Abstract
Pancreatic ductal adenocarcinoma (PDAC) represents a biggest challenge in clinic oncology due to its invasiveness and lack of targeted therapeutics. Our recent study showed that schizophrenia susceptibility factor dysbindin exhibited significant higher level in serum of PDAC patients. However, the functional relevance of dysbindin in PDAC is still unclear. Here, we show that dysbindin promotes tumor growth both in vitro and in vivo by accelerating the G1/S phase transition in cell cycle via PI3K/AKT signaling pathway. Mechanistically, dysbindin interacts with PI3K and stimulates the kinase activity of PI3K. Moreover, overexpression of dysbindin in PDAC is correlated with clinicopathological characteristics significantly, such as histological differentiation (P = 0.011) and tumor size (P = 0.007). Kaplan–Meier survival curves show that patients with high dysbindin expression exhibit poorer overall survival, compared to those with low dysbindin expression (P < 0.001). Multivariate analysis reveals that dysbindin is an independent prognostic factor for pancreatic ductal adenocarcinoma (P = 0.001). Thus, our findings reveal that dysbindin is a novel PI3K activator and promotes PDAC progression via stimulation of PI3K/AKT. Dysbindin therefore represents a potential target for prognosis and therapy of PDAC.
Keywords: pancreatic ductal adenocarcinoma, dysbindin, cell cycle, PI3K, prognostic factor
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal malignancy and is the leading cause of cancer-related deaths worldwide (Costello and Neoptolemos, 2011; Ryan et al., 2014). In most cases, PDAC is diagnosed at advanced disease stages, when radical pancreatic resection is not possible. The 5-year survival rate is only 1%–5% after the second stage, and the median survival time is <6 months if the disease is left untreated (Carrato et al., 2015; Le et al., 2016). Despite tremendous research efforts, the mechanism underlying disease progression in PDAC remains largely unclear due to the heterogeneity and complex nature of this malignancy (Garcia et al., 2014; Karandish and Mallik, 2016). Molecular pathological studies have demonstrated that PDAC is driven by a sequence of complex deviations at the molecular level, as gene expression alterations are frequently observed and are crucial for pancreatic ductal adenocarcinoma development (Jones et al., 2008; Lindberg et al., 2014).
Dysbindin, which is mainly expressed in the central nervous system, has emerged as a putative schizophrenia susceptibility factor (Benson et al., 2001; Straub et al., 2002; Kircher et al., 2009). Evidences indicate that dysbindin is involved in neurotransmission transmission and neurodevelopment and is associated with cognitive deficit phenotypes within schizophrenia (Golimbet, 2008; Mullin et al., 2015). Genetic variations in dysbindin may result in heightened susceptibility to schizophrenia, which are considered as the most promising risk factor in the pathogenesis of schizophrenia (Bray et al., 2005; Cheah et al., 2015). Meanwhile, several studies imply that dysbindin may be overexpressed in cancer tissues, since the transcript and alterations of dysbindin have been identified in various cancers (Higo et al., 2005; Cerami et al., 2012; Gao et al., 2013). Little information is available for the role of dysbindin in human cancers. Recently, our study disclosed that serum dysbindin levels were increased in PDAC patients and suggested that dysbindin may be a novel potential serum biomarker for PDAC (Guo et al., 2016). However, the role of dysbindin in pancreatic cancer progression remains confused.
PI3K/AKT signaling is commonly deregulated in pancreatic cancer, which is considered as the second well-characterized RAS effector pathway (Baer et al., 2015; Mann et al., 2016). Accumulating evidence has uncovered that PI3K/AKT signaling is involved in many cellular processes, such as proliferation, suppression of apoptosis, metastasis, and angiogenesis, implying an essential role of PI3K/AKT signaling in pancreatic cancer (Ma et al., 2010; Sun et al., 2016; Wang et al., 2016). Previous study has demonstrated that dysbindin can activate PI3K/AKT signaling pathway (Numakawa et al., 2004), but the underlying mechanism remains to be elucidated.
In this study, we provided the first evidence that dysbindin facilitated tumor growth in vitro and in vivo by promoting the G1/S phase transition. We then elucidated the underlying mechanism that dysbindin upregulated the PI3K/AKT signaling by interacting with PI3K and modulating the kinase activity of PI3K. Further evidences illustrated that overexpression of dysbindin in PDAC was significantly correlated with clinicopathologic characteristics and poor prognosis of patients.
Results
Dysbindin promotes tumor growth in vitro and in vivo
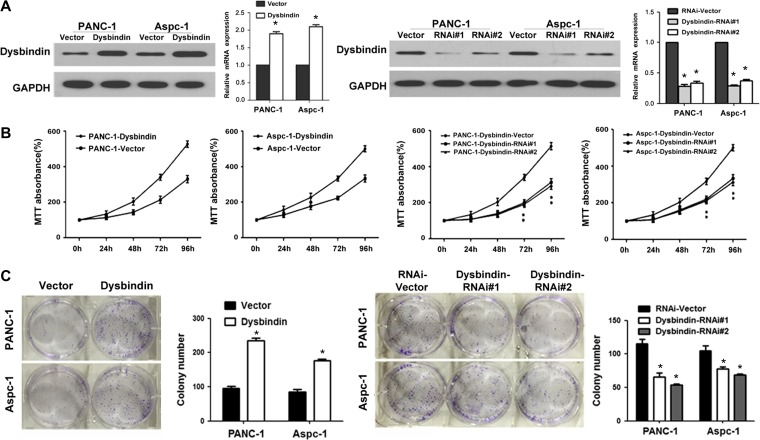
As overexpression of dysbindin is commonly noted in germinal cells of mouse embryos, which is suggested to play a role in cell proliferation (Higo et al., 2005), we investigated the effect of dysbindin on pancreatic tumor growth. PANC-1 and Aspc-1 cell lines with dysbindin overexpression and dysbindin knockdown were established, respectively (Figure 1A). As determined by MTT assays, cell viability was enhanced in dysbindin-overexpressing cells. Conversely, dysbindin knockdown was dramatically associated with a significant decrease in cell viability (Figure 1B). We further examined the effects of dysbindin on the colony formation ability of pancreatic cancer cells. As shown in Figure 1C, dysbindin-overexpressing cells exhibited higher colony formation ability, as the size and number of cell colonies were increased compared to control cells. Parallel results were observed in dysbindin-knockdown cells, as the size and number of cell colonies were decreased compared to control cells.
Figure 1.
Dysbindin promotes cell proliferation in vitro. (A) Western blotting analysis and real-time PCR for validation of dysbindin-overexpressing and dysbindin-knockdown PANC-1 and Aspc-1 cells. (B) MTT assays of dysbindin-overexpressing and dysbindin-knockdown cells. (C) Colony formation assay of dysbindin-overexpressing and dysbindin-knockdown cells. Data are expressed as mean ± SEM of three independent experiments in triplicate. Quantitative analyses are representative results from three independent experiments. *P < 0.05.
