Abstract
Signal transduction pathways alter the gene expression program in response to extracellular or intracellular cues. Mitogen-activated protein kinases (MAPKs) govern numerous cellular processes including cell growth, stress response, apoptosis, and differentiation. In the past decade, MAPKs have been shown to regulate the transcription machinery and associate with chromatin-modifying complexes. Moreover, recent studies demonstrate that several MAPKs bind directly to chromatin at target genes. This review highlights the recent discoveries of MAPK signaling in regard to histone modifications and chromatin regulation. Evidence suggesting that further unknown mechanisms integrate signal transduction with chromatin biology is discussed.
Signal transduction pathways alter the gene expression program in response to extracellular or intracellular cues. Mitogen-activated protein kinases (MAPKs) govern numerous cellular processes including cell growth, stress response, apoptosis, and differentiation. Over the past decade, MAPK-signaling studies have shown that phosphorylation of transcription factors by MAPKs is a trigger for the recruitment of transcriptional machinery, including RNA Polymerase II (Pol II), chromatin-remodeling complexes, histone acetyltransferase complexes, and histone deacetylases to target genes (Suganuma and Workman, 2011). Moreover, recent studies demonstrate that several MAPKs bind directly to chromatin at target genes. This Perspective reviews the recent discoveries of MAPK signaling in regard to histone modifications and chromatin regulation. Evidence suggesting that further unknown mechanisms integrate signal transduction with chromatin biology is discussed.
Cells have elaborate systems in place for survival under extracellular stress conditions. For example, MAPKs are responsible for amplification of cell wall integrity (CWI) signaling in Saccharomyces cerevisiae and regulate both gene activation and transcriptional elongation. CWI signaling occurs in response to cell wall stressors, such as elevation of growth temperature (Levin, 2005). Such stress activates MAPKs (Mpk1 or Mlp1), which bind to the transcription factor SBF (SCB [Swi4,6-dependent cell cycle box] binding factor). SBF is comprised of the Swi4 and Swi6 proteins. Phosphorylated Mpk1 and Mlp1 kinases bind to the CWI signaling target gene, FKS2, in response to heat stress. Phosphorylated Mpk1 has also been shown to function in transcription elongation on the FKS2 gene via association with the RNA Pol II-associated complex (Paf1C), consisting of the Paf1, Cdc73m Rtf1, Ctr9, and Leo1 proteins. The association of Paf1 with the FKS2 gene is required for SBF occupancy of the promoter (Kim and Levin, 2011). However, the binding of phosphorylated Mpk1 to Paf1C-Pol II on the coding region is not dependent on SBF, since SBF binds only to the promoter of FKS2. In addition, the association of Mapk1-Paf1-Pol II with FKS2 prevents recruitment of the Sen1 complex (Sen1–Nrd1–Nab3), which has been shown to terminate, specifically, short transcripts including small nucleolar RNA (snoRNA) cryptic unstable transcripts (Arigo et al., 2006). The influence of FKS2 on transcription elongation is lost in the paf1-4A mutant, which is defective in its interaction with Mpk1 but still binds RNA Pol II. This transcriptional suppression is reduced in the sen-1 mutant. Therefore, the repression of FKS2 transcription by dissociation of Paf1 with Mpk1 depends on loss of Sen1 recruitment to FKS2 gene (Kim and Levin, 2011). Hence, Mpk1 links to transcription elongation through its interaction with Paf1; however, the regulation of intermolecular switching between Mpk2 and Sen1 complex with Paf1 on the coding region of the FKS2 gene is unclear.
MAPKs have been found to interact directly with target genes. Upon osmotic stress, the Hog1 kinase translocates into the nucleus (Pelet et al., 2011). Chromatin immunoprecipitation experiments coupled with microarrays (ChIP-chip) have shown that Hog1 binds to 36 different genes at the onset of osmotic stress (Pokholok et al., 2006).
