Ellison et al. provide the first evidence that the genomic region bearing the Or gene is involved in the presence of carotenoids in carrot. Using a global collection of diverse carrot accessions, they identify 12 genomic regions...
Keywords: Daucus carota, GWAS, population structure, carotenoids, domestication, selective sweep
Abstract
Carrots are among the richest sources of provitamin A carotenes in the human diet, but genetic variation in the carotenoid pathway does not fully explain the high levels of carotenoids in carrot roots. Using a diverse collection of modern and historic domesticated varieties, and wild carrot accessions, an association analysis for orange pigmentation revealed a significant genomic region that contains the Or gene, advancing it as a candidate for carotenoid presence in carrot. Analysis of sequence variation at the Or locus revealed a nonsynonymous mutation cosegregating with carotenoid content. This mutation was absent in all wild carrot samples and nearly fixed in all orange domesticated samples. Or has been found to control carotenoid presence in other crops but has not previously been described in carrot. Our analysis also allowed us to more completely characterize the genetic structure of carrot, showing that the Western domesticated carrot largely forms one genetic group, despite dramatic phenotypic differences among market classes. Eastern domesticated and wild accessions form a second group, which reflects the recent cultivation history of carrots in Central Asia. Other wild accessions form distinct geographic groups, particularly on the Iberian peninsula and in Northern Africa. Using genome-wide Fst, nucleotide diversity, and the cross-population composite likelihood ratio, we analyzed the genome for regions putatively under selection during domestication and identified 12 regions that were significant for all three methods of detection, one of which includes the Or gene. The Or domestication allele appears to have been selected after the initial domestication of yellow carrots in the East, near the proposed center of domestication in Central Asia. The rapid fixation of the Or domestication allele in almost all orange and nonorange carrots in the West may explain why it has not been found with less genetically diverse mapping populations.
CARROT domestication and modern breeding have been driven by selection for large roots containing abundant carotenoids, which are responsible for orange pigmentation in the taproot. The presence of carotenoids in root tissues is unlikely to confer an advantage for natural selection, but is meaningful in a domesticated context (Iorizzo et al. 2016) due to their visual appeal and the role of dietary provitamin A compounds in human health (Arscott and Tanumihardjo 2010). Carrots are among the richest sources of provitamin A carotenes in the human diet (Simon et al. 2009) and significant breeding effort has focused on increasing root carotenoid levels (Simon 2000; Simon and Goldman 2007; Simon et al. 2008). Although carotenoid biosynthetic genes have been mapped (Just et al. 2007), they do not fully explain the presence of high levels of carotenoids in carrot roots, leaving much of that mechanism unknown (Iorizzo et al. 2016; Ellison et al. 2017).
While carrot is known as a bright orange root crop, the original carrots domesticated in Central Asia ca. 900 CE were purple and yellow (Banga 1963) (Figure 1, A and B). There is some evidence for orange carrots earlier in history (Stolarczyk and Janick 2011), but it was not until six centuries after domestication that orange roots appeared consistently in the historical record. Wild carrot is indigenous to Europe, North Africa, and Western Asia, with its center of diversity in present day Afghanistan (Vavilov and Dorofeev 1992). Based on most historical records, the first evidence of carrot being cultivated as a food crop was in the Iranian Plateau and Persia in the 10th century (Banga 1957a,b, 1963; Brothwell and Brothwell 1969), and molecular evidence supports a Central Asian origin of domesticated carrot (Iorizzo et al. 2013). Carrot cultivation spread westward to North Africa and Europe, and eastward to Asia. Orange roots appeared in Spain and Germany in the 16th century (Stolarczyk and Janick 2011), and quickly became the predominant color (Figure 1, C and D).
Figure 1.
Carrot accessions illustrating the stages of carrot domestication and improvement. Carrots shown are fully mature (100 days). (A) Wild, (B) Eastern landrace, (C) Western historic open pollinated, and (D) modern hybrids (left: processing type and right: imperator type). Photo courtesy of Matthew Mirkes.
