The molecular mechanisms of sleep are not fully understood. Huang et al. demonstrate that loss of Caenorhabditis elegans UNC-7 or UNC-9 innexins dramatically reduces sleep during L4/A lethargus and that those innexins are partially required...
Keywords: Caenorhabditis elegans sleep, gap junction, cGMP-dependent kinase, NCA channel
Abstract
An essential characteristic of sleep is heightened arousal threshold, with decreased behavioral response to external stimuli. The molecular and cellular mechanisms underlying arousal threshold changes during sleep are not fully understood. We report that loss of UNC-7 or UNC-9 innexin function dramatically reduced sleep and decreased arousal threshold during developmentally timed sleep in Caenorhabditis elegans. UNC-7 function was required in premotor interneurons and UNC-9 function was required in motor neurons in this paradigm. Simultaneous transient overexpression of UNC-7 and UNC-9 was sufficient to induce anachronistic sleep in adult animals. Moreover, loss of UNC-7 or UNC-9 suppressed the increased sleep of EGL-4 gain-of-function animals, which have increased cyclic-GMP–dependent protein kinase activity. These results suggest C. elegans gap junctions may act downstream of previously identified sleep regulators. In other paradigms, the NCA cation channels act upstream of gap junctions. Consistent with this, diminished NCA channel activity in C. elegans robustly increased arousal thresholds during sleep bouts in L4-to-adult developmentally timed sleep. Total time in sleep bouts was only modestly increased in animals lacking NCA channel auxiliary subunit UNC-79, whereas increased channel activity dramatically decreased sleep. Loss of EGL-4 or innexin proteins suppressed UNC-79 loss-of-function sleep and arousal defects. In Drosophila, the ion channel narrow abdomen, an ortholog of the C. elegans NCA channels, drive the pigment dispersing factor (PDF) neuropeptide release, regulating circadian behavior. However, in C. elegans, we found that loss of the PDF receptor PDFR-1 did not suppress gain-of-function sleep defects, suggesting an alternative downstream pathway. This study emphasizes the conservation and importance of neuronal activity modulation during sleep, and unequivocally demonstrates that gap junction function is critical for normal sleep.
DURING sleep, animals have heightened arousal thresholds due to decreased response and desensitization of specific neurons (Campbell and Tobler 1984; Schwarz et al. 2011; Nichols et al. 2017). How does this transient and profound state change occur at a molecular level? Previous work has revealed multiple players, including neurotransmitters, neuropeptides, receptors, and kinases (Cirelli 2009; Allada et al. 2017). However, genes regulating sleep act in diverse pathways, suggesting that additional critical genes are not yet known and that understanding the relationship between these genes will be critical. Conserved genes regulating sleep can be identified in many model organisms, including Caenorhabditis elegans (Trojanowski and Raizen 2016).
C. elegans sleeps spontaneously at specific times or under specific conditions (Van Buskirk and Sternberg 2007; Ghosh and Emmons 2008; Raizen et al. 2008; Hill et al. 2014). The best characterized sleep states are developmentally timed sleep and stress-induced sleep. Molecular and genetic pathways required for sleep in these paradigms do not completely overlap, although commonalities have been identified (Trojanowski et al. 2015). Here, we focus almost exclusively on developmentally timed sleep, which occurs during a 2.5-hr period, called lethargus, coincident with cuticle molting during diapause. During lethargus, C. elegans exhibits interspersed bouts of sleep and motion that vary in duration, lasting from seconds to minutes. During sleep bouts, animals spontaneously cease locomotion, assume a specific posture, and show diminished response to external stimuli. During motion bouts, animals resume sinusoidal locomotion and respond robustly to external stimuli (Raizen et al. 2008; Singh et al. 2011; Iwanir et al. 2013). The mechanisms underlying this state change remain elusive.
Three major components of sleep behavior can be defined: circadian regulation, sleep homeostasis, and the state of sleep itself (Campbell and Tobler 1984). At this point, circadian regulation is well described, but the other two are less understood. The molecular pathways that regulate these three components of sleep are likely interlinked and complex. Using an evolutionary and ethological strategy, it may be possible to disentangle these behaviors by studying species that have fewer circadian behaviors. While the state of sleep and sleep homeostasis are conserved in C. elegans, many aspects of circadian biology are not conserved. There is no evidence for an endogenous, genetically encoded 24-hr internal clock. C. elegans lacks classical proteins involved in light entrainment, like Cryptochrome. However, proteins intimately involved in circadian rhythms in other species, like Period and timeless, have C. elegans orthologs that play essential roles in the developmental timing of diapause and lethargus, but not circadian behavior (Banerjee et al. 2005; Monsalve et al. 2011). Thus far, endogenous C. elegans circadian behaviors have not been described, although cyclic gene expression changes can be artificially entrained (van der Linden et al. 2010). Other components of sleep are well conserved and a decade of C. elegans sleep research has established the utility of this model organism in the field. There is deep conservation of genes and pathways regulating sleep and sleep homeostasis between C. elegans and other species (Singh et al. 2014; Trojanowski and Raizen 2016). Studies in C. elegans have contributed to a growing list of genes affecting sleep, including egl-4/cyclic-GMP (cGMP)–dependent kinase, lin-12/glp-1/Notch receptors, aptf-1/AP2-family transcription factor, and gpb-2/Gβ protein (Raizen et al. 2008; Singh et al. 2011; Huang et al. 2017b).
Here, we synthesize previous work in the sleep field with the results of an unbiased forward genetic screen for sleep genes in C. elegans, revealing a hierarchy of genes involved in sleep and arousal. In a classic genetic screen, we identify a new role for UNC-7 and UNC-9 innexins in sleep and find that these gap junction proteins are likely required downstream of the C. elegans cGMP-dependent protein kinase G (PKG), EGL-4. We also find that the NCA cation channels, identified in a mouse genetic screen for sleep genes (Funato et al. 2016), regulate arousal and sleep in C. elegans. Finally, we determine that EGL-4 PKG, but not C. elegans pigment dispersing factor (PDF) signaling, is required downstream of NCA channels to regulate sleep.
Materials and Methods
C. elegans culture and strain information
C. elegans were cultured on standard nematode growth media (NGM) seeded with OP50 E. coli. Animals were grown at 25° and assayed at 22° unless noted otherwise. Strains used are listed in Supplemental Material, Table S1.
Microfluidic chamber-based assessment of L4/A sleep
The microfluidic, chamber-based sleep assay was adapted from previous studies (Singh et al. 2011). Briefly, kanamycin-treated OP50 was resuspended as described in NGM (without agar) as a food source for the animals in the chambers. Mid-L4 stage animals, L4 substages L4.3–L4.4 (Mok et al. 2015), were loaded into each chamber and covered with a glass coverslip, which was sealed to the chip-containing chambers using molten 2% agar. Images were recorded every 10 sec for 12 hr and analyzed using a MATLAB (MathWorks, Natick, MA) script for image subtraction (Singh et al. 2011) and a custom Python script for calculating total time in sleep bouts (Huang et al. 2017a).
Arousal threshold assessment (touch and light)
Arousal threshold assays with blue light stimulation were conducted as previously described (Huang et al. 2017b). Briefly, L4/A lethargus animals were picked to freshly made assay plates with food and allowed to recover for 15 min. During the assay, the plates remained unperturbed with the lids off. A blue light laser pointer was used to stimulate the animal over the head region. Response latency for animals during both sleep bouts and motion bouts was recorded. Examination of unc-7(e5) animals suggested possible defects in their response to blue light stimulation during motion bouts. Therefore, arousal thresholds determinations for unc-7 and unc-9 studies tested latency to respond to posterior body gentle touch (Singh et al. 2011) in both sleep bouts and motion bouts. For all arousal determinations, at least eight animals per genotype and per condition (asleep or in motion) were tested in each trial. At least three independent trials were conducted. The experimenter was blinded to genotype.
PLM imaging
PLM GCaMP6 imaging in roaming animals was done with a custom-made system published earlier (Venkatachalam et al. 2016). One L4 animal, either before lethargus or during lethargus, was picked onto each assay plate (NGM in 100 mm plate seeded with 10 μl OP50). Animals were covered with a 48 × 65 mm #1 coverslip and left unperturbed for between 10 and 20 min before imaging. When imaging, the agar plate was fit into a groove and secured with a custom-made metal plate screwed down on the plate. An automated tapping device will exert controlled tapping when triggered. To avoid desensitization, the interval between tapping for an individual animal was at least 10 min.
Plasmids and transgenic strains
Transgenic strains were generated by microinjecting plasmids into nematodes using standard methods (Evans 2006). Plasmids used and transgene concentrations are listed in Table S2. Phenotypic rescue studies showed dose dependence. For unc-7 transgenes, high concentrations caused paralysis. For results herein, only transgene concentrations that did not cause paralysis were used.
