Baker’s yeast bearing incompatible MLH1 and PMS1 mismatch repair alleles are mutators that can adapt more rapidly to stress—but do so at the cost of long-term fitness...
Keywords: DNA mismatch repair, genetic incompatibility, mutation rate, natural yeast isolates, adaptation, Saccharomyces cerevisiae
Abstract
Laboratory baker’s yeast strains bearing an incompatible combination of MLH1 and PMS1 mismatch repair alleles are mutators that can adapt more rapidly to stress, but do so at the cost of long-term fitness. We identified 18 baker’s yeast isolates from 1011 surveyed that contain the incompatible MLH1-PMS1 genotype in a heterozygous state. Surprisingly, the incompatible combination from two human clinical heterozygous diploid isolates, YJS5845 and YJS5885, contain the exact MLH1 (S288c-derived) and PMS1 (SK1-derived) open reading frames originally shown to confer incompatibility. While these isolates were nonmutators, their meiotic spore clone progeny displayed mutation rates in a DNA slippage assay that varied over a 340-fold range. This range was 30-fold higher than observed between compatible and incompatible combinations of laboratory strains. Genotyping analysis indicated that MLH1-PMS1 incompatibility was the major driver of mutation rate in the isolates. The variation in the mutation rate of incompatible spore clones could be due to background suppressors and enhancers, as well as aneuploidy seen in the spore clones. Our data are consistent with the observed variance in mutation rate contributing to adaptation to stress conditions (e.g., in a human host) through the acquisition of beneficial mutations, with high mutation rates leading to long-term fitness costs that are buffered by mating or eliminated through natural selection.
LOSS of DNA mismatch repair (MMR) functions is often seen in bacteria grown in stressful environments. MMR-defective bacteria display an increased mutation supply and adapt to stress by acquiring beneficial mutations; however, their high mutation rate ultimately results in the accumulation of deleterious mutations that reduce fitness (Chao and Cox 1983; LeClerc et al. 1996; Taddei et al. 1997; Boe et al. 2000; Denamur et al. 2000; Giraud et al. 2001; Tanaka et al. 2003; Townsend et al. 2003). Bacteria defective in MMR can overcome fitness costs associated with high mutation rate by regaining MMR functions through horizontal gene transfer (Denamur et al. 2000). In baker’s yeast, loss of MMR functions in the laboratory provides an adaptive advantage to stress (Thompson et al. 2006; Raynes et al. 2011; Bui et al. 2015). However, there is no evidence that baker’s yeast defective in MMR can undergo horizontal transfer in the wild (Liti and Louis 2005; Ruderfer et al. 2006; Nishant et al. 2010). We and others hypothesized that MMR-defective baker’s yeast isolates in the wild could potentially mate with MMR-proficient isolates to become nonmutators, and thus eliminate long-term fitness costs [reviewed in Bui et al. (2017)].
Saccharomyces cerevisiae are usually nonpathogenic and are infectious only in immunocompromised individuals, causing opportunistic infections (Perez-Torrado and Querol 2015; Hall and Noverr 2017). Phylogenetic analyses of 16 human clinical isolates indicated that these isolates were not derived from a common ancestor or single strain, but may be a consequence of multiple independent origins arising from chance events involving the opportunistic colonization of human tissues by different strains (Schacherer et al. 2009; Strope et al. 2015; Zhu et al. 2016; Skelly et al. 2017). Yeast strains that infect humans must survive unfavorable growth conditions such as elevated temperature and exposure to antifungal agents. Interestingly, recent studies have suggested that ploidy changes and chromosome aneuploidy can promote phenotypic diversity; in some cases, such events are thought to increase the rate of acquisition of adaptive mutations in the face of stress and have been seen at higher frequencies in clinical isolates (Selmecki et al. 2009, 2015; Pavelka et al. 2010; Rancati and Pavelka 2013; Zörgö et al. 2013; Zhu et al. 2014, 2016; Cromie and Dudley 2015; Gerstein and Berman 2015; Hose et al. 2015; Sirr et al. 2015; Sunshine et al. 2015). At present, it is unclear if baker’s yeast strains associated with humans can be MMR defective; however, two recent reports have identified Cryptococcus fungal human pathogens that are defective in MMR and are mutators (Billmyre et al. 2017; Boyce et al. 2017).
We showed previously that the MLH1 and PMS1 MMR genes from the S288c and SK1 yeast strains can display negative epistasis; strains bearing the S288c MLH1-SK1 PMS1 genotype are mutators (Heck et al. 2006). Single-amino acid changes in each protein, MLH1 D761 from S288c and PMS1 K818/822 from SK1, were sufficient to cause this incompatibility (Supplemental Material, Figure S1; Heck et al. 2006). The MLH1-PMS1 combinations are compatible in the S288c and SK1 group strains, as well as in the inferred ancestral strain from which they diverged (Figure S1, Heck et al. 2006; Bui et al. 2015). The incompatible combination of S288c MLH1-SK1 PMS1 in laboratory strains (abbreviated as cMLH1-kPMS1) confers an ∼100-fold increase in the mutation rate in an assay in which null mutants display a 10,000-fold higher rate (Heck et al. 2006; Bui et al. 2015). Incompatible strains have an adaptive advantage in high-salt stress (Bui et al. 2015). However, they also display a long-term fitness cost due to the accumulation of deleterious mutations. This was demonstrated by a fitness decline of incompatible cells in competition experiments between compatible and incompatible cells in rich media (Bui et al. 2017). To our knowledge, the cMLH1-kPMS1 genotype is the only incompatibility involving MMR genes that has been characterized. However, recessive mutations in MMR genes MSH2 and MSH6 found in colon cancer patients were postulated to be due to a compound effect of mutations in the two genes, which could reflect negative epistasis (Kariola et al. 2003).
In a search for yeast bearing the incompatible cMLH1-kPMS1 combination, we screened a collection of 1011 naturally occurring worldwide isolates of S. cerevisiae (Bui et al. 2017; Peter et al. 2018). Only YJM523, a human clinical isolate of S. cerevisiae, is homozygous for cMLH1-kPMS1 incompatibility (Strope et al. 2015; Bui et al. 2017; Skelly et al. 2017). A spore clone of YJM523, YJM555, was a nonmutator that had accumulated multiple suppressors of the incompatibility phenotype (Bui et al. 2017; Skelly et al. 2017). To our knowledge, baker’s yeast isolates that are mutators have not been identified in the wild (Bui et al. 2017; Skelly et al. 2017). However, yeast heterozygous for the cMLH1-kPMS1 incompatibility, a recessive trait (Heck et al. 2006), might have an advantage because they can be nonmutators in a heterozygous diploid background. Under stress conditions, the diploids can sporulate and give rise to mutator spore clones. These mutators would be able to provide a transient adaptive advantage to stress, and can potentially mate back to compatible strains and become nonmutators to prevent long-term fitness costs.
In eukaryotes, a complete loss of MMR functions would likely have dramatic effects on organism fitness. In changing environments, as would likely be the case for yeast growing in a human host, adaptation to stress would involve a compromise between efficient growth and long-term survival. One way to deal with such variable stress is for progeny of an organism to display variations in mutation supply that can yield adaptation phenotypes and thus prevent the population from undergoing extinction under stress [see Richard and Yvert (2014)]. We examined two human clinical diploid isolates, YJS5845 and YJS5885, that are each heterozygous for cMLH1-kPMS1 incompatibility polymorphisms. In both isolates, one of the two copies of MLH1 is identical in amino acid sequence to S288c MLH1, and one of two copies of PMS1 is identical to SK1 PMS1. The isolates were nonmutators but derived spore clones displayed a 340-fold range of mutation rates in a DNA slippage assay, with cMLH1-kPMS1 incompatibility being the major contributor to increased mutation. In contrast, the difference in mutation rates between laboratory-compatible and -incompatible strains was only 11-fold, indicating that the human isolate backgrounds contained suppressors and enhancers of mutation rate. Curiously, high levels of aneuploidy were observed in the spore clones from YJS5845, suggesting additional mechanisms that may contribute to the modification of mutation rate and/or adaptation. These findings suggest that there is a variation in mutation supply that is balanced with associated fitness costs. Such variation could provide a greater chance for yeast to survive stress conditions present in a human host.
Materials and Methods
S. cerevisiae strains and media
The natural isolates YJS5845, YJS5885, YJS4806, YJS4810, YJS5882, YJS5678, YJS5512, and YJS4970 (Table 1) were obtained from the 1011 Yeast Genomes Project (Peter et al. 2018). YJM521, YJM523, and YJM555 were obtained from the Fungal Genetics Stock Center (FGSC) collection (Strope et al. 2015; http://www.fgsc.net/). Isolates were sporulated on plates by streaking cells from frozen stocks onto YPD (2% peptone, 1% yeast extract, and 2% dextrose) media. Cells were grown for 1 day at 30°, after which they were patched onto sporulation medium [1% (g/liter) potassium acetate and 2% (g/liter) agar] and incubated at 30° for 3 days. Tetrads were dissected and germinated on minimal complete media (Rose et al. 1990). Isolates were sporulated in liquid by growing overnights in YPD. Next, 50 μl of each overnight was inoculated into 5 ml YPA (2% peptone, 1% yeast extract, and 2% potassium acetate) media and grown with shaking at 30° for 16 hr, after which each culture was washed with 3 ml of sterile water, and then transferred to 1% potassium acetate and incubated at 30° with shaking for 3 days. To test for the growth of spore clones in lactate media, spore clones were patched on YPL plates (2% peptone, 1% yeast extract, 2% lactate, and 2% agar) and grown for 4 days at 30° (Table S5).
