Abstract
Triacylglycerol (TAG) is the most important caloric source with respect to energy homeostasis in animals. In addition to its evolutionarily conserved importance as an energy source, TAG turnover is crucial to the metabolism of structural and signaling lipids. These neutral lipids are also key players in development and disease. Here, we review the metabolism of TAG in the Drosophila model system. Recently, the fruit fly has attracted renewed attention in research due to the unique experimental approaches it affords in studying the tissue-autonomous and interorgan regulation of lipid metabolism in vivo. Following an overview of the systemic control of fly body fat stores, we will cover lipid anabolic, enzymatic, and regulatory processes, which begin with the dietary lipid breakdown and de novo lipogenesis that results in lipid droplet storage. Next, we focus on lipolytic processes, which mobilize storage TAG to make it metabolically accessible as either an energy source or as a building block for biosynthesis of other lipid classes. Since the buildup and breakdown of fat involves various organs, we highlight avenues of lipid transport, which are at the heart of functional integration of organismic lipid metabolism. Finally, we draw attention to some “missing links” in basic neutral lipid metabolism and conclude with a perspective on how fly research can be exploited to study functional metabolic roles of diverse lipids.
Keywords: Drosophila fat, triacylglycerol, lipogenesis, lipolysis, lipases, lipid transport, FlyBook
ALL animals are faced with large fluctuations in energy intake and demand on diurnal to seasonal scales. To maintain energy homeostasis during nonfeeding developmental stages or upon metabolic stress, animals have developed the ability to store a surplus of calories, which serve as flexible on-demand energy depots. The quantitatively most important energy reservoirs in many metazoans, including those in the model invertebrate Drosophila melanogaster, include the carbohydrate glycogen and neutral triacylglycerol (TAG) lipids. TAG represents the most concentrated form of chemically bound energy. On the one hand, the carbon atoms of TAG are in more reduced states than the carbon atoms of carbohydrates. This results in a greater caloric yield upon complete oxidation of TAG. Moreover, the weight to energy content ratio of TAG is favorable due to its apolar nature, which unlike glycogen allows its anhydrous storage in specialized intracellular organelles called lipid droplets (LDs) (Thiam et al. 2013; Zechner et al. 2017). Importantly, the metabolic function of TAG is not limited to energy storage. Fatty acid (FA) moieties stored in the form of TAG may also serve as building blocks for structural membrane lipids or signaling molecules. TAG also represents an essential metabolic sink that sequesters excess carbon units from dietary sugar or fat, which stress cells via their conversion to lipo- or glucotoxic metabolic intermediates (Garrido et al. 2015; Welte and Gould 2017; Zechner et al. 2017). These principal biological functions of TAG have been highly conserved during evolution, and are similar in Drosophila and higher metazoans. This makes the fruit fly an increasingly popular model to study the general (patho)physiology of TAG metabolism [reviewed in Baker and Thummel (2007), Owusu-Ansah and Perrimon (2014), and Musselman and Kühnlein (2018)].
Reflecting its central role in metabolic homeostasis, dynamic changes in TAG levels characterize all stages of Drosophila development (Carvalho et al. 2012; Guan et al. 2013). The accumulation and catabolism of TAG stores are essential for successful oogenesis and embryogenesis, respectively (Buszczak et al. 2002; Grönke et al. 2005). Embryos are provided with maternal TAG stores, which are largely depleted during embryogenesis (Carvalho et al. 2012; Guan et al. 2013; Tennessen et al. 2014). This catabolic phase is followed by a massive buildup of TAG stores during larval growth that ceases in the postfeeding stages of metamorphosis (Carvalho et al. 2012; Guan et al. 2013). In larval and adult stages, TAG stores represent an essential reservoir for surviving starvation (Grönke et al. 2003, 2007; Gutierrez et al. 2006; Bi et al. 2012). When food is abundant, complex diet–genotype interactions influence TAG storage in adult Drosophila, and the dietary history of parental or grandparental generations can influence the TAG phenotype of the offspring through epigenetic mechanisms (Öst et al. 2014; Palu et al. 2017). Fluctuations in energy intake are integrated by an endocrine system that coordinates catabolic and anabolic branches of TAG metabolism between several organ systems, which are central to the organismal lipid metabolism. Endocrine signals cause alterations in the activity of protein factors, such as transporters, enzymes, and scaffolding or regulatory proteins, which regulate the trafficking and metabolism of TAG at the molecular level. The enzymatic and transport machineries that govern TAG metabolism, and the principal regulatory mechanisms controlling these enzymatic activities in Drosophila, are the focus of this review. We begin with a brief overview of the major endocrine pathways and organ systems implicated in TAG metabolism, before we describe the proteins orchestrating the synthesis, degradation, and transport of TAG in detail.
Hormonal Control of Drosophila TAG Metabolism
The past decade has yielded a substantial increase in knowledge about endocrine systems that regulate TAG metabolism in Drosophila. A comprehensive description of these endocrine pathways is the subject of a number of recent excellent reviews (Rajan and Perrimon 2013; Lehmann 2018). Here, we briefly summarize the implications of some major hormonal pathways in the regulation of TAG storage that include insulin, adipokinetic hormone (Akh), juvenile hormone (JH), and ecdysone signaling.
Insulin signaling is a central nutrient-sensing pathway regulating growth, aging, stress responses, reproduction, and metabolism. In Drosophila, multiple insulin-like peptides (Dilps) serve as ligands for a single insulin receptor (InR), which relays the signal via a conserved cascade to downstream intracellular effectors such as kinases and transcription factors. Stimulation of these pathways translates into alterations in the expression and activities of key metabolic enzymes [for review, see Teleman (2010), Nässel and Broeck (2015), and Nässel et al. (2015)]. Binding of Dilps to InR is relayed to initial activation of the Drosophila ortholog of phosphatidylinositol-3-phosphate kinase (PI3K) Pi3K92E (Leevers et al. 1996; Oldham et al. 2002) and, subsequently, to Akt1 kinase (Verdu et al. 1999; Cho et al. 2001). Akt1 phosphorylates and regulates multiple protein targets including other kinases (Papadopoulou et al. 2004; DiAngelo and Birnbaum 2009), metabolic enzymes, and transcription factors (Jünger et al. 2003; Puig et al. 2003; Wang et al. 2008; Bolukbasi et al. 2017). Akt1 promotes phosphorylation and cytoplasmic retention of the transcription factor Forkhead box subgroup O (Foxo) and decreases its transcriptional output (Puig et al. 2003). Accordingly, Foxo plays a central role in connecting insulin signaling to TAG metabolism. Conversely, a reduction in insulin signaling, e.g., upon starvation, promotes nuclear translocation and transcriptional activity of Foxo, which in turn alters expression of a wide range of metabolic genes (Puig et al. 2003; Alic et al. 2011). Here, Foxo promotes the expression of lipases and increases the enzymatic breakdown of TAG (Vihervaara and Puig 2008; Wang et al. 2011). Consequently, transcriptional repression of lipolysis by the insulin/Foxo axis is a critical tissue-autonomous determinant of TAG homeostasis in the Drosophila fat body (DiAngelo and Birnbaum 2009; Wang et al. 2011). However, systemic defects in insulin signaling cause a complex array of growth-related, developmental, and metabolic phenotypes in Drosophila that typically includes increased organismal TAG stores (Böhni et al. 1999; Oldham et al. 2002; Werz et al. 2009; Grönke et al. 2010; Slack et al. 2010; Song et al. 2010).
The glucagon-like peptide Akh was named after its stimulatory effect on TAG mobilization and was first described in migratory locusts (Mayer and Candy 1969). The hormone is produced by the neuroendocrine cells of the corpora cardiaca and is released on demand to the hemolymph to bind to its cognate G protein-coupled Akh receptor (AkhR) on fat body cells. AkhR translates the Akh signal to downstream effectors via two distinct second-messenger systems (Park et al. 2002; Staubli et al. 2002). In the first system, it activates the adenylate cyclase-catalyzed production of cyclic adenosine-3′,5′-monophosphate (cAMP), an allosteric activator of protein kinase A (Pka), which promotes the phosphorylation of several downstream effectors including transcription factors, kinases, and LD proteins (Patel et al. 2005; Lee et al. 2018). In the second system, it activates a phospholipase C that converts phosphatidylinositol 4,5-bisphosphate (PIP2) to inositol-1,4,5-trisphosphate (IP3), which, in turn, triggers activation of endoplasmic reticulum (ER)-resident IP3 receptors and increases cytosolic concentrations of the second messenger Ca2+ [for a review, see Gäde and Auerswald (2003)]. This Ca2+ response transmits the Akh signal via currently unknown mechanisms to a gene expression signature that favors TAG catabolism (described in more detail below; Baumbach et al. 2014a). Although many components of the Akh pathway were first characterized in other insects, research on Drosophila has been fundamental in determining the genetic basis of this process. Consistent with the “adipokinetic” nature of the pathway, ablation of either the AkhR or the Akh gene impairs TAG mobilization in Drosophila (Grönke et al. 2007; Bharucha et al. 2008; Gáliková et al. 2015; Sajwan et al. 2015). Further, Drosophila genetic studies have also confirmed the crucial role of Ca2+ in transmitting the Akh signal and identified the G proteins Gγ1, Gαq, and the phospholipase C Plc21C as signal transducers along this axis (Baumbach et al. 2014b). In addition, several proteins that either promote or antagonize intracellular Ca2+ levels, such as the ER sensor Stromal interaction molecule (Stim), the plasma membrane channel Olf186-F, the ER Ca2+ channels sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) and Itp-R83a, and the Ca2+-binding protein calmodulin (Cam), have been linked to TAG homeostasis in Drosophila (Baumbach et al. 2014a; Bi et al. 2014; Moraru et al. 2017). Notably, the Akh pathway intersects with several other endocrine axes including insulin signaling (Choi et al. 2015; Kim and Neufeld 2015; Rajan et al. 2017; Song et al. 2017).
JH is the umbrella term for a group of acyclic sesquiterpenoids that are produced by the cells of the corpora allata, and that regulate diverse insect traits such as development, reproduction, and aging [for a review, see Flatt et al. (2005)]. JHs likely act by directly binding to transcription factors such as Methoprene-tolerant (Met) and germ cell-expressed bHLH-PAS (gce; bHLH-PAS stands for basic helix-loop-helix-PER-ARNT-SIM) to regulate expression of JH-responsive genes in target tissues (Charles et al. 2011; Jindra et al. 2015). Recent studies implicate JH signaling in the regulation of TAG metabolism (Baumann et al. 2013; Kang et al. 2017). For example, flies lacking corpora allata cells or lacking Met show reduced TAG levels, whereas pharmacological treatment with the JH analog methoprene increases TAG levels in adult flies. Moreover, loss-of-function alleles of the JH effector Kruppel homolog 1 (Kr-h1) have been shown to affect lipogenic and lipolytic gene expression, and TAG homeostasis in Drosophila larvae. The delineation of physical and genetic interactions between Kr-h1 and the insulin effector Foxo suggests an intersection of both pathways in the regulation of TAG homeostasis (Kang et al. 2017).
Ecdysteroids are a group of polyhydroxylated steroid hormones that are essential for molting and metamorphosis in Drosophila [reviewed in Tennessen and Thummel (2011)]. The “prototypical” ecdysteroid, ecdysone, is synthesized from cholesterol through several enzymatic steps in the prothoracic gland of Drosophila larvae and in currently unknown tissues in adults [reviewed in Schwedes and Carney (2012)]. It is converted to the active metabolite 20-hydroxyecdysone in several target tissues, and signals through a heterodimeric nuclear hormone receptor complex comprised of Ecdysone receptor (EcR) and Ultraspiracle (Koelle et al. 1991; Yao et al. 1992, 1993; Thomas et al. 1993). Activation of this heterodimer regulates the expression of ecdysone-responsive genes by binding to specific promoter sequences. More recently, ecdysone signaling has been implicated in the regulation of TAG metabolism in various developmental stages. EcR is required in the central nervous system for sexually dimorphic feeding behavior and TAG accumulation in mature female flies (Sieber and Spradling 2015). EcR also regulates lipid accumulation during oogenesis, a process that is essential for oocyte maturation (Carney and Bender 2000; Sieber and Spradling 2015). Likewise, ecdysone signaling has been shown to promote TAG accumulation during metamorphosis (Francis et al. 2010), whereas its role in third-instar larval TAG homeostasis is controversial (Wang et al. 2010; Kamoshida et al. 2012). Notably, ecdysone antagonizes insulin signaling and promotes nuclear translocation of the insulin-responsive transcription factor Foxo (Rusten et al. 2004; Colombani et al. 2005). Conversely, insulin signaling dampens ecdysone signaling by controlling the expression of the transcriptional coactivator Diabetes and obesity regulated (encoded by DOR), suggesting extensive cross talk between both pathways in the regulation of TAG metabolism (Francis et al. 2010).
Physiology of Drosophila TAG Metabolism at a Glance
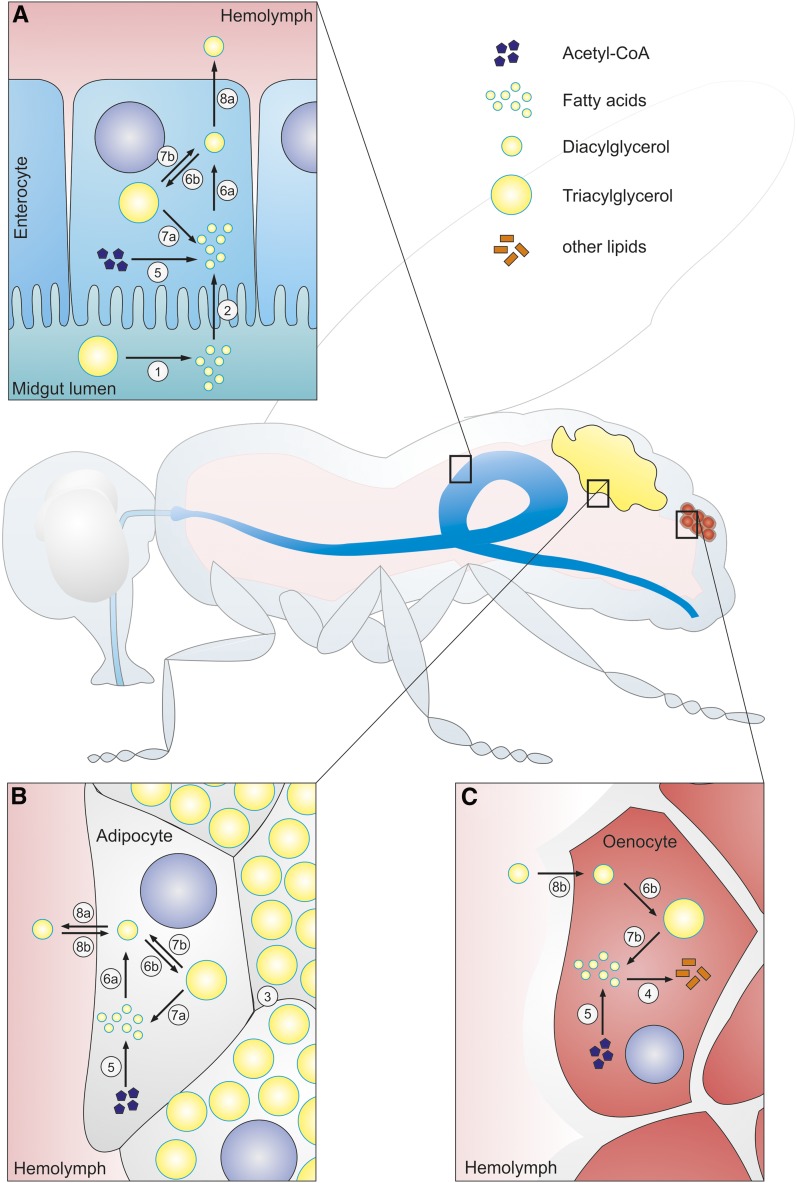
Figure 1 summarizes the TAG metabolism in major fly organs of glycerolipid absorption, storage, and utilization/processing. TAG is a natural component of many standard laboratory diets used for the cultivation of Drosophila. After ingestion, dietary TAG is hydrolyzed by digestive lipase(s) into free FAs, glycerol, and/or other acylglycerol intermediates in the midgut lumen. These TAG-derived metabolites are then absorbed by enterocytes and reincorporated into complex lipids (Canavoso et al. 2001; Sieber and Thummel 2009; Carvalho et al. 2012). Enterocytes convert dietary FA and glycerol moieties to the acylglycerol intermediate diacylglycerol (DAG), the major transport form of neutral lipid in the hemolymph (Palm et al. 2012). Excess dietary FA is also converted to TAG and deposited in intracellular LDs for transient storage (Carvalho et al. 2012; Palm et al. 2012). In addition, midgut enterocytes convert acetyl-CoA units derived from the metabolism of ingested carbohydrates into FAs (a process termed de novo lipogenesis), which are also incorporated into DAG or TAG for transport into other tissues or for transient storage, respectively (Figure 1) (Song et al. 2014). Hemolymph circulates through an open circulatory system, and transports nutrients and humoral factors between tissues. DAG, exported into the hemolymph by the midgut, exists in the form of specific lipoprotein complexes that serve as reusable shuttles for lipid transport between tissues (Palm et al. 2012). The hemolymph exports lipids to the brain, oocytes, oenocytes, and imaginal discs, in addition to other tissues and organs (Parra-Peralbo and Culi 2011; Palm et al. 2012; Parvy et al. 2012). Most midgut-derived DAG is directed to the prime energy storing tissue, the fat body, which has a uniquely high capacity for storage of TAG. It utilizes ingested lipids and de novo synthesized FAs for TAG storage (Canavoso et al. 2001; Palm et al. 2012; Parvy et al. 2012). In times of starvation, fat body TAG reserves are mobilized by enzymatic hydrolysis in a process termed lipolysis and supply DAG to other tissues (Figure 1) (Chino and Gilbert 1964; Arrese and Wells 1997; Grönke et al. 2005, 2007). One prominent example is the oenocyte, which acquires neutral fat from fat body lipolysis specifically upon fasting and possesses unique biochemical pathways for processing FA units into cuticle structural lipids, some of which serve as pheromones (Gutierrez et al. 2006; Chatterjee et al. 2014; Makki et al. 2014) (Figure 1). Although the fat body is the primary TAG storage organ, most other tissues maintain autonomous TAG pools and typically express specific sets of enzymes for the synthesis of TAG (Chintapalli et al. 2007; Kühnlein 2011). Likewise, enzymatic machineries that catabolize TAG (and DAG) to FA moieties, which are required for further catabolism and resultant energy production, are believed to be a general feature of all fly tissues and organs.
