Abstract
Body size is a tightly regulated phenotype in metazoans that depends on both intrinsic and extrinsic factors. While signaling pathways are known to control organ and body size, the downstream effectors that mediate their effects remain poorly understood. In the nematode Caenorhabditis elegans, a Bone Morphogenetic Protein (BMP)-related signaling pathway is the major regulator of growth and body size. We investigated the transcriptional network through which the BMP pathway regulates body size and identified cuticle collagen genes as major effectors of growth control. We demonstrate that cuticle collagens can act as positive regulators (col-41), negative regulators (col-141), or dose-sensitive regulators (rol-6) of body size. Moreover, we find a requirement of BMP signaling for stage-specific expression of cuticle collagen genes. We show that the Smad signal transducers directly bind conserved Smad-binding elements in regulatory regions of col-141 and col-142, but not of col-41. Hence, cuticle collagen genes may be directly and indirectly regulated via the BMP pathway. Our work thus connects a conserved signaling pathway with its critical downstream effectors, advancing insight into how body size is specified. Since collagen mutations and misregulation are implicated in numerous human genetic disorders and injury sequelae, understanding how collagen gene expression is regulated has broad implications.
Keywords: growth, body size, collagen, BMP, C. elegans
THE Transforming Growth Factor-β (TGF-β) superfamily encompasses > 30 ligands including Bone Morphogenetic Proteins (BMPs), Activin, and Nodal. BMPs play essential roles in development and have been studied in a variety of contexts (Wu and Hill 2009). However, the roles of BMPs in growth and body size regulation are relatively unexplored. This deficit is partly due to body size being expressed as a complex trait with continuous variation. The existence of monogenic variants with discrete body size changes provides us with an entry point into growth- and size-regulating mechanisms. We have capitalized on the major visible effect of BMP signaling on growth regulation in the nematode Caenorhabditis elegans to uncover transcriptional targets acting as effectors of body size regulation.
BMPs are highly conserved among animal species along with the components of their signaling pathways. BMP homologs are present in Drosophila [e.g., Decapentaplegic (Dpp)] and C. elegans [e.g., Dpp/BMP-like-1 (DBL-1)] (Wu and Hill 2009), genetically tractable organisms in which Smads, the intracellular components of the TGF-β signaling pathway, were first identified (Sekelsky et al. 1995; Savage et al. 1996). Receptor-regulated Smads (R-Smads) are directly phosphorylated by the TGF-β receptors on the C-terminus. This phosphorylation allows formation of the heterotrimeric Smad complex with co-Smads that accumulate in the nucleus to regulate target genes of the pathway (Wrana et al. 1992; Estevez et al. 1993; Liu et al. 1995; Lagna et al. 1996; Zhang et al. 1996; Abdollah et al. 1997; Souchelnytskyi et al. 1997; Qin et al. 2001; Wu et al. 2001; Chacko et al. 2004; Nicolas et al. 2004). Smads are known to bind a 4-bp GTCT Smad-Binding Element (SBE); furthermore, R-Smads for BMP ligands associate with GC-rich sequences (GC-SBEs) (Kim et al. 1997; Rushlow et al. 2001; Gao et al. 2005). While much detailed knowledge has been obtained in studies of a few direct Smad target genes, fewer studies have addressed the direct and indirect transcriptional programs required to mediate biological functions in intact organisms.
In C. elegans, the DBL-1/BMP signaling pathway is the major regulator of growth and body size. DBL-1 ligand is secreted by neurons and body wall muscle, and is necessary for body size regulation, and other developmental and homeostatic processes (Suzuki et al. 1999). The small body size phenotype is the result of a reduction in cell size rather than cell number. Previous work has pinpointed the hypodermis, the outermost multinucleated epithelium, as the main target tissue of DBL-1 signaling. In dbl-1 mutants, different tissue sizes are reduced to different extents, with hypodermal tissue reduced in proportion to body size (Nagamatsu and Ohshima 2004). Furthermore, we have previously shown by tissue-specific expression of SMA-3 (R-Smad) in a sma-3 mutant background that activation of the DBL-1 pathway in the hypodermis is necessary and sufficient for normal body size (Wang et al. 2002). Similar conclusions have been drawn in experiments with the DBL-1 receptors (Inoue and Thomas 2000; Yoshida et al. 2001). To identify the transcriptional targets of the DBL-1 pathway that may function in body size regulation, we performed microarray analysis on dbl-1 mutants (Liang et al. 2007). The functions of putative target genes were analyzed by RNA interference (RNAi) knockdowns, leading us to concentrate on a group of cuticle collagen genes.
The cuticle serves as the skin of the worm and its main component is collagen, encoded by > 170 genes in the cuticular collagen multigene family (Johnstone 2000). The cuticle is synthesized and secreted by the underlying hypodermis, and polymerizes on the external surface forming a flexible barrier. At the end of each larval stage, the cuticle molts revealing a newly formed cuticle underneath. Some cuticle collagen genes are expressed in specific larval stages while others are expressed in each cuticle synthesis period (Jackson et al. 2014). In spite of extensive study, only a few of the cuticle collagen genes have been described to have visible mutant phenotypes; these abnormalities include body morphology defects such as DumPY (Dpy), ROLler (Rol), BLIster (Bli), SQuaT (Sqt), and LONg (Lon) (Page and Johnstone 2007). We reasoned that altered expression of cuticle collagen genes could contribute to the small body size phenotype of DBL-1 pathway mutants.
We show here that cuticle collagen genes are direct and possibly indirect transcriptional targets of the DBL-1 pathway. We demonstrate SMA-4 (co-Smad) binding in the intergenic region between col-141 and col-142. This result is the first demonstration that the DBL-1 pathway Smads bind DNA in vitro, further validating the functional conservation of Smads in the nematode model. We find that other collagen genes are likely indirect targets of the DBL-1 pathway. Moreover, we observe loss of stage-specific expression of cuticle collagens in the absence of DBL-1 signaling. Lastly, we demonstrate through collagen loss-of-function mutants, RNAi, and overexpression studies that cuticle collagens are the effectors of body size regulation by the DBL-1 pathway.
Materials and Methods
Strains
C. elegans strains were grown at 20° using standard methods (Brenner 1974). All experiments were performed at 20°. In addition to strains generated in this study, the following strains were used: N2 (wild-type), CS24 sma-3(wk30), and MT2709 rol-6(e187n1270).
Body size measurements
Worms were grown at 20°, and collected at the given time point (Figure 1A) or developmental stage (all other experiments). Animals were photographed using QC Capture 2.73.0. The length of each worm was determined by drawing a segmented line along the midline from the head to the tail, and the length was measured using Image-Pro Express 5.1.0.12 software.
Figure 1.