Table 1. Yeast isolates analyzed in this study.
| Lab, standard, and isolate identifier | Origin | Ploidy | SNPs (singletons) | Spore formation (%) (n) | Three or four spores (%) | Two spores (%) | Viable spores (tetrads) (%) |
|---|---|---|---|---|---|---|---|
| YJS5845, CIC, Ponton 11 | Human, mouth, Spain | 2 | 68,564 (825) | 93 (147) | 53 | 39 | 20 (40) |
| YJS5845 + CHR XIV | 96 (174) | 60 | 35 | 19 (39) | |||
| YJS5885, CKN, CLI_19 | Human, feces, France | 2 | 76,104 (955) | 57 (139) | 31 | 24 | 30 (28) |
| YJM521 | Human clinical, CA | 2 | 53,947 (22) | 78 (221) | 69 | 5.9 | 94 (10) |
| YJS4806, CFI, WLP013 | Beer, UK | 4* | nt | 15 (10) | |||
| YJS4810, CFN, WLP006 | Beer, unknown | 4* | nt | 5.0 (10) | |||
| YJS5882, CKK, CLI_16 | Human clinical, France | 4 | nt | 72 (8) | |||
| YJS5678, CBF, SD-15 | Bakery, Italy | 4* | nt | 70 (10) | |||
| YJS5512, BML, NCYC_2780 | Human clinical, Belgium | 4* | nt | nt | |||
| YJS4970, CGC, UCD_06-645 | Fruit, Davis, CA | 4 | no spores |
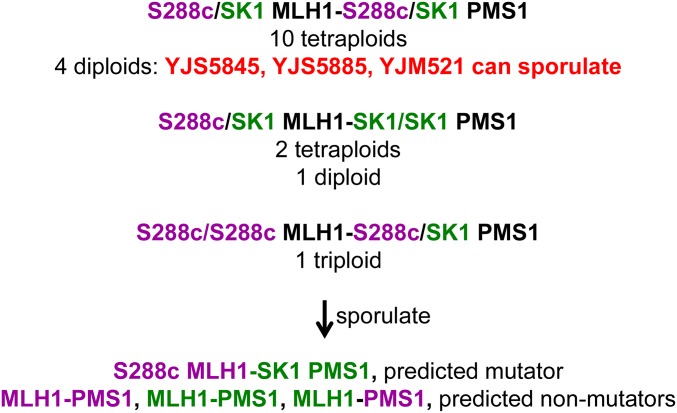
Nineteen of 1011 baker’s yeast isolates contain the incompatible MLH1-PMS1 genotype in heterozygous (18 isolates) or homozygous (YJM523) combinations (Bui et al. 2017). One isolate is triploid and 12 are tetraploid (six are shown here), two of which cannot sporulate. The remaining six are diploid, and the four diploid isolates that sporulate, YJS5845, YJS5885, YJM521, and YJM523, all belong to the MR3 mosaic clade (113 members) that are admixed with ancestry from two or more populations. In the MR3 clade, the mean number of singletons was 721 (± 1150 SD, ± 108 SE, and median = 163). The diploid isolates YJM521 (Clemons et al. 1997) and YJS5845 are homothallic, and YJS5885 appears to be functionally heterothallic (see text). The percentage of cells forming at least one spore (n is the cells examined) is shown under spore formation, and the distribution of these cells into 3, 4, or 2 spore asci is shown, along with the % spore viability seen in dissected tetrads (n = number dissected). Our original stock of YJS5845 contained euploid and aneuploid (additional copy of chromosome XIV) cells. The euploid and aneuploid isolates were analyzed for spore formation and viability separately. *, contains chromosomal aneuploidy (Peter et al. 2018); nt, not tested.
The S288c-derived strains EAY1365 (MATa, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2::insE-A14, mlh1Δ::KanMX4, pms1Δ::KanMX4), EAY1369 (MATα, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2::insE-A14, cPMS1::HIS3, cMLH1), EAY1370 (MATα, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2::insE-A14, kPMS1::HIS3, cMLH1), EAY1372 (MATa, ura3-52, leu2Δ1, trp1Δ63, lys2::insE-A14, msh2Δ::hisG), FY90 (MATa, ade8; Winston et al. 1995), and EAY4087 (MATa, ade8, mlh1Δ::KanMX), and the natural isolates YJS5845, YJS5885, and their spore clone derivatives, were analyzed for mutator phenotypes (see below for details). Transformation of plasmids into strains, isolates, and spore clones was performed as described previously (Gietz and Schiestl 2007). In this manuscript, genes derived from the S288c background are designated with a “c” (e.g., cMLH1) and those derived from SK1 with a “k” (e.g., kMLH1).
Plasmids
pEAA213 (cMLH1, ARSH4 CEN6, LEU2) and pEAA214 (kMLH1, ARSH4 CEN6, LEU2) were described previously (Argueso et al. 2003; Heck et al. 2006). The cMLH1 gene from YJM521 was cloned into pEAA213 as described previously (Bui et al. 2017). All of the constructs expressed MLH1 via the S288c MLH1 promoter. pEAA238 (cPMS1, ARSH4, CEN6, HIS3) and pEAA239 (kPMS1, ARSH4, CEN6, HIS3) were described previously (Argueso et al. 2003; Heck et al. 2006). The kPMS1 gene from YJM521 was cloned into pEAA238 as described previously (Bui et al. 2017). All of the constructs expressed PMS1 via the S288c PMS1 promoter.
pEAA613 (ARS-CEN, NATMX) contains the URA3 promoter-KanMX::insE-A14 reversion reporter (Bui et al. 2017). This reporter is expressed via the URA3 promoter (−402 to the ATG start site). A 55-bp sequence containing a +1 frameshift in the 14-bp homopolymeric A run (insE-A14; Tran et al. 1997) was inserted immediately after the URA3 ATG, followed by codons 18–269 of the KANMX open reading frame derived from pFA6-KANMX. pEAA611 (ARS-CEN, NATMX) contains the URA3 promoter-KanMX::insE-A10 in-frame reporter (Bui et al. 2017).
Genotyping analysis
The 2.3-kb MLH1 and 2.6-kb PMS1 open reading frames from YJM521, YJS5845, YJS5885, YJS4806, YJS4810, YJS5882, YJS5678, YJS5512, YJS4970, and derived spore clones were determined by sequencing PCR-amplified DNA from chromosomal DNA (Tables S1 and S2; Hoffman and Winston 1987) using Expand High Fidelity Polymerase (Roche Life Sciences). Primers AO324 (5′-ATAGTGTAGGAGGCGCTG-3′) and AO821 (5′-AACTTTGCGGCCGCGGATCCAGCCAAAACGTTTTAAAGTTA-3′) were used to amplify the MLH1 open reading frame, and primers AO481 (5′-CCACGTTCATATTCTTAATGGCTAAGC-3′) and AO548 (5′-CGATTCTAATACAGATTTTAATGACC-3′) were used to amplify the PMS1 open reading frame. PCR products were sequenced by the Sanger method in the Cornell Biotechnology Resource Center.
The diploid isolates YJM521, YJS5845, and YJS5885 were shown previously to be heterozygous for genetic information at both the MLH1 and PMS1 incompatibility sites (Gly/Asp at amino acid 761 in MLH1 and Arg/Lys at amino acid 818/822 in PMS1; Bui et al. 2017; Skelly et al. 2017). For each diploid isolate, the sequences of the two MLH1 and PMS1 alleles were determined by sequencing the MLH1 and PMS1 genes from two ancestral (MLH1 Gly 761 and PMS1 Arg 818/822) and two incompatible (MLH1 Asp 761 and PMS1 Lys 818/822) haploid spore clones. In all cases, the DNA sequences of the two different spore clone isolates of the same MLH1-PMS1 genotype were identical, thus allowing us to assign the MLH1 and PMS1 sequences present in each parental chromosome (Table S2).
To determine the compatibility genotype (Gly/Asp at amino acid 761 in MLH1 and Arg/Lys at amino acid 818/822 in PMS1; Figure 1 and Figure S1) of YJM521, YJS5845, and YJS5885 spore clones, MLH1 and PMS1 open reading frame PCR products were sequenced with primers AO328 (5′-GACGAGTTAAATGACGATGCTTCC-3′) and AO485 (5′-AAAGTATCTGACGTTAACAGTTTC-3′), respectively. To test for the presence of the proline 271 suppressor polymorphism in MLH1-incompatible spore clones (Demogines et al. 2008) in YJS5845 and YJS5885, MLH1 PCR products were also sequenced using primer AO325 (5′-CATGTGGCAACAGTCACAGTAACG-3′). None of the spore clones (10 from YJS5845 and 11 from YJS5885) contained this polymorphism, displaying instead the leucine residue.
Figure 1.
Isolates containing heterozygous MLH1-PMS1 genotypes predicted to form mutator spore progeny. Eighteen isolates were identified from the 1011 yeast genome project that are heterozygous for the MLH1 761 and/or PMS1 818/822 genotypes, and are predicted to yield incompatible spore progeny (Bui et al. 2017). In Figure S1, a model is presented in which ancestral isolates bearing MLH1 Gly 761 and PMS1 Arg 818/822 alleles acquire neutral or beneficial mutations that lead to the derived S288c (purple, Asp 761, Arg 818/822) and SK1 (green, Gly 761, Lys 818/822) group isolates.
