanier and his colleagues Michael Doudoroff and Edward Adelberg referred to the observed differences between eukaryotic and prokaryotic cells as the “greatest single discontinuity to be found in the present‐day living world” 1. Indeed, most simple eukaryotic cells do possess an intricate level of complexity, a fact that is well illustrated by microsporidian fungi. These obligate intracellular parasites possess highly reduced genomes, have uniquely lost some pathways, and present genome‐lacking mitochondria, called mitosomes 2. A more extreme example is provided by Monocercomonoides sp., a flagellated excavate, living in the intestines of chinchillas. This unicellular eukaryote seems to have completely lost mitochondria, representing the first case of a eukaryote convincingly shown to lack this organelle 3. Yet, these organisms, and many others with reduced forms of mitochondria, represent relatively recent adaptations to particular lifestyles or niches, where a varying repertoire of mitochondrial functions became dispensable 4. Although the loss of mitochondria may seem an extreme reduction of complexity, these organisms retain an otherwise complex intracellular compartmentation, with a cell nucleus, cytoskeleton, endoplasmic reticulum, and other vesicles. In comparison, the most complex forms of prokaryotic cells look overly simplistic. Perhaps, the most sophisticated prokaryotic cell could be represented by the Planctomycetes, a group of Gram‐negative bacteria. Species of this clade, of which some were originally described as eukaryotes, show a convoluted membrane organization which was proposed to be unique among prokaryotes and to closely resemble those of eukaryotes 5. However, recent research has shown that these apparent compartmentalization results from invaginations of the cytoplasmic membrane, rather than by independent membrane‐bounded organelles 6. Thus, ever if our exploration efforts are widening the range of cellular complexities in both eukaryotes and prokaryotes, a fundamental gap still remains. Elucidating how this discontinuity could have been bridged during evolution has occupied the minds of many researchers and has been the matter of many heated debates. A historical account of the developments on the different theories for the origin of eukaryotes (a process also known as eukaryogenesis) has been nicely summarized elsewhere 7.
One of the most revolutionizing ideas of how eukaryotes could have been formed from preexisting simpler cells was put forward by Lynn Margulis (Lynn Sagan by then) in her famous 1967 article 8. The idea that certain cellular organelles could have been the result of symbiotic relationships with once free‐living bacteria was certainly not new 9. However, Margulis not only revived those ideas but also put them into a broader and more specific theoretical framework: The Serial Endosymbiosis Theory. In Margulis’ view, the origin of eukaryotes was driven by the need to survive in a newly formed oxygen‐rich atmosphere resulting from the advent of photosynthetic prokaryotes 8. The engulfment of an aerobic prokaryotic microbe (the protomitochondrion) by a heterotrophic anaerobe would have allowed the survival of the latter and marked the origin of eukaryotes. The sum of simpler forms gives rise to more complex forms and can do so in nonadditive ways that bring about emerging properties. In this regard, Margulis herself proposed that mitochondrial endosymbiosis triggered the formation of the nucleus and the endoplasmic reticulum, enabled by the acquisition of pathways for phospholipid and steroid synthesis and the availability of larger amounts of energy 8. Subsequent symbiotic events would have brought, according to Margulis, flagella by endosymbiosis with spirochaetes and plastids by endosymbiosis with cyanobacteria; this latter event restricted to the common ancestor of archaeplastida. Heavily resisted at the beginning, the idea of endosymbiotic origins of some organelles finally pervaded the research community and is now broadly accepted although not for all organelles 10, 11, 12. Indeed, the findings that mitochondria and plastids harbor genomes whose sequences resemble specific prokaryotic clades—alpha‐proteobacteria for mitochondria and cyanobacteria for plastids—were most persuasive. Other proposed endosymbiotic origins such as that of flagella have been discarded in the light of the absence of any evidence 12. The broad acceptance of the endosymbiotic origin of mitochondria and plastids has likely influenced endosymbiotic hypotheses for other ubiquitous eukaryotic organelles, such as the peroxisome 13, 14. However, nowadays hypotheses purporting symbiotic origins of peroxisomes have been discarded 15, 16, 17 and replaced by alternative ones, involving endogenous scenarios 18, 19, 20.
Among all organelles, the mitochondrion occupies a central position in hypothetical eukaryogenesis scenarios 9. This organelle plays a central metabolic role in most eukaryotic organisms. Most notably but not exclusively, and also not necessarily (as we will see below), mitochondria can be responsible for the production of most of the energy in a cell 21. This organelle is also a clear ancient eukaryotic feature, inferred to be present in the last common ancestor of all eukaryotes, the so‐called last eukaryotic common ancestor (LECA), 22, and it is also the only such ancient structure for which there is evidence for an endosymbiotic origin 23. Hence, the establishment of mitochondrial endosymbiosis is a clear hallmark of eukaryotic evolution. Undoubtedly, mitochondrial endosymbiosis must have had a radical impact on the cellular metabolism of both the host and the endosymbiont. This is a central idea that has accompanied eukaryogenesis hypotheses since the times of Margulis 8. Although most hypotheses agree on the central role of mitochondrial endosymbiosis, there is disagreement on whether this event occurred early or late during the process of eukaryogenesis 24. Although there is a plethora of proposed hypothetical scenarios for the origin of eukaryotes, differing in many details other than the timing of mitochondrial endosymbiosis, in this article I will collectively discuss them as mito‐early or mito‐late, based solely on this temporal aspect.