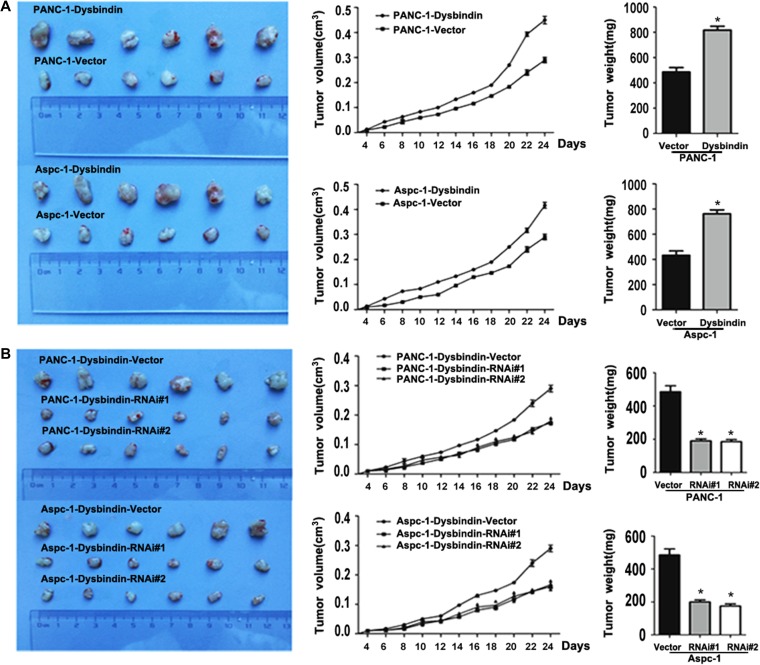
To investigate the role of dysbindin in tumorigenesis in vivo, xenograft tumor models were performed in nude mice by subcutaneously inoculating dysbindin-overexpressing and dysbindin-knockdown cells in the dorsal aspect of mice. We found that tumors formed in mice inoculated with dysbindin-overexpressing cells grew significantly faster than the control group at the indicated time points and presented increased tumor weight. Consistently, dysbindin-knockdown groups exhibited smaller tumor volumes than the control group at the indicated time points and the tumor weight was significantly decreased (Figure 2). Hence, we suggested that dysbindin could promote tumor tumorigenesis in pancreatic cancer.
Figure 2.
Dysbindin facilitates tumor growth in vivo. (A) Xenografted tumors of dysbindin-overexpressing and control groups. Left: tumor size; middle: tumor volumn detected at the indicating time points; right: tumor weight. (B) Xenografted tumors of dysbindin-knockdown and control groups. Left: tumor size; middle: tumor volume detected at the indicating time points; right: tumor weight. *P < 0.05.
Dysbindin accelerates the G1/S phase transition and regulates cell cycle-related factors
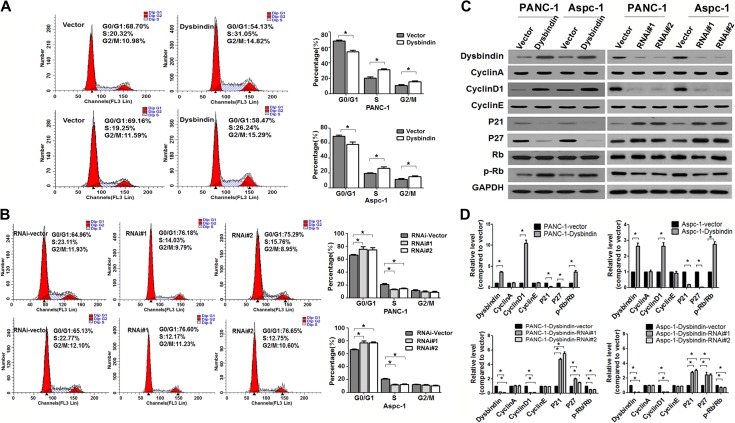
As shown in Figure 3A, flow cytometry data indicated that dysbindin overexpression significantly increased the percentage of cells in the S and G2 phases and decreased the percentage of cells in the G0/G1 phase. Conversely, dysbindin knockdown increased the percentage of cells in the G0/G1 phase and decreased the percentage of cells in the S phase (Figure 3B). This result indicated that dysbindin accelerated the G1/S phase transition in pancreatic cancer cells.
Figure 3.
Dysbindin accelerates the G1/S phase transition and regulates the expression of cell cycle-related factors. (A) Flow cytometry of dysbindin-overexpressing and control cells. Data are expressed as mean ± SEM of three independent experiments in triplicates. *P < 0.05. (B) Flow cytometry of dysbindin-knockdown and control cells. Data are expressed as mean ± SEM of three independent experiments in triplicate. *P < 0.05. (C) Western blotting analysis of proteins involved in cell cycle. (D) Quantification of C from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05.
Given the above changes in cell cycle, we subsequently measured the expression of cyclin A, cyclin D1, and cyclin E, which may regulate the G1/S phase transition. The expression of cyclin D1 was exclusively increased in dysbindin-overexpressing cells and was exclusively reduced in dysbindin-knockdown cells, while the expression of cyclin A and cyclin E remained unaltered. The expression of Rb protein, a downstream target of cyclin D1, did not change; however, the expression of p-Rb was increased in dysbindin-overexpressing cells and was reduced in dysbindin-knockdown cells. Moreover, the expression of p21Clip1 and p27Kip1, the cell cycle inhibitors, was suppressed by dysbindin (Figure 3C and D). These data confirmed that dysbindin regulated the G1/S phase transition in pancreatic cancer cells, consistent with those results of the cell cycle analysis.