Figure 1.
Triacylglycerol (TAG) metabolism in major fly organs of glycerolipid absorption, storage, and utilization/processing. (A) The midgut serves as major site of TAG digestion and lipid absorption. Dietary TAG is enzymatically processed to lipolytic products such as fatty acids (FAs) by digestive lipases prior to absorption by the enterocytes of the midgut (1–2). Enterocytes produce additional FAs through de novo synthesis from acetyl-CoA (5). FAs from both sources are converted to diacylglycerol (DAG) via enzymatic pathways (6a). DAG is exported to the hemolymph for lipid transport to other tissues (8a). Alternatively, DAG can be esterified with excessive FAs to TAG for storage (6b). TAG can be remobilized to FA and DAG by intracellular lipolysis (7a–b). (B) The fat body serves as a major energy reservoir that stores bulk TAG (3). Similar metabolic pathways as in the midgut direct the synthesis and turnover of FA, DAG, and TAG (5–7). Adipocytes control lipid transport via the hemolymph by the export and import of DAG (8a–b). (C) Oenocytes are an exemplary site of lipid utilization. FAs derived from de novo lipogenesis (5) or TAG lipolysis (7b) are converted to structural or signaling lipids (4). Oenocytes also convert surplus DAG acquired from the hemolymph (8b) to TAG for storage (6b). Note that TAG metabolism is not restricted to the organs displayed but is an inherent property of all fly tissues. In particular, the central nervous system, as well as cardiac and skeletal muscles, play important roles in organismal TAG homeostasis integration and/or show pathophysiological phenotypes in response to TAG dysregulation.
De Novo Lipogenesis and Dietary Lipid Digestion
FA moieties that are stockpiled in TAG are either derived from the diet or synthesized de novo through a cascade of enzymatic reactions. The basic pathway of de novo FA formation requires two enzymes. First, Acetyl-CoA-carboxylase (ACC) catalyzes the formation of malonyl-CoA from acetyl-CoA. Second, the multienzyme complex FA synthase (FAS, encoded by FASN) sequentially condenses malonyl-CoA units with acetyl-CoA to produce long-chain FAs (Smith et al. 2003; Barber et al. 2005). The Drosophila genome encodes a single ACC gene and three distinct FASN genes (Parvy et al. 2012). ACC is ubiquitously expressed, with highest levels in oenocytes and fat body cells (Parvy et al. 2012). Three FASN-related genes—FASN1 (CG3523), FASN2 (CG3524), and FASN3 (CG17374)—show distinct expression patterns and are differentially expressed in the fat body, oenocytes, midgut, and muscles (Garrido et al. 2015; Wicker-Thomas et al. 2015). Both, ACC and FASN functions are essential for development as genetic loss of the single Drosophila ACC gene, or a combined deletion of FASN1 and FASN2, results in embryonic and larval lethality, respectively (Parvy et al. 2012; Garrido et al. 2015). These developmental deficits are in part due to oenocyte-specific functions of de novo FA synthesis (Parvy et al. 2012). The long-chain and very long-chain FA pools generated in oenocytes are further converted to cuticular hydrocarbons and other lipids, which are essential for the barrier functions of the cuticle and the trachea (Chung et al. 2014; Wicker-Thomas et al. 2015). Notably, the tissue-autonomous TAG pool of larval oenocytes accumulates even in the absence of ACC function, suggesting that tissue-autonomous storage lipid synthesis in oenocytes does not contribute to the developmental arrest evoked by defects in de novo FA synthesis (Parvy et al. 2012). Instead, ACC and FASN contribute to storage lipid synthesis mainly in two other tissues, the fat body and the midgut (Palm et al. 2012; Parvy et al. 2012). In the latter tissue, de novo lipogenesis contributes to the formation of DAG for export to the hemolymph and for transport of FA moieties to other tissues (Palm et al. 2012; described below). In the fat body, de novo lipogenesis generates a quantitatively important fraction of storage lipids (Parvy et al. 2012; Garrido et al. 2015; Wicker-Thomas et al. 2015). Both, fat body and midgut lipogenesis have important tissue-autonomous functions in maintaining whole-body energy homeostasis. Animals deficient in de novo lipogenesis due to combined mutations in FASN1 and FASN2 store less TAG in the larval as well as adult states, suggesting a continuous contribution of de novo FA synthesis to fat body TAG storage throughout development (Wicker-Thomas et al. 2015). This blockade of fat body lipogenesis renders larvae sensitive to excess sugar, and suggests a dual role of de novo lipogenesis in bulk energy storage and in sugar detoxification (Garrido et al. 2015). Sugar-stressed FASN1/2 mutant cells display a cell-autonomous decrease in size, and accumulate advanced glycation end products that form by covalent bonding between amine and carbonyl groups of sugars, or their α-oxalaldehyde derivatives (Garrido et al. 2015). However, FASN1 knockdown did not result in increased sugar sensitivity in another study (Havula et al. 2013), suggesting that specific features of the diet or genetic background might modulate this phenotype.
It is notable that genetic blockade of lipogenesis in skeletal muscle tissues impairs muscle function, and translates into defects in larval motility and dietary-induced hyperactivity in adults (Katewa et al. 2012; Garrido et al. 2015). In adult flies, dietary restriction has been shown to increase steady-state whole-body TAG levels as well as overall TAG turnover rates. These metabolic adaptions, as well as life-extending effects of dietary restriction, were compromised by muscle-specific knockdown of ACC (Katewa et al. 2012). Unrestricted lipogenesis via ACC and FASN gene functions was identified as a pathogenic mechanism involved in cardiac dysfunction of flies with mutations in the easily shocked (eas) gene, which encodes an ethanolamine kinase (Lim et al. 2011). Eas mutant hearts exhibit elevated lipogenic gene expression and increased TAG content, likely due to a derepression of the transcription factor sterol regulatory element-binding protein (SREBP, discussed in further detail below). The increased cardiac TAG levels and cardiac dysfunction could be corrected by genetically interfering with the cardiac expression of lipogenic genes. However, it is currently not known if the phenotypes evoked by dysregulated lipogenesis in skeletal or cardiac muscle directly involve tissue-autonomous changes of TAG metabolism, or changes in the metabolism of other lipids. Notably, a recent study demonstrated that ceramide accumulation causes lipotoxic cardiomyopathy in flies and identified FASN1 as a ceramide-interacting protein (Walls et al. 2018). Thus, ectopic TAG accumulation in cardiac muscle may be a metabolic signature, rather than a mechanistic cause, of FA-induced stress. In summary, de novo lipogenesis contributes a quantitatively important source of FAs for TAG storage in various developmental stages of Drosophila, but also provides tissue-specific pools of FAs for essential, as well as potentially lipotoxic, downstream metabolites that are unrelated to TAG.
The diet constitutes an alternative source of FA for the synthesis of structural and storage lipids. The Drosophila midgut—the main site of digestion and nutrient absorption—is composed of defined subregions with distinct morphological, histological, physiological, and genetic properties (Buchon et al. 2013; Lemaitre and Miguel-Aliaga 2013). Digestive enzymes are frequently organized in genomic clusters that may have arisen by gene duplication followed by functional divergence. The corresponding gene products are often sequentially expressed in various gut regions, suggesting a tight regulation of nutrient digestion in each section of the digestive tract. The resorption of dietary lipids requires the digestion of complex lipids into simple building blocks by the action of dietary lipases to yield FAs, glycerol, and acylglycerol intermediates. The Drosophila genome encodes about a dozen genes with predicted lipase function whose expression in the midgut qualifies them as putative digestive enzymes (Horne et al. 2009; Sieber and Thummel 2009). However, relatively little is known about the biochemistry and enzymology of lipid digestion in Drosophila. Alterations in dietary status or food composition cause distinct expression responses for these putative midgut lipases (Zinke et al. 2002; Chintapalli et al. 2007; Sieber and Thummel 2009; Karpac et al. 2013). For example, expression of the pancreatic lipase-related genes CG6277 and CG6283 is repressed by dietary sugar, whereas expression of the gastric lipase-related magro gene is induced by feeding, and suppressed by starvation and cholesterol (Grönke et al. 2005; Horner et al. 2009; Chng et al. 2014; Mattila et al. 2015). Although the large number of putative midgut lipases may suggest substantial functional redundancy in dietary lipid breakdown, Magro has been ascribed a major function in lipid digestion (Sieber and Thummel 2009). Magro exhibits TAG and cholesterol ester hydrolase activity in vitro, and is expressed at high levels in the proventriculus from where it is secreted into the gut lumen. Knockdown of magro expression or pharmacological inhibition of midgut lipase activity decreases midgut TAG lipase activity and assimilation of dietary TAG, which manifests in decreased whole-body TAG storage and starvation sensitivity (Sieber and Thummel 2009, 2012).
Transcriptional Regulation of De Novo Lipogenesis and Digestion of Dietary TAG
The enzymatic control of de novo lipogenesis and lipid digestion is governed by changes in the dietary status of the animal, which are sensed and transmitted by transcription factors that respond to distinct dietary or hormonal cues. Among them, the SREBPs have an evolutionarily conserved function in controlling lipogenic gene expression (Dobrosotskaya et al. 2002; Seegmiller et al. 2002; McKay et al. 2003; Walker et al. 2011). SREBPs regulate transcription of a variety of genes involved in de novo lipogenesis and membrane biosynthesis, including ACC and FASN, and are targeted by numerous hormonal and dietary signals. SREBPs are membrane-bound bHLH leucine zipper (bHLH-Zip) transcription factors that are activated by proteolytic cleavage at specific sites that, in turn, facilitate the release and nuclear translocation of the active bHLH-Zip domains. A lipid-sensing chaperone termed SREBP cleavage-activating protein (SCAP), and specific proteases including site-specific proteases 1 and 2 (S1P and S2P) constitute the basic molecular machinery that controls SREBP activation. This machinery senses membrane lipids such as sterols and phospholipids, which act in a feedback mode to inhibit SREBP processing and activation [for reviews, see Horton (2002), Rawson (2003), and Osborne and Espenshade (2009)]. The Drosophila genome encodes single orthologs of SREBP, SCAP, S1P, and S2P (Seegmiller et al. 2002). Consistent with its lipogenic function, SREBP is mainly expressed in the midgut, fat body, and oenocytes (Kunte et al. 2006). Drosophila SREBP null mutants are FA auxotrophic and die as second-instar larvae unless supplemented with dietary FAs. Mutant larvae show decreased expression of lipogenic genes such as ACC and FASN (Kunte et al. 2006). Although TAG storage has not been investigated in SREBP mutant larvae, the lean phenotype of Drosophila mutants lacking components of the SREBP-processing machinery (SCAP or S2P) suggests that SREBP function is required for the buildup of organismal TAG stores (Matthews et al. 2010). Moreover, manipulations of SREBP expression in the larval midgut by ectopic expression or RNA interference (RNAi)-mediated knockdown have been shown to reciprocally affect midgut lipid stores and body TAG levels (Song et al. 2014). Thus, the impairments in TAG storage observed upon manipulations of SREBP expression largely recapitulate the phenotypes of ACC or FASN deficiency described above. At present, it is unclear as to what extent the developmental defects displayed by SREBP mutants are related to the reduced capacity to build up energy reserves or to defects in the synthesis of other lipids required for development (Kunte et al. 2006). The tissue-autonomous function of lipogenesis in larval oenocytes that generates essential TAG-unrelated lipids suggests that a similar mechanism may account for the SREBP mutant phenotype (Parvy et al. 2012). However, results of genetic rescue experiments argue against an oenocyte-autonomous role for SREBP, but rather point to essential functions of this protein in the midgut and fat body (Kunte et al. 2006). In accordance with the evolutionarily conserved function of SREBP proteins in sensing membrane lipids, the processing of Drosophila SREBP is inhibited by synthesis of the major Drosophila phospholipid phosphatidylethanolamine (PE) (Dobrosotskaya et al. 2002). RNAi-mediated interference with PE synthesis in the adult fat body causes enhanced ACC and FASN gene expression and obesity (Baumbach et al. 2014a). Likewise, defective PE synthesis in eas mutants has been linked to elevated lipogenic gene expression and tissue-autonomous ectopic TAG deposition in cardiac muscle (Lim et al. 2011). In addition to membrane lipids, SREBP activity is also regulated by hormones. Enterocyte SREBP activity is induced by JH signaling in response to mating and is required for optimal fecundity (Reiff et al. 2015). Conversely, tachykinin, secreted from enteroendocrine cells upon fasting, represses midgut SREBP activity via activation of Pka (Song et al. 2014). Notably, SREBP control by the PI3K/Akt/TOR (target of rapamycin) pathway [reviewed by Krycer et al. (2010)] appears to be evolutionarily highly conserved among flies and mammals, exhibiting similar effects on cell growth in these organisms (Porstmann et al. 2008). Lipogenic gene expression is also controlled by the heterodimeric transcription factor Mondo/Bigmax (also abbreviated Mio/Mlx), which coordinates transcriptional responses to dietary sugar [reviewed in Mattila and Hietakangas (2017)]. bigmax mutant larvae have decreased ACC and FASN gene expression, and store less TAG. Upon increased dietary sugar, Mondo/Bigmax induces expression of the transcription factor Sugarbabe, which acts in a feedforward loop to transcriptionally activate lipogenic gene expression (Mattila et al. 2015). Notably, Mondo/Bigmax is also required for the sugar-dependent repression of certain genes. These include the putative gut lipases CG6283 and CG6277, suggesting that Mondo/Bigmax coordinates the expression of digestive enzymes with dietary needs and coordinates sugar metabolism, de novo lipogenesis, and lipid digestion (Mattila et al. 2015).
In addition to Mondo/Bigmax, several other transcription factors have been implicated in the regulation of dietary lipid digestion. The Hormone receptor-like in 96 (Hr96, also known as DHR96) acts in the midgut as a transcriptional regulator of genes involved in dietary lipid breakdown and uptake (Horner et al. 2009; Sieber and Thummel 2009, 2012). Hr96 is required for expression of the major digestive TAG lipase Magro. Hr96 mutants are lean and genetic rescue experiments support the conclusion that Magro acts as main effector downstream of Hr96 to control organismal TAG storage (Sieber and Thummel 2009). Magro expression is highly sensitive to alterations in food intake. Genetic models of hyperphagia in flies show increased magro expression and intestinal lipase activity (Subramanian et al. 2013). In contrast, the suppression of magro expression upon starvation is controlled by the insulin-responsive transcription factor Foxo (Karpac et al. 2013). Interference with intestinal foxo expression in adult flies increases magro expression and organismal TAG stores. Conversely, increased Foxo activity in the aging gut has been causally linked to decreased magro expression and compromised organismal TAG stores (Karpac et al. 2013).
