Abstract
Insects are unable to synthesize essential amino acids (EAAs) de novo, thus rely on dietary or symbiotic sources for them. Wood is a poor resource of nitrogen in general, and EAAs in particular. In this study, we investigated whether gut microbiota of the Asian longhorned beetle, Anoplophora glabripennis (Motschulsky), a cerambycid that feeds in the heartwood of healthy host trees, serve as sources of EAAs to their host under different dietary conditions. δ 13 C-stable isotope analyses revealed significant δ 13 C-enrichment (3.4 ± 0.1‰; mean ± SEM) across five EAAs in wood-fed larvae relative to their woody diet. δ 13 C values for the consumers greater than 1‰ indicate significant contributions from non-dietary EAA sources (symbionts in this case). In contrast, δ 13 C-enrichment of artificial diet-fed larvae (controls) relative to their food source was markedly less (1.7 ± 0.1‰) than was observed in wood-fed larvae, yet still exceeded the threshold of 1‰. A predictive model based on δ 13 C EAA signatures of five EAAs from representative bacterial, fungal, and plant samples identified symbiotic bacteria and fungi as the likely supplementary sources of EAA in wood-fed larvae. Using the same model, but with an artificial diet as the dietary source, we identified minor supplementary bacterial sources of EAA in artificial diet-fed larvae. This study highlights how microbes associated with A. glabripennis can serve as a source of EAAs when fed on nutrient-limited diets, potentially circumventing the dietary limitations of feeding on woody substrates.
Keywords: Anoplophora glabripennis, δ 13 C fingerprint , gut microbiota, xylophagy, EAA
Acquiring sufficient protein and essential amino acids (EAAs) from suboptimal diets is a major nutritional obstacle for terrestrial herbivores, especially xylophagous (wood-feeding) insects, owing to the low nutritional content of their diets and the importance of EAAs for growth and development ( Mattson 1980 ). As insects cannot synthesize these amino acids de novo, they are proposed to overcome this limitation by utilizing gut microbial sources ( Dillon and Dillon 2004 ). Although EAA provisioning has been a suggested function of many insect gut microbiota ( Douglas 2009 ), it remains a relatively underexplored line of inquiry. Advanced molecular and genomic approaches have identified amino acid provisioning pathways of multiple obligate endosymbionts in a variety of insects, providing support for their ability to provision their hosts with EAA. For example, the obligate endosymbionts of pea aphids ( MacDonald et al. 2012 ), sharpshooters ( Wu et al. 2006 ), and cockroaches ( Sabree et al. 2009 , Tokuda et al. 2013 ) and the bacterial endosymbionts of obligate termite-associated protists ( Hongoh et al. 2008 ), as well as gut microbiome of Anoplophora glabripennis ( Scully et al. 2014 ), have been shown to possess EAA biosynthetic pathways that purportedly supplement their insect host's nutritional requirements.
Some challenges inherent in investigating EAA provisioning by insect gut microbes can largely be ascribed to the difficulties in the nature, design, and execution of these types of experiments ( Douglas 2013 , Engel and Moran 2013 ). The diversity and plasticity of facultative microbial members of many insect gut communities constitute another obstacle to assigning EAA provisioning functions, as specific members in complex communities are difficult to identify as being responsible for a given proposed function ( Engel and Moran 2013 ). It is possible, however, to overcome some of these limitations in delineating symbiotic EAA provisioning if these functions are viewed as being provided by a “gut microbiome” (including bacteria and fungi) as opposed to the function of specific member(s) of the community, particularly as insects may derive EAA from their gut microbiota via direct assimilation or incorporation following indiscriminate digestion of these microbes ( Douglas 2009 ).
One approach to investigating microbial origins of EAA is by means of stable isotope analysis. This approach has been used to confirm insect gut microbial functions such as nitrogen fixation in the Eastern subterranean termite ( Reticulitermes flavipes (Kollar); Bentley 1984 ), leaf-cutter ants ( Atta sp. (F.)), and Acromyrmex sp. (Mayr; Pinto-Tomas et al. 2009 ), as well as nitrogenous waste recycling by gut microbes in the cerambycid A. glabripennis (Motschulsky; Ayayee et al. 2014 ) and the pine bark beetle, Dendroctonus valens (LeConte; Morales-Jimenez et al. 2013 ). A particularly promising stable isotope technique for investigating gut microbial EAA provisioning in insects is stable carbon isotope fingerprinting of amino acids, which is based on how bacteria, fungi, and plants fractionate carbon differentially during EAA biosynthesis, resulting in unique and distinct 13 C/ 12 C ratios in the EAAs (δ 13 C EAA ; Larsen et al. 2009 ). This approach has subsequently been used in several other ecological studies ( Larsen et al. 2011 , Larsen et al. 2013 , Vokhshoori, McCarthy & Larsen 2014 ). The δ-notation in the term δ 13 C EAA above is defined as [(R sample EAA /R standard EAA )-1] × 1,000 (per mille, ‰), where R is the ratio of heavy ( 13 C) to light ( 12 C) isotopes in a particular EAA of the sample R sample EAA and the standard R standard EAA ( Coleman and Fry 1991 ).