Although mito‐early hypotheses argue that mitochondria came early in the process of eukaryogenesis, perhaps as its first event, mito‐late hypotheses view mitochondria as an important step that happened after some complexity was acquired. In addition, although mito‐early and mito‐late hypotheses agree on the nature of the mitochondrial ancestor (an alphaproteobacterium), they differ in the nature of the host that engulfed it. Typically, mito‐early hypotheses assume a prokaryotic host (an archaeon), whereas mito‐late hypotheses tend to assume a host with already some eukaryotic features, sometimes referred to as a “proto‐eukaryote” 9, 25. The level of complexity of this purported proto‐eukaryote can vary widely among different scenarios. The initial mito‐late hypothesis put forward by Cavalier‐Smith known as the “archezoa hypothesis,” purported amitochondriate eukaryotes, such as the microsporidians described earlier, as primitive 26. In his view, eukaryotes would have developed most of the complexity present in modern cells before acquiring mitochondria. Supporting this view, early molecular phylogenies placed amitochondriate eukaryotes as early branching lineages in the tree of eukaryotes. However, this placement was later found to be a phylogenetic artifact, and the archezoa hypothesis was abandoned in favor of mito‐early hypotheses of the time 27. However, it is important to note that the falsification of the primitive nature of currently known amitochondriate organisms does not necessarily imply that mitochondria were the first acquired complex feature in eukaryotes. Hence, we return to the same basic problem, most hallmark features of eukaryotes are mapped to the same common ancestor, without providing any information about the relative order in which they originated. In discussing mito‐early and mito‐late hypotheses, I will intentionally move away from comparing specific scenarios such as the archezoa 26 or the hydrogen 28 hypotheses, which represent the two extremes in terms of temporal acquisition of mitochondria. I will instead discuss the types of existing data that can be assessed to ponder on the time of the acquisition of mitochondria relative to the arrival of other protein families to the lineage leading to LECA.
THE IDENTITY OF THE MITOCHONDRIAL ANCESTOR
The mitochondrion is one of the necessary pieces of the eukaryogenesis puzzle, and the one for which we have more certainties, making it one of the few solid anchor points in the line of events that lead to the origin of eukaryotes. In many eukaryotes, mitochondria play a central role in energy metabolism, having been described as the cell's energy factory. However, this is an overly simplistic view of the cellular roles that can be adscribed to mitochondria. Mitochondria can play important roles in many other cellular processes ranging from biosynthesis of coenzymes to signaling and apoptosis 21. Indeed, mitochondria can entirely lose their ability of producing energy and still be an essential organelle in many organisms 4. Having a look at the broad diversity of mitochondria in the major eukaryotic lineages, particularly at those which have been reduced to the bare minimum, the key role that seems to be the minimal common denominator is the synthesis of Iron–Sulfur clusters. Indeed, the recent acquisition, via horizontal gene transfer, of a bacterial derived pathway for the synthesis of iron–sulfur clusters seems to have been the definitive step that enabled the complete vanishing of the mitochondrion in the already introduced amitochondriate Micromonas 3. However, which were the original functions of mitochondria, those that enabled the establishment of an initial symbiosis?
Initial phylogenies pointed to an alpha‐proteobacterial nature of the proto‐mitochondrion, with different studies disagreeing on which specific lineage of alpha‐proteobacteria 4, 11, 23. A recent phylogenomic analysis, which includes many metagenomic derived alpha‐proteobacterial sequences, suggests an earlier branching of the mitochondrial lineage, being a sister group to all sequenced alpha‐proteobacteria 29. As alpha‐proteobacteria are highly versatile in terms of their genome composition and metabolic properties and lifestyles, knowing that the proto‐mitochondrion was an alpha‐proteobacterium tells us little about the possible metabolic bases of the endosymbiotic interactions. One way to narrow down the key pathways is to search for the alpha‐proteobacterial proteins that were retained in LECA. This has been done using a phylogenetic approach to reconstruct the potential proto‐mitochondrial metabolism 30, 31. Such reconstructions of the alpha‐proteobacterial derived content in eukaryotes show that the heritage from the proto‐mitochondrion goes beyond mitochondrial metabolism, extending to pathways that are now found in several compartments such as the cytoplasm or the peroxisomes. That several, somehow unrelated, metabolic pathways have been retained from this ancestral symbiont suggests a complex symbiotic relationship with multifaceted benefits for the symbiotic partners. Such broad proto‐mitochondrial heritage is difficult to explain by models based on a simple benefit of the symbiosis such as those based on the exchange of a single metabolic product. The other side of the coin is represented by the perhaps uncomfortable finding that only a minor fraction of the mitochondrial proteome can be traced back to alpha‐proteobacteria 31, 32. Although part of this could be explained by insufficient phylogenetic signal, it is now clear that the mitochondrial proteome has been shaped by the incorporation of proteins of non‐alphaproteobacterial proteins 33. As we will see below, we face the same problem when we trace the origins of bacterial derived proteins that were present in LECA, which is perhaps an even more uncomfortable finding.
COMPLEX, CHIMERIC NATURE OF LECA
The nature of the host that engulfed the mitochondrion is one of the main conundrums regarding eukaryogenesis. To approach the question we must first resort to describe what we know about LECA, a descendant of that mysterious host that postdates the mitochondrial acquisition. We are having an increasingly resolved picture of how the proteome of LECA looked like based on comparisons from extant genomic sequences. Although there are several procedures to reconstruct ancestral proteomes 34, most approaches rely on finding protein families that are sufficiently widespread among major eukaryotic lineages and for which a vertical inheritance from LECA can be assessed 22, 25. We have already discussed that the inspection of these reconstructed proteomes, as well the distribution of traits among major eukaryotic lineages, points to LECA being already a full‐fledged eukaryote not missing any of the hallmark traits of eukaryotes. We will now focus on the evolutionary origins of the eukaryotic protein families that have been inferred to have been present in LECA. Where do they come from? The answer is not a simple one.
