Abstract
Objective The objective of the Strategic Health IT Advanced Research Project area four (SHARPn) was to develop open-source tools that could be used for the normalization of electronic health record (EHR) data for secondary use—specifically, for high throughput phenotyping. We describe the role of Intermountain Healthcare’s Clinical Element Models ([CEMs] Intermountain Healthcare Health Services, Inc, Salt Lake City, Utah) as normalization “targets” within the project.
Materials and Methods Intermountain’s CEMs were either repurposed or created for the SHARPn project. A CEM describes “valid” structure and semantics for a particular kind of clinical data. CEMs are expressed in a computable syntax that can be compiled into implementation artifacts. The modeling team and SHARPn colleagues agilely gathered requirements and developed and refined models.
Results Twenty-eight “statement” models (analogous to “classes”) and numerous “component” CEMs and their associated terminology were repurposed or developed to satisfy SHARPn high throughput phenotyping requirements. Model (structural) mappings and terminology (semantic) mappings were also created. Source data instances were normalized to CEM-conformant data and stored in CEM instance databases. A model browser and request site were built to facilitate the development.
Discussion The modeling efforts demonstrated the need to address context differences and granularity choices and highlighted the inevitability of iso-semantic models. The need for content expertise and “intelligent” content tooling was also underscored. We discuss scalability and sustainability expectations for a CEM-based approach and describe the place of CEMs relative to other current efforts.
Conclusions The SHARPn effort demonstrated the normalization and secondary use of EHR data. CEMs proved capable of capturing data originating from a variety of sources within the normalization pipeline and serving as suitable normalization targets.
Keywords: semantics, information storage and retrieval, vocabulary, controlled, electronic health records/standards, health information systems/standards
BACKGROUND AND SIGNIFICANCE
The Clinical Element Model ([CEM] Intermountain Healthcare Health Services, Inc, Salt Lake City, Utah) 1,2 is Intermountain Healthcare’s formalism for authoring Detailed Clinical Models. 3–5 Models authored under this formalism are called CEMs. A CEM provides a specification of “valid data” for a particular clinical element—the valid attributes for the element (and the valid values for those attributes), the valid data type, the valid physiologic range, etc.
The Strategic Health IT Advanced Research Project (SHARP) 6 was a program funded by the Office of the National Coordinator for Health Information Technology. Its focus was enhancing the quality, safety, and efficiency of health care through information technology. The authors participated in “area four” of the SHARP program, which specifically focused on data normalization. 7 Thus, SHARP area four was nicknamed “SHARPn,” the “n” reflecting “normalization.”
OBJECTIVE
Traditionally, a patient’s clinical data (medical history, examination data, hospital visits, and physician notes, etc) are stored inconsistently and in multiple locations, both electronically and nonelectronically. SHARPn’s task was to develop a “normalization framework” of open-source components that could be dynamically configured to accept electronic health record (EHR) data in various formats—eg, HL7 Version 2 8 (Health Level Seven International, Ann Arbor, Michigan) messages, text documents, and HL7 Consolidated Clinical Document Architecture ([C-CDA] Health Level Seven International, Ann Arbor, Michigan) 9 documents—and normalize them into common, standards-conformant structures and codes that would, as a result, contain comparable information suitable for secondary uses. High throughput phenotyping was the secondary use that drove requirements.
CEMs were chosen as the normalization targets for SHARPn. A framework was developed to accept data in various formats and converted them to CEM instances. While the overall SHARPn project has been described elsewhere, 10,11 this paper focuses particularly on the CEM modeling and terminology aspect of the project.
MATERIALS AND METHODS
Clinical Element Models
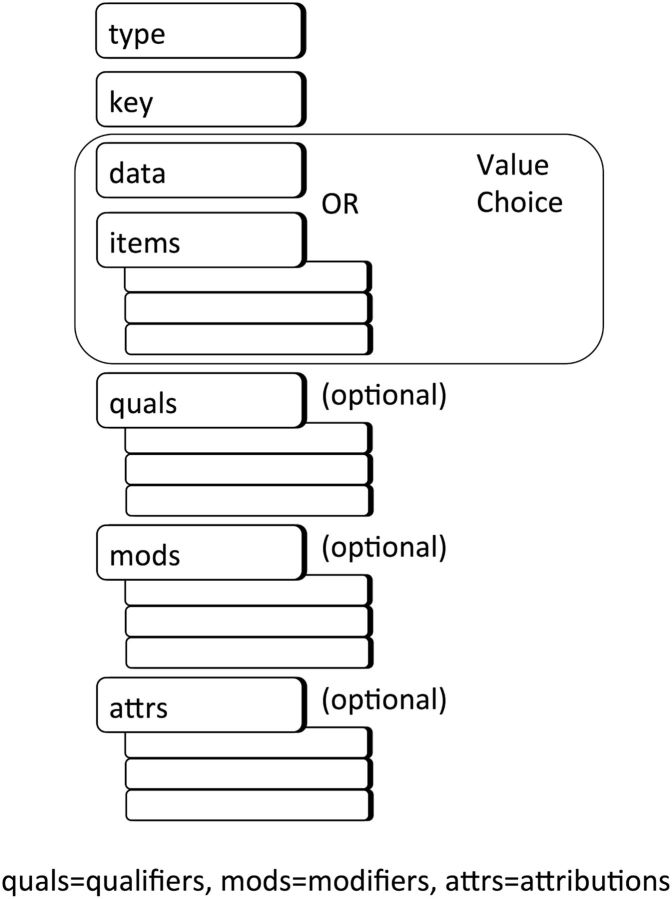
The CEM strategy is based on a very generic Abstract Instance Model, shown in figure 1 . This model constitutes the general structure for every model (and hence every instance of data). It declares that every model has a “type” (an identifier for the model), a “key” (the “real world” concept that is the focus of the model, often mapping to a code in a standard terminology), and a “value choice” (either a single “data” or a collection of “items”). Values are defined using HL7 Version 3 data types (with some practical constraints). A CEM may optionally contain qualifiers (which amend the meaning of the data, eg, the body location from which a heart rate measurement was taken), modifiers (which drastically change the meaning of the instance, eg, the heart rate measurement pertains to a subject of “fetus” instead of to the patient of record), and attributions (which capture the “who,” “when,” “where,” and “why” of an action, such as “observed,” “documented,” etc, related to an instance of data). Items, qualifiers, modifiers, and attributions are themselves defined as CEMs.