As the gut microbial community of insects is generally composed of bacteria and/or fungi ( Dillon and Dillon 2004 , Engel and Moran 2013 ), contributions of EAAs from gut microbes can be determined relative to EAAs from an insect’s diet using the δ 13 C EAA fingerprinting approach. The hypothesis behind this approach is that larval δ 13 C EAA values will match their dietary sources closely with little δ 13 C EAA fractionation when these amino acids are incorporated directly from the diet ( McMahon et al. 2010 , Newsome et al. 2011 ). In contrast, larval δ 13 C EAA values are not expected to match dietary δ 13 C EAA values when the host is supplemented with de novo synthesized EAAs by microbial symbionts. We can determine whether a consumer directly sources EAAs from its diet or from alternate sources by quantifying δ 13 C isotopic differences between the consumer and the diet for the amino acids of interest. This difference/discrimination (Δ 13 C) between consumer and dietary δ 13 C EAA is calculated as: δ 13 C Consumer EAA − δ 13 C Dietary EAA . Any significant Δ 13 C deviation beyond an analytical error of 1‰ implies alternate or additional sources of EAAs apart from dietary sources ( Newsome et al. 2011 ). Discrimination factors (Δ 13 C or Δ 15 N), however, are known to exhibit considerable variation, and are dependent on the diet of a consumer, as well as the particular consumer tissue examined ( Caut et al. 2008 ), and serve to provide important metrics in trophic ecology studies.
In this study, we investigated EAA provisioning by the gut microbiota of A. glabripennis , a xylophagous (wood-feeding) beetle in the family Cerambycidae. A. glabripennis subsists on a woody diet with limited bioavailable nitrogen by harboring a diverse and relatively plastic gut microbial community consisting of bacteria, yeasts, and a soft-rot fungus Fusarium solani species complex 6 (FSSC 6; Geib et al. 2012 , Scully et al. 2012 ). Newly emerged A. glabripennis females feed on host trees for about two weeks, during which time they mature sexually and develop an egg load prior to mating ( Haack et al. 2010 ); fertilized eggs, which are laid individually under the bark of trees, usually hatch in about 14 d. The time to complete development from larva to adult ranges from one to two years, depending on environmental temperature, with the insect overwintering as a larva ( Haack et al. 2010 ). A. glabripennis larvae may be particularly susceptible to nutritional limitations because they develop deep in the wood of apparently healthy trees (not predigested by wood-decay fungi). The larvae are thus hypothesized to require microbial contributions to overcome EAA limitations in their dietary substrate.
The gut microbiota of A. glabripennis has previously been demonstrated to contribute to host nitrogen economy and ecology through biological nitrogen fixation and urea recycling using bulk tissue amino acid δ 15 N analyses ( Ayayee et al. 2014 ). Recycled nitrogen ( 15 N) was routed into EAAs and non-EAAs in insect biomass. The incorporation of fixed and recycled 15 N into EAA provided empirical evidence of microbial provisioning and insect assimilation of provisioned nitrogen; however, these results lacked verification of the biosynthetic origin or source of the EAA carbon skeletons in the larvae. The microbial community of A. glabripennis has a broad range of metabolic capabilities, including degradation of plant cells walls, detoxification of xenobiotics, and synthesis of EAAs, all of potential use to the insect host ( Scully et al. 2013a , 2014 ).
Given the limited availability of EAA in the beetles' host trees, we hypothesized that A. glabripennis larvae obtain supplementary EAA from symbiotic sources (bacteria and fungi) during their development on Acer spp. (maple) host trees. We also hypothesized that symbiotic EAA supplementation would differ (be lower) on a nutritionally optimal artificial diet (control), compared with supplementation in larvae extracted from naturally infested host trees.
Materials and Methods
Sources of Insects
A. glabripennis: red maple feeding group
A. glabripennis larvae feeding in naturally infested red maple ( Acer rubrum L . ) were collected from the USDA-regulated infestation site in Bethel, OH, in 2013. Third to fourth instars were removed from the host tree and prepared for analysis (referred to hereafter as wood-fed larvae). Bark and wood from these red maple logs were also sampled.
A. glabripennis: artificial diet feeding group
A. glabripennis larvae used in this experiment were removed from Norway maple logs ( Acer platanoides L . ) as neonates and placed on a nutritionally optimal artificial diet for 2 mo before being used in the experiment; colony rearing of this species in quarantine routinely begins with oviposition into logs to obtain neonates that are then transferred to an artificial diet to complete development ( Keena 2005 ). The artificial diet contains casein (25%) and torula yeast (75%) as the main protein sources, as well as the food preservative agents sodium propionate, sorbic acid, and methyl paraben (the latter two act as antifungal agents). Sterility of the artificial diet was ensured by autoclaving and by UV surface exposure for 2–3 h.
Samples from the Ascomycota fungus F. solani , which is consistently found associated with wood-feeding A. glabripennis, were cultured and submitted for 13 C-stable isotope analysis. Briefly, two carboxymethylcellulose agar plates (CMC; Geib et al. 2009 ) were inoculated using a frozen, pure culture of F. solani (ATCC MYA 4552) that had been grown on infested barley. Samples from these plates were subjected to PCR followed by sequencing of ITS2 amplicons to confirm the identity of the fungus (data not shown). Two F. solani hyphal samples were obtained by scraping with a sterilized spatula from the surface of the CMC plates, making sure to collect only fungal materials, and put into 1.5-ml eppendorf tubes. The samples were similarly prepared and sent for analysis at the University of California, Davis Stable Isotope Facility. To account for potential inter-lab differences, we quantified δ 13 C EAA from single bacterial ( Burkholderia xenovorans) , fungal ( Aureobasidium pullulans ), and plant ( Quercus robur) reference samples present in the Larsen data set at the UC Davis Stable Isotope Facility, under the same conditions as the samples described in this study. The offset values following calibration were applied to the original Larsen data set prior to linear discriminant function analysis (LDA).