All studies agree that LECA's proteome was chimeric, with a proteome fraction whose origin can be traced back to archaea, another one that has bacterial sequences as the closest non‐eukaryotic relatives, and yet a third fraction, dubbed eukaryote‐specific, for which no prokaryotic homologs can be found 22, 25. This chimeric nature is compatible with all symbiosis‐based hypotheses for the origin of eukaryotes that involve at least one archaeal and one bacterial partner. I want to stress the “at least” of the preceding sentence, which means the chimeric nature of LECA is not only compatible with binary symbiogenic hypothesis, that is, involving just two partners, but also with those that involve more than two partners. Most studies also agree in that the fraction of archaeal and bacterial derived proteins is not balanced neither in terms of their relative abundance nor their functionalities. With regards to their functionalities, the archaeal derived component tends to have informational or structural roles, whereas metabolic functions abound in the bacterial derived component. Although these generalities describe the main trend, this is by no means a dichotomy, and there are examples of bacterial derived structural genes and archaeal derived metabolic genes. Nevertheless, these functional differences point to different and complementary roles of the two main fractions of the proteome, as it would be expected from a symbiotic interaction. With regard to their sizes, the bacterial derived fraction is much larger, usually two or three times larger than the archaeal derived content. This may be considered at odds with scenarios proposing an archaeal host and a single bacterial endosymbiont, as it would imply an extreme streamlining of the host in comparison to the endosymbiont. In contrast to this, most examples from extant endosymbiotic relationships show extreme reductions in the endosymbiont side 35. However, in these cases the endosymbiont is usually complementing a single requirement of the host (e.g., the synthesis of an essential amino acid), and it may well be that the ancestral eukaryogenesis event was of a different sort.
There is another observation regarding the chimeric LECA proteome that is more intriguing. Both blast‐based and phylogeny‐based methods to assign ancestry to LECA protein families can provide more specific taxonomic assignments beyond the archaea/bacterial divide. When this is done, a diversity of taxonomic assignments is found, particularly among the bacterial derived component 36, 37. Most surprisingly, only a small fraction of the bacterial derived component can be traced back to alpha‐proteobacteria. This heterogeneity of phylogenetic signals has been constant across studies, but its relevance has been usually disregarded based on (i) the known pitfalls of taxonomy assigning methods, and the noisy nature of phylogenetic signal and/or (ii) the known propensity of prokaryotic species to exchange genes through horizontal gene transfer, which can be attributable to the proto‐mitochondrion. These two factors are certainly blurring the ancestry signal in LECA's proteome and can be the source of a fraction—if not all—of the observed phylogenetic disparity. However, these two processes imply some expectations in the type of signal distortions that they would produce. For instance, phylogenetic noise would not result in phylogenetic affiliations distributed randomly across the prokaryotic phylogeny, but rather one would expect misplacements to be more often resulting in assignments to lineages closer to the true ancestral lineages—i.e., other proteobacterial lineages—than to distantly related ones. However, only about one‐third of the phylogenetic assignments correspond to proteobacterial lineages, and other, distantly related lineages have a surprisingly high number of assignments. Horizontal gene transfer among prokaryotes would produce alternative signals, even in distantly related lineages. However, if such transfers are assumed to have occurred before the endosymbiotic event, one must conclude that they were lost from any sampled sister lineage of the proto‐mitochondrion; otherwise, the ancestry of proto‐mitochondrial protein would be mapped to that sister lineage. This leaves a narrow window of time to acquire a surprisingly high amount of non‐proteobacterial genes in LECA. That is, under such assumption the proto‐mitochondrial ancestor would be predicted to have a larger fraction of foreign genes than genes derived vertically from its direct proteobacterial ancestors. Moreover, these genes should have been gained in a relatively short time span since the divergence of the closest sequenced sister lineage and the endosymbiotic event. Furthermore, as I discuss below, there are other considerations that, in my view, compromise such scenarios.
THE (PALM) TREE OF EUKARYOTES, ITS ELUSIVE ROOT, AND ITS LENGTH‐CHANGING STEM
Extant diversity of eukaryotes can be broadly divided into several main supergroups whose monophily is relatively well established 38. These include Unikonts (uniting opisthokonts and amoebozoa), Archaeplastida (including green plants, red algae, and glaucophytes), Excavates (including Discoba and Metamonada), and the so‐called SAR supergroup that joins Stramenopiles, Alveolates and Rhizaria. In addition, a growing number of “orphan” lineages seem to branch early within the eukaryotic tree of life but have no clear affiliation to any of these supergroups or may be even form novel supergroups, including, among others, Cryptophytes, Picozoa, Haptophytes, or Collodictyon. Great efforts have been put to resolve the earliest branches of eukaryotic tree of life, including the placement of its root 38, 39, 40, 41, 42. Resolving the root and the early branching pattern of the tree of eukaryotes is a daunting task, mostly because the main eukaryotic groups are separated by very short branches, and thus the phylogenetic signal is scarce.
When prokaryotic sequences are used to root the trees, however, relatively long branches separate the eukaryotes from their prokaryotic counterparts. I usually refer to this feature with the metaphor of the palm tree of eukaryotes to emphasize the fact that the main extant eukaryotic groups seem to have followed a fast radiation from LECA (Fig. 1). A relatively long stem without ramifications precedes the appearance of LECA—the trunk of the palm tree—and separates it from its closest prokaryotic relatives. The palm tree shape is reminiscent of an ancient radiation probably accompanied by massive extinctions of lineages that diverged earlier. Certainly, this is something suggestive of a major evolutionary innovation in the surviving lineage or, alternatively, a major existing feature in this lineage that allowed it to survive major changes in the environment. Similar “palm tree” structures are observable at other lineages (for instance, in birds). Here too, most bird lineages radiated very rapidly in a relatively short time 44, and the extinction of earlier branching lineages such as dinosaurs leaves a naked stem in molecular phylogenies separating the last common ancestor of extant birds to its closest extant sister group, represented by crocodiles 45. This stem is as long, in terms of substitutions per site, as the distance from this last common bird ancestor to one of its extant species (i.e., chicken).
Figure 1.