Enzymatic Synthesis of TAG
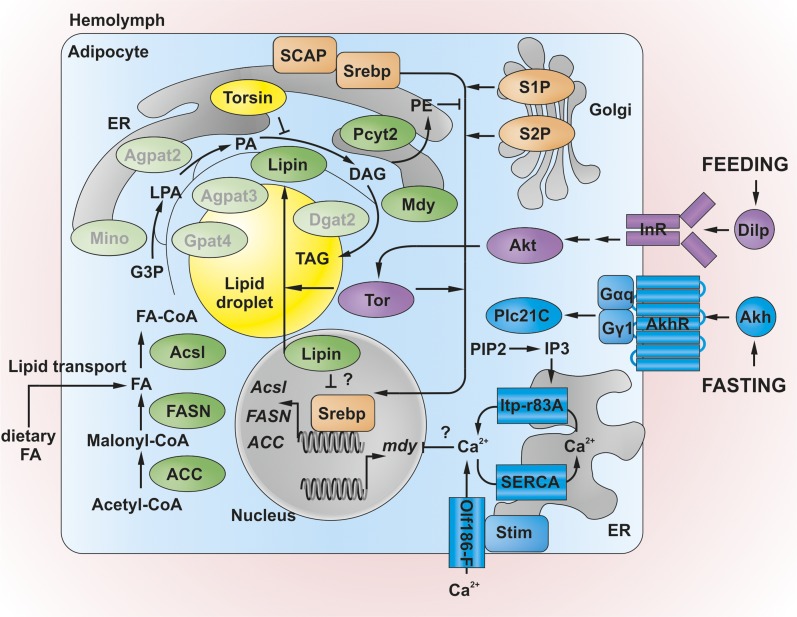
Figure 2 summarizes the enzymatic steps and subcellular topology of TAG synthesis and its regulation. Excess FAs acquired via the diet or synthesized de novo need to be converted to complex lipids for transport or storage. This conversion involves the sequential esterification of FA moieties to a glycerol backbone. The canonical glycerol-3-phosphate (G3P) pathway (also called the Kennedy pathway) of TAG synthesis is initiated by G3P-O-acyltransferase (GPAT), which catalyzes the transfer of an FA unit from acyl-CoA to G3P [for review, see Bell and Coleman (1980), Liu et al. (2012), and Wang et al. (2017)]. The resultant lysophosphatidic acid (LPA, also termed 1-acylglycero-3-phosphate) is converted to phosphatidic acid (PA) in a second acyltransferase reaction by LPA acyltransferase (LPAAT; also termed 1-acylglycero-3-phosphate-O-acyltransferase, AGPAT). PA is converted by PA phosphatase (PAP) to DAG, which is the major transport unit of FA in the hemolymph of Drosophila (Palm et al. 2012). An alternative pathway of DAG synthesis involves the acylation of monoacylglycerol (MAG) by MAG-O-acyltransferase (MGAT). However, unlike the precursors of the G3P pathway, MAG is not synthesized de novo, but derived from the breakdown of complex lipids. Both the G3P and MGAT pathways converge at DAG (Liu et al. 2012). Finally, DAG-O-acyltransferase (DGAT) transfers FA from acyl-CoA to DAG, resulting in the formation of TAG. Each step is catalyzed by isoenzymes that differ in tissue expression and subcellular localization (Figure 2) (Chintapalli et al. 2007; Wilfling et al. 2013; Wang et al. 2017). TAG synthesis competes with several other biochemical pathways that consume FA, e.g., FA oxidation or membrane lipid synthesis. The synthesis routes of glycerophospholipids share the initial steps with TAG, and branch off at the level of PA or DAG. Notably, the analysis of the Drosophila third-instar larval lipidome indicates a selective enrichment of medium-chain FAs in DAG and TAG as compared to membrane glycerophospholipids (Palm et al. 2012). It is presently unclear if this diversification involves the “channeling” of different FAs along de novo synthesis pathways toward either TAG or glycerophospholipids, e.g., via different isoenzymes, or selective remodeling reactions after synthesis.
Figure 2.
Triacylglycerol (TAG) synthesis in Drosophila: biochemistry and main regulatory pathways. Fatty acids (FAs) derived from the diet or synthesized de novo by Acetyl-CoA-carboxylase (ACC) and the FA synthase complex (FASN) are activated to FA-CoA by Acsl, and are esterified consecutively by glycerol-3-phosphate acyltransferases (GPATs) (possibly Gpat4 or Mino) to lysophosphatidic acid (LPA) and by LPA acyltransferases (possibly Agpat3, Agpat2) to phosphatidic acid (PA). Lipin dephosphorylates PA to diacylglycerol (DAG), which is further esterified by DAG acyltransferases (Mdy and possibly Dgat2) to TAG and stored in lipid droplets (LDs). Acyltransferases show differential localization to the endoplasmic reticulum (ER) or LDs, respectively. Lipin shuttling between the cytoplasm and the nucleus is controlled by the Tor kinase, which in turn is subject to insulin pathway regulation via Akt under feeding conditions. Lipin is also under the inhibitory control of Torsin. The transcription factor sterol regulatory element-binding protein (SREBP) promotes FA-CoA synthesis via positive control of ACC, FASN, and Acsl expression. The canonical pathway of SREBP activation requires its interaction with the escort factor SREBP cleavage-activating protein (SCAP), and its proteolytic processing via Golgi-resident S1P and S2P. SREBP activation is promoted by target of rapamycin (Tor) kinase and inhibited by the membrane lipid PE, which is synthesized from DAG by Pcyt2. Nuclear Lipin may also inhibit SREBP activity. Under fasting conditions, mdy expression is suppressed in a Ca2+-dependent manner under the control of adipokinetic hormone (Akh) signaling. Akh receptor (AkhR) activation by Akh triggers store-operated entry of extracellular Ca2+ via small G proteins, IP3 signaling under the control of phospholipase C, and ER Ca2+ store depletion, which promotes interaction of Stim with the plasma membrane Ca2+ channel Olf186-F to result in extracellular Ca2+ influx until the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) restores ER Ca2+ levels. The proximal mechanisms of Ca2+-dependent mdy repression are currently unknown. See main text for details and definitions of abbreviations. Proteins shown in faded green with gray lettering are based on homology-based activity predictions and await experimental validation. Note the antagonistic action of the lipoanabolic insulin/Tor pathway (in magenta) under feeding vs. the lipocatabolic Akh pathway (in blue) under fasting conditions.
The Drosophila genome encodes three putative GPAT isoenzymes that belong to two different protein families and are structurally related to bona fide mammalian GPATs. The gene product of minotaur (mino), an ortholog of mammalian GPAT1 and GPAT2, has been named after its localization in the labyrinthine fusome network that connects nurse cells of Drosophila ovaries (Vagin et al. 2013). Interfering with mino function in ovaries causes defects in piwi-interacting RNA biosynthesis by a mechanism that is unrelated to its predicted enzymatic function. Consistent with an additional role in TAG formation, ectopic expression of mino increased fat storage in larval salivary glands (Tian et al. 2011; Pagac et al. 2016). However, the specific contribution of Mino to TAG synthesis has not been investigated in flies deficient for mino. The genes CG15450 and CG3209/Gpat4 encode structurally related proteins with homology to mammalian GPAT3 and GPAT4, respectively. Whereas CG15450 expression is restricted to the testis, Gpat4 is ubiquitously expressed (Chintapalli et al. 2007) and the protein localizes to LDs in Drosophila S2 cells (Wilfling et al. 2013). Drosophila Gpat4 mutants exhibit a semilethal phenotype. More than 80% of Gpat4 mutant animals do not develop into larval stages. Animals that escape early death exhibit severe metabolic defects and developmental delay, but give rise to adults of normal size (Yan et al. 2015). Of note, TAG levels were unaltered in surviving mutants, suggesting that the developmental phenotypes of flies lacking Gpat4 are unrelated to storage lipid metabolism. Depletion of Gpat4 expression by RNAi reduced LDs in glial cells of Drosophila larvae (Bailey et al. 2015) and reduced TAG formation of embryonic S2 cells (Wilfling et al. 2013), suggesting a cell type-specific contribution of GPAT4 to TAG synthesis, which awaits further characterization.
As for the initial step of the G3P pathway, the second step has also been attributed to a set of various isoenzymes with putative LPAAT/AGPAT function. The Drosophila genome encodes four genes of the LPAAT/AGPAT family including CG4729/Agpat3, CG4753/Agpat4, fu12/Agpat2, and CG3812/Agpat1. No mutant alleles of these genes have been characterized so far. The function of these genes is largely inferred by homology to bona fide mammalian AGPATs and lacks biochemical evidence. A specific role of Agpat3 in TAG storage metabolism has been suggested by a cell-based study using in vitro-cultured S2 cells in which the protein was localized to LDs and contributed to FA-induced TAG formation (Wilfling et al. 2013). In addition, in vivo knockdown of Agpat3 gene expression reduced LD formation in glial cells (Bailey et al. 2015). Whether this protein controls TAG formation in other cells or acts redundantly to other AGPATs remains to be investigated.
The third step in TAG formation is the dephosphorylation of PA to DAG. PA is a substrate for type I and type II PAP enzymes that are discriminated by their requirement for Mg2+ (Zhang and Reue 2017). The Mg2+-dependent type I PAP activity is encoded by Lipin genes that have been assigned major roles in glycerolipid and TAG synthesis in diverse phyla (Péterfy et al. 2001; Han et al. 2006; Donkor et al. 2007). Apart from their enzymatic function in glycerolipid synthesis, Lipins have been shown to act as transcriptional cofactors in the nucleus (Finck et al. 2006; Peterson et al. 2011). Drosophila has a single Lipin gene (Lpin) that encodes at least three different protein isoforms with tissue- and developmental stage-specific expression patterns (Valente et al. 2010). Lpin mutants exhibit a plethora of phenotypes including developmental delay, pupal semilethality, and reduced fertility (Ugrankar et al. 2011). Consistent with a key function of Lipin in TAG formation, mutant larvae present with severe lipodystrophy, reduced organismal TAG levels, and decreased LD size (Ugrankar et al. 2011). Conversely, TAG levels and LD size are increased in the fat body of young larvae mutant for the Lipin repressor Torsin, which affects Lipin localization (Grillet et al. 2016). This fat body phenotype is not limited to cellular TAG content, but also manifests as altered cell morphology, including fragmented nuclei, aberrant organelle structure, and altered cell shape (Ugrankar et al. 2011). Specifically, loss of Lpin is associated with a cell-autonomous reduction in insulin signaling and fat cell size (Schmitt et al. 2015). This phenotype is further enhanced upon genetic impairment of insulin and Tor signaling, suggesting that Lpin function is required for the normal insulin responsiveness of fat cells (Schmitt et al. 2015).
The last and only committed step in TAG synthesis is the conversion of DAG to TAG by DGAT. Eukaryotic DGAT isoenzymes belong to two unrelated protein families, membrane-bound O-acyltransferase (MBOAT) and DGAT2, respectively. Although the Drosophila genome encodes for proteins of both families, a single enzyme of the MBOAT family, Midway (Mdy), has been assigned a major role in Drosophila TAG metabolism. Midway is structurally related to mammalian DGAT1 and has been shown to possess DGAT activity in vitro (Buszczak et al. 2002). Mutations in the mdy gene were initially identified in a genetic screen for female sterile mutants (Schüpbach and Wieschaus 1991). Mdy mutants display reduced oocyte lipid stores and degeneration of egg chambers during midoogenesis (Buszczak et al. 2002). Since TAG serves as an energy resource of the developing embryo, it has been suggested that defects in TAG synthesis are causative for egg chamber degeneration in mdy mutants (Buszczak et al. 2002). Likewise, mdy lack-of-function mutations in adult fat-storing tissues cause a strong reduction in organismal TAG stores, arguing for a principle role of this enzyme in TAG synthesis in Drosophila (Beller et al. 2010; Baumbach et al. 2014a). The Drosophila genome contains three DGAT2 family members encoded by the uncharacterized genes CG1941, CG1942/Dgat2, and CG1946. The associated proteins are structurally related to mammalian DGAT2 and MGAT enzymes. Although recombinant CG1942/Dgat2 protein has been shown to localize to LDs in Drosophila S2 cells, a reduction in CG1942/Dgat2 expression did not compromise TAG formation (Wilfling et al. 2013). Moreover, and in contrast to Midway, the enzymatic activities of Drosophila DGAT2 family members have not been characterized. Thus, it remains to be shown if Drosophila DGAT2 enzymes represent bona fide DGATs or, rather, function in the MGAT pathway of TAG synthesis. Notably, MGAT activity has been measured in fat body samples of the tobacco hornworm Manduca sexta, further arguing for the presence of this pathway in insects (Arrese et al. 1996).
Regulation of TAG Synthesis
Although data from genome-wide transcriptomics suggests nutritional regulation of several acyltransferases (Zinke et al. 2002; Harbison et al. 2005; Mattila et al. 2015) implicated in TAG synthesis, little is known about the underlying molecular mechanisms of these transcriptional responses or the physiological significance for storage lipid metabolism. Mdy gene expression levels decrease in response to genetic manipulations that increase cytosolic Ca2+ levels in fat body cells, such as knockdown of the ER Ca2+ pump SERCA (Bi et al. 2014). Conversely, depletion of cytosolic Ca2+ levels in the fat body tissue of adult flies by knockdown of the ER calcium sensor Stim, or by knockdown of the IP3 receptor gene Itp-r83A, increase mdy gene expression (Figure 2) (Baumbach et al. 2014a). This response is in part due to a tissue-nonautonomous effect on activation of the orexigenic brain short neuropeptide F (sNPF) and ensuing hyperphagia. Notably, a similar change in mdy transcript expression was also elicited by genetic manipulations that compromise Akh signaling in the fat body, which is in line with the observation that Ca2+ is an important second messenger mediating Akh responses in insects (Arrese et al. 1999; Baumbach et al. 2014a). Conversely, genetic activation of Akh signaling suppressed mdy expression. However, the molecular mechanisms that relay Akh/Ca2+ signaling to altered mdy transcript levels remain elusive (Figure 2). Unlike many other anabolic genes, transcriptional expression of Lpin is induced upon starvation, compared to the ad libitum fed state (Harbison et al. 2005; Ugrankar et al. 2011). In addition, fasting alters the subcellular localization of Lipin and increases its abundance in the nucleus (Figure 2). Although the nuclear functions of Drosophila Lipin are not fully understood, studies with mammalian orthologs suggest that nuclear Lipin acts as a transcriptional activator of genes involved in FA utilization and as a repressor of lipogenic genes (Finck et al. 2006; Peterson et al. 2011). Insulin- and TOR complex 1 (TORC1)-dependent phosphorylation of mammalian Lipins prevents nuclear translocation (Harris et al. 2007; Peterson et al. 2011). Likewise, nuclear translocation of Drosophila Lipin is suppressed by TORC1 signaling, whereas altered insulin pathway activity did not change Lipin localization in vivo (Schmitt et al. 2015). Although these observations suggest a dual role for Lipin in TAG synthesis and the regulation of gene expression, knowledge about putative transcriptional targets of Drosophila Lipin is currently lacking.