Figure 1:
The Abstract Instance Model
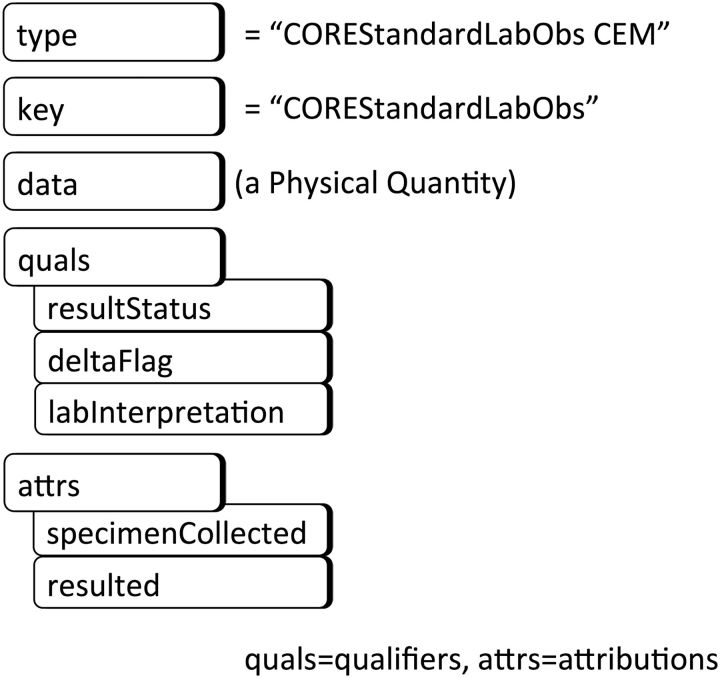
An abbreviated depiction of the CEM “COREStandardLabObs,” representing a standard laboratory test result, is provided in figure 2 as illustration of a CEM conforming to the Abstract Instance Model. (The prefix “CORE” will be explained later during the discussion of specialization.) In this CEM, the type is a code that identifies this model, ie, a code that means “the CEM for a standard laboratory test result.” The CEM’s key is bound to a value set of LOINC ([Logical Observation Identifier Names and Codes] Regenstrief Institute, Indianapolis, Indiana) codes representing the various laboratory tests used in clinical care. The value choice in the case of this CEM is a single data value. Its data type is “physical quantity,” which consists of a number and a unit of measure. The CEM’s valid qualifiers are “result status,” “delta flag,” and “lab interpretation,” and its valid attributions are structures for capturing the who/when/where information about the specimen collection and results of the laboratory test.
Figure 2:
An example CEM as a constraint on the Abstract Instance Model
Controlled terminologies play a fundamental role in the CEMs. Valid codes for the keys and in many cases the data values are selected from standard terminologies wherever possible. Hence, not just the structure but the semantics of the normal CEMs are definitively declared.
Regardless of the type of information source (text document, HL7 message, etc) or of the particular system (Intermountain Healthcare’s EHR, Mayo Clinic’s laboratory system, etc), all laboratory test results would be normalized to instances of this model. Those laboratory test results would then be available in a common form to phenotyping algorithms.
A syntax for expressing the CEM (ie, the constraints) is required. During the SHARPn project, we used the GE Healthcare-developed Constraint Definition Language (CDL) to represent the CEMs. (GE Healthcare [General Electric Company, Fairfield, Connecticut] has not published documentation of the CDL language. The language is GE Healthcare-owned, but the CEM artifacts expressed in CDL for the SHARPn project are publicly free for use under the Intermountain license.) An example of that language is shown in figure 3 . We used an Eclipse-based plugin to author in CDL and a Subversion (SVN) repository to store the models.
Figure 3:
An example of a CEM in CDL
Operating mechanisms
The SHARPn phenotyping subteam dictated modeling requirements, requesting the models that they anticipated would be needed by their phenotyping algorithms. Specifically, they planned to support 2 National Quality Forum (NQF) Quality Measures—NQF 0018 NCQA: Controlling High Blood Pressure 12 and NQF 0064 NCQA: Diabetes: LDL Management & Control (NQF endorsement since removed). 13 Once presented with the requirements, representatives from the Intermountain Healthcare modeling team, the Mayo Clinic/AgileX team building the SHARPn normalization pipeline and the CEM database, and the project principal and coprincipal investigators followed an agile approach to developing and implementing the CEMs: (1) the participants would discuss requirements; (2) the Intermountain team would create new models or modify existing ones; (3) the Mayo/AgileX participants would implement them in XSDs; and (4) iterative refinement would take place as necessary.