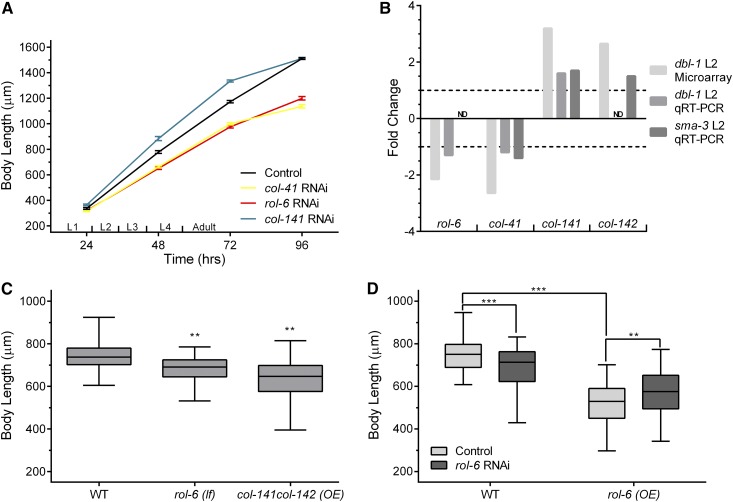
Cuticle collagen gene inhibition and overexpression leads to body size phenotypes. (A) RNAi inhibition of col-41, rol-6, and col-141 led to changes in body size. Animals were measured at indicated time points following embryo collection. n = 30 for each data point. Error bars show SE. (B) Fold-change in expression levels of cuticle collagen genes. Data from dbl-1 mutants at L2 stage from microarray analysis is compared with qRT-PCR results from sma-3 and/or dbl-1 mutants at L2 stage. (C) Comparing body size phenotype of rol-6(e187n1270) mutant and overexpressing col-141col-142 worms relative to wild-type. Body length was measured at the L4 stage. An independently generated transgenic line gave equivalent results. (D) Overexpression of rol-6 leads to smaller body size. rol-6 RNAi in the rol-6 overexpression line alleviates small body size. Body length was measured at the L4 stage. An independently generated transgenic line gave equivalent results. OE, overexpression; Control, empty vector. Box plots show second and third quartiles; whiskers show maximum and minimum. Statistical significance was determined using Student’s t-test: * P < 0.05, ** P < 0.005, and *** P < 0.0005. ND, not determined. qRT-PCR, quantitative reverse transcriptase PCR; RNAi, RNA interference.
RNAi
RNAi by feeding was performed as described (Kamath and Ahringer 2003; Liang et al. 2013). RNAi clones that target the unique (non-Gly-X-Y repeat) regions of collagen genes were generated by cloning desired PCR fragments into L4440. The following primers were used to PCR amplify respective genes:
Electrophoretic mobility shift assay
Protein purification:
GST-tagged versions of the MH1 domains of SMA-2, SMA-3, and SMA-4 were cloned in the pGEX-4T2 vector. All three proteins were isolated using BL21 competent cells, grown in a starter culture overnight at 37°. From the starter culture, a fresh culture of 100 ml was started and grown at 37° until OD of 500 was reached. Expression was induced using IPTG at a final concentration of 1 mM in 100 ml cultures and grown at 28° for 6–8 hr. Standard batch purification was performed using GST beads, and eluted in 30 mM reduced glutathione and 50 mM Tris buffer, pH 8.0. Three elutions were taken; each of the primary elutions were run on 4–12% Bis-Tris gels (Life Technology) for size separation under denaturing conditions.
Gel shifts:
Double-stranded probes were labeled with 32P and incubated with protein in binding buffer at room temperature for 20 min. Binding buffer was composed of 50% glycerol, 100 mM HEPES, 15 mM DTT, 0.5 mg/ml BSA, 0.5 M KCl, 50 mM ZnSO4, and 50 µg/ml dIdC (deoxyinosine:deoxycytidine), samples were run on 5% native acrylamide gel for 1–1.5 hr, dried for 1 hr at 65°, and developed for 1 hr at −80°. Probe 1: 5′-TTCAAAATAAGACAACACAGAAAGTAGGGT-3′; probe 2: 5′-TGACCTTTTCATGATCATAAGACCCGGTTT-3′; probe 3: 5′-ACGGTTTCAAGGTCTGTCTCCTCGAACACG-3′; and probe 4: 5′-GGTGAGACAAGCAATGAGAATAGACACACA-3′.
Quantitative RT-PCR
Worms were grown at 20° until a large number of eggs were observed on plates. Worms were then washed off using M9 buffer and the remaining eggs were allowed to hatch for 4 hr. Worms were then collected and placed on new plates, and grown at 20° until late second larval (L2) or adult stage, collected, and mRNA was extracted using the RNeasy mini kit (QIAGEN, Valencia, CA). Since rol-6(OE) is an extrachromosomal array, for analysis of rol-6(OE) and rol-6(RNAi), we used the following protocol from http://groups.molbiosci.northwestern.edu/morimoto/research/Protocols/IX.%20C.%20elegans/B.%20Extraction/2.%20TotalRNA.pdf. Ten L4 worms from L4440 control, rol-6(OE) on L4440, and rol-6(RNAi) were picked into 20 μl of M9 buffer. Next, 250 μl of Trizol was added. Samples were vortexed intermittently and kept on ice, then frozen in liquid nitrogen. Samples were thawed, and extraction performed with 50 μl chloroform two times. RNA was precipitated with 125 μl isopropanol and washed with 70% ethanol. DNA was removed using Ambion Turbo DNAse. RT-PCR and quantitative reverse transcriptase PCR (qRT-PCR) were performed as previously described (Yin et al. 2015). The following primers were used for qRT-PCR:
Chromatin immunoprecipitation sequencing
Chromatin immunoprecipitation sequencing (ChIP-seq) was performed by Michelle Kudron (Valerie Reinke Model Organism ENCyclopedia Of DNA Elements and model organism Encyclopedia of Regulatory Networks group) on the sma-3(wk30);Is[GFP::SMA-3] strain at late L2 stage (Gerstein et al. 2010). Data are available at encodeproject.org.
Genome editing and transgenic strains
To obtain a col-141 knock-in line via clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9, we followed the Dickinson (Dickinson et al. 2015) protocol and used the selection excision cassette pDD282 vector. All constructs were assembled using a New England Biolabs (Beverly, MA) Hi-fi DNA Assembly kit. All other strains were constructed by microinjections, myo-2::gfp (20 ng/μl), transgene of interest (100 ng/μl), and carrier DNA (80 ng/μl). col-141 and col-142 overexpression lines were created via microinjection (50 ng/μl each, myo-2::gfp at 12 ng/μl). rol-6 overexpression lines were made via injection of rol-6 (50 ng/μl) and myo-2::gfp (10 ng/μl). The col-p::2xNLSmCherry vector was constructed via subcloning the intergenic region (XbaI/KpnI) into the p702 vector containing 2xNLSmCherry and confirmed via sequencing. The mutated col-p region was synthesized by Integrated DNA Technologies and subcloned into the p702 vector.
Imaging
All fluorescence images were taken via a Zeiss ([Carl Zeiss], Thornwood, NY) Apotome microscope. For fluorescence quantification, images were taken at 63× and, utilizing ImageJ software, circles were drawn around the fluorescent nuclei and quantified.
Data availability
Strains and plasmids are available upon request. All data necessary for confirming the conclusions of the article are present within the article or in supplemental files available at FigShare. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6305780.
Results
Cuticle collagen genes are transcriptional targets of the DBL-1 pathway and are required for normal body size
We sought to connect the DBL-1 transcriptional program to its downstream effectors of body size regulation. In previous work, we identified five putative target genes with body size phenotypes (Liang et al. 2007), but all of these genes encode signaling molecules or transcription factors, so they are not the downstream effectors of growth control. It has been proposed that changes in hypodermal polyploidy contribute to the reduced growth of dbl-1 mutant adults (Lozano et al. 2006), although these changes cannot account for differences in size during larval stages. We tested the functions of putative target genes encoding DNA licensing factors and cyclins that could regulate polyploidization, but knockdown of these genes by RNAi did not produce body size phenotypes (Supplemental Material, Figure S1). We next analyzed the functions of target genes involved in fat metabolism, but discovered that they function independently of body size regulation (Clark et al. 2018). Finally, we turned our attention to four cuticle collagen genes that we identified as putative transcriptional targets of the DBL-1 pathway. We performed qRT-PCR to verify transcriptional changes in cuticle collagen genes. We compared expression in dbl-1 and sma-3 mutants to wild-type controls at the L2 stage, the same stage in which the microarray analysis had been conducted. In the absence of DBL-1 signaling, rol-6 and col-41 are downregulated while col-141 and col-142 are upregulated (Figure 1B), in agreement with the microarray studies.