Efficiency of plating
EAY1369 (cMLH1-cPMS1, compatible) and EAY1370 (cMLH1-kPMS1, incompatible) strains, and YJM521, YJM555, YJS5845, and YJS5885 isolates/spore clone derivatives, were transformed with pEAA611 (URA3 promoter-KanMX::insE-A10 in-frame reporter; Bui et al. 2017) and grown on YPD media containing clonNAT (100 µg/ml). Independent transformants were grown overnight on YPD-clonNAT (50 µg/ml). Next, 10 µl of 100, 10−1, 10−2, 10−3, 10−3, and 10−4 dilutions were plated on YPD-clonNAT (50 µg/ml) and YPD-clonNAT (50 µg/ml) geneticin (200 µg/ml) plates. Growth on YPD-clonNAT and YPD-clonNAT geneticin plates was compared, and strains that grew to the same extent on both plates (within a little fold variation) were considered to have an efficiency of plating of ∼1. Reversion assays were performed only on spore clones showing an efficiency of plating of ∼1 (Figure S3).
kanMX::insE-A14 reversion assay
The S288c strains EAY1369 (cMLH1-cPMS1, compatible) and EAY1370 (cMLH1-kPMS1, incompatible), and YJS5845 and YJS5885 isolates and spore clone derivatives, were transformed with pEAA613 and grown on YPD media containing clonNAT (100 µg/ml). Independent transformants were subsequently grown overnight in YPD + clonNAT, and then plated on to YPD + clonNAT (50 µg/ml) and YPD + clonNAT (50 µg/ml) with geneticin (200 µg/ml). These strains were analyzed for reversion to resistance to geneticin using methods described previously (Tran et al. 1997; Zubko and Zubko 2014; Bui et al. 2015, 2017). Previously, we measured resistance to geneticin (G418) for EAY1369 and YJM555 lacking the kanMX::insE-A14 reporter plasmid (Bui et al. 2017). We estimated the mutation rates to be < 2 × 10−10, indicating that spontaneous reversion to G418r would not interfere with the detection of G418r using the pEAA613 kanMX::insE-A14 reporter plasmid. The 95% C.I.s were determined as described by Dixon and Massey (1969). Mann–Whitney U-tests were performed using Prism (GraphPad Prism 7.00 for Mac OS X; GraphPad Software, La Jolla, CA, www.graphpad.com) to determine the significance of the differences in median reversion rates (Wilcoxon 1945; Mann and Whitney 1947).
Sequencing of homopolymeric A repeats in kanMX::insE-A14 G418r clones
A 2.2-kb region of the plasmid pEAA613 containing kanMX::insE-A14 was PCR amplified from total DNA (Hoffman and Winston 1987) isolated from revertant colonies (NATr, G418r) or nonrevertant controls (NATr, G418s) using Phusion Hot Start II DNA polymerase (Thermo Fisher Scientific) and primers AO3879 (5′-CTCGTTTTCGACACTGGATGGC-3′) and AO3880 (5′-GCGTGAGCTATGAGAAAGCGC-3′). Primer AO3838 (5′-TGGTCGGAAGAGGCATAAATTC-3′) was used to sequence the PCR product in the region surrounding the homopolymeric A sequence.
lys2-A14 reversion assay
Independent colonies of EAY1365 (relevant genotype lys2-A14) containing the ARS-CEN, MLH1 and ARS-CEN, PMS1 plasmids presented in Table 2 were inoculated into YPD liquid media, grown overnight at 30°, and then plated onto LYS, HIS, LEU dropout and HIS, LEU dropout synthetic plates. These strains were analyzed for reversion to Lys+ as described previously (Tran et al. 1997; Bui et al. 2015). The 95% C.I.s were determined as described by Dixon and Massey (1969). The Mann–Whitney U-test (Mann and Whitney 1947) was performed to determine the significance of the differences in median reversion rates.
Table 2. Mutation rates in an S288c strain containing MLH1 and PMS1 gene combinations identical in amino acid sequence to those present in S288c, SK1, YJS5845, YJS5885, and YJM521.
| MLH1-PMS1 genotype | Lys+ reversion rate (10−7), (95% C.I.) | Relative rate | (n) |
|---|---|---|---|
| S288c-S288c, compatible | 4.1 (1.7–13.8) | 1 | 13 |
| S288c-SK1, incompatible | 311 (111–919)a | 75 | 16 |
| YJM521c-YJM521k | 92 (78.4–690)b | 22 | 29 |
| YJM521c-SK1 | 80 (47–182)b | 19 | 15 |
| mlh1Δ, pms1Δ | 45,300 (13,170–126,800)a | 10,970 | 10 |
EAY1365 (relevant genotype mlh1Δ::KanMX4, pms1Δ::KanMX4) was transformed with ARS-CEN plasmids containing the MLH1 and PMS1 genes obtained from the indicated strains and isolates. Independent cultures (n) were examined for reversion to Lys+. Median mutation rates are presented with 95% C.I.s, and relative mutation rates compared to the wild-type strain are shown. Data for S288c-S288c compatible, S288c-SK1 incompatible, and mlh1Δ, pms1Δ were reported previously (Bui et al. 2017). Note that MLH1 open reading frames in YJS5845c and YJS5885c are identical to S288c MLH1, and the PMS1 open reading frames in YJS5845k and YJS5885k are identical to SK1 PMS1 (Table S2).
Significantly different from S288c-S288c (P < 0.001, Mann–Whitney U-test).
Significantly different from S288c-S288c, compatible (P < 0.01, Mann–Whitney U-test).
5-fluoroorotic acid resistance assay
Resistance to 5-fluoroorotic acid (5-FOA) was measured in FY90, EAY4087 (mlh1Δ derivative of FY90), and haploid spore clones of YJS5885 using a protocol similar to that of Lang and Murray (2008). Single colonies from a synthetic complete media were grown overnight in 2 ml liquid synthetic complete media (0.7% yeast nitrogen base, 0.087% complete amino acid mix, and 2% dextrose), diluted 1:500 into synthetic complete media, and grown in 5 ml for 2 days at 30° with shaking. Appropriate dilutions were then plated onto 5-FOA (0.1% 5-FOA, 0.7% yeast nitrogen base, 0.087% amino acid mix without uracil, 0.005% uracil, 2% dextrose, and 2% agar) and synthetic complete plates, and mutation rates were determined using previously published methods (Tran et al. 1997; Zubko and Zubko 2014; Bui et al. 2015). Next, 95% C.I.s were determined as described by Dixon and Massey (1969). Mann–Whitney U-tests were performed using Prism (GraphPad Prism 7.00 for Mac OS X; GraphPad Software; www.graphpad.com) to determine the significance of the differences in median reversion rates (Wilcoxon 1945; Mann and Whitney 1947).
The URA3 open reading frame was PCR amplified from genomic DNA of independent 5-FOA-resistant (FOAr) spore clones using Phusion Hot Start II DNA polymerase (Thermo Fisher Scientific), and primers AO1115 (5′-AGAAGAGTATTGAGAAGGGCAA-3′) and AO3784 (5′-TTAGTTTTGCTGGCCGCATC-3′). Primers AO3156 (5′-GGTGAAGGATAAGTTTTGACCATCAAAGAA-3′) and AO3788 (5′-CTGGAGTTAGTTGAAGCATTAGGTC-3′) were used to sequence the PCR product.
Whole-genome sequencing
Single colonies of isolates, spore clones, and transformants were grown overnight in 2 ml YPD media and genomic DNA was isolated using the YeaStar Genomic DNA kit (Zymo Research). DNA was barcoded using Illumina Nextera XT and high-throughput DNA sequencing was performed on an Illumina NextSeq500 at the Cornell Biotechnology Resource Center, achieving a 50-fold mean coverage. Sequences were aligned to the reference S288c genome sequence (SGD: https://www.yeastgenome.org/) using HISAT2 to create SAM (sequence alignment map) files. SAM files were converted to binary version BAM files. BAM files were sorted and indexed, and duplicates were removed using SAMTools-1.7 (http://samtools.sourceforge.net/). Aneuploidy plots (Figure 4B and Figure S6) were constructed using a custom-made script (provided by V. P. Ajith, IISER Trivandrum); read counts were tabulated for 5000-bp windows using the GenomicRanges and GenomicAlignments Bioconductor packages in a custom R-script. SnpEff was used to annotate variants using sequence information for YJS5845 and YJS5885 as described in Peter et al. (2018).
Figure 4.
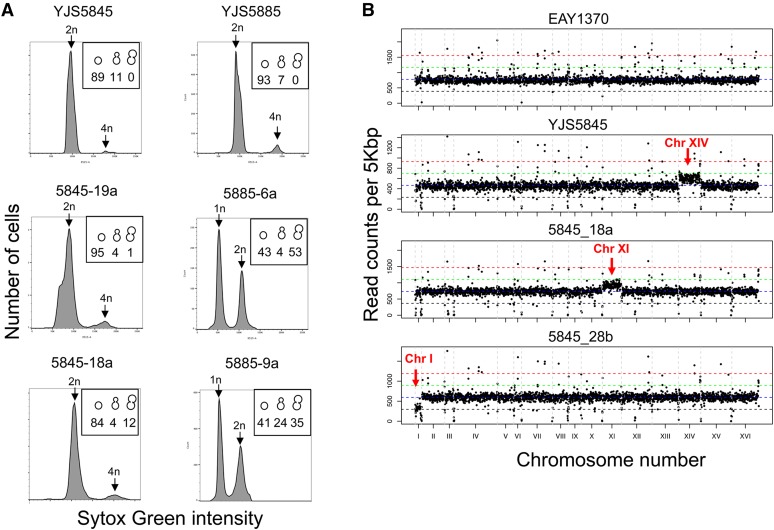
Ploidy of isolates and spore clones. (A) Representative flow cytometry plots of YJS5845 (left), YJS5885 (right), and derived spore clones. All spore clones of YJS5845 and YJS5885 tested were diploid and haploid, respectively. The black arrows show the position of 1n, 2n, and 4n DNA content. Inset shows percentage of single cells, small-budded cells, and large-budded cells assessed by light microscopy. (B) Whole-genome sequencing was performed for YJS5845 and spore clones (Materials and Methods). YJS5845, and spore clones 5845-18a and 5845-28b, displayed aneuploidy for chromosomes (Chr) XIV, XI, and I, respectively.