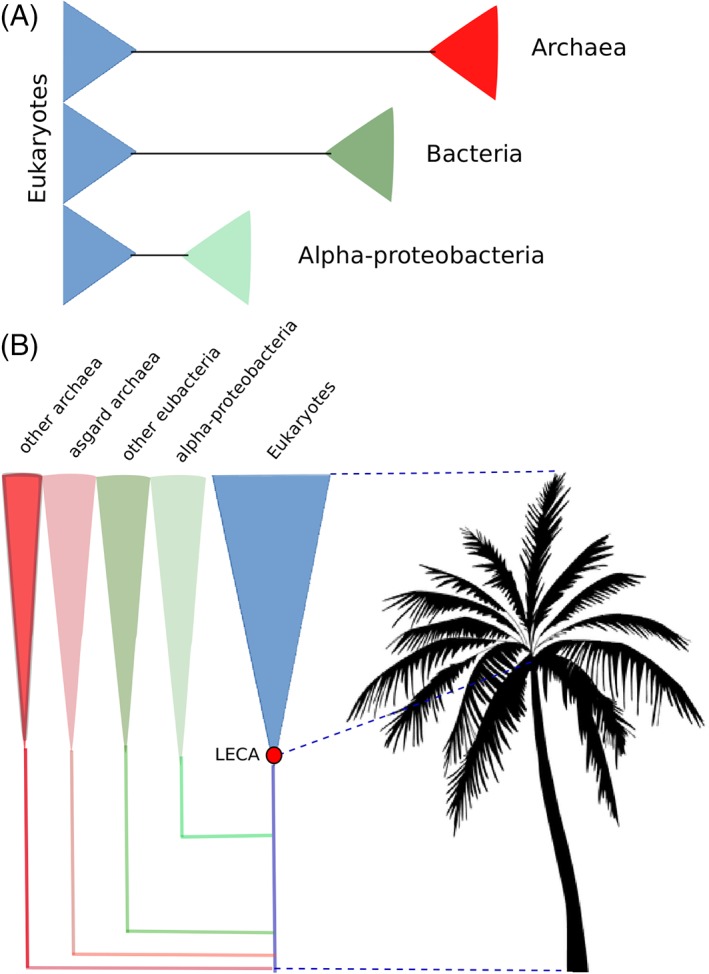
The diversity of stem lengths and the palm tree of eukaryotes. (A) The different nature of the outgroups used to root the tree of eukaryotes results in distinct distances to eukaryotes. Schematic trees depicted differences in branch lengths from the last common ancestor of eukaryotes to the last common ancestor of the used prokaryotic outgroups. Branch lengths have been normalized relative to the distance from LECA to the amoebozoan Dictyostelium discoideum, as measured in Bayesian trees (CAT‐GTR + Γ4) from recent analyses 40, 43. (B) The radiation of the major groups of eukaryotes shortly after LECA and the absence of close pre‐LECA lineages can be idealized with the aspect of a palm tree, where a crown of branches stems out from a single node (LECA), which is subtended by a relative long trunk (right). The length of this stem can vary depending on the subset of proteins used to build the tree (left).
Given the absence of clear fossil records, it is difficult to estimate the amount of time represented by the stem preceding LECA. In most phylogenetic trees, particularly those based on informational genes of archaeal origin 46, 47, the branch separating the eukaryotic homologs from their prokaryotic counterparts is relatively long and comparable in length to branches connecting the most basal eukaryotic nodes with extant sequences (Fig. 1). This indicates that, at least in terms of molecular change (substitutions per site, as the typical unit measure in molecular phylogenies), the stem phase in the evolution toward LECA is not negligible, and only somewhat shorter to the phase spanning since LECA until the extant species. One important observation is that the length of this stem depends on the prokaryotic outgroup that is used in the reconstruction, with archaeal outgroups generally producing the longer stems. Nevertheless, even taking the outgroup producing the shortest stems, alpha‐proteobacteria, this stem length is not negligible, representing, in some trees, about two‐thirds of the length of the distance spanning from LECA to extant eukaryotic species such as the amoebozoan Dictyostelium discoideum.
VARIABLE STEM LENGTHS IN THE TREE OF EUKARYOTES
An important observation regarding the length of the stem subtending LECA is that, as mentioned above, different outgroups provide different lengths relative to intra‐LECA branch lengths (Fig. 1). Branch lengths are represented as number of substitutions per analyzed site, which is the result of both evolutionary rates (how fast sequences change) and time span (how much time the sequences have had to accumulate changes). Different proteins evolve at different rates, and even if one speaks about relative distances, rates are not necessarily kept constant over time. As we will see below, this is one of the central aspects around which the discussions on the information provided by molecular phylogenies are centered. There are several radically different interpretations of the observation that the length of the stem varies depending on the used outgroups. In one interpretation different outgroups provide different lengths because they diverged from eukaryotes at different times, so that the archaeal component of eukaryotes branched out from archaea before the bacterial component of eukaryotes. An alternative interpretation would stress rate differences as the main driver of the stem length. Molecular phylogenies are based on concatenation of a relatively small sample of widespread genes. Hence, one could argue that informational genes such as those encoding ribosomal proteins would have suffered an acceleration of their evolutionary rates before LECA, to then slow down. Or, alternatively, that bacterial derived genes, and particularly alpha‐proteobacterial genes, would have radically accelerated their evolutionary rates after LECA. On the other hand, observed differences in branch length between asgard and non‐asgard archaeal roots or between alpha‐proteobacteria and non‐alphaproteobacteria bacterial roots agree with recently obtained topologies in molecular phylogenies that place alpha‐proteobacteria and asgard as the closest archaeal and bacterial sister groups to the eukaryotic archaeal derived component and to mitochondria, respectively 29, 48. As branching order correlates with stem lengths, this would seem to support the interpretation that divergence time is indeed determining differences in stem lengths, although one could counterargue that differences in rates may have an effect on topologies as well. A third possible interpretation is that biases in sampling of genomes across the different taxonomic group may affect these stem lengths, as one may have been lucky to sample a closer sister group among bacteria than among archaea. This possibility is indeed a real one as shown by the recent discovery of asgard archaea 48, which were revealed as the closest sister group to eukaryotes, and indeed shortened the stem of trees based on archaeal derived proteins (Fig. 1). As genome sequences accumulate, particularly from metagenomics studies, we are more likely to exhaust extant lineages, by having at least a sampled representative. However, we will never be certain of that. In addition, extinct ancient lineages cannot be sampled in molecular studies, and thus the possibility that a closer prokaryotic sister‐clade to eukaryotes went extinct cannot be excluded. All these issues complicate interpretations of observable differences in reconstructed phylogenies and should always be considered. These challenges notwithstanding molecular phylogenies remain a useful piece of information.