TAG Storage: Formation of LDs
Although the Drosophila fat body has a unique capacity to synthesize and store TAG, most other tissues harbor the protein machinery of TAG synthesis and storage. TAG storage has been observed in the midgut, brain, oenocytes (Gutierrez et al. 2006; Palm et al. 2012; Bailey et al. 2015), imaginal discs, ovaries (Parra-Peralbo and Culi 2011), and salivary glands (Thiel et al. 2013). At the cellular level, fat storage is accomplished by deposition of lipid esters into discrete globular organelles termed LDs (Welte 2015). The evolutionarily conserved organization of LDs is characterized by a hydrophobic core of storage lipid esters (largely TAG and sterol ester), surrounded by a closed phospholipid monolayer to which proteins are attached (Welte 2015). Proteomic studies of LDs from Drosophila embryos, larvae, or in vitro-cultured cells have identified specific sets of LD proteins with functions in metabolism, membrane trafficking, signaling, and protein turnover (Beller et al. 2006; Cermelli et al. 2006; Krahmer et al. 2013). Although the general organization of LDs is highly conserved between distant phyla such as fungi, plants, or animals, there is morphological and functional heterogeneity between LDs, even within single cells or tissues (Wilfling et al. 2013; Thul et al. 2017). Although the exact mechanisms of LD biogenesis are incompletely understood, a prevalent model suggests that LDs arise through the deposition of neutral lipids between the leaflets of the ER lipid bilayer, developing nascent droplets that finally bud off from the ER membrane after reaching a critical size [reviewed in Walther et al. (2017)]. This process is regulated and assisted by distinct proteins whose dysfunction compromises normal LD morphology and function. Genome-wide RNAi screens in Drosophila cells were indispensable to the comprehensive identification of proteins required for normal LD structure (Beller et al. 2008; Guo et al. 2008). A prominent example is Seipin, which was initially identified as being deficient in human patients with congenital lipodystrophy (Magré et al. 2001). Orthologous proteins in diverse eukaryotic model organisms such as yeast, plants, and insects have shaped the view that Seipin has a conserved function in establishing and maintaining LD morphology (Grippa et al. 2015; Wang et al. 2016; Taurino et al. 2018). The single Drosophila Seipin ortholog is expressed mainly in the fat body, but also in the salivary gland, midgut, and muscle (Tian et al. 2011). Drosophila Seipin localizes at contact sites between the ER and LDs, and promotes distinct steps in LD maturation in Drosophila S2 cells (Wang et al. 2016). Seipin mutant flies display fat body-autonomous reductions in LD size and organismal TAG levels (Tian et al. 2011). Notably, this deficit in TAG storage has been linked to impaired activity of the ER Ca2+ pump SERCA (Bi et al. 2014) and subsequent mitochondrial dysfunction, which leads to reduced lipogenesis (Ding et al. 2018). The Seipin mutant also represents a striking example of ectopic fat accumulation in salivary glands, which is unrelated to the function of Seipin in the fat body. Genetic and biochemical studies have linked ectopic LD formation in Seipin mutant salivary glands to the accumulation of PA, and aberrant TAG synthesis via Lipin and Mdy, rather than to defective fat breakdown (Tian et al. 2011). Notably, lipodystrophy, as well as ectopic fat accumulation, are also major pathological manifestations of loss-of-function mutations in mammalian Seipin (Magré et al. 2001; Chen et al. 2012), highlighting the value of Drosophila as a model providing novel insights into human LD-associated pathologies.
TAG Mobilization by Lipolysis
During nonfeeding developmental stages, periods of increased energy demand, or nutrient deprivation, insects mobilize their energy stores. The mobilization of fat body TAG requires hydrolysis of the ester bonds in TAG by the action of lipases. The prevailing model of insect fat body lipolysis is that TAG hydrolysis directly generates DAG, which is exported into the hemolymph for transport into other tissues (Arrese and Wells 1997). A major executor of fat body lipolysis in Drosophila is the TAG hydrolase Brummer (Bmm), a member of the patatin-like domain-containing family of proteins (Grönke et al. 2005). Orthologous proteins in diverse phyla such as plants (Eastmond 2006), yeast (Athenstaedt and Daum 2003; Kurat et al. 2005), and mammals (Zimmermann et al. 2004) have been assigned analogous functions in lipid mobilization, highlighting the ancestral role of Bmm-related proteins in TAG breakdown. Drosophila bmm mutant flies accumulate excessive TAG during adulthood and display a reduced rate of TAG breakdown upon starvation. Due to the excessive fat depots and delayed TAG consumption, bmm mutants substantially outlive wild-type flies during nutrient deprivation, suggesting impaired, though still functional, lipid mobilization (Grönke et al. 2005). An alternative lipolytic system in Drosophila depends on Akh signaling that controls the expression of one or more currently uncharacterized TAG lipase(s), which complement Bmm lipase function (Grönke et al. 2007). The bmm gene is broadly expressed and abundant in several other tissues than the fat body, including the gut, salivary glands, heart, and oenocytes (Chintapalli et al. 2007). In line with a general function of Bmm in TAG breakdown, bmm mutants accumulate excessive TAG in peripheral tissues such as the midgut (Palm et al. 2012) and in the Malpighian tubules (P. Hehlert and R. P. Kühnlein, in preparation). Notably, bmm transcripts are maternally provided and expressed throughout embryogenesis, suggesting an essential function of Bmm during embryonic TAG mobilization. In line with this hypothesis, a lack of zygotic and maternal bmm function results in embryonic lethality (Grönke et al. 2005).
DAG that has been liberated from fat body TAG stores needs to be further hydrolyzed before FA can enter β-oxidative or anabolic pathways. Brummer lacks DAG lipase activity in vitro (Grönke et al. 2005) and is, therefore, unlikely to provide the activity necessary to catabolize DAG. A candidate enzyme for this function is the Drosophila hormone-sensitive lipase (Hsl), which has been named because of its homology to mammalian Hsl. Mammalian Hsl is a hydrolase with broad substrate specificity and acts as a key DAG lipase in adipose tissue, testis, and muscle (Haemmerle et al. 2002). Drosophila Hsl has been implicated in TAG mobilization in parallel with Bmm lipase in the larval fat body (Bi et al. 2012). As the enzymatic properties of Drosophila Hsl have not yet been characterized, it remains to be shown if Drosophila Hsl acts as a DAG lipase in peripheral tissues and to what extent it contributes to complete acylglycerol breakdown for energy production.
Regulation of Lipolysis
Consistent with its major physiological function with respect to energy metabolism, TAG mobilization in Drosophila is nutritionally regulated both at the transcriptional and post-translational levels. For example, bmm transcription is suppressed by feeding and induced upon nutrient withdrawal. An important transcriptional inducer of bmm expression is the insulin-responsive transcription factor Foxo (Figure 3) (Wang et al. 2011). Nuclear Foxo promotes transcription of a plethora of fasting-induced genes while it suppresses others [Karpac et al. (2013) and see above]. The nuclear abundance of Foxo is regulated by post-transcriptional modifications, including phosphorylation and acetylation, that control its retention in the cytosol. In addition to the canonical inhibitory Foxo phosphorylation by Akt1, Salt-inducible kinase 3 (Sik3) acts as an important negative regulator of Foxo and indirectly controls its sequestration in the cytosol (Puig et al. 2003; Wang et al. 2011; Choi et al. 2015). Sik3 phosphorylates histone deacetylase 4 (HDAC4), which sequesters HDAC4 in the cytosol, rendering it unable to interact with and activate nuclear Foxo. This results in decreased Foxo activity in the nucleus and, consequently, decreased bmm expression (Figure 3) (Wang et al. 2011; Choi et al. 2015). The failure to suppress bmm expression causes reduced TAG levels and increased starvation sensitivity in Sik3 mutant animals (Wang et al. 2011). Although the insulin pathway likely directly engages Sik3 by Akt1-dependent phosphorylation (Wang et al. 2011), Sik3 is also a direct target of the Drosophila ortholog of mammalian liver kinase B1 (Lkb1), which acts through an insulin-independent pathway, suggesting that various pathways converge at Sik3 to control bmm expression via Foxo (Choi et al. 2015). Additionally, Foxo-dependent bmm expression is repressed by the zinc finger transcription factor Kr-h1, a central effector of JH signaling (Kang et al. 2017). Notably, transcriptional control of bmm by Foxo is suggested to be evolutionarily highly conserved, as the mammalian Brummer ortholog adipose triglyceride lipase is subject to FoxO1 regulation (Chakrabarti and Kandror 2009). Bmm transcript levels show an antagonistic response to disturbed Akh and fat body Ca2+ homeostasis, as opposed to the lipogenesis gene mdy (Baumbach et al. 2014a). Genetic manipulations that deplete cytosolic Ca2+ or that result in a loss-of-function of AkhR reduce bmm transcript concentrations (Baumbach et al. 2014a), suggesting that Akh signaling via Ca2+ promotes bmm expression (Figure 3) (Baumbach et al. 2014a; Choi et al. 2015). As described above, hyperphagia driven by orexigenic sNPF contributes, in part, to this response, indicating a yet unknown nonautonomous tissue regulatory circuit controlled by fat body Ca2+. Recently, Akh signaling via Ca2+ and Calcium/Cam-dependent protein kinase II (CaMKII) has been demonstrated to inhibit secretion of the adipokine Unpaired 2 (Upd2) from the fat body (Rajan et al. 2017). Upd2 triggers systemic insulin signaling from the central brain (Rajan and Perrimon 2012) and impairs TAG mobilization in the fat body, possibly via repressing Foxo-dependent bmm transcription (Rajan and Perrimon 2012; Rajan et al. 2017). Although not formally proven, this model implies that alterations in Upd2 and systemic insulin signaling underlie Akh/Ca2+-dependent regulation of bmm expression. Similar to bmm, the expression of Hsl transcripts is induced upon starvation, but the underlying molecular mechanisms are currently unknown (Bi et al. 2012). Post-transcriptional lipase regulation has been less studied in Drosophila, but two important mechanisms include Bmm transport to the LD surface, and the functional interaction of both Bmm and Hsl with LD coat proteins. Genome-wide RNAi screens in cultured Drosophila cell lines identified the Coat protein complex I (COPI) retrograde vesicle trafficking machinery as a key regulator of LD structure and TAG storage (Beller et al. 2008; Guo et al. 2008). Further studies identified defective trafficking of Bmm or orthologous proteins to the LD surface as a likely cause of the TAG overstorage phenotype provoked by defective COPI function (Beller et al. 2008; Wilfling et al. 2014). Notably, fat body-specific knockdown of the COPI-associated small GTPase adenosine diphosphate ribosylation factor at 79F (Arf79F) recapitulates the overstorage phenotype observed in cultured cells in adult flies (Baumbach et al. 2014a).
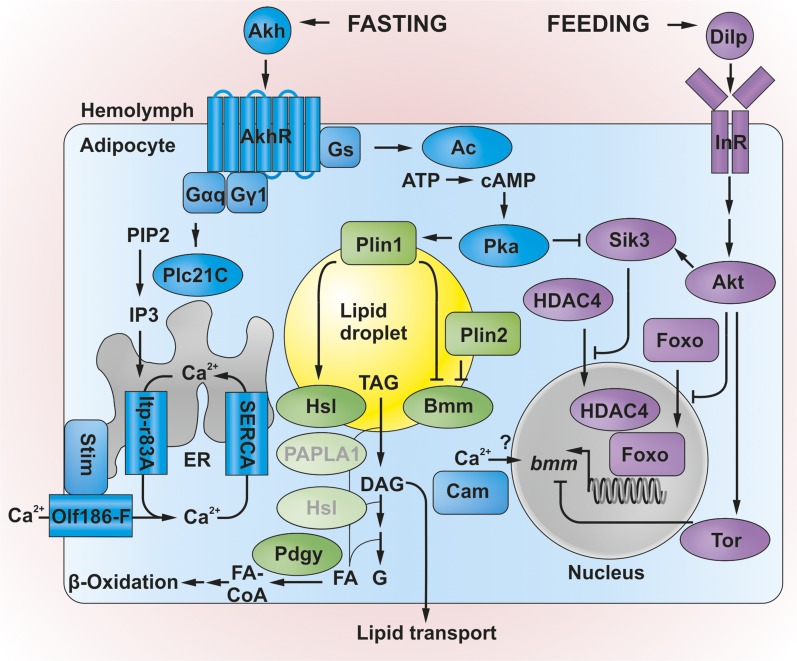
Figure 3.
Triacylglycerol (TAG) lipolysis in Drosophila: biochemistry and main regulatory pathways. Mobilization of lipid droplet (LD) fat stores in adipocytes requires TAG hydrolysis by Bmm and Hsl [possibly also by PAPLA1 (Phosphatidic Acid Phospholipase A1)] to fatty acids (FAs) and diacylglycerol (DAG), which become exported to peripheral tissues, or are subjected to further lipolytic cleavage (possibly by Hsl) to release glycerol (G) and FAs. The latter can be catabolized via β-oxidation after activation to FA-CoA by Pdgy. LD coat proteins Plin1 and Plin2 restrict Bmm activity. Plin1 phosphorylation by protein kinase A (Pka) may also promote adipokinetic hormone (Akh)-stimulated TAG mobilization by facilitating the activity of Hsl and/or other currently unknown lipases. Transcription of bmm is under antagonistic control of the Akh pathway during fasting and under the insulin signaling pathway during feeding conditions. Insulin signaling operates via activation of Akt, which represses bmm expression by promoting phosphorylation and cytoplasmic retention of the transcription factor Foxo. In parallel, Akt activates Sik3, which promotes the phosphorylation and cytoplasmic retention of the Foxo-activator HDAC4. Target of rapamycin (Tor) kinase, which is positively regulated by insulin signaling, represses bmm transcript levels by a currently unknown mechanism. The insulin and Akh pathways converge at Sik3, which is suppressed by Pka. Pka, in turn, depends on the cyclic AMP (cAMP) branch of Akh/AkhR (Akh receptor) signaling, which operates via Gs and adenylate cyclase (Ac). Lipolysis also involves the Ca2+ branch of AkhR activation (for details see Figure 2 legend and main text). Genetic data support the idea that proteins such as Cam relay the rise in intracellular Ca2+, resulting in bmm expression by unknown mechanisms. Proteins shown in faded green with gray lettering are based on homology-based activity predictions and await experimental validation. See main text for definitions of protein name abbreviations. Note the antagonistic action of the lipoanabolic insulin/Tor pathway (in magenta) under feeding vs. the lipocatabolic Akh pathway (in blue) under fasting conditions.
Perilipins are evolutionarily conserved LD-associated proteins with key functions in regulating LD structure and catabolism (Bickel et al. 2009). Studies of perilipin function in diverse phyla uncovered two canonical functions of this protein family by (1) limiting basal access of lipases to the LD surface, and (2) promoting lipase activity and LD access upon hormonal signaling [for review see Bickel et al. (2009), Kimmel and Sztalryd (2016), and Sztalryd and Brasaemle (2017)]. The Drosophila genome encodes two perilipin-related proteins termed Plin1 (also called Lipid storage droplet-1, Lsd-1) and Plin2 (also called Lipid storage droplet-2, Lsd-2), both of which contribute to the regulation of lipocatabolism (Figure 3). Plin2 acts in an antilipolytic way and opposes bmm function in a gene dosage-dependent manner (Grönke et al. 2003, 2005). Consequently, plin2 mutants are lean and starvation-sensitive, whereas transgenic flies with increased plin2 expression are obese and starvation-resistant (Grönke et al. 2003). Although the underlying molecular mechanism has not been formally addressed in Drosophila, these observations are in line with the canonical function of perilipins in limiting basal access of lipases to the LD surface, and preventing interactions between lipase and lipase substrates (Sztalryd and Brasaemle 2017). In contrast to Plin2, the second Drosophila perilipin Plin1 has been attributed with both pro- and antilipolytic functions. Genetic deficiency of plin1 results in obesity of larvae and adult flies, and impaired yet functional lipid mobilization (Beller et al. 2010; Bi et al. 2012). A double deficiency in plin1 and bmm further accentuated this phenotype resulting in flies completely lacking lipid mobilization competence (Beller et al. 2010). Loss of Plin1 impaired the starvation-induced enrichment of Hsl at larval LDs, suggesting that it acts as a docking interface for lipolytic factors (Bi et al. 2012). However, further genetic interaction studies demonstrated that Plin1 also contributes to limit basal Bmm activity (Beller et al. 2010). Both Plins are phosphorylated (Welte et al. 2005; Arrese et al. 2008; Beller et al. 2010), although the significance and dynamics of these modifications for the regulation of lipolysis in Drosophila are not completely understood (see below). A recent report linked ectopic TAG accumulation in skeletal muscle to defective proteolytic degradation of Plin2, suggesting that post-translational regulation of Plins contributes to TAG homeostasis (Yan et al. 2017). As noted above, Akh signaling influences the expression of key lipolytic/lipogenic effectors such as bmm and mdy, but also acts acutely via a second, currently unknown lipolytic effector (Grönke et al. 2007). The bmm and Akh/AkhR lipocatabolic systems show a strong genetic interaction and double mutants are extremely obese, unable to mobilize lipid stores, and highly starvation-sensitive (Grönke et al. 2007). Although the molecular events that relay Akh signaling to acute alterations in lipolytic output are understudied in Drosophila, other insect models have contributed important mechanistic insights into this process. In the M. sexta fat body, Akh signaling promotes elevated cAMP levels and subsequent activation of Pka, which phosphorylates the Manduca homolog of Plin1 (Arrese et al. 1999; Patel et al. 2005). This phosphorylation event renders Plin1-coated LDs susceptible to lipolytic activity by a cytosolic TAG lipase termed triglyceride lipase (TGL) (Patel et al. 2005; Arrese et al. 2006). Hsl and PA Phospholipase A1 (PAPLA1) are among the candidate Akh-dependent lipases in Drosophila (Figure 3). Whereas Hsl, together with Bmm, has been shown to coordinate TAG mobilization in larval stages, its function in adults remains unclear (Bi et al. 2012). Drosophila PAPLA1 is an ortholog of Manduca TGL and has been shown to possess TAG hydrolase activity in vitro (Arrese et al. 2006). PAPLA1 mutant animals show a complex spectrum of phenotypes including impaired fecundity and fertility, decreased photoreceptor sensitivity, and metabolic abnormalities (Kunduri et al. 2014; Gáliková et al. 2017). However, TAG storage and mobilization are comparable in PAPLA1 mutants and wild-type animals (Gáliková et al. 2017), which makes the hypothesis of in vivo relevance of PAPLA1 in Drosophila TAG mobilization controversial.