Flow cytometry
Cells were prepared using a protocol modified from Rosebrock (2017). Cells were grown overnight in rich medium, and washed and fixed in 70% ethanol at −20° for 24 hr. Fixed cells were washed, resuspended in 50 mM sodium citrate pH 7.4, and allowed to rehydrate. RNase A (Thermo Fisher Scientific) was added to a final concentration of 0.2 mg/ml and incubated at 37° overnight. Cells were then treated with proteinase K (New England Biolabs, Beverly, MA) to a final concentration of 16 units/ml at 37° for 1 hr, and then sonicated for 10 sec at 30% power. Cells were then pelleted, supernatant was removed, and fresh 50 mM sodium citrate pH 7.4 was added, followed by staining of cells using Sytox Green at a final concentration of 1 μM. Flow cytometry was performed on a BD FACS Aria at the Cornell University Flow Cytometry Core Laboratory. YJS5885 spore clones showed multiple peaks even after gating out cell clumps, therefore the peaks were sorted and visualized under the microscope to determine the composition of cells under each peak. Peak one had single cells, peak two had a combination of single cells and small-budded cells, and peaks three and four had large-budded cells or multiple-budded cells. The populations from the last two peaks were removed during analysis, because they were present as cell clumps and were thus not a correct representation of the DNA content in each cell, using FlowJo 10.4.2 software. The unsorted cells were counted under the microscope to determine the fraction of single cells, small-budded cells, and large-budded cells. The buds with diameter less than one-half of mother cell were classified as small-budded cells (Chan and Botstein 1993).
Data availability
Strains and plasmids are available upon request, and the entire DNA sequences of the MLH1 and PMS1 genes from 1011 isolates are presented in Bui et al. (2017). Supporting information contains all detailed descriptions of supplemental files. All wet laboratory experiments presented (lys2-A14 reversion, kanMX::insE-A14 reversion, 5-FOA resistance) were repeated on at least 2 separate days. The following figures and tables can be found in the Genetics Society of America Figshare portal:
Figure S1: incompatibility involving the MLH1 and PMS1 MMR genes.
Figure S2: DNA sequence, as shown by chromatogram traces, of the MLH1 incompatibility site (bp 2282, Gly or Asp at amino acid 761) in the indicated isolates and spore clones.
Figure S3: efficiency of plating of strains transformed with pEAA611, comparing growth on clonNAT and clonNAT + G418 plates.
Figure S4: sequencing analysis of G418-resistant revertants and sensitive control colonies.
Figure S5: flow cytometry of spore clones.
Figure S6: ploidy of YJS5845 and YJS5885 isolates, and their spore clones.
Table S1: genotyping of spore clones obtained by dissection of isolate tetrads.
Table S2: genotyping of MLH1 and PMS1 loci in YJM and YJS isolates, and derived spore clones.
Table S3: analysis of HO, PHO80, and STP22 genes in YJS5845 and YJS5885 for variants using SnpEff.
Table S4: analysis of resistance to 5-FOA in YJS5885 spore clones.
Table S5: sporulation and lactate growth phenotype.
Table S6: assigning MLH1 polymorphisms found in heterozygous genotypes onto the MLH1 structure–function map.
Table S7: amino acid heterozygosities identified in MLH1 in 107 yeast isolates.
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7233104.
Results
Genotyping of yeast isolates containing incompatible cMLH1-kPMS1 combinations
Among 1011 yeast isolates, we identified one clinical isolate (YJM523) that was homozygous and 18 that were heterozygous for an S288c MLH1-SK1 PMS1 incompatibility that confers a mutator phenotype in laboratory strains (Materials and Methods; Figure 1, Figure S1, and Table S1; Bui et al. 2017). This phenotype is abbreviated as cMLH1-kPMS1 and was assigned based on the amino acid at position 761 in MLH1 and 818/822 in PMS1. For YJM523, analysis of a derived spore clone (YJM555), and DNA sequencing and phylogeny analysis, suggested that it is homothallic and homozygous for MLH1 and PMS1 sequence information (Table S2; Strope et al. 2015). Genetic analysis indicated that YJM555 is not a mutator but contains multiple mutations that suppress and enhance the cMLH1-kPMS1 incompatibility (Bui et al. 2017; Skelly et al. 2017). Of the 18 isolates heterozygous for cMLH1-kPMS1 incompatibility, five are diploid, three of which can sporulate (Figure 1 and Table 1). The remainder are triploid or tetraploid. Several tetraploids were genotyped (Figure S2 and Table S1), showing a variety of cMLH1:kMLH1 genotypes (4:0, 3:1, and 2:2).
Characterizing the yeast isolates that contain incompatible cMLH1-kPMS1 combinations
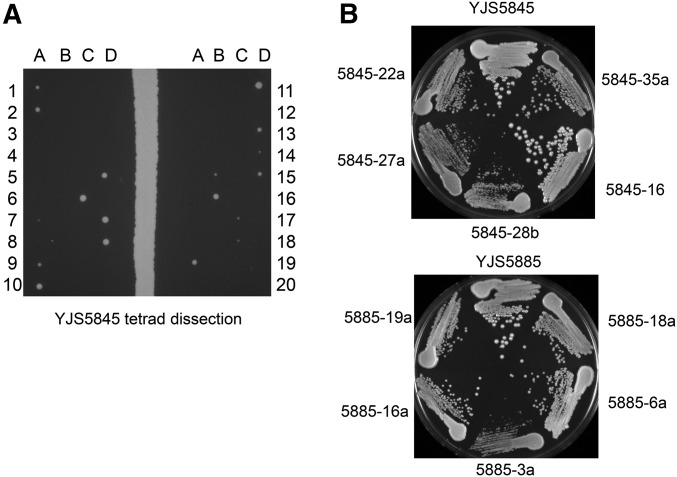
Three heterozygous diploid clinical isolates (YJM521, YJS5845, and YJS5885) displayed rapid vegetative growth and formed colonies from single cells on YPD plates in 24 hr. They also showed a high efficiency of sporulation, with variable spore viability (20–94%) and spore clone growth (Figure 2 and Table 1). At least one of the YJS5845 spore clones genotyped displayed what appeared to be invasive growth properties on YPD. We also saw some deviation from 1:1:1:1 for MLH1 and PMS1 genotypes in YJS5845 and YJS5885, which we had not seen in the high spore viability isolate YJM521 (Table S1). We sporulated some of the tetraploids with the goal of identifying homozygous incompatible spore clones. However, as shown in Figure S2 and Table S1, such clones were not identified.
Figure 2.
YJS5845, YJS5885, and spore clones have different colony sizes/growth properties. (A) YJS5845 was sporulated and then tetrad-dissected on minimal complete plates. Germinated spore clones were photographed after a 48-hr incubation at 30°. (B) YJS5845 and YJS5885 isolates, and representative spore clones, were struck to single colonies on YPD media and photographed after a 48-hr incubation at 30°.
YJS5845, YJS5885, and YJM521 are all human clinical isolates, belong to an admixture clade, and have different geographic locations (Table 1). For MLH1, YJS5845 and YJS5885 contain the exact amino acid sequences for the S288c MLH1 allele on one chromosome and the exact amino sequences for the YJM521 MLH1 k-allele on the other chromosome (Table S2). For PMS1, YJS5845 and YJS5885 contain the exact amino acid sequences for the SK1 PMS1 allele on one chromosome. The other PMS1-bearing chromosome in both isolates contains the S288c PMS1 K818 allele; however, this chromosome contains unique variants for the two isolates (Table S2). YJM521 is homozygous for the MLH1-P271 suppressor allele (Table 2 and Table S2).
Mutator phenotypes exhibited by incompatible S288c MLH1-SK1 PMS1 combinations of the YJM521, YJS5845, and YJS5885 isolates in the S288c strain background
The incompatible combinations of MLH1 and PMS1 present in YJS5845, YJS5885, and YJM521 were tested for their ability to confer a mutator phenotype in the S288c background (Table 2). The incompatible combination from YJM521 was cloned; the incompatible combinations in YJS5845 and YJS5885 are represented by cMLH1-kPMS1 because the amino acid sequences are identical to the cMLH1-kPMS1 combination. The cMLH1-kPMS1 combination representing YJS5845 and YJS5885 conferred an incompatible mutator phenotype (75-fold higher than compatible), while the cMLH1-kPMS1 combination from YJM521 displayed a suppressed incompatible mutator phenotype (19-fold higher than compatible) that was expected because YJM521 is homozygous for the MLH1-P271 suppressor allele (Demogines et al. 2008).
Spore clones from YJS5845 and YJS5885 display a wide range of mutator phenotypes, with the MLH1-PMS1 genotype being the major contributor to mutation rate
We examined the mutation rate in isolates and spore clone derivatives that had an ∼100% efficiency of growth in YPD media containing geneticin when transformed with the in-frame plasmid pEAA611 (Table 3). This was done to remove spore clones that were highly sensitive to aminoglycoside antibiotics (often observed in yeast isolates) (Ernst and Chan 1985; Wickert et al. 1998). It is important to note that as a result of this analysis we assayed mutator phenotypes in roughly half of the spore clones obtained; the other half displayed poor efficiencies of plating on YPD media containing geneticin (Figure S3; Materials and Methods; no linkage was seen between efficiency of plating and MLH1, PMS1 compatible or incompatible genotypes). Using SNPeff (Materials and Methods), we confirmed the sequences of previously identified genes involved in antibiotic sensitivity, STP22 and PHO80, in YJS5845 and YJS5885, but did not find any disruptions or predicted deleterious alleles (Table S3; Ernst and Chan 1985; Wickert et al. 1998).