Dysbindin upregulates PI3K/Akt signaling pathway by promoting PI3K kinase activity
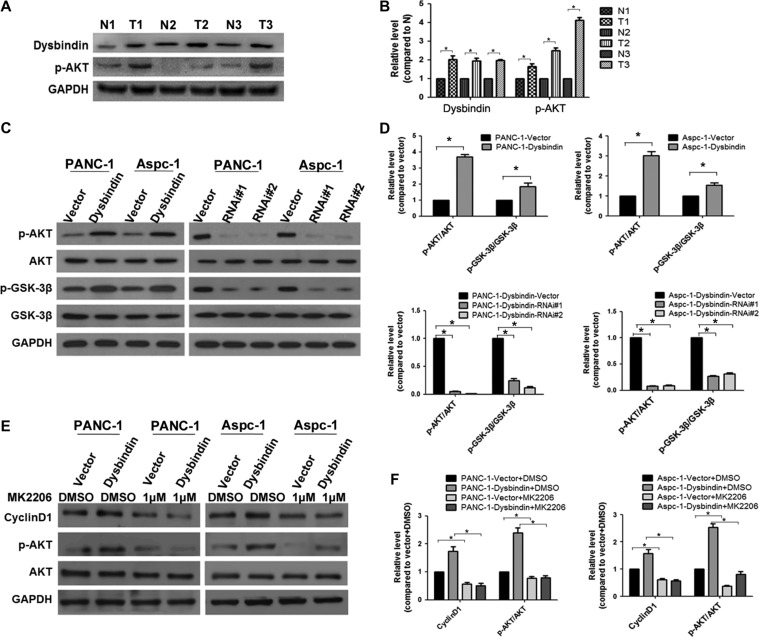
As cyclin D1 was regulated by the AKT/GSK-3β signaling pathway (Zeng et al., 2012), we questioned whether dysbindin influences cell cycle by regulating the AKT signaling pathway. First, we detected the phosphorylation level of AKT in pancreatic cancer to verify whether the AKT signaling pathway was activated in pancreatic cancer. We found that the phosphorylation level of AKT was much higher in tumor specimens than in adjacent noncancerous specimens, consistent with the increased expression of dysbindin in cancerous specimens (Figure 4A and B). We then detected AKT phosphorylation levels in dysbindin-overexpressing or dysbindin-knockdown cells by western blotting analysis. Dysbindin overexpression increased the level of phosphorylated AKT, while dysbindin knockdown decreased the level of phosphorylated AKT. The level of p-GSK-3β was also increased in dysbindin-overexpressing cells and reduced in dysbindin-knockdown cells, consistent with the expression of cyclin D1 in these cells (Figure 4C and D). MK2206, a specific AKT inhibitor to block AKT signaling, could significantly reduce the expression of cyclin D1 induced by dysbindin, following the abrogation of AKT activity (Figure 4E and F).
Figure 4.
Dysbindin promotes tumor cell growth via activation of AKT. (A) Western blotting analysis of dysbindin and phosphorylated AKT in PDAC tissue samples. N: adjacent noncancerous tissues, T: tumor tissues. (B) Quantification of A from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05. (C) Western blotting analysis of the AKT/GSK-3β pathway in stable cells. (D) Quantification of C from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05. (E) Western blotting analysis of AKT activity in dysbindin-overexpressing cells treated with MK2206 at a concentration of 1 μM for 24 h. (F) Quantification of E from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05.
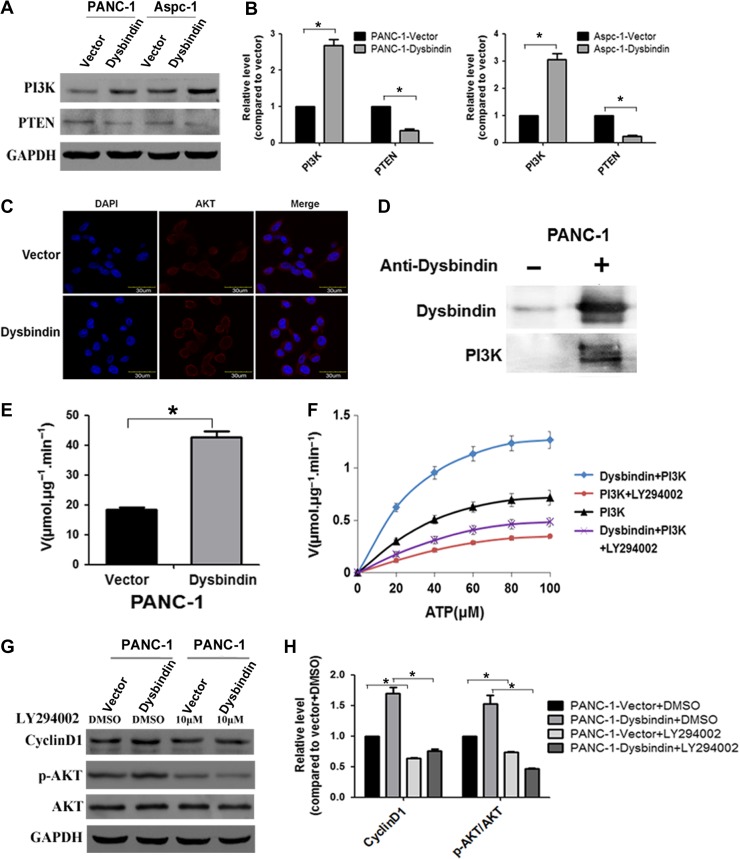
PI3K can recruit and phosphorylate AKT via the production of PIP3, and PTEN is a major negative regulator of PI3K/AKT signaling. In this study, we found that PI3K was upregulated and PTEN was downregulated in dysbindin-overexpressing cells, suggesting that dysbindin could upregulate the PI3K/AKT pathway with a suppression of PTEN activity (Figure 5A and B). Immunofluorescence showed that the level of AKT located in the plasma membrane was increased in dysbindin-overexpressing cells, compared to control cells (Figure 5C), suggesting that increased activity of PI3K by dysbindin promoted the recruitment of AKT to the membrane. To identify the protein interacting with dysbindin in PI3K/AKT pathway, we performed immunoprecipitation experiment by which we discovered the dysbindin−PI3K interaction in PANC-1 cells, demonstrated by western blotting analysis (Figure 5D). To test whether dysbindin modulates PI3K activity, an in vitro kinase assay as recently described was carried out (Mo et al., 2016). Specifically, aliquots (100 ng) of recombinant active PI3 kinase from Sigma was incubated with 10 μM PIP2 and 100 M ATP in the presence or absence of purified dysbindin (100 ng), followed by standard kinase assay as described (Mo et al., 2016). As shown in Figure 5E, dysbindin significantly increased the PI3K kinase activity. As reflected by PIP2 phosphorylation, dysbindin stimulated the kinase activity of PI3K directly (Figure 5F; P < 0.01). Importantly, this dysbindin-elicited increase in PI3K activity was abolished upon an addition of PI3K inhibitor LY294002, confirming that the stimulation of kinase activity was due to PI3 kinase other than the potential catalytic activities co-purified with dysbindin. Furthermore, blockage of PI3K signaling using LY294002 significantly reduced the level of phosphorylated AKT and cyclin D1 in dysbindin-overexpressing cells (Figure 5G and H). These results demonstrated that dysbindin directly stimulated the kinase activity of PI3K in vitro and in cultured PDAC cells.
Figure 5.
