In vitro and in silico studies of the CaaX-type prenyl transferases suggest a wider array of prenylatable sequences than those determined in vivo. Berger and Kim et al. investigate whether this disconnect is due to use of...
Keywords: CaaX, farnesyl transferase, isoprenylation, Hsp40, thermotolerance
Abstract
Protein isoprenylation targets a subset of COOH-terminal Cxxx tetrapeptide sequences that has been operationally defined as a CaaX motif. The specificity of the farnesyl transferase toward each of the possible 8000 combinations of Cxxx sequences, however, remains largely unresolved. In part, it has been difficult to consolidate results stemming from in vitro and in silico approaches that yield a wider array of prenylatable sequences relative to those known in vivo. We have investigated whether this disconnect results from the multistep complexity of post-translational modification that occurs in vivo to CaaX proteins. For example, the Ras GTPases undergo isoprenylation followed by additional proteolysis and carboxymethylation events at the COOH-terminus. By contrast, Saccharomyces cerevisiae Hsp40 Ydj1p is isoprenylated but not subject to additional modification. In fact, additional modifications are detrimental to Ydj1p activity in vivo. We have taken advantage of the properties of Ydj1p and a Ydj1p-dependent growth assay to identify sequences that permit Ydj1p isoprenylation in vivo while simultaneously selecting against nonprenylatable and more extensively modified sequences. The recovered sequences are largely nonoverlapping with those previously identified using an in vivo Ras-based yeast reporter. Moreover, most of the sequences are not readily predicted as isoprenylation targets by existing prediction algorithms. Our results reveal that the yeast CaaX-type prenyltransferases can utilize a range of sequence combinations that extend beyond the traditional constraints for CaaX proteins, which implies that more proteins may be isoprenylated than previously considered.
THE CaaX proteins are abundant eukaryotic proteins with diverse biological functions. They are operationally defined by a COOH-terminal CaaX motif that is subject to an ordered series of post-translational modifications involving covalent attachment of a C15 (farnesyl) or C20 (geranylgeranyl) isoprenoid to cysteine, endoproteolysis to remove aaX, and carboxymethylation (Figure 1A) (Wang and Casey 2016; Hampton et al. 2018). More complexity can occur in the form of additional modification to certain CaaX proteins (e.g., palmitoylation). The multistep canonical modification of CaaX proteins has been extensively studied in vivo using relatively few CaaX proteins, with Ras-related GTPases being the most often cited. Modifications modulate CaaX protein function and/or localization, and there is intense interest on developing therapeutic inhibitors for all steps of the pathway (e.g., prenyltransferase inhibitors, protease inhibitors, methyltransferase inhibitors) (Silvius 2002; Winter-Vann and Casey 2005; Konstantinopoulos et al. 2007; Berndt et al. 2011; Cox et al. 2015; Wang and Casey 2016).
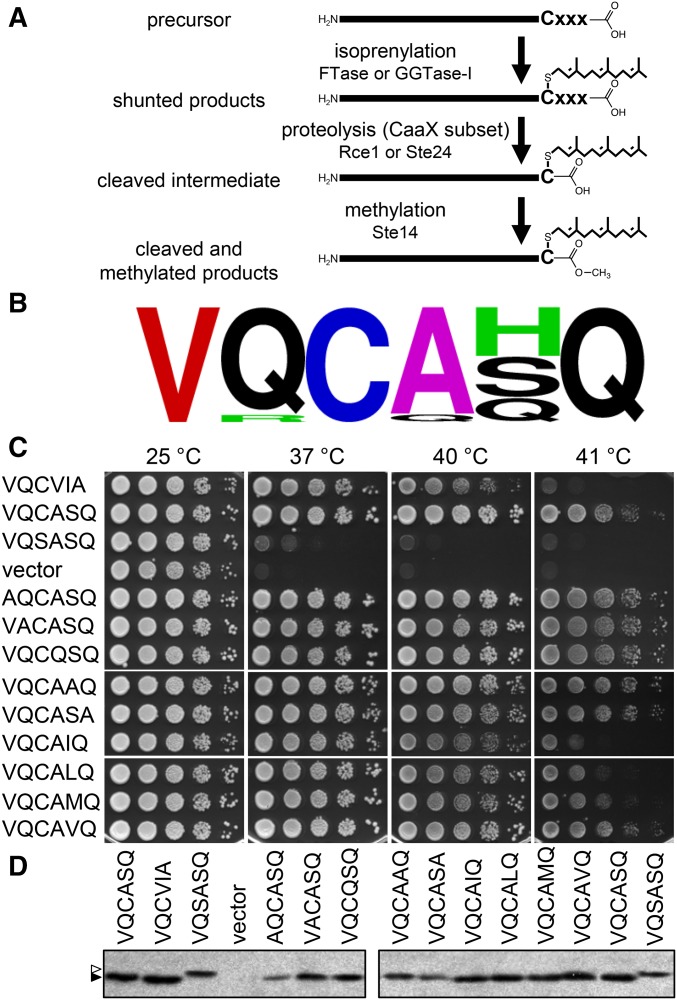
Figure 1.
Site-directed mutation of the Ydj1p Cxxx motif reveals flexibility in sequence requirements for functional levels of isoprenylation. (A) The Cxxx motif directs protein isoprenylation. Both farnesyl (C15) and geranylgeranyl (C20) can be added to Cxxx proteins; only C15 addition is shown for clarity. The isoprenylated species is either the endpoint modification (e.g., Ydj1p; shunted proteins) or an intermediate that is additionally modified by proteolysis and carboxymethylation (e.g., K-Ras4b; traditional CaaX proteins). Not shown are more extensive modifications that can also occur, such as palmitoylation (e.g., H- and N-Ras) or distal proteolysis (e.g., lamin A; yeast a-factor). (B) WebLogo frequency analysis of the last seven amino acids associated with 14 Ydj1p homologs retrieved from the Homologene Database (http://www.ncbi.nlm.nih.gov/homologene). Color scheme is as described in Materials and Materials and Methods. See Figure S1 for specific sequence details. (C) Ydj1p mutants were evaluated for their ability to support high-temperature yeast growth. yWS304 yeast (ydj1∆) expressing the indicated plasmid-encoded Ydj1p mutant were cultured in selective SC-uracil media and pinned as 10-fold serial dilutions onto nonselective YPD; the leftmost spot in each panel is undiluted. Plates were incubated at the indicated temperature as described in Materials and Methods. (D) The Ydj1p mutants indicated in C were expressed in yWS304 (ydj1∆) and cell lysates evaluated by anti-Ydj1p immunoblot. The specific plasmids used for Figure 1 are listed in Table S2. Unmodified Ydj1p (open triangle) migrates at a larger apparent kDa than prenylated Ydj1p (solid triangle).
The isoprenylation step of the canonical modification pathway has received the most investigative attention. The two isoprenoid transferases targeting CaaX sequences are the farnesyl transferase (FTase) and geranylgeranyl transferase (GGTase-I). Their specificities toward CaaX motifs clearly involve sequence determinants, but these are not yet fully resolved despite intensive investigations using in vivo, in vitro, and in silico methods. Historically, the determinants have been defined as a cysteine (required), followed by two aliphatic amino acids, and one of several amino acids at the last position. While CaaX sequences can be recognized by both FTase and GGTase-I, geranylgeranylation is reportedly enhanced for sequences ending Leu or Phe (Finegold et al. 1991; Moores et al. 1991; Yokoyama et al. 1995; Hartman et al. 2005; Maurer-Stroh and Eisenhaber 2005; Krzysiak et al. 2010). The aliphatic requirement at a1 and a2 positions of the CaaX motif should not be viewed as rigid, however, because prenylatable sequences clearly fall outside the traditional consensus. Examples include yeast Ydj1p (CASQ) and human Stk11/Lkb1 (CKQQ), among many others (Caplan et al. 1992b; Sapkota et al. 2001). Increasing evidence indicates that noncanonical sequences are modified by isoprenylation but are not cleaved and carboxymethylated (i.e., shunted products) (Hildebrandt et al. 2016).
Comparisons of known prenylated sequences and subsequent systematic amino acid substitution analysis of associated CaaX sequences initially contributed to the development of rules for prenyltransferase selectivity (Moores et al. 1991; Reiss et al. 1991; Yokoyama et al. 1991; Trueblood et al. 1993, 1997; Caplin et al. 1994; Omer and Gibbs 1994; Fu and Casey 1999; Roskoski 2003). This in vivo work, much of it originally performed using the yeast system, suggested enrichment of aliphatic residues at a1 and a2, more so at a2, and significant variation at X, leading to the canonical definition now widely accepted. A recent study utilizing a high-throughput yeast genetic approach and a Ras reporter revealed a similar bias for aliphatic residues at a1 and a2, again more so at a2 (Stein et al. 2015). Additional rules have derived from studies involving mammalian FTase mutants with altered selectivity (Hougland et al. 2012), computational modeling of substrates within the mammalian FTase active site (Reid et al. 2004; London et al. 2011), and reactivity of mammalian FTase against arrayed peptide sets (Hougland et al. 2009, 2010; Krzysiak et al. 2010; Wang et al. 2014). Collectively, these ex vivo approaches suggest that reactive sequences can significantly deviate from the historical CaaX definition. The rules proposed to govern FTase selectivity have been incorporated into predictive algorithms (Maurer-Stroh and Eisenhaber 2005; London et al. 2011). These algorithms often fail to predict, however, noncanonical motifs found on well-documented prenylated CaaX proteins (e.g., CKQQ present on human Stk11/Lkb1, human Nap1L1, and yeast Pex19p; CASQ present on yeast Ydj1p).
A striking difference between in vivo and ex vivo studies of FTase specificity is the strong enrichment of branched chain amino acids (BCAs) (i.e., Ile, Leu, Val) at the a2 position in many in vivo studies. We hypothesize that the results of in vivo studies are inherently biased because reporters used in those studies (e.g., a-factor; Ras and Ras-related GTPases) use the canonical prenylation pathway and are subject to multiple post-translational modifications (i.e., Ras-like modifications). In such situations, outputs are governed not only by isoprenylation efficiency but also proteolysis and carboxymethylation efficiencies. At least two in vivo approaches are being developed that minimize the effect of proteolysis and methylation on specificity studies. One involves the combined use of prenylation probes (e.g., alkyne farnesyl analogs) and mass spectrometry methods to identify prenylated proteins (i.e., the prenylome) (Kho et al. 2004; Onono et al. 2010; Suazo et al. 2016; Wang and Distefano 2016). Application of such technology has begun to confirm known and identify novel prenylated proteins, but direct detection of the prenyl group itself on these proteins remains a key challenge. Moreover, there may not be exact equivalency of chemical probes and farnesyl diphosphate when used by the FTase (Jennings et al. 2016). A second approach that we describe in this study involves the use of the yeast Hsp40 Ydj1p chaperone as a genetic reporter (Caplan et al. 1992b; Flom et al. 2008). An advantage of Ydj1p over previously used in vivo reporters is that it is an uncleaved CaaX protein (Hildebrandt et al. 2016). It is thus useful for identifying prenylatable sequences without concern for proteolysis and methylation, which are actually detrimental to Ydj1p activity (Hildebrandt et al. 2016). We report the use of Ydj1p-based screening to recover sequences that support prenylation of Ydj1p, which upon evaluation largely fail to match the operationally defined CaaX consensus, supporting a broader specificity than anticipated for the yeast CaaX-type prenyltransferases.