Carotenoid levels have doubled due to plant breeding over the past 60 years (Simon 1990). Hence, there has been substantial effort to understand the mechanisms of carotenoid accumulation and regulation. Allelic variation at two genes, Y and Y2, accounts for most of the distinctive color and carotenoid accumulation differences observed in orange, yellow, and white carrot roots (Buishand and Gabelman 1979). However, carotenoid biosynthesis genes in carrot do not map near enough to Y or Y2 to be responsible for these differences (Just et al. 2007). The popularity of orange carrot likely fixed many of the alleles responsible for carotenoid presence in roots in domesticated populations. Researchers have therefore looked outside the biosynthetic pathway to regulatory and other modifying genes for explanation. Iorizzo et al. (2016) used two mapping populations and the newly assembled carrot genome to identify a candidate outside of the carotenoid biosynthetic pathway for the Y gene, DCAR_032551, that regulates photosystem development and conditions a portion of carotenoid presence in carrot roots.
In cauliflower, the Orange (Or) gene accounts for elevated levels of carotenoids (Li et al. 2001). The Or gene is responsible for both the biogenesis of chromoplasts, where carotenoids are stored, and post-transcriptional regulation of Phytoene Synthase (PSY), an enzyme necessary for carotenoid biosynthesis (Lu et al. 2006; Yuan et al. 2015; Zhou et al. 2015). Mutations in the Or gene have been associated with the presence of carotenoids in nonleaf tissue through the differentiation of noncolored plastids into chromoplasts in Arabidopsis, cauliflower, and sweet potato (Yuan et al. 2015). Maass et al. (2009) noted that the presence of large amounts of β-carotene in the form of crystals in carrot is strikingly similar to that found in the cauliflower Or mutant (Maass et al. 2009). The Or gene has not previously been associated with carotenoid presence in carrot. Previous carotenoid studies have focused either on biparental populations derived from crosses among domesticated carrot (Buishand and Gabelman 1979), or on crosses between wild carrot from North America and domesticated carrot (Santos and Simon 2002; Just et al. 2007; Ellison et al. 2017). Previous studies were also limited in their ability to detect significant associations by population size and marker density (Iorizzo et al. 2013).
In this study, we use 674 globally distributed domesticated and wild carrot accessions to conduct a genome-wide association study (GWAS) for carrot root pigmentation. We also analyze the population structure that developed during carrot dispersal and domestication. We sampled germplasm from all major global regions where carrot originated or was domesticated. Previous studies have identified three major genetic groups—Western, Eastern, and wild—but with limited numbers of accessions and low marker density (Iorizzo et al. 2013). Our analysis enabled the identification of both new and previously characterized regions of the carrot genome that were likely involved in selective sweeps during domestication, and we present the first indication of the Or gene playing a role in carotenoid presence in carrot.
Materials and Methods
Plant materials and phenotypic evaluation
Included were 154 wild and 520 domesticated carrot (Daucus carota L.) samples. Samples 1–144 were sown on certified organic land at Tipi Organic Produce in Evansville, WI and Elderberry Hill Farm in Waunakee, WI in 2013 and 2014. Samples 43XXX and 53XXX were grown at the University of Wisconsin–Madison West Madison Agricultural Research Station (Verona, WI) in 2014 and 2015 (X stands for any digit 0–9). Samples 30XXX and 32XXX were grown at the University of Wisconsin–Madison Hancock Agricultural Research Station (Hancock, WI) in 2013, and samples labeled GHXXXX, DH, and 493 were grown at the Walnut Street Greenhouse (Madison, WI) in 2013. Two samples of Daucus syrticus (Ames 29096 and Ames 29108) were used as an outgroup species (Arbizu et al. 2014). Passport data for the 674 accessions can be found in Supplemental Material, Table S1 (available at Figshare: 10.6084/m9.figshare.6177110).