Transcription activation of the Hog1-driven genes, such as pSTL1, increased linearly with increased osmotic stress at high doses of salt (from 0.4 to 0.8 M NaCl). However, it was found that expression of the pSTL1 gene is bimodal under weak osmotic stress conditions (0.15–0.2 M NaCl) through single-cell analysis for fluorescent-expressed proteins and transcription reporter probing by flow cytometry (Pelet et al., 2011). Interestingly, histone H3 quantitative ChIP data showed that bimodal Hog1-driven gene expression is reflected by the degree of histone eviction at the promoter, when the cell is exposed to osmotic stress up to 0.2 M. Thus, the bimodality of expression was attributed to a threshold in activation, resulting from chromatin remodeling at target genes. Interpretively, some cells pass this threshold at low levels of osmotic stress, while others do not. Consistent with this observation, mutations that might impair chromatin remodeling raised this threshold for H3 eviction and increased the bimodality of expression. Hog1 is known to recruit the RSC (remodels the structure of chromatin) nucleosome remodeling complex to stress-response genes. Loss of Hog1 resulted in loss of histone eviction at the promoter. Mutation of SAGA subunits (gcn5, ada2, or spt3) or the RSC subunit (rsc9) reduced histone eviction. Loss of SAGA reduced bimodal expression in low osmotic stress, but loss of RSC had a smaller effect on bimodal gene expression (Pelet et al., 2011). These results suggest that the chromatin-remodeling threshold requires histone acetylation by SAGA (Figure 1). Hog1 phosphorylates the Sko1 transcription factor, which then recruits SAGA to Hog1-activated genes. It has been shown that subunits of RSC contain bromodomains, which interact with acetylated nucleosomes during remodeling (Kasten et al., 2004). Thus, a possible scenario is that Hog1 at the pSTL1 gene promoter recruits SAGA through Sko1 phosphorylation, and SAGA-acetylated histones at the promoter may induce RSC remodeling (Figure 1). However, any of these steps could be rate-limiting, leading to the bimodal gene expression patterns observed at low levels of osmotic stress. It has also been suggested that SAGA may not be required for chromatin remodeling, but its HAT activity plays several important roles in gene activation (Biddick et al., 2008). Hence, other components required for chromatin remodeling are also likely to be involved; for example, histone chaperones accepting the evicted histones. It is still unknown how nucleosomes are rearranged during and after cellular exposure to osmotic stress. Regardless of which exact step is limiting, the requirement for chromatin remodeling imposes a transcriptional threshold in the response of individual cells to linear-signaling gradients (Pelet et al., 2011).
Figure 1.
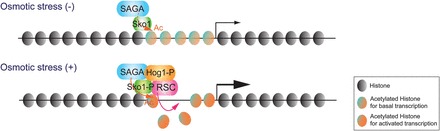
MAPK signals for recruitment of chromatin modifiers in response to stress. The transcription of Hog1 target genes is maintained at a low level in the yeast cells in the absence of stress (Osmotic stress (−)). When cells are exposed to osmotic stress with ∼0.2 M NaCl (Osmotic stress (+)), phosphorylated Hog1 (Hog1-P) efficiently translocates into the nucleus, recruits RSC, and phosphorylates Sko1. Sko1 recruits SAGA onto the nucleosomes of Hog1 target gene promoters. The histones acetylated by SAGA at the promoters are evicted by RSC. Ac indicates acetylation.
Extracellular signal-related kinases (ERK) 1/2 bind directly to the insulin gene promoter in pancreatic beta cells in response to glucose (Lawrence et al., 2008). The binding of ERK1/2 to the insulin or the fos gene promoter in response to glucose is required for ERK1/2 kinase activities. In interferon-gamma-induced genes, the ERK2 kinase binds to sequence-specific sites in the promoters and represses transcription (Hu et al., 2009). The DNA consensus sequence G/CAAAG/C interacts directly with recombinant ERK2, and the binding of ERK2 to this motif suppresses gene expression. Expression of 78 of 82 genes containing this consensus sequence was increased in ERK2 knock-down cells. Moreover, suppression of gene expression by ERK2 did not depend on its kinase activity (Hu et al., 2009). Hence, it would be interesting to study how kinase-activity-dependent transcription activation is coupled with catalytic activity-independent kinase repression of target gene expression.
Histones bound to downstream target genes can be direct targets of MAPK signaling. Progesterone activates the Src/Ras/ERK/MSK1 cascade in T47D-MTVL breast cancer cells. Progesterone receptors ERK and MSK1 are rapidly recruited to the mouse mammary tumor virus (MMTV) promoter, where MSK1 phosphorylates histone H3 serine 10 (H3S10) 5 min after hormone treatment, leading to displacement of the repressor, heterochromatin protein 1 gamma (HP1 γ) (Vicent et al., 2006). Hormone treatment of T47D-MTVL cells also leads to the recruitment of Brg1, a component of the BAF (SWI/SNF) chromatin-remodeling complex and the PCAF (SAGA) histone acetyltransferase, which acetylates histone H3K14 on the promoter (Vicent et al., 2009). Non-histone proteins, including several MAPKs, are known to be reversibly phosphorylated and dephosphorylated. However, phosphorylation of histones on the target genes implies that MAPK may epigenetically control target nucleosomes.