Materials and Methods
Yeast strains
Strains used in this study are listed in Supplemental Material, Table S1. Most have been previously described, several were isolated from a commercial MATa haploid genomic deletion library, and several were created for this study (Michaelis and Herskowitz 1988; Chen et al. 1997; Brachmann et al. 1998; Hildebrandt et al. 2016). Plasmids were introduced into strains via a lithium acetate-based transformation procedure (Elble 1992).
yWS2542 (MATa his3leu2met15ura3ydj1::NATRram1::KANR) was created by replacing the YDJ1 open reading frame with the nourseothricin resistance cassette (NATR) in yWS1632 (MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ram1::KANR). This was accomplished by transformation of the strain with an extensive digestion of pWS1623 (BamHI, HindIII, PvuII) and selection on YPD containing 100 µg/ml nourseothricin. yWS2544 (MATa his3leu2met15ura3ydj1::NATR) was made in similar fashion using BY4741 as the parent strain. The gene replacements were confirmed by PCR and Western blot against Ydj1p.
yWS304 (MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ydj1::KANR) and yWS1635 (MATa his3∆1 leu2∆0 met15∆0 ura3∆0 ydj1::KANRste14::KANR) were used for unbiased selection screens to identify Ydj1p-Cxxx mutants that could support thermotolerance. Unless otherwise noted, strains were routinely propagated at 30° or room temperature if temperature sensitive (23–25°) on either YPD or selective media as appropriate.
Plasmids
The plasmids used in this study, other than those recovered by screening, are listed in Table S2. Plasmids were either previously reported or constructed by standard molecular methods. All new plasmids created for this study were analyzed by restriction digest and DNA sequencing (Genewiz, South Plainfield, NJ or Eurofins Genomics, Louisville, KY) to verify proper sequence of the entire open reading frame (MFA1) or the 3′ end encoding the Cxxx sequence (YDJ1; ∼900 bp 5′ from stop codon). Plasmids encoding Ydj1p and a-factor with specific Cxxx sequences were derived by the same approach using pWS1132 (CEN URA3YDJ1-SASQ) and pWS610 (CEN LEU2MFA1), respectively, as the parent plasmids. Mutagenic oligonucleotides were designed to encode the desired Cxxx sequences and used to produce PCR products compatible with PCR-directed, plasmid-based recombination methods (Oldenburg et al. 1997) (Table S3). The PCR products have homology to the appropriate parent plasmid in regions flanking the intended mutation site. The parent plasmids were readied for recombination-based gap repair by digestion with NheI (pWS1132) or MluI and SphI (pWS610). Following cotransformation of digested plasmid and PCR product into yeast and appropriate selection (i.e., SC-uracil or SC-leucine), plasmids were recovered from individual yeast colonies, evaluated by restriction enzyme mapping, and sequenced to confirm the identity of recovered plasmids. pWS1623 was made in similar fashion using an NheI and BsaBI digestion of pWS1132 and a PCR product encoding NATR to replace the entirety of the YDJ1 ORF.
Plasmids encoding Ydj1p with random Cxxx sequences for the purpose of thermotolerance selection were also created by PCR-directed, plasmid-based recombination. In this case, the mutagenic oligonucleotide used for PCR was synthesized to contain random nucleotides for the xxx codons (oWS986). The COOH-terminal sequences associated with Ydj1p mutants recovered by the selection scheme are listed in Table S4.
Thermotolerance selection
Yeast deficient for YDJ1 (ydj1∆) were cultured to late log phase, harvested, and cotransformed with NheI-linearized pWS1132 and PCR product; control transformations with each DNA component alone were also prepared. For the cotransformed condition, multiple replicates were prepared. A portion of one transformation mix (10%) was plated onto SC-uracil and incubated at room temperature (23–25°). This allowed for an estimation of the number of recombinant plasmids created by the procedure. The remaining portion of the transformation mix (90%) and replicate transformation mixes (100%) were plated onto YPD and incubated at 40° for 48 hr followed by growth at room temperature (24–36 hr) to facilitate better visual identification of colonies. Surviving colonies were amplified as liquid cultures in SC-uracil, and cell pellets used for isolation of plasmids via sequential yeast and Escherichia coli DNA miniprep protocols. Several selection rounds were performed over the course of the study to accumulate the plasmids. In these experiments, both yWS304 and yWS1635 were used as the ydj1∆ background.
We used the above selection method to directly assess the effect of high temperature on transformation and recombination efficiencies by plating out equal portions of a single transformation mix in replicate onto SC-uracil (23–25°) and YPD (40° incubation). The transformation mix was prepared using yWS304, NheI-linearized pWS1132, and a PCR product designed to encode a thermotolerant Ydj1p mutant (AQCASQ). The fraction of colonies observed at 40° over 23–25° was calculated and applied as a correction factor to the initial estimate of colonies screened.
Temperature sensitivity assay and scoring of Ydj1p Cxxx variants
Thermotolerance assays were performed as previously described, with minor modifications (Hildebrandt et al. 2016; Blanden et al. 2018). In brief, plasmid-transformed strains expressing Ydj1p Cxxx variants were cultured to saturation (25°, 24–30 hr) in SC-uracil liquid media, serially diluted into H2O (10-fold dilutions), and replica-pinned in duplicate onto YPD solid media. For assays involving temperature and time optimizations, saturated cultures were diluted at fixed dilutions into YPD and spotted using a multichannel pipettor; the dilutions are specified in the associated figure legend. Strains expressing unmodified Ydj1p (SASQ), cleaved and carboxymethylated Ydj1p (CVIA), and shunted Ydj1p (CASQ) were typically included as controls, although the combination of controls pinned/spotted onto YPD plates varied between sets of mutants evaluated; shunted refers to CaaX motifs that are isoprenylated but not cleaved and carboxymethylated (Hildebrandt et al. 2016). Plates were typically incubated at various temperatures after an appropriate time of growth: 25° for 72 hr, 37° for 48 hr, 40° and 41° for 72 hr plus 24 hr at nonrestrictive temperature to allow better visualization of microcolonies; alterations to these temperature and time schedules are noted in appropriate figure legends.
On average, 4.25 replicates were evaluated for each strain expressing a Ydj1 mutant under the conditions described above; the range was three to eight replicates. Four independent observers scored the qualitative growth phenotype of replicates using a 1–5 score range in single-blind fashion. The scores were determined relative to reference strains expressing Ydj1p (SASQ), Ydj1p (CVIA), and Ydj1p (CASQ), which were standardly assigned scores of 1 (no growth at 40 and 41°), 3 (weak growth at 40 and 41°), and 5 (strong growth at 40 and 41°), respectively. Intermediate phenotypic scores of 2 and 4 were allowed for Ydj1p mutants. An average thermotolerance (T) score and standard deviation were calculated for each reference and mutant from the indicated number of replicates (n) reported with each T-score. Because reference strains were typically included in each experiment, the number of replicates for these is much higher than that of mutants examined.
Yeast lysate preparations for SDS-PAGE analysis and immunoblotting
Yeast were cultured to log phase (A600 0.75–1.0) in selective SC-uracil at 25° unless otherwise noted. Cell pellets of equal mass were harvested by centrifugation, washed with water, and processed by alkaline hydrolysis and trichloroacetic acid precipitation (Kim et al. 2005). Total cell precipitates were resuspended in urea-containing Sample Buffer (250 mM Tris, 6 M urea, 5% β-mercaptoethanol, 4% SDS, 0.01% bromophenol blue; pH 8), and analyzed by SDS-PAGE and immunoblotting. Blots were processed according to standard protocols using appropriate dilutions of rabbit anti-Ydj1p (courtesy of Dr. Avrom Caplan) and HRP-conjugated donkey or goat anti-rabbit antibodies (GE Healthcare, Little Chalfont, UK; Kindle Biosciences, Greenwich, CT). Antibody dilutions were prepared using TBST (10 mM Tris, 150 mM NaCl, 0.1% Tween-20; pH 7.5) containing 1% milk (w/v). Immune complexes on blots were detected using X-ray film after treatment with HyGLO development solution (Denville Scientific, South Plainfield, NJ) or using a KwikQuant Imager at multiple exposure times after treatment with the KwikQuant Western Blot Detection Kit (Kindle Biosciences).
Yeast mating assay
Qualitative and quantitative yeast mating assays were performed as previously described except that mating mixtures were pinned instead of spotted with a multichannel pipettor (Kim et al. 2005; Alper et al. 2006; Hildebrandt et al. 2016). In brief, MATa strains expressing various a-factor mutants were cultured to saturation SC-leucine liquid media; the MATα strain (IH1793) was cultured in parallel in YPD liquid media. Saturated cultures were normalized to an A600 value 1.0 ± 0.05 using appropriate fresh media, and normalized cell suspensions were mixed 1:9 (MATa:MATα) in individual wells of a 96-well plate. The mating mixtures were further subject to 10-fold serial dilution within the 96-well plate using the normalized MATα cell suspension as the diluent. For qualitative analysis, each diluted series was pinned in duplicate onto SC-lysine and synthetic dextrose minimal (SD) solid media. For quantitative analysis, equivalent volumes of empirically identified mixtures from the dilution series were spread onto SC-lysine and synthetic dextrose minimal plates in duplicate, such that individual colony density was projected to be 50–200 colonies per plate; the dilution mixture used varied between all samples. The SC-lysine cell count reports on the total number of mating competent cells (i.e., MATa haploid cells), while the synthetic dextrose minimal media cell count reports on the number of mating competent cells that underwent mating events (i.e., diploids). To cross-compare the values obtained, the colony counts were mathematically corrected for the dilution evaluated to estimate the number of colony forming units (CFUs) in each undiluted sample. The CFU values were used to determine mating efficiency (CFUSD/CFUSC-lysine) relative to a positive control expressing wild-type a-factor that was operational defined as having 100% mating efficiency (i.e., SM2331 transformed with pWS610; Tables S1 and S2). For both qualitative and quantitative analyses, plates were typically incubated 3 days at 30°.
Digital imaging of yeast plates and immunoblots
A Cannon flat-bed scanner was used to image plates and X-ray films (300 dpi, grayscale, .TIFF format). Plates were scanned face down without lids using a black background; films were scanned using a white background. Some immunoblot images were captured using a KwikQuant Imager system (.TIFF format). Digitized images of plates and immunoblots were imported into Photoshop for minor adjustments (i.e., image rotation, contrast, cropping, etc.), then copied to PowerPoint for final figure assembly. Contrast settings were adjusted within Photoshop to be identical for all plate images and to maximize dynamic range of signal; contrast settings for film-based immunoblot images were subject to Photoshop’s autocontrast function; the contrast settings for KwikQuant-based images were unaltered.