We hypothesized that regulated expression of these cuticle collagen genes contributes to body size regulation. An alternative hypothesis is that the altered expression of cuticle collagen genes is a response to, rather than a cause of, the altered size of dbl-1 mutants (Roberts et al. 2010). To address the role of these cuticle collagen genes in body size regulation, we generated gene-specific RNAi clones targeting the genes. We performed RNAi of col-41, rol-6, and col-141, and measured body length at different time points during larval and adult growth (Figure 1A). RNAi of col-41 and rol-6 led to a decrease in body size at all time points. Conversely, col-141(RNAi) led to a transient increase in body size in the larval stages, suggesting that cuticle collagens can act as negative as well as positive regulators of body size. In these experiments, we did not observe any detectable differences in developmental timing. Nevertheless, in subsequent experiments, we determined body size at defined developmental stages based on vulval and gonadal developmental milestones. We verified our RNAi results for rol-6 using an available loss-of-function mutation in rol-6. This rol-6(lf) mutant also has a small body size, as illustrated by body length measured at the L4 stage (Figure 1B).
If cuticle collagen expression is truly instructive rather than permissive for body size regulation, we would expect that overexpression of respective collagens should also lead to body size changes. Therefore, we performed overexpression of col-141 and col-142, by increasing copy number in transgenics carrying a genomic fragment that contains both genes, which are adjacent in the genome. Overexpression of col-141 and col-142 resulted in decreased body size relative to wild-type worms (Figure 1B), confirming these genes as negative regulators of growth. We performed a similar experiment to overexpress rol-6. Surprisingly, overexpression of rol-6 led to a decrease, rather than an increase, in body size (Figure 1C). This result suggests that excess ROL-6 can cause a reduction in body size. Since the overexpression lines have a smaller body size than rol-6 loss-of-function, we were able to test this hypothesis by using RNAi to inactivate rol-6 in these lines. RNAi inhibition of rol-6 expression reduced the severity of the growth defect in the overexpressing lines, bringing their phenotypes closer to the observed size of rol-6(RNAi) or rol-6(lf) (Figure 1C; similar results were obtained with an independent line). Finally, we verified expression changes in rol-6(OE) and rol-6(RNAi) by qRT-PCR; RNAi reduced rol-6 expression to 0.01 of the level of L4440 empty vector control, while rol-6(OE) increased expression by 18% to 1.18 the level of control. These results suggest that rol-6 is dose-sensitive and must be expressed in a particular range, otherwise aberrant body size is observed.
Direct regulation of cuticle collagen genes by DBL-1 Smads
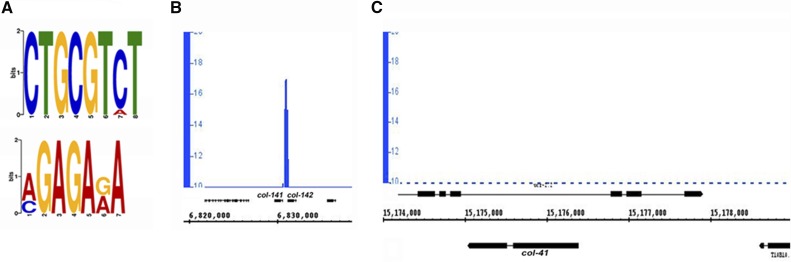
To identify which transcriptional targets of DBL-1 signaling were potentially directly regulated by Smad binding, genome-wide ChIP-seq was performed using a strain expressing functional GFP::SMA-3 from the endogenous sma-3 promoter in a sma-3(wk30) background (encodeproject.org). Using the online Galaxy analysis software (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010), we extracted genomic sequences from the coordinates obtained via ChIP-seq. MEME-ChIP software (Bailey et al. 2009) was used to discover enriched motifs and generate logo plots. The top two enriched motifs were the CTGCGTCT SBE and GAGA (Figure 2A). The presence of GTCT as an enriched motif indicates an increased presence of SMA-3 at canonical Smad-binding sites. The GAGA motif is bound by the transcription factor EOR-1 in C. elegans, and is associated with open chromatin regions during larval development (Daugherty et al. 2017). Among the cuticle collagen genes that we established as transcriptional targets, the intergenic region between col-141 and col-142 had strong Smad binding (Figure 2B). No additional SMA-3 recruitment sites were identified in this genomic region, suggesting that the intergenic region serves to regulate both of these genes. Therefore, we chose to study this region, which we will refer to hereafter as col-p. In contrast to col-141 and col-142, SMA-3 ChIP-seq peaks were not found in the genomic regions surrounding col-41 (Figure 2C).
Figure 2.
SMA-3 associates with Smad-binding elements (SBEs) in the intergenic region between col-141 and col-142 (col-p). (A) SMA-3 chromatin immunoprecipitation sequencing revealed SBE as a highly enriched motif via MEME-ChIP motif discovery analysis. The e value for CTGCGTCT motif is 3.6E−101; the e value for AGAGAGA motif is 7.6E−083. (B) SMA-3 associates with the intergenic region between col-141 and col-142 (col-p). (C) No peaks of SMA-3 are detected in the genomic region surrounding col-41. The nearest peaks are located distal to the neighboring coding sequences T10B10.3 and ucr-2.2. We were unable to analyze binding of SMA-3 to the rol-6 genomic region due to multiple copies of rol-6(d) present as a marker on the transgene in the strain used for this experiment.