Dysbindin interacts with PI3K and upregulates its kinase activity. (A) Western blotting analysis of the PI3K and PTEN in dysbindin-overexpressing and control cells. (B) Quantification of A from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05. (C) Immunofluorescent images representative of AKT (red) in the dysbindin-overexpressing and control cells. Original magnification: 120×. (D) Immunoprecipitation analysis for the interaction of dysbindin and PI3K in PANC-1 cells. (E) The PI3K kinase activity in dysbindin-overexpressing cells was measured by fluorometry-based kinase assay. (F) Hela cells expressing FLAG-dysbindin were synchronized by nocodazole (100 ng/ml), and protease inhibitor cocktail and phosphatase inhibitor cocktail were added to cells for 0.5 h in the presence of MG132. Immuno-isolated dysbindin was subjected to an in vitro kinase assay with PI3K. The kinetics curves of dysbindin were generated by fluorometry-based kinase assay. (G) Western blotting analysis of AKT/Cyclin D1 in dysbindin-overexpressing and control cells treated with LY294002 at a concentration of 10 μM for 24 h. (H) Quantification of G from three independent experiments. Data are expressed as mean ± SEM. *P < 0.05.
Dysbindin expression correlates with clinicopathologic characteristics of PDAC patients
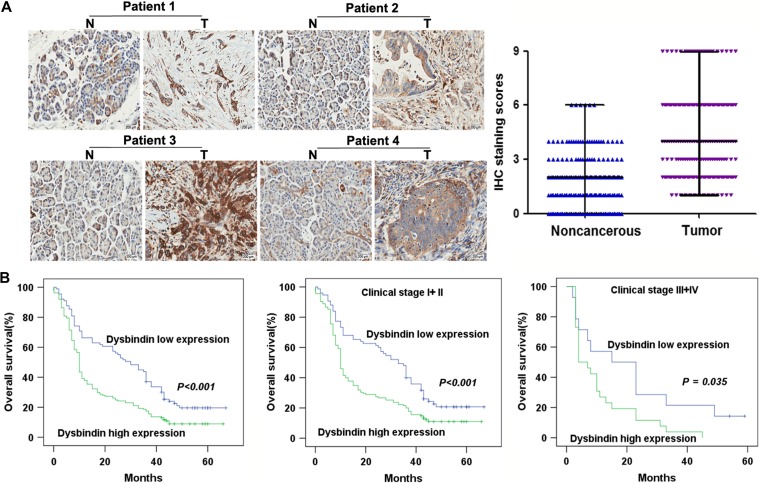
To examine whether dysbindin is involved in PDAC development we detected dysbindin expression in a tissue microarray. We found that dysbindin staining was much stronger in tumor lesions (T) than in paired adjacent noncancerous tissues (N) (P < 0.001) (Figure 6A). The clinicopathological characteristics of the patients subjected to the tissue microarray and the IHC staining results are presented in Table 1. More than half of PDAC tissues (n = 161, 64.4%) exhibited high dysbindin expression (score ≥4); however, 35.6% of samples (n = 89) exhibited low dysbindin expression (score < 4). Dysbindin expression was correlated with histological differentiation (P = 0.011) and tumor size (P = 0.007). However, dysbindin expression was not correlated with age, gender, tumor location, lymph node invasion, or clinical stage (P > 0.05).
Figure 6.
The expression of dysbindin is increased in PDAC and correlated with overall survival of PDAC patients. (A) IHC for dysbindin and quantification of staining intensity in PDAC and adjacent noncancerous tissues. N, noncancerous tissues; T, tumor tissues. Original magnification, 200×. (B) Overall survival of PDAC patients with high and low expression levels of dysbindin.
Table 1.
Correlations between dysbindin expression and clinical characteristics in patients with PDAC.
| Parameters | Patient number (n = 250) | Dysbindin expression | P-value (χ2 test) | |
|---|---|---|---|---|
| High (n = 161) | Low (n = 89) | |||
| Age (years) | ||||
| ≤60 | 120 | 72 | 48 | |
| >60 | 130 | 89 | 41 | 0.187 |
| Gender | ||||
| Male | 131 | 89 | 42 | |
| Female | 119 | 72 | 47 | 0.236 |
| Tumor location | ||||
| Head | 39 | 28 | 11 | |
| Body/tail | 211 | 133 | 78 | 0.364 |
| Histological differentiation | ||||
| Well | 23 | 9 | 14 | |
| Moderate/poor | 227 | 152 | 75 | 0.011* |
| Size | ||||
| ≤2 cm | 40 | 18 | 22 | |
| >2 cm | 210 | 143 | 67 | 0.007* |
| Lymph node invasion | ||||
| Absent | 148 | 90 | 58 | |
| Present | 102 | 71 | 31 | 0.179 |
| Clinical stage | ||||
| I + II | 210 | 135 | 75 | |
| III + IV | 40 | 26 | 14 | 0.931 |
*Statistically significant.
P-values <0.05 are in bold.
Dysbindin expression is associated with overall survival in PDAC patients
Kaplan–Meier survival curves and log-rank test survival analyses indicated that the overall survival of patients with high dysbindin expression was significantly poorer than that of patients with low dysbindin expression (P < 0.001), as well as in early stage (P < 0.001) and advanced stage of PDAC (P = 0.035), respectively (Figure 6B). The parameters related to patient prognosis were then evaluated using univariate and multivariate analyses, as shown in Table 2. Univariate analysis showed that dysbindin expression (HR: 1.788, 95% CI: 1.339–2.387, P < 0.001), histological differentiation (HR: 1.712, 95% CI: 1.055–2.779, P = 0.030), tumor size (HR: 1.975, 95% CI: 1.325–2.942, P = 0.001), lymph node invasion (HR: 2.044, 95% CI: 1.553–2.690, P < 0.001), and clinical stage (HR: 1.649, 95% CI: 1.159–2.346, P = 0.005) were prognostic factors for overall survival in PDAC patients. Furthermore, multivariate analysis using Cox proportional hazards model showed that dysbindin expression (HR: 1.668, 95% CI: 1.240–2.244, P = 0.001), lymph node invasion (HR: 2.265, 95% CI: 1.699–3.020, P < 0.001), and clinical stage (HR: 1.894, 95% CI: 1.303–2.755, P = 0.001) were significant and independent prognostic factors for overall survival in PDAC patients. These results indicated that dysbindin expression was associated with the prognosis of PDAC patients.
Table 2.
Univariate and multivariate analyses of prognostic parameters for survival in PDAC patients.