Amino acid frequency analysis
For our analyses, the entire set of Ydj1p-based sequences was always evaluated (n = 153). For the Ras-based sequences, the high-probability sequences (i.e., enrichment score >3) were culled to eliminate low-confidence sequences as suggested by the authors of the original study, which created a reduced set of high-probability sequences (n = 369) (Stein et al. 2015). For WebLogo-based analyses, appropriate groupings of Ydj1p- and Ras-based sequences were uploaded to the WebLogo server (http://weblogo.berkeley.edu/logo.cgi) and analyzed for amino acid frequency using a custom color scheme (Crooks et al. 2004). Cys was set to blue, polar charged amino acids were set to green (Asp, Arg, Glu, His, and Lys), polar uncharged residues were set to black (Asn, Gln, Ser, Thr, and Tyr), branched chain amino acids (BCAs) were set to red (Ile, Leu, and Val), and all other residues were set to purple (Ala, Gly, Met, Phe, Pro, and Trp). For bar graphs, the number of occurrences of a specific amino acid at each position of the Ydj1p-based sequences was normalized to the number of codons for that particular amino acid. Normalization was not applied to Ras-based sequences because this issue was addressed by the study design. High- and low-frequency amino acids for both sets of sequences were defined as those with normalized frequencies outside a 95% confidence level relative to the mean frequency for all amino acids at each position.
Prenylation predictions
For analyses using the Prenylation Prediction Suite algorithm (PrePS; http://mendel.imp.ac.at/PrePS), each Cxxx sequence was evaluated in the context of a 26 amino acid window representing the COOH terminus of a protein (Maurer-Stroh and Eisenhaber 2005). While PrePS requires a minimal length of 15 amino acids for analysis, we used 26 to be somewhat consistent with our previous amino acid frequency analysis of the COOH-terminal region of Ydj1p homologs where a window of 25 amino acids was evaluated (Hildebrandt et al. 2016). Ydj1p-derived sequences were evaluated in the context of Ydj1p (i.e., RASRGGANYDSDEEEQGGEGVQCxxx). Ras-derived sequences and the full set of 8000 Cxxx sequences were evaluated in the context of human H-Ras (i.e., RQHKLRKLNPPDESGPGCMSCKCxxx). The Cxxx sequences associated with the set of 89 yeast Cxxx proteins identified in the Saccharomyces Genome Database (SGD; https://www.yeastgenome.org) were evaluated in the context of the parental protein. Sequences were binned into groups based on their PrePS values for predicted probability of farnesylation: highly probable (scores >0), ambiguous (−2 to 0), or weakly predicted (< −2) (Maurer-Stroh and Eisenhaber 2005). For predictions using FlexPepBind scores, each Ydj1p- and Ras-based sequence was associated with its FlexPepBind score, and sequences binned into groups based on predicted probability of farnesylation: highly probable (scores < −1.1), ambiguous (−1.1 to −0.4), or weakly predicted (> −0.4) (London et al. 2011).
Our in-house prenylation and cleavage prediction rules were based on a simple point system involving assessment of the amino acid at each position of the Cxxx sequence. Prenylation potential was scored using a negative point scale where disfavored amino acids were counted. One negative point was assigned when the amino acid was low frequency in both Ydj1p- and Ras-based sets of sequences (e.g., Phe at x1); one extra negative point was assigned when it was absent in both (e.g., Lys at x2) (see Table 2). The potential range of scores with this method was 0 to −5. Sequences with scores of 0 were categorized as having a strong probability of prenylation, those with scores of −1 were categorized as having ambiguous potential, and those scoring −2 points or less were considered to have weak prenylation potential. Cleavage potential was scored using a mixture of positive and negative point scales where only the Ras-based data set was considered. One positive point was assigned when the amino acid was high frequency (e.g., Ala at x1). Negative points were assigned as for the prenylation rule (e.g., Phe at x1), also including the extra count for absent residues (e.g., Lys at x2). The potential range of scores with this method was 3 to −5. Sequences with scores of 1 or more were categorized as having a strong probability of cleavage, and all other sequences were categorized as having weak probability.
Table 2. High- and low-frequency amino acids in Ydj1p- and Ras-based data sets.
| Position | Reporter | Above CILa | Within CIL | Below CIL |
|---|---|---|---|---|
| x1 | Ydj1p | A T V D G S | N R E I K | F H Q W Y L M P; C (absent) |
| Ras | A T V I L | N R C M S P | F H Q W Y D E G K | |
| x2 | Ydj1p | V A G H N Q S Y | T I L M P | K R W (all absent); D E C F |
| Ras | V I L M | T C F | K R W G H N P S (all absent); D E A Q Y | |
| x3 | Ydj1p | Q S G H I N V | T A D M W | K P (all absent); E R Y C F L |
| Ras | Q S A C M | T F I L N V | K P R W (all absent); E Y D G H |
The number of instances that each amino acid was observed at a particular position within the population of sequences was determined. For Ydj1-based sequences (n = 153), the number of occurrences for each amino acid was normalized to adjust for codon bias (i.e., there are more Leu than Met codons). For Ras2-based sequences (n = 369), normalization was not needed due to study design. An SD and 95% confidence interval level (CIL) was determined for each population at each position that was used to determine amino acids above or below the interval. Amino acids that are in both the Ydj1- and Ras-based data sets for an indicated frequency group are bold.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7075025.
Results
Multiple Cxxx sequences can sustain Ydj1p-dependent thermotolerance
Farnesylation of Ydj1p is required for high-temperature growth of yeast (i.e., thermotolerance) and mitochondrial import (Atencio and Yaffe 1992; Caplan et al. 1992a, Caplan, 1992b). Replacing the normally uncleaved prenylation motif of Ydj1p (CASQ) with cleavable motifs (CTLM or CVIA) alters yeast thermotolerance, the ability to overexpress Ydj1p, and Ydj1p subcellular localization (Hildebrandt et al. 2016). These effects are correlated with the COOH-terminal cleavage state of the reporter rather than altered protein expression, stability, or farnesylation. It remains unclear why optimal Ydj1p thermotolerance function requires an uncleaved COOH-terminus. Nevertheless, Ydj1p serves as a unique reporter to investigate protein prenylation because it does not require subsequent proteolytic and methylation modifications associated with the canonical modification pathway (i.e., Ras-like modifications).
The last six residues of Ydj1p are conserved across species (Hildebrandt et al. 2016) (Figure 1B and Figure S1). The last four amino acids of the sequence form the CaaX motif that we operationally refer to as a Cxxx motif in this study. To initially determine whether any of the residues in this region, besides cysteine, contributed to the thermotolerance function of Ydj1p, substitution mutations within the conserved COOH-terminal region were created and thermotolerance profiles examined after plasmid-based reintroduction of the mutants into ydj1∆ yeast. For one set of mutants, alanine was substituted at every conserved position, except at x1 of the Cxxx sequence where glutamine was used to replace the naturally occurring alanine at that position. In a second set of mutants, the x2 position was varied with aliphatic amino acids isoleucine, leucine, methionine, and valine in an effort to make the sequence more canonical (i.e., aliphatic residues at both x1 and x2). The substitution mutants all supported thermotolerance behavior at 40°, although subtle growth pattern differences were observed at 41° (Figure 1C). Importantly, the mutants were all more thermotolerant at higher temperatures than nonfarnesylated Ydj1p (i.e., SASQ) or Ydj1p that was fully modified in a manner typically associated with traditional CaaX proteins (i.e., CVIA). For the latter, colonies were typically smaller at higher temperatures and colony growth was less robust and reproducible at 41° between experiments.
With the exception of the nonfarnesylated Ydj1p mutant (i.e., SASQ), Ydj1p substitution mutants appeared fully isoprenylated as judged by a gel-shift assay (Figure 1D). In this gel-shift assay, farnesylated Ydj1p migrates faster than nonprenylated Ydj1p generated through either mutation of the Cxxx motif (i.e., SASQ) or expression of Ydj1p in a farnesylation-defective yeast background (i.e., ram1∆). These results were interpreted to indicate that multiple Cxxx motifs can promote Ydj1p farnesylation and support Ydj1p-dependent thermotolerance. It remains unclear why Ydj1p family members have a conserved COOH-terminal sequence, but we speculate that these residues provide optimal functionality for at least one of the many roles attributed to Ydj1p (e.g., protein translocation, protein folding, prion clearance, etc.) (Caplan et al. 1992a; Lu and Cyr 1998a,b; Flom et al. 2008; Summers et al. 2009).
Unbiased identification of sequences that support Ydj1p-based thermotolerance
The ability of Ydj1p to support thermotolerance and be isoprenylated in the context of different Cxxx sequences suggested that many sequences might be able to promote this behavior. This lead us to hypothesize that the thermotolerance profile of Ydj1p could be used as a genetic reporter to identify the breadth of Cxxx sequences capable of supporting thermotolerance. Moreover, we predicted that carefully selecting thermotolerance conditions would allow for genetic enrichment of shunted Cxxx motifs (i.e., prenylated but not cleaved and carboxymethylated) over nonprenylated and fully modified sequences (e.g., SASQ and CVIA, respectively). We therefore designed a high-temperature selection strategy to enrich for shunted Ydj1p Cxxx mutants. At the highest temperatures applied, the selection strategy prevented growth of nonprenylated Ydj1p (i.e., SASQ) and forced slow growth of fully modified Ydj1p (i.e., CVIA and CTLM) relative to shunted Ydj1p (i.e., CASQ) (Figure 2, A and B).
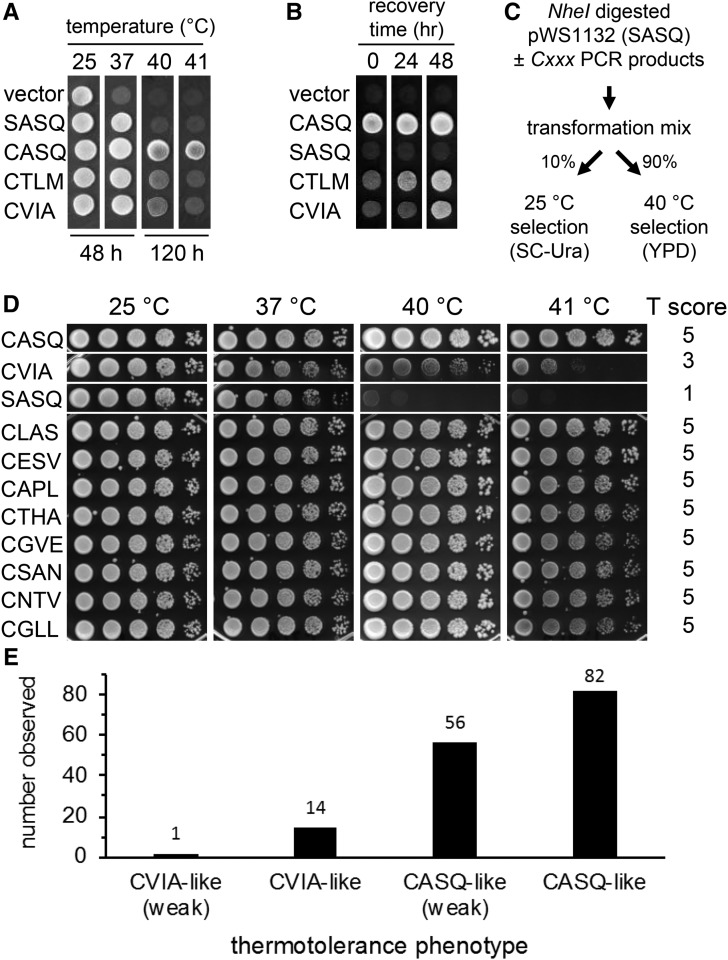
Figure 2.