RESULTS
The place of models in the framework
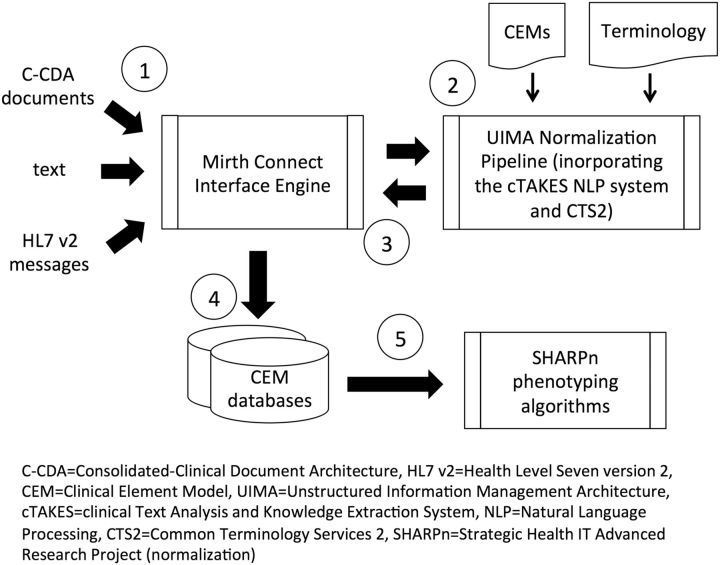
The SHARPn framework and the place of CEMs and terminology in the framework are shown in figure 4 . At step 1, the framework used Mirth (Quality Systems, Inc, Irvine, California), 14 an open-source health care integration engine, to accept the various input data (HL7 Version 2 messages, C-CDA documents, and text) from a file system or from external entities via the Nationwide Health Information Network and channel them to components of the normalization “pipeline” (step 2). The pipeline was based on the Apache Unstructured Information Management Architecture 15 (The Apache Software Foundation, Forest Hill, Maryland) platform. The component responsible for normalizing text was the Apache clinical Text Analysis and Knowledge Extraction System 16 (The Apache Software Foundation, Forest Hill, Maryland) natural language processing (NLP) system. The pipeline incorporated Common Terminology Services 2, 17 services to access terminology content. The SHARPn CEMs were compiled into EXtensible Markup Language Schema Definitions (XSDs), and the pipeline normalized the input patient data to XSD-conformant Extensible Markup Language (XML) documents. The resulting XML documents/CEM instances were then sent back to Mirth (step 3), which serialized them and routed them to a database designed to store CEM instances (step 4). Several databases were incorporated in the framework, including MySQL and CouchDB. The normalized CEM instances were then available to be retrieved by the algorithms that executed phenotyping logic (step 5). During the execution phase, 273 patients’ EHR data were normalized and a phenotyping algorithm was executed over those data. One thousand medication order messages were also normalized. Pathak et al 10 describes the execution and its results in more detail.
Figure 4:
The SHARPn Framework
Tooling
We developed a web-based browser (clinicalelement.com) from which CEMs could be viewed and downloaded. This tool allowed all SHARPn team members to view existing models and also allowed the modeling team to deliver new models developed for the project. We also developed a web-based request tool from which team members could request new models or modifications to models and with which the modeling team could track the requests.
The models
As a result of the phenotyping team’s requests, 28 “statement” models (analogous to “classes”) and numerous “component” models (analogous to “attributes”) were created or repurposed. Table 1 lists the statement models. As mentioned, when model requirements were identified, existing models were first examined. Those models marked with an “R” are models that were created for other projects but that we found could be repurposed for SHARPn. Those models marked with an “N” in the table were created especially for the project because no existing model satisfied the requirements.