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Dysbindin expression (high vs. low) | 1.788 | 1.339–2.387 | <0.001* | 1.668 | 1.240–2.244 | 0.001* |
| Age (≤60 vs. >60) | 1.141 | 0.872–1.491 | 0.336 | |||
| Gender (male vs. female) | 0.944 | 0.722–1.234 | 0.671 | |||
| Tumor location (head vs. body/tail) | 1.097 | 0.753–1.598 | 0.628 | |||
| Histological differentiation (well vs. moderate/poor) | 1.712 | 1.055–2.779 | 0.030* | 1.464 | 0.889–2.410 | 0.134 |
| Size (≤2 cm vs. >2 cm) | 1.975 | 1.325–2.942 | 0.001* | 1.492 | 0.991–2.245 | 0.055 |
| Lymph node invasion (absent vs. present) | 2.044 | 1.553–2.690 | <0.001* | 2.265 | 1.699–3.020 | <0.001* |
| Clinical stage (I + II vs. III + IV) | 1.649 | 1.159–2.346 | 0.005* | 1.894 | 1.303–2.755 | 0.001* |
Univariate analysis and multivariate analyses were analyzed by Cox proportional hazards model. HR, hazard ratio; 95% CI, 95% confidence interval.
*Statistically significant.
P-values <0.05 are in bold.
Discussion
Our previous study provided the first line of evidence that dysbindin as a novel serum biomarker candidate may be valuable for the screenings for PDAC (Guo et al., 2016). However, the functional role of dysbindin in pancreatic cancer has not been characterized. As an essential component of biogenesis in lysosome-related organelles complex-1 (BLOC-1), dysbindin is recognized as a synaptic and microtubular protein that binds to brain snapin (Talbot et al., 2006; Mullin et al., 2011). In the central nervous system, dysbindin enacts its chief functions of facilitating neurite outgrowth and regulating synaptic activity (Liao and Chen, 2004; Ma et al., 2011). Moreover, two following binding sites for transcription factors were identified in the promotor region of dysbindin: nuclear factor-1 (NF-1) and specificity protein-1 (SP-1), both of which may regulate cell growth and embryogenesis (Liao and Chen, 2004; Plachez et al., 2008; Zhao et al., 2013; Beketaev et al., 2016). Transcription factor E2F1, which regulates cell growth and proliferation, can bind to the dysbindin promoter and cause a 10-fold increase in dysbindin expression (Iwanaga et al., 2006). These results collectively suggested that dysbindin may be involved in cell viability; however, the role of dysbindin in cell growth and proliferation has not been verified. As a result of this investigation, we found that dysbindin promotes pancreatic tumor growth via PI3K pathway. Since majority of solid tumor cells exhibit their invasiveness in PI3K-dependent manner, it would be of great interest to examine whether the PI3K-dependent effect of dysbindin is generally conserved in solid tumors. Thus, our demonstration of dysbindin-elicited tumor growth is by all means not unique for PDCA at this point.
It is well known that cell cycle control is crucial for cell proliferation, and cell cycle dysregulation is a hallmark of tumor cells (Oshima and Campisi, 1991; Stewart et al., 2003; Golias et al., 2004). As we demonstrated that dysbindin may promote cell growth, we then analyzed the changes in cell cycle and found that dysbindin induced an increased fraction of cells in the S phase at the expense of the G0/G1 phase, suggesting that dysbindin may promote the G1/S phase transition in cell cycle. Growth regulatory factors are generally confirmed to function at specific cell cycle stages, such that cyclin A and cyclin E govern the G1/S phase transition and S-phase progression (Mishra, 2013) and cyclin B is required for the G2/M phase transition (Sanchez and Dynlacht, 2005). Cyclin D1 has been verified as a vital protein with respect to the regulation of cell cycle, especially for exiting from the G1 phase and passing through the G1/S phase transition (Sanchez and Dynlacht, 2005; Zhang et al., 2012). A recent study suggested that dysbindin may directly bind to the C-terminal of cyclin D1 and modulate the homeostasis of cyclin D1 in HEK293 cells (Ito et al., 2016). In this study, we noticed that dysbindin upregulated the levels of phosphorylated Rb and cyclin D1, accompanied with decreased expression of the CDK inhibitors p21 and p27. However, cyclin A and cyclin E did not significantly change. Thus, we reason that dysbindin promotes the G1/S phase transition probably via upregulating cyclin D1.
Furthermore, AKT is an important serine/threonine kinase involved in various biological processes. Activated AKT has been demonstrated to upregulate cyclin D1 by phosphorylating and inactivating GSK-3β (Diehl et al., 1998; Parrales et al., 2011). PI3K can phosphorylate and activate AKT by recruiting AKT to the plasma membrane (King et al., 2015). As we found that dysbindin upregulated the phosphorylated level of AKT, we then examined whether PI3K was involved in dysbindin-mediated process. PI3K is an upstream of AKT, with both serine/threonine kinase activity and phosphatidylinositol kinase activity. Previous studies predicted that dysbindin may contain several serine/threonine phosphorylation sites, implying the potential interaction between dysbindin and serine/threonine kinase. Our results showed that dysbindin could interact with PI3K and directly upregulate the kinase activity of PI3K, which may result in AKT recruitment to the plasma membrane. These findings indicated that dysbindin promoted cell proliferation via the PI3K/AKT signaling pathway, consistent with a previous study reporting dysbindin-mediated promotion of cell viability (Numakawa et al., 2004). However, the underlying mechanism by which dysbindin activates PI3K in cancer cells needs further investigations.
Despite great efforts in investigations of molecular markers for PDAC, none have been successfully integrated into clinical practice (Winter et al., 2013). To date, serum CA-199 remains the routinely used biomarker approved by the FDA for PDAC; however, the clinical use of CA-199 has limited values (Winter et al., 2013). Considering the role of dysbindin in tumor growth, we evaluated the possibility of dysbindin as a potential biomarker in predicting the clinical outcome of PDAC patients. In this study, we detected the expression of dysbindin in PDAC and investigated the correlations between dysbindin expression and PDAC progression. We found that dysbindin expression in PDAC tissues was markedly correlated with histological differentiation and tumor size. Moreover, high dysbindin expression was also significantly associated with poorer overall survival of patients. Our previous study revealed that serum dysbindin was effective in screening of PDAC (Guo et al., 2016) and this study suggested that dysbindin may serve as an independent prognostic factor for PDAC, demonstrating a novel potential role of dysbindin in PDAC management.
Taken together, our study shows that dysbindin promotes tumor growth by activating the PI3K/AKT signaling pathway and overexpression of dysbindin is associated with poor prognosis of PDAC. Our findings indicate that dysbindin is a novel prognostic indicator and a potential therapeutic target for PI3K-active tumors such as PDAC. Further studies are needed to evaluate whether activity of dysbindin can be manipulated to inhibit PDAC progression and delineate the mechanism of action underlying dysbindin-elicited PI3K activation in PDAC.