Transient overexpression using heat shock
Transgenic animals with transgenes under the control of heat shock promoter were picked onto fresh plates at the young adult stage to induce transgene expression. Plates were sealed with Parafilm (Bemis Company) and heat shocked for 75 min in a 33.5° water bath, agar side down. Parafilm was removed and animals allowed to recover in a 20° incubator for 60 min. Then, animals were scored in the next 20-min interval. To avoid disturbing animals, plate lids were kept closed and plates were placed on the dissection scope agar side up (to avoid condensation on the inner surface of the plate lid when scoring). Animals were scored as Ans (in anachronistic sleep) if they did not move spontaneously or pump for 5 sec; either pharyngeal pumping or spontaneous locomotion was sufficient to score an animal as nonAns (not in anachronistic sleep).
To score locomotion and pharyngeal pumping separately, animals were treated as described above. Animals were scored as not moving if they did not move spontaneously for 5 sec, independent of the pumping status. Animals were scored as not pumping if they did not pump for 5 sec, regardless of locomotion. For all heat shock studies, at least 10 animals per genotype were tested for each trial and at least three independent trials were conducted. The experimenter was blinded to genotype.
Statistics
Statistical analyses used Student’s t-test for most of the studies. Mann–Whitney–Wilcoxon tests were performed for analyses of arousal thresholds with blue light because of the non-normally distributed data.
Data and reagent availability
Strains and plasmids are available upon request, unless otherwise stated. Full data tables are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7187531.
Results
Loss of UNC-7 or UNC-9 gap junction proteins dramatically decreases sleep bouts during lethargus
To identify conserved genes required for C. elegans developmentally timed sleep during the transition to adulthood (L4/A lethargus), we previously undertook a forward genetic screen (Huang et al. 2017b). Subsequently, we determined that the unc-7(rt212) mutant allele, a premature stop mutation (W365Opal, isoform a), caused decreased total time in sleep bouts during L4/A lethargus (total sleep time, Figure S1A). The unc-7 gene encodes a C. elegans gap junction/innexin protein with at least 10 alternative splice forms, differing at the N terminus. The unc-7(rt212) missense allele is predicted to prematurely truncate all unc-7 splice forms. unc-7(rt212) decreased endogenous L4/A lethargus sleep defects (Figure S1A) and suppressed adult anachronistic sleep induced by Notch coligand OSM-11 overexpression (Figure S1B).
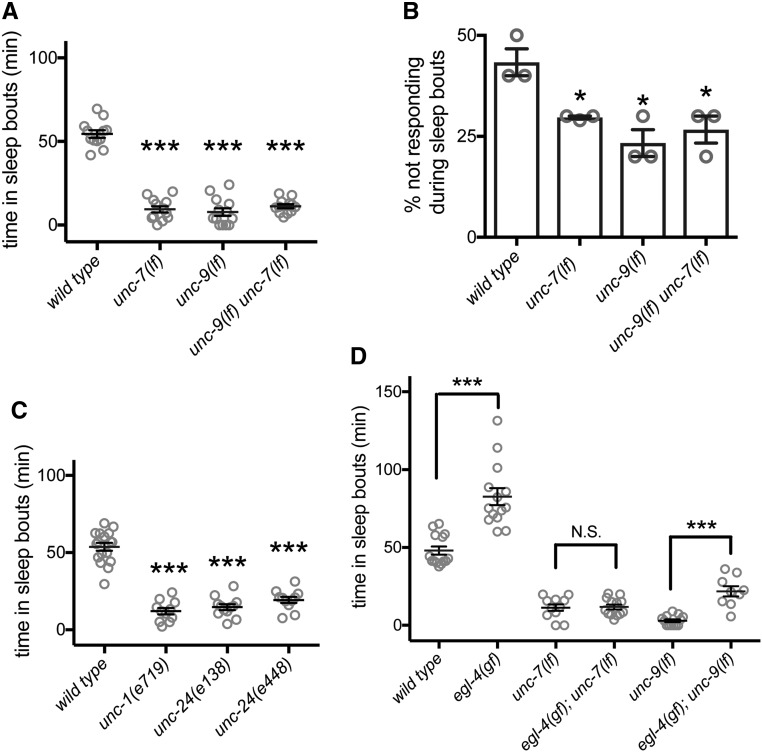
UNC-7 normally functions with UNC-9 to form gap junctions between cells. Loss of either UNC-7 or UNC-9 causes the animals to have uncoordinated locomotion with inappropriate posture predominantly during forward locomotion, but activity levels of adult animals are not decreased (Starich et al. 1993, 1996, 2009). We found that unc-7(e5) loss-of-function or unc-9(e101) loss-of-function animals had dramatically reduced L4/A lethargus total sleep (Figure 1A) and decreased lethargus sleep bout duration (Figure S1C). Introduction of a transgene containing the unc-9 promoter driving unc-9 complementary DNA (cDNA) almost completely restored total sleep quantity in unc-9(e101) animals (Figure S1D). To corroborate the sleep defects observed based on motion detection, we also examined another important aspect of sleep: altered arousal thresholds. During C. elegans L4/A lethargus sleep bouts, response to a variety of external stimuli is diminished (Raizen et al. 2008; Singh et al. 2011; Huang et al. 2017b); animals carrying mutations that perturb sleep quantity usually also have perturbed arousal thresholds during sleep bouts (Singh et al. 2014). Consistent with this general rubric, we found arousal defects in unc-7(lf) and unc-9(lf) animals. While animals lacking unc-7 or unc-9 always responded to gentle touch outside of lethargus or during lethargus motion bouts, loss of either unc-7 or unc-9 decreased arousal thresholds specifically during sleep bouts (Figure 1B, response to gentle touch). Additionally, we undertook a small pilot study and found that changes in stimulus-evoked, intracellular calcium signals in mechanosensory neurons that are characteristic of lethargus (Schwarz and Bringmann 2013; Cho and Sternberg 2014; Nichols et al. 2017) were not seen in unc-9(lf) animals (Figure S1E). Consistent with reduced time in sleep bouts, decreased arousal thresholds suggests a poor quality of sleep together with loss of UNC-7 or UNC-9 function. Double mutant unc-9(lf) unc-7(lf) animals did not have additive defects (Figure 1, A and B), suggesting that UNC-7 and UNC-9 likely function together. Combined, these results demonstrate that UNC-7 and UNC-9 function is important for modulating sensory neuron response, setting arousal thresholds during sleep bouts, and normal sleep quantity during lethargus.
Figure 1.
UNC-7 and UNC-9 gap junction proteins are required for normal L4/A developmentally timed sleep and arousal and are downstream of EGL-4 PKG. (A) Total time in sleep bouts for wild-type, unc-7(e5lf), unc-9(e101lf), and unc-9(e101lf) unc-7(e5lf) animals during L4 to adult lethargus. (B) Percent of wild-type, unc-7(e5lf), unc-9(e101lf), and unc-9(e101lf) unc-7(e5lf) animals not responding to gentle touch during L4 to adult sleep bouts. Because of the response defect of unc-7(e5lf) and unc-9(e101lf) animals to blue light stimuli, the arousal threshold was measured by gentle touch instead. Three independent trials were tested. n = 10 per genotype for each trial. (C) Total time in sleep bouts for wild-type, unc-1(e719lf), unc-24(e138lf), and unc-24(e138lf) animals during L4 to adult lethargus. (D) Total time in sleep bouts for egl-4(ad450gf), unc-7(e5lf), egl-4(ad450gf); unc-7(e5lf), unc-9(e101lf), and egl-4(ad450gf); unc-9(e101lf) animals during L4 to adult lethargus. Error bars show the SEM. * P < 0.05, *** P < 0.001. All error bars represent standard error of the mean.
Gap junction accessory proteins are also required for normal lethargus sleep bouts
The stomatin-like proteins UNC-1 and UNC-24 are thought to regulate UNC-7/UNC-9 gap junction function via an unknown mechanism (Chen et al. 2007); unc-24(lf) and unc-1(lf) animals have severe locomotion defects that are almost identical to unc-7(lf) and unc-9(lf) animals. Both of the unc-24(lf) alleles examined resulted in decreased total sleep time during L4/A lethargus (Figure 1C). Additionally, we tested the loss-of-function allele unc-1(e719) and the dominant gain-of-function allele unc-1(e1598) (Park and Horvitz 1986). unc-1 loss of function reduced total sleep time and unc-1 gain of function increased total sleep time (Figure 1C and Figure S1F). Combined, these results confirm that reduced gap junction accessory protein function results in decreased developmentally timed sleep.
Gap junction proteins likely act downstream of EGL-4 PKG
Orthologs of the C. elegans EGL-4 PKG, regulate sleep in vertebrates and invertebrates (Raizen et al. 2008; Langmesser et al. 2009). In C. elegans, loss of EGL-4 function decreases sleep quantity and lowers arousal thresholds during developmentally timed sleep. Further, increased EGL-4 activity increases both sleep quantity and arousal thresholds (Raizen et al. 2008). We confirmed these changes in sleep quantity and arousal thresholds for egl-4(n479) and egl-4(ad450), which are loss- and gain-of-function alleles, respectively. To our knowledge, no downstream targets of EGL-4 PKG pertinent to sleep have been identified in any species. To determine if C. elegans gap junction proteins might act downstream, we examined double mutant animals, as the downstream gene usually suppresses in this context (Huang and Sternberg 1995). We found that the increased sleep of egl-4(ad450) gain-of-function animals was completely suppressed by loss of unc-7 and that egl-4(ad450) increased sleep was almost completely suppressed by loss of unc-9 (Figure 1D). This suggests that EGL-4 PKG acts upstream in a genetic pathway and that EGL-4 PKG requires UNC-7 gap junction function, and is heavily dependent on UNC-9 function, to increase sleep.