Smads associate with Smad-binding elements in col-p
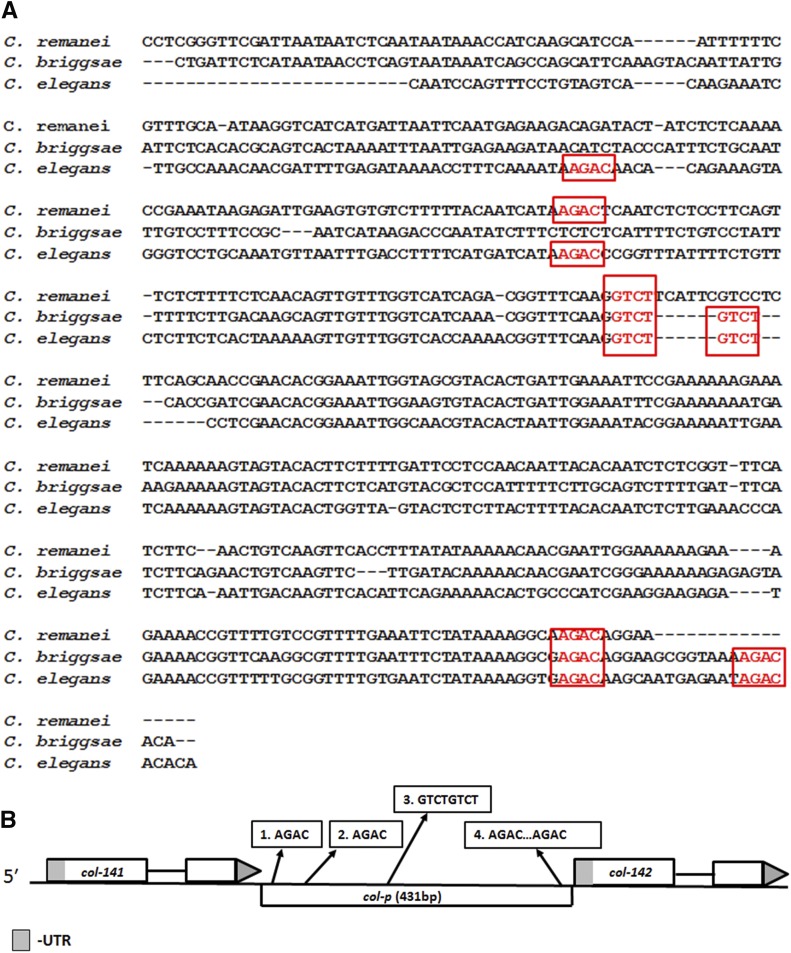
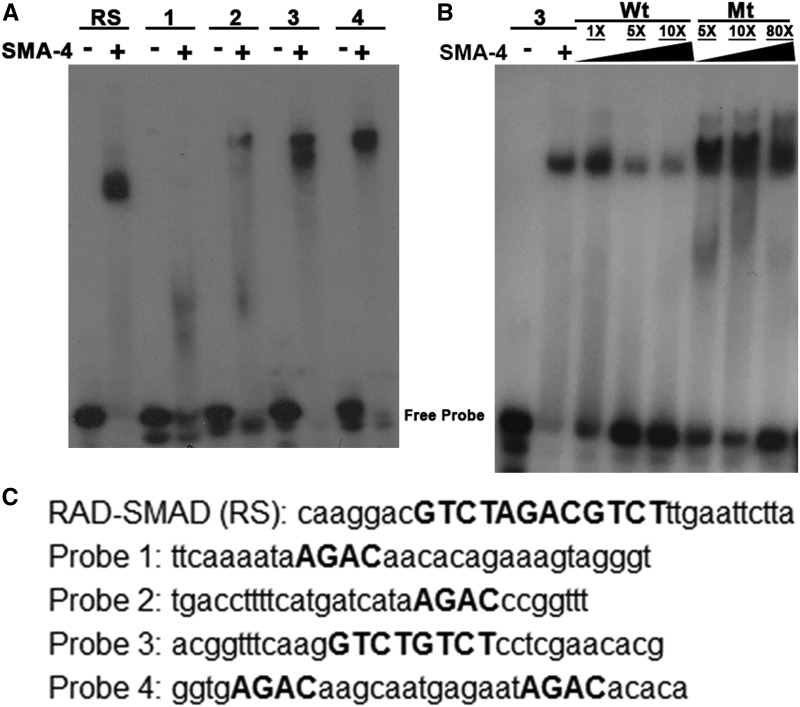
We generated a multiple sequence alignment between col-p sequences from three related nematode species (Figure 3A). This analysis of col-p revealed multiple SBEs, some of which are conserved between two or three analyzed species. To elucidate the molecular requirements for Smad recruitment, we used the electrophoretic mobility shift assay (EMSA) to test R-Smad SMA-3 and co-Smad SMA-4 binding to putative regulatory sequences. The MH1 Smad DNA-binding domains were expressed as GST fusion proteins in bacteria. We used an artificial Smad-binding sequence based on the well-characterized RAD-SMAD reporter (Tian et al. 2010) as a positive control. The SMA-4 MH1 domain binds strongly to this probe (Figure 4A). We then generated four probes from col-p based on sequence alignment and the presence of GTCT sequences (Figure 4C). The SMA-4 MH1 domain strongly and specifically bound to probes 3 and 4 (Figure 4, A and B). To determine if the conserved SBEs are necessary for SMA-4 binding, we introduced two single-base pair substitutions in the SBEs, changing GTCT to ATCT. Unlabeled wild-type and mutant probes were used in a competition assay with labeled wild-type probe 3. Unlike wild-type probes, mutant probes with ATCT are unable to compete for SMA-4 binding (Figure 4B). Similar results were obtained with mutated probe 4 (Figure S2). Therefore, SMA-4 binding depends on intact GTCT sequences. Overall, our EMSA results indicate that SMA-4 can bind at least two specific sites in col-p (in probes 3 and 4). Additional binding sites may also be present, because we did not test the entire intergenic region. SMA-3 MH1 domain did not bind to any of the tested probes, possibly because BMP R-Smads prefers GC-SBEs to canonical SBEs (see Discussion).
Figure 3.
Multiple conserved Smad-binding elements (SBEs) are present in the intergenic region between col-141 and col-142. (A) Five out of the six putative SBE sites are conserved among multiple Caenorhabditis species in the intergenic region between col-141 and col-142. Nucleotide alignments were made using ClustalW. (B) Schematic of the location of SBEs in the intergenic region between col-141 and col-142.
Figure 4.
Smads bind directly to Smad-binding elements (SBEs) in col-p. (A) SMA-4 strongly binds to probes 3 and 4 containing SBEs. (B) SMA-4 binds specifically to an SBE (2xGTCT) in probe 3. (C) Probes from col-p region used to test binding of Smad MH1 DNA-binding domains. SBEs are capitalized. In (B), mutated probe 3 had sequence 5′-ACGGTTTCAAGATCTATCTCCTCGAACACG-3′, in which GTCT motifs were mutated to ATCT. Mt, mutated; RS, RAD-SMAD; Wt, wild-type.
Identification of genomic sequences required for col-141 and col-142 expression
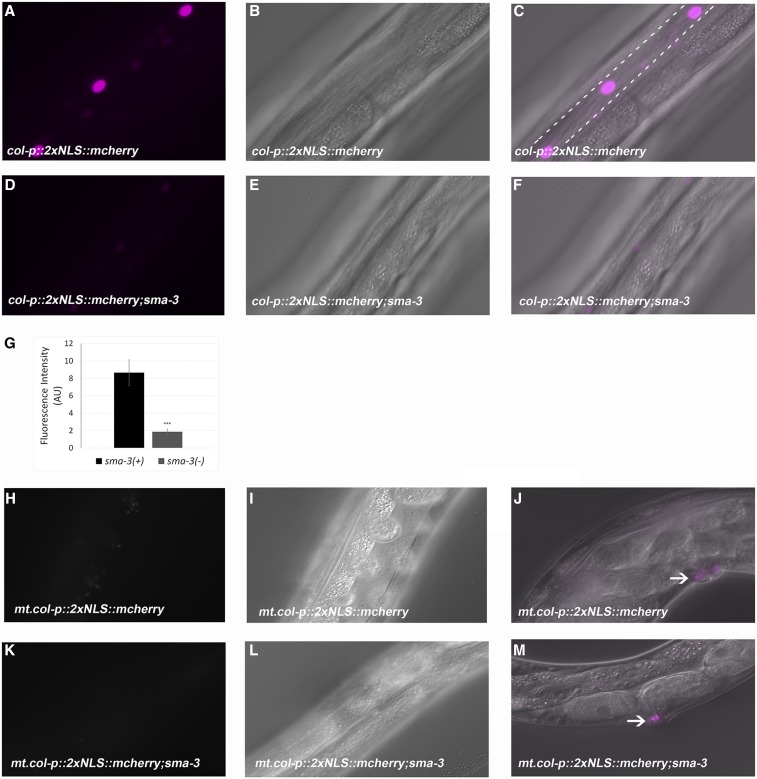
Having identified Smad-binding sites in col-p, we further aimed to determine whether these sites are relevant for gene expression in vivo. Hence, we created a construct with col-p driving expression of 2× nuclear localized mCherry and obtained multiple lines via microinjection. As expected, we found that these sequences drive expression in epidermal tissues. The main C. elegans epidermal tissue is a large multinucleate syncytium known as hyp7. Lateral epidermal cells known as seam cells act as stem cells that divide asymmetrically, giving rise to one daughter cell that joins the hypodermis. Ventral epidermal cells either join the hypodermal syncytium or contribute to the vulva (Sulston and Horvitz 1977). mCherry expression driven by col-p was observed in the seam cells and in the vulva at the adult stage (Figure 5, A–C). Fluorescence was detectable in the adult stage only. This pattern is consistent with previous genome-wide studies (Jackson et al. 2014) that showed peak col-141 and col-142 expression at the adult stage. To test if the DBL-1 pathway would regulate mCherry expression, we further crossed these lines into the sma-3 mutant background. We observed little to no expression of mCherry expression at all stages (Figure 5, D–F). We quantitated fluorescence intensity and found that it was significantly reduced (Figure 5G). This result did not align with our microarray and qRT-PCR results, which showed increased rather than decreased expression of col-141 and col-142 in DBL-1 pathway mutants. However, those assays were conducted at the L2 stage, at which time fluorescence of our reporter is undetectable. As described below, qRT-PCR experiments in adult animals are consistent with our reporter results.