Pigmentation analysis was conducted within 5 weeks of carrot harvest. Roots were sliced in cross section 5–10 cm from the root tip and root phloem color was classified as orange, purple, red, white, or yellow. Visual assessment data can be found in Table S2. Carotenoid presence/absence and root color classes are highly heritable, and not strongly influenced by environmental effects, with evidence of discrete inheritance of total carotenoids in crosses of white carrots with carrots of other colors (Buishand and Gabelman 1979; Fernandes Santos and Simon 2006). However, carotenoid concentration can be influenced by environmental conditions and so samples used for quantifying carotenoids were all taken from the Hancock Agricultural Research Station. Carotenoid content was quantified using lyophilized root tissue for HPLC analysis, as modified from Simon and Wolff (1987) and Simon et al. (1989). Briefly, 0.1 g of lyophilized carrot root tissue was crushed and then soaked in 2.0 ml of petroleum ether at 4°. After 12–16 hr, 300 μl of the petroleum ether extract was added to 700 μl of methanol, eluted through a Rainin Microsorb-MV column, and analyzed on a Millipore (Bedford, MA) Waters 712 WISP HPLC system. Synthetic β-carotene (Sigma [Sigma Chemical], St. Louis, MO) was used in each independent run as a reference standard for calibration. Lutein, α-carotene, and β-carotene were quantified by absorbance at 450 nm. Concentrations are described in μg g−1 dry weight. HPLC data can be found in Table S3.
Genotyping, SNP production, and filtering
Total genomic DNA of individual plants was isolated from ∼0.1 g of lyophilized leaves of 4-week-old plants following the 10% CTAB protocol described by Murray and Thompson (1980), with modifications by Boiteux et al. (1999). All DNA was quantified using the Quantus PicoGreen dsDNA Kit (Life Technologies, Grand Island NY) and normalized to 10 ng/μl. Genotyping-by-sequencing (GBS), as described by Elshire et al. (2011), was carried out at the University of Wisconsin–Madison Biotechnology Center (WI) with minimal modification and half-sized reactions. Briefly, DNA samples were digested with ApeKI, barcoded, and pooled for sequencing, and 80–95 pooled samples were run per single Illumina (San Diego, CA) HiSeq 2000 lane, using 100-nt reads and v3 Sequencing by Synthesis reagents (Illumina). Images were analyzed using CASAVA 1.8.2. and bcl2fastq-1.8.4.
The TASSEL-GBS pipeline version 5.2.26 was used to call SNPs as described by Bradbury et al. (2007) and Glaubitz et al. (2014) using the carrot reference genome (GenBank accession LNRQ01000000.1; Iorizzo et al. 2016). SNPs were filtered into two data sets. D1 had < 30% missing data for genotype and marker, a 5% minor allele frequency, no more than two alleles, and at least 5× depth per marker. Markers were further filtered to set heterozygous markers with an allele ratio < 0.3 or > 0.7 to missing, leaving 39,710 SNPs in 674 samples. Missing genotype calls in D1 were imputed using Beagle 4.1 with n iterations = 10 (Browning and Browning 2016). The average SNP distribution across the carrot genome was ∼54 SNPs per 500 kb bin or 1 SNP per 10 kb (Figure S1) with an average 18× coverage per SNP. To reduce potential confounding effects of population structure and differentiation, we excluded 21 wild samples from Portugal (D1-noPT).
The same filtering parameters were used for data set D2 except that SNPs were filtered using 10% missing data and were not imputed. Additionally, two samples from the outgroup D. syrticus were included for a total of 676 individuals and 32,128 SNPs. The SNP distribution for D2 was 43 SNPs per 500 kb (Figure S2) and 20× coverage per SNP. Additional information about SNP filtering can be found in Figure S3. A subsample of D2 was created to exclude samples with > 30% admixture (D2-lowAd) as determined by STRUCTURE.
SNP density across chromosomes, using 500,000-nt bins for D1 and D2, can be found in Figures S1 and S2. Filtering parameters for each SNP data set can be found in Figure S3. SNP data sets are in Data D1 and Data D2.
Linkage disequilibrium
TASSEL 5 (Bradbury et al. 2007) was used to calculate linkage disequilibrium (LD) for the full matrix of SNPs for data set D1-noPT. Reported values of LD decay use an r2 cutoff of 0.1 and 0.2 for filtered SNPs (P < 0.01) (Vos et al. 2017). The half-distance of LD decay was calculated as when the LD decay curve intersected with half the maximum LD value. Genome-wide sliding window analysis of LD was conducted for both wild and domesticated samples using VCFtools with the parameters –geno-r2 –ldwindow 100 (51). r2 values with fewer than 95 SNPs per bin were removed. Sliding window analysis was visualized using qqman in R studio (Wickham 2009).