UNC-7 and UNC-9 act in interneurons and motor neurons, respectively
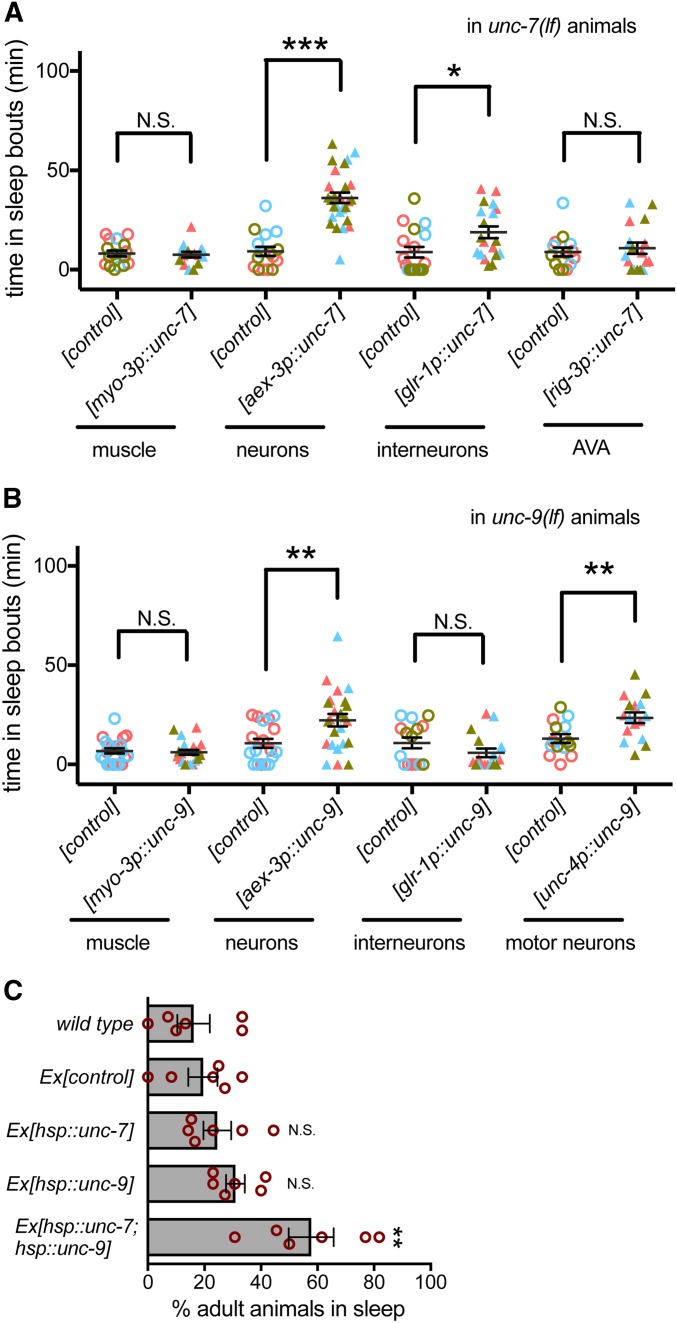
UNC-7 and UNC-9 are broadly expressed in C. elegans neurons and body wall muscles (Starich et al. 2009). To determine where these genes are required for sleep, we drove the expression of unc-7 or unc-9 cDNAs in mutant animals using tissue-specific promoters. Expression of UNC-7 or UNC-9 in all neurons, rather than body wall muscle, ameliorated sleep defects in unc-7(lf) or unc-9(lf) animals, respectively (Figure 2, A and B). These and rescue studies presented below did not restore wild-type level sleep; this may be a consequence of chimeric extrachromosomal array expression.
Figure 2.
UNC-7 and UNC-9 is partially required in premotor interneurons and motor neurons, respectively, during sleep. (A) Total time in sleep bouts for unc-7(e5lf) animals in L4 to adult lethargus rescued with myo-3 promoter, aex-3 promoter, glr-1 promoter, and rig-3 promoter driven unc-7 and their corresponding promoter driven GFP as controls. Colors represent three independent extrachromosomal arrays tested for each condition (strain information in Table S2). (B) Total time in sleep bouts for unc-9(e101lf) animals in L4 to adult lethargus rescued with myo-3 promoter, aex-3 promoter, glr-1 promoter, and unc-4 promoter driven unc-9 and their corresponding promoter driven GFP as controls. Colors represent two or three independent extrachromosomal arrays tested for each condition (strain information in Table S2). (C) Percent of animals with anachronistic sleep in wild-type animals, animals overexpressing hsp::gfp ([control]), hsp::unc-7, hsp::unc-9, and both hsp::unc-7 and hsp::unc-9. Data from three independent extrachromosomal arrays were pooled for each genotype. At least three independent trials were tested. n = 10 per genotype for each trial. * P < 0.05, ** P < 0.01, *** P < 0.001. All error bars represent standard error of the mean.
UNC-7 function is required in premotor AVA interneurons for coordinated locomotion (Kawano et al. 2011). To determine if UNC-7 function is also required in AVA for normal sleep, we expressed UNC-7 using the glr-1 promoter, which drives expression in 17 classes of interneurons, including AVA interneurons (Hart et al. 1995). This partially rescued sleep defects (Figure 2A). Expression of UNC-7 in AVA neurons alone using the rig-3 promoter did not rescue sleep. Combined, these results suggest that UNC-7 function is required in neurons, and that UNC-7 function in a subset of glr-1-expressing neurons plays a role in developmentally timed sleep.
UNC-9 function is required in A-class motor neurons for coordinated locomotion (Kawano et al. 2011). Consistent with this, expression of UNC-9 cDNA using the glr-1 promoter did not restore sleep (Figure 2B). However, expression of UNC-9 in cholinergic neurons, which includes the A-class motor neurons, partially restored sleep. These results suggest that UNC-7/UNC-9 gap junctions may function between premotor interneurons and motor neurons to promote developmentally timed sleep, but their function in other neurons is likely also important. UNC-7 and UNC-9 act, at least in part, in interneurons and motor neurons to promote lethargus sleep bouts. The partial rescue of lethargus sleep bouts observed here may suggest that these gap junction proteins act at other sites to facilitate activity changes associated with sleep.
Simultaneous expression of both UNC-7 and UNC-9 induces anachronistic sleep in adult animals
UNC-7 and UNC-9 are required for developmentally timed sleep, but is their activation sufficient to induce sleep? We tested this by inducing expression of unc-7, unc-9, or both in adult animals using the heat-inducible hsp-16.2 promoter to drive cDNA expression. After heat shock, C. elegans enter stress-induced quiescence, but in our hands, most wild-type animals resume locomotion and pharyngeal pumping 1 hr after heat shock ends. However, when UNC-7 and UNC-9 were simultaneously expressed, anachronistic sleep bouts were observed in adult animals 1 hr after heat shock (Figure 2C). Expression of either UNC-7 or UNC-9 was not sufficient, demonstrating that increasing UNC-7 and UNC-9 gap junctions is sufficient to induce anachronistic sleep in adult C. elegans and that both are required. UNC-7 and UNC-9 can function as heterotypic gap junction channels or as hemichannels; homomeric channels have been observed in ectopic expression systems (Starich et al. 2009; Bouhours et al. 2011; Meng et al. 2016). Heterotypic channels may be critical for the anachronistic sleep observed here.
The relationship between lethargus sleep and anachronistic sleep observed in adult animals after heat shock induced expression of unc-7 and unc-9 may be complicated by contribution of stress-induced sleep, ectopic overexpression, and uncoupling of this behavior from the developmental context of diapause. Nevertheless, we were able to examine the consequences of egl-4 PKG loss of function in anachronistic sleep caused by UNC-7 and UNC-9 ectopic overexpression. Loss of egl-4 did not significantly alter the fraction of animals not pumping after UNC-7/UNC-9 ectopic overexpression (Figure S2A), suggesting that EGL-4 is irrelevant or acts upstream in this paradigm. However, egl-4 loss did suppress the locomotion changes caused by UNC-7/UNC-9 ectopic overexpression (Figure S2B), suggesting that EGL-4 may act downstream in this anachronistic paradigm. We are hesitant to draw conclusions about developmentally timed sleep from anachronistic sleep, but unc-7 and unc-9 may act downstream of egl-4 for cessation of pumping in anachronistic sleep.