With notable exceptions, the steps of DAG breakdown to FA, and its subsequent degradation by mitochondrial or peroxisomal β-oxidation, have not been comprehensively characterized in the fly with respect to the involved genes and their corresponding regulatory mechanisms. However, the starvation-responsive nuclear receptor HNF4 is a critical regulator of lipid catabolism because it coordinates numerous genes involved in larval lipolysis, FA activation, and β-oxidation (Palanker et al. 2009). HNF4-induced genes include acyl-CoA synthetase (ACS) genes such as bubblegum (bgm). In the absence of bgm gene function, larvae accumulate lipids in the gastric caecae (Sivachenko et al. 2016). The importance of FA activation by acyl-CoA formation in global TAG control is further emphasized by the involvement of the Foxo-dependent ACS gene pudgy (pdgy) in TAG homeostasis. Pudgy mutants accumulate excessive storage fat and display compromised TAG mobilization (Xu et al. 2012). In contrast, mutations of the carnitine pamitoyltransferases 1 and 2, which transport activated FA into mitochondria, have comparably mild phenotypes. These mutants are starvation-sensitive, and store ectopic lipids in brain and flight muscles without an effect on global body TAG content (Schulz et al. 2015). In contrast, flies deficient for the mitochondrial trifunctional protein α or β subunits, which assemble the enzyme complex responsible for the last three steps in β-oxidation, accumulate increased body fat stores (Kishita et al. 2012). Collectively, single-gene analyses to date reveal complex roles for β-oxidation genes with respect to fly TAG homoeostasis, which might, in part, be due to functional redundancies among multiple gene family members [for an overview see Palanker et al. (2009)]. Notably, adult flies incapable of TAG mobilization are viable on a regular, carbohydrate-rich diet, but are starvation-hypersensitive (Grönke et al. 2007). This finding predicts that dietary challenge might reveal more severe consequences of genetic β-oxidation impairment.
Lipid Transport
The interorgan transport of Drosophila lipids via hemolymph involves specific lipoprotein carriers. Upon feeding, this system carries dietary or de novo-synthesized lipids from the midgut to tissues as energy supply, or to the fat body for storage. Upon starvation, the same protein system delivers lipids from the fat body to peripheral tissues (Palm et al. 2012). Like de novo lipogenesis, hemolymph lipid transport is essential, and deficiency of lipid transport components leads to developmental arrest in preadult stages (Palm et al. 2012). Hemolymph contains distinct lipoproteins of various densities that have various qualitative and quantitative roles in lipid transport. Lipophorin (Lpp) is the major lipoprotein in fly hemolymph and carries > 95% of total hemolymph lipids in feeding third-instar larvae (Palm et al. 2012). The protein fraction of Lpp consists of the two apoproteins ApoLI and ApoLII, which derive from proteolytic cleavage of a single Apolipophorin (Apolpp) precursor (encoded by the apolpp gene) and assemble into lipoprotein complexes in a 1:1 M ratio (Sundermeyer et al. 1996; Canavoso et al. 2001; Panáková et al. 2005; Palm et al. 2012). This protein component binds a mixture of polar and neutral lipids, depending on the developmental and dietary status of the animal (Palm et al. 2012). DAG is the primary neutral lipid of Drosophila Lpp and serves as the major transport form of FA in hemolymph. In contrast, only trace amounts of TAG and free FA have been identified in hemolymph. The remaining Lpp lipids include PE, free sterol, phosphatidylcholine (PC), sphingolipids, and hydrocarbons (Fernando-Warnakulasuriya and Wells 1988; Carvalho et al. 2012). Lpp-associated DAG in feeding third-instar larvae is markedly enriched in medium-chain FA chains, which are typically not found in PE or other phospholipids, suggesting channeling of specific FAs in glycerolipid synthesis pathways (Palm et al. 2012). Although the structure of Drosophila Lpp is currently unknown, studies in other insect species suggest that Lpp comprises a spherical particle with a hydrophobic core of neutral lipids and a surface covered by phospholipids, apolipoproteins, and a small fraction of DAG (Kawooya et al. 1991). Genetic studies in Drosophila third-instar larvae indicate that the fat body is the major site of Lpp synthesis and secretion (Palm et al. 2012). This observation confirms earlier biochemical studies in other insect species that demonstrate Lpp synthesis and release from isolated fat body explants (Prasad et al. 1986; Weers et al. 1992). Although the midgut is the dedicated site of lipid absorption, de novo synthesis, and export to the circulatory system, it lacks the capacity to synthesize lipoproteins, but relies on lipoprotein secretion from other tissues, principally the fat body (Palm et al. 2012). However, recent data suggest that, in response to nutritional challenges such as high fat levels, cardiac muscle cells deliver an additional pool of Lpp to the hemolymph (Lee et al. 2017).
In the fat body of feeding Drosophila third-instar larvae, Lpp is assembled as a PE-rich, but DAG-poor, precursor particle (Palm et al. 2012). The maturation of Lpp requires the presence of microsomal TAG transfer protein (Mtp), an ER-resident lipid transport protein with an evolutionarily conserved chaperoning function in the secretion of apoB-containing lipoproteins (Sellers et al. 2003). Accordingly, RNAi-mediated depletion of Mtp results in the accumulation of immature Apolpp precursors in the fat body and a deficiency of mature Lpp in hemolymph (Palm et al. 2012). Unlike its mammalian ortholog, Drosophila Mtp lacks TAG transfer activity, but displays phospholipid transfer activity in vitro consistent with its function in the biogenesis of the PE-enriched precursor Lpp (Rava and Hussain 2007). After its secretion by the fat body or cardiac muscle, Lpp is recruited to the midgut, where it is loaded with diet-derived DAG and sterols that are then redistributed to other tissues. As a consequence, RNAi-mediated interference of the hemolymph Lpp system manifests in a massive accumulation of LDs in the midgut of feeding third-instar larvae (Panáková et al. 2005; Palm et al. 2012), and in a concomitant depletion of lipid in other tissues such as the brain or imaginal discs (Palm et al. 2012; Rodríguez-Vázquez et al. 2015). Biochemical studies in non-Drosophila insects indicate that lipid delivery to tissues via Lpp occurs without concomitant degradation of ApoLI/II proteins, suggesting that Lpp acts as a reusable lipid shuttle (Downer and Chino 1985; van Heusden et al. 1991; Arrese et al. 2001).
Several molecular components are required for the interaction of Lpp with target tissues and/or lipid transfer. Lipid transfer particle (LTP, encoded by the Apoltp gene) is synthesized in the fat body as an ApoLTP precursor and, analogous to Lpp, becomes processed to ApoLTP-I and ApoLTP-II via proteolytic cleavage before secretion into hemolymph. As for Lpp, the maturation and secretion of LTP depend on the activity of Mtp (Ryan et al. 1988; Palm et al. 2012). Although circulating LTP is associated with only 1% of hemolymph lipids, it is required for bulk lipid transport by facilitating lipid transfer between Lpp and tissues. Genetic depletion of LTP in Drosophila third-instar larvae compromises Lpp recruitment to the midgut and its loading with DAG, resulting in ectopic LD accumulation in the midgut epithelium (Palm et al. 2012). LTP activity has also been implicated in lipid transfer from lipid-loaded Lpp to the ovaries and wing discs (Rodríguez-Vázquez et al. 2015).
In addition to LTP, lipid transport between hemolymph and tissues depends on members of the evolutionarily conserved low-density lipoprotein receptor family of proteins. The Drosophila genome encodes seven members of this family, of which Lpp receptors 1 and 2 (LpR1 and LpR2) share the highest sequence homology to LpR from Locusta migratoria, and to the mammalian very low-density lipoprotein-like receptor (Parra-Peralbo and Culi 2011). LpR2 directs the recruitment and extracellular stabilization of both LTP and Lpp to target tissues, suggesting that LpRs transiently form ternary complexes to facilitate lipid transport (Parra-Peralbo and Culi 2011; Rodríguez-Vázquez et al. 2015). Through the use of alternative promoters and alternative splicing mechanisms, the Drosophila LpR1 and LpR2 genes are expressed as multiple isoforms, with tissue-specific expression patterns and various capacities for lipid transfer (Parra-Peralbo and Culi 2011; Parvy et al. 2012). LpR2 is the major receptor for lipid uptake into nurse cells and oocytes during vitellogenesis, with minor contributions of LpR1 to this process (Parra-Peralbo and Culi 2011; Sieber and Spradling 2015). Loss of both receptors results in oocyte degeneration during midoogenesis. This phenotype is only partially tissue-autonomous as germline-specific deficiency causes reduced oocyte storage lipid content, but not midoogenesis degeneration (Parra-Peralbo and Culi 2011). In addition, LpR2 has also been assigned a predominant role in lipid uptake into larval oenocytes upon fasting, whereas both LpR1 and LpR2 act redundantly in mediating lipid acquisition of the wing disc (Parvy et al. 2012; Rodríguez-Vázquez et al. 2015). Notably, whole-body TAG content and mobilization were unaffected by genetic deficiency of LpR1 and LpR2, suggesting that the major routes of lipid transport are not grossly impaired. In LpR1 LpR2 double mutants, LTP failed to accumulate at nurse cell plasma membranes or wing imaginal discs, but was still recruited to many other tissues, suggesting that LpR1/LpR2-independent lipid transport pathways contribute to lipid transport (Parra-Peralbo and Culi 2011; Rodríguez-Vázquez et al. 2015).
Despite the identification of key molecular components of intertissue lipid transport in Drosophila, little is known about the molecular mechanisms of this process. Biochemical studies in other insects suggest that LTP acquires limited amounts of lipid from a donor lipoprotein or a tissue, and transfers it to a receptor (Blacklock et al. 1992). Consistent with this hypothesis, depletion of the Lpp acceptor by RNAi increases the density of LTP particles (Palm et al. 2012). Notably, the ability of LTP to transfer lipids to Lpp appears to be tissue-selective. For example, LTP transfers lipids from the explanted midgut, but not from the explanted fat body of feeding Drosophila third-instar larvae to Lpp (Palm et al. 2012). Although Lpp receptors have been shown to induce Lpp endocytosis in cells and insect tissues (Dantuma et al. 1999; Van Hoof et al. 2002, 2003; Callejo et al. 2008), the functional relevance of this observation for lipid transport mechanisms is unclear and may again be tissue-specific. Genetic experiments that interfered with the endocytosis machinery suggest that lipid transport to the oocyte is endocytosis-independent (Rodríguez-Vázquez et al. 2015), whereas Lpp loading at the midgut of third-instar larvae partly depends on functional endocytosis (Palm et al. 2012). How lipid gets transported across the endosomal or plasma membranes, and if this process involves lipid processing (e.g., by lipoprotein or lysosomal lipases), is currently unknown.
Future Perspectives
The preponderance of human lipid storage-associated disorders such as diabetes, obesity, and cardiovascular disease has spurred efforts to investigate TAG metabolism in genetically tractable model organisms. Metabolic phenotyping of Drosophila mutants, in combination with the use of genetic screens, have been fundamental tools in the identification of evolutionarily conserved regulators of TAG storage (Pospisilik et al. 2010; Reis et al. 2010; Baumbach et al. 2014a). The past decades of research on Drosophila TAG metabolism have shaped the view that the molecular processes of TAG formation, degradation, and storage are remarkably similar between Drosophila and higher organisms. Nevertheless, a comprehensive understanding of the molecular, cellular, and organismal principles of Drosophila TAG metabolism requires continued efforts among the steadily growing research community active in this field. Several important directions in this area are likely to fill those knowledge gaps and contribute novel insights into TAG metabolism, with relevance to basic and translational research.
Characterizing the basic enzymology and biochemistry of Drosophila TAG metabolism is a promising field. Genetic evidence indicates the presence of yet unknown enzymatic effectors in TAG formation and degradation in flies. However, functional annotations of enzymes are often based on homology to mammalian proteins and require confirmatory biochemical evidence. Moreover, enzyme isoforms may differentially or redundantly contribute to tissue-autonomous TAG metabolism and/or act during specific developmental stages. With clustered regularly interspaced short palindromic repeats/Cas9-based genome editing tools, the study of redundant genetic control by multiple genes is now accessible to functional analyses. Complementation of such in vivo functional analysis with the comprehensive enzymatic characterization of Drosophila TAG metabolism promises to be a solid framework for addressing open fundamental questions in lipid biology. Some of the important issues to be addressed include: which isoenzymes catalyze TAG formation or breakdown in the fat body and peripheral tissues, how do they incorporate specific FAs into DAG and TAG as opposed to glycerophospholipids, and what is the contribution of lysosomes to the breakdown of lipoproteins or cytosolic LDs?
FAs are precursors for both structural and storage lipids. Accordingly, metabolic pathways of TAG are not independent from the effects of the presence of structural lipids such as glycerophospholipids or sphingolipids, which may interact with TAG metabolism at multiple levels. A prominent example of this interconnection is the repression of SREBP activity by the major Drosophila membrane lipid PE, which leads to tissue-autonomous or global alterations in de novo lipogenesis and TAG storage (Lim et al. 2011; Meltzer et al. 2017; Ziegler et al. 2017). Likewise, PC, the second most abundant glycerophospholipid in Drosophila membranes, has been identified as a critical modulator of TAG storage. PC acts as a structural component of the phospholipid monolayer surrounding LDs, and depletion of PC synthesis enzymes favors LD coalescence, and increases LD size and TAG levels (Krahmer et al. 2011; Moessinger et al. 2014). In analogy to glycerophospholipid metabolic pathways, sphingolipid formation and breakdown have also been implicated in TAG storage (Bauer et al. 2009; Kohyama-Koganeya et al. 2011; Nirala et al. 2013; Walls et al. 2013; Sociale et al. 2018). Reciprocal to the concept that membrane lipids affect the storage lipid pools, FAs liberated from TAG stores may also affect synthesis of sphingolipids or glycerophospholipids, with profound effects on membrane structure and function. This dual role of TAG lipolysis in providing FAs for β-oxidation, as well as providing structural lipids, might be particularly relevant during nonfeeding stages of development or during tissue remodeling as, for example, during embryogenesis or metamorphosis. The advent of lipidomic and metabolic labeling techniques in Drosophila developmental research provides unique tools for future studies of these processes (Carvalho et al. 2012; Guan et al. 2013; Musselman et al. 2013).
The regulation of TAG homeostasis in a constantly changing environment requires elaborate endocrine mechanisms that orchestrate intertissue communication. Research in Drosophila has contributed significantly to answer the question of how organs sense and communicate fluctuations in energy status (Colombani et al. 2003; Géminard et al. 2009). These studies have recently highlighted the role of cytokines and other humoral factors, which are released in tissue-specific patterns to communicate changes in the nutrient status and adjust TAG homeostasis, often via cross talk with insulin or Akh signaling pathways (Rajan and Perrimon 2012; Agrawal et al. 2016; Delanoue et al. 2016; Song et al. 2017; Zhao and Karpac 2017). However, how these pathways relay the information to direct changes in the expression or activity of enzymatic effectors that control TAG storage and mobilization is still poorly understood. One attractive hypothesis of coordinated, environmental-responsive gene regulation involves microRNAs (miRs). Notably, starvation reduces the levels of miR machinery components, implying the possible coupling of miR effectors to energy status (Barrio et al. 2014). Several miRs have been implicated in TAG storage control, e.g., mir-278 (Teleman et al. 2006) and mir-14 (Xu et al. 2003; Varghese et al. 2010), which both modulate energy homeostasis via insulin signaling. Whether or not miRs control TAG metabolism more directly at the level of enzymes is an important issue to be resolved in future research.