Population structure
We used data set D2 and conducted eight replications of the Bayesian clustering program STRUCTURE version 2.3.4 (Pritchard et al. 2000) with populations (K value) ranging from 1 to 14, with a burn-in length of 20,000 and 50,000 Monte Carlo iterations, respectively. An admixture model with no previous population information was included; all other parameters were set to default values. STRUCTURE results were processed in the software STRUCTURE HARVESTER 0.6.94 with parameter–evanno (Earl and vonHoldt 2012), to detect the most likely number of clusters by using the rate of change in the log probability between successive values of K (ΔK) (Evanno et al. 2005). Population structure was visualized using distruct software version 1.1 (Rosenberg 2004).
Principal component analysis
An eigenvalue decomposition of the SNP covariance matrix was performed using TASSEL 5 using default parameters for D2 and D2-lowAd. All individuals’ loadings were plotted along the first and second principal components (PCs) using ggplot in R. Individuals were colored according to their STRUCTURE group identity.
Maximum-likelihood tree
Using data sets D2 and D2-lowAd, maximum likelihood analyses were conducted with the GTR+G nucleotide substitution model using RAxML version 8.2.9 (Stamatakis 2014). GATK HaplotypeCaller (McKenna et al. 2010) with parameters –genotyping_mode GENOTYPE_GIVEN_ALLELES was used to call SNPs for the two outgroup accessions, D. syrticus SRR2147152 and SRR2147153 (Arbizu et al. 2016). FigTree (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize phylogenetic trees.
Pairwise Fst
Weir and Cockerham’s method for calculating pairwise Fst (Weir and Cockerham 1984) was implemented within the genet.dist function of the R package hierfstat (Goudet 2005). Pairwise values were calculated on all K = 6 subpopulations using data sets D2 and D2-lowAd. The data set was first converted to a genind object using the df2genind command of the R package adegenet using default parameters.
Sliding window analysis of nucleotide diversity, Fst, and cross-population composite likelihood ratio
Selective sweep detection analyses used data set D1-noPT. VCFtools was used to calculate genetic diversity (π) in 500-kb windows across the carrot genome (–window-pi 500000) for wild and domesticated carrot samples. Genomic bins were set at 500-kb regions. Each bin was analyzed for reduced nucleotide diversity, elevated Fst, and high cross-population composite likelihood ratio (XP-CLR) scores. Potential sweeps corresponded to each bin; however, the start and stop points of the sweep are very likely different from those of the bin. Bin sizes were selected to contain an average of 50 SNPs. Potential selective sweep regions were found by calculating the difference between wild and domesticated nucleotide diversity bins, and selecting bins in the top 5% of values (P > 1.578). The population differentiation statistic, Fst, was estimated between wild and domesticated samples in VCFtools in 500-kb windows with 100-kb steps (-weir -fst-pop -fst-window-size 500000-fst-window-step 100000) (Danecek et al. 2011). Potential sweep regions were defined as the top 5% of values A third method, XP-CLR, was implemented to test for selective sweeps (Chen et al. 2010). The XP-CLR software was run with parameters: -w1 0.005 50 100 1 -p1 0.9 for each chromosome. Sweeps were detected unidirectionally from wild to domesticated. The genetic distances between SNPs were interpolated according to their physical distances in a high-density genetic map (Iorizzo et al. 2016). Mean XP-CLR scores were tabulated in nonoverlapping 10-kb windows across the genome. Windows with the top 1% of XP-CLR values (11.93) were selected and placed in corresponding bins from the Fst and nucleotide diversity analyses. Genome-wide sliding window analyses were plotted using the R package qqman (Turner 2014). Overlapping genomic regions in the top 5% for nucleotide diversity and Fst, and top 1% for XP-CLR scores, were identified as the most likely selective sweeps. The physical positions of four previously identified Diversity Arrays Technology (DArT) markers (crPT9895272, crPT9891905, crPT9890532, and crPT9885102; Grzebelus et al. 2014), which flank previously identified domestication signatures, were located in the carrot genome (GenBank accession LNRQ01000000.1; Iorizzo et al. 2016) using BLASTn.