Sample Preparation and Amino Acid Stable Isotope Analysis (δ 13 C AA )
A. glabripennis larvae were surface-sterilized by rinsing once in 10% Coverage Plus (Steris Corporation, Mentor, OH) and twice in deionized water. Whole larval guts were removed and discarded to ensure that microbial δ 13 C EAA were eliminated from the quantification of δ 13 C EAA from host tissues. Larval carcasses were subsequently lyophilized for 72 h. Bark and wood samples from host trees were milled and lyophilized as well for 72 h. All insects, wood and artificial diet samples were ball-ground and sent to the Stable Isotope Facility at UC Davis for sample preparation and EAA δ 13 C analysis. To determine if there were any host tree-specific differences in EAA provisioning, wood and bark samples from silver maple ( Acer saccharinum L), another suitable host tree for this beetle, were collected and prepared in the same manner as the red maple samples.
For δ 13 C AA analysis, the samples were acid-hydrolyzed and derivatized in a solution of methanol (Thermo Fisher Scientific, Waltham, MA, USA), pyridine, and methyl chloroformate (Sigma Aldrich, St. Louis, MO, USA) using a one-step rapid derivatization method ( Walsh et al. 2014 ). Approximately 0.35-µl aliquots of derivatized samples were injected into a splitless liner at 250°C with a helium flow rate of 2.8 ml/min. Conditions and optimization during derivatization and analysis were performed as per protocol ( Chen et al. 2010 , Walsh et al. 2014 ). Compound-specific isotope 13 C-amino acid analysis (δ 13 C EAA ) was performed using the trace gas chromatograph (Thermo Electron, Bremen Germany) coupled to a Delta V Advantage isotope ratio mass spectrometer via the GC Combustion Interface III (Thermo Electron, Bremen, Germany) using the high polar VF-23ms column (Agilent Technologies). Combustion and reduction furnace temperatures were 950 and 650°C, respectively.
For each sample, the 13 C/ 12 C ratio was determined and calibrated to the international reference standard, Vienna Pee Dee Belemnite scale (V-PDB), and expressed as the δ−notation (‰) δ 13 C EAA , as shown above. We prepared and analyzed each biological sample in duplicate and corrected for the addition of carbon during derivatization after analysis ( Walsh et al. 2014 ). This correction was based on the number of moles of carbon in the derivatized amino acid of the sample of interest and an empirically determined correction factor based on the amino acid of derivatized and underivatized reference amino acid samples. The carbon-corrected δ 13 C EAA values of the EAAs leucine (Leu), isoleucine (Ile), lysine (Lys), phenylalanine (Phe), and valine (Val) are shown in Supplementary Data (online only). The inter-lab calibrations carried out on the EAAs from the additional bacterial, fungal, and plant samples are shown in Supplementary Data (online only).
Statistical Analyses and Model Generation
Mixed model analysis using JMP 10 (SAS Inc. NC, USA) was performed with amino acid, treatment group, and amino acid × group interactions as independent variables, and raw δ 13 C EAA values of the five EAAs measured in this study as the dependent variable. Treatment groups included the larvae and the diets they were fed ( n = 4). Analyses were carried out separately for the A. glabripennis : red maple and the A. glabripennis : artificial diet feeding groups. Means separations were performed using Tukey’s HSD on the δ 13 C EAA values. Individual amino acid discriminations (Δ 13 C EAA ; δ 13 C-enrichments or depletions) in larvae relative to their diets were determined as Δ 13 C EAAx = (δ 13 C LarvalEAAx – δ 13C Diet EAAx ), where EAAx is a given EAA.
LDA was used to create a classification model based on δ 13 C EAA values from a training data set consisting of three classifiers, fungi, bacteria, and host tree ( A. rubrum and A. saccharinum ) samples, to predict biosynthetic origins of larval EAA ( Supplementary Data [online only]). Reference bacterial and fungal data were obtained from a previously published data set ( Larsen et al. 2013 ). The LDAs were run using R package MASS (R version 3.03; http://www.R-project.org ).
Ethic approval
All applicable institutional and/or national guidelines for the care and use of animals were followed.
Results
A. glabripennis Larvae Are 13 C-Enriched Relative to Their Diets
There were no significant differences in δ 13 C EAA values between red maple bark and red maple wood, thus these data were pooled, and are hereafter referred to as red maple. In the A. glabripennis larvae: red maple feeding group, the mean δ 13 C EAA of wood-fed larvae across all five EAAs (−25.4 ± 0.4‰; mean ± SE) was significantly different from that of the red maple host tree δ 13 C EAA (–28.9 ± 0.3‰; F = 48; df = 1, 68; P < 0.0001; Table 1 ). There was also significant 13 C-enrichment in wood-fed larvae (Δ 13 C EAA = δ 13 C Larval EAA – δ 13 C Red maple EAA ) of 3.4 ± 0.07‰ relative to red maple ( Table 1 ), which indicates the presence of non-host tree sources of EAA. Of the five EAAs examined in wood-fed larvae, leucine and lysine were the most 13 C-enriched relative to the red maple host tree (Δ 13 C EAA ), followed by phenylalanine and valine, with isoleucine being the least 13 C-enriched ( Fig. 1 A).