Effect of temperature and time on yeast thermotolerance. (A) Yeast expressing the indicated Ydj1p variants were cultured to saturation in selective media, diluted into YPD, spotted using a multichannel pipettor onto YPD solid media, and plates incubated at indicated temperatures and times. A 1:20 dilution was the source for spots incubated at 25°, and a 1:2 dilution was used for spots incubated at other temperatures. (B) Yeast were cultured and processed as described for A, using the 40° condition with the following alterations: the 1:2 dilution was into YPD, incubation at 40° was for 72 hr followed by recovery at 25° for the indicated times. (C) Flow diagram of screen used to identify thermotolerant yeast expressing Ydj1p Cxxx variants in either the ydj1∆ or ydj1∆ ste14∆ background. (D) Examples of thermotolerance profiles observed and thermotolerance (T) scores assigned when expressed in ydj1∆ background; the controls and panel of mutants are replicated in Figure S2. Yeast thermotolerance assays were performed as described for Figure 1. (E) The thermotolerance profiles of all recovered mutants were scored relative to yeast expressing Ydj1p with a SASQ, CVIA, or CASQ motif, where the controls were assigned scores of 1, 3, and 5, respectively. The mutants were then binned according to their scores; see Materials and Methods for details on binning.
We next devised a strategy to create and evaluate thermotolerance of a library of Ydj1p Cxxx mutants in vivo. In brief, ydj1∆ yeast was cotransformed with a linearized yeast expression vector encoding nonprenylated Ydj1p (i.e., SASQ) and a library of PCR products encoding Cxxx sequences. This resulted in recombination events between the DNA fragments, leading to the formation of a library of Ydj1p mutants having the potential to encode all 8000 possible COOH-terminal tetrapeptide combinations of the form Cxxx, where C is Cys and “x” is any amino acid (Figure 2C) (Oldenburg et al. 1997). Importantly, this strategy allowed for immediate selection of colonies with thermotolerant properties. We initially estimated that we evaluated ∼480,000 recombination events, which was enough to achieve near complete coverage of all Cxxx permutations (estimated 99.9% completeness; ∼1% probability that all Cxxx sequences were sampled) (Firth and Patrick 2008). This value was revised to ∼67,200 recombination events (93.5% completeness; ∼0% probability of full coverage) when it was determined that the number of colonies capable of forming at higher temperature was ∼14% that observed at room temperature, which we infer is due to reductions in transformation and/or recombination efficiency.
Our selection strategy yielded 172 thermotolerant colonies from which plasmids were recovered and subject to DNA sequencing. One of the plasmids encoded wild-type Ydj1p (i.e., CASQ) thus validating the design of the screen. A set of 153 plasmids encoding unique Ydj1p Cxxx sequences, inclusive of the wild-type sequence, was defined by eliminating a small number of sequences recovered multiple times. The unique set of plasmids was reintroduced into ydj1∆ yeast for more detailed analyses of thermotolerance (Figure 2D and Figure S2). This analysis yielded a T-score for each Ydj1p Cxxx variant, where higher scores were associated with better thermotolerant behavior (see Materials and Methods for details on the scoring rubric). We binned the Cxxx variants based on their T-scores (Figure 2E and Table 1). The CASQ-like group formed the largest cohort and displayed phenotypic growth similar to that of wild-type Ydj1p (CASQ) (54% of hits; T-score 4.5–5.0). These Cxxx variants supported strong thermotolerance (i.e., growth). The CASQ-like (weak) group was next largest (37%; T-score 3.5–4.5.). These Cxxx variants were phenotypically less thermotolerant than wild-type Ydj1p but more tolerant than cleaved Ydj1p (i.e., CVIA). The CVIA-like group accounted for a small percentage of total hits (9%; T-score 2.5–3.5). These Cxxx variants behaved much like cleaved Ydj1p (i.e., CVIA). The CVIA-like (weak) group was formed by a single Cxxx variant (0.6%; T-score 2.47). Overall, our scoring analysis indicated that 90% of Cxxx variants were categorized as having thermotolerance profiles better than that of cleaved Ydj1p (i.e., CVIA).
Table 1. Summary of thermotolerance phenotypes observed and associated prenylation prediction scores.
| Categorya | Number observed | Average T-scoreb | Average FPB score | Average RRS E-score | Average PrePS score |
|---|---|---|---|---|---|
| All unique sequences | 153 | 4.36 ± 0.57 | −0.18 ± 1.41 | 0.62 ± 2.26 | −3.31 ± 2.19 |
| CASQ-like | 82 | 4.76 ± 0.15 | −0.26 ± 1.20 | 0.31 ± 1.62 | −3.55 ± 1.99 |
| CASQ-like (weak) | 56 | 4.15 ± 0.22 | 0.20 ± 1.58 | 0.24 ± 0.79 | −3.47 ± 1.97 |
| CVIA-like | 14 | 3.04 ± 0.24 | −1.18 ± 1.35 | 3.24 ± 5.02 | −1.61 ± 5.02 |
| CVIA-like (weak)c | 1 | 2.47 | −1.02 | 9.98 | 1.35 |
| CASQ-like (x2 = ILV)c | 10 | 4.66 ± 0.12 | −1.87 ± 0.50 | 1.63 ± 4.60 | −1.93 ± 2.45 |
| Ydj1p/RRS overlapc | 8 | 3.30 ± 0.66 | −1.83 ± 0.81 | 9.37 ± 4.23 | 0.94 ± 0.54 |
Specific sequences and groupings are listed in Table S4.
Thermotolerance (T) score averages were calculated using data from Figure S2. FlexPepBind (FPB) scores were derived from London et al. (2011); a score of −1.1 or less is predicted to have a high probability of prenylation. RRS Enrichment (E) scores were derived from Stein et al. (2015); a score of three or more is predicted to have a high probability of prenylation. PrePS scores were retrieved from the PrePS server; a score above 0 is predicted to have a high probability of prenylation.
The scores reported for the single CVIA-like (weak) sequence are for that sequence alone. Scores for CASQ-like (x2 = ILV) are for the subset of CASQ-like sequences with BCAs at x2. Scores for Ydj1p/RRS overlap are for those sequences recovered in both the Ydj1p and RRS screens.
Weaker thermotolerance phenotypes are associated with prenylation defects and enhanced cleavage propensity
To investigate the extent of prenylation associated with sequences conferring the strongest thermotolerance phenotype, we randomly identified 10% of the Ydj1p Cxxx variants in each of the CASQ-like and CASQ-like (weak) phenotypic categories and assessed prenylation by gel-shift assay (Figure 3A). An online algorithm (Research Randomizer; https://www.randomizer.org/) was used to randomly identify the 10% subsets (eight and six sequences per category, respectively) (Urbaniak and Plous 2013). Most of these sequences (9 out of 14 evaluated) presented as a single band with the same mobility as farnesylated Ydj1p. Several sequences presented as doublet bands, indicative of incomplete prenylation. The prenylated species (i.e., lower band) appeared qualitatively stronger than that of the non-prenylated species (i.e., upper band) in most instances, with the exception of CDFI where the bands were qualitatively assessed to be about equal intensity. Thus, partial prenylation of 50% or greater appears to be sufficient to support Ydj1p-dependent thermotolerance. We also performed gel-shift studies on the 15 sequences conferring the weakest thermotolerance phenotypes: CVIA-like and CVIA-like (weak) (Figure 3B). The CWGG mutant was the only mutant in this set to present with a doublet pattern, with the nonprenylated species being the major band. Thus, incomplete prenylation most likely explains the weak thermotolerance profile of this sequence. Considering all our data, however, it appears that the completeness of isoprenylation cannot account for the range of thermotolerance behaviors observed for mutant sequences.
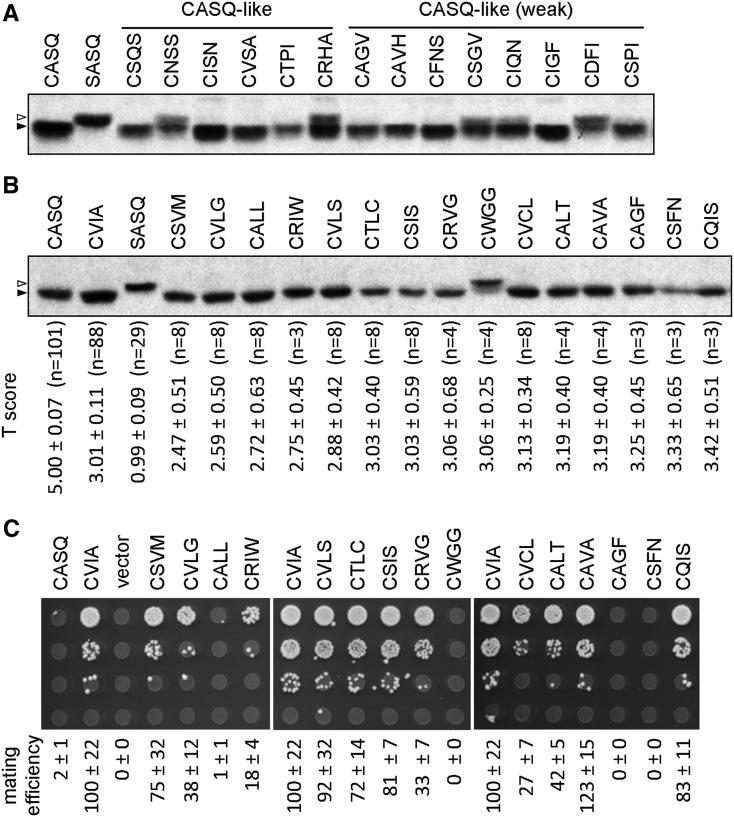
Figure 3.
Isoprenylation and cleavage properties of Ydj1p Cxxx mutants identified through thermotolerance screening. (A and B) Yeast expressing the indicated Ydj1p mutants were evaluated by a gel-shift assay as described in Figure 1C. The sequences evaluated were either randomly identified from (A) the CASQ-like and CASQ-like (weak) groups or (B) reflect the combined set of sequences presenting with CVIA-like and CVIA-like (weak) thermotolerance phenotypes. Reference controls included on the blots are: farnesylated and uncleaved Ydj1p (CASQ); farnesylated, cleaved, and carboxymethylated Ydj1p (CVIA); unmodified Ydj1p (SASQ). Unmodified Ydj1p (open triangle) migrates at a larger apparent kDa than prenylated Ydj1p (closed triangle). The values in B reflect the thermotolerance scores observed for the corresponding mutant. (C) Yeast expressing the indicated a-factor Cxxx variants in a mfa1∆ mfa2∆ background were assessed for mating competence using a serial dilution mating assay (panel) and quantitative mating assay (values). Values represent mating efficiency relative to wild-type CVIA, which was set to 100%; values were determined using four or more replicates from two or more individual experiments. The panel is representative of one of the replicates.