Normal NCA channel activity is required during C. elegans sleep bouts
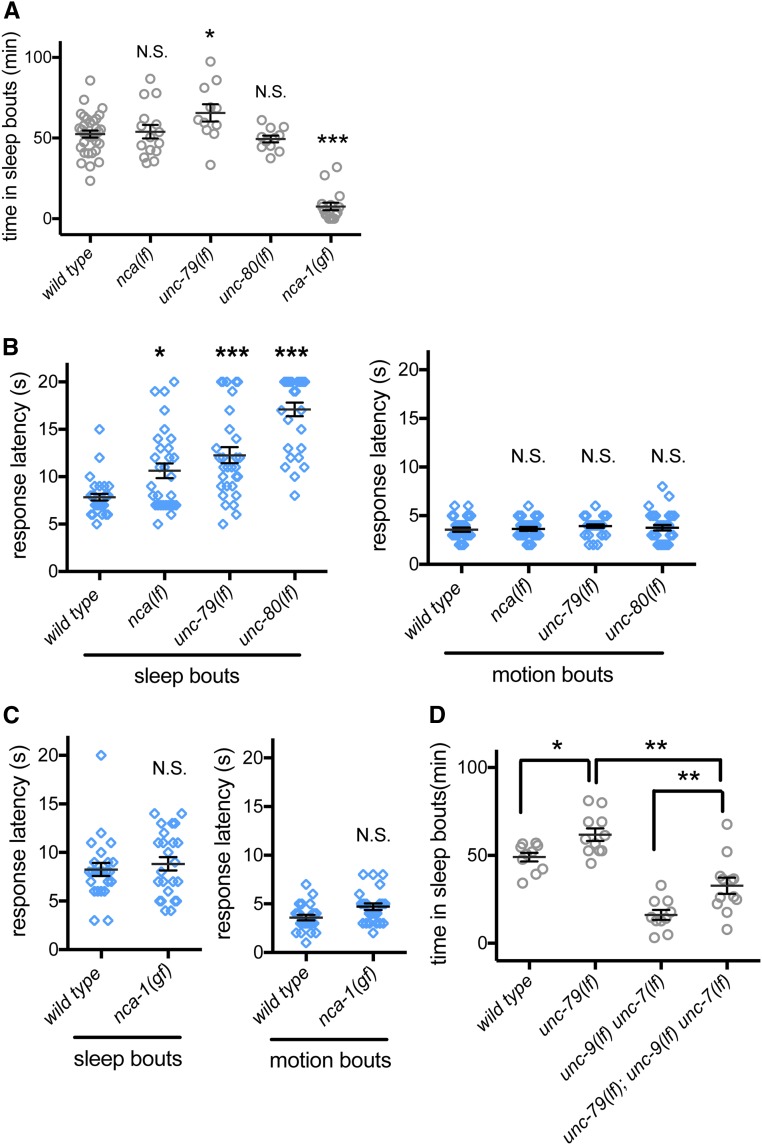
In C. elegans, UNC-7 and UNC-9 act downstream of NCA cation channels to modulate response to anesthetics (Sedensky and Meneely 1987). The NCA ion channels (NALCN channels in mammals) are conserved cation channels that are a member of the voltage-gated sodium and calcium channel family (Lee et al. 1999; Ren 2011; Liebeskind et al. 2012). The NALCN/NCA channels used to be considered sodium leak channels, although more functions have been proposed for these channels. (Senatore et al. 2013; Senatore and Spafford 2013; Boone et al. 2014) The NALCN/NCA channels have been implicated in various aspects of sleep in flies and mice (Lear et al. 2005; Funato et al. 2016). In flies, antiphase cycles in resting potassium and sodium conductance in clock neurons drive daily rhythms of activity, arousal, and sleep. This rhythmic sodium conductance is driven by sodium leak channel activity, which regulates circadian pacemaker neuron activity (Flourakis et al. 2015). We hypothesized that, in C. elegans, the NCA cation channels might also regulate arousal and sleep. The C. elegans NCA channel comprises the functionally redundant α1 subunits NCA-1 and NCA-2, which are encoded by nca-1 and nca-2 genes, hence the name NCA channels. NCA-1 and NCA-2 act redundantly to regulate resting membrane potential (Gao et al. 2015). Increased NCA channel function causes exaggerated body bends during locomotion, while NCA channel loss causes short transient pauses in C. elegans locomotion (Yeh et al. 2008). To assess the effect of NCA channels on C. elegans sleep during L4/A lethargus, we first examined the gain-of-function allele of nca-1(e625): sleep bouts during L4/A lethargus were almost completely eliminated in these animals (Figure 3A), suggesting that heightened NCA channel activity disturbs sleep.
Figure 3.
Normal NCA cation channel activity is required for proper response latency during sleep bouts. (A) Total time in sleep bouts for wild-type, nca-1(gk5lf); nca-1(gk9lf) [simplified as nca(lf)], unc-79(ec1lf), unc-80(e1272lf), and nca-1(e625gf) animals during L4 to adult lethargus. (B) Response latency of wild-type, nca(lf), unc-79(ec1lf), and unc-80(e1272lf) animals to blue light stimulation during sleep bouts (left panel) and motion bouts (right panel) in L4 to adult lethargus. (C) Response latency of nca-1(e625gf) animals to blue light stimulation during sleep bouts (left panel) and motion bouts (right panel) in L4 to adult lethargus. (D) Total time in sleep bouts for wild-type, unc-79(ec1lf), unc-9(e101lf) unc-7(e5lf), and unc-79(ec1lf); unc-9(e101lf) unc-7(e5lf) animals during L4 to adult lethargus. All sleep bouts and motions bouts were assessed during L4 to adult lethargus. * P < 0.05, ** P < 0.01, *** P < 0.001. All error bars represent standard error of the mean.
Animals lacking both nca-1 and nca-2 are referred to as nca(lf) animals in the literature (Yeh et al. 2008) and we adopt that convention here. We found that nca(lf) animals had normal total time in sleep bouts during L4/A lethargus (Figure 3A). Other aspects of sleep/motion bout timing and duration were also normal when compared to wild-type animals (Figure S3, A and B). This discordance led us to more closely examine the role of NCA channels and associated proteins in C. elegans sleep.
Functional NCA channels require two conserved auxiliary subunits, called UNC-79 and UNC-80 in C. elegans. These two genes do not function redundantly, as loss of either unc-79 or unc-80 perturbs NCA channel function with defects equal to nca(lf) animals (Yeh et al. 2008). When we examined L4/A lethargus sleep in unc-79(lf) animals, we observed a small increase in total sleep time for two different mutant allele strains (Figure 3A and Figure S3C). However, total sleep time and other measures of L4/A sleep were normal in unc-80(lf) animals (Figure 3A and Figure S3, A–C). Given the dramatic effect of nca(gf) on sleep, we considered the possibility that nca(lf) altered other aspects of L4/A lethargus sleep.
NCA loss-of-function animals had normal total sleep time, allowing us to assess arousal thresholds during sleep bouts. We previously established that blue light can be used to wake C. elegans from sleep bouts and that time to respond (response latency) is an effective measure of arousal threshold (Huang et al. 2017b). Loss-of-function alleles of NCA channel genes, including nca(lf), unc-79(lf), and unc-80(lf), caused increased latency to respond during sleep bouts (Figure 3B, left panel). To rule out the possibility that NCA channel function is required for response to blue light, we also examined latency to respond during L4/A motion bouts. Time to respond to blue light was normal in nca(lf), unc-79(lf), and unc-80(lf) animals (Figure 3B, right panel). Assessing arousal thresholds in nca-1(gf) animals was more challenging. Sleep bouts are rare and the average sleep bout duration in these animals was roughly 50% shorter (Figure S3B). With these caveats, we found that arousal thresholds in nca-1(gf) animals were not different from wild-type animals (Figure 3C). Combined, these results suggest that appropriate C. elegans NCA activity is critical for normal sleep.
We also examined sleep and arousal in animals defective in nlf-1 function; NLF-1 is an endoplasmic reticulum-associated protein that promotes, but is not absolutely required for, appropriate C. elegans NCA channel localization and function (Xie et al. 2013). The locomotion defects in adult nlf-1(lf) animals were not as severe as those of unc-79(lf), unc-80(lf), or nca(lf) animals (Xie et al. 2013). We did not observe changes in L4/A lethargus sleep quantity or in arousal thresholds for either sleep or motion bouts (Figure S3, D and E). Combined, all of these results confirm the conserved role of NCA channels in sleep-associated behaviors extends to C. elegans. Loss of NCA function leads to aberrantly increased arousal thresholds only during sleep bouts and increased NCA function prevents sleep bouts. Regulation of NCA channel activity might be critical for entry into or maintenance of the sleep state with appropriately increased arousal thresholds.
We undertook rescue experiments to pinpoint the site of action for NCA channels by expressing the channel pan-neuronally, in premotor-interneurons, and in different classes of motor neurons in nca(lf) animals. We found that expressing NCA in any of these neuron populations rescued the arousal defects during lethargus (Figure S4C). These results are reminiscent of the distributed sites of action for C. elegans egl-4 in sleep bout arousal (Raizen et al. 2008) and consistent with the hypothesis that arousing factors can act at various locations to awaken animals.