Figure 5.
Identification of genomic sequences required for col-141 expression. (A–C) col-p::2xNLS::mCherry leads to adult stage-specific expression in seam cell nuclei. Dotted lines outline the approximate positions of the seam cells. (D–F) mCherry expression is severely depleted in the sma-3 mutant background. (G) Quantification of mCherry expression in wild-type and sma-3 mutant background. Error bars show SE; AU, arbitrary units. * P < 0.05, ** P < 0.005, and *** P < 0.0005. (H–J) Mutating Smad-binding element (SBE) 3 from GTCT to ATCA, and SBE 4 from AGAC to TGAT, leads to loss of mCherry expression. (K–M) Crossing of mt.col-p::2xNLS::mCherry into sma-3 mutant background does not change the ablated mCherry expression. (A, D, H, and K) Fluorescent images; (B, E, I, and L) Nomarski images; (C and F) merged images; and (J and M) merged images showing expression in the vulval region.
To test if SBEs present in col-p are relevant for mCherry expression, we made transgenic animals with mutated SBEs (mt.col-p) driving 2xNLS::mCherry. Expression of mCherry became undetectable in the hypodermis, but was retained in the vulva (Figure 5J), indicating that Smad binding at these SBEs is required for gene expression in the hypodermis (Figure 5, H and I). To test if mCherry expression was the same in the presence and absence of sma-3, we crossed these worms into the sma-3 mutant background (Figure 5, K–M). Again, we detected no fluorescence in the hypodermis.
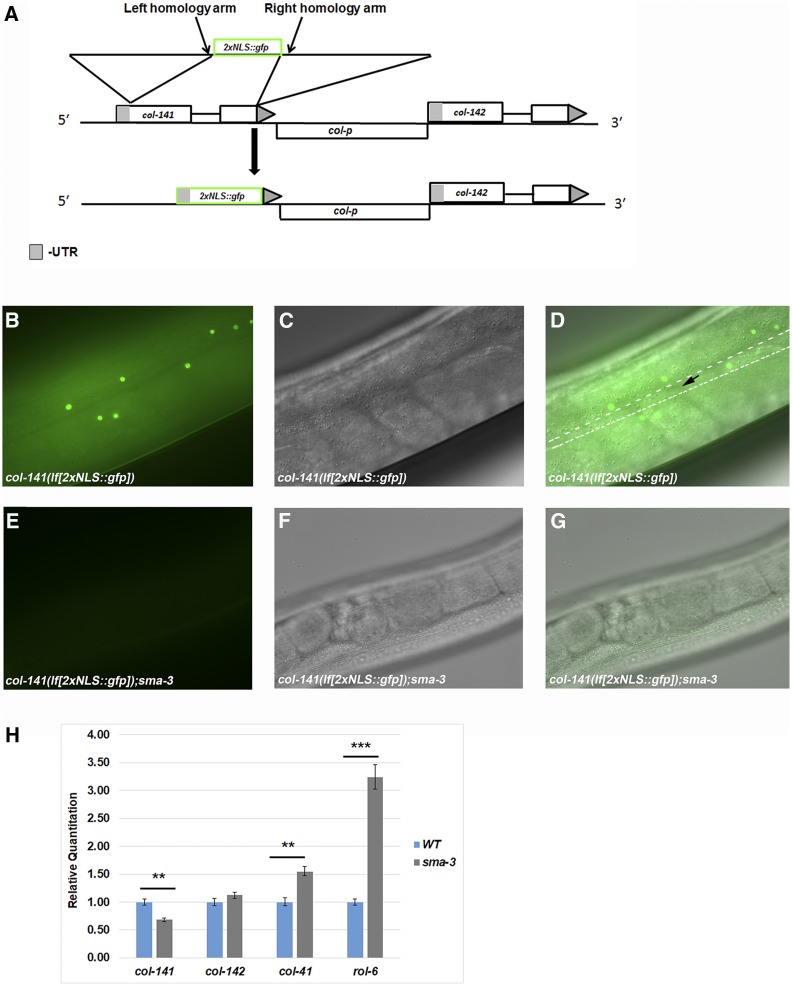
To verify the results from our extrachromosomal reporter, we wanted to study the expression of col-141 in its endogenous genomic environment. Utilizing CRISPR-Cas9, we knocked in 2xNLS::GFP to replace col-141, making a transcriptional reporter and a loss-of-function simultaneously (Figure 6A). We characterized the reporter and observed GFP fluorescence specifically in epidermal nuclei at the adult stage (Figure 6, B–D). Little to no expression was observed in larval stages, consistent with the result from the extrachromosomal reporter. These two constructs showed some differences in spatial expression pattern. While the extrachromosomal reporter showed expression in the seam cells and the vulva, expression from the endogenous promoter was primarily seen in hyp7, and absent from seam cells (black arrow in Figure 6D) and the vulva, suggesting that endogenous sequences outside of col-p contribute to spatial specificity. To confirm that DBL-1 signaling regulates GFP transcription from the col-141 locus, we crossed the col-141 CRISPR reporter into a sma-3 mutant background (Figure 6, E–G). We observed a loss of GFP expression, replicating the results of the extrachromosomal col-p::2xNLSmCherry transgenic animals (Figure 5, A and D).
Figure 6.
An endogenous clustered regularly interspaced short palindromic repeats (CRISPR)-generated reporter verifies requirements for col-141 expression. (A) Utilizing CRISPR-Cas9 genome editing to knock out and replace col-141 with 2xNLS::gfp, creating a loss-of-function mutant and transcriptional reporter simultaneously. (B–D) GFP expression in col-141(lf) mutant. Dotted lines outline the approximate positions of the seam cells. Black arrow denotes a seam cell nucleus lacking GFP expression. (E–G) Loss of GFP expression in the sma-3 mutant background. (B and E) Fluorescent images; (C and F) Nomarski images; and (D and G) merged images. (H) Regulation of cuticle collagen gene expression by sma-3 in adult stages determined by quantitative reverse transcriptase PCR mirrors the transcriptional reporter. Results show the mean from four independent biological replicates with two technical replicates for each sample. Statistical analysis was performed by student’s t-test. ** P < 0.005 and *** P < 0.0005. WT, wild-type.