We determined that the majority of the 15 CVIA-like and CVIA-like (weak) mutants were fully prenylated because they presented as a single band with mobility similar to prenylated Ydj1p. We thus hypothesized that the reduced thermotolerance associated with these mutants was likely due to the presence of cleavable sequences (i.e., Ras-like). To assess cleavage status, we first evaluated the thermotolerant behavior of ydj1∆ ste14∆ yeast expressing the 15 Ydj1p Cxxx variants. This genetic background lacks the isoprenylcysteine carboxymethyl transferase and improves the thermotolerance profiles of yeast expressing cleavable Ydj1p Cxxx variants (Hildebrandt et al. 2016). Indeed, thermotolerance profiles improved for all but Ydj1p CWGG (Figure S3). To examine cleavage status more directly, we next evaluated the sequences in the context of the yeast a-factor mating pheromone (Figure 3C). The biological activity of a-factor in vivo requires it to be farnesylated, cleaved, and carboxymethylated (He et al. 1991; Hrycyna et al. 1991; Boyartchuk et al. 1997; Chen et al. 1997). Defects in any one of these steps reduce bioactivity as measured through a pheromone-based biological mating assay. Through both qualitative and quantitative mating assays, we observed that the bioactivities of the a-factor Cxxx mutants could be categorized into two groups. Category I mutants formed the largest group and had substantial bioactivity relative to wild-type a-factor (i.e., 10% or greater). This observation indicates that these sequences are susceptible to cleavage. We suspect that the varied bioactivities of category I mutants reflect differences in their cleavage efficiencies or perhaps alternative geranylgeranylation of these sequences. Fourfold reduced bioactivity has been reported for synthetic geranylgeranylated a-factor, which can also be produced in vivo in the context of the CVIL sequence (Caldwell et al. 1994). Category II mutants had very limited or no bioactivity (<1%) and were represented by CALL, CWGG, CAGF, and CSFN. Whereas the absence of bioactivity associated with CWGG is most likely due to a prenylation defect, this does not readily explain the loss of bioactivity observed for the other category II sequences. These sequences appear to be fully prenylated and cleaved in the context of Ydj1p, thus we speculate that they may have a prenylation or cleavage defect in the context of a-factor. Such defects would negatively affect multiple downstream steps required for pheromone bioactivity that depend on efficient initial farnesylation and cleavage (e.g., Ste14p-mediated carboxymethylation; Ste6p-dependent export; Ste3p receptor interaction).
Ydj1p-based thermotolerant behavior may involve alternative isoprenylation
Yeast thermotolerance depends on the farnesylation of Ydj1p. It is unclear whether geranylgeranylation can substitute in this capacity. We were especially interested in sequences ending in Leu or Phe, which reportedly confer GGTase-I reactivity (Finegold et al. 1991; Moores et al. 1991; Yokoyama et al. 1995; Hartman et al. 2005; Maurer-Stroh and Eisenhaber 2005; Krzysiak et al. 2010; Gangopadhyay et al. 2014). To address this potential, we investigated whether the 10 Ydj1 CxxL and CxxF variants recovered through screening could be geranylgeranylated, and promote thermotolerance in the absence of FTase activity (Figure 4A).
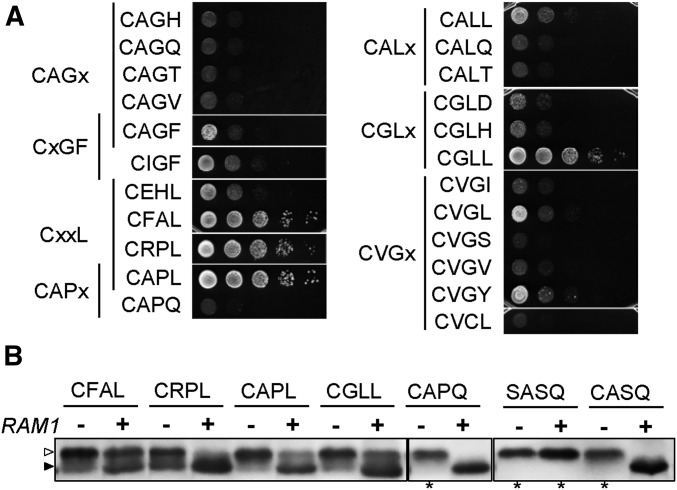
Figure 4.
Investigations of the geranylgeranylation potential of Ydj1p. (A) The indicated Ydj1p Cxxx variants were assessed for their ability to restore thermotolerance to a ydj1∆ ram1∆ (yWS2542) background as described for Figure 1, except that the 40° incubation was ∼80 hr without recovery. (B) Lysates for gel-shift assays were prepared from mid-log cultures incubated at either 40° or 25° (the latter are marked with an asterisk). The strain backgrounds used were ydj1∆ RAM1 (+; yWS2544) and ydj1∆ ram1∆ (−; yWS2542). Of note, the ram1∆ cultures were slow growing relative to RAM1 cultures at 40°, taking 36 or more hours instead of 18–24 hr to achieve the same cell density. Unmodified Ydj1p (open triangle) migrates at a larger apparent kDa than prenylated Ydj1p (closed triangle).
The two CxxF sequences, CAGF and CIGF, both displayed weak thermotolerance. We also analyzed several sequences similar to CAGF that were recovered by screening (i.e., CAGx), and these did not demonstrate thermotolerance. More substantial thermotolerance was observed for CxxL sequences, including CRPL and CAPL that only differ at the x1 position. By contrast to CAPL, the very similar CAPQ sequence recovered by screening did not support thermotolerance. The remaining CxxL sequences were part of subsets CALx, CGLx, and CVGx. Among these subsets, those with Leu or Tyr at x3 generally displayed minor thermotolerance. Leu at the x3 position, however, was not always a positive predictor of thermotolerance in the absence of FTase activity (e.g., CVCL). Collectively, these results indicate that Ydj1p Cxx(L/F/Y) sequences, where Leu is potentially more favorably than Phe or Tyr, are susceptible to prenylation in the absence of FTase, consistent with previous reports of GGTase-I selectivity being influenced by the x3 position (Hartman et al. 2005). But, it is not clear under our screening conditions whether geranylgeranylation is competing with farnesylation for these sequences or only occurs when farnesylation is genetically disrupted (i.e., ram1∆ background-specific). Thus, we cannot exclude the possibility that some sequences recovered by thermotolerance screening are indeed geranylgeranylated.
We also examined the properties of potentially geranylgeranylated Ydj1p Cxxx variants by gel-shift assay using lysates prepared from yeast cultured at elevated temperature (Figure 4B). GGTase-I activity (Cdc43p/Ram2p) is essential, so we could not express variants in a strain background lacking this activity. Instead, we expressed variants in the FTase-defective background (i.e., ram1∆) and reasoned that any shifted bands still present in this background were a consequence of GGTase-I activity. We focused on the subset of four Ydj1p Cxxx variants that supported robust growth of the ydj1∆ ram1∆ strain on solid media because yeast expressing other Ydj1p Cxxx variants did not grow well at elevated temperature. The four Ydj1p Cxxx variants were also evaluated in a background with normal FTase activity (i.e., RAM1). In the presence of FTase, a shifted protein band indicative of prenylated Ydj1p was present in each case. This species was estimated to be at least 50% or more of the total Ydj1p present in each of the lysates. In the absence of FTase, the shifted protein band was present but represented a reduced amount of the total Ydj1p in the sample. We interpret these observations to indicate that low levels of prenylation can occur to Ydj1p Cxxx variants in the absence of FTase. We also evaluated several other Ydj1p Cxxx variants, including CASQ, SASQ and CAPQ. Yeast expressing these variants were cultured at room temperature when expressed in the ram1∆ background because of their inability to grow at elevated temperature. None of these variants displayed a shifted band in the absence of FTase.
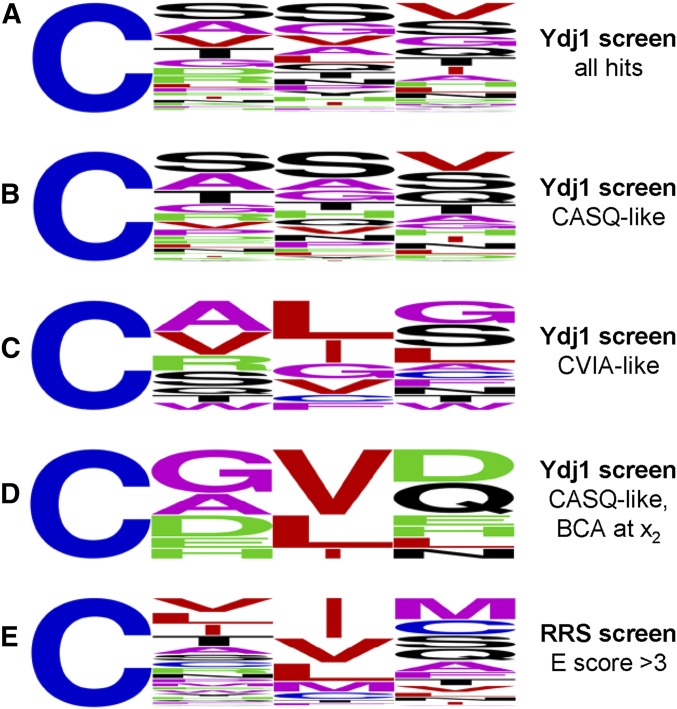
Frequency analyses of recovered sequences
The Ydj1p-based selection strategy recovered numerous sequence combinations. WebLogo frequency analysis of all the identified sequences revealed no obvious enrichment of any particular amino acid at the x1, x2, or x3 position (Figure 5A). Analysis of sequences grouped by thermotolerance scores also failed to reveal an enrichment pattern for the variants that performed phenotypically most similarly to shunted Ydj1p (i.e., CASQ-like) (Figure 5B). While the small subset of variants that performed similarly to cleaved Ydj1p (i.e., CVIA-like) also lacked obvious enrichment of any type of amino acid at x1 or x3, they were enriched for BCAs at x2 (Figure 5C). The presence of a BCAs at x2 alone, however, was not a good predictor of weaker thermotolerance. Frequency analysis of CASQ-like sequences with x2 BCAs revealed that charged residues (i.e., Arg, Asp, Glu, His, or Lys) were present at x1 or x3 in 7 out of 10 instances (Figure 5D); the outliers were CAVQ, CGLL, and CGVQ. Similar analysis of CASQ-like (weak) sequences revealed charged residues at x1 or x3 in 10 out of 11 instances; the outlier was CAVG.
Figure 5.
Frequency analysis of sequences identified by thermotolerance screening. Sequences were categorized by their associated thermotolerance scores (see Figure S2). For each grouping, a WebLogo analysis was performed (Crooks et al. 2004). Groupings were (A) all identified sequences (n = 153), (B) those most like CASQ (i.e., CASQ-like; T-score range 4.5–5; n = 82), (C) those most like CVIA (i.e., CVIA-like; T-score range 2.50–3.49; n = 14), (D) the subset of sequences from B that had a branched chain amino acid at x2 (n = 10), and (E) sequences identified by a Ras-based strategy as having high likelihood of prenylation (enrichment score >3; n = 369) (Stein et al. 2015).
We also conducted frequency analysis of sequences predicted to be prenylated in yeast that were obtained using a Ras-based in vivo reporter (Ras Recruitment System screen; RRS screen) (Stein et al. 2015). The Ras-based strategy used an enrichment scoring system to identify sequences having a high probability of prenylation (n = 496). For our analyses, we culled the high-probability sequences to eliminate low-confidence sequences as suggested by the authors, which created a reduced set of sequences (n = 369). This reduced set was enriched for BCAs at x2, while a wide range of amino acids were present x1 and x3 (Figure 5E). Interestingly, most of the sequences identified through Ydj1p-based screening were scored as low-probability sequences in the Ras-based screen (Figure S4A). Among the overlapping set of sequences identified in both screens, six of the eight sequences presented phenotypically as CVIA-like or CVIA-like (weak) in the Ydj1p-based thermotolerance test; the two outliers were CAVQ (CASQ-like) and CVTS [CASQ-like (weak)].