Table 1.
Means comparisons of five δ 13 C EAA (ile, leu, phe, lys, and val) for wood-fed larvae and host tree in the A. glabripennis larvae: red maple feeding group, and for artificial diet-fed larvae and artificial diet in the A. glabripennis larvae: artificial diet feeding group
| A. glabripennis larvae: red maple experiment | δ 13 C EAA (±SE) (‰) |
|---|---|
| Red maple | − 28.9 ± 0.3a |
| Wood-fed larvae | −25.4 ± 0.4b |
| F (1,68) = 48, P < 0.0001 | |
| A. glabripennis larvae: artificial diet experiment | δ 13 C EAA (±SE) (‰) |
| Artificial diet | −26.8 ± 0.2a |
| Artificial diet-fed larvae | −25.1 ± 0.1b |
| F (1,34) = 78, P < 0.0001 |
Shown are mean values for three replicates per larval group, four replicates for red maple tree (bark + wood), and one replicate for the artificial diet control. Different letters represent a statistically significant difference at P < 0.05 within each feeding group.
Fig. 1.
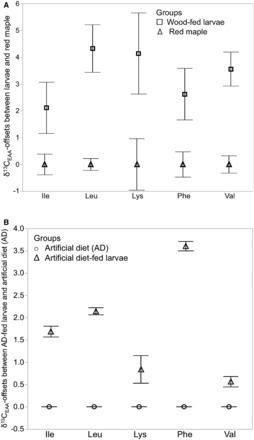
(A) δ 13 C discrimination (Δ 13 C EAA ; enrichment or depletion) of essential amino acids in wood-fed larvae normalized to essential amino acids in the red maple host tree; Δ 13 C EAA = (δ 13 C wood-fed larvae EAA – δ 13 C red maple EAA ). F(1,68) = 48.04, P < 0.0001. Shown are mean values for three replicates per larval group and four replicates for host tree. Error bars represent standard errors of the mean. (B) δ 13 C discrimination (Δ 13 C EAA enrichment or depletion) of essential amino acids (Δ 13 C EAA ) in artificial diet-fed larvae normalized to the artificial diet essential amino acids; Δ 13 C EAA = (δ 13 C larvae EAA – δ 13 C Artificial diet EAA ). F(10, 77) = 12, P < 0.0001. Shown are mean values for three replicates per larval group and one replicate for the artificial diet food source.
In the A. glabripennis larvae: artificial diet feeding group, the mean δ 13 C EAA of artificial diet-fed larvae across all five EAAs (−25.1 ± 0.1‰) was significantly different from that of the artificial diet (−26.8 ± 0.2‰; F = 78; df = 1, 34; P < 0.0001; Table 1 ). On average, artificial diet-fed larvae were minimally Δ 13 C-enriched by 1.7 ± 0.12‰ relative to the artificial diet itself ( Table 1 ). Individual EAA enrichments in artificial diet-fed larvae were comparatively lower than those in wood-fed larvae relative to their respective diets ( Fig. 1 B).
Validation of Predictive Model
We used LDA to determine which δ 13 C EAA values discriminate between bacteria, fungi, and host tree. To assess classification of the training data samples, we used jacknifed (i.e., leave one out) predictions ( Larsen et al. 2009 ). The 95% confidence limits for each group are depicted as ellipses and the decision boundaries between the groups as lines ( Fig. 2 ). After establishing the discrimination model, we then predicted posterior probabilities, i.e., the probability that a particular sample belonged to one or another of the three groups. The greater the distance of a particular consumer from the centroid of a classification group, i.e., potential EAA source, the greater the probability that mixing of EAA sources occurred. Given the distinct discrimination scores between the classification groups, we interpreted discriminant scores of consumers falling outside the 95% confidence limits of their food sources as a strong indication of microbial EAA supplementation. All three classifiers (bacteria, fungi, and host tree) were correctly assigned to their respective groups by the training model ( Supplementary Data [online only]). In addition, in the predictive model, the two F. solani samples were correctly classified as fungi using bacteria, fungi, and Acer spp. as the classifiers, indicating the validity of the model for subsequent use in determining group assignment of larval δ 13 C EAA data.
Fig. 2.