DBL-1 signaling is required for stage-specific regulation of cuticle collagen genes
We sought to resolve our discrepant results on the direction of regulation of col-141 and col-142 by the DBL-1 pathway. Initial qRT-PCR performed at the L2 stage, as well as the original microarray experiments (Liang et al. 2007), indicated an increase in col-141 and col-142 expression in DBL-1 pathway mutants (Figure 1B). Meanwhile, the col-p::2xNLSmCherry and col-141(lf[2xNLS::gfp]) strains showed a reduction of expression at the adult stage. To confirm that loss of fluorescence expression in col-141(lf[2xNLS::gfp]) and col-p::2xNLSmCherry replicated col-141 and col-142 expression in the sma-3 mutant background, we performed qRT-PCR on sma-3 mutant adults. We observed a significant decrease in col-141 levels in sma-3 mutant adults (Figure 6H), corroborating the observations from our transcriptional reporters. In some experiments, we saw a decrease in col-142 levels in sma-3 mutant adults, but this decrease did not reach statistical significance. We extended these observations by determining the expression levels of col-41 and rol-6 in sma-3 mutant adults. In these experiments, we observed an increase in col-41 and rol-6 levels (Figure 6H), again reversing the direction of regulation in comparison with the L2 stage. We note that an independent microarray experiment also identified col-41 as upregulated and col-142 as downregulated in sma-2 mutants at the L4 stage (Luo et al. 2010), similar to our results in adults. These results suggest that DBL-1 signaling is required for the appropriate temporal stage-specific expression of cuticle collagen genes.
Discussion
Body size regulation has been investigated in multiple organisms, including humans (Le Goff and Cormier-Daire 2012; Lui et al. 2015) mice, dogs, and Drosophila (Baker et al. 1993; Oldham et al. 2000; Shingleton 2005; Sutter et al. 2007). In these organisms, signaling pathways have been identified that contribute to body size regulation; however, the effector genes for body size regulation through those pathways are poorly characterized. We have capitalized on the genetic tractability of C. elegans to study how the conserved DBL-1 BMP-related signaling pathway regulates body size. In our work, we elucidated the transcriptional network initiated by DBL-1 to explain how body size is modulated by DBL-1 activity. We have shown that cuticle collagen genes are transcriptional targets of the DBL-1 pathway via microarray analysis and qRT-PCR. Moreover, we showed that DBL-1 is required for stage-specific expression of cuticle collagen genes. Through RNAi, mutant, and overexpression studies, we discovered a positive regulator (col-41), a dose-sensitive regulator (rol-6), and a negative regulator (col-141) of body size. We showed association of Smads in the intergenic region between col-141 and col-142 via ChIP-seq, and using EMSA we showed in vitro binding of SMA-4 MH1 domain to conserved SBE sites in this region.
Since cuticle collagen genes are encoded by such a large gene family in C. elegans, it is reasonable to ask why so many genes are required. One possible explanation is that the large number of genes allows rapid synthesis of the cuticle during the discrete cuticle synthesis periods of the worm’s lifecycle. Our work supports the notion that there is also a degree of functional specificity. Consistent with this hypothesis, > 60% of cuticle collagen genes have been described as having stage-specific expression patterns (Jackson et al. 2014). In particular, col-41 and rol-6 have peaks of expression in the L2 stage, while col-141 and col-142 have highest expression in adults. In addition to our own work, two published microarray analyses identified cuticle collagen genes as transcriptional targets of the DBL-1 pathway (Liang et al. 2007; Luo et al. 2010; Roberts et al. 2010). Furthermore, specific subsets of cuticle collagen genes have been described as transcriptional targets of Wnt and insulin signaling pathways in C. elegans (Jackson et al. 2014; Ewald et al. 2015). Both DBL-1 and Wnt pathway mutants have been shown to have cuticle defects (Jackson et al. 2014; Schultz et al. 2014), demonstrating that there are functional consequences to the alterations in collagen gene expression. Furthermore, Ewald et al. (2015) demonstrated that specific cuticle collagens (which include col-141) contribute to the regulation of longevity by the insulin signaling pathway.
Interestingly, previous work has demonstrated that the DBL-1 pathway can also regulate the cuticle post-transcriptionally. For example, LON-3 is a cuticle collagen that acts downstream of DBL-1 to regulate body size (Nystrom et al. 2002; Suzuki et al. 2002), but it is regulated post-transcriptionally by the DBL-1 pathway (Suzuki et al. 2002). Like COL-141 and possibly COL-142, LON-3 is a negative regulator of growth for which loss-of-function produces longer animals and overexpression results in shorter animals (Nystrom et al. 2002). Based on all of this evidence, we conclude that cuticle collagen genes are major effectors of body size through the DBL-1 pathway. We speculate that the specific collagen isoforms deposited in the cuticle contribute to the characteristics of that cuticle, due to differences in how they cross-link with other components. COL-41, as a positive regulator of growth, may contribute to a more elastic, expandable cuticle. COL-141, COL-142, and LON-3, as negative regulators of growth, may contribute to a more rigid, less easily expanded cuticle. C. elegans cuticle collagens have been categorized into six families based on the patterns of cysteine residues flanking the Gly-X-Y repeats (Yang and Kramer 1994). Collagens in the same family can potentially form heterotrimers cross-linked through these cysteine residues. Interestingly, COL-41, COL-141, and COL-142 all belong to the same family, so it is possible that the relative proportion of these subunits changes the characteristics of the cuticle. ROL-6 and LON-3 belong to another family (Nystrom et al. 2002). It is possible that the small body size seen with ROL-6 overexpression is a consequence of excessive incorporation of the growth-limiting collagen LON-3.
In humans, collagens constitute ∼30% of the protein mass and are the most abundant protein. Collagens function in a diverse range of processes such as mechanical properties of tissue, ligands for receptors, cell growth, differentiation, cell migration, bone formation, skeletogenesis, and the integrity and function of the epidermis (Kadler et al. 2007; Matsuo et al. 2008; Hynes 2009). Collagen expression levels must be tightly controlled during wound healing, and inappropriate expression of collagens can lead to fibrosis (Andrews et al. 2015; Prabhu and Frangogiannis 2016). Mutations in collagens have been recognized in rare heritable diseases such as osteogenesis imperfecta, several subtypes of Ehlers–Danlos syndrome, various chondrodysplasias, X-linked forms of Alpoty syndrome, Ullrich muscular dystrophy, corneal endothelial dystrophy, and Knobloch syndrome (Myllyharju and Kivirikko 2004). Collagen mutations, as seen in osteogenesis imperfecta, can result in increased TGF-β signaling (Grafe et al. 2014). Therefore, collagens and their regulation have significant implications for human health. Our work further adds to the functional importance of collagens in the context of growth regulation.
In addition to highlighting the functional significance of collagen genes, our work sheds light on the mechanisms of action of the DBL-1 Smads. Three Smads act in this pathway: the R-Smads SMA-2 and SMA-3, and the co-Smad SMA-4. All three Smads are necessary for pathway function, and they presumably form a heterotrimeric complex. We performed genome-wide ChIP-seq analysis on GFP::SMA-3 and identified GTCT, the canonical SBE, as enriched at sites of SMA-3 occupancy. In Drosophila, Mad, the Dpp R-Smad, binds GC-rich Smad-binding sites termed GC-SBE (GGAGCC) (Morikawa et al. 2011), while Medea, the Dpp co-Smad, binds canonical SBEs (GTCT). In our analyses of targets of the DBL-1 pathway, we observed Smad binding to the intergenic region between col-141 and col-142. Therefore, we tested for direct binding of isolated MH1 domains of Smads SMA-3 and SMA-4 to SBEs in this region. Consistent with findings in Drosophila, the MH1 domain of co-Smad SMA-4, but not that of R-Smad SMA-3, was able to bind the SBE’s directly in vitro. Similarly, the co-Smad of the C. elegans DAF-7/TGF-β pathway, DAF-3, has previously been shown to bind canonical GTCT sequences (Thatcher et al. 1999). Since there is no evidence for direct binding of R-Smad SMA-3 to GTCT sequences, the enrichment of this motif at SMA-3 genomic binding sites may be due to SMA-3 being recruited as part of the Smad complex, with co-Smad SMA-4 making the direct contact with this site.