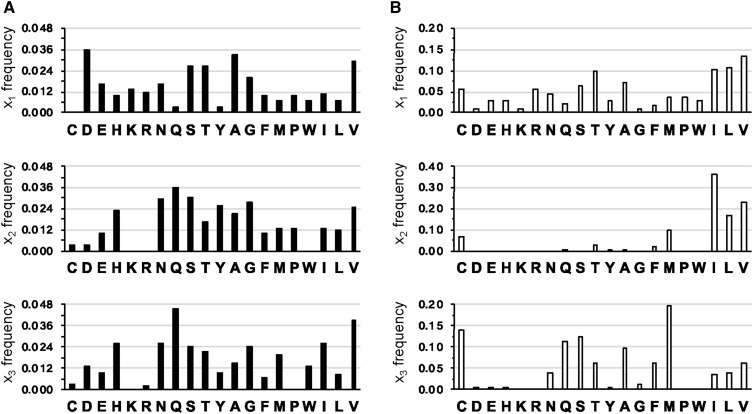
Initial evaluation of the Ydj1p- and Ras-based sequences revealed that some amino acids were absent or at low frequency in one, the other, or both sets. Some amino acids were also very common. For example, Ser was observed over 20 times at each position in Ydj1p-based sequences. This over-representation likely reflects codon bias in the degenerate oligo used to create Cxxx permutations (i.e., Ser is encoded by six codons). To better understand the frequency occurrence of each amino acid in the Ydj1p set without codon bias, we normalized the occurrence of each residue based on the number of potential codons for that amino acid (Figure 6A). We built high- and low-frequency groups by identifying amino acids with normalized frequencies that were outside a 95% confidence interval level relative to the average frequency of all amino acids (Table 2). We built the same groups for the reduced set of Ras-based sequences, but normalization was not needed due the nature of the experimental strategy used to identify those sequences (Figure 6B and Table 2) (Stein et al. 2015). Few amino acids were consistently present at high frequency independent of reporter: x1 (Ala, Thr, Val), x2 (Val), and x3 (Gln and Ser); these results are similar to those observed in vitro with mammalian FTase (Hartman et al. 2005; Hougland et al. 2009, 2010). We view these amino acids as extremely favorable for farnesylation. There were additional high-frequency amino acids identifiable in the context of one or the other reporter, indicating that reporter-specific effects need to be considered when evaluating the data sets. Considering the combined set of high-frequency amino acids, eight were strongly favored at x1, 11 at x2, and 10 at x3. We interpret this observation to indicate that the yeast FTase can tolerate many different amino acids at these individual positions, similar to the reported behavior of mammalian FTase in vitro. Of note, charged amino acids were generally excluded as high-frequency amino acids from both data sets, with the exception of Asp (x1) and His (x2 and x3) that were highly enriched in Ydj1p sequences. By contrast to high-frequency amino acids, more amino acids were consistently present at low frequency independent of reporter: x1 (His, Gln, Phe, Trp, Tyr), x2 (Arg, Asp, Glu, Lys, Trp), and x3 (Arg, Glu, Lys, Pro, Tyr). We view these low-frequency amino acids as incompatible with efficient farnesylation when present at the indicated positions, although their incompatibility may be context specific (i.e., dependent on residues at other positions) (Hougland et al. 2009, 2010). Considering the combined set of low-frequency amino acids, 13 were disfavored at x1, 13 at x2, and 12 at x3. Some of the disfavored amino acids from the RRS set may be incompatible with proteolysis rather than farnesylation. Thus, the low-frequency amino acids common to both the Ydj1p and RRS data sets are likely the extent of disfavored amino acids: x1 (His),; x2 (Arg, Asp, Glu, Lys), and x3 (Arg, Glu, Lys).
Figure 6.
Amino acid frequency in hits recovered in Ydj1p- and Ras-based screens. The frequency occurrences of each amino acid at the x1, x2 and x3 positions were calculated for the set of (A) Ydj1p and (B) Ras-based sequences. The complete set of Ydj1p-based sequences (n = 153) and the reduced-size set of Ras-based sequences (n = 369) were used for the analysis. Frequency values for Ydj1p-based sequences were normalized for codon bias (e.g., Leu codons are over-represented 6× relative to the Met codon); Ras-based sequences did not need normalization due to study design. Amino acids are clustered as reported for WebLogo analyses. Note that y-axis scales differ for the three panels in B.
Overall, our frequency analyses established that membership in a frequency group was clearly influenced by the reporter utilized in the screen. Of additional note, the Ydj1p-based selection recovered CASQ as an independent isolate whereas the RRS screen did not identify this sequence as significantly enriched. Collectively, our observations indicate that sequences recovered by Ydj1p- and Ras-based approaches yield largely nonoverlapping sets of sequences, which we hypothesize reflect shunted and cleaved sequences, respectively. Additively, the two sets of sequences likely represent a comprehensive spectrum of prenylatable sequences, although it is possible that additional Ydj1p-based sequences have gone unidentified. For example, we did not recover the Pex19p CKQQ motif, whose prenylation is well documented (Götte et al. 1998). This sequence is associated with CASQ-like behavior in the context of Ydj1p (ER Hildebrandt and WK Schmidt, unpublished results).
Evaluation of Ydj1p-based sequences using prenylation prediction algorithms
We evaluated our Ydj1p-based hits in the context of several prenylation prediction algorithms, as it was unclear how efficiently these noncanonical sequences would be scored by methods trained using learning sets based on different measures of prenylation reactivity. These included recently developed FlexPepBind, which takes into account structure-based constraints derived from the conserved features in solved FTase structures, the web-hosted PrePS algorithm, and an in-house algorithm that used disfavored amino acids identified by our study as the guide to predict likelihood of farnesylation (Figure S4, B and C and Table 3) (Maurer-Stroh and Eisenhaber 2005; London et al. 2011). It should be noted that FlexPepBind and PrePS were optimized for mammalian FTase, potentially limiting their utility for predicting modification of yeast Cxxx sequences.
Table 3. Results of isoprenylation prediction algorithms.
| Isoprenylation prediction (%) | ||||
|---|---|---|---|---|
| Test set | Method | High | Ambiguous | Low |
| Ydj1-based (n = 153) | FlexPepBind | 27a | 20 | 54 |
| PrePS | 7 | 25 | 68 | |
| in-house | 86 | 14 | 0 | |
| Ras-based (n = 369) | FlexPepBind | 60 | 15 | 25 |
| PrePS | 56 | 32 | 12 | |
| in-house | 86 | 14 | 0 | |
| Cxxx (n = 8000) | FlexPepBind | 17 | 12 | 71 |
| PrePS | 5 | 14 | 80 | |
| in-house | 42 | 28 | 30 | |
| SGD (n = 89)b | FlexPepBind | 28 | 27 | 46 |
| PrePS | 26 | 8 | 66 | |
| in-house | 61 | 16 | 24 | |
The summed value of the three categories for some sets exceeds 100% due to rounding errors.
This test set was identified using appropriate sequence patterns and the Pattern Match search function associated with the Saccharomyces Genome Database (SGD). The set was culled of sequences annotated as dubious open reading frames.
Relatively few Ydj1p-based sequences were identified as having a high probability of prenylation by FlexPepBind and PrePS (27 and 7%, respectively), with the majority of sequences judged to have a low probability (54 and 68%, respectively). By contrast, a majority of sequences were judged to have a high probability of prenylation by in-house rules (86%), and none were judged to have a low probability. Considering the Ras-based-sequences, in-house rules also identified more high probability sequences (86%) than FlexPepBind and PrePS (60 and 56%, respectively). We also evaluated the full set of 8000 Cxxx combinations using our in-house rules and the FlexPepBind and PrePS algorithms. In-house rules identified many high probability sequences (42%) whereas fewer were identified by FlexPepBind and PrePS (17, and 5%, respectively). A similar pattern was observed for a set of Cxxx proteins identified in the SGD. The initially identified set of sequences (n = 108) was culled to a smaller set (n = 89) by eliminating those annotated as dubious open reading frames. In-house rules identified a majority of sequences as having a high probability of prenylation (61%), whereas FlexPepBind and PrePS identified fewer (28 and 26%, respectively) (Table S5 and Table 3).
Discussion
There are a limited number of genetic reporters suitable for investigations of prenyltransferase specificity. Past studies in yeast have relied on the Ras GTPase and a-factor mating pheromone, which both require full COOH-terminal modification for their optimal activities. By contrast, Ydj1p only requires isoprenylation for its optimal activity, and lack thereof or more extensive Ras-like modification of its COOH-terminus are actually detrimental to its activity in promoting yeast thermotolerance. The specific reason that shunted Ydj1p is required for optimal thermotolerance remains undefined. We hypothesize that Ydj1p uses its farnesylated COOH-terminal region for physical interactions with key client proteins required for an effective heat stress response. This is akin to the chaperone Pex19p where its farnesylation stabilizes conformations that promote association with cargo proteins (Emmanouilidis et al. 2017). Full modification of Ydj1p would thus be expected to alter the biophysical properties of its COOH-terminus, potentially disrupting client interactions. A free charged COOH terminus may be needed for client interactions, which can be provided by any sequence that can be prenylated but resists cleavage (i.e., shunted). Alternatively, it may be that the canonical multistep modification of Ydj1p enhances its hydrophobicity and promotes increased association with membranes, limiting its accessibility to cytosolic client proteins. Regardless of the reason that Ydj1p-based thermotolerance requires shunting, we were able to successfully take advantage of this phenotype to develop Ydj1p as a novel reporter for sampling the prenylation potential of sequence space. We did this to specifically test our prediction that the multistep complexity of post-translational modification that occurs in vivo to Ras and a-factor may limit interpretation of prenyltransferase specificity.
Through a Ydj1p-based genetic thermotolerance selection strategy, we recovered many sequences that did not conform to the CaaX motif typically associated with prenylproteins. Interestingly, we observed that the sequences recovered using Ydj1p were mostly nonoverlapping with those obtained using Ras-based methods, suggesting that the combined set of sequences represents a more comprehensive set of prenylatable motifs that is larger than previously appreciated. The combined set suggests that a large combination of amino acids is tolerated by the prenyltransferases across all positions of the motif. Because early investigations into the specificity of the prenyltransferases were often limited to a small subset of prenylproteins (e.g., Ras and Ras-related GTPases), this led to the use of the traditional CaaX consensus often described in the literature (Trueblood et al. 1993, 1997; Omer and Gibbs 1994; Roskoski 2003), although “Cxxx” is a more accurate description of the consensus in light of our findings and others (Stephenson and Clarke 1990; Kinsella et al. 1991; Hrycyna and Clarke 1992).