Although cells need to mount a significant transcriptional response to MAPK signaling, their recovery from stress signaling is important. Transcriptional co-repressor complexes have been shown to antagonize transcription activation by MAPK signaling. The methyl-CpG-binding domain protein 3 (MBD3), containing the nucleosome remodeling and histone deacetylation (NuRD) repressor complex, interacts with unphosphorylated c-Jun but not phosphorylated c-Jun (Aguilera et al., 2011). This interaction recruits NuRD onto the activator protein 1 (AP-1) recognition site of the c-Jun gene in HCT116 cells. Acetylation of histone H3K9 and H3K14 was found at a region 5′ of the AP-1 site on the c-Jun gene in the absence of stress. This acetylation required JNK (cJun-NH2-terminal Kinase) activity, indicating a basal level of JNK signaling in the absence of stress. However, under this condition, NuRD suppresses the levels of H3K9 and K14 acetylation (Aguilera et al., 2011). Hence, NuRD may serve to suppress acetylation levels at c-Jun and c-Jun transcription under non-stress conditions.
In Drosophila, the activation transcription factor-2 (ATF2) contributes to heterochromatin formation, maintaining H3K9me2 and HP1 occupancy (Seong et al., 2011). Heat shock induces Mekk1-p38 kinase to phosphorylate ATF2, which then relocalizes to specific loci in euchromatin, leading to disruption of heterochromatin and reduction of H3K9me2 and HP1 occupancy. This stress-induced heterochromatin disruption via ATF2 phosphorylation is inherited in subsequent generations (Seong et al., 2011). Since MAPK activation responds to several different stimuli, it will be important to define how MAPK-induced modifications to chromatin are inherited. Why do suppression mechanisms for recovery from stress signaling not recover the original chromatin configurations? To address the epigenetic effects from stress signaling, it will be important to determine and reproduce the physiologically normal range of heat shock stimulation in animals.
MAPK signaling affects chromatin modifications in multiple ways through phosphorylation of transcription factors, which recruit chromatin-modifying complexes and through direct phosphorylation of histones. Exploring the prevalence and interplay between these pathways will be an important step toward understanding signaling, chromatin, and epigenetics.
[This research was supported by the Stowers Institute for Medical Research. We thank Joanne Chatfield in the Stowers Institute for Medical Research for editing of this manuscript.]
References
- Aguilera C., Nakagawa K., Sancho R., et al. c-Jun N-terminal phosphorylation antagonises recruitment of the MBD3/NuRD repressor complex. Nature. 2011;469:231–235. doi: 10.1038/nature09607. [DOI] [PubMed] [Google Scholar]
- Arigo J.T., Carroll K.L., Ames J.M., et al. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell. 2006;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Biddick R.K., Law G.L., Chin K.K., et al. The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. J. Biol. Chem. 2008;283:33101–33109. doi: 10.1074/jbc.M805258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Xie Z., Onishi A., Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten M., Szerlong H., Erdjument-Bromage H. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.Y., Levin D.E. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.C., McGlynn K., Shao C., Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in beta-cells. Proc. Natl Acad. Sci. USA. 2008;105:13315–13320. doi: 10.1073/pnas.0806465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelet S., Rudolf F., Nadal-Ribelles M., Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science. 2011;332:732–735. doi: 10.1126/science.1198851. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Zeitlinger J., Hannett N.M., Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Seong K.H., Li D., Shimizu H., Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–1061. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- Suganuma T., Workman J.L. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Vicent G.P., Ballare C., Nacht A.S., Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell. 2006;24:367–381. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Vicent G.P., Zaurin R., Ballare C., Erk signaling and chromatin remodeling in MMTV promoter activation by progestins. Nucl. Recept. Signal. 2009;7:e008. doi: 10.1621/nrs.07008. [DOI] [PMC free article] [PubMed] [Google Scholar]