Table 3. Reversion assay using the URA3 promoter-KanMX::insE-A14 plasmid.
| Strain, isolate, or spore clone | Genotype MLH1-PMS1 | Incompatible/compatible | Rate* G418r (10−7), (95% C.I.), n | Rate** G418r (10−7), (95% C.I.), n | Relative rate |
|---|---|---|---|---|---|
| EAY1369 | c-c | C | 5.2 (3.3–7.9), 15 | 1 | |
| EAY1370 | c-k | I | 57 (38–89), 19a | 11 | |
| EAY1372 | msh2Δ | Not applicable | 9,540 (6,640–24,800), 10a,b | 1840 | |
| YJS5845 | c/k, c/k | Parental (C/I) | 2.6 (2.07–2.8), 16a,b | 0.50 | |
| 5845-7a | k-c | C | 0.99 (0.56–1.1), 10c,d | 0.19 | |
| 5845-16 | k-k | C | 2.1 (1.6–2.7), 10a,b | 0.40 | |
| 5845-27a | k-k | C | 2.1 (1.2–3.7), 10a,b | 0.41 | |
| 5845-35a | k-k | C | 5.8 (3.1–14), 15b | 1.1 | |
| 5845-19a | k-k | C | 6.0 (3.8–7.4), 15b | 1.2 | |
| 5845-22a | k-k | C | 7.2 (5.2–9.4), 15b | 1.3 | |
| 5845-21a | c-k | I | 2.7 (0.53–10), 16b,c | 0.38 (0.32–0.94), 36b,c | 0.51 |
| 5845-20a | c-k | I | 3.6 (0.9–8.4), 11b | 0.69 | |
| 5845-30a | c-k | I | 16 (7.6–26), 16a,b | 3.2 | |
| 5845-19 | c-k | I | 141 (86–320), 16a,b | 27 | |
| 5845-18a | c-k | I | 330 (253–1,300), 10a,b | 64 | |
| YJS5885 | c/k, c/k | Parental (C/I) | 2.6 (1.4–6.5), 15b,c | 0.51 | |
| 5885-1a | k-c | C | 0.74 (0.54–1.2), 15a,b | 0.14 | |
| 5885-14a | c-c | C | 1.1 (0.44–3.4), 10a,b | 0.21 | |
| 5885-20b | k-c | C | 1.5 (1–1.6), 13a,b | 0.29 | |
| 5885-5b | k-k | C | 1.6 (0.53–2.2), 15a,b | 0.31 | |
| 5885-10a | k-k | C | 2.1 (1.2–3.5), 25a,b | 1.6 (0.65–2.6), 34a,b | 0.41 |
| 5885-19b | k-c | C | 2.5 (1.4–3.2), 25a,b | 1.4 (0.75–2.6), 34a,b | 0.48 |
| 5885-15b | c-c | C | 13 (8.6–18), 25a,b | 8.6 (5.8–13), 36b,c | 2.5 |
| 5885-6a | c-c | C | 62 (46–97), 10a | 12 | |
| 5885-9a | c-k | I | 17 (11–23), 15a,b | 3.3 | |
| 5885-4b | c-k | I | 24 (16–29), 15a,b | 4.6 | |
| 5885-16a | c-k | I | 85 (37–222), 10a | 16 | |
| 5885-19a | c-k | I | 86 (20–130), 10a | 17 |
The isolates YJS5845 and YJS5885, and derived spore clones, were transformed with pEAA613 (ARS–CEN URA3 promoter-kanMX::insE-A14). Four to six independent cultures of each transformant were examined for reversion to geneticin resistance as described in the Materials and Methods. Median mutation rates are presented with 95% C.I.s and relative mutation rates compared to EAY1369 (S288c compatible) are shown. n = number of independent repetitions. The genotype of the isolates and spore clones is presented with respect to the S288c (c) or SK1 (k) amino acid position in amino acid 761 in Mlh1 and 818/822 in Pms1 (Figure 1). Heterozygous genotypes are indicated by the “/.” In this nomenclature, c-c = S288c genotype, k-k = SK1, k-c = ancestral, and c-k = incompatible. **For 5845-21a, 5845-41a, 5885-10a, 5885-15b, and 5885-19b spore clones there were four (of eight tested), four (of eight tested), two (of seven tested), two (of seven tested), and two (of six tested) transformants, respectively, that gave low reversion rates (see Materials and Methods for details). The rates of reversion to G418r in this column include the data from the low-reversion transformants; however, the column marked with * does not include these transformants, nor does the relative rate column. C, compatible; I, incompatible.
Significantly different from EAY1369 (P < 0.001, Mann–Whitney U-test).
Significantly different from EAY1370 (P < 0.001, Mann–Whitney U-test).
Significantly different from EAY1369 (P < 0.01, Mann–Whitney U-test).
Significantly different from EAY1370 (P < 0.01, Mann–Whitney U-test). YJS5845 compatible and incompatible spore clones (P < 0.001), and YJS5885 compatible and incompatible spore clones (P < 0.001), are significantly different from each other (Mann–Whitney U-test).
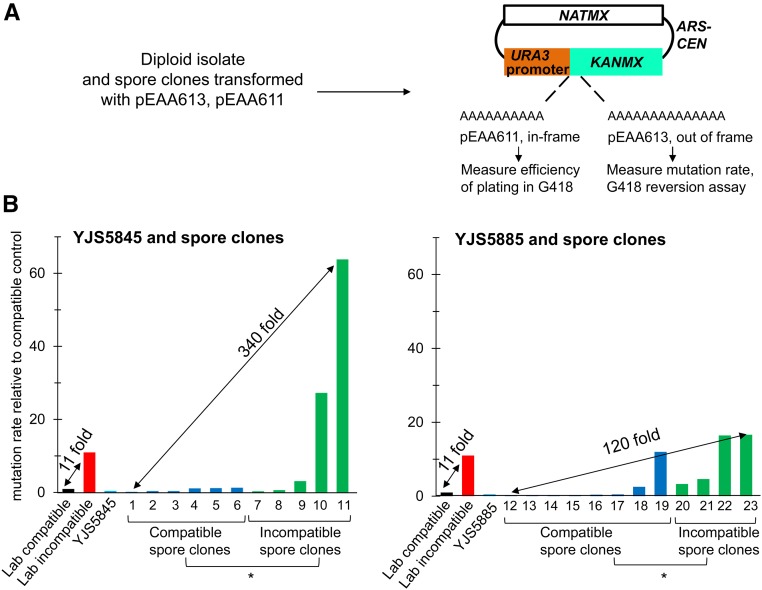
To measure mutation rate, isolates and spore clones were transformed with pEAA613, a mutation rate reporter vector that measures reversion of an A14 sequence inserted into the KANMX gene (Figure 3A). We confirmed that in these isolates and spore clones, resistance to G418 resulted in frameshift mutations in the A14 sequence that restored the KANMX reading frame (Figure S4). Mutation rates in EAY1369 (compatible) and EAY1370 (incompatible) controls were highly reproducible when measured with independent transformations and repetitions on different days. YJS5885 and YJS5845 were nonmutators but yielded spore clones with a range of mutation rates in a DNA slippage assay that varied over ∼340-fold (Figure 3B and Table 3). Mutation rates of incompatible spore clones were significantly different from compatible spore clones in both YJS5845 and YJS5885, as determined by the Mann–Whitney U-test (P < 0.001, Figure 3B and Table 3). This indicates that the cMLH1-kPMS1 genotype is the major contributor to high mutation rates, though there were examples where spore clones compatible for MLH1-PMS1 showed mutation rates similar to that observed in incompatible strains and vice versa (for example, see analyses of spore clones 5885-6a and 5845-21a in Figure 3B and Table 3). Several compatible and incompatible spore clones had significantly different mutation rates compared to EAY1369 (laboratory compatible) and EAY1370 (laboratory incompatible). As shown in Figure 3B and Table 3, up to a sevenfold lower mutation rate was seen between compatible spore clones and strain EAY1369, and up to a sixfold higher mutation rate was seen between incompatible spore clones and strain EAY1370. These data indicate that modifiers are present in the isolates that impact mutation rate (Figure 3B and Table 3).
Figure 3.