With respect to yeast prenyltransferase specificity, frequency analysis of both Ydj1p and Ras data sets suggests strongly preferred and disfavored amino acids within the Cxxx motif. At x1, eight amino acids were frequently observed. These included expected aliphatic amino acids (Ala, Gly, Ile, Leu, Val), small polar uncharged residues (Ser, Thr), and an unexpected polar charged residue (Asp). At x2, 11 amino acids were frequently observed. These included expected aliphatic amino acids (Ala, Gly, Ile, Leu, Met, Val), unexpected polar uncharged residues (Asn, Gln, Ser, Tyr), and a polar/weakly charged residue (His; the charged state of His depends on local environment; only 5–10% of free His is charged at physiological pH). At x3, 10 amino acids were frequently observed. These included aliphatic amino acids (Ala, Gly, Ile, Met, Val), polar uncharged residues (Asn, Cys, Gln, Ser), and a polar/weakly charged residue (His). The above reflects amino acids most highly enriched relative to other amino acids (i.e., above a 95% confidence interval), so the number of amino acids tolerated at each position is actually greater when considering amino acids with average to above average frequency occurrences. We expect that future studies involving modeling or cocrystallization of atypical peptide sequences within the prenyltransferase active sites will help establish how these enzymes can accommodate such a variety of side chains at each position. We also expect that multivariate analysis will lead to a better understanding of the substrate features being recognized, but such studies will likely require more Ydj1p-based sequences than we have presently collected.
Perhaps more telling for yeast prenyltransferase specificities are the residues that were infrequently recovered in the combined data sets, and in particular those that were reporter-independent. While each position of the Cxxx motif seems to have a disfavored set of amino acids, poor prenylation outcomes were common when the x1 position had a bulky amino acid, and x2 and x3 positions had either charged or bulky residues; notably, mammalian FTase also has steric restrictions at these positions (Reid et al. 2004; Hougland et al. 2009). The negative constraint for Pro at x3 is interesting in that it suggests that the conformation restrictions introduced by Pro at this position may eliminate critical interactions with the prenyltransferase and/or lead to unfavorable contacts within the substrate binding site.
We initiated our study expecting that Ydj1p could serve as a reporter for identifying shunted Cxxx sequences (i.e., CASQ-like). Indeed, the vast majority of recovered sequences behave phenotypically as if they are not cleaved. It remains to be determined, however, whether each identified sequence is actually shunted. Such an analysis would require either individual purification and mass-spectrometry analysis of the COOH-terminus of each Ydj1p Cxxx variant or indirectly assessing cleavage through other reporters such as a-factor (i.e., mating assays) or Ras (i.e., localization assays). It also remains to be determined how well each sequence is prenylated and by which prenyl group. While we did not evaluate all Cxxx variants, our investigations with subsets of sequences indicate that many are substantially farnesylated, often to completion.
Besides information on yeast prenyltransferase specificity, our Ydj1p-based data also hints at specificity determinants related to Cxxx cleavage. While our screen was not primarily intended to identify cleaved Cxxx sequences, we did recover a small set of sequences that behave phenotypically as if they are cleaved (i.e., CVIA-like). Not surprising, this phenotype is generally favored when aliphatic amino acids are present at x2, except in instances where charged residues flank this position or prenylation is otherwise disfavored. The limited number of CVIA-like sequences in our Ydj1p-based data set limits our ability to analyze cleavage specificity. We propose that sequences recovered through Ras-based screening offer a better set for such analyses. Because the Ras-based sequences are largely nonoverlapping with the sequences obtained by Ydj1p-based screening, the sets of sequences clearly differ in some capacity. The most parsimonious explanation is that the Ras-based sequences are cleaved while the Ydj1p-based sequences are shunted. Analysis of Ras-based sequences indicates that they are also enriched for aliphatic residues at x2. This suggests that Cxax (a = aliphatic) may be a more precise consensus for cleavage by the yeast CaaX proteases. It remains to be determined whether cleavage in these instances is due to Rce1p or Ste24p, the two proteases identified as cleaving CaaX proteins (Boyartchuk et al. 1997).
Overall, we interpret our analyses to indicate that many sequences have the potential to be prenylated in yeast (i.e., Cxxx), and that a subset have characteristics that make them susceptible to cleavage (i.e., Cxax). Using our in-house rules for prenylation, and additional rules to predict cleavage potential, we categorized the 89 Cxxx proteins identifiable in the SGD (Table S5). Over one-third of the sequences (36%; n = 32) were predicted to be prenylated and cleaved (i.e., Ras-like). This group includes the Ras and Rho GTPases (RAS2, RHO1, RHO2, etc.), a Gγ subunit (STE18), and a-factor (MFA1, MFA2). Other members of this group have not been investigated with respect to their Cxxx modifications (e.g., ABC transporter Atr1; ureidoglycolate lyase Dal3; Hsp40 Xdj1) and are interesting candidates to investigate as part of future studies on protein prenylation. About one-quarter of sequences (24%; n = 22) were predicted to be prenylated and not cleaved (i.e., shunted). This group included Ydj1p, as expected, and notables Pex19p and Nap1p. The CKQQ motif associated with Pex19p is farnesylated, and this motif is also present on the mammalian tumor suppressor STK11/Lkb1, for which there is evidence of farnesylation and shunting (James et al. 1994; Götte et al. 1998; Sapkota et al. 2001). A similar CKQx motif is present on Nap1p (yeast, human, and plant orthologs). The farnesylation status of yeast Nap1p has not yet been specifically investigated, but both human and plant Nap1 are farnesylated (Kho et al. 2004; Galichet and Gruissem 2006; Onono et al. 2010); cleavage status is unknown. Of the remaining sequences, about one-fifth (16%; n = 14) had ambiguous prenylation predictions with varying cleavage potential, and about one-quarter (23%; n = 21) were not predicted to be farnesylated. None of the proteins in these sets are known to be farnesylated (e.g., CUP1-1, HMG1, etc.). The proper binning of many proteins into what are appropriate categories provides confidence that our in-house prediction methods have potential applicability. Our prediction methods can be improved in future studies by incorporating additional discrimination parameters. For example, the accessibility of Cxxx sequences to the cytosolic prenyltransferases was not considered, so some proteins may be false positives within the canonical or shunted sequence groups.
The activities of the prenyltransferases and CaaX proteases have received much attention over the years, but their specificities have been hard to resolve despite a combination of in vivo, in vitro, and in silico methodologies. Here we used a genetic approach to identify a large set of prenylatable sequences in yeast that are not predicted to be modified by existing prediction methods. While this data set provides strong evidence for broader yeast prenyltransferase specificity, it has also allowed for reinterpretation of pre-existing data in a manner that informs on cleavage specificity. Beyond studies of the yeast enzymes, we envision that the specificities of other prenyltransferases and CaaX proteases could be investigated using our methods by heterologous expression of desired enzymes (e.g., yeast expressing human FTase instead of yeast enzyme). Intriguingly, considerable evidence suggests conserved specificity between yeast and human prenyltransferases, suggesting that the specificities observed in our study may ultimately hold true for the human enzymes (Gomez et al. 1993; Omer et al. 1993; Wang et al. 2014).
Acknowledgments
We thank Avrom Caplan (City College of New York) for anti-Ydj1p primary antibody, Ora Furman-Schueler (Hebrew University of Jerusalem) for sharing FlexPepBind scores, and Wayland Yeung (University of Georgia) for help with PrePS scores. We also thank members of the Schmidt laboratory for critical discussions and comments on this manuscript, and the following laboratory members for technical assistance: Alona Botnar, Manuel Fierro, and Ryan Peppenhorst, and Rajani Ravishankar. This work was funded by a grant from the National Institutes of Health to W.K.S. (GM117148).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7075025.
Communicating editor: S. Fields
Literature Cited
- Alper B. J., Nienow T. E., Schmidt W. K., 2006. A common genetic system for functional studies of pitrilysin and related M16A proteases. Biochem. J. 398: 145–152. 10.1042/BJ20060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atencio D., Yaffe M., 1992. MAS5, a yeast homolog of DnaJ involved in mitochondrial protein import. Mol. Cell. Biol. 12: 283–291. 10.1128/MCB.12.1.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N., Hamilton A. D., Sebti S. M., 2011. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11: 775–791. 10.1038/nrc3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanden M. J., Suazo K. F., Hildebrandt E. R., Hardgrove D. S., Patel M., et al. , 2018. Efficient farnesylation of an extended C-terminal C(x)3X sequence motif expands the scope of the prenylated proteome. J. Biol. Chem. 293: 2770–2785. 10.1074/jbc.M117.805770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyartchuk V. L., Ashby M. N., Rine J., 1997. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275: 1796–1800. 10.1126/science.275.5307.1796 [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Caldwell G. A., Wang S.-H., Naider F., Becker J., 1994. Consequences of altered isoprenylation targets on a-factor export and bioactivity. Proc. Natl. Acad. Sci. USA 91: 1275–1279. 10.1073/pnas.91.4.1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. J., Cyr D. M., Douglas M. G., 1992a. Ydj1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71: 1143–1155. 10.1016/S0092-8674(05)80063-7 [DOI] [PubMed] [Google Scholar]
- Caplan A. J., Tsai J., Casey P. J., Douglas M. G., 1992b. Farnesylation of Ydj1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J. Biol. Chem. 267: 18890–18895. [PubMed] [Google Scholar]
- Caplin B. E., Hettich L. A., Marshall M. S., 1994. Substrate characterization of the Saccharomyces cerevisiae protein farnesyltransferase and type-I protein geranylgeranyltransferase. Biochim. Biophys. Acta 1205: 39–48. 10.1016/0167-4838(94)90089-2 [DOI] [PubMed] [Google Scholar]
- Chen P., Sapperstein S. K., Choi J. D., Michaelis S., 1997. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J. Cell Biol. 136: 251–269. 10.1083/jcb.136.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. D., Der C. J., Philips M. R., 2015. Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin. Cancer Res. 21: 1819–1827. 10.1158/1078-0432.CCR-14-3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R., 1992. A simple and efficient procedure for transformation of yeasts. Biotechniques 13: 18–20. [PubMed] [Google Scholar]