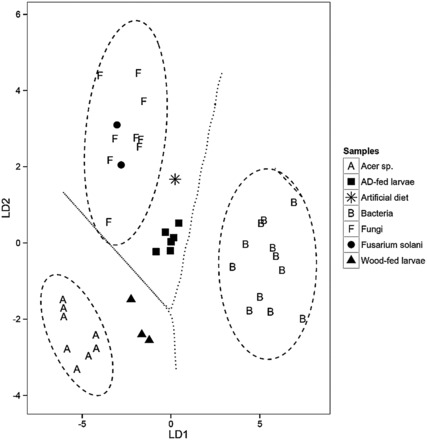
A linear discriminant function analysis (LDA) using δ 13 C EAA from (A) wood-fed and artificial diet-fed larvae with three classifier groups: maple trees ( Acer sp .; n = 8), fungi ( n = 9), and bacteria ( n = 12), and four predictor groups: A. glabripennis wood-fed larvae ( n = 3), A. glabripennis artificial diet-fed larvae (AD-fed larvae; n = 6), the artificial diet food source (AD; n = 1), and the fungal symbiont F. solani ( n = 2). Classification along LD1 (83.5%) accounts for the separation between Acer sp. , bacteria, and fungi. The classification along LD2 (16.5%) accounts for the separation between Acer sp. and fungi (Wilks’ Lambda = 0.01, F = 49.2, P < 0.0001). Letters indicate classifiers groups, while shapes indicate consumers. The ellipses signify the 95% confidence limits for each classifier group, and the dashed lines between classifiers signify the decision boundaries for each classifier group. The essential amino acids used in both plots were isoleucine, leucine, lysine, phenylalanine, and valine.
Wood-Fed Larvae Derive EAA From Fungal and Bacterial Associates
All wood-fed larval samples fell outside the 95% confidence limit of their woody diets, Acer spp. ( Fig. 2 ; Wilks’ lambda = 0.01, F = 49.2, P < 0.0001). Based on the classification criterion (outside the 95% confidence limit of) and, on the offset between these three wood-feed larval samples and the wood diet, fungal and bacterial sources of EAA were suggested as the most likely sources ( Fig. 2 ).
Artificial Diet-Fed Larvae Derive EAA From Gut Bacteria
Maple EAAs were used in the predictive model for the artificial diet-fed group because the larvae were collected from Norway maple logs as neonates prior to being placed on the artificial diet, which is standard operating procedure for colony rearing of this species. We assessed potential carryover of plant δ 13 C signatures following assimilation of host plant-derived EAAs during the initial growth period in the host trees by using host plants ( Acer spp.) in the supervised model. There was no evidence of host plant contributions to larval EAA under these conditions, suggesting complete turnover of any host tree-derived EAAs. In the predictive model, the artificial diet and artificial diet-fed larval samples were outside the 95% confidence limit of the fungal classifier ( Fig. 2 ; Wilks’ lambda = 0.01, F = 49.2, P < 0.0001). The offset of the larval samples from the fungal classifier as well as the artificial diet classifier indicated that the EAA input was primarily bacterial ( Fig. 2 ).
Discussion
Diet quality is known to affect gut microbial composition of insects ( Colman et al. 2012 ), including A. glabripennis ( Geib et al. 2009 ), and in turn influence utilization of microbiota as sources of EAAs. Our results supports the hypothesis that gut microbiota (mixture of bacteria and fungi) harbored by A. glabripennis are responsible for provisioning EAA to the host insect. The positive mean EAA discriminations (Δ 13 C) of 3.4‰ for wood-fed larvae ( Fig. 1 A) and 1.7‰ for the artificial diet-fed larvae relative to their diets were the first indications in this study of larval utilization of non-dietary sources of EAAs, albeit the lower non-dietary contribution of EAA in artificial diet-fed larvae compared with that of tree-fed larvae. These positive discriminations deviate from the expected 1‰ ( Newsome et al. 2011 ) that would be expected if EAA inputs were solely from dietary sources.
Larval A. glabripennis are known to harbor a diverse gut bacterial community as well as several fungi and yeasts with varied metabolic capacities ( Geib et al. 2009 , Scully et al. 2014 ). Consistent with the results reported herein, previous studies implicated provisioning of EAA by the gut microbiome in this system. A. glabripennis gut transcriptomic studies uncovered the presence of complete bacterial and fungal biosynthetic pathways for several EAAs in the A. glabripennis microbiome, as well as complementary insect- and host-derived biosynthetic pathways for the synthesis of non-EAAs using by-products from cellulose degradation ( Scully et al. 2014 ); no evidence of insect-derived genes or transcripts for biosynthesis of EAAs were detected ( Scully et al. 2013a , b ). Other studies detected the presence of fungal and bacterial transcripts predicted to encode ABC transporters, which may be involved in the displacement of EAAs into the gut lumen from microbial cells ( Scully et al. 2014 ), as well as A. glabripennis -derived transcripts encoding protein transporters ( Scully et al. 2013b ), which may be involved in EAA uptake from the gut lumen through a cation co-transport mechanism, which is a mechanism whereby mammalian and insect epithelial cells take up EAAs ( Castagna et al. 1997 ).
Some A. glabripennis microbiome members play key roles in lignin ( Geib et al. 2008 ) and cellulose ( Geib et al. 2009 ) degradation, making available carbon moieties, for metabolic processes, such as EAA biosynthesis. Detection of 15 N-labeled metabolites in larval biomass following microbial 15 N 2 gas fixation and 15 N-urea hydrolysis indicated that A. glabripennis larvae benefit from these processes via gut-associated bacteria ( Ayayee et al. 2014 ). In addition, the recycled 15 N stable isotopes were found to be routed into both EAAs and non-EAAs in larval biomass ( Ayayee et al. 2014 ).