Table 1:
CEMs used in the SHARPn project
| Model Name | Description | New (N) or Repurposed (R) |
|---|---|---|
| Statement CEMs: | “Statements” are independent facts in a patient’s record. Heart rate measurements, diagnoses, procedure records, documentation of allergies, etc are all statements. | |
| GenericStatement | A generic structure that serves as the parent of the other statement models. | N |
| AdministrativeDiagnosis | Statement of a diagnosis for administrative purposes. | R |
| AdministrativeProcedure | Statement of a procedure for administrative purposes. | N |
| AllergyIntolerance * | Statement that a patient has an allergy or intolerance. | R |
| Assertion * | Statement of a symptom or finding that exists in a patient. The value is a code representing the symptom or finding, eg, “nausea,” “fever,” etc. | R |
| Assessment * (and its 3 data type-specific variants) | Statement of a symptom or finding that follows an attribute/value pattern, eg, heart rate/numeric heart rate value, hair color (blonde, black, brown, etc), APGAR pulse score (“0,” “1,” “2”), etc. The data type may be a code, a numeric, or an ordinal. | R |
| DiseaseDisorder * | Statement of a disease/disorder existing in a patient. | R |
| NotedDrug * | Statement of a medication the patient is noted to have taken or be taking. | N |
| Procedure * | Statement of a procedure performed on the patient. | R |
| StandardLabObs * (and its 6 data type-specific variants) | Statement of a standard laboratory result. The 6 variants of the model support the result value being coded, quantitative, ordinal, string, interval, or titer. | R |
| Patient CEM: | A “Patient,” because of its central position in the health care record, is a special type of CEM. | |
| Patient * | CEM for capturing the identity and demographics of a patient. | R |
| Links: | A “Link,” or “Semantic Link,” is an association between 2 instances. Just as is the case for all other data instances, semantic link instances must conform to a CEM. | |
| ComponentToComponentLink | A CEM defining the structure of a link between 2 instances of component CEMs. | N |
| StatementToComponentLink | A CEM defining the structure of a link between an instance of a statement CEM and an instance of a component CEM. | N |
| StatementToStatementLink | A CEM defining the structure of a link between 2 instances of statement CEMs. | N |
| Collections: | “Collections” are loose groupings of CEM instances; all instances are related to the same patient but not necessarily recorded at the same time, by the same provider, etc. | |
| AllergyIntoleranceConcern * | A collection of allergy/intolerance instances. | N |
| NotedDrugList * | A collection of notedDrug instances. | N |
| ProblemConcern * | A collection of problem instances. | N |
| Panels: | “Panels” are strong groupings of CEM instances; all instances are related to the same patient and derived from the same source (specimen or instrument) and associated with the same or closely-related timestamp. | |
| StandardLabPanel * | A panel of StandardLabObs instances. | R |
| VitalSignPanel * | A panel of Assessment instances where the assessments represent vital sign measurements. | R |
| Components: | “Components” are analogous to “attributes” in traditional modeling/programming; they are only used within other CEMs and cannot stand by themselves. They themselves are CEMs, eg, the “StartTime” component within the “NotedDrug” CEM is itself defined by a separate CEM. | |
| A number of “component” models were also created or repurposed for SHARP. |
Abbreviations: SHARPn, Strategic Health IT Advanced Research Project area four; APGAR, appearance, pulse, grimace, activity, respiration; CEM, Clinical Element Model.
*Both a “Core” version and a “Secondary Use” version of these models were created, as explained in the section “Context differences and core models.”
For most models, a “core” version and a “secondary use” version were created. Those models are indicated by an asterisk after the model name. The notion of a core version was an attempt to support multiple contexts. Models intended for specific contexts could start with the same core model and constrain the model according to its specific needs. Context-specific model requirements are addressed further in the Discussion section. Where no context-specific differences were anticipated, no separate core and secondary use versions were created.
“Metadata” was modeled outside the logical CEM models, in “reference classes.” The reference class attributes captured context and provenance information. For example, “patientExternalId” and “clinicalEncounterId” captured the patient and encounter context to which the data pertained, respectively. Since SHARPn’s objective was to normalize data coming from EHRs for secondary use purposes, provenance information identifying the source system of the data and the kind of process that normalized the data (NLP, messaging interface, etc) were important to capture. The “sourceDataInfo” and “normalized” structures captured these data. The reference class attributes used for most SHARPn CEMs are shown in table 2 .
Table 2:
Reference class attributes used in most SHARPn CEMs
| Attribute | Definition |
|---|---|
| attribRecordedTime | The time this CEM instance was stored in the CEM instance database. |
| clinicalEncounterId | External (business) identifier(s) for the clinical encounter associated with this CEM instance. |
| externalId | External (business) identifier(s) for this CEM instance. |
| instanceId | Internal identifier for this CEM instance. |
| normalized | A structure containing information related to the process of normalizing input data into this CEM instance. |
| patientExternalId | External (business) identifier(s) for the patient associated with this CEM instance. |
| patientGloballyUniqueId | An identifier for this CEM instance that is not duplicated in any internal or external system. |
| sourceDataInfo | A structure containing information about the original data before normalization into this CEM instance. |
| sourceSystemId | An identifier for the system that originated the original data normalized to this CEM instance. |
| status | The status of this CEM instance. |
Abbreviations: SHARPn, Strategic Health IT Advanced Research Project area four; CEM, Clinical Element Model.
These reference classes were “added” to a CEM when it was compiled. The separation of these reference classes from the CEMs forces the separation between logical model and implementation, and it allows different implementations to add their own implementation-specific attributes to a CEM.
Terminology content
Coded terminology plays an essential role in normalization, providing semantics to the models and their attributes. The SHARPn project’s approach was to directly support the terminology standards declared by the US government. Guided by prescribed or de facto standards, we selected a particular code system for each clinical area. The selected code systems are shown in table 3 .
Table 3:
Code systems referenced by the SHARPn models
| Domain | Standard Terminology |
|---|---|
| Drug names | RxNorm |
| Clinical drugs | RxNorm |
| Ingredients | RxNorm |
| Forms | RxNorm |
| Routes | HL7 |
| Units | SNOMED CT a |
| Frequencies | SNOMED CT |
| Disease/disorder | SNOMED CT |
| Alleviating/exacerbating factor | SNOMED CT |
| Laboratory test results | LOINC |
Abbreviations: SHARPn, Strategic Health IT Advanced Research Project area four; HL7, Health Level Seven International; SNOMED CT, Systematized Nomenclature of Medicine–Clinical Terms; LOINC, Logical Observation Identifiers Names and Codes.
a SNOMED CT (The International Health Terminology Standards Development Organisation, Copenhagen, Denmark)
A number of codes could not be found in the standard terminologies, such as codes for “permanent” (for an address use), “significant change up” (for a measure of change in a laboratory value), and “resulted” (for a type of attribution). A SHARP code system was created to maintain these codes.