The function of TAG synthesis and TAG deposition in LDs is not limited to energy storage, and may be highly tissue-specific. One of the most active fields in fly lipid research currently addresses the roles of TAG metabolism in neurodegeneration. Early molecular characterization of Drosophila neurodegeneration mutants, such as swiss cheese (sws) (Kretzschmar et al. 1997) and bubblegum (bgm) (Min and Benzer 1999), has already indicated the importance of lipid metabolism in neuronal integrity with relevance to human neurodegenerative disorders [reviewed in Lessing and Bonini (2009)]. More recent research has demonstrated that LDs fulfill a developmental function in brain glial cells by serving as a sink to protect unsaturated FAs, which are instrumental for neuroblast function, from excessive lipid peroxidation (Bailey et al. 2015). Similarly, transient LD accumulation in adult brain glia has been suggested to act neuroprotectively by scavenging peroxidated lipids under conditions of mitochondrial dysfunction in neurons (Liu et al. 2015). In this context, glial LDs participate in an evolutionarily conserved glia–neuron lactate shuttle (Liu et al. 2017). Under stress conditions, neurons use glia-derived lactate for the production of lipids, which are subject to peroxidation and become exported to build up glial LDs. Conditional ectopic LD formation is not restricted to the nervous systems, as exemplified by the larval salivary gland. In this tissue, ectopic LDs appear upon genetically compromised function of Seipin or of the cytidine 5′-diphosphate-DAG synthase gene Cds. In these cases, ectopic LD formation has been proposed to result from rerouting of glycerophospholipid precursors such as PA into TAG (Tian et al. 2011; Liu et al. 2014). In line with the concept of “lipotoxicity,” these studies suggest a role of TAG synthesis and LD formation as a tissue-specific metabolic buffer for excess lipid. Yet, ectopic LD formation is highly dependent on the (patho)physiological circumstances, and on the cellular context. Collectively, the role of ectopic LDs as a trigger, bystander, or buffer of cellular stress is still controversial, which makes them an active and promising area of TAG metabolism research in the fly.
Acknowledgments
The authors thank the peer reviewers for constructive criticisms on the manuscript and Lisa Pichler for copy editing. They apologize to those colleagues whose important contributions to this active research field are not adequately covered due to the scope of this review. The authors acknowledge the support of the University of Graz.
Footnotes
Communicating editor: C. Thummel
Literature Cited
- Agrawal N., Delanoue R., Mauri A., Basco D., Pasco M., et al. , 2016. The Drosophila TNF eiger is an adipokine that acts on insulin-producing cells to mediate nutrient response. Cell Metab. 23: 675–684. 10.1016/j.cmet.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Alic N., Andrews T. D., Giannakou M. E., Papatheodorou I., Slack C., et al. , 2011. Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol. Syst. Biol. 7: 502 10.1038/msb.2011.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese E. L., Wells M. A., 1997. Adipokinetic hormone-induced lipolysis in the fat body of an insect, Manduca sexta: synthesis of sn-1,2-diacylglycerols. J. Lipid Res. 38: 68–76. [PubMed] [Google Scholar]
- Arrese E. L., Rojas-Rivas B. I., Wells M. A., 1996. Synthesis of sn-1,2-diacylglycerols by monoacylglycerol acyltransferase from Manduca sexta fat body. Arch. Insect Biochem. Physiol. 31: 325–335. [DOI] [PubMed] [Google Scholar]
- Arrese E. L., Flowers M. T., Gazard J. L., Wells M. A., 1999. Calcium and cAMP are second messengers in the adipokinetic hormone-induced lipolysis of triacylglycerols in Manduca sexta fat body. J. Lipid Res. 40: 556–564. [PubMed] [Google Scholar]
- Arrese E. L., Canavoso L. E., Jouni Z. E., Pennington J. E., Tsuchida K., et al. , 2001. Lipid storage and mobilization in insects: current status and future directions. Insect Biochem. Mol. Biol. 31: 7–17. 10.1016/S0965-1748(00)00102-8 [DOI] [PubMed] [Google Scholar]
- Arrese E. L., Patel R. T., Soulages J. L., 2006. The main triglyceride-lipase from the insect fat body is an active phospholipase A1: identification and characterization. J. Lipid Res. 47: 2656–2667. 10.1194/jlr.M600161-JLR200 [DOI] [PubMed] [Google Scholar]
- Arrese E. L., Rivera L., Hamada M., Mirza S., Hartson S. D., et al. , 2008. Function and structure of lipid storage droplet protein 1 studied in lipoprotein complexes. Arch. Biochem. Biophys. 473: 42–47. 10.1016/j.abb.2008.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G., 2003. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278: 23317–23323. 10.1074/jbc.M302577200 [DOI] [PubMed] [Google Scholar]
- Bailey A. P., Koster G., Guillermier C., Hirst E. M., MacRae J. I., et al. , 2015. Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell 163: 340–353. 10.1016/j.cell.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. D., Thummel C. S., 2007. Diabetic larvae and obese flies—emerging studies of metabolism in Drosophila. Cell Metab. 6: 257–266. 10.1016/j.cmet.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M. C., Price N. T., Travers M. T., 2005. Structure and regulation of acetyl-CoA carboxylase genes of metazoa. Biochim. Biophys. Acta 1733: 1–28. 10.1016/j.bbalip.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Barrio L., Dekanty A., Milán M., 2014. MicroRNA-mediated regulation of Dp53 in the Drosophila fat body contributes to metabolic adaptation to nutrient deprivation. Cell Rep. 8: 528–541. 10.1016/j.celrep.2014.06.020 [DOI] [PubMed] [Google Scholar]
- Bauer R., Voelzmann A., Breiden B., Schepers U., Farwanah H., et al. , 2009. Schlank, a member of the ceramide synthase family controls growth and body fat in Drosophila. EMBO J. 28: 3706–3716. 10.1038/emboj.2009.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A. A., Benoit J. B., Michalkova V., Mireji P. O., Attardo G. M., et al. , 2013. Juvenile hormone and insulin suppress lipolysis between periods of lactation during tsetse fly pregnancy. Mol. Cell. Endocrinol. 372: 30–41. 10.1016/j.mce.2013.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J., Hummel P., Bickmeyer I., Kowalczyk K. M., Frank M., et al. , 2014a. A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metab. 19: 331–343. 10.1016/j.cmet.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Baumbach J., Xu Y., Hehlert P., Kühnlein R. P., 2014b. Gαq, Gγ1 and Plc21C control Drosophila body fat storage. J. Genet. Genomics 41: 283–292. 10.1016/j.jgg.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Bell R. M., Coleman R. A., 1980. Enzymes of glycerolipid synthesis in eukaryotes. Annu. Rev. Biochem. 49: 459–487. 10.1146/annurev.bi.49.070180.002331 [DOI] [PubMed] [Google Scholar]
- Beller M., Riedel D., Jänsch L., Dieterich G., Wehland J., et al. , 2006. Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics 5: 1082–1094. 10.1074/mcp.M600011-MCP200 [DOI] [PubMed] [Google Scholar]
- Beller M., Sztalryd C., Southall N., Bell M., Jäckle H., et al. , 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6: e292 10.1371/journal.pbio.0060292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M., Bulankina A. V., Hsiao H.-H., Urlaub H., Jäckle H., et al. , 2010. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12: 521–532. 10.1016/j.cmet.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Bharucha K. N., Tarr P., Zipursky S. L., 2008. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J. Exp. Biol. 211: 3103–3110. 10.1242/jeb.016451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Xiang Y., Chen H., Liu Z., Grönke S., et al. , 2012. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J. Cell Sci. 125: 3568–3577. 10.1242/jcs.101329 [DOI] [PubMed] [Google Scholar]
- Bi J., Wang W., Liu Z., Huang X., Jiang Q., et al. , 2014. Seipin promotes adipose tissue fat storage through the ER Ca2+-ATPase SERCA. Cell Metab. 19: 861–871. 10.1016/j.cmet.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Bickel P. E., Tansey J. T., Welte M. A., 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta 1791: 419–440. 10.1016/j.bbalip.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklock B. J., Smillie M., Ryan R. O., 1992. Insect lipid transfer particle can facilitate net vectorial lipid transfer via a carrier-mediated mechanism. J. Biol. Chem. 267: 14033–14037. [PubMed] [Google Scholar]
- Böhni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., et al. , 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97: 865–875. 10.1016/S0092-8674(00)80799-0 [DOI] [PubMed] [Google Scholar]
- Bolukbasi E., Khericha M., Regan J. C., Ivanov D. K., Adcott J., et al. , 2017. Intestinal fork head regulates nutrient absorption and promotes longevity. Cell Rep. 21: 641–653. 10.1016/j.celrep.2017.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Osman D., David F. P., Fang H. Y., Boquete J. P., et al. , 2013. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 3: 1725–1738 (erratum: Cell Rep. 3: 1755) 10.1016/j.celrep.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Lu X., Segraves W. A., Chang T. Y., Cooley L., 2002. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics 160: 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callejo A., Culi J., Guerrero I., 2008. Patched, the receptor of Hedgehog, is a lipoprotein receptor. Proc. Natl. Acad. Sci. USA 105: 912–917. 10.1073/pnas.0705603105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavoso L. E., Jouni Z. E., Karnas K. J., Pennington J. E., Wells M. A., 2001. Fat metabolism in insects. Annu. Rev. Nutr. 21: 23–46. 10.1146/annurev.nutr.21.1.23 [DOI] [PubMed] [Google Scholar]
- Carney G. E., Bender M., 2000. The Drosophila ecdysone receptor (EcR) gene is required maternally for normal oogenesis. Genetics 154: 1203–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M., Sampaio J. L., Palm W., Brankatschk M., Eaton S., et al. , 2012. Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 8: 600–617. 10.1038/msb.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermelli S., Guo Y., Gross S. P., Welte M. A., 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16: 1783–1795. 10.1016/j.cub.2006.07.062 [DOI] [PubMed] [Google Scholar]
- Chakrabarti P., Kandror K. V., 2009. FoxO1 controls insulin-dependent adipose triglyceride lipase (ATGL) expression and lipolysis in adipocytes. J. Biol. Chem. 284: 13296–13300. 10.1074/jbc.C800241200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles J.-P., Iwema T., Epa V. C., Takaki K., Rynes J., et al. , 2011. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc. Natl. Acad. Sci. USA 108: 21128–21133. 10.1073/pnas.1116123109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D., Katewa S. D., Qi Y., Jackson S. A., Kapahi P., et al. , 2014. Control of metabolic adaptation to fasting by dILP6-induced insulin signaling in Drosophila oenocytes. Proc. Natl. Acad. Sci. USA 111: 17959–17964. 10.1073/pnas.1409241111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Chang B., Saha P., Hartig S. M., Li L., et al. , 2012. Berardinelli-seip congenital lipodystrophy 2/seipin is a cell-autonomous regulator of lipolysis essential for adipocyte differentiation. Mol. Cell. Biol. 32: 1099–1111. 10.1128/MCB.06465-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chino H., Gilbert L. I., 1964. Diglyceride release from insect fat body: a possible means of lipid transport. Science 143: 359–361. 10.1126/science.143.3604.359 [DOI] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A. T., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Chng W.-B. A., Sleiman M. S. B., Schüpfer F., Lemaitre B., 2014. Transforming growth factor β/Activin signaling functions as a sugar-sensing feedback loop to regulate digestive enzyme expression. Cell Rep. 9: 336–348. 10.1016/j.celrep.2014.08.064 [DOI] [PubMed] [Google Scholar]
- Cho K. S., Lee J. H., Kim S., Kim D., Koh H., et al. , 2001. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc. Natl. Acad. Sci. USA 98: 6144–6149. 10.1073/pnas.101596998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Lim D.-S., Chung J., 2015. Feeding and fasting signals converge on the LKB1–SIK3 pathway to regulate lipid metabolism in Drosophila. PLoS Genet. 11: e1005263 10.1371/journal.pgen.1005263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H., Loehlin D. W., Dufour H. D., Vaccarro K., Millar J. G., et al. , 2014. A single gene affects both ecological divergence and mate choice in Drosophila. Science 343: 1148–1151. 10.1126/science.1249998 [DOI] [PubMed] [Google Scholar]
- Colombani J., Raisin S., Pantalacci S., Radimerski T., Montagne J., et al. , 2003. A nutrient sensor mechanism controls Drosophila growth. Cell 114: 739–749. 10.1016/S0092-8674(03)00713-X [DOI] [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., et al. , 2005. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310: 667–670. 10.1126/science.1119432 [DOI] [PubMed] [Google Scholar]
- Dantuma N. P., Potters M., De Winther M. P., Tensen C. P., Kooiman F. P., et al. , 1999. An insect homolog of the vertebrate very low density lipoprotein receptor mediates endocytosis of lipophorins. J. Lipid Res. 40: 973–978. [PubMed] [Google Scholar]
- Delanoue R., Meschi E., Agrawal N., Mauri A., Tsatskis Y., et al. , 2016. Drosophila insulin release is triggered by adipose Stunted ligand to brain Methuselah receptor. Science 353: 1553–1556. 10.1126/science.aaf8430 [DOI] [PubMed] [Google Scholar]
- DiAngelo J. R., Birnbaum M. J., 2009. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol. Cell. Biol. 29: 6341–6352. 10.1128/MCB.00675-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Yang X., Tian H., Liang J., Zhang F., et al. , 2018. Seipin regulates lipid homeostasis by ensuring calcium-dependent mitochondrial metabolism. EMBO J. 37: e97572 10.15252/embj.201797572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrosotskaya I. Y., Seegmiller A. C., Brown M. S., Goldstein J. L., Rawson R. B., 2002. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296: 879–883. 10.1126/science.1071124 [DOI] [PubMed] [Google Scholar]
- Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K., 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282: 3450–3457. 10.1074/jbc.M610745200 [DOI] [PubMed] [Google Scholar]
- Downer R., Chino H., 1985. Turnover of protein and diacylglycerol components of lipophorin in insect haemolymph. Insect Biochem. 15: 627–630. 10.1016/0020-1790(85)90124-6 [DOI] [Google Scholar]
- Eastmond P. J., 2006. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675. 10.1105/tpc.105.040543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando-Warnakulasuriya G. J. P., Wells M. A., 1988. Isolation and characterization of lipophorin from Drosophila melanogaster larvae. Arch. Insect Biochem. Physiol. 8: 243–248. 10.1002/arch.940080405 [DOI] [Google Scholar]
- Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., et al. , 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 4: 199–210. 10.1016/j.cmet.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Flatt T., Tu M.-P., Tatar M., 2005. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. BioEssays 27: 999–1010. 10.1002/bies.20290 [DOI] [PubMed] [Google Scholar]
- Francis V. A., Zorzano A., Teleman A. A., 2010. dDOR is an EcR coactivator that forms a feed-forward loop connecting insulin and ecdysone signaling. Curr. Biol. 20: 1799–1808. 10.1016/j.cub.2010.08.055 [DOI] [PubMed] [Google Scholar]
- Gäde G., Auerswald L., 2003. Mode of action of neuropeptides from the adipokinetic hormone family. Gen. Comp. Endocrinol. 132: 10–20. 10.1016/S0016-6480(03)00159-X [DOI] [PubMed] [Google Scholar]
- Gáliková M., Diesner M., Klepsatel P., Hehlert P., Xu Y., et al. , 2015. Energy homeostasis control in Drosophila adipokinetic hormone mutants. Genetics 201: 665–683. 10.1534/genetics.115.178897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáliková M., Klepsatel P., Münch J., Kühnlein R. P., 2017. Spastic paraplegia-linked phospholipase PAPLA1 is necessary for development, reproduction, and energy metabolism in Drosophila. Sci Rep. 7: 46516 10.1038/srep46516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Rubin T., Poidevin M., Maroni B., Le Rouzic A., et al. , 2015. Fatty acid synthase cooperates with glyoxalase 1 to protect against sugar toxicity. PLoS Genet. 11: e1004995 10.1371/journal.pgen.1004995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géminard C., Rulifson E. J., Léopold P., 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10: 199–207. 10.1016/j.cmet.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Grillet M., Dominguez Gonzalez B., Sicart A., Pöttler M., Cascalho A., et al. , 2016. Torsins are essential regulators of cellular lipid metabolism. Dev. Cell 38: 235–247. 10.1016/j.devcel.2016.06.017 [DOI] [PubMed] [Google Scholar]
- Grippa A., Buxó L., Mora G., Funaya C., Idrissi F.-Z., et al. , 2015. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211: 829–844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Beller M., Fellert S., Ramakrishnan H., Jäckle H., et al. , 2003. Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 13: 603–606. 10.1016/S0960-9822(03)00175-1 [DOI] [PubMed] [Google Scholar]