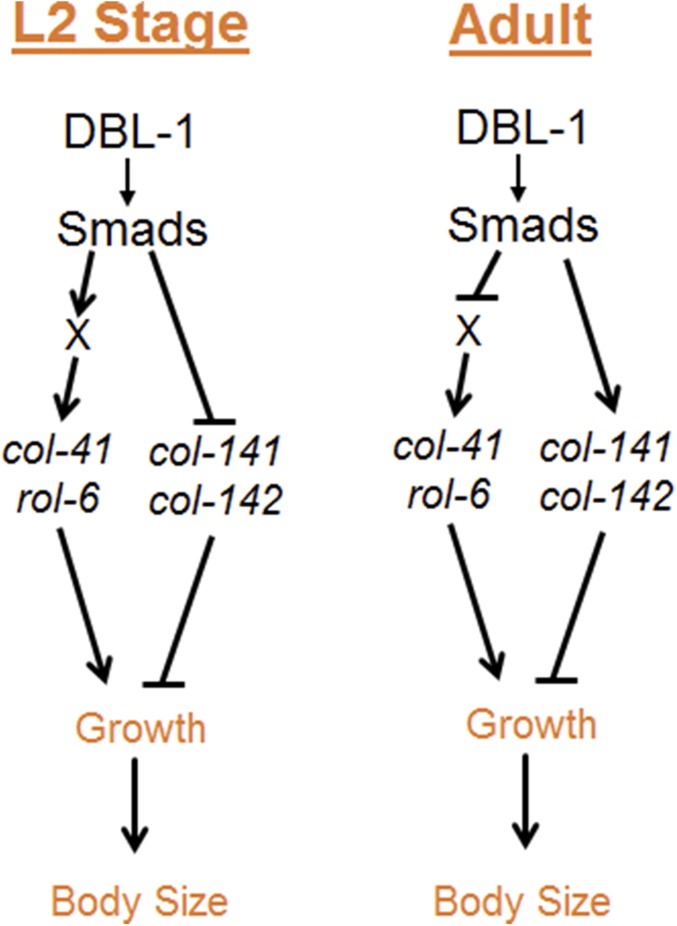
In conclusion, our work elucidates the transcriptional network for body size regulation through the DBL-1/BMP pathway in C. elegans. We propose a model (Figure 7) in which DBL-1 signaling leads to Smad activation causing direct and indirect regulation of specific cuticle collagen genes. This regulation occurs in a stage-specific manner to initiate the correct temporal program of collagen gene expression. Inappropriate expression of cuticle collagen genes leads to aberrant body size. This work thus provides the first mechanistic link between BMP signaling and effectors of body size regulation.
Figure 7.
DBL-1 regulation of growth. Our model proposes stage-specific regulation of cuticle collagen genes to contribute to the growth and body size of C. elegans.
Acknowledgments
We thank Michelle Kudron with the Model Organism ENCyclopedia Of DNA Elements and model organism Encyclopedia of Regulatory Networks projects for performing chromatin immunoprecipitation sequencing; Randy Hui for technical assistance with electrophoretic mobility shift assays; Amy Park for RNA interference (RNAi) experiments; and Emma Ciccarelli for constructing the col-142 RNAi clone. We are grateful to Jun (Kelly) Liu and Geraldine Seydoux for sharing protocols, and Hannes Buelow for providing clones. This work was carried out in partial fulfillment of the requirements for the Ph.D. degree from the Graduate Center of City University of New York (U.M.). Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD-010440). This work was supported by PSC-CUNY ENHC-45-57, NIH grants R15 GM-112147 and R15 G-097692 to C.S.-D., and by NIH grant GM-63024 to C.A.R.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6305780.
Communicating editor: B. Goldstein
Literature Cited
- Abdollah S., Macias-Silva M., Tsukazaki T., Hayashi H., Attisano L., et al. , 1997. TGF-β RI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 272: 27678–27685. 10.1074/jbc.272.44.27678 [DOI] [PubMed] [Google Scholar]
- Andrews J. P., Marttala J., Macarak E., Rosenbloom J., Uitto J., 2015. Keloid pathogenesis: potential role of cellular fibronectin with the EDA domain. J. Invest. Dermatol. 135: 1921–1924. 10.1038/jid.2015.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J., Liu J. P., Robertson E. J., Efstratiadis A., 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75: 73–82. 10.1016/S0092-8674(05)80085-6 [DOI] [PubMed] [Google Scholar]
- Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., et al. , 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19: Unit 19.10.1–Unit 19.10.21. 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko B. M., Qin B. Y., Tiwari A., Shi G., Lam S., et al. , 2004. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol. Cell 15: 813–823. 10.1016/j.molcel.2004.07.016 [DOI] [PubMed] [Google Scholar]
- Clark J. F., Meade M., Ranepura G., Hall D. H., Savage-Dunn C., 2018. Caenorhabditis elegans DBL-1/BMP regulates lipid accumulation via interaction with insulin signaling. G3 (Bethesda) 8: 343–351. 10.1534/g3.117.300416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty A. C., Yeo R. W., Buenrostro J. D., Greenleaf W. J., Kundaje A., et al. , 2017. Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Genome Res. 27: 2096–2107. 10.1101/gr.226233.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D., Goldstein B., 2015. Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200: 1035–1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M., Attisano L., Wrana J. L., Albert P. S., Massague J., et al. , 1993. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365: 644–649. 10.1038/365644a0 [DOI] [PubMed] [Google Scholar]
- Ewald C. Y., Landis J. N., Porter Abate J., Murphy C. T., Blackwell T. K., 2015. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519: 97–101. 10.1038/nature14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Steffen J., Laughon A., 2005. Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J. Biol. Chem. 280: 36158–36164. 10.1074/jbc.M506882200 [DOI] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., et al. , 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15: 1451–1455. 10.1101/gr.4086505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J., Nekrutenko A., Taylor J., Galaxy T., 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11: R86 10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafe I., Yang T., Alexander S., Homan E. P., Lietman C., et al. , 2014. Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nat. Med. 20: 670–675. 10.1038/nm.3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O., 2009. The extracellular matrix: not just pretty fibrils. Science 326: 1216–1219. 10.1126/science.1176009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Thomas J. H., 2000. Targets of TGF-β signaling in Caenorhabditis elegans dauer formation. Dev. Biol. 217: 192–204. 10.1006/dbio.1999.9545 [DOI] [PubMed] [Google Scholar]
- Jackson B. M., Abete-Luzi P., Krause M. W., Eisenmann D. M., 2014. Use of an activated beta-catenin to identify Wnt pathway target genes in Caenorhabditis elegans, including a subset of collagen genes expressed in late larval development. G3 (Bethesda) 4: 733–747. 10.1534/g3.113.009522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone I. L., 2000. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 16: 21–27. 10.1016/S0168-9525(99)01857-0 [DOI] [PubMed] [Google Scholar]
- Kadler K. E., Baldock C., Bella J., Boot-Handford R. P., 2007. Collagens at a glance. J. Cell Sci. 120: 1955–1958. 10.1242/jcs.03453 [DOI] [PubMed] [Google Scholar]
- Kamath R., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kim J., Johnson K., Chen H. J., Carroll S., Laughon A., 1997. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 388: 304–308. 10.1038/40906 [DOI] [PubMed] [Google Scholar]
- Lagna G., Hata A., Hemmati-Brivanlou A., Massague J., 1996. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature 383: 832–836. 10.1038/383832a0 [DOI] [PubMed] [Google Scholar]
- Le Goff C., Cormier-Daire V., 2012. From tall to short: the role of TGFβ signaling in growth and its disorders. Am. J. Med. Genet. C. Semin. Med. Genet. 160C: 145–153. 10.1002/ajmg.c.31337 [DOI] [PubMed] [Google Scholar]
- Liang J., Yu L., Yin J., Savage-Dunn C., 2007. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev. Biol. 305: 714–725. 10.1016/j.ydbio.2007.02.038 [DOI] [PubMed] [Google Scholar]
- Liang J., Xiong S., Savage-Dunn C., 2013. Using RNA-mediated interference feeding strategy to screen for genes involved in body size regulation in the nematode C. elegans. J. Vis. Exp. 72: e4373. 10.3791/4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Ventura F., Doody J., Massague J., 1995. Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol. Cell. Biol. 15: 3479–3486. 10.1128/MCB.15.7.3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano E., Sáez A. G., Flemming A. J., Cunha A., Leroi A. M., 2006. Regulation of growth by ploidy in Caenorhabditis elegans. Curr. Biol. 16: 493–498. 10.1016/j.cub.2006.01.048 [DOI] [PubMed] [Google Scholar]