Materials and methods
Cell culture and retroviral infection
The human pancreatic cancer cell lines PANC-1 and Aspc-1 were purchased from American Type Culture Collection (ATCC) and cultured at 37°C in 5% CO2 in DMEM/high glucose medium (Hyclone) supplemented with 10% heat-inactivated fetal bovine serum (Biological Industries). The dysbindin expression construct was generated by cloning PCR-amplified human dysbindin cDNA into a pMSCV plasmid. Two human dysbindin-targeting siRNA sequences were cloned to generate dysbindin-shRNA(s) with the following sequences: RNAi#1 (GCAGATGGACCTGATGGACAT) and RNAi#2 (GCCTGAATCCAGTACCTGTCA) (synthesized by GenePharma). Retroviral production and infection were conducted according to the manufacturer’s instructions. Stable cell lines were generated upon selection with 0.5 μg/ml puromycin for 10 days. MK-2206 was obtained from Selleck Chemicals, and dissolved with DMSO. Cells were treated with MK-2206 at the concentration of 1 μM for 24 h (Deng et al., 2014). LY294002 was obtained from Sigma, and dissolved with DMSO. Cells were treated with LY294002 at the concentration of 10 μM for 24 h (Shi et al., 2014).
Flow cytometry
The harvested cells were fixed in 70% ethanol overnight and rehydrated in phosphate-buffered saline (PBS). Then, the cells were treated with RNase A (1 mg/ml) for 30 min and stained with propidium iodide (10 μg/ml) for 5 min. Finally, the cells were analyzed using a FACScan analyzer (Becton Dickinson).
MTT assay
The cells were seeded in 96-well plates at a density of 1 × 103 cells/well and incubated overnight. Next, 20 μl of MTT solution (5 mg/ml in PBS) were added to each well, and the cells were incubated for an additional 4 h at 37°C. Then, 150 μl of DMSO (Sigma-Aldrich) was added to each well. The optical density (OD) was measured at a wavelength of 490 nm. Three experimental replicates were performed.
Colony formation assay
Transfected cells and control cells were plated in 6-well plates (5 × 102 cells/plate) and cultured for 10 days. The colonies were stained with 1% crystal violet for 30 sec after fixation with 10% formaldehyde for 15 min.
Xenograft tumor models
Balb/c nude male mice (6–8 weeks of age) were obtained from Weitonglihua Experimental Animal Technical Co., LTD. The animals were maintained under specific pathogen-free conditions, with food and water supplied ad libitum. The Balb/c nude mice were randomly divided into 10 groups (n = 6/group). Stable cells were injected subcutaneously on the dorsal aspect of each mouse at a concentration of 1 × 106/ml. The tumors were examined once weekly. Length, width, and thickness measurements were performed using calipers, and tumor volumes were calculated from these measurements. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
RNA extraction and real-time PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Reverse transcription reactions were performed using a high capacity cDNA reverse transcription kit (Promega). Real-time PCR was performed using a Bio-Rad instrument (Roche) with the SYBR Premix Ex Taq TM (TaKaRa) according to the manufacturer’s instruction. All reactions were performed in a 25 μl reaction volume in triplicates. Dysbindin and GAPDH primers were obtained from Sangon Biotech as follows: dysbindin, (forward) 5′-GTACCTGTCCACTGGCTAC-3′ and (reverse) 5′-CCTCCTGGTCCGATATGTC-3′; GAPDH, (forward) 5′-CTGACTTCAACAGCGACACC-3′ and (reverse) 5′-TGCTGTAGCCAAATTCGTTG-3′ (Huang et al., 2005). The fold change in the expression of each target gene was calculated by the 2−△△CT method, and the relative amount of target gene mRNA was normalized to the house-keeping gene GAPDH.
Western blotting
Proteins extracted either from tissues or from cells were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes, which were incubated with anti-dysbindin, anti-GAPDH, anti-p21, anti-p27, anti-Cyclin A, anti-cyclin D1, anti-Cyclin E, anti-p-Rb, anti-Rb, anti-p-GSK-3β, anti-GSK-3β (Abcam), anti-PI3K, anti-p-AKT, anti-AKT (Millipore), and anti-PTEN (Abmart). Peroxidase-conjugated anti-mouse or anti-rabbit antibodies were used as secondary antibodies, and the final bands were visualized by enhanced chemiluminescence assay (Thermo Scientific). The quantification of western blot was performed using ImageJ software, and the statistical analyses were conducted from three independent experiments. The density of each band was normalized to GAPDH, and the cells transfected with vector or treated with DMSO were employed as the control.
Immunoprecipitation
The experiments were performed as previously described (Liu et al., 2016). PANC-1 cells (1 × 106) were lysed in 1 ml of lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride). Lysates were clarified by centrifugation at 16000 g for 10 min at 4°C. For each immunoprecipitation, 0.4 ml lysate was incubated with 0.5 mg anti-dysbindin antibody or control IgG for 2 h. Sepharose beads were washed three times with 1 ml lysis buffer containing 0.25 M NaCl. The precipitates were analyzed by standard immunoblot procedures.
Immunofluorescent staining and confocal microscopy
Cells were washed three times with cold phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 30 min. After blocked with 10% goat serum, cells were incubated with anti-AKT antibody (Millipore) overnight in 4°C. After PBS washes, cells were incubated with Cy3-conjugated secondary antibody and counterstained with DAPI. Laser confocal microscopy was performed, using an Olympus Fluoview FV 1000 confocal laser microscope (Olympus). Images of AKT staining were captured using the red channel at 120× magnification.
Enzymatic assay and characterization of kinetics
Dysbindin was purified from HEK293T cells, PI3K was purchased from Abcam, and PIP2 was purchased from J&K Chemical. ADP assay were performed using Universal Fluorometric Kinase Assay Kits (Sigma). For the kinetics of dysbindin, dysbindin (10 nM) was incubated with PI3K (10 nM) and P1P2 (10 μM) in 20 μl kinetics reaction solution (60 mM HEPES, pH 7.5, 3 mM MgCl2, 3 mM MnCl2, and 3 μM Na-orthovanadate) in the presence of 0–100 μM ATP for 0.5 h at 37°C. To determine the effect of dysbindin on kinase activity, dysbindin-overexpressing cells were lysed, and aliquots of cell lysates were incubated in 20 μl kinetics reaction solution for 0.5 h at 37°C. Then the ADP sensor buffer and sensor solution were added, and the assay mixture was incubated for another 15 min at room temperature. The fluorescence intensities were monitored and quantified as a 540/590 nm excitation/emission ratio. An ADP standard curve was obtained for quantification purposes, and the Km and kcat values were calculated according to the Michaelis–Menten equation. To validate the dysbindin-elicited PI3K activity, we include PI3K inhibitor in addition.