- Grönke S., Mildner A., Fellert S., Tennagels N., Petry S., et al. , 2005. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1: 323–330. 10.1016/j.cmet.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Grönke S., Müller G., Hirsch J., Fellert S., Andreou A., et al. , 2007. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 5: e137 10.1371/journal.pbio.0050137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Clarke D.-F., Broughton S., Andrews T. D., Partridge L., 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6: e1000857 10.1371/journal.pgen.1000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X. L., Cestra G., Shui G., Kuhrs A., Schittenhelm R. B., et al. , 2013. Biochemical membrane lipidomics during Drosophila development. Dev. Cell 24: 98–111. 10.1016/j.devcel.2012.11.012 [DOI] [PubMed] [Google Scholar]
- Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., et al. , 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453: 657–661. 10.1038/nature06928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E., Wiggins D., Fielding B., Gould A. P., 2006. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445: 275–280. 10.1038/nature05382 [DOI] [PubMed] [Google Scholar]
- Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., et al. , 2002. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J. Biol. Chem. 277: 4806–4815. 10.1074/jbc.M110355200 [DOI] [PubMed] [Google Scholar]
- Han G.-S., Wu W.-I., Carman G. M., 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218. 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S. T., Chang S., Kamdar K. P., Mackay T. F. C., 2005. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 6: R36 10.1186/gb-2005-6-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., et al. , 2007. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282: 277–286. 10.1074/jbc.M609537200 [DOI] [PubMed] [Google Scholar]
- Havula E., Teesalu M., Hyötyläinen T., Seppälä H., Hasygar K., et al. , 2013. Mondo/ChREBP-Mlx-regulated transcriptional network is essential for dietary sugar tolerance in Drosophila. PLoS Genet 9: e1003438 10.1371/journal.pgen.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne I., Haritos V. S., Oakeshott J. G., 2009. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem. Mol. Biol. 39: 547–567. 10.1016/j.ibmb.2009.06.002 [DOI] [PubMed] [Google Scholar]
- Horner M. A., Pardee K., Liu S., King-Jones K., Lajoie G., et al. , 2009. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 23: 2711–2716. 10.1101/gad.1833609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. D., 2002. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem. Soc. Trans. 30: 1091–1095. 10.1042/bst0301091 [DOI] [PubMed] [Google Scholar]
- Jindra M., Uhlirova M., Charles J.-P., Smýkal V., Hill R. J., 2015. Genetic evidence for function of the bHLH-PAS protein Gce/Met as a juvenile hormone receptor. PLoS Genet. 11: e1005394 10.1371/journal.pgen.1005394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jünger M. A., Rintelen F., Stocker H., Wasserman J. D., Végh M., et al. , 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2: 20 10.1186/1475-4924-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoshida Y., Fujiyama-Nakamura S., Kimura S., Suzuki E., Lim J., et al. , 2012. Ecdysone receptor (EcR) suppresses lipid accumulation in the Drosophila fat body via transcription control. Biochem. Biophys. Res. Commun. 421: 203–207. 10.1016/j.bbrc.2012.03.135 [DOI] [PubMed] [Google Scholar]
- Kang P., Chang K., Liu Y., Bouska M., Birnbaum A., et al. , 2017. Drosophila Kruppel homolog 1 represses lipolysis through interaction with dFOXO. Sci. Rep. 7: 16369 10.1038/s41598-017-16638-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpac J., Biteau B., Jasper H., 2013. Misregulation of an adaptive metabolic response contributes to the age-related disruption of lipid homeostasis in Drosophila. Cell Rep. 4: 1250–1261. 10.1016/j.celrep.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa S. D., Demontis F., Kolipinski M., Hubbard A., Gill M. S., et al. , 2012. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab. 16: 97–103. 10.1016/j.cmet.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawooya J. K., van der Horst D. J., van Heusden M. C., Brigot B. L., van Antwerpen R., et al. , 1991. Lipophorin structure analyzed by in vitro treatment with lipases. J. Lipid Res. 32: 1781–1788. [PubMed] [Google Scholar]
- Kim J., Neufeld T. P., 2015. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat. Commun. 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A. R., Sztalryd C., 2016. The perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu. Rev. Nutr. 36: 471–509. 10.1146/annurev-nutr-071813-105410 [DOI] [PubMed] [Google Scholar]
- Kishita Y., Tsuda M., Aigaki T., 2012. Impaired fatty acid oxidation in a Drosophila model of mitochondrial trifunctional protein (MTP) deficiency. Biochem. Biophys. Res. Commun. 419: 344–349. 10.1016/j.bbrc.2012.02.026 [DOI] [PubMed] [Google Scholar]
- Koelle M. R., Talbot W. S., Segraves W. A., Bender M. T., Cherbas P., et al. , 1991. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67: 59–77. 10.1016/0092-8674(91)90572-G [DOI] [PubMed] [Google Scholar]
- Kohyama-Koganeya A., Nabetani T., Miura M., Hirabayashi Y., 2011. Glucosylceramide synthase in the fat body controls energy metabolism in Drosophila. J. Lipid Res. 52: 1392–1399. 10.1194/jlr.M014266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., et al. , 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14: 504–515. 10.1016/j.cmet.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N., Hilger M., Kory N., Wilfling F., Stoehr G., et al. , 2013. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics 12: 1115–1126. 10.1074/mcp.M112.020230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar D., Hasan G., Sharma S., Heisenberg M., Benzer S., 1997. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J. Neurosci. 17: 7425–7432. 10.1523/JNEUROSCI.17-19-07425.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krycer J. R., Sharpe L. J., Luu W., Brown A. J., 2010. The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol. Metab. 21: 268–276. 10.1016/j.tem.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Kühnlein R. P., 2011. The contribution of the Drosophila model to lipid droplet research. Prog. Lipid Res. 50: 348–356. 10.1016/j.plipres.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Kunduri G., Yuan C., Parthibane V., Nyswaner K. M., Kanwar R., et al. , 2014. Phosphatidic acid phospholipase A1 mediates ER–Golgi transit of a family of G protein–coupled receptors. J. Cell Biol. 206: 79–95. 10.1083/jcb.201405020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte A. S., Matthews K. A., Rawson R. B., 2006. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 3: 439–448. 10.1016/j.cmet.2006.04.011 [DOI] [PubMed] [Google Scholar]
- Kurat C. F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., et al. , 2005. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281: 491–500. 10.1074/jbc.M508414200 [DOI] [PubMed] [Google Scholar]
- Lee P.-T., Lin G., Lin W.-W., Diao F., White B. H., et al. , 2018. A kinase-dependent feedforward loop affects CREBB stability and long term memory formation. Elife 7: e33007 10.7554/eLife.33007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Bao H., Ishikawa Z., Wang W., Lim H.-Y., 2017. Cardiomyocyte regulation of systemic lipid metabolism by the apolipoprotein B-containing lipoproteins in Drosophila. PLoS Genet. 13: e1006555 10.1371/journal.pgen.1006555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leevers S. J., Weinkove D., MacDougall L. K., Hafen E., Waterfield M. D., 1996. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 15: 6584–6594. 10.1002/j.1460-2075.1996.tb01049.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., 2018. Endocrine and physiological regulation of neutral fat storage in Drosophila. Mol. Cell. Endocrinol. 461: 165–177. 10.1016/j.mce.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Miguel-Aliaga I., 2013. The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 47: 377–404. 10.1146/annurev-genet-111212-133343 [DOI] [PubMed] [Google Scholar]
- Lessing D., Bonini N. M., 2009. Maintaining the brain: insight into human neurodegeneration from Drosophila melanogaster mutants. Nat. Rev. Genet. 10: 359–370. 10.1038/nrg2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. Y., Wang W., Wessells R. J., Ocorr K., Bodmer R., 2011. Phospholipid homeostasis regulates lipid metabolism and cardiac function through SREBP signaling in Drosophila. Genes Dev. 25: 189–200. 10.1101/gad.1992411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang K., Sandoval H., Yamamoto S., Jaiswal M., et al. , 2015. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 160: 177–190. 10.1016/j.cell.2014.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., MacKenzie K. R., Putluri N., Maletić-Savatić M., Bellen H. J., 2017. The glia-neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab. 26: 719–737.e6. 10.1016/j.cmet.2017.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Siloto R. M. P., Lehner R., Stone S. J., Weselake R. J., 2012. Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 51: 350–377. 10.1016/j.plipres.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang W., Shui G., Huang X., 2014. CDP-diacylglycerol synthetase coordinates cell growth and fat storage through phosphatidylinositol metabolism and the insulin pathway. PLoS Genet. 10: e1004172 10.1371/journal.pgen.1004172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magré J., Delépine M., Khallouf E., Gedde-Dahl T., Van Maldergem L., et al. , 2001. Identification of the gene altered in Berardinelli–Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28: 365–370. 10.1038/ng585 [DOI] [PubMed] [Google Scholar]
- Makki R., Cinnamon E., Gould A. P., 2014. The development and functions of oenocytes. Annu. Rev. Entomol. 59: 405–425. 10.1146/annurev-ento-011613-162056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. A., Ozdemir C., Rawson R. B., 2010. Activation of sterol regulatory element binding proteins in the absence of Scap in Drosophila melanogaster. Genetics 185: 189–198. 10.1534/genetics.110.114975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J., Hietakangas V., 2017. Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 207: 1231–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J., Havula E., Suominen E., Teesalu M., Surakka I., et al. , 2015. Mondo-mlx mediates organismal sugar sensing through the gli-similar transcription factor sugarbabe. Cell Rep. 13: 350–364. 10.1016/j.celrep.2015.08.081 [DOI] [PubMed] [Google Scholar]
- Mayer R. J., Candy D. J., 1969. Control of haemolymph lipid concentration during locust flight: an adipokinetic hormone from the corpora cardiaca. J. Insect Physiol. 15: 611–620. 10.1016/0022-1910(69)90259-5 [DOI] [Google Scholar]
- McKay R. M., McKay J. P., Avery L., Graff J. M., 2003. C elegans: a model for exploring the genetics of fat storage. Dev. Cell 4: 131–142. 10.1016/S1534-5807(02)00411-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer S., Bagley J. A., Perez G. L., O’Brien C. E., DeVault L., et al. , 2017. Phospholipid homeostasis regulates dendrite morphogenesis in Drosophila sensory neurons. Cell Rep. 21: 859–866. 10.1016/j.celrep.2017.09.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K. T., Benzer S., 1999. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science 284: 1985–1988. 10.1126/science.284.5422.1985 [DOI] [PubMed] [Google Scholar]
- Moessinger C., Klizaite K., Steinhagen A., Philippou-Massier J., Shevchenko A., et al. , 2014. Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 15: 43 10.1186/s12860-014-0043-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraru A., Cakan-Akdogan G., Strassburger K., Males M., Mueller S., et al. , 2017. THADA regulates the organismal balance between energy storage and heat production. Dev. Cell 41: 72–81.e6 (erratum: Dev. Cell 41: 450) 10.1016/j.devcel.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman L. P., Kühnlein R. P., 2018. Drosophila as a model to study obesity and metabolic disease. J. Exp. Biol. 221: jeb163881 10.1242/jeb.163881 [DOI] [PubMed] [Google Scholar]
- Musselman L. P., Fink J. L., Ramachandran P. V., Patterson B. W., Okunade A. L., et al. , 2013. Role of fat body lipogenesis in protection against the effects of caloric overload in Drosophila. J. Biol. Chem. 288: 8028–8042. 10.1074/jbc.M112.371047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R., Vanden Broeck J., 2015. Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides. Cell. Mol. Life Sci. 73: 271–290. 10.1007/s00018-015-2063-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R., Liu Y., Luo J., 2015. Insulin/IGF signaling and its regulation in Drosophila. Gen. Comp. Endocrinol. 221: 255–266. 10.1016/j.ygcen.2014.11.021 [DOI] [PubMed] [Google Scholar]
- Nirala N. K., Rahman M., Walls S. M., Singh A., Zhu L. J., et al. , 2013. Survival response to increased ceramide involves metabolic adaptation through novel regulators of glycolysis and lipolysis. PLoS Genet. 9: e1003556 10.1371/journal.pgen.1003556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S., Stocker H., Laffargue M., Wittwer F., Wymann M., et al. , 2002. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development 129: 4103–4109. [DOI] [PubMed] [Google Scholar]
- Osborne T. F., Espenshade P. J., 2009. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 23: 2578–2591. 10.1101/gad.1854309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öst A., Lempradl A., Casas E., Weigert M., Tiko T., et al. , 2014. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159: 1352–1364. 10.1016/j.cell.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E., Perrimon N., 2014. Modeling metabolic homeostasis and nutrient sensing in Drosophila: implications for aging and metabolic diseases. Dis. Model. Mech. 7: 343–350. 10.1242/dmm.012989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac M., Cooper D. E., Qi Y., Lukmantara I. E., Mak H. Y., et al. , 2016. SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17: 1546–1559. 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L., Tennessen J. M., Lam G., Thummel C. S., 2009. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 9: 228–239. 10.1016/j.cmet.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W., Sampaio J. L., Brankatschk M., Carvalho M., Mahmoud A., et al. , 2012. Lipoproteins in Drosophila melanogaster–assembly, function, and influence on tissue lipid composition. PLoS Genet. 8: e1002828 10.1371/journal.pgen.1002828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palu R. A. S., Praggastis S. A., Thummel C. S., 2017. Parental obesity leads to metabolic changes in the F2 generation in Drosophila. Mol. Metab. 6: 631–639. 10.1016/j.molmet.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panáková D., Sprong H., Marois E., Thiele C., Eaton S., 2005. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 435: 58–65. 10.1038/nature03504 [DOI] [PubMed] [Google Scholar]
- Papadopoulou D., Bianchi M. W., Bourouis M., 2004. Functional studies of shaggy/glycogen synthase kinase 3 phosphorylation sites in Drosophila melanogaster. Mol. Cell. Biol. 24: 4909–4919. 10.1128/MCB.24.11.4909-4919.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Kim Y.-J., Adams M. E., 2002. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc. Natl. Acad. Sci. USA 99: 11423–11428 (erratum: Proc. Natl. Acad. Sci. USA 99: 13961) 10.1073/pnas.162276199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Peralbo E., Culi J., 2011. Drosophila lipophorin receptors mediate the uptake of neutral lipids in oocytes and imaginal disc cells by an endocytosis-independent mechanism. PLoS Genet 7: e1001297 10.1371/journal.pgen.1001297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvy J.-P., Napal L., Rubin T., Poidevin M., Perrin L., et al. , 2012. Drosophila melanogaster Acetyl-CoA-carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLoS Genet. 8: e1002925 10.1371/journal.pgen.1002925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. T., Soulages J. L., Hariharasundaram B., Arrese E. L., 2005. Activation of the lipid droplet controls the rate of lipolysis of triglycerides in the insect fat body. J. Biol. Chem. 280: 22624–22631. 10.1074/jbc.M413128200 [DOI] [PubMed] [Google Scholar]
- Péterfy M., Phan J., Xu P., Reue K., 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27: 121–124. 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., et al. , 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146: 408–420. 10.1016/j.cell.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T., Santos C. R., Griffiths B., Cully M., Wu M., et al. , 2008. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8: 224–236. 10.1016/j.cmet.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik J. A., Schramek D., Schnidar H., Cronin S. J. F., Nehme N. T., et al. , 2010. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of Brown vs. White adipose cell fate. Cell 140: 148–160. 10.1016/j.cell.2009.12.027 [DOI] [PubMed] [Google Scholar]
- Prasad S. V., Fernando-Warnakulasuriya G. J., Sumida M., Law J. H., Wells M. A., 1986. Lipoprotein biosynthesis in the larvae of the tobacco hornworm, Manduca sexta. J. Biol. Chem. 261: 17174–17176. [PubMed] [Google Scholar]