Genome-wide association analysis
A GWAS was performed for carrot root pigmentation using data set D1 by implementing the EGSCORE function in the GenABLE R package (Aulchenko et al. 2007). The following parameters were used: naxes = 2, times = 1, quiet = FALSE, bcast = 0, clamda = T, and propsPs = 1. No fixed effects were included as covariates. The kinship matrix was calculated using the ibs command in GenABLE with the weight parameter set to “freq.” The diagonal of the kinship matrix was replaced with the default option of the variance of the average homozygosity within each individual to account for the level of inbreeding of an individual with itself. Manhattan and qqplots were drawn using the R package qqman (Turner 2014). While there may be some confounding of population structure and polymorphisms due to the orange phenotype being absent in wild carrot accessions but common in domesticated accessions, when accounting for kinship, the Q-Q plot does not show inflation of observed -values below expected -values of at least 2.5 (Figure S4). This indicates that any potential confounding of the root pigmentation phenotype with the underlying population structure was adequately controlled in our model. The most significant SNP in our GWAS, located on chromosome 3 at position 5,224,824, was used to analyze allele frequency distribution according the groupings identified by STRUCTURE at K = 6, PC1 and PC2, domestication or wild status, and orange or nonorange status.
Observed heterozygosity, homozygosity, and gene diversity
Observed heterozygosity Ho, within population gene diversity (HS), overall gene diversity (Ht), and overall Fst were calculated using the basic.stats function in the R package hierfstat (Goudet 2005) using data sets D1, D2, and D2 lowAd. Data sets were first converted to genind objects using the df2genind command of the R package adegenet using default parameters. Sample homozygosity was calculated using data set D1 by implementing the hom() function in the GenABLE R package (Aulchenko et al. 2007). Differences in mean homozygosity among STRUCTURE populations were ascertained via pairwise Student’s t-tests with pooled SDs and Bonferroni correction using the pairwise.t.test command in R.
Candidate gene sequence analysis
Thirteen previously resequenced carrot samples (Table S4) were surveyed for any sequence variation within the open reading frame of the Or gene (DCAR_009172). One SNP was identified between low- and high-carotenoid genotypes within exon 5. A transition of T to C at position 3350 resulted in a change of the codon TTG to TCG, causing a missense mutation of leucine to serine. This SNP is located on chromosome 3, position 5,197,361. To genotype carrot samples for T3350C, primers that flank the SNP were generated (Table S5). PCR-based sequencing was performed on 197 domesticated and 82 wild carrot samples. Sanger sequencing was performed on an ABI 3730xl DNA Analyzer by the University of Wisconsin Biotechnology Center DNA Sequence Facility. A gene model for Or was generated from the website http://wormweb.org/exonintron. Phenotypic differences for lutein, α-carotene, and β-carotene were analyzed for the three Or genotypic classes. For each trait, significance between different genotypic classes was determined by using the aov and TukeyHSD functions in R. A genotyping workflow can be found in Figure S5.
Data availability
Carrot sequences used for the Or gene alignment and D. syrticus samples used as an outgroup for phylogenetic analysis are available under the National Center for Biotechnology Information Bioproject accession PRJNA291976. All supplementary tables and data sets necessary to reproduce the analyses for this manuscript are available at Figshare: 10.6084/m9.figshare.6177110.
Results
Rapid decay of LD in carrot
LD analysis of wild carrot accessions demonstrated a very rapid genome-wide decay between 100 bp and 1 kb , and a rapid decay of 400 bp and 13 kb in domesticated accessions. This rapid decay was further supported by estimates of wild and domesticated samples having an LD half-life of 67 and 6544 bp, respectively (Figure S6). Determination of LD decay distances does not have a consensus method in the literature, with both thresholds (0.1 and 0.2) and half-life methods used (Vos et al. 2017). Half-life methods may be more robust to differences in minor allele frequencies and have been used in a number of species (Kim et al. 2007; Lam et al. 2010; Branca et al. 2011; Zhao et al. 2011; Vos et al. 2017). This is the first report of genome-wide LD in carrot. LD decay rates appear slower in domesticated samples around regions putatively under selection, such as the Y region (Iorizzo et al. 2016) and the cult region (Macko-Podgorni et al. 2017), as well as several carotenoid biosynthesis genes (Clotault et al. 2010; Soufflet-Freslon et al. 2013).