Patients and tissue samples
PDAC tissue samples and paired adjacent noncancerous samples were obtained from 250 patients, who underwent surgery in Xijing Hospital of the Fourth Military Medical University from August 2010 to August 2013. None of the patients had received any treatments prior to undergoing the investigational surgery, such as surgery, chemotherapy, or radiotherapy. Primary cancer tissues were verified independently by three pathologists. Pancreatic cancer histological classification and staging were performed using the TNM (tumor, node, metastasis) criteria of the International Union Against Cancer and the Guidelines for Clinical and Pathological Studies of PDAC. This study was approved by the ethics committee of the Xijing Hospital of the Fourth Military Medical University, and written informed consent was obtained from all patients.
Tissue microarray construction and immunohistochemistry
Tissue microarray (TMA) construction was performed as described previously (Ke et al., 2009). Immunohistochemistry (IHC) was performed on paraffin sections on the TMA, according to the manufacturer’s protocols. Briefly, the sections were heated at 60°C for 30 min and washed with xylene to de-wax. The samples were then rehydrated with different grade concentrations of ethanol. Antigen was retrieved by autoclaving the tissue sections for 5 min in 10 mM citrate buffer (pH 6.0). After the sections had cooled to room temperature, endogenous peroxidase was blocked by 3% H2O2 for 20 min, and the sections were incubated in goat serum for 60 min to reduce non-specific reactions. Subsequently, the samples were incubated with a rabbit monoclonal antibody against dysbindin (1:200 dilution; Abcam) overnight at 4°C. Then, the appropriate secondary antibody was used. Finally, the TMA slides were stained with diaminobenzidine and counterstained with hematoxylin.
Evaluation of immunostaining intensity
The IHC results were evaluated independently by two pathologists, and dysbindin staining was quantified based on the proportion of positively stained tumor cells (staining area) and the staining intensity. The indicated staining scores were applied for the corresponding intensities (0, no staining; 1, light yellow; 2, yellow brown; 3, strong brown color), and the indicated scores were applied for the corresponding proportions of positive tumor cells (0, 0% positive tumor cells; 1, 0%–10% positive tumor cells; 2, 10%–50% positive tumor cells; 3, 50%–100% positive tumor cells). The final immunoreactivity score (IS) for grouping was the product of the staining area score and the staining intensity. For the statistical analysis, the scores were grouped in two categories: scores of 0–3 were considered as low expression and 4–9 as high expression (Moss et al., 2008).
Statistical analysis
Statistical analyses were conducted using SPSS 17.0 for Windows. Comparisons between two categories were performed using Mann–Whitney tests, Student’s t-test, and correlations between dysbindin expression and clinicopathological characteristics were evaluated using the Chi-square test. Survival curves were plotted by the Kaplan–Meier method using the log-rank test. Prognostic value with respect to predicting overall survival was assessed by univariate and multivariate Cox proportional hazards regression analysis. P < 0.05 was considered statistically significant.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81372607, 31430054, 31320103904, and 31621002), National Key Research and Development Program of China (2017YFC1308600), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19040000), the Ministry of Science and Technology of China (2017YFA0503600 and 2016YFA0100500), and MOE Innovative Team (IRT_17R102).
Conflict of interest: none declared.
Acknowledgements
The authors are grateful to Yueyun Ma (Department of Clinical Laboratory Medicine, Xijing Hospital, Fourth Military Medical University), Yu Xue (Huazhong University of Science and Technology), and Quan Bai (Northwest University, China) for technical help.
References
- Baer R., Cintas C., Therville N., et al. (2015). Implication of PI3K/Akt pathway in pancreatic cancer: when PI3K isoforms matter? Adv. Biol. Regul. 59, 19–35. [DOI] [PubMed] [Google Scholar]
- Beketaev I., Zhang Y., Weng K.C., et al. (2016). cis-Regulatory control of Mesp1 expression by YY1 and SP1 during mouse embryogenesis. Dev. Dyn. 245, 379–387. [DOI] [PubMed] [Google Scholar]
- Benson M.A., Newey S.E., Martin-Rendon E., et al. (2001). Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 276, 24232–24241. [DOI] [PubMed] [Google Scholar]
- Bray N.J., Preece A., Williams N.M., et al. (2005). Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum. Mol. Genet. 14, 1947–1954. [DOI] [PubMed] [Google Scholar]
- Carrato A., Falcone A., Ducreux M., et al. (2015). A systematic review of the burden of pancreatic cancer in Europe: real-world impact on survival, quality of life and costs. J. Gastrointest. Cancer 46, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah S.Y., Lawford B.R., Young R.M., et al. (2015). Dysbindin (DTNBP1) variants are associated with hallucinations in schizophrenia. Eur. Psychiatry 30, 486–491. [DOI] [PubMed] [Google Scholar]
- Costello E., and Neoptolemos J.P. (2011). Pancreatic cancer in 2010. New insights for early intervention and detection. Nat. Rev. Gastroenterol. Hepatol. 8, 71–73. [DOI] [PubMed] [Google Scholar]
- Deng X., Hu J., Ewton D.Z., et al. (2014). Mirk/dyrk1B kinase is upregulated following inhibition of mTOR. Carcinogenesis 35, 1968–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl J.A., Cheng M., Roussel M.F., et al. (1998). Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.N., Grasso D., Lopez-Millan M.B., et al. (2014). IER3 supports KRAS(G12D)-dependent pancreatic cancer development by sustaining ERK1/2 phosphorylation. J. Clin. Invest. 124, 4709–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golias C.H., Charalabopoulos A., and Charalabopoulos K. (2004). Cell proliferation and cell cycle control: a mini review. Int. J. Clin. Pract. 58, 1134–1141. [DOI] [PubMed] [Google Scholar]
- Golimbet V.E. (2008). Molecular genetics of cognitive deficit in schizophrenia. Mol. Biol. 42, 830–839. [PubMed] [Google Scholar]
- Guo X., Lv X., Fang C., et al. (2016). Dysbindin as a novel biomarker for pancreatic ductal adenocarcinoma identified by proteomic profiling. Int. J. Cancer 139, 1821–1829. [DOI] [PubMed] [Google Scholar]
- Higo M., Uzawa K., Kouzu Y., et al. (2005). Identification of candidate radioresistant genes in human squamous cell carcinoma cells through gene expression analysis using DNA microarrays. Oncol. Rep. 14, 1293–1298. [PubMed] [Google Scholar]