What precedes LECA? Most authors agree that LECA does not correspond to the first eukaryote and that it must have been preceded by an evolutionary period that comprises all major changes 22, 25. Somewhere in this period, we could place the first eukaryotic common ancestor (FECA), an idealized predecessor of LECA that would possess just the minimum set of features to be considered a eukaryote. This definition is sufficiently ambiguous to allow for almost any imaginary intermediate. One could, for instance, use the presence of a nucleus as the defining feature of eukaryotes, and thus the origin of this feature would mark the birth of FECA. However, other authors could consider the presence of any membranous intracellular compartment as the hallmark of eukaryotes. For instance, some hypotheses consider that the endosymbiosis of mitochondria marks the origin of eukaryotes and preceded the origin of the nucleus. This implicitly defines FECA as a proto‐mitochondrion‐bearing cell that lacks a nucleus. As this issue is mostly semantical, we will be content with the idea that FECA is a predecessor of LECA that is sufficiently distinct to other prokaryotic lineages and that could be considered already a eukaryote. The period of time between FECA and LECA has been dubbed the stem phase 25. If FECA only displays one or few of the distinctive features of eukaryotes, this stem phase must have comprised the establishment of most major eukaryotic features. The duration of the stem phase and the order in which the major eukaryotic features emerged is unknown.
TOWARD MORE COMPLEX MODELS FOR EUKARYOGENESIS
Aiming to shed some light into the contentious issue of the timing of mitochondrial endosymbiosis, we undertook a phylogenetic analysis of the LECA proteome in which we assessed both the nature of the sister group to eukaryotic protein families and the length of the branch subtending the eukaryotic sequences 36. The analysis of the sister‐clade provided similar heterogeneous taxonomic assignments as previous studies, with only a relatively small fraction of proteins being assigned to alpha‐proteobacteria and even proteobacteria. The most important result consisted in that the lengths of the stems for the different bacterial‐derived proteins in LECA were different depending on the taxonomic nature of the sister‐branch. Importantly, alpha‐proteobacterial derived proteins had significantly shorter stem lengths than proteins of other bacterial origins. Considering that differences were observed independently of the current function of the protein family, we suggested that the differences in stem length result from differences in the time of acquisition of the different families, which implies a later origin of the alpha‐proteobacterial content relative to proteins of other bacterial origins.
As mentioned above, branch lengths result from a composite of evolutionary rates and time, which cannot be disentangled, and this has been a point of discussion regarding what interpretations can result from our observations 4, 49, 50. We observed similar differences when stem lengths were considered as raw values or when they were normalized by the median lengths of the branches going from LECA to the tips represented by extant eukaryotes. This suggests that, overall, variations in rates among families are not determining the observed pattern. Moreover, our results were consistent when protein families were binned by function (e.g., operational or metabolic). As discussed above, rate shifts across time could also explain the observed patterns if they differentially affected genes of different inferred phylogenetic origins. However, in the absence of a specific hypothesis that would explain why genes of proto‐mitochondrial origin and different functions would have been affected in this particular way, I consider that differences in time are the most plausible scenario to explain the observation. Under this assumption, our results would point to a relatively late acquisition of the mitochondrion by a host that possessed a chimeric proteome, already containing proteins of alternative bacterial origins.
Where do these LECA protein families which are assigned to a non‐alphaproteobacterial bacterial origin come from? I have already given some arguments against the idea that all these proteins are the result of horizontal gene transfer (HGT) to the ancestor of mitochondria, their differences in branch lengths provide another one. HGT‐derived proteins from other groups should display a similar or even shorter distance to their sister groups (i.e., the putative donor group), and only convoluted scenarios implying massive losses in the donor group following the acquisition of the gene could explain a longer branch length 36. In contrast, the simpler assumption that these proteins were in the LECA lineage before the arrival of the alpha‐proteobacterial component directly predicts longer stem lengths, which is compatible with the mentioned observations.
Although resisted by some, the idea that the host that engulfed the proto‐mitochondrion was already a chimeric organism with archaeal and bacterial content is compatible with our knowledge of extant prokaryotic communities and symbiotic associations. Firstly, although HGT to the proto‐mitochondrion does not provide an explanation to the overall observed patterns in branch lengths, HGT from bacterial sources to the archaeal host could provide. Indeed, acquisitions of bacterial genes from various sources by the archaeal host occurred before the establishment of the proto‐mitochondrial endosymbiosis would result in this type of phylogenetic footprint: heterogeneous phylogenetic origins, each having different relative branch length distributions. Supporting this scenario is the finding that genomes from asgard archaea seem to display rather high fractions of putatively transferred genes from bacteria 51. It is thus plausible to assume that the archaeal ancestor of LECA was similarly prone to acquire foreign genes. The recurrent acquisition of genes from particular groups of bacteria could have been favored by the establishment of stable ecological associations with particular groups of bacteria, as those existing in extant bacterial–archaeal syntrophic communities 52.