Population structure dynamics among wild and domesticated carrot
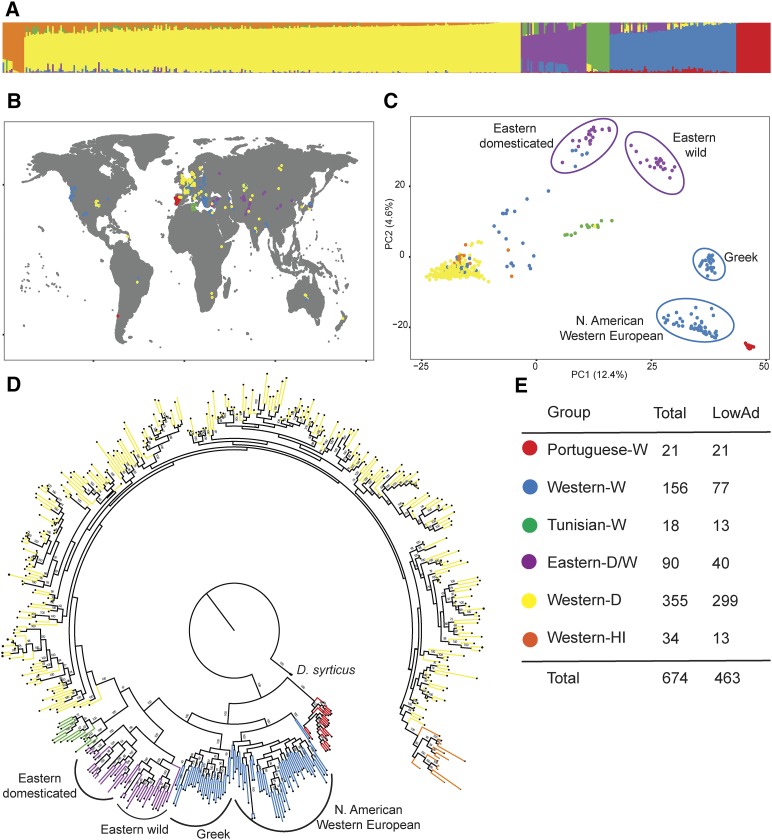
An examination of population structure was carried out using STRUCTURE software with K 6 as the number of groups strongly supported by the Evanno method (Evanno et al. 2005) (Figure 2A and Figures S7–S10). The support for K 6 was slightly stronger than K 4 or K 5, and as we are interested in understanding population structure in carrot, we chose to work with the largest K value strongly supported by the data.
Figure 2.
Population structure of 463 carrot accessions with admixture (D2-LowAd). (A) STRUCTURE groups. Percentage of membership (q) for each group identified at K6. (B) Geographic distribution of accessions, each represented by a point on the map colored according to STRUCTURE group. Current commercial varieties not shown. (C) Principal component (PC) analysis plot of the first two principal components. PC1 and PC2 account for 12.4 and 4.6% of the total variation, respectively. (D) Maximum-likelihood tree of carrot accessions. Numbers on the branches indicate bootstrap support. Black branch represents outgroup D. syrticus. (E) Color key. Total number of accessions in each STRUCTURE Group. LowAd, low admixture.
To maximize cluster separation, a low-admixture group (D2-LowAd) of 463 accessions was created by only including samples when the proportion of inferred ancestry was > 70 % (Figure 2E and Table S1). Clustering with STRUCTURE indicated divisions between Western domesticated, Western wild, and all Eastern samples, as well as emergent subclusters corresponding to geographic origin, including samples from Tunisia, which were mostly wild with a few domesticated samples, and wild accessions from Portugal (Figure 2A). An additional cluster formed for Western Imperator hybrids (Western-HI) (Figure 1D and Figure 2A). Imperator carrots are one of the major commercial market classes in the US and cultivars are primarily F1 hybrids. This market class has been the focus of much of US carrot breeding efforts in recent decades (Simon 2000). The Q matrix of individual accessions is reported in Table S1. In the presentation and discussion of results, we use W to refer to wild accessions, D to refer to domesticated accessions, and HI to refer to Hybrid Imperator accessions.