Mutation rate assay. (A) Isolates and spore clones were transformed with pEAA613, an ARS-CEN, URA3 promoter-KanMX::insE-A14 plasmid used to measure mutation rates in natural yeast isolates that contains a NatMX selectable marker and a frameshift reporter in which the insE-A14 sequence from Tran et al. (1997) was inserted immediately after methionine 17 in the KanMX open reading frame. pEAA611 is in-frame for KanMX whereas pEAA613 contains a +1-frameshift mutation that disrupts KanMX function. Frameshift mutation events (e.g., a −1 deletion in the homopolymeric A run) are detected on YPD plates containing clonNAT and geneticin (G418). Spore clones were first screened for efficiency of plating in G418 by transforming with pEAA611 that contains KanMX::insE-A10. (B) Mutation rates of YJS5845, YJS5885, and their spore clones in a G418 reversion assay relative to the compatible S288c-derived strain EAY1369. The parental isolates YJS5845 and YJS5885 are in light blue, compatible spore clones are in blue, incompatible spore clones are in green, the EAY1369 compatible laboratory strain is in black, and the EAY1370 incompatible laboratory strain is in red. Compatible and incompatible spore clones from YJS5845 and YJS5885 were significantly different from each other, as determined by Mann–Whitney U-test (* P < 0.001). The spore clones are named as follows: 1, 5845-7a; 2, 5845-16; 3, 5845-27a; 4, 5845-35a; 5, 5845-19a; 6, 5845-22a; 7, 5845-21a; 8, 5845-20a; 9, 5845-30a; 10, 5845-19; 11, 5845-18a; 12, 5885-1a; 13, 5885-14a; 14, 5885-20b; 15, 5885-5b; 16, 5885-10a; 17, 5885-19b; 18, 5885-15b; 19, 5885-6a; 20, 5885-9a; 21, 5885-4b; 22, 5885-16a; and 23, 5885-19a
YJM521 displayed poor efficiency of plating with pEAA611 (a plasmid containing an in-frame homopolymeric A10 run in the KANMX gene; Figure S3). We also found that the previously characterized isolate YJM555 (Bui et al. 2017), a spore clone of YJM523, also displayed inefficient plating with pEAA611. Thus, it was not possible to accurately measure mutation rates in these isolates or their spore clones using pEAA613 (Table 3). However, YJM521 is unlikely to be a strong mutator because it is homozygous for the MLH1-P271 suppressor allele (Table 2), and YJM555 was shown not to be a mutator using a 5-FOA-based reversion assay (Skelly et al. 2017) and had acquired suppressor mutations (Bui et al. 2017).
We also measured the rate of resistance to 5-FOA in haploid YJS5885 spore clones (see FACS analysis below). This resistance results primarily from recessive base substitution mutations in the URA3 gene that confer loss-of-function (Lang and Murray 2008). Zeyl and DeVisser (2001) observed a 152-fold difference in the rate of 5-FOAr between wild-type and msh2 haploid strains, and Thompson et al. (2006) reported a 10-fold difference between wild-type and mutator strains. We observed a 20-fold difference between the haploid wild-type in the S288c background (7.9 × 10−8) and an mlh1Δ derivative (1.6 × 10−6; Table S4). Interestingly, we observed a 13-fold range in mutation rate in a set of seven YJS5885 spore clones (4.8 × 10−8 – 6.4 × 10−7; Table S4). We sequenced the URA3 open reading frame from 13 5-FOA-resistant colonies (Table S4). Ten independent URA3 mutations were detected: three missense, five nonsense, and two single-base deletions. There were no mutations identified in three mutants. Similar percentages of 5-FOAr colonies containing mutations in the URA3 gene were seen in our study (77%) and an earlier one (87%; Lang and Murray 2008). The rank order of mutation rates for the YJS5885 spore clones in the 5-FOAr and kanMX::insE-A14 reversion assays did not precisely correlate (Table 3 and Table S4). This result is not surprising because the two assays measure different mutation spectra, with the kanMX::insE-A14 reversion assay detecting DNA slippage events in homopolymeric repeats and the 5-FOAr assay detecting primarily base substitutions (Lang and Murray 2008).
YJS5845 and YJS5885 spore clones are diploid and haploid, respectively
Wild yeasts are primarily homothallic, indicating that daughter cells can switch mating type and mate with mother cells (Mortimer 2000; Butler et al. 2004). YJM521 and YJM523 are homothallic (Clemons et al. 1997), and we hypothesized that YJS5845 is also homothallic because it yielded spore clones (12 of 15) that can sporulate (Table S5). It appears that YJS5885 is functionally heterothallic because most of its spore clones (24 of 26) were unable to sporulate. We tested the ploidy of YJS5845 and YJS5885 spore clones, for which we measured mutation rates by flow cytometry, and showed that YJS5845 spore clones were diploid while those of YJS5885 were haploid (Figure 4A and Figure S5). There are no disruptions in the open reading frame of the HO gene, which codes for the endonuclease involved in mating type switching in both YJS5845 and YJS5885 (Nasmyth 1993; Table S3). However, there is a predicted deleterious variant in the HO gene of YJS5845 but not in YJS5885 (Table S3). To determine if defects in sporulation correlated to growth deficiency or defects in mitochondria (normal mitochondrial function is necessary for meiosis; Gorsich and Shaw 2004), spore clones from YJS5845 and YJS5885 were analyzed for their ability to grow on media containing lactate as a carbon source. Three of 14 YJS5845 spore clones and 9 of 19 YJS5885 spore clones failed to grow with lactate as a carbon source (Table S5). This explains why two of the YJS5845 spore clones were unable to sporulate, but we believe that YJS5885 is functionally heterothallic because its spore clones did not diploidize (Figure 4A and Figure S5).
Spore clones from saturated cultures were prepared for flow cytometry and subsequently examined by light microscopy to determine the populations of single, small-budded, and large-budded cells in each sample. We observed that YJS5885 spore clones had a higher proportion of large-budded cells (34–87% large-budded cells as compared to < 1% large-budded cells in the isolate YJS5885; Figure 4A and Figure S5), suggesting a possible activation of a G2/M checkpoint or a cell division defect. Some cells of spore clones were much larger than the cells from the original isolate (5885-9a, 20b, and 5b), which might be due to higher DNA content in the spore clones or the possible defects outlined above. Spore clones of YJS5845 were primarily single unbudded cells and appeared similar to the parental isolate.
Spore clones of YJS5845 display chromosome gain and loss
Whole-genome sequence analyses of YJS5845 and YJS5885, and their spore clones, indicated that most of the spore clones are euploid (Figure 4B and Figure S6). Interestingly, YJS5845 and 2 of 16 spore clones analyzed, 5845-18a and 5845-28b, displayed aneuploidy, with gains of a chromosome in YJS5845 (XIV) and 5845-18a (XI), and a loss of a chromosome in 5845-28b (I). The YJS5845 stock that we obtained is a mixture of euploid and aneuploid cells (trisomy in chromosome XIV that contains PMS1), as determined by sequencing the PMS1 gene from several single colonies struck out from our original stock. Based on the heterozygosity in the PMS1 sequence, we determined that of the 14 single YJS5845 colonies sequenced for PMS1, six had three copies (indicating aneuploidy) and eight had two copies of PMS1 (indicating euploidy). However, the sporulation efficiency and viability of euploid and aneuploid YJS5845 were very similar (Table 1). The majority of the spore clones (except for 5845-7a, 16, and 19) were isolated from the euploid YJS5845. YJS5885 is euploid and all spore clones derived from YJS5885 were also euploid (Figure S6).
Discussion
Spore clones of human clinical isolates yield a wide range of mutator phenotypes
Mutator phenotypes can be challenging to analyze in natural isolates because they lack genetic markers, display differences in growth, show colony variation, and can have differential resistance to antibiotics. We overcame these hurdles to measure mutation rates in YJS5845 and YJS5885 isolates, and their spore clone derivatives, using a recently developed frameshift mutation reporter. We found that while the isolates themselves were nonmutators, they sporulated at high efficiency and their spore clones displayed a wide range of mutation rates, with a 340-fold difference between the lowest and highest mutator using a DNA slippage assay. cMLH1-kPMS1 incompatibility was the major contributor of the high mutation rate (Figure 3 and Table 3). This study provides a practical and efficient strategy to characterize the phenotypic properties of human clinical yeast isolates. Other strategies, such as bottleneck analysis of large numbers of independent isolates, are also possible but are time consuming and would require a significant effort to analyze a large amount of whole-genome sequencing data.
Opposing forces of genetic drift and natural selection ensure that most individuals in a population evolve to a basal nonzero mutation rate [reviewed in Lynch et al. (2016)]. For the natural isolates analyzed in this study, we hypothesize that stress can induce sporulation, giving rise to individuals that display high deviations from the basal mutation rate. Under unpredictable and changing stress conditions in the human host, it is likely to be valuable to produce spores with a broad range of mutation rates that will ensure population survival, but such a range might not be optimal for individual cells. Spore clones with a high mutation rate might rapidly gain beneficial mutations but will also acquire deleterious mutations. Thus, having a range of mutation rates gives the population an opportunity to survive changing stress conditions. To prevent long-term fitness costs, these strains can either acquire suppressor mutations to become nonmutators, or mate back with the compatible strains or outcross to become diploid nonmutators. The compatible mutants surviving stress would also have a higher mutation rate, but it is reasonable to assume that diploids resulting from mating of compatible and incompatible spore clones would have complementary recessive mutations, and thus a nonmutator phenotype (as seen for the low spore viability in YJS5845 and YJS5885).
Outcrossing with different isolates is thought to occur at a low frequency in the wild, once in every 50,000 generations (Ruderfer et al. 2006; Magwene et al. 2011). Stressful environments likely influence this rate, with levels estimated to be as high as one in every 100 to one in every two generations (Marsit and Dequin 2015). While such estimates are valuable, we hypothesize that the YJS5845 and YJS5885 isolates sporulated in stress conditions could yield mutator clones that are capable of mating with siblings, and thus do not require outcrossing to become compatible.
Modifiers of incompatibility genotypes in clinical isolates
The mutation rates of compatible and incompatible spore clones of YJS5845 and YJS5885 vary significantly from their corresponding laboratory strains (Figure 3 and Table 3). This finding indicates the presence of enhancers and suppressors of mutation rate in the YJS5885 and YJS5845 isolate backgrounds. Previously we identified both intragenic (MLH1-P271; Demogines et al. 2008) and extragenic modifiers (Bui et al. 2017) of the cMLH1-kPMS1 incompatibility. In the case of the homozygous incompatible spore clone, YJM555, we observed what appeared to be multiple intergenic modifiers of the cMLH1-kPMS1 incompatibility (Bui et al. 2017; Skelly et al. 2017).