Alternatively, or in addition, the proto‐mitochondrion may have not been the first endosymbiont of this host but only the latest one. The presence of multiple obligate or facultative symbionts within the same host, or the evolutionary replacement of symbionts, has been documented in several extant eukaryotic lineages, particularly in insects 53, 54. Endosymbiosis generally leads to extreme genome reductions and can be considered an evolutionary bottleneck, often resulting in extinction. If the proto‐miochondrion host was able to engulf an alpha‐proteobacterium, it may have as well established earlier endosymbiotic interactions with other unrelated bacteria. The attractiveness of this idea is that it provides a broader temporal framework for a stepwise appearance of the cellular innovations that allowed the establishment of the mitochondrial endosymbiosis (e.g., organellar targeting and import machinery). These earlier endosymbiosis may have favored early developments in the host directed to retain and control the endosymbionts. The earlier endosymbionts would have been victims of the evolutionary ratchet that leads to genome reduction and eventual extinction of the endosybionts, particularly when genes from the endosymbiont performing the key biochemical pathways supporting the symbiotic relationships were transferred to the host genome. The host, already primed to live in association with an endosymbiont, may have established newer symbioses with other organisms until, eventually, the proto‐mitochondrial endosymbiosis would have been established. In contrast to these putative previous endosymbionts, the proto‐mitochondrion was never lost, perhaps because one or some of the key pathways supporting its symbiosis could never be transferred to the host genome (e.g., key subunits of some respiratory complexes) or because some key pathways could not efficiently perform their function outside the endosymbiont compartment (e.g., oxidative phosphorylation or assembly of iron–sulfur clusters).
These two symbiotic scenarios—microbial communities and serial endosymbiotic interactions—serve to explain waves of acquisition of genes from different sources and at different times. Hence, they could explain the observed differences in stem lengths among the bacterial derived component in LECA (Fig. 2). I consider them plausible as they are based in currently existing ecological interactions. I refer to this scenario as the “pre‐mitochondrial symbioses” hypothesis to underscore the fact that multiple symbioses with different partners and perhaps of multiple types (i.e., endo‐ or ecto‐symbiosis) preceded the advent of mitochondrial endosymbiosis. In contrast to the recently proposed pre‐endosymbiont hypothesis 55, which postulates that the host that engulfed the proto‐mitochondrion already possessed an endogenously generated membrane‐bound compartment, the above‐mentioned symbiosis‐based processes do not necessarily require the endogenous development of a compartment to accommodate the symbionts. However, it also does not preclude that possibility. In addition, the proposed evolutionary framework is compatible, although does not necessarily provide any further support, with any hypothesis that explains the origin of any eukaryotic innovation as a response to an endosymbiotic interaction. Rather, it simply opens the possibility that the trigger endosymbiont was a different one than the proto‐mitochondrion. Thus the pre‐mitochondrial symbioses hypothesis opens the door for the possibility of the development of symbiont‐induced cellular structures before the advent of mitochondrial symbiosis. Admittedly, this hypothesis adds complexity to eukaryogenesis scenarios. However, it does so to accommodate the empirically observed complexity in terms of heterogeneous bacterial origins and phylogenetic distances of the reconstructed LECA proteome.
Figure 2.
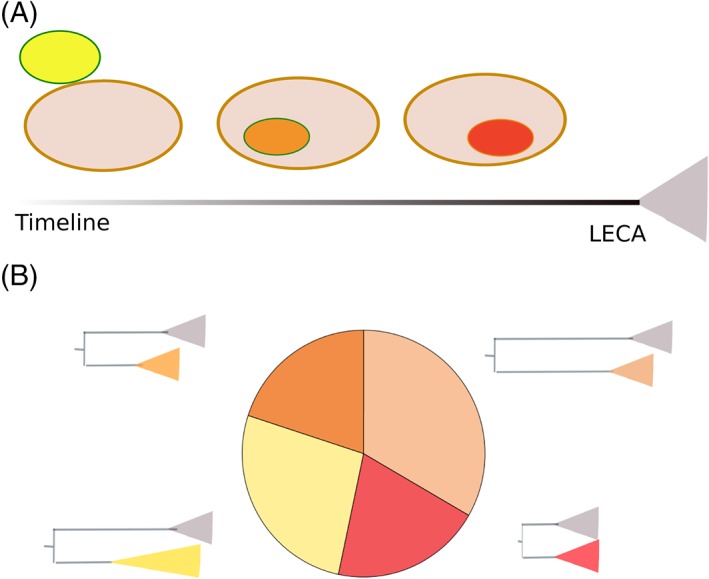
The pre‐mitochondrion symbioses hypothesis. (A) Putative ecto‐ (yellow oval) or endo‐symbioses (orange oval) could have been established by the host (large oval) before the advent of the proto‐mitochondrion (red oval). For simplicity, only one of each of these types of symbioses is represented but the hypothesis is not binding to any particular number of symbiosis or their specific type. (B) If these earlier symbiosis involved symbionts of different phylogenetic origins and they promoted the transfer of genes to the increasingly chimeric host genome, these would result in particular phylogenetic patterns of the LECA proteome in which (i) the phylogenetic nature of the LECA protein would be heterogeneous, containing more than two distinct types of phylogenetic origins and (ii) protein families of different phylogenetic origins would tend to show different stem lengths.
CONCLUDING REMARKS
The origin of eukaryotes remains one of the most difficult questions in evolutionary biology. Given the scarcity of data, most proposed scenarios for the origin of eukaryotes are necessarily highly speculative. Phylogenomic analysis of sequenced genomes of extant species provide one of the few types of empirical data that we can use to support or challenge the different proposed scenarios. In addition, observations of currently existing ecological or evolutionary processes serve us to build possible alternative scenarios that fit the newly obtained empirical data. Phylogenomic analysis of the reconstructed LECA proteomes indicates that the bacterial derived repertoire is heterogeneous, both in terms of the inferred ancestry and the distance to the closest bacterial relatives. Furthermore, these two properties are not independent, with phylogenetic distance to the ancestors being significantly shorter for alpha‐proteobacterial derived proteins, as compared to alternative bacterial origins. To reconcile these observations, we need to go beyond simple models that involve a single bacterial endosymbiont engulfed by an archaeal ancestor. The proposed pre‐mitochondrial symbioses scenarios propose that symbiotic interactions with different partners preceded the mitochondrial endosymbiosis which promoted different waves of acquisition of genetic material. Rather than providing a specific ecological or metabolic scenario, the proposed new framework can be translated to several of the previously proposed scenarios as long as they can contemplate other symbiotic interactions preceding the mitochondrial endosymbiosis.