Model mappings
For the models to be used as normalization targets, mappings from HL7 messages, NLP structures, and C-CDA documents to the CEMs were required. These mappings were created in spreadsheets and diagrams. The logic was then transferred into the pipeline code. The pipeline was designed in a modular fashion such that new mappings could be “plugged in” as needed.
CEMs were also successfully mapped to the NQF’s Quality Data Model (QDM), 18 since phenotyping algorithms executed the logic of QDM-based clinical quality measures. 19
Terminology mappings
We also needed to develop mappings from local codes that would be received in messages to the normalized terminology content referenced in the CEMs. An example of a terminology mapping is shown in figure 5 . The figure shows a local code “2” for a patient’s race mapped to the normal CEM code “2106-3” (“white”). When a message contains “2,” the pipeline would call terminology mapping services that would normalize the code to “2106-3,” and that code would be stored in the SHARP CEM instance database. The benefit of normalized codes is that secondary use applications such as the phenotyping algorithms need to only understand 1 structure and code system for medications rather than myriad possibilities. The drawback is potential information loss, as terminology mappings are sometimes lossy. This is ameliorated by the fact that the original code is preserved in the CEM’s data type.
Figure 5:
An example of a SHARPn terminology mapping
DISCUSSION
Context differences and core models
SHARPn’s secondary use context had slightly different model requirements from the EHR context for which the CEMs were initially created. For example, in an EHR, the “reporting priority” and “updated time” are necessary to track the progress of the result through its lifecycle. On the other hand, these attributes are useless in the secondary use context because by the time the result reaches a secondary use database, it is essentially static data—it has already undergone its lifecycle. Similarly, an EHR needs to record such patient demographic information as “religious congregation” and “disabilities” because it impacts the accommodations given to a patient during care. While such demographics might also be useful in secondary use, we made the simplifying assumption that such demographic attributes would not be needed for our particular phenotyping requirements, and we omitted them from the SHARPn models.
Given these discussions, it appeared that creation of a single, context-independent model for each element would be unachievable. The question, then, was how to create models for different contexts yet facilitate as much interoperability between them as possible. One obvious solution would be a traditional object-oriented extension approach—create parent models that contain only the attributes that are common across all contexts, extending them with context-specific models that add context-specific attributes. The other obvious approach would be a constraint strategy in which a common parent model contains all attributes needed by any and all contexts while the child context-specific models constrain out of the common model those attributes not pertinent to the context. A disadvantage of this latter approach is that whenever any new attribute is identified as necessary (for any context), it needs to be added to the parent superset. The maintainers of all the constraining context models would then need to assess whether the new attribute would be applicable to their contexts and if not, constrain it out. The advantage is that tighter control would be possible; in the extension approach, maintainers of child context models could add attributes to their models unbeknownst to the other maintainers of child models. They could add similar attributes in slightly different ways, diluting interoperability. And the proliferation of different hierarchies of classes could easily become unwieldy and difficult to understand.
Consequently, since interoperability is a prime objective, we decided on the restriction approach even though it would require more review and oversight. We surmised that the required review and oversight would even prove beneficial to the process, as stakeholders would need to communicate and collaborate.
Under this approach, in cases where a model that would meet SHARPn requirements already existed in the Intermountain set, we copied it and saved it as a core SHARPn CEM. We then created a secondary use SHARPn model that constrained out of the core model those attributes that the SHARPn team deemed not applicable to secondary use. Asterisks after the model names in table 1 indicate those models for which this core/secondary use strategy was implemented. Figure 6 shows an example. The top of this figure shows a core model (for a numeric standard laboratory test result). The attributes that were constrained out to achieve a secondary use model have been struck out. The bottom of the figure shows the resulting secondary use (numeric standard laboratory test result) model.
Figure 6:

A diagrammatic example of a secondary use constraint on a core model
Other context differences called for different models, as opposed to a common model whose attributes were constrained differently. For example, SHARPn had a requirement for a “medication” model. For its EHR context, Intermountain had developed a medication order model and anticipated developing a medication administration model also. In SHARPn discussions, we realized that what was needed for secondary use was neither an order nor an administration but rather a level of abstraction higher than the EHR models—something to note that the patient was taking “drug X,” the amount taken, and whether the information was known via a clinical note, a prescription, an order, or an administration. In the case of secondary use of EHR data, summary information such as average daily dose of a medication and daily dose count of a medication are very useful rather than the information regarding individual administrations or orders that would be found in the EHR. Hence, a new model called “noted drug” was created.
Model granularity
In the EHR context, Intermountain modeled CEMs very specifically. We created models for “heart rate measurement,” “nausea,” “serum glucose result,” “serum potassium result,” etc. We were very specific in order to capture all detail needed to specify valid data. For example, the serum glucose and serum potassium models are very similar, both containing a “reference range” attribute, a “status” attribute, etc. The only differences of note are that the serum glucose model specifies that the valid unit of measure for a serum glucose measurement is “mg/dL” with a “physiological limit” of “0–3000,” while the potassium result model specifies “mEq/L” for the unit of measure and “0–10” for the physiologic limit.