Is there evidence for SNPs in MLH1 conferring a mutator phenotype or acting as intragenic modifiers of the cMLH1-kPMS1 incompatibility? Of the 1011 yeast isolates analyzed by Bui et al. (2017), 107 display heterozygosities in the MLH1 and PMS1 genes. This information, and the fact that the yeast MLH1 gene has been extensively mutagenized through alanine scanning, random mutagenesis, and site-specific mutagenesis (both in known domains and in homology modeling to Hereditary nonpolyposis colorectal cancer (HNPCC) alleles; see Table S6), encouraged us to determine if any MMR alleles exist in heterozygous isolates that could confer a deleterious phenotype. As shown in Tables S6 and S7, we mapped heterozygosities present in 107 (including a newly identified one) isolates onto the MLH1 structure–function map and found that some cluster to regions in MLH1 predicted to affect function. These observations provide support for the idea that mutators in yeast grown in stressful conditions could be obtained through incompatibilities as well as the presence of recessive deleterious alleles.
Yeast growing in a human environment are likely to encounter multiple stresses
Human clinical isolates of the fungal pathogen Cryptococcus have been identified that display a mutator phenotype due to mutations in the MSH2 MMR gene (Billmyre et al. 2017; Boyce et al. 2017). The authors of these studies suggest that pathogens undergo a significant change in environment when entering a human host and that challenges specific to the human host, such as survival in the presence of antifungals, provide an adaptive advantage for Cryptococcus isolates that are mutators (Billmyre et al. 2017; Boyce et al. 2017). In a human environment, S. cerevisiae is likely to deal with challenges that are analogous to those faced by Cryptococcus, including growth at high temperature and exposure to fungicides. In fact, several causative alleles were identified in clinical and laboratory yeast strains that provide growth advantages at high temperature (Steinmetz et al. 2002; Sinha et al. 2008). As summarized in the Introduction, many human clinical isolates of S. cerevisiae are mosaics (including YJS5845 and YJS5885) that contain a mixture of alleles from different subpopulations (Liti et al. 2009; Perez-Torrado and Querol 2015; Strope et al. 2015; Skelly et al. 2017).
Our study of YJS5845 and YJS5885 highlights how isolates can adapt to stressful human environments. The incompatible alleles do not confer a mutator phenotype in these heterozygotes but may provide an advantage, as they appear to be poised to adapt to stress through the variable mutation rates in the progeny (Figure 3B and Table 3). This variation in mutation rates is due to the presence of modifiers in the background of these strains as a consequence of being heterozygotes. Furthermore, one of the isolates and its spore clones show evidence of losing and gaining chromosomes at high frequency (Figure 4B), which could also be highly beneficial for rapid adaptation in the clinical setting by affecting mutation rates. Variance in mutation rate, at least for baker’s yeast, is likely to provide only a transient advantage due to fitness costs, and changes in environment would need to be accompanied by the acquisition of suppressor mutations, mating with nearby spore clones, return to euploidy, or by outcrossing to become nonmutators.
Ploidy, mutation rate, and adaptation
Chromosome aneuploidy and increases-in-ploidy have been shown to provide faster routes to adaptation, by increasing mutation rate and increasing the likelihood of gaining a beneficial mutation (Selmecki et al. 2009; Pavelka et al. 2010; Rancati and Pavelka 2013; Zörgö et al. 2013; Zhu et al. 2014; Gerstein and Berman 2015). For example, compared to haploid and diploid baker’s yeast, tetraploid yeasts display more rapid adaptation, suggesting that increased ploidy impacts the rate of adaptation by providing a broad spectrum of adaptive mutations (Selmecki et al. 2015; Scott et al. 2017). Consistent with this observation, diploid mutators display a growth advantage over diploid and haploid nonmutators, and haploid mutators, in several stress conditions (Thompson et al. 2006). In our study, we identified both haploid (YJS5885-derived) and diploid (YJS5845-derived) spore clone progeny, suggesting that ploidy may also play a role in regulating mutation rates of spore clones.
Zhu et al. (2014, 2016) analyzed 132 clinical isolates of S. cerevisiae by whole-genome sequencing and found that roughly one-third had higher levels of ploidy (3n and 4n genome copy number), one-quarter had partial chromosome copy-number variations, and one-third were aneuploid. We observed that our YJS5845 stock consisted of a mix of euploid and aneuploid cells. Furthermore, euploid YJS5845 yielded aneuploid spore clones, either through meiotic or mitotic chromosome segregation defects (Figure 4B). Interestingly, YJS5845 belongs to an admixture clade; thus, it is reasonable to assume that the chromosome segregation defects seen in this isolate are due to incompatibilities in processes involving a large number of components. While we do not have direct evidence that the aneuploidy seen in YJS5845 impacts mutation rate, work from the studies referenced above suggest that it is likely to play a significant role.
Acknowledgments
We thank members of the Alani laboratory for their helpful comments and advice, V. P. Ajith and K. T. Nishant at Indian Institute of Science Education and Research Trivandrum for sharing the aneuploidy script and for their help in analyzing whole-genome sequencing data, Christopher Donahue at the Flow Cytometry Core Laboratory for his help in running the flow cytometry and sorting experiments, and Peter Schweitzer and Jeff Glaubitz at the Biotechnology Resource Center sequencing facility for performing whole-genome sequencing. J.S. is a member of the Institut Universitaire de France. V.R., D.T.B., N.A.-S., and E.A. were supported by National Institutes of Health (NIH) grant GM-053085. C.F.A. was supported by NIH grant GM-095793. The 1011 Yeast Genomes Project (J.S. and A.F.) was funded by France Génomique (ANR-10-INBS-09-08). D.T.B. was a fellow of the Vietnam Education Foundation. N.A.-S. was supported by a scholarship from the Saudi Arabian Cultural Mission. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the NIH.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7233104.
Communicating editor: J. Nickoloff
Literature Cited
- Argueso J. L., Kijas A. W., Sarin S., Heck J., Waase M., et al. , 2003. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol. 23: 873–886. 10.1128/MCB.23.3.873-886.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre R. B., Clancey S. A., Heitman J., 2017. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. Elife 6: e28802 10.7554/eLife.28802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L., Danielsen M., Knudsen S., Petersen J. B., Maymann J., et al. , 2000. The frequency of mutators in populations of Escherichia coli. Mutat. Res. 448: 47–55. 10.1016/S0027-5107(99)00239-0 [DOI] [PubMed] [Google Scholar]
- Boyce K. J., Wang Y., Verma S., Shakya V. P. S., Xue C., et al. , 2017. Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. MBio 8: e00595-17 10.1128/mBio.00595-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui D. T., Dine E., Anderson J. B., Aquadro C. F., Alani E. E., 2015. A genetic incompatibility accelerates adaptation in yeast. PLoS Genet. 11: e1005407 10.1371/journal.pgen.1005407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui D. T., Friedrich A., Al-Sweel N., Liti G., Schacherer J., et al. , 2017. Mismatch repair incompatibilities in diverse yeast populations. Genetics 205: 1459–1471. 10.1534/genetics.116.199513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Kenny C., Fagan A., Kurischko C., Gaillardin C., et al. , 2004. Evolution of the MAT locus and its HO endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 101: 1632–1637. 10.1073/pnas.0304170101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Botstein D., 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135: 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L., Cox E. C., 1983. Competition between high and low mutating strains of Escherichia coli. Evolution 37: 125–134. 10.1111/j.1558-5646.1983.tb05521.x [DOI] [PubMed] [Google Scholar]
- Clemons K. V., Park P., McCusker J. H., McCullough M. J., Davis R. W., et al. , 1997. Application of DNA typing methods and genetic analysis to epidemiology and taxonomy of Saccharomyces isolates. J. Clin. Microbiol. 35: 1822–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G. A., Dudley A. M., 2015. Aneuploidy: tolerating tolerance. Curr. Biol. 25: R771–R773. 10.1016/j.cub.2015.06.056 [DOI] [PubMed] [Google Scholar]
- Demogines A., Wong A., Aquadro C., Alani E., 2008. Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLoS Genet. 4: e1000103 10.1371/journal.pgen.1000103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E., Lecointre G., Darlu P., Tenaillon O., Acquaviva C., et al. , 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103: 711–721. 10.1016/S0092-8674(00)00175-6 [DOI] [PubMed] [Google Scholar]
- Dixon W. J., Massey F. J., 1969. Introduction to Statistical Analysis. McGraw-Hill, New York. [Google Scholar]
- Ernst J. F., Chan R. K., 1985. Characterization of Saccharomyces cerevisiae mutants supersensitive to aminoglycoside antibiotics. J. Bacteriol. 163: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A. C., Berman J., 2015. Shift and adapt: the costs and benefits of karyotype variations. Curr. Opin. Microbiol. 26: 130–136. 10.1016/j.mib.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., 2007. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 38–41. 10.1038/nprot.2007.15 [DOI] [PubMed] [Google Scholar]
- Giraud A., Matic I., Tenaillon O., Clara A., Radman M., et al. , 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291: 2606–2608. 10.1126/science.1056421 [DOI] [PubMed] [Google Scholar]
- Gorsich S. W., Shaw J. M., 2004. Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell 15: 4369–4381. 10.1091/mbc.e03-12-0875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. A., Noverr M. C., 2017. Fungal interactions with the human host: exploring the spectrum of symbiosis. Curr. Opin. Microbiol. 40: 58–64. 10.1016/j.mib.2017.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J. A., Argueso J. L., Gemici Z., Reeves R. G., Bernard A., et al. , 2006. Negative epistasis between natural variants of the Saccharomyces cerevisiae MLH1 and PMS1 genes results in a defect in mismatch repair. Proc. Natl. Acad. Sci. USA 103: 3256–3261. 10.1073/pnas.0510998103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F., 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. 10.1016/0378-1119(87)90131-4 [DOI] [PubMed] [Google Scholar]