In this study, to determine the origins of non-dietary EAAs, we carried out supervised predictive modeling using previously reported δ 13 C EAA data from bacteria and fungi ( Supplementary Data ; Larsen et al. 2013 ), and dietary sources ( Acer spp. or artificial diet). The various biosynthetic processes leading to the synthesis of EAAs in fungi, bacteria, and plants generate distinct signatures ( Larsen at al. 2009 ), which were utilized in this study, to determine bacterial and fungal input. In wood-fed larvae, the model placed larval samples outside of the 95% confidence limit of the host tree classifier, Acer spp. ( Fig. 2 ), lowering the confidence that the larvae acquired EAA from their woody diet. According to the classification criterion in the predictive model, the greater the distance of a particular consumer from the centroid of a classification group, i.e., potential EAA source, the greater the probability that mixing of EAA sources occurred. Given the distinct distances/placement of the wood-fed larvae from the centroid of the wood diet classification group (outside the 95% confidence interval decision region), we interpreted our results as indicative of acquisition of EAAs of non-dietary origin. Furthermore, based on location of wood-fed larval samples toward the decision boundaries of bacterial and fungal classifiers ( Fig. 2 ) and the determined 13 C-discrimination factor of 3.4‰ between the woody diet and the wood-fed larvae, non-dietary sources of EAAs in wood-fed larvae were attributed to a mixture of gut-associated bacteria and fungi.
Under ideal circumstances, a more direct approach to investigating the microbial origin of EAAs in wood-fed larvae would be to eliminate or drastically reduce the gut microbiome, followed by comparative analysis of the source(s) of EAA between antibiotic-treated and non-treated wood-fed larval samples. Unfortunately, this approach was not possible to perform because, unless using an artificial diet, A. glabripennis larvae will only feed in woody tissue in intact larval galleries and cannot be treated and then reinserted into a living tree without significant mortality and fitness costs to the larvae that survive. Nonetheless, the location of the larval samples outside of the Acer spp . decision region, together with the 13 C- offset results between the wood-fed larvae and their wood diet ( Fig. 1 ), provide support for our assertion that the origin of EAA in wood-fed larvae is microbial.
In the artificial diet and artificial diet-fed larval feeding group, both the diet and the larval EAAs were classified outside of the 95% confidence limit of the fungal classifier as well as that of the diet itself ( Fig. 2 ). These offsets indicate that the larval EAAs in these diet-fed samples likely originated from bacteria, although the contributions from bacteria were not as great as the contributions from microbiota to the wood-fed larvae. This is not unexpected, given that the artificial diet is a complete diet. We suspect that Lactobacillus is the bacterial source of EAA in artificial diet-fed larvae, as this bacterium was the dominant member of the gut community in diet-fed larvae in a previous study ( Geib et al. 2009 ).
Utilization of gut microbes as sources of EAAs reported herein for A. glabripennis larvae have been similarly reported in collembolans under comparable circumstances ( Larsen et al. 2011 ). Collembolans were found to be more dependent on gut bacterial sources of EAAs when fed on nitrogen-limited diets than on optimum diets. Larvae may use gut microbes differently on diets that vary in quality, thereby obtaining different types of amino acids or amounts of EAA from bacteria and fungi under different conditions. Fungi associated with A. glabripennis larvae represent a more immediate source of EAA, mainly because they can directly utilize larger, partially degraded products from lignocellulose digestion (e.g., hemicellulose, xylose) for EAA biosynthesis. Bacteria, on the other hand, may require smaller, completely degraded products (e.g., glucose, xylose) as starting material for EAA biosynthesis. This would be consistent with the time-dependent EAA synthesis schema following lignocellulose degradation by larger cellulolytic protists prior to utilization of by-products by smaller cellulolytic bacteria in the hindguts of termites ( Brune 2014 , Tokuda et al. 2014 ).
In this study, we avoided the confounding factors of non-consumer input to the calculated Δ 13 C by using only larval carcasses for analysis after removing the entire digestive tract to eliminate microbial influences on the consumer δ 13 C data. Additionally, the issue of multiple dietary sources and mixing of 13 C signatures in the artificial diet was avoided by using artificial diets as predictors and not as classifiers in the supervised model. Although there are limitations in the numbers and ecologically relevant members of representative bacterial and fungal groups in the δ 13 C EAA fingerprint data set, the data set and the model adequately classified known F. solani samples used in this study as fungi, and provided sufficient resolution of group membership of larval EAA. Further additions to the data set, and use of cultivated insect host-derived microbial isolates might improve the classification power of the prediction model.
In conclusion, the results in this study, along with results from previous studies investigating A. glabripennis and its associated gut microbes, provide a framework for conceptualizing the nutrient provisioning functions of gut-associated bacteria and fungi in A. glabripennis larvae. Briefly, cellulose ingested by larvae is degraded by a combination of larval and microbial processes ( Geib et al. 2009 ), yielding intermediary metabolites that are subsequently utilized for other metabolic processes by both the host insect and its associated microbes. In A. glabripennis, gut bacteria fix and recycle nitrogen, which can contribute nitrogen in the form of NH 4+ that is then routed into intermediary metabolites and their end-products, such as EAAs and non-EAAs ( Ayayee et al. 2014 ). Complementary microbial and host processes lead to the production of non-EAAs from the intermediary metabolites, and microbe-only processes lead to the production of EAAs from the intermediary metabolites ( Scully et al. 2014 ). Larvae can then acquire the EAAs following synthesis by the microbiome. Although this study does not definitively determine how larvae acquire EAAs from microbial sources, a combination of digestion (of gut microbes) as well as larval uptake of EAAs secreted by gut microbes into the lumen are likely routes of provisioning. Further studies are required to definitively verify or ascertain which of these processes dominate. Finally, this study highlights the utility of 13 C-fingerprinting in studying the metabolic basis of insect–microbe interactions, and exemplifies some of the technical and experimental challenges that need to be optimized for widespread use of this technique.