Similarly, a model for a heart rate measurement and a systolic blood pressure measurement share much the same structure. However, the valid (or at least the common) values captured for a heart rate measurement’s measurement location differ from those of a systolic blood pressure measurement. The same is true for the method/device. And, perhaps most significantly, the 2 are clinically very different measurements. Consequently, in the EHR context, we created different models for the heart rate measurement and systolic blood pressure measurement.
In the EHR context, CEMs would govern data storage. All incoming data (whether from applications, NLP, messages, or structured documents) would be submitted to validation against the CEMs. If data instances do not pass validation, the intent is to store the data in an alternate form and alert the modeling team. The team would then investigate whether a new model or modifications to the existing model would be needed and make changes if necessary. Depending on the kind of data, the stored instance may be migrated to the new or modified model.
In contrast, in the secondary use of EHR data, the data would be generated without surveillance and immediate reaction as in the EHR case. The focus would be more on normalizing structure and high-level semantics rather than the very fine-grained validation of the EHR context. Consequently, for the SHARPn project, we created models at a higher abstraction level. We created models such as “assertion” (used to assert the existence of a condition/disease/disorder in a patient) instead of “nausea” and “diabetes”; “assessment” (used for observations that have a value, whether it be numeric or categorical) instead of “heart rate measurement” and “urine description”; and “standard lab result” instead of “glucose result” and “hematocrit result.” We even created a “generic statement” model to support cases in which the pipeline received data that could not even be distinguished as one of the aforementioned types.
We acknowledged that the type of granular specification available in our EHR CEMs would only improve the normalization and hence the interoperability of the data, but we left this level of normalization to a future exercise. With more attention towards high-quality EHR data, we envisioned that that type of granular normalization could take place as a second phase of processing. The very specific CEMs could be used in that phase.
CEMs vs other modeling efforts
There is a number of health care information modeling projects currently underway or recently completed 20–28 as well as a number of projects whose principal effort was not modeling but during which model creation was a necessary component. 29–32 The Clinical Information Modeling Initiative (CIMI) 33 is an effort to unify such modeling efforts, and representatives of many of the aforementioned efforts are participating. CIMI’s objective is to provide an open collection of information models in an agreed-upon format. Intermountain Healthcare intends to migrate its CEMs to that format at the point CIMI models are mature. A major objective of CIMI is to move beyond the level of modeling addressed by SHARPn, creating not just higher-level models akin to the SHARPn models but also very specific models as Intermountain has done. Had the CIMI higher-level models been available, the SHARPn project would likely have used those as normalization targets instead of CEMs. Instead, SHARPn chose CEMs because of Intermountain’s experience in developing them for EHR data representation and converting them into computable artifacts.
Two efforts presently prominent in the US are the QDM and C-CDA. As part of the SHARPn project, both were successfully mapped to the SHARPn CEMs. A valid question would be why 1 of them was not used as the normalization targets for the project. C-CDA has been designed to represent documents, and the QDM has been designed to facilitate expression of quality measures. In contrast, we designed the CEMs to be implementable as discrete EHR data instances, and thus felt they were better suited for the pipeline and instance database. Using CEMs also gave us the freedom to design the models according to pipeline implementation requirements. For example, in some cases, C-CDA is more granular than the pipeline context and does not address the use case of receiving unknown elements. In retrospect, we could have patterned the CEMs more closely to 1 of these efforts. Which of the 2 (or of the other modeling efforts cited above) to select, however, would have been a somewhat arbitrary choice. We look to the CIMI effort to provide a set of common logical models that then can be mapped to C-CDA, QDM, or any of the other contexts.
Iso-semantic models
Ideally, we would create just 1 model for each clinical element to be stored. However, that proves to be a difficult proposition. We have mentioned, for example, that the SHARPn models were more generic than the Intermountain CEMs originally created. The consequence is that we now have 2 models under which a data element such as heart rate measurement might be stored—1 specific and the other generic. Many more situations of needing to create iso-semantic 34 models will arise, especially as we consider more granular models. Interoperability with models from other projects further compounds the problem. The conclusion is that mechanisms for mapping between these iso-semantic models are critical to a coherent normalization strategy.
Expertise required
The SHARPn project’s contribution of open-source software components is significant to the normalization effort. However, normalization software alone will not completely solve the problem. We observed that implementation of the components in a multi-institution environment would still require a fair amount of manual labor involving content expertise. For example, mapping the Mayo Clinic’s medication order messages (which sent First Databank codes in HL7 Version 2 message structures) to CEM attributes (which reference RxNorm codes) requires a deep understanding of HL7 Version 2, the CEMs, RxNorm, and FDB. It is highly unlikely that a typical institution will have human resources that possess the necessary content knowledge or the wherewithal to train and hire such resources. To overcome this barrier to adoption, an extremely beneficial follow-on to the SHARPn project would be to develop open-source mapping tools with “embedded intelligence”—tools that “understand” the structures and semantics of the input sources and the normalization targets, and that can guide and assist users as they create structural and semantic mappings. They could use sophisticated lexical and probabilistic matching algorithms, incorporating large amounts of an institution’s historical data to assist them in suggesting matches, and leverage creative visualization techniques (heat maps, bubble charts, etc) to present possible matches.