- Huang H., Regan K.M., Wang F., et al. (2005). Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl Acad. Sci. USA 102, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Morishita R., and Nagata K. (2016). Schizophrenia susceptibility gene product dysbindin-1 regulates the homeostasis of cyclin D1. Biochim. Biophys. Acta 1862, 1383–1391. [DOI] [PubMed] [Google Scholar]
- Iwanaga R., Komori H., Ishida S., et al. (2006). Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene 25, 1786–1798. [DOI] [PubMed] [Google Scholar]
- Jones S., Zhang X., Parsons D.W., et al. (2008). Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandish F., and Mallik S. (2016). Biomarkers and targeted therapy in pancreatic cancer. Biomark. Cancer 8, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke A.W., Shi G.M., Zhou J., et al. (2009). Role of overexpression of CD151 and/or c-Met in predicting prognosis of hepatocellular carcinoma. Hepatology 49, 491–503. [DOI] [PubMed] [Google Scholar]
- King D., Yeomanson D., and Bryant H.E. (2015). PI3King the lock: targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J. Pediatr. Hematol. Oncol. 37, 245–251. [DOI] [PubMed] [Google Scholar]
- Kircher T., Markov V., Krug A., et al. (2009). Association of the DTNBP1 genotype with cognition and personality traits in healthy subjects. Psychol. Med. 39, 1657–1665. [DOI] [PubMed] [Google Scholar]
- Le N., Sund M., and Vinci A. (2016). Prognostic and predictive markers in pancreatic adenocarcinoma. Dig. Liver Dis. 48, 223–230. [DOI] [PubMed] [Google Scholar]
- Liao H.M., and Chen C.H. (2004). Mutation analysis of the human dystrobrevin-binding protein 1 gene in schizophrenic patients. Schizophr. Res. 71, 185–189. [DOI] [PubMed] [Google Scholar]
- Lindberg J.M., Newhook T.E., Adair S.J., et al. (2014). Co-treatment with panitumumab and trastuzumab augments response to the MEK inhibitor trametinib in a patient-derived xenograft model of pancreatic cancer. Neoplasia 16, 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Liu X., Zhou T., et al. (2016). IRE1-RACK1 axis orchestrates ER stress preconditioning-elicited cytoprotection from ischemia/reperfusion injury in liver. J. Mol. Cell Biol. 8, 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Sawai H., Matsuo Y., et al. (2010). IGF-1 mediates PTEN suppression and enhances cell invasion and proliferation via activation of the IGF-1/PI3K/Akt signaling pathway in pancreatic cancer cells. J. Surg. Res. 160, 90–101. [DOI] [PubMed] [Google Scholar]
- Ma X., Fei E., Fu C., et al. (2011). Dysbindin-1, a schizophrenia-related protein, facilitates neurite outgrowth by promoting the transcriptional activity of p53. Mol. Psychiatry 16, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Mann K.M., Ying H., Juan J., et al. (2016). KRAS-related proteins in pancreatic cancer. Pharmacol. Ther. 168, 29–42. [DOI] [PubMed] [Google Scholar]
- Mishra R. (2013). Cell cycle-regulatory cyclins and their deregulation in oral cancer. Oral Oncol. 49, 475–481. [DOI] [PubMed] [Google Scholar]
- Mo F., Zhuang X., Liu X., et al. (2016). Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12, 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss S.F., Lee J.W., Sabo E., et al. (2008). Decreased expression of gastrokine 1 and the trefoil factor interacting protein TFIZ1/GKN2 in gastric cancer: influence of tumor histology and relationship to prognosis. Clin. Cancer Res. 14, 4161–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin A.P., Gokhale A., Larimore J., et al. (2011). Cell biology of the BLOC-1 complex subunit dysbindin, a schizophrenia susceptibility gene. Mol. Neurobiol. 44, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin A.P., Sadanandappa M.K., Ma W., et al. (2015). Gene dosage in the dysbindin schizophrenia susceptibility network differentially affect synaptic function and plasticity. J. Neurosci. 35, 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numakawa T., Yagasaki Y., Ishimoto T., et al. (2004). Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum. Mol. Genet. 13, 2699–2708. [DOI] [PubMed] [Google Scholar]
- Oshima J., and Campisi J. (1991). Fundamentals of cell proliferation: control of the cell cycle. J. Dairy Sci. 74, 2778–2787. [DOI] [PubMed] [Google Scholar]
- Parrales A., Lopez E., and Lopez-Colome A.M. (2011). Thrombin activation of PI3K/PDK1/Akt signaling promotes cyclin D1 upregulation and RPE cell proliferation. Biochim. Biophys. Acta 1813, 1758–1766. [DOI] [PubMed] [Google Scholar]
- Plachez C., Lindwall C., Sunn N., et al. (2008). Nuclear factor I gene expression in the developing forebrain. J. Comp. Neurol. 508, 385–401. [DOI] [PubMed] [Google Scholar]
- Ryan D.P., Hong T.S., and Bardeesy N. (2014). Pancreatic adenocarcinoma. N. Engl. J. Med. 371, 1039–1049. [DOI] [PubMed] [Google Scholar]
- Sanchez I., and Dynlacht B.D. (2005). New insights into cyclins, CDKs, and cell cycle control. Semin. Cell Dev. Biol. 16, 311–321. [DOI] [PubMed] [Google Scholar]
- Shi P., Yin T., Zhou F., et al. (2014). Valproic acid sensitizes pancreatic cancer cells to natural killer cell-mediated lysis by upregulating MICA and MICB via the PI3K/Akt signaling pathway. BMC Cancer 14, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart Z.A., Westfall M.D., and Pietenpol J.A. (2003). Cell-cycle dysregulation and anticancer therapy. Trends Pharmacol. Sci. 24, 139–145. [DOI] [PubMed] [Google Scholar]
- Straub R.E., Jiang Y., MacLean C.J., et al. (2002). Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 71, 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wu C., Ma J., et al. (2016). Toll-like receptor 4 promotes angiogenesis in pancreatic cancer via PI3K/AKT signaling. Exp. Cell Res. 347, 274–282. [DOI] [PubMed] [Google Scholar]
- Talbot K., Cho D.S., Ong W.Y., et al. (2006). Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum. Mol. Genet. 15, 3041–3054. [DOI] [PubMed] [Google Scholar]
- Wang M.C., Jiao M., Wu T., et al. (2016). Polycomb complex protein BMI-1 promotes invasion and metastasis of pancreatic cancer stem cells by activating PI3K/AKT signaling, an ex vivo, in vitro, and in vivo study. Oncotarget 7, 9586–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J.M., Yeo C.J., and Brody J.R. (2013). Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J. Surg. Oncol. 107, 15–22. [DOI] [PubMed] [Google Scholar]
- Zeng Z., Lin H., Zhao X., et al. (2012). Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin. Cancer Res. 18, 4059–4069. [DOI] [PubMed] [Google Scholar]
- Zhang Y.Y., Xu Z.N., Wang J.X., et al. (2012). G1/S-specific cyclin-D1 might be a prognostic biomarker for patients with laryngeal squamous cell carcinoma. Asian Pac. J. Cancer Prev. 13, 2133–2137. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhang W., Guo Z., et al. (2013). Inhibition of the transcription factor Sp1 suppresses colon cancer stem cell growth and induces apoptosis in vitro and in nude mouse xenografts. Oncol. Rep. 30, 1782–1792. [DOI] [PubMed] [Google Scholar]