Supplementary Material
Acknowledgments
The Alphawood Foundation, Chicago, IL, funded this project, and USDA National Institute of Food and Agriculture (NIFA) Grant 2015-67013-23287 provided funding for this research to KH and CR. Thomas Larsen was supported by the DFG-funded Cluster of Excellence “The Future Ocean”. We thank Liz McCarthy for culturing the F. solani isolates and acknowledge the staff of the University of California, Davis Stable Isotope Facility for sample preparation and analysis.
References Cited
- Ayayee P., Rosa C., Ferry J. G., Felton G., Saunders M., Hoover K. . 2014. . Gut microbes contribute to nitrogen provisioning in a wood-feeding cerambycid . Environ. Entomol. 43 : 903 – 912 . [DOI] [PubMed] [Google Scholar]
- Bentley B. L. 1984. . Nitrogen fixation in termites: Fate of newly fixed nitrogen . J. Insect Physiol. 30 : 653 – 655 . [Google Scholar]
- Brune A. 2014. . Symbiotic digestion of lignocellulose in termite guts . Nat. Rev. Microbiol. 12 : 168 – 180 . [DOI] [PubMed] [Google Scholar]
- Castagna M., Shayakul C., Trotti D., Sacchi V. F. . 1997. . Molecular characteristics of mammalian and insect amino acid transporters: Implications for amino acid homeostasis . J. Expt. Biol. 200 : 269 – 286 . [DOI] [PubMed] [Google Scholar]
- Caut S., Angulo E., Courchamp F. . 2008. . Discrimination factors (delta 15N & delta 13C) in an omnivorous consumer: Effect of diet isotopic ratio . Funct.Ecol. 22 : 255 – 263 . [Google Scholar]
- Chen W. P., Yang X. H., Hegeman A. D., Gray W. M., Cohen J. D. . 2010. . Microscale analysis of amino acids using gas chromatography–mass spectrometry after methyl chloroformate derivatization . J. Chromatogr. B 878 : 2199 – 2208 . [DOI] [PubMed] [Google Scholar]
- Coleman D. C., Fry B. . 1991. . Carbon isotope techniques . Academic Press; , San Diego, CA: . [Google Scholar]
- Colman D. R., Toolson E. C., Takacs-Vesbach C. D. . 2012. . Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 21 : 5124 – 5137 . [DOI] [PubMed] [Google Scholar]
- Dillon R., Dillon V. M. . 2004. . The gut bacteria of insects: Non-pathogenic interactions . Annu. Rev. Entomol. 49 : 71 – 92 . [DOI] [PubMed] [Google Scholar]
- Douglas A. E. 2013. . Microbial brokers of insect-plant interactions revisited . J. Chem. Ecol. 39 : 952 – 961 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. E. 2009. . The microbial dimension in insect nutritional ecology . Funct. Ecol. 23 : 38 – 47 . [Google Scholar]
- Engel P., Moran N. A. . 2013. . The gut microbiota of insects - diversity in structure and function . FEMS Microbiol. Rev. 37 : 699 – 735 . [DOI] [PubMed] [Google Scholar]
- Geib S. M., Filley T. R., Hatcher P. G., Hoover K., Carlson J. E., Jimenez-Gasco M. D., Nakagawa-Izumi A., Sleighter R. L., Tien M. . 2008. . Lignin degradation in wood-feeding insects . Proc. Natl. Acad. Sci. USA 105 : 12932 – 12937 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib S. M., Jimenez-Gasco M. D., Carlson J. E., Tien M., Hoover K. . 2009. . Effect of host tree species on cellulase activity and bacterial community composition in the gut of larval asian longhorned beetle . Environ. Entomol. 38 : 686 – 699 . [DOI] [PubMed] [Google Scholar]
- Geib S. M., Scully E. D., Jimenez-Gasco M., Carlson H. E., Tien M., Hoover K. . 2012. . Phylogenetic analysis of Fusarium solani associated with the asian longhorned beetle Anoplophora glabripennis . Insects 3 : 141 – 160 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack R. A., Herard F., Sun J., Turgeon J. J. . 2010. . Managing invasive populations of asian longhorned beetle and citrus longhored beetle: A worldwide perspective . Annu. Rev. Entomol. 55 : 521 – 546 . [DOI] [PubMed] [Google Scholar]
- Hongoh Y., Hattori M., Ohkuma M., Sharma V. K., Prakash T., Noda S., Toh H., Taylor T. D., Kudo T., Sakaki Y., Toyoda A., Hattori M., et al. . 2008. . Genome of an endosymbiont coupling N 2 fixation to cellulolysis within protist cells in termite gut . Science 322 : 1108 – 1109 . [DOI] [PubMed] [Google Scholar]
- Keena M. A. 2005. . Pourable artificial diet for rearing Anoplophora glabripennis (coleoptera: Cerambycidae) and methods to optimize larval survival and synchronize development . Annu. Entomol. Soc. Am. 90 : 536 – 547 . [Google Scholar]
- Larsen T., Larsen J., Ventura M., O'Brien D. M., Magid J., Lomstein B. A. . 2011. . Contrasting effects of nitrogen limitation and amino acid imbalance on carbon and nitrogen turnover in three species of collembola . Soil Biol. Biochem. 43 : 749 – 759 . [Google Scholar]