- Puig O., Marr M. T., Ruhf M. L., Tijan R., 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A., Perrimon N., 2012. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151: 123–137 [corrigenda: Cell 152: 1197 (2013)] 10.1016/j.cell.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A., Perrimon N., 2013. Of flies and men: insights on organismal metabolism from fruit flies. BMC Biol. 11: 38 10.1186/1741-7007-11-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A., Housden B. E., Wirtz-Peitz F., Holderbaum L., Perrimon N., 2017. A mechanism coupling systemic energy sensing to adipokine secretion. Dev. Cell 43: 83–98.e6. 10.1016/j.devcel.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rava P., Hussain M. M., 2007. Acquisition of triacylglycerol transfer activity by microsomal triglyceride transfer protein during evolution. Biochemistry 46: 12263–12274. 10.1021/bi700762z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson R. B., 2003. The SREBP pathway--insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4: 631–640. 10.1038/nrm1174 [DOI] [PubMed] [Google Scholar]
- Reiff T., Jacobson J., Cognigni P., Antonello Z., Ballesta E., et al. , 2015. Endocrine remodelling of the adult intestine sustains reproduction in Drosophila. Elife 4: e06930 10.7554/eLife.06930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis T., Van Gilst M. R., Hariharan I. K., 2010. A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLoS Genet. 6: e1001206 10.1371/journal.pgen.1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Vázquez M., Vaquero D., Parra-Peralbo E., Mejía-Morales J. E., Culi J., 2015. Drosophila lipophorin receptors recruit the lipoprotein LTP to the plasma membrane to mediate lipid uptake. PLoS Genet. 11: e1005356 10.1371/journal.pgen.1005356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T. E., Lindmo K., Juhasz G., Sass M., Seglen P. O., et al. , 2004. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell 7: 179–192. 10.1016/j.devcel.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Ryan R. O., Haunerland N. H., Bowers W. S., Law J. H., 1988. Insect lipid transfer particle catalyzes diacylglycerol exchange between high-density and very-high-density lipoproteins. Biochim. Biophys. Acta 962: 143–148. 10.1016/0005-2760(88)90105-1 [DOI] [PubMed] [Google Scholar]
- Sajwan S., Sidorov R., Stašková T., Žaloudíková A., Takasu Y., et al. , 2015. Targeted mutagenesis and functional analysis of adipokinetic hormone-encoding gene in Drosophila. Insect Biochem. Mol. Biol. 61: 79–86. 10.1016/j.ibmb.2015.01.011 [DOI] [PubMed] [Google Scholar]
- Schmitt S., Ugrankar R., Greene S. E., Prajapati M., Lehmann M., 2015. Drosophila Lipin interacts with insulin and TOR signaling pathways in the control of growth and lipid metabolism. J. Cell Sci. 128: 4395–4406. 10.1242/jcs.173740 [DOI] [PubMed] [Google Scholar]
- Schulz J. G., Laranjeira A., Van Huffel L., Gärtner A., Vilain S., et al. , 2015. Glial β-oxidation regulates Drosophila energy metabolism. Sci. Rep. 5: 7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T., Wieschaus E., 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedes C. C., Carney G. E., 2012. Ecdysone signaling in adult Drosophila melanogaster. J. Insect Physiol. 58: 293–302. 10.1016/j.jinsphys.2012.01.013 [DOI] [PubMed] [Google Scholar]
- Seegmiller A. C., Dobrosotskaya I., Goldstein J. L., Ho Y. K., Brown M. S., et al. , 2002. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev. Cell 2: 229–238. 10.1016/S1534-5807(01)00119-8 [DOI] [PubMed] [Google Scholar]
- Sellers J. A., Hou L., Athar H., Hussain M. M., Shelness G. S., 2003. A Drosophila microsomal triglyceride transfer protein homolog promotes the assembly and secretion of human apolipoprotein B. Implications for human and insect transport and metabolism. J. Biol. Chem. 278: 20367–20373. 10.1074/jbc.M300271200 [DOI] [PubMed] [Google Scholar]
- Sieber M. H., Spradling A. C., 2015. Steroid signaling establishes a female metabolic state and regulates SREBP to control oocyte lipid accumulation. Curr. Biol. 25: 993–1004. 10.1016/j.cub.2015.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber M. H., Thummel C. S., 2009. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 10: 481–490. 10.1016/j.cmet.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber M. H., Thummel C. S., 2012. Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the Drosophila LipA homolog magro. Cell Metab. 15: 122–127. 10.1016/j.cmet.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachenko A., Gordon H. B., Kimball S. S., Gavin E. J., Bonkowsky J. L., et al. , 2016. Neurodegeneration in a Drosophila model of adrenoleukodystrophy: the roles of the Bubblegum and Double bubble acyl-CoA synthetases. Dis. Model. Mech. 9: 377–387. 10.1242/dmm.022244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C., Werz C., Wieser D., Alic N., Foley A., et al. , 2010. Regulation of lifespan, metabolism, and stress responses by the Drosophila SH2B protein, Lnk. PLoS Genet. 6: e1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Witkowski A., Joshi A. K., 2003. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 42: 289–317. 10.1016/S0163-7827(02)00067-X [DOI] [PubMed] [Google Scholar]
- Sociale M., Wulf A.-L., Breiden B., Klee K., Thielisch M., et al. , 2018. Ceramide synthase Schlank is a transcriptional regulator adapting gene expression to energy requirements. Cell Rep. 22: 967–978. 10.1016/j.celrep.2017.12.090 [DOI] [PubMed] [Google Scholar]
- Song W., Ren D., Li W., Jiang L., Cho K. W., et al. , 2010. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 11: 427–437. 10.1016/j.cmet.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Veenstra J. A., Perrimon N., 2014. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 9: 40–47. 10.1016/j.celrep.2014.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Cheng D., Hong S., Sappe B., Hu Y., et al. , 2017. Midgut-derived activin regulates glucagon-like action in the fat body and glycemic control. Cell Metab. 25: 386–399. 10.1016/j.cmet.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staubli F., Jorgensen T. J. D., Cazzamali G., Williamson M., Lenz C., et al. , 2002. Molecular identification of the insect adipokinetic hormone receptors. Proc. Natl. Acad. Sci. USA 99: 3446–3451. 10.1073/pnas.052556499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M., Metya S. K., Sadaf S., Kumar S., Schwudke D., et al. , 2013. Altered lipid homeostasis in Drosophila InsP3 receptor mutants leads to obesity and hyperphagia. Dis. Model. Mech. 6: 734–744. 10.1242/dmm.010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeyer K., Hendricks J. K., Prasad S. V., Wells M. A., 1996. The precursor protein of the structural apolipoproteins of lipophorin: cDNA and deduced amino acid sequence. Insect Biochem. Mol. Biol. 26: 735–738. 10.1016/S0965-1748(96)00060-4 [DOI] [PubMed] [Google Scholar]
- Sztalryd C., Brasaemle D. L., 2017. The perilipin family of lipid droplet proteins: gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862: 1221–1232. 10.1016/j.bbalip.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurino M., Costantini S., De Domenico S., Stefanelli F., Ruano G., et al. , 2018. SEIPIN proteins mediate lipid droplet biogenesis to promote pollen transmission and reduce seed dormancy. Plant Physiol. 176: 1531–1546. 10.1104/pp.17.01430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A. A., 2010. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425: 13–26. 10.1042/BJ20091181 [DOI] [PubMed] [Google Scholar]
- Teleman A. A., Maitra S., Cohen S. M., 2006. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 20: 417–422. 10.1101/gad.374406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Thummel C. S., 2011. Coordinating growth and maturation - insights from Drosophila. Curr. Biol. 21: R750–R757. 10.1016/j.cub.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Bertagnolli N. M., Evans J., Sieber M. H., Cox J., et al. , 2014. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 4: 839–850. 10.1534/g3.114.010652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam A. R., Farese R. V., Jr., Walther T. C., 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14: 775–786. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel K., Heier C., Haberl V., Thul P. J., Oberer M., et al. , 2013. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J. Cell Sci. 126: 2198–2212. 10.1242/jcs.120493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. E., Stunnenberg H. G., Stewart A. F., 1993. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature 362: 471–475. 10.1038/362471a0 [DOI] [PubMed] [Google Scholar]
- Thul P. J., Tschapalda K., Kolkhof P., Thiam A. R., Oberer M., et al. , 2017. Targeting of the Drosophila protein CG2254/Ldsdh1 to a subset of lipid droplets. J. Cell Sci. 130: 3141–3157. 10.1242/jcs.199661 [DOI] [PubMed] [Google Scholar]
- Tian Y., Bi J., Shui G., Liu Z., Xiang Y., et al. , 2011. Tissue-autonomous function of Drosophila seipin in preventing ectopic lipid droplet formation. PLoS Genet. 7: e1001364 10.1371/journal.pgen.1001364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrankar R., Liu Y., Provaznik J., Schmitt S., Lehmann M., 2011. Lipin is a central regulator of adipose tissue development and function in Drosophila melanogaster. Mol. Cell. Biol. 31: 1646–1656. 10.1128/MCB.01335-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V. V., Yu Y., Jankowska A., Luo Y., Wasik K. A., et al. , 2013. Minotaur is critical for primary piRNA biogenesis. RNA 19: 1064–1077. 10.1261/rna.039669.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente V., Maia R. M., Vianna M. C. B., Paçó-Larson M. L., 2010. Drosophila melanogaster lipins are tissue-regulated and developmentally regulated and present specific subcellular distributions. FEBS J. 277: 4775–4788. 10.1111/j.1742-4658.2010.07883.x [DOI] [PubMed] [Google Scholar]
- van Heusden M. C., van der Horst D. J., Kawooya J. K., Law J. H., 1991. In vivo and in vitro loading of lipid by artificially lipid-depleted lipophorins: evidence for the role of lipophorin as a reusable lipid shuttle. J. Lipid Res. 32: 1789–1794. [PubMed] [Google Scholar]
- Van Hoof D., Rodenburg K. W., Van der Horst D. J., 2002. Insect lipoprotein follows a transferrin-like recycling pathway that is mediated by the insect LDL receptor homologue. J. Cell Sci. 115: 4001–4012. 10.1242/jcs.00113 [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Rodenburg K. W., van der Horst D. J., 2003. Lipophorin receptor-mediated lipoprotein endocytosis in insect fat body cells. J. Lipid Res. 44: 1431–1440. 10.1194/jlr.M300022-JLR200 [DOI] [PubMed] [Google Scholar]
- Varghese J., Lim S. F., Cohen S. M., 2010. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 24: 2748–2753. 10.1101/gad.1995910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu J., Buratovich M. A., Wilder E. L., Birnbaum M. J., 1999. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1: 500–506. 10.1038/70293 [DOI] [PubMed] [Google Scholar]
- Vihervaara T., Puig O., 2008. dFOXO regulates transcription of a Drosophila acid lipase. J. Mol. Biol. 376: 1215–1223. 10.1016/j.jmb.2007.12.042 [DOI] [PubMed] [Google Scholar]
- Walker A. K., Jacobs R. L., Watts J. L., Rottiers V., Jiang K., et al. , 2011. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147: 840–852. 10.1016/j.cell.2011.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls S. M., Jr., Attle S. J., Brulte G. B., Walls M. L., Finley K. D., et al. , 2013. Identification of sphingolipid metabolites that induce obesity via misregulation of appetite, caloric intake and fat storage in Drosophila. PLoS Genet 9: e1003970 10.1371/journal.pgen.1003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls S. M., Cammarato A., Chatfield D. A., Ocorr K., Harris G. L., et al. , 2018. Ceramide-protein interactions modulate ceramide- associated lipotoxic cardiomyopathy. Cell Rep. 22: 2702–2715. 10.1016/j.celrep.2018.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C., Chung J., Farese R. V., 2017. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33: 491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Goode J., Best J., Meltzer J., Schilman P. E., et al. , 2008. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 7: 434–444. 10.1016/j.cmet.2008.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Moya N., Niessen S., Hoover H., Mihaylova M. M., et al. , 2011. A hormone-dependent module regulating energy balance. Cell 145: 596–606. 10.1016/j.cell.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Becuwe M., Housden B. E., Chitraju C., Porras A. J., et al. , 2016. Seipin is required for converting nascent to mature lipid droplets. Elife 5: e16582 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Airola M. V., Reue K., 2017. How lipid droplets “TAG” along: glycerolipid synthetic enzymes and lipid storage. Biochim. Biophys. Acta 1862: 1131–1145. 10.1016/j.bbalip.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu S., Liu H., Wang J., Zhou S., et al. , 2010. 20-hydroxyecdysone reduces insect food consumption resulting in fat body lipolysis during molting and pupation. J. Mol. Cell Biol. 2: 128–138. 10.1093/jmcb/mjq006 [DOI] [PubMed] [Google Scholar]
- Weers P. M., van der Horst D. J., van Marrewijk W. J., van den Eijnden M., van Doorn J. M., et al. , 1992. Biosynthesis and secretion of insect lipoprotein. J. Lipid Res. 33: 485–491. [PubMed] [Google Scholar]
- Welte M. A., 2015. Expanding roles for lipid droplets. Curr. Biol. 25: R470–R481. 10.1016/j.cub.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M. A., Gould A. P., 2017. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862: 1260–1272. 10.1016/j.bbalip.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M. A., Cermelli S., Griner J., Viera A., Guo Y., et al. , 2005. Regulation of lipid-droplet transport by the perilipin homolog LSD2. Curr. Biol. 15: 1266–1275. 10.1016/j.cub.2005.06.062 [DOI] [PubMed] [Google Scholar]
- Werz C., Köhler K., Hafen E., Stocker H., 2009. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 5: e1000596 10.1371/journal.pgen.1000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker-Thomas C., Garrido D., Bontonou G., Napal L., Mazuras N., et al. , 2015. Flexible origin of hydrocarbon/pheromone precursors in Drosophila melanogaster. J. Lipid Res. 56: 2094–2101. 10.1194/jlr.M060368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., et al. , 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24: 384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Thiam A. R., Olarte M.-J., Wang J., Beck R., et al. , 2014. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 3: e01607 10.7554/eLife.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy S. Y., Guo M., Hay B. A., 2003. The Drosophila MicroRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795. 10.1016/S0960-9822(03)00250-1 [DOI] [PubMed] [Google Scholar]
- Xu X., Gopalacharyulu P., Seppänen-Laakso T., Ruskeepää A.-L., Aye C. C., et al. , 2012. Insulin signaling regulates fatty acid catabolism at the level of CoA activation. PLoS Genet. 8: e1002478 10.1371/journal.pgen.1002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Wang H., Chen H., Lindström-Battle A., Jiao R., 2015. Ecdysone and insulin signaling play essential roles in readjusting the altered body size caused by the dGPAT4 mutation in Drosophila. J. Genet. Genomics 42: 487–494. 10.1016/j.jgg.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Yan Y., Wang H., Hu M., Jiang L., Wang Y., et al. , 2017. HDAC6 suppresses age-dependent ectopic fat accumulation by maintaining the proteostasis of PLIN2 in Drosophila. Dev. Cell 43: 99–111.e5. 10.1016/j.devcel.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Yao T. P., Segraves W. A., Oro A. E., McKeown M., Evans R. M., 1992. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71: 63–72. 10.1016/0092-8674(92)90266-F [DOI] [PubMed] [Google Scholar]
- Yao T. P., Forman B. M., Jiang Z., Cherbas L., Chen J. D., et al. , 1993. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 366: 476–479. 10.1038/366476a0 [DOI] [PubMed] [Google Scholar]
- Zechner R., Madeo F., Kratky D., 2017. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18: 671–684. 10.1038/nrm.2017.76 [DOI] [PubMed] [Google Scholar]
- Zhang P., Reue K., 2017. Lipin proteins and glycerolipid metabolism: roles at the ER membrane and beyond. Biochim. Biophys. Acta 1859: 1583–1595. 10.1016/j.bbamem.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Karpac J., 2017. Muscle directs diurnal energy homeostasis through a myokine-dependent hormone module in Drosophila. Curr. Biol. 27: 1941–1955.e6. 10.1016/j.cub.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A. B., Thiele C., Tenedini F., Richard M., Leyendecker P., et al. , 2017. Cell-autonomous control of neuronal dendrite expansion via the fatty acid synthesis regulator SREBP. Cell Rep. 21: 3346–3353. 10.1016/j.celrep.2017.11.069 [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., et al. , 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306: 1383–1386. 10.1126/science.1100747 [DOI] [PubMed] [Google Scholar]
- Zinke I., Schütz C. S., Katzenberger J. D., Bauer M., Pankratz M. J., 2002. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 21: 6162–6173. 10.1093/emboj/cdf600 [DOI] [PMC free article] [PubMed] [Google Scholar]