- Hose J., Yong C. M., Sardi M., Wang Z., Newton M. A., et al. , 2015. Dosage compensation can buffer copy-number variation in wild yeast. Elife 4: e05462 10.7554/eLife.05462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariola R., Otway R., Lonnqvist K. E., Raevaara T. E., Macrae F., et al. , 2003. Two mismatch repair gene mutations found in a colon cancer patient–which one is pathogenic? Hum. Genet. 112: 105–109. 10.1007/s00439-002-0866-4 [DOI] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178: 67–82. 10.1534/genetics.107.071506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc J. E., Li B., Payne W. L., Cebula T. A., 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274: 1208–1211. 10.1126/science.274.5290.1208 [DOI] [PubMed] [Google Scholar]
- Liti G., Louis E. J., 2005. Yeast evolution and comparative genomics. Annu. Rev. Microbiol. 59: 135–153. 10.1146/annurev.micro.59.030804.121400 [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Ackerman M. S., Gout J. F., Long H., Sung W., et al. , 2016. Genetic drift, selection and the evolution of the mutation rate. Nat. Rev. Genet. 17: 704–714. 10.1038/nrg.2016.104 [DOI] [PubMed] [Google Scholar]
- Magwene P. M., Kayikci O., Granek J. A., Reininga J. M., Scholl Z., et al. , 2011. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 1987–1992. 10.1073/pnas.1012544108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H. B., Whitney D. R., 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18: 50–60. 10.1214/aoms/1177730491 [DOI] [Google Scholar]
- Marsit S., Dequin S., 2015. Diversity and adaptive evolution of Saccharomyces wine yeast: a review. FEMS Yeast Res. 15: fov067 10.1093/femsyr/fov067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., 2000. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 10: 403–409. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., 1993. Regulating the HO endonuclease in yeast. Curr. Opin. Genet. Dev. 3: 286–294. [DOI] [PubMed] [Google Scholar]
- Nishant K. T., Wei W., Mancera E., Argueso J. L., Schlattl A., et al. , 2010. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 6: e1001109 10.1371/journal.pgen.1001109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N., Rancati G., Zhu J., Bradford W. D., Saraf A., et al. , 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Torrado R., Querol A., 2015. Opportunistic strains of Saccharomyces cerevisiae: a potential risk sold in food products. Front. Microbiol. 6: 1522 10.3389/fmicb.2015.01522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J., De Chiara M., Friedrich A., Yue J. X., Pflieger D., et al. , 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G., Pavelka N., 2013. Karyotypic changes as drivers and catalyzers of cellular evolvability: a perspective from non-pathogenic yeasts. Semin. Cell Dev. Biol. 24: 332–338. 10.1016/j.semcdb.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Raynes Y., Gazzara M. R., Sniegowski P. D., 2011. Mutator dynamics in sexual and asexual experimental populations of yeast. BMC Evol. Biol. 11: 158 10.1186/1471-2148-11-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Yvert G., 2014. How does evolution tune biological noise? Front. Genet. 5: 374 10.3389/fgene.2014.00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Winston F., Hieter P., 1990. Methods in Yeast Genetics–A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rosebrock, A. P., 2017 Analysis of the budding yeast cell cycle by flow cytometry. Cold Spring Harb. Protoc. 2017: [DOI] [PubMed]
- Ruderfer D. M., Pratt S. C., Seidel H. S., Kruglyak L., 2006. Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38: 1077–1081. 10.1038/ng1859 [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345. 10.1038/nature07670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A. L., Richmond P. A., Dowell R. D., Selmecki A. M., 2017. The influence of polyploidy on the evolution of yeast grown in a sub-optimal carbon source. Mol. Biol. Evol. 34: 2690–2703. 10.1093/molbev/msx205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A. M., Dulmage K., Cowen L. E., Anderson J. B., Berman J., 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5: e1000705 10.1371/journal.pgen.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A. M., Maruvka Y. E., Richmond P. A., Guillet M., Shoresh N., et al. , 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352. 10.1038/nature14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha H., David L., Pascon R. C., Clauder-Munster S., Krishnakumar S., et al. , 2008. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180: 1661–1670. 10.1534/genetics.108.092932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirr A., Cromie G. A., Jeffery E. W., Gilbert T. L., Ludlow C. L., et al. , 2015. Allelic variation, aneuploidy, and nongenetic mechanisms suppress a monogenic trait in yeast. Genetics 199: 247–262. 10.1534/genetics.114.170563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly D. A., Magwene P. M., Meeks B., Murphy H. A., 2017. Known mutator alleles do not markedly increase mutation rate in clinical Saccharomyces cerevisiae strains. Proc. Biol. Sci. 284: 20162672 10.1098/rspb.2016.2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. 10.1038/416326a [DOI] [PubMed] [Google Scholar]
- Strope P. K., Skelly D. A., Kozmin S. G., Mahadevan G., Stone E. A., et al. , 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25: 762–774. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine A. B., Payen C., Ong G. T., Liachko I., Tan K. M., et al. , 2015. The fitness consequences of aneuploidy are driven by condition-dependent gene effects. PLoS Biol. 13: e1002155 10.1371/journal.pbio.1002155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F., Radman M., Maynard-Smith J., Toupance B., Gouyon P. H., et al. , 1997. Role of mutator alleles in adaptive evolution. Nature 387: 700–702. 10.1038/42696 [DOI] [PubMed] [Google Scholar]
- Tanaka M. M., Bergstrom C. T., Levin B. R., 2003. The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics 164: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. A., Desai M. M., Murray A. W., 2006. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Curr. Biol. 16: 1581–1590. 10.1016/j.cub.2006.06.070 [DOI] [PubMed] [Google Scholar]
- Townsend J. P., Nielsen K. M., Fisher D. S., Hartl D. L., 2003. Horizontal acquisition of divergent chromosomal DNA in bacteria: effects of mutator phenotypes. Genetics 164: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H. T., Keen J. D., Kricker M., Resnick M. A., Gordenin D. A., 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17: 2859–2865. 10.1128/MCB.17.5.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickert S., Finck M., Herz B., Ernst J. F., 1998. A small protein (Ags1p) and the Pho80p-Pho85p kinase complex contribute to aminoglycoside antibiotic resistance of the yeast Saccharomyces cerevisiae. J. Bacteriol. 180: 1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxon F., 1945. Individual comparisons by ranking methods. Biom. Bull. 1: 80–83. 10.2307/3001968 [DOI] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288c. Yeast 11: 53–55. 10.1002/yea.320110107 [DOI] [PubMed] [Google Scholar]
- Zeyl C., DeVisser J. A., 2001. Estimates of the rate and distribution of fitness effects of spontaneous mutation in Saccharomyces cerevisiae. Genetics 157: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. O., Siegal M. L., Hall D. W., Petrov D. A., 2014. Precise estimates of mutation rate and spectrum in yeast. Proc. Natl. Acad. Sci. USA 111: E2310–E2318. 10.1073/pnas.1323011111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. O., Sherlock G., Petrov D. A., 2016. Whole genome analysis of 132 clinical Saccharomyces cerevisiae strains reveals extensive ploidy variation. G3 (Bethesda) 6: 2421–2434. 10.1534/g3.116.029397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörgö E., Chwialkowska K., Gjuvsland A. B., Garré E., Sunnerhagen P., et al. , 2013. Ancient evolutionary trade-offs between yeast ploidy states. PLoS Genet. 9: e1003388 10.1371/journal.pgen.1003388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko E. I., Zubko M. K., 2014. Deficiencies in mitochondrial DNA compromise the survival of yeast cells at critically high temperatures. Microbiol. Res. 169: 185–195. 10.1016/j.micres.2013.06.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request, and the entire DNA sequences of the MLH1 and PMS1 genes from 1011 isolates are presented in Bui et al. (2017). Supporting information contains all detailed descriptions of supplemental files. All wet laboratory experiments presented (lys2-A14 reversion, kanMX::insE-A14 reversion, 5-FOA resistance) were repeated on at least 2 separate days. The following figures and tables can be found in the Genetics Society of America Figshare portal:
Figure S1: incompatibility involving the MLH1 and PMS1 MMR genes.
Figure S2: DNA sequence, as shown by chromatogram traces, of the MLH1 incompatibility site (bp 2282, Gly or Asp at amino acid 761) in the indicated isolates and spore clones.
Figure S3: efficiency of plating of strains transformed with pEAA611, comparing growth on clonNAT and clonNAT + G418 plates.
Figure S4: sequencing analysis of G418-resistant revertants and sensitive control colonies.
Figure S5: flow cytometry of spore clones.
Figure S6: ploidy of YJS5845 and YJS5885 isolates, and their spore clones.
Table S1: genotyping of spore clones obtained by dissection of isolate tetrads.
Table S2: genotyping of MLH1 and PMS1 loci in YJM and YJS isolates, and derived spore clones.
Table S3: analysis of HO, PHO80, and STP22 genes in YJS5845 and YJS5885 for variants using SnpEff.
Table S4: analysis of resistance to 5-FOA in YJS5885 spore clones.
Table S5: sporulation and lactate growth phenotype.
Table S6: assigning MLH1 polymorphisms found in heterozygous genotypes onto the MLH1 structure–function map.
Table S7: amino acid heterozygosities identified in MLH1 in 107 yeast isolates.
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7233104.