Scalability and sustainability
The models evolved as we practiced an agile development approach. For instance, we added a “drug vehicle” attribute to the noted drug model when the phenotyping team anticipated that need. We also made changes to the models to make them more robust to the variety of system capabilities, such as adding a precoordinated “clinical drug” attribute to the existing postcoordinated attributes in the noted drug model to accommodate systems that might not support representing medication information in postcoordinated fashion. We also made changes to support future requirements, such as creating the ability to add “extension” attributes to models. Consequently, by the time the last pipeline executions took place, the models were capable of serving as targets for most/all data typically stored in an EHR and no errors attributable to the models were encountered.
This initial set of 28 models fulfilled the phenotyping team’s narrow requirements for the project’s algorithms. Certainly, more classes of models (eg, “allergy,” “radiology order,” etc) will be needed. (In fact, many CEMs not used in the SHARPn project already exist.) And it may not be feasible to represent more complex, presently-less-understood areas (eg, -omics data) in present CEM structures. But within the realm of typically stored EHR data, CEMs appear to be a promising and scalable option.
The issues discussed in the “Iso-semantic models” and “Expertise required” sections, however, are hindrances to scalability and sustainability. Even if the CEMs themselves were robust and scalable, implementations would need to map their systems to the CEMs. The systems’ models and the CEMs constitute iso-semantic models that need to be mapped. The terminologies used by the systems may need to be mapped to the CEMs’ terminologies, requiring effort and expertise. As stated, robust, intelligent tools to aid mapping and hiding complexity are a critical need.
A library of models will evolve and expand over time. It will require governance and financial and human resources. It is likely that the CIMI library will supplant the CEMs, curated under the auspices of a yet-to-be-determined standards organization, but its maintenance will likely be a community effort. As has been discussed, the CIMI library will include models several orders of magnitude more than this SHARPn set at a higher level of granularity. Its maintenance will present a challenge to scalability and sustainability, but the development effort will settle over time (as the greater part of common EHR are represented); and with adequate governance structures and creative community incentivizing, the challenge should not be insurmountable.
CONCLUSION
We have described the role of CEMs in the SHARPn project, which provided structures to which data from HL7 Version 2 messages, NLP outputs, and C-CDA documents could be normalized. They were also shown to be mappable to QDM-based quality measures for high throughput phenotyping. Use of standardized terminology enhanced the normalization. Very specific semantic normalization via very granular CEMs was left to a future exercise.
Recognition of context differences stimulated different model constraints and granularity. Mechanisms for mapping between iso-semantic models and how to reduce the amount of content expertise needed to implement and use the normalization pipeline remain issues to be solved by the community.
FUNDING
This manuscript was made possible by funding from the Strategic Health IT Advanced Research Projects (SHARP) Program (90TR002) administered by the Office of the National Coordinato r for Health Information Technology. The contents of the manuscript are solely the responsibility of the authors.
COMPETING INTERESTS
None
ACKNOWLEDGEMENTS
The authors wish to acknowledge Teresa Conway, Nathan Davis, Doug Mitchell, and Cyndalynn Tilley for their valuable contributions to Intermountain’s modeling efforts during the project.
REFERENCES
- 1. Coyle JF, Rossi Mori A, Huff SM . Standards for detailed clinical models as the basis for medical data exchange and decision support . Int J Med Inf 2003. ; 69 ( 2–3 ): 157 – 174 . [DOI] [PubMed] [Google Scholar]
- 2. Coyle JF . The Clinical Element Model Detailed Clinical Models [dissertation] . Salt Lake City: : The University of Utah; ; 2013. . [Google Scholar]
- 3. Huff SM, Rocha RA, Coyle JF, et al. . Integrating detailed clinical models into application development tools . Stud Health Technol Inform 2004. ; 107 ( Pt 2 ): 1058 – 1062 . [PubMed] [Google Scholar]
- 4. Goossen W, Goossen-Baremans A, van der Zel M . Detailed clinical models: a review . Healthc Inform Res 2010. ; 16 ( 4 ): 201 – 214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parker CG, Rocha RA, Campbell JR, et al. . Detailed clinical models for sharable, executable guidelines . Stud Health Technol Inform 2004. ; 107 ( Pt 1 ): 145 – 148 . [PubMed] [Google Scholar]
- 6. Office_of_the_National_Coordinator for Health Information Technology (ONC) . Final report: assessing the SHARP experience. http://www.healthit.gov/sites/default/files/sharp_final_report.pdf . Updated 29 Aug 2014. Accessed March 20, 2015 . [Google Scholar]
- 7. Strategic Health IT Advanced Research Projects (SHARP): Research Focus Area 4 – Secondary Use of EHR Data. SHARPn Website. http://sharpn.org . Updated 18 Dec 2014. Accessed March 19, 2015.
- 8. HL7 standards product brief - HL7 Version 2 product suite . Health Level Seven International Website. http://www.hl7.org/implement/standards/product_brief.cfm?product_id=185 . Updated 11 Feb 2015. Accessed March 19, 2015 . [Google Scholar]
- 9. HL7 standards product brief - HL7 implementation guide for CDA release 2: IHE health story consolidation, release 1.1 - US realm. Health Level Seven International Website. http://www.hl7.org/implement/standards/product_brief.cfm?product_id=258 . Updated 14 Aug 2014. Accessed March 19, 2015.