The observed population substructure was supported by phylogenetic analysis, PC analysis (PCA), and pairwise Fst. Using D. syrticus as an outgroup (Arbizu et al. 2014), the maximum-likelihood analysis identified the same six strongly supported clades (bootstrap ): Portuguese-W, Western-W, Eastern-W/D, Tunisian-W, Western-D, and Western-HI carrots (Figure 2D). PCA revealed a clear separation between wild and domesticated carrots along the first PC (12.4% of variation explained), and between Eastern and Western samples along the second PC (4.6% of variation explained, Figure 2C). Pairwise Fst calculations further supported differentiation between the six subclusters (Table S6). The Portuguese-W samples were the most strongly divergent of all the STRUCTURE groups, forming a very distinct subpopulation separate from other wild carrot accessions. Observed heterozygosity (Ho) for accessions in data set D2-lowAd was 0.18 (Table S7). All analyses and results were also confirmed on data set D2, without removal of high-admixture samples (Figure S8, Table S6, and Table S7).
GWAS analysis identifies Or as a candidate gene for carotenoid presence in carrot
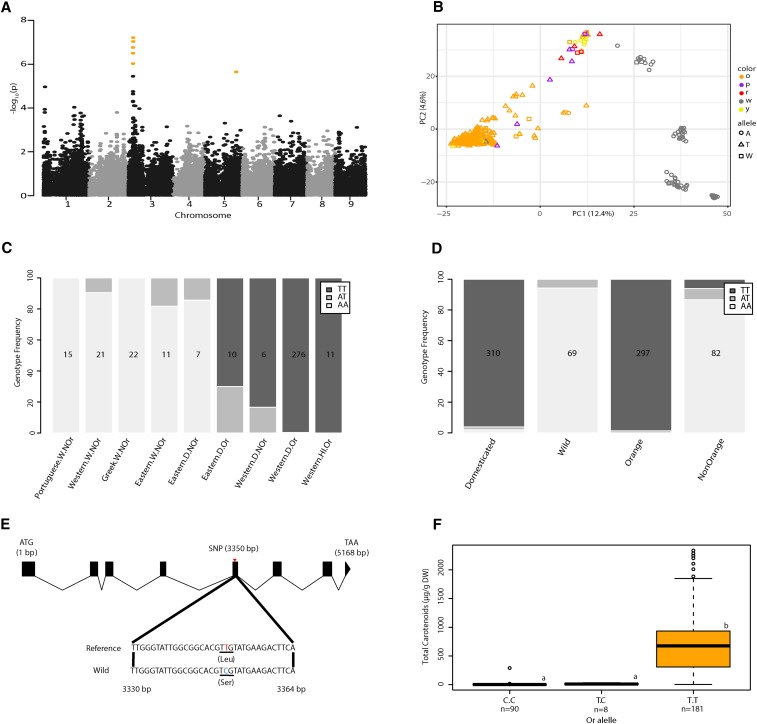
To identify genomic regions potentially related to carotenoid presence, we performed a GWAS for orange pigmentation in carrot root using data set D1 (Figure S3). We found a previously unidentified significant 143-kb GWAS signal on chromosome 3 containing 17 annotated genes (Figure 3A and Table S8). Or, a gene associated with carotenoid biosynthesis regulation and chromoplast formation (Lu et al. 2006; Li et al. 2012; Zhou et al. 2015), is in the middle of the 143-kb region encompassing the most significant SNPs in our GWAS analysis. No other genes in the 143-kb region are known to be associated with carotenoid presence. We selected the most significant GWAS SNP, located on chromosome 3 at position 5,224,824, to serve as a proxy to analyze the Or allele frequency distribution with respect to PC1 and PC2 of our PCA (Figure 3B), STRUCTURE populations (Figure 3C), domestication status (wild and domesticated), and pigmentation status (orange and nonorange) (Figure 3D). The wild-type allele (A) appears nearly fixed in all wild STRUCTURE groups as well as the eastern domesticated nonorange samples. The domestication allele (T) was found at high frequency in the eastern domesticated orange samples as well as western domesticated nonorange samples, and is fixed in all western domesticated orange samples. Further, we observe the domestication genotype (TT) to be absent in all wild samples and the wild genotype (AA) to be absent in orange samples.
Figure 3.