Gap junction protein function contributes to NCA channel modulation of sleep
Either NCA channel gain of function or gap junction loss of function results in decreased sleep. We examined the possibility that these genes act in the same pathway for sleep, as observed in other contexts (Sedensky and Meneely 1987; Bouhours et al. 2011). For this analysis, we used unc-79(lf) animals, which have increased sleep, and introduced loss-of-function alleles for unc-7 and unc-9. Loss of UNC-7, UNC-9, or simultaneous loss of both gap junction proteins suppressed unc-79(lf) defects; total sleep time in triple mutant animals was not as low as sleep time in unc-9(lf) unc-7(lf) animals (Figure 3D), but was dramatically lower than the quantity of sleep observed in wild-type animals. We found that loss of only unc-7 or unc-9 also decreased sleep in unc-79(lf) animals (Figure S3F), to levels comparable to the triple mutant in Figure 3D. The incomplete suppression means either that these genes function in independent pathways or that gap junctions act genetically downstream of NCA channels in a pathway critical for lethargus sleep, along with other effectors.
C. elegans NCA channel loss of function leads to “fainting bouts,” which cause transient cessation of locomotion in adult animals (Yeh et al. 2008). We examined the consequences of nca-1(gf) in anachronistic sleep caused by UNC-7 and UNC-9 ectopic overexpression in adult animals. nca-1(gf) did not suppress pumping or locomotion changes observed in animals ectopically overexpressing UNC-7 and UNC-9 (Figure S2, A and B), suggesting that NCA channels either act upstream of gap junctions or are irrelevant in this paradigm.
EGL-4 PKG may act downstream of NCA channels in C. elegans sleep
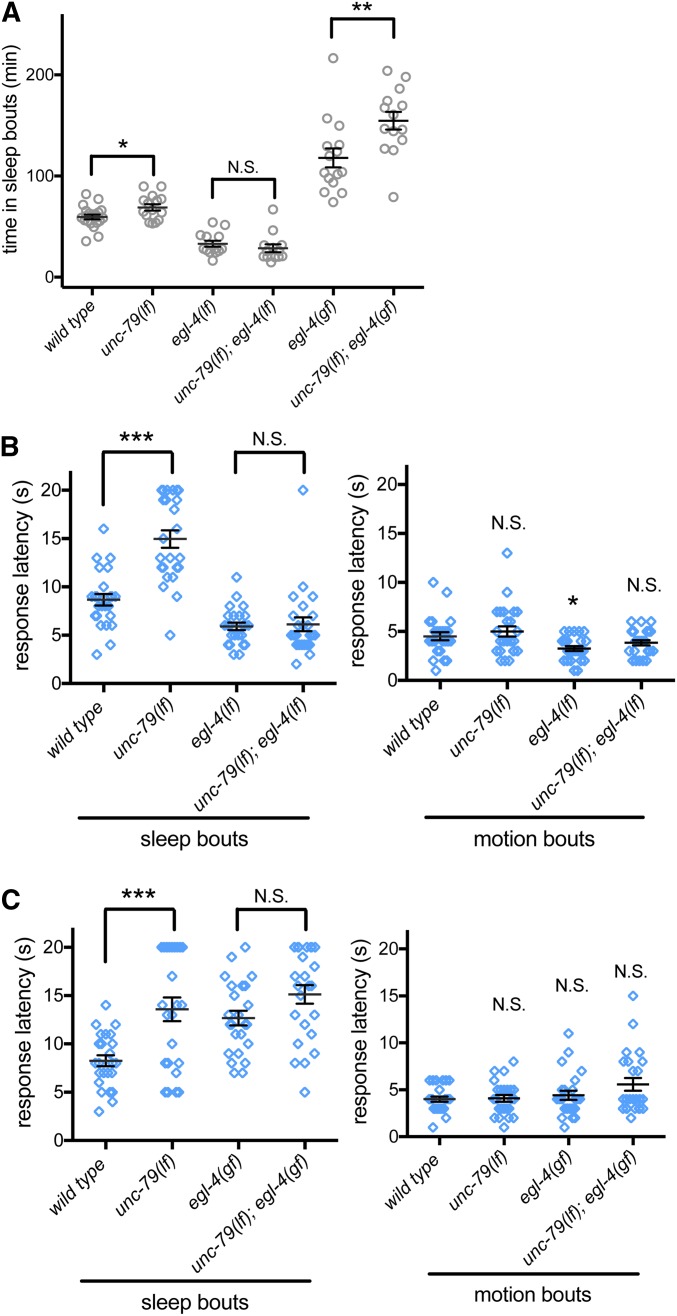
We also examined the relationship between EGL-4 and NCA channels during C. elegans L4/A lethargus, focusing initially on sleep quantity. We found that the increased sleep of unc-79(ec1) loss-of-function animals was completely eliminated by loss of egl-4; total time in sleep bouts of animals lacking both unc-79 and egl-4 was indistinguishable from animals with loss of egl-4 function alone (Figure 4A). Therefore, egl-4 likely acts downstream of unc-79 in a genetic pathway that regulates sleep quantity. Next, we examined the arousal thresholds in the double mutant animals. Again, unc-79(ec1) arousal changes during sleep bouts were completely suppressed by loss of egl-4 (Figure 4B). Combined, these results suggest that EGL-4 likely acts downstream of NCA channels for both sleep bouts and arousal threshold changes in a single pathway.
Figure 4.
EGL-4 PKG is downstream of NCA channels during L4/A lethargus. (A) Total time in sleep bouts for wild-type, unc-79(ec1lf), egl-4(n479lf), unc-79(ec1lf); egl-4(n479lf), egl-4(ad450gf), and unc-79(ec1lf); egl-4(ad450gf). (B) Response latency of wild-type, unc-79(ec1lf), egl-4(n479lf), and unc-79(ec1lf); egl-4(n479lf) animals to blue light stimulation during sleep bouts (left panel) and motion bouts (right panel). (C) Response latency of wild-type, unc-79(ec1lf), egl-4(ad450gf), and unc-79(ec1lf); egl-4(ad450gf) animals to blue light stimulation during sleep bouts (left panel) and motion bouts (right panel). All sleep bouts and motions bouts were assessed during L4 to adult lethargus. * P < 0.05, ** P < 0.01, *** P < 0.001. All error bars represent standard error of the mean.
Sometimes, additional information can be gained by examining gain-of-function alleles in epistasis studies. Therefore, we constructed double mutant unc-79(lf); egl-4(gf) animals. There was no additivity in arousal changes between unc-79(lf) and egl-4(gf) animals (Figure 4C), consistent with EGL-4 and NCA acting in the same pathway for arousal threshold changes during L4/A sleep. However, we noted that unc-79(lf); egl-4(gf) double animals had increased sleep compared with unc-79(lf) and egl-4(gf) single mutant animals (Figure 4A). Because the increased sleep in double mutant animals is more than additive, interpretation is difficult, but not inconsistent with these two genes acting in the same pathway. Loss of NCA channel function may increase downstream EGL-4 kinase activity. Alternatively, NCA channels may act through EGL-4, as well as other mechanisms. Combined, these genetic epistasis studies suggest that NCA channels likely act through the EGL-4 PKG to regulate sleep and arousal.
NCA channels act downstream or in parallel to PDF signaling
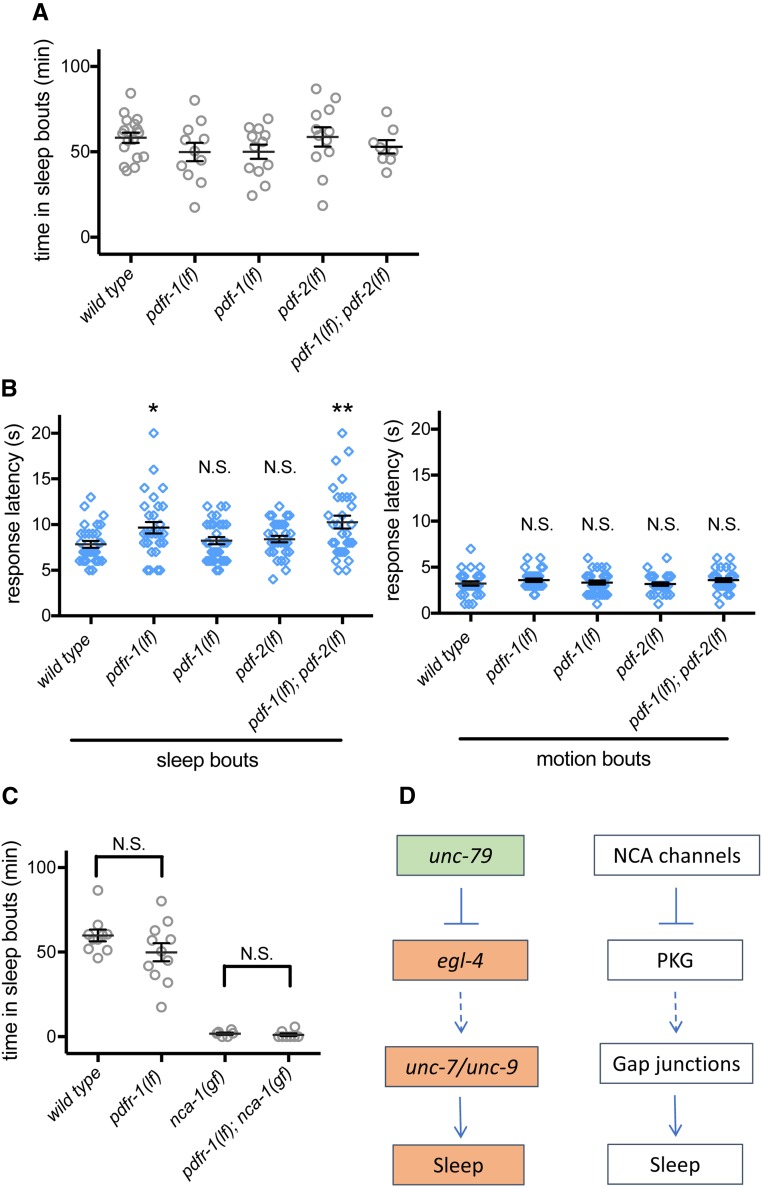
Previous studies in Drosophila have suggested that NCA channels and PDF receptors act coordinately in DN1p circadian neurons to regulate circadian behavior (Zhang et al. 2010). In C. elegans, there are two PDF neuropeptide-encoding genes (pdf-1 and pdf-2) and one gene encoding a receptor (pdfr-1). Previous work in C. elegans has shown in animals lacking neuropeptide Y signaling, loss of PDF-1 or the PDFR-1 receptor restores L4/A lethargus sleep quantity (Choi et al. 2013). PDF-1 or PDFR-1 loss also decreases motility in adult C. elegans (Meelkop et al. 2012; Choi et al. 2013). However, a requirement for PDF signaling in L4/A lethargus sleep and arousal has not been reported previously.