- Lui J. C., Garrison P., Baron J., 2015. Regulation of body growth. Curr. Opin. Pediatr. 27: 502–510. 10.1097/MOP.0000000000000235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Kleemann G. A., Ashraf J. M., Shaw W. M., Murphy C. T., 2010. TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell 143: 299–312. 10.1016/j.cell.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N., Tanaka S., Yoshioka H., Koch M., Gordon M. K., et al. , 2008. Collagen XXIV (Col24a1) gene expression is a specific marker of osteoblast differentiation and bone formation. Connect. Tissue Res. 49: 68–75. 10.1080/03008200801913502 [DOI] [PubMed] [Google Scholar]
- Morikawa M., Koinuma D., Tsutsumi S., Vasilaki E., Kanki Y., et al. , 2011. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 39: 8712–8727. 10.1093/nar/gkr572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J., Kivirikko K. I., 2004. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 20: 33–43. 10.1016/j.tig.2003.11.004 [DOI] [PubMed] [Google Scholar]
- Nagamatsu Y., Ohshima Y., 2004. Mechanisms for the control of body size by a G-kinase and a downstream TGFbeta signal pathway in Caenorhabditis elegans. Genes Cells 9: 39–47. 10.1111/j.1356-9597.2004.00700.x [DOI] [PubMed] [Google Scholar]
- Nicolas F. J., De Bosscher K., Schmierer B., Hill C. S., 2004. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 117: 4113–4125. 10.1242/jcs.01289 [DOI] [PubMed] [Google Scholar]
- Nystrom J., Shen Z. Z., Aili M., Flemming A. J., Leroi A., et al. , 2002. Increased or decreased levels of Caenorhabditis elegans lon-3, a gene encoding a collagen, cause reciprocal changes in body length. Genetics 161: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S., Bohni R., Stocker H., Brogiolo W., Hafen E., 2000. Genetic control of size in Drosophila. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 945–952. 10.1098/rstb.2000.0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A. P., and I. L. Johnstone, 2007 The cuticle (March 19, 2007), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.138.1, http://www.wormbook.org.
- Prabhu S. D., Frangogiannis N. G., 2016. The biological basis for cardiac repair after myocardial infarction. From inflammation to fibrosis. Circ. Res. 119: 91–112. 10.1161/CIRCRESAHA.116.303577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B. Y., Chacko B. M., Lam S. S., de Caestecker M. P., Correia J. J., et al. , 2001. Structural basis of Smad1 activation by receptor kinase phosphorylation. Mol. Cell 8: 1303–1312. 10.1016/S1097-2765(01)00417-8 [DOI] [PubMed] [Google Scholar]
- Roberts A. F., Gumienny T. L., Gleason R. J., Wang H., Padgett R. W., 2010. Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans. BMC Dev. Biol. 10: 61 10.1186/1471-213X-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow C., Colosimo P. F., Lin M., Xu M., Kirov N., 2001. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev. 15: 340–351. 10.1101/gad.861401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C., Das P., Finelli A. L., Townsend S. R., Sun C. Y., et al. , 1996. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc. Natl. Acad. Sci. USA 93: 790–794. 10.1073/pnas.93.2.790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz R. D., Bennett E. E., Ellis E. A., Gumienny T. L., 2014. Regulation of extracellular matrix organization by BMP signaling in Caenorhabditis elegans. PLoS One 9: e101929 [corrigenda: PLoS One 10: e0118036 (2015)] 10.1371/journal.pone.0101929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., Newfeld S. J., Raftery L. A., Chartoff E. H., Gelbart W. M., 1995. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics 139: 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton A. W., 2005. Body-size regulation: combining genetics and physiology. Curr. Biol. 15: R825–R827. 10.1016/j.cub.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Souchelnytskyi S., Tamaki K., Engstrom U., Wernstedt C., ten Dijke P., et al. , 1997. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J. Biol. Chem. 272: 28107–28115. 10.1074/jbc.272.44.28107 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sutter N. B., Bustamante C. D., Chase K., Gray M. M., Zhao K., et al. , 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316: 112–115. 10.1126/science.1137045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Yandell M. D., Roy P. J., Krishna S., Savage-Dunn C., et al. , 1999. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126: 241–250. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Morris G. A., Han M., Wood W. B., 2002. A cuticle collagen encoded by the lon-3 gene may be a target of TGF-β signaling in determining Caenorhabditis elegans body shape. Genetics 162: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher J. D., Haun C., Okkema P. G., 1999. The DAF-3 Smad binds DNA and represses gene expression in the Caenorhabditis elegans pharynx. Development 126: 97–107. [DOI] [PubMed] [Google Scholar]
- Tian C., Sen D., Shi H., Foehr M. L., Plavskin Y., et al. , 2010. The RGM protein DRAG-1 positively regulates a BMP-like signaling pathway in Caenorhabditis elegans. Development 137: 2375–2384. 10.1242/dev.051615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tokarz R., Savage-Dunn C., 2002. The expression of TGF-β signal transducers in the hypodermis regulates body size in C. elegans. Development 129: 4989–4998. [DOI] [PubMed] [Google Scholar]
- Wrana J., Attisano L., Carcamo J., Zentella A., Doody J., et al. , 1992. TGF-β signals through a heteromeric protein kinase receptor complex. Cell 71: 1003–1014. 10.1016/0092-8674(92)90395-S [DOI] [PubMed] [Google Scholar]
- Wu J. W., Hu M., Chai J., Seoane J., Huse M., et al. , 2001. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol. Cell 8: 1277–1289. 10.1016/S1097-2765(01)00421-X [DOI] [PubMed] [Google Scholar]
- Wu M. Y., Hill C. S., 2009. TGF-β superfamily signaling in embryonic development and homeostasis. Dev. Cell 16: 329–343. 10.1016/j.devcel.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Yang J., Kramer J. M., 1994. In vitro mutagenesis of Caenorhabditis elegans cuticle collagens identifies a potential subtilisin-like protease cleavage site and demonstrates that carboxyl domain disulfide bonding is required for normal function but not assembly. Mol. Cell. Biol. 14: 2722–2730. 10.1128/MCB.14.4.2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Madaan U., Park A., Aftab N., Savage-Dunn C., 2015. Multiple cis elements and GATA factors regulate a cuticle collagen gene in Caenorhabditis elegans. Genesis 53: 278–284. 10.1002/dvg.22847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Morita K., Mochii M., Ueno N., 2001. Hypodermal expression of Caenorhabditis elegans TGF-beta type I receptor SMA-6 is essential for the growth and maintenance of body length. Dev. Biol. 240: 32–45. 10.1006/dbio.2001.0443 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng X., We R., Derynck R., 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature 383: 168–172. 10.1038/383168a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. All data necessary for confirming the conclusions of the article are present within the article or in supplemental files available at FigShare. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6305780.