- 10. Pathak J, Bailey KR, Beebe CE, et al. . Normalization and standardization of electronic health records for high-throughput phenotyping: The SHARPn consortium . J Am Med Inform Assoc 2013. ; 20 ( e2 ): e341 – e348 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rea S, Pathak J, Savova G, et al. . Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project . J Biomed Inform 2012. ; 45 ( 4 ): 763 – 771 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Controlling high blood pressure . National Quality Forum Website. http://www.qualityforum.org/QPS/0018 . Updated 1 Dec 2013. Accessed March 19, 2015 . [Google Scholar]
- 13. Comprehensive diabetes care: LDL-C control <100 mg/dL. National Quality Forum Website. http://www.qualityforum.org/QPS/0064 . Updated 4 Sept 2014. Accessed March 19, 2015.
- 14. Mirth BJ . Standards-based open source healthcare interface engine . Technol Innovation Manage Rev 2008. . http://timreview.ca/article/205 . Accessed October 29, 2013 . [Google Scholar]
- 15. Ferrucci D, Lally A . Building an example application with the unstructured information management architecture . IBM Syst J 2004. ; 43 ( 3 ): 455 – 475 . [Google Scholar]
- 16. Savova G, Masanz J, Ogren P, et al. . Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications . J Am Med Inform Assoc 2010. ; 17 ( 5 ): 507 – 513 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. CTS2 . Mayo Clinic Website. http://informatics.mayo.edu/cts2/index.php/Main_Page . Updated 17 Jan 2014. Accessed March 19, 2015 . [Google Scholar]
- 18. Healthcare IT knowledge base: QDM. Quality Data Model. National Quality Forum Website. http://public.qualityforum.org/hitknowledgebase/Pages/QDM.aspx . Updated 14 Jan 2013. Accessed March 19, 2015.
- 19. Li D, Endle CM, Murthy S, et al. . Modeling and executing electronic health records driven phenotyping algorithms using the NQF Quality Data Model and JBoss Drools Engine . Proc AMIA Annu Fall Symp 2012. : 532 – 541 . [PMC free article] [PubMed] [Google Scholar]
- 20. HL7 . FHIR ballot reconciliation master. Health Level Seven International FHIR Website. http://hl7.org/implement/standards/fhir/index.html . Updated 29 Sept 2014. Accessed March 19, 2015 . [Google Scholar]
- 21. Goossen WT . Using detailed clinical models to bridge the gap between clinicians and HIT . In: De Clercq, De Moor G, Bellon J , eds. Collaborative Patient Centred eHealth . Amsterdam, The Netherlands: : IOS Press; ; 2008. : 3 – 10 . [PubMed] [Google Scholar]
- 22. European Committee for Standardization (CEN) . ISO/CEN 13606: Health Informatics-Electronic Health Record Communication . Brussels, Belgium: : European Committee for Standardization; ; 2010. . [Google Scholar]
- 23. Beale T . Archetypes and the EHR . Stud Health Technol Inform 2003. ; 96 : 238 – 244 . [PubMed] [Google Scholar]
- 24. Beale T. Archetypes: constraint-based domain models for future-proof information systems . In: Baclawski K, Kilov H , eds. Eleventh OOPSLA Workshop on Behavioral Semantics: Serving the Customer, 2002 . Evanston, IL: : Northeastern University; ; 2002. : 16 – 32 . [Google Scholar]
- 25. Hoy D, Hardiker NR, McNicoll IT, et al. . A feasibility study on clinical templates for the National Health Service in Scotland . Stud Health Technol Inform 2007. ; 129 : 770 – 774 . [PubMed] [Google Scholar]
- 26. Ahn SJ, Kim Y, Yun JH, et al. . Clinical contents model to ensure semantic interoperability of clinical information . J KIISE: Software Appl 2010. ; 37 ( 12 ): 871 – 881 . [Google Scholar]
- 27. Regan-Udall Foundation for the Food and Drug Administration . Common data model. Observational Medical Outcomes Partnership Website. http://omop.org/CDM . Updated 18 Feb 2015. Accessed March 19, 2015 . [Google Scholar]
- 28. Reisinger SJ, Ryan PB, O’Hara DJ, et al. . Development and evaluation of a common data model enabling active drug safety surveillance using disparate healthcare databases . J Am Med Inform Assoc 2010. ; 17 : 652 – 662 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bradshaw RL, Matney S, Livne OE, et al. . Architecture of a federated query engine for heterogeneous resources . Proc AMIA Annu Fall Symp 2009. : 70 – 74 . [PMC free article] [PubMed] [Google Scholar]
- 30. Narus SP, Srivastava R, Gouripeddi R, et al. . Federating clinical data from six pediatric hospitals: process and initial results from the PHIS+ consortium . Proc AMIA Annu Fall Symp 2011. : 994 – 1003 . [PMC free article] [PubMed] [Google Scholar]
- 31. Friedman C . Towards a comprehensive medical language processing system: methods and issues . Proc AMIA Annu Fall Symp 1997. : 595 – 599 . [PMC free article] [PubMed] [Google Scholar]
- 32. Friedman C, Hripcsak G, DuMouchel W, et al. . Natural language processing in an operational clinical information system . Nat Lang Eng 1995. ; 1 ( l ): 83 – 108 . [Google Scholar]
- 33. Clinical Information Modeling Initiative Website. http://www.opencimi.org/ . Accessed March 19, 2015.
- 34. Oniki TA, Coyle JF, Parker CG, et al. . Lessons learned in detailed clinical modeling at Intermountain Healthcare . J Am Med Inform Assoc 2014. ; 21 ( 6 ): 1076 – 1081 . [DOI] [PMC free article] [PubMed] [Google Scholar]