To directly determine the role of PDF signaling in lethargus sleep, we measured the total time in sleep bouts in pdfr-1(ok3425), pdfr-1(lst34), pdf-1(tm1996), or pdf-2(tm4393) loss-of-function animals and in pdf-1(lf); pdf-2(lf) double mutant animals. All of these mutant strains had normal total sleep during L4/A lethargus (Figure 5A and Figure S4A). We also examined arousal thresholds during L4/A lethargus sleep and motion bouts. Both pdfr-1(lf) animals and pdf-1(lf); pdf-2(lf) double mutant animals had slightly increased arousal thresholds only during sleep bouts (Figure 5B and Figure S4B). Response latency was normal during motion bouts for all genotypes. pdf-1(lf) and pdf-2(lf) may act redundantly in this scenario, through the pdfr-1 receptor. While the effect of altered PDF signaling was less dramatic than that of NCA channel perturbations, these results suggest that PDF neuropeptide signaling contributes to arousal during lethargus sleep bouts. Next, we examined the relationship between PDF signaling and NCA channels based on genetic epistasis. We generated pdfr-1(lf); nca-1(gf) double mutant animals. PDFR-1 loss did not ameliorate the sleep defects of nca-1(gf) animals (Figure 5C). Combined, these results suggest that PDF signaling is not required for NCA channel modulation of C. elegans lethargus sleep and that loss of PDF signaling has a modest effect on arousal threshold during lethargus sleep bouts.
Figure 5.
PDF signaling acts upstream or in parallel to NCA channels. (A) Total time in sleep bouts for wild-type, pdfr-1(ok3425lf), pdf-1(tm1996lf), pdf-2(tm4393lf), and pdf-1tm1996(lf); pdf-2(tm4393lf) animals. (B) Response latency of wild-type, pdfr-1(ok3425lf), pdf-1(tm1996lf), pdf-2(tm4393lf), and pdf-1(tm1996lf); pdf-2(tm4393lf) animals to blue light stimulation during sleep bouts (left panel) and motion bouts (right panel). (C) Total time in sleep bouts for wild-type, pdfr-1(ok3425lf), nca-1(e625gf), and pdfr-1(ok3425lf); nca-1(e625gf) animals. (D) Model for genetic pathways of the conserved cation channel and gap junction proteins regulating sleep across species. Dashed arrows represent possible parallel pathway. All sleep bouts and motions bouts were assessed during L4 to adult lethargus. * P < 0.05, ** P < 0.01. All error bars represent standard error of the mean.
In summary, we have unequivocally established a requirement for UNC-7 and UNC-9 gap junctions in L4/A lethargus sleep and arousal thresholds. We also find that NCA cation channels and EGL-4 PKG likely act upstream of UNC-7 and UNC-9 gap junction proteins in this behavior. NCA and PKG play critical roles in sleep and arousal across species, including C. elegans, Drosophila, and mice. We suggest that a pathway including these proteins and gap junctions may play a conserved role regulating sleep across all animal species (Figure 5D).
Discussion
Here, we report that loss of C. elegans UNC-7 or UNC-9 innexin gap junction proteins resulted in dramatically reduced time in sleep bouts during L4/A lethargus, decreased bout duration, and decreased arousal thresholds during these sleep bouts. UNC-7 function was partially required in premotor interneurons and UNC-9 function was partially required in motor neurons during sleep. Moreover, transient, simultaneous ectopic overexpression of UNC-7 and UNC-9 induced anachronistic sleep in adult animals. Based on genetic epistasis studies in lethargus sleep, unc-7 and unc-9 acted downstream of egl-4, which encodes the C. elegans PKG. Additionally, we found that loss of the C. elegans NCA channel increased arousal thresholds during L4/A lethargus sleep bouts and increased cation channel activity decreased sleep in this context. NCA channels acted upstream of egl-4 and gap junctions in lethargus sleep. Loss of C. elegans PDF neuropeptides or receptors had no effect on sleep quantity during lethargus, but their loss slightly increased arousal thresholds during these sleep bouts. Our results suggest that in C. elegans lethargus sleep, PDF signaling likely does not act downstream of NCA channels. This unequivocally demonstrates a requirement for gap junction proteins in sleep and demonstrates that EGL-4 PKG regulation of sleep is dependent on gap junction function.
A recent forward genetic screen in mice for genes involved in sleep, identified a dominant, missense mutation in murine NALCN, which increases channel activity and leads to decreased rapid eye movement sleep (Funato et al. 2016). Murine Nalcn encodes the direct ortholog of C. elegans nca-1 and nca-2. Our work in C. elegans finds that increased NCA channel activity decreases sleep bout quantity during lethargus, which is consistent with a requirement for NALCN channels in murine sleep (Funato et al. 2016). We speculate that if these channels act in wake-promoting neurons or sensory neurons, their loss might increase arousal thresholds during sleep. In Drosophila, PDF signaling and cation channels coordinately regulate Drosophila circadian behavior, based on activity and entrainment to light/dark cycles (Zhang et al. 2010). Drosophila PDF-expressing neurons play critical roles in activating neurons that express NCA channels, which in turn helps drive circadian behaviors. In C. elegans, PDF signaling regulates activity levels in adult animals and suppresses sleep quantity defects in animals lacking neuropeptide Y signaling (Meelkop et al. 2012; Choi et al. 2013). Our work confirms that loss of PDF signaling does not change C. elegans lethargus sleep quantity (Choi et al. 2013), but reveals that PDF signaling regulates arousal thresholds during sleep bouts, confirming a cross-species role for PDF signaling in arousal and sleep. We note that loss of nlf-1, which encodes an ER-localized protein required for NCA channel trafficking, did not perturb sleep or arousal thresholds (Figure S3, D and E). As the fainting locomotion defects in nlf-1 loss-of-function animals are less severe than those seen in nca loss-of-function animals (Xie et al. 2013), NLF may also interact with or regulate additional, unidentified channels that affect both locomotion and sleep, antagonizing NCA channel function. Combined, these studies in disparate animal species highlight the conserved requirement for NACLN channels in sleep, although the molecular mechanisms underlying sleep regulation may require further study.
Invertebrate gap junctions comprise innexin proteins, while vertebrate gap junctions comprise connexin proteins. Despite their analogous function in electrically coupling adjacent cells, innexin and connexin protein amino acid sequences cannot be easily aligned, which makes drawing conclusions about orthology or orthologous function difficult (Abascal and Zardoya 2013). Vertebrates also have genes that encode innexin protein orthologs, called pannexins. Innexin and pannexin proteins share sequence similarity and both can function as hemichannels (Bruzzone et al. 2003; Bouhours et al. 2011), which are nonspecific transmembrane channels.
Prior work is not inconsistent with a conserved role for gap junctions in sleep. Rapid eye movement sleep in rats is disrupted by quinine, a nonspecific blocker of gap junctions and various other channels and solute pumps (Franco-Pérez and Paz 2009; Connors 2012). Further, mice lacking Pannexin 1 function show increased wake and decreased slow-wave sleep, possibly due to the depleted extracellular adenosine (Kovalzon et al. 2017). Also, Connexin 43 knockout mice show excessive sleepiness and fragmented wakefulness, which may be attributed to connexin loss in astrocytes causing impaired lactate shuttling that impairs orexin neuron function (Clasadonte et al. 2017). Additionally, Connexin 36 knockout mice show dampened circadian activity rhythms that may be attributed to loss of synchronized spiking in the suprachiasmatic nucleus (Long et al. 2005). Finally, a recent study in Drosophila has demonstrated requirement for innexin 6 activity in dorsal fan-shaped body neurons that regulate arousal during both sleep and awaking states (Troup et al. 2018). Our work here demonstrates that gap junction function is required for normal C. elegans lethargus sleep and that these gap junctions act genetically downstream of EGL-4 PKG. We suggest that increased gap junction activity may increase coupling between neurons and the consequent shunting may serve to decrease neuronal excitability in circuits, allowing normal C. elegans sleep. Or, the synchronized neuronal activity characteristic of sleep in many species may require gap junction function. Additional studies are clearly required to understand how gap junction proteins act to regulate sleep and arousal across species.