- Larsen T., Taylor D. L., Leigh M. B., O'Brien D. M. . 2009. . Stable isotope fingerprinting: A novel method for identifying plant, fungal, or bacterial origins of amino acids . Ecology 90 : 3526 – 3535 . [DOI] [PubMed] [Google Scholar]
- Larsen T., Ventura M., Andersen N., O'Brien D. A., Piatkowski U., McCarthy M. D. . 2013. . Tracing carbon sources through aquatic and terrestrial food webs using amino acid stable isotope fingerprinting: E73441 . PLoS One 8 : 1 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald S. J., Lin G. G., Russell C. W., Thomas G. H., Douglas A. E. . 2012. . The central role of the host cell in symbiotic nitrogen metabolism . Proc. R. Soc. Biol. Sci. 279 : 2965 – 2973 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson W. J. 1980. . Herbivory in relation to plant nitrogen content . Ann. Rev. Ecol. Syst. 11 : 119 – 161 . [Google Scholar]
- McMahon K. W., Fogel M. L., Elsdon T. S., Thorrold S. R. . 2010. . Carbon isotope fractionation of amino acids in fish muscle reflects biosynthesis and isotopic routing from dietary protein . J. Anim. Ecol. 79 : 1132 – 1141 . [DOI] [PubMed] [Google Scholar]
- Morales-Jimenez J., de Leon A. V. P., Garcia-Dominguez A., Martinez-Romero E., Zuniga G., Hernandez-Rodriguez C. . 2013. . Nitrogen-fixing and uricolytic bacteria associated with the gut of Dendroctonus rhizophagus and Dendroctonus valens (curculionidae: Scolytinae) . Microb. Ecol. 66 : 200 – 210 . [DOI] [PubMed] [Google Scholar]
- Newsome S. D., Fogel M. L., Kelly L., del Rio C. M. . 2011. . Contributions of direct incorporation from diet and microbial amino acids to protein synthesis in nile tilapia . Funct. Ecol. 25 : 1051 – 1062 . [Google Scholar]
- Pinto-Tomas A. A., Anderson M. A., Suen G., Stevenson D. M., Chu F.S.T., Cleland W. W., Weimer P. J., Currie C. R. . 2009. . Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants . Science 326 : 1120 – 1123 . [DOI] [PubMed] [Google Scholar]
- Sabree Z. L., Kambhampati S., Moran N. A. . 2009. . Nitrogen recycling and nutritional provisioning by Blattabacterium , the cockroach endosymbiont . Proc. Natl. Acad. Sci. USA 106 : 19521 – 19526 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E. D., Carlson J., Hoover K., Tien M., Geib S. M. . 2013b. . Midgut transcriptome profiling of Anoplophora glabripennis a lignocellulose degrading cerambycid beetle . BMC Genomics 14 : 1 – 26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E. D., Carlson J., Hoover K., Tien M., McKenna D., Geib S. M. . 2014. . Functional genomics and microbiome profiling of the asian longhorned beetle ( Anoplophora glabripennis ) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles . BMC Genomics 15 : 1 – 21 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E. D., Geib S. M., Hoover K., Tien M., Tringe S. G., Barry K. W., del Rio T. G., Chovatia M., Herr J. R., Carlson J. E. . 2013a. . Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle . PLoS One 8 : e73827 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E. D., Hoover K., Carlson J., Tien M., Geib S. M. . 2012. . Proteomic analysis of Fusarium solani isolated from the longhorned beetle, Anoplophora glabripennis . PLoS One 7 : e32990 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda G., Bandi C., Paulsen I. T., Watanabe H. . 2013. . Maintenance of essential amino acid synthesis pathways in the Blattabacterium cuenoti symbiont of a wood-feeding cockroach . Biol. Lett. 9 : 20121153 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda G., Suboi Y., Kihara K., Saitou S., Moriya S., Lo N., Kikuchi J. . 2014. . Metabolomic profiling of 13 C-labelled cellulose digestion in a lower termite: Insights into gut symbiont function . Proc. R. Soc. Biol. Sci. 281 : 20140990 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokhshoori N. L., McCarthy M. D., Larsen T. . 2014. . Reconstructing 13 C isoscapes of phytoplankton production in a coastal upwelling system with amino acid isotope values of littoral mussels . Mar. Ecol. Prog. Ser. 504 : 59 – 72 . [Google Scholar]
- Walsh R. G., He S., Yarnes C. T. . 2014. . Compound-specific 13 C and 15 N analysis of amino acids: A rapid, chloroformate-based method for ecological studies . Rap. Commun. Mass Spectrom. 28 : 96 – 108 . [DOI] [PubMed] [Google Scholar]
- Wu D., Daugherty S. C., Van Aken S. E., Pai G. H., Watkins K. L., Khouri H., Tallon L. J., Zaborsky J. M., Dunbar H. E., Tran P. L., et al. . 2006. . Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters . PLoS Biol. 4 : 1079 – 1092 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