- Emmanouilidis L., Schutz U., Tripsianes K., Madl T., Radke J., et al. , 2017. Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. Nat. Commun. 8: 14635 10.1038/ncomms14635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold A. A., Johnson D. I., Farnsworth C. C., Gelb M. H., Judd S. R., et al. , 1991. Protein geranylgeranyltransferase of Saccharomyces cerevisiae is specific for Cys-Xaa-Xaa-Leu motif proteins and requires the CDC43 gene product but not the DPR1 gene product. Proc. Natl. Acad. Sci. USA 88: 4448–4452. 10.1073/pnas.88.10.4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A. E., Patrick W. M., 2008. GLUE-IT and PEDEL-AA: new programmes for analyzing protein diversity in randomized libraries. Nucleic Acids Res. 36: W281–W285. 10.1093/nar/gkn226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G. A., Lemieszek M., Fortunato E. A., Johnson J. L., 2008. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol. Biol. Cell 19: 5249–5258. 10.1091/mbc.e08-04-0435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. W., Casey P. J., 1999. Enzymology and biology of CaaX protein prenylation. Recent Prog. Horm. Res. 54: 315–342. [PubMed] [Google Scholar]
- Galichet A., Gruissem W., 2006. Developmentally controlled farnesylation modulates AtNAP1;1 function in cell proliferation and cell expansion during Arabidopsis leaf development. Plant Physiol. 142: 1412–1426. 10.1104/pp.106.088344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangopadhyay S. A., Losito E. L., Hougland J. L., 2014. Targeted reengineering of protein geranylgeranyltransferase type I selectivity functionally implicates active-site residues in protein-substrate recognition. Biochemistry 53: 434–446. 10.1021/bi4011732 [DOI] [PubMed] [Google Scholar]
- Gomez R., Goodman L. E., Tripathy S. K., O’Rourke E., Manne V., et al. , 1993. Purified yeast protein farnesyltransferase is structurally and functionally similar to its mammalian counterpart. Biochem. J. 289: 25–31. 10.1042/bj2890025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte K., Girzalsky W., Linkert M., Baumgart E., Kammerer S., et al. , 1998. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol. Cell. Biol. 18: 616–628. 10.1128/MCB.18.1.616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton S. E., Dore T. M., Schmidt W. K., 2018. Rce1: mechanism and inhibition. Crit. Rev. Biochem. Mol. Biol. 53: 157–174. 10.1080/10409238.2018.1431606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H. L., Hicks K. A., Fierke C. A., 2005. Peptide specificity of protein prenyltransferases is determined mainly by reactivity rather than binding affinity. Biochemistry 44: 15314–15324. 10.1021/bi0509503 [DOI] [PubMed] [Google Scholar]
- He B., Chen P., Chen S. Y., Vancura K. L., Michaelis S., et al. , 1991. RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc. Natl. Acad. Sci. USA 88: 11373–11377. 10.1073/pnas.88.24.11373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E. R., Cheng M., Zhao P., Kim J. H., Wells L., et al. , 2016. A shunt pathway limits the CaaX processing of Hsp40 Ydj1p and regulates Ydj1p-dependent phenotypes. eLife 5: e15899 10.7554/eLife.15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland J. L., Lamphear C. L., Scott S. A., Gibbs R. A., Fierke C. A., 2009. Context-dependent substrate recognition by protein farnesyltransferase. Biochemistry 48: 1691–1701. 10.1021/bi801710g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland J. L., Hicks K. A., Hartman H. L., Kelly R. A., Watt T. J., et al. , 2010. Identification of novel peptide substrates for protein farnesyltransferase reveals two substrate classes with distinct sequence selectivities. J. Mol. Biol. 395: 176–190. 10.1016/j.jmb.2009.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland J. L., Gangopadhyay S. A., Fierke C. A., 2012. Expansion of protein farnesyltransferase specificity using “tunable” active site interactions: development of bioengineered prenylation pathways. J. Biol. Chem. 287: 38090–38100. 10.1074/jbc.M112.404954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna C. A., Clarke S., 1992. Maturation of isoprenylated proteins in Saccharomyces cerevisiae. J. Biol. Chem. 267: 10457–10464. [PubMed] [Google Scholar]
- Hrycyna C. A., Sapperstein S. K., Clarke S., Michaelis S., 1991. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and RAS proteins. EMBO J. 10: 1699–1709. 10.1002/j.1460-2075.1991.tb07694.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G. L., Goldstein J. L., Pathak R. K., Anderson R. G., Brown M. S., 1994. PxF, a prenylated protein of peroxisomes. J. Biol. Chem. 269: 14182–14190. [PubMed] [Google Scholar]
- Jennings B. C., Danowitz A. M., Wang Y. C., Gibbs R. A., Distefano M. D., et al. , 2016. Analogs of farnesyl diphosphate alter CaaX substrate specificity and reactions rates of protein farnesyltransferase. Bioorg. Med. Chem. Lett. 26: 1333–1336. 10.1016/j.bmcl.2015.12.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho Y., Kim S. C., Jiang C., Barma D., Kwon S. W., et al. , 2004. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. USA 101: 12479–12484. 10.1073/pnas.0403413101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lapham A., Freedman C., Reed T., Schmidt W., 2005. Yeast as a tractable genetic system for functional studies of the insulin-degrading enzyme. J. Biol. Chem. 280: 27481–27490. 10.1074/jbc.M414192200 [DOI] [PubMed] [Google Scholar]
- Kinsella B. T., Erdman R. A., Maltese W. A., 1991. Carboxyl-terminal isoprenylation of ras-related GTP-binding proteins encoded by rac1, rac2, and ralA. J. Biol. Chem. 266: 9786–9794. [PubMed] [Google Scholar]
- Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G., 2007. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6: 541–555. 10.1038/nrd2221 [DOI] [PubMed] [Google Scholar]
- Krzysiak A. J., Aditya A. V., Hougland J. L., Fierke C. A., Gibbs R. A., 2010. Synthesis and screening of a CaaL peptide library vs. FTase reveals a surprising number of substrates. Bioorg. Med. Chem. Lett. 20: 767–770. 10.1016/j.bmcl.2009.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Lamphear C. L., Hougland J. L., Fierke C. A., Schueler-Furman O., 2011. Identification of a novel class of farnesylation targets by structure-based modeling of binding specificity. PLoS Comput. Biol. 7: e1002170 10.1371/journal.pcbi.1002170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Cyr D. M., 1998a. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273: 5970–5978. 10.1074/jbc.273.10.5970 [DOI] [PubMed] [Google Scholar]
- Lu Z., Cyr D. M., 1998b. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273: 27824–27830. 10.1074/jbc.273.43.27824 [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S., Eisenhaber F., 2005. Refinement and prediction of protein prenylation motifs. Genome Biol. 6: R55 10.1186/gb-2005-6-6-r55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S., Herskowitz I., 1988. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell. Biol. 8: 1309–1318. 10.1128/MCB.8.3.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores S. L., Schaber M. D., Mosser S. D., Rands E., O’Hara M. B., et al. , 1991. Sequence dependence of protein isoprenylation. J. Biol. Chem. 266: 14603–14610. [PubMed] [Google Scholar]
- Oldenburg K. R., Vo K. T., Michaelis S., Paddon C., 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25: 451–452. 10.1093/nar/25.2.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A., Gibbs J. B., 1994. Protein prenylation in eukaryotic microorganisms: genetics, biology and biochemistry. Mol. Microbiol. 11: 219–225. 10.1111/j.1365-2958.1994.tb00302.x [DOI] [PubMed] [Google Scholar]
- Omer C. A., Kral A. M., Diehl R. E., Prendergast G. C., Powers S., et al. , 1993. Characterization of recombinant human farnesyl-protein transferase: cloning, expression, farnesyl diphosphate binding, and functional homology with yeast prenyl-protein transferases. Biochemistry 32: 5167–5176. 10.1021/bi00070a028 [DOI] [PubMed] [Google Scholar]
- Onono F. O., Morgan M. A., Spielmann H. P., Andres D. A., Subramanian T., et al. , 2010. A tagging-via-substrate approach to detect the farnesylated proteome using two-dimensional electrophoresis coupled with Western blotting. Mol. Cell. Proteomics 9: 742–751. 10.1074/mcp.M900597-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T. S., Terry K. L., Casey P. J., Beese L. S., 2004. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J. Mol. Biol. 343: 417–433. 10.1016/j.jmb.2004.08.056 [DOI] [PubMed] [Google Scholar]
- Reiss Y., Seabra M. C., Armstrong S. A., Slaughter C. A., Goldstein J. L., et al. , 1991. Nonidentical subunits of p21H-ras farnesyltransferase. Peptide binding and farnesyl pyrophosphate carrier functions. J. Biol. Chem. 266: 10672–10677. [PubMed] [Google Scholar]
- Roskoski R., Jr., 2003. Protein prenylation: a pivotal posttranslational process. Biochem. Biophys. Res. Commun. 303: 1–7. 10.1016/S0006-291X(03)00323-1 [DOI] [PubMed] [Google Scholar]
- Sapkota G. P., Kieloch A., Lizcano J. M., Lain S., Arthur J. S., et al. , 2001. Phosphorylation of the protein kinase mutated in Peutz-Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90(RSK) and cAMP-dependent protein kinase, but not its farnesylation at Cys(433), is essential for LKB1 to suppress cell vrowth. J. Biol. Chem. 276: 19469–19482. 10.1074/jbc.M009953200 [DOI] [PubMed] [Google Scholar]
- Silvius J. R., 2002. Mechanisms of Ras protein targeting in mammalian cells. J. Membr. Biol. 190: 83–92. 10.1007/s00232-002-1026-4 [DOI] [PubMed] [Google Scholar]
- Stein V., Kubala M. H., Steen J., Grimmond S. M., Alexandrov K., 2015. Towards the systematic mapping and engineering of the protein prenylation machinery in Saccharomyces cerevisiae. PLoS One 10: e0120716 10.1371/journal.pone.0120716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson R. C., Clarke S., 1990. Identification of a C-terminal protein carboxyl methyltransferase in rat liver membranes utilizing a synthetic farnesyl cysteine-containing peptide substrate. J. Biol. Chem. 265: 16248–16254. [PubMed] [Google Scholar]
- Suazo K. F., Schaber C., Palsuledesai C. C., Odom John A. R., Distefano M. D., 2016. Global proteomic analysis of prenylated proteins in Plasmodium falciparum using an alkyne-modified isoprenoid analogue. Sci. Rep. 6: 38615 10.1038/srep38615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. W., Douglas P. M., Ren H. Y., Cyr D. M., 2009. The type I Hsp40 Ydj1 utilizes a farnesyl moiety and zinc finger-like region to suppress prion toxicity. J. Biol. Chem. 284: 3628–3639. 10.1074/jbc.M807369200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C., Ohya Y., Rine J., 1993. Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 4260–4275. 10.1128/MCB.13.7.4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C. E., Boyartchuk V. L., Rine J., 1997. Substrate specificity determinants in the farnesyltransferase beta- subunit. Proc. Natl. Acad. Sci. USA 94: 10774–10779. 10.1073/pnas.94.20.10774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak G., Plous S., 2013. Research randomizer. Available at: https://www.randomizer.org/. Accessed: January 2018.
- Wang M., Casey P. J., 2016. Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17: 110–122. 10.1038/nrm.2015.11 [DOI] [PubMed] [Google Scholar]
- Wang Y. C., Distefano M. D., 2016. Synthetic isoprenoid analogues for the study of prenylated proteins: fluorescent imaging and proteomic applications. Bioorg. Chem. 64: 59–65. 10.1016/j.bioorg.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. C., Dozier J. K., Beese L. S., Distefano M. D., 2014. Rapid analysis of protein farnesyltransferase substrate specificity using peptide libraries and isoprenoid diphosphate analogues. ACS Chem. Biol. 9: 1726–1735. 10.1021/cb5002312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter-Vann A. M., Casey P. J., 2005. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer 5: 405–412. 10.1038/nrc1612 [DOI] [PubMed] [Google Scholar]
- Yokoyama K., Goodwin G. W., Ghomashchi F., Glomset J. A., Gelb M. H., 1991. A protein geranylgeranyltransferase from bovine brain: implications for protein prenylation specificity. Proc. Natl. Acad. Sci. USA 88: 5302–5306. 10.1073/pnas.88.12.5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K., McGeady P., Gelb M. H., 1995. Mammalian protein geranylgeranyltransferase-I: substrate specificity, kinetic mechanism, metal requirements, and affinity labeling. Biochemistry 34: 1344–1354. 10.1021/bi00004a029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7075025.