Do C. elegans UNC-7 and UNC-9 proteins function as hemichannels or as gap junctions proteins in the context of C. elegans sleep? Previous work has established that C. elegans UNC-7 may function as a hemichannel in some contexts, but must function as a gap junction for normal locomotion (Bouhours et al. 2011). We do not directly test if UNC-7 acts as a hemichannel in the context of sleep. However, our overexpression data showed that only by overexpressing both UNC-7 and UNC-9 together were we able to induce sleep in adult animals. This suggests that either collaboration of UNC-7 and UNC-9 in heterotypic gap junctions is required for normal sleep, or that in the ectopic overexpression paradigm, high levels of both UNC-7 and UNC-9 hemichannels are required.
During lethargus, C. elegans stop feeding and pharyngeal pumping ceases. Why pumping normally ceases during lethargus is unclear. Previous work has demonstrated a requirement for UNC-7 in EGF-signaling induced pumping cessation during sleep bouts (Van Buskirk and Sternberg 2007). Further, a recent report found that optical silencing of body wall muscles can induce pumping inhibition in adult animals via UNC-7 gap junctions between the I1 neuron and RIP neurons, the only intra- and extrapharyngeal neural connection (Takahashi and Takagi 2017). However, even in animals lacking unc-7 function, we observed that pharyngeal pumping ceases during lethargus, suggesting that other factors are at work. We note that lethargus-specific decreases in pharyngeal muscle excitability may play a major role in suppressing pharyngeal pumping and feeding during lethargus, as optical stimulation of pharyngeal muscles cannot drive contraction (Trojanowski et al. 2016). Gap junction proteins may contribute to cessation of feeding during lethargus, but other regulatory pathways are likely major players.
Results presented here suggest that gap junctions function downstream of other proteins known to regulate C. elegans sleep. Indeed, C. elegans gap junctions function downstream of the cGMP-dependent protein kinase EGL-4 in sleep, suggesting a model in which EGL-4 directly or indirectly regulates gap junction function. Gap junction activity could be regulated at multiple levels, including protein local expression, membrane protein endocytosis and exocytosis, and/or protein modifications that might alter gap junction permeability (Kjenseth et al. 2010; Axelsen et al. 2013; Pogoda et al. 2016). Phosphorylation and redox changes can regulate connexin activity and trafficking (Pogoda et al. 2016), but a connection between gap junction proteins and cGMP-dependent kinase activity has not been examined to our knowledge. The only exception is one in vitro study showing that nitric oxide can lead to PKG phosphorylation of connexins (Patel et al. 2006). Further work examining how gap junctions are regulated might provide novel insights into how cGMP-dependent protein kinases regulate sleep and arousal across species.
Acknowledgments
We thank Mei Zhen (Lunenfeld–Tanenbaum Research Institute) and Dong Yan (Duke University) for numerous constructs and strains, Gary Silverman and Stephen Pak (Washington University in St. Louis) and Barry Connors (Brown University) for manuscript discussion, and Aravinthan Samuel (Harvard University) for critical advice and support. Some strains were provided by the Caenorhabditis Genetics Center, funded by National Institutes of Health Office of Research Infrastructure Programs (grant P40 OK010440). We acknowledge support from the Brown Institute for Brain Science/Norman Prince Neurosciences Institute (BIBS/NPNI) Postdoctoral Fellowship in Translational Neuroscience (to H.H.), Karen T. Romer Undergraduate Teaching and Research Awards (to D.J.H. and L.L.S.), Burroughs Wellcome Fund Career Award (to V.V.) and National Institute of Neurological Disorders and Stroke (NINDS) grant NS055813 (to A.C.H.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7187531.
Communicating editor: M. Sundaram
Literature Cited
- Abascal F., Zardoya R., 2013. Evolutionary analyses of gap junction protein families. Biochim. Biophys. Acta 1828: 4–14. 10.1016/j.bbamem.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Allada R., Cirelli C., Sehgal A., 2017. Molecular mechanisms of sleep homeostasis in flies and mammals. Cold Spring Harb. Perspect. Biol. 9 10.1101/cshperspect.a027730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen L. N., Calloe K., Holstein-Rathlou N. H., Nielsen M. S., 2013. Managing the complexity of communication: regulation of gap junctions by post-translational modification. Front. Pharmacol. 4: 130 10.3389/fphar.2013.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D., Kwok A., Lin S. Y., Slack F. J., 2005. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev. Cell 8: 287–295. 10.1016/j.devcel.2004.12.006 [DOI] [PubMed] [Google Scholar]
- Boone A. N., Senatore A., Chemin J., Monteil A., Spafford J. D., 2014. Gd3+ and calcium sensitive, sodium leak currents are features of weak membrane-glass seals in patch clamp recordings. PLoS One 9: e98808 10.1371/journal.pone.0098808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhours M., Po M. D., Gao S., Hung W., Li H., et al. , 2011. A co-operative regulation of neuronal excitability by UNC-7 innexin and NCA/NALCN leak channel. Mol. Brain 4: 16 10.1186/1756-6606-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone R., Hormuzdi S. G., Barbe M. T., Herb A., Monyer H., 2003. Pannexins, a family of gap junction proteins expressed in brain. Proc. Natl. Acad. Sci. USA 100: 13644–13649. 10.1073/pnas.2233464100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. S., Tobler I., 1984. Animal sleep: a review of sleep duration across phylogeny. Neurosci. Biobehav. Rev. 8: 269–300. 10.1016/0149-7634(84)90054-X [DOI] [PubMed] [Google Scholar]
- Chen B., Liu Q., Ge Q., Xie J., Wang Z. W., 2007. UNC-1 regulates gap junctions important to locomotion in C. elegans. Curr. Biol. 17: 1334–1339. 10.1016/j.cub.2007.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. Y., Sternberg P. W., 2014. Multilevel modulation of a sensory motor circuit during C. elegans sleep and arousal. Cell 156: 249–260. 10.1016/j.cell.2013.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Chatzigeorgiou M., Taylor K. P., Schafer W. R., Kaplan J. M., 2013. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron 78: 869–880. 10.1016/j.neuron.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., 2009. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat. Rev. Neurosci. 10: 549–560. 10.1038/nrn2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasadonte J., Scemes E., Wang Z., Boison D., Haydon P. G., 2017. Connexin 43-mediated astroglial metabolic networks contribute to the regulation of the sleep-wake cycle. Neuron 95: 1365–1380.e5. 10.1016/j.neuron.2017.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors B. W., 2012. Tales of a dirty drug: carbenoxolone, gap junctions, and seizures. Epilepsy Curr. 12: 66–68. 10.5698/1535-7511-12.2.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, T. C., 2006 Transformation and microinjection (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.108.1, http://www.wormbook.org. [Google Scholar]
- Flourakis M., Kula-Eversole E., Hutchison A. L., Han T. H., Aranda K., et al. , 2015. A conserved bicycle model for circadian clock control of membrane excitability. Cell 162: 836–848. 10.1016/j.cell.2015.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Pérez J., Paz C., 2009. Quinine, a selective gap junction blocker, decreases REM sleep in rats. Pharmacol. Biochem. Behav. 94: 250–254. 10.1016/j.pbb.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Funato H., Miyoshi C., Fujiyama T., Kanda T., Sato M., et al. , 2016. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539: 378–383. 10.1038/nature20142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Xie L., Kawano T., Po M. D., Guan S., et al. , 2015. The NCA sodium leak channel is required for persistent motor circuit activity that sustains locomotion. Nat. Commun. 6: 6323 (erratum: Nat. Commun. 6: 7191) 10.1038/ncomms7323 [DOI] [PubMed] [Google Scholar]
- Ghosh R., Emmons S. W., 2008. Episodic swimming behavior in the nematode C. elegans. J. Exp. Biol. 211: 3703–3711. 10.1242/jeb.023606 [DOI] [PubMed] [Google Scholar]
- Hart A. C., Sims S., Kaplan J. M., 1995. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 378: 82–85. 10.1038/378082a0 [DOI] [PubMed] [Google Scholar]
- Hill A. J., Mansfield R., Lopez J., Raizen D. M., Van Buskirk C., 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol. 24: 2399–2405. 10.1016/j.cub.2014.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Singh K., Hart A. C., 2017a. Measuring Caenorhabditis elegans sleep during the transition to adulthood using a microfluidics-based system. Bio Protoc. 7: e2174 10.21769/BioProtoc.2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Zhu C. T., Skuja L. L., Hayden D. J., Hart A. C., 2017b. Genome-wide screen for genes involved in Caenorhabditis elegans developmentally timed sleep. G3 (Bethesda) 7: 2907–2917. 10.1534/g3.117.300071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. S., Sternberg P. W., 1995. Genetic dissection of developmental pathways. Methods Cell Biol. 48: 97–122. 10.1016/S0091-679X(08)61385-0 [DOI] [PubMed] [Google Scholar]