ACKNOWLEDGEMENTS
TG acknowledges support from the Spanish Ministry of Economy, Industry, and Competitiveness (MEIC) for the EMBL partnership, and grants ‘Centro de Excelencia Severo Ochoa 2013‐2017’ SEV‐2012‐0208, and BFU2015‐67107 cofounded by European Regional Development Fund (ERDF); from the CERCA Programme/Generalitat de Catalunya; from the Catalan Research Agency (AGAUR) SGR857, and grant from the European Union's Horizon 2020 research and innovation programme under the grant agreement ERC‐2016‐724173 the Marie Sklodowska‐Curie grant agreement no. H2020‐MSCA‐ITN‐2014‐642095.
REFERENCES
- 1. Stanier, R. , Doudoroff, M. & Adelberg, E. , 1965. The Microbial World 2nd edn, Upper Saddle River, NJ: Prentice Hall. [Google Scholar]
- 2. Keeling, P. J. , and Fast, N. M. (2002) Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 56, 93–116. [DOI] [PubMed] [Google Scholar]
- 3. Karnkowska, A. , Vacek, V. , Zubáčová, Z. , Treitli, S. C. , Petrželková, R. , et al. (2016) A eukaryote without a mitochondrial organelle. Curr. Biol. 26(10), 1274–1284. [DOI] [PubMed] [Google Scholar]
- 4. Roger, A. J. , Muñoz‐Gómez, S. A. , and Kamikawa, R. (2017) The origin and diversification of mitochondria. Curr. Biol. 27(21), R1177–R1192. [DOI] [PubMed] [Google Scholar]
- 5. Fuerst, J. A. , and Sagulenko, E. (2011) Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9(6), 403–413. [DOI] [PubMed] [Google Scholar]
- 6. Boedeker, C. , Schüler, M. , Reintjes, G. , Jeske, O. , van Teeseling, M. C. F. , et al. (2017) Determining the bacterial cell biology of planctomycetes. Nat. Commun. 8, 14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Archibal, J. (2011) One Plus One Equals One: Symbiosis and the Evolution of Complex Life. Report of the National Center for Science Education, (November 1859). pp. 1–3, Oxford University Press, Oxford. [Google Scholar]
- 8. Sagan, L. (1967) On the origin of mitosing cells. J. Theoret. Biol. 14(3), 255–274. [DOI] [PubMed] [Google Scholar]
- 9. Martin, W. F. , Garg, S. , and Zimorski, V. (2015) Endosymbiotic theories for eukaryote origin. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 370(1678), 20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gabaldón, T. , and Pittis, A. A. (2015) Origin and evolution of metabolic sub‐cellular compartmentalization in eukaryotes. Biochimie 119, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gray, M. W. (2017) Lynn Margulis and the endosymbiont hypothesis: 50 years later. Mol. Biol. Cell 28(10), 1285–1287. Available at: http://www.molbiolcell.org/lookup/doi/10.1091/mbc.E16-07-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin, W. F. (2017) Physiology, anaerobes, and the origin of mitosing cells 50 years on. J. Theoret. Biol. 434, 2–10. [DOI] [PubMed] [Google Scholar]
- 13. de Duve, C. (1969) Evolution of the peroxisome. Annals N. Y. Acad. Sci. 168(2), 369–381. [DOI] [PubMed] [Google Scholar]
- 14. De Duve, C. (2007) The origin of eukaryotes: a reappraisal. Nat. Rev. Genet. 8(5), 395–403. [DOI] [PubMed] [Google Scholar]
- 15. Gabaldón, T. , and Capella‐Gutiérrez, S. (2010) Lack of phylogenetic support for a supposed actinobacterial origin of peroxisomes. Gene 465(1–2), 61–65. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20600706. [DOI] [PubMed] [Google Scholar]
- 16. Gabaldón, T. , Snel, B. , Zimmeren, F. . , Hemrika, W. , Tabak, H. , et al. (2006) Origin and evolution of the peroxisomal proteome. Biol. Direct 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schlüter, A. , et al. (2006) The evolutionary origin of peroxisomes: an ER‐peroxisome connection. Mol. Biol. Evol. 23(4), 838–845. [DOI] [PubMed] [Google Scholar]
- 18. Gabaldón, T. (2014a) A metabolic scenario for the evolutionary origin of peroxisomes from the endomembranous system. Cell. Mol. Life Sci. 71(13), 2373–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gabaldón, T. (2014b) Evolutionary considerations on the origin of peroxisomes from the endoplasmic reticulum, and their relationships with mitochondria. Cell. Mol. Life Sci. 71(13), 2379–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Speijer, D. (2017) Evolution of peroxisomes illustrates symbiogenesis. Bio Essays 39(9), 1700050. [DOI] [PubMed] [Google Scholar]
- 21. Friedman, J. R. , and Nunnari, J. (2014) Mitochondrial form and function. Nature 505, 335 Available at: 10.1038/nature12985–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koumandou, V. L. , Wickstead, B. , Ginger, M. L. , van der Giezen, M. , Dacks, J. B. , et al. (2013) Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit. Rev. Biochem. Mol. Biol. 48(4), 373–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray, M. W. , Burger, G. , and Lang, B. F. (1999) Mitochondrial evolution. Science 283(5407), 1476–1481. [DOI] [PubMed] [Google Scholar]
- 24. Poole, A. M. , and Gribaldo, S. (2014) Eukaryotic origins: how and when was the mitochondrion acquired? Cold Spring Harbor Perspect. Biol. 6(12), a015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koonin, E. V. (2010) The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11(5), 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cavalier‐Smith, T. (1987) Eukaryotes with no mitochondria. Nature 326(6111), 332–333. [DOI] [PubMed] [Google Scholar]
- 27. Embley, T. M. , and Martin, W. (2006) Eukaryotic evolution, changes and challenges. Nature 440(7084), 623–630. [DOI] [PubMed] [Google Scholar]
- 28. Martin, W. , and Müller, M. (1998) The hydrogen hypothesis for the first eukaryote. Nature 392(6671), 37–41. [DOI] [PubMed] [Google Scholar]