- Iwanir S., Tramm N., Nagy S., Wright C., Ish D., et al. , 2013. The microarchitecture of C. elegans behavior during lethargus: homeostatic bout dynamics, a typical body posture, and regulation by a central neuron. Sleep (Basel) 36: 385–395. 10.5665/sleep.2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T., Po M. D., Gao S., Leung G., Ryu W. S., et al. , 2011. An imbalancing act: gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron 72: 572–586. 10.1016/j.neuron.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Kjenseth A., Fykerud T., Rivedal E., Leithe E., 2010. Regulation of gap junction intercellular communication by the ubiquitin system. Cell. Signal. 22: 1267–1273. 10.1016/j.cellsig.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Kovalzon V. M., Moiseenko L. S., Ambaryan A. V., Kurtenbach S., Shestopalov V. I., et al. , 2017. Sleep-wakefulness cycle and behavior in pannexin1 knockout mice. Behav. Brain Res. 318: 24–27. 10.1016/j.bbr.2016.10.015 [DOI] [PubMed] [Google Scholar]
- Langmesser S., Franken P., Feil S., Emmenegger Y., Albrecht U., et al. , 2009. cGMP-dependent protein kinase type I is implicated in the regulation of the timing and quality of sleep and wakefulness. PLoS One 4: e4238 10.1371/journal.pone.0004238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear B. C., Lin J. M., Keath J. R., McGill J. J., Raman I. M., et al. , 2005. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48: 965–976. 10.1016/j.neuron.2005.10.030 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Cribbs L. L., Perez-Reyes E., 1999. Cloning of a novel four repeat protein related to voltage-gated sodium and calcium channels. FEBS Lett. 445: 231–236. 10.1016/S0014-5793(99)00082-4 [DOI] [PubMed] [Google Scholar]
- Liebeskind B. J., Hillis D. M., Zakon H. H., 2012. Phylogeny unites animal sodium leak channels with fungal calcium channels in an ancient, voltage-insensitive clade. Mol. Biol. Evol. 29: 3613–3616. 10.1093/molbev/mss182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. A., Jutras M. J., Connors B. W., Burwell R. D., 2005. Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat. Neurosci. 8: 61–66. 10.1038/nn1361 [DOI] [PubMed] [Google Scholar]
- Meelkop E., Temmerman L., Janssen T., Suetens N., Beets I., et al. , 2012. PDF receptor signaling in Caenorhabditis elegans modulates locomotion and egg-laying. Mol. Cell. Endocrinol. 361: 232–240. 10.1016/j.mce.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Meng L., Chen C. H., Yan D., 2016. Regulation of gap junction dynamics by UNC-44/ankyrin and UNC-33/CRMP through VAB-8 in C. elegans neurons. PLoS Genet. 12: e1005948 10.1371/journal.pgen.1005948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok D. Z., Sternberg P. W., Inoue T., 2015. Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev. Biol. 15: 26 10.1186/s12861-015-0076-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve G. C., Van Buskirk C., Frand A. R., 2011. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21: 2033–2045. 10.1016/j.cub.2011.10.054 [DOI] [PubMed] [Google Scholar]
- Nichols A. L. A., Eichler T., Latham R., Zimmer M., 2017. A global brain state underlies C. elegans sleep behavior. Science 356: eaam6851 10.1126/science.aam6851 [DOI] [PubMed] [Google Scholar]
- Park E. C., Horvitz H. R., 1986. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics 113: 821–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel L. S., Mitchell C. K., Dubinsky W. P., O’Brien J., 2006. Regulation of gap junction coupling through the neuronal connexin Cx35 by nitric oxide and cGMP. Cell Commun. Adhes. 13: 41–54. 10.1080/15419060600631474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda K., Kameritsch P., Retamal M. A., Vega J. L., 2016. Regulation of gap junction channels and hemichannels by phosphorylation and redox changes: a revision. BMC Cell Biol. 17: 11 10.1186/s12860-016-0099-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen D. M., Zimmerman J. E., Maycock M. H., Ta U. D., You Y. J., et al. , 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451: 569–572 (erratum: Nature 453: 952) 10.1038/nature06535 [DOI] [PubMed] [Google Scholar]
- Ren D., 2011. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron 72: 899–911. 10.1016/j.neuron.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J., Bringmann H., 2013. Reduced sleep-like quiescence in both hyperactive and hypoactive mutants of the Galphaq gene egl-30 during lethargus in Caenorhabditis elegans. PLoS One 8: e75853 10.1371/journal.pone.0075853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J., Lewandrowski I., Bringmann H., 2011. Reduced activity of a sensory neuron during a sleep-like state in Caenorhabditis elegans. Curr. Biol. 21: R983–R984. 10.1016/j.cub.2011.10.046 [DOI] [PubMed] [Google Scholar]
- Sedensky M. M., Meneely P. M., 1987. Genetic analysis of halothane sensitivity in Caenorhabditis elegans. Science 236: 952–954. 10.1126/science.3576211 [DOI] [PubMed] [Google Scholar]
- Senatore A., Spafford J. D., 2013. A uniquely adaptable pore is consistent with NALCN being an ion sensor. Channels (Austin) 7: 60–68. 10.4161/chan.23981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A., Monteil A., van Minnen J., Smit A. B., Spafford J. D., 2013. NALCN ion channels have alternative selectivity filters resembling calcium channels or sodium channels. PLoS One 8: e55088 10.1371/journal.pone.0055088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Chao M. Y., Somers G. A., Komatsu H., Corkins M. E., et al. , 2011. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr. Biol. 21: 825–834. 10.1016/j.cub.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Ju J. Y., Walsh M. B., DiIorio M. A., Hart A. C., 2014. Deep conservation of genes required for both Drosophila melanogaster and Caenorhabditis elegans sleep includes a role for dopaminergic signaling. Sleep (Basel) 37: 1439–1451. 10.5665/sleep.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich T. A., Herman R. K., Shaw J. E., 1993. Molecular and genetic analysis of unc-7, a Caenorhabditis elegans gene required for coordinated locomotion. Genetics 133: 527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich T. A., Lee R. Y., Panzarella C., Avery L., Shaw J. E., 1996. eat-5 and unc-7 represent a multigene family in Caenorhabditis elegans involved in cell-cell coupling. J. Cell Biol. 134: 537–548. 10.1083/jcb.134.2.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich T. A., Xu J., Skerrett I. M., Nicholson B. J., Shaw J. E., 2009. Interactions between innexins UNC-7 and UNC-9 mediate electrical synapse specificity in the Caenorhabditis elegans locomotory nervous system. Neural Dev. 4: 16 10.1186/1749-8104-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Takagi S., 2017. Optical silencing of body wall muscles induces pumping inhibition in Caenorhabditis elegans. PLoS Genet. 13: e1007134 10.1371/journal.pgen.1007134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski N. F., Raizen D. M., 2016. Call it worm sleep. Trends Neurosci. 39: 54–62. 10.1016/j.tins.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski N. F., Nelson M. D., Flavell S. W., Fang-Yen C., Raizen D. M., 2015. Distinct mechanisms underlie quiescence during two Caenorhabditis elegans sleep-like states. J. Neurosci. 35: 14571–14584. 10.1523/JNEUROSCI.1369-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski N. F., Raizen D. M., Fang-Yen C., 2016. Pharyngeal pumping in Caenorhabditis elegans depends on tonic and phasic signaling from the nervous system. Sci. Rep. 6: 22940 10.1038/srep22940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troup M., Yap M. H., Rohrscheib C., Grabowska M. J., Ertekin D., et al. , 2018. Acute control of the sleep switch in Drosophila reveals a role for gap junctions in regulating behavioral responsiveness. eLife 7: e37105 10.7554/eLife.37105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C., Sternberg P. W., 2007. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat. Neurosci. 10: 1300–1307. 10.1038/nn1981 [DOI] [PubMed] [Google Scholar]
- van der Linden A. M., Beverly M., Kadener S., Rodriguez J., Wasserman S., et al. , 2010. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 8: e1000503 10.1371/journal.pbio.1000503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam V., Ji N., Wang X., Clark C., Mitchell J. K., et al. , 2016. Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 113: E1082–E1088. 10.1073/pnas.1507109113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Gao S., Alcaire S. M., Aoyagi K., Wang Y., et al. , 2013. NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron 77: 1069–1082. 10.1016/j.neuron.2013.01.018 [DOI] [PubMed] [Google Scholar]
- Yeh E., Ng S., Zhang M., Bouhours M., Wang Y., et al. , 2008. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 6: e55 10.1371/journal.pbio.0060055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chung B. Y., Lear B. C., Kilman V. L., Liu Y., et al. , 2010. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr. Biol. 20: 591–599. 10.1016/j.cub.2010.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]