- 29. Martijn, J. , Vosseberg, J. , Guy, L. , Offre, P. , and Ettema, T. J. G. (2018) Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557(7703), 101–105. [DOI] [PubMed] [Google Scholar]
- 30. Gabaldón, T. , and Huynen, M. A. (2003) Reconstruction of the proto‐mitochondrial metabolism. Science 301(5633), 609. [DOI] [PubMed] [Google Scholar]
- 31. Gabaldón, T. , and Huynen, M. A. (2007) From endosymbiont to host‐controlled organelle: the hijacking of mitochondrial protein synthesis and metabolism. PLoS Comput. Biol. 3(11), 2209–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurland, C. , and Andersson, S. (2000) Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64(4), 786–820. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=11104819%5Cnpapers2://publication/uuid/706B4447-6118-4CE9-BBEE-B9EA7D29FB4F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gabaldón, T. , and Huynen, M. A. (2004) Shaping the mitochondrial proteome. Biochim. Biophys. Acta Bioenerget. 1659, 212–220. [DOI] [PubMed] [Google Scholar]
- 34. Gabaldón, T. , and Huynen, M. A. (2008) Reconstruction of ancestral proteomes In Ancestral Sequence Reconstruction. Oxford University Press, Oxford. [Google Scholar]
- 35. Lane, C. E. (2007) Bacterial endosymbionts: genome reduction in a hot spot. Curr. Biol. 17(13), R508–R510. [DOI] [PubMed] [Google Scholar]
- 36. Pittis, A. A. , and Gabaldón, T. (2016) Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531(7592), 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rochette, N. C. , Brochier‐Armanet, C. , and Gouy, M. (2014) Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes. Mol. Biol. Evol. 31(4), 832–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Burki, F. (2014) The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harbor Perspect. Biol. 6(5), a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Derelle, R. , and Lang, B. F. (2012) Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol. Biol. Evol. 29(4), 1277–1289. [DOI] [PubMed] [Google Scholar]
- 40. Derelle, R. , Torruella, G. , Klimeš, V. , Brinkmann, H. , Kim, E. , et al. (2015) Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl. Acad. Sci. 112(7), E693–E699. Available at: http://www.pnas.org/lookup/doi/10.1073/pnas.1420657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He, D. , Fiz‐Palacios, O. , Fu, C. J. , Fehling, J. , Tsai, C. C. , et al. (2014) An alternative root for the eukaryote tree of life. Curr. Biol. 24(4), 465–470. [DOI] [PubMed] [Google Scholar]
- 42. Simpson, A. G. B. , and Roger, A. J. (2002) Eukaryotic evolution: getting to the root of the problem. Curr. Biol. 12(20), R691–R693. [DOI] [PubMed] [Google Scholar]
- 43. Raymann, K. , Brochier‐Armanet, C. , and Gribaldo, S. (2015) The two‐domain tree of life is linked to a new root for the archaea. Proc. Natl. Acad. Sci. 112, 6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jarvis, E. D. , Mirarab, S. , Aberer, A. J. , Li, B. , Houde, P. , et al. (2014) Whole‐genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215), 1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Green, R. E. , Braun, E. L. , Armstrong, J. , Earl, D. , Nguyen, N. , et al. (2014) Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346(6215), 1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hug, L. A. , Baker, B. J. , Anantharaman, K. , Brown, C. T. , Probst, A. J. , et al. (2016) A new view of the tree of life. Nat. Microbiol. 1(5), 1254449. [DOI] [PubMed] [Google Scholar]
- 47. Williams, T. A. , Foster, P. G. , Nye, T. M. W. , Cox, C. J. , and Embley, T. M. (2012) A congruent phylogenomic signal places eukaryotes within the Archaea. Proc. R. Soc. B Biolog. Sci. 279(1749), 4870–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zaremba‐Niedzwiedzka, K. , Caceres, E. F. , Saw, J. H. , Bäckström, D. , Juzokaite, L. , et al. (2017) Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541(7637), 353–358. [DOI] [PubMed] [Google Scholar]
- 49. Martin, W. F. , Roettger, M. , Ku, C. , Garg, S. G. , Nelson‐Sathi, S. , et al. (2017) Late mitochondrial origin is an artifact. Genome Biol. Evol. 9(2), 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pittis, A. A. , and Gabaldon, T. (2016) On phylogenetic branch lengths distribution and the late acquistion of mitochondria. bioRxiv. 10.1101/064873. [DOI] [Google Scholar]
- 51. Spang, A. , Saw, J. H. , Jørgensen, S. L. , Zaremba‐Niedzwiedzka, K. , Martijn, J. , et al. (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521(7551), 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kouzuma, A. , Kato, S. , and Watanabe, K. (2015) Microbial interspecies interactions: recent findings in syntrophic consortia. Front. Microbiol. 6, (MAY), 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koga, R. , Bennett, G. M. , Cryan, J. R. , and Moran, N. A. (2013) Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ. Microbiol. 15(7), 2073–2081. [DOI] [PubMed] [Google Scholar]
- 54. Smith, W. A. , Oakeson, K. F. , Johnson, K. P. , Reed, D. L. , Carter, T. , et al. (2013) Phylogenetic analysis of symbionts in feather‐feeding lice of the genus Columbicola: evidence for repeated symbiont replacements. BMC Evol. Biol. 13(1), 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gray, M. W. (2014) The pre‐endosymbiont hypothesis: a new perspective on the origin and evolution of mitochondria. Cold Spring Harbor Perspect. Biol. 6(3), a016097. [DOI] [PMC free article] [PubMed] [Google Scholar]