Genome-wide association analysis of orange pigmentation and identification of the candidate gene Or on chromosome 3. (A) Manhattan plot for orange carrot root color. SNPs with empirically-adjusted P-values < 0.05, were defined as significant and are colored orange. (B) Allele frequency of SNP S3_5228434 and pigmentation classification superimposed on the principal component analysis from Figure 2C. o, orange; p, purple; r, red; w, white; y, yellow. (C) SNP S3_5228434 genotype frequency separated by STRUCTURE classification. Or and NOr indicates orange or nonorange root pigmentation. W, D, or HI is wild, domesticated, or Hybrid Imperator, respectively. (D) SNP S3_5228434 genotype frequency separated by root pigmentation and domestication status. Quantities indicate the number of samples in each group. (E) Open reading frame of Or and the nonsynonymous mutation in exon 5 at position 3350 (T3350C). (F) Box plots for total carotenoids for the three Or genotypes (C/C, T/C, and TT) at position 3350. Center line median, box limits upper and lower quartiles, whiskers = 1.5× the interquartile range, dots outliers. Different letters indicate significant differences between genotypes (P < 0.05, Tukey’s honest significant difference). Reported values are in dry weight.
To better characterize the association of carotenoid presence and the Or gene, we looked for mutations cosegregating between five high- and eight low-carotenoid accessions that had been previously resequenced (Iorizzo et al. 2016), and found a nonsynonymous mutation at position 3350 in exon 5, causing a serine to leucine amino acid change (Figure 3B). An additional 198 domesticated and 81 wild samples were phenotyped for total carotenoid content ( dry weight) using HPLC and genotyped at Or. Those samples with the T/T genotype had significantly higher amounts of total carotenoid content than those that were heterozygous (C/T) or homozygous wild-type (C/C) (Figure 3F and Table S3). This is the first report of an association between Or and carotenoid presence in carrot.
Identification of selection signatures during carrot domestication
During crop domestication, highly favorable alleles undergo intensive selection and reach fixation rapidly, resulting in reduced variation in neighboring genomic regions and thereby creating a signature of a selective sweep. We used three measures to analyze sweeps: reduced nucleotide diversity (π) (Nei and Li 1979) in domesticated samples as compared to wild ones, high population differentiation (Fst) (Wright 1951) between wild and domesticated samples, and allele frequency differentiation between populations (XP-CLR) (Chen et al. 2010). To reduce potential confounding effects of population structure and differentiation, we removed the 21 Portuguese-W samples from the selective sweep analyses (data set D1-noPT, Figure S3).
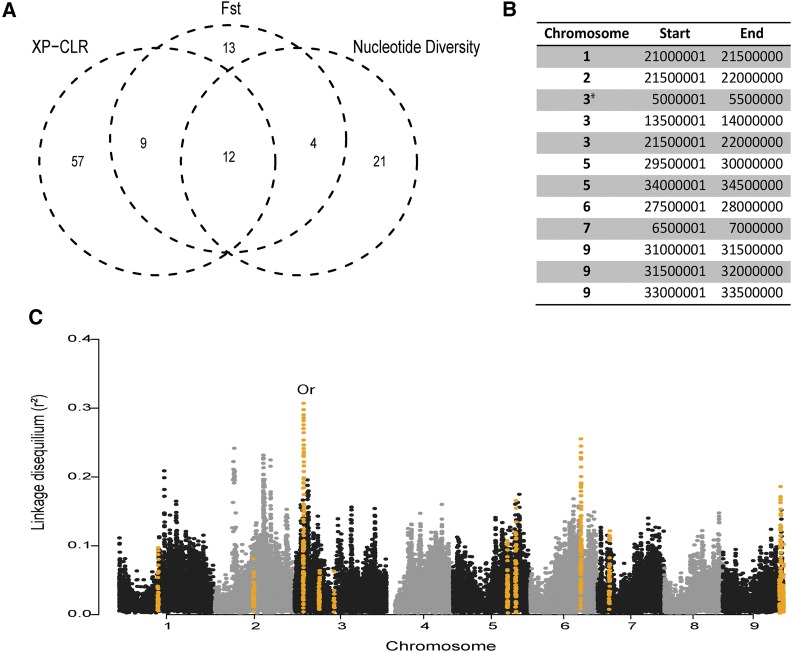
Overall, we found little reduction in genetic diversity in all domesticated carrot compared to all wild carrot averaged across the whole genome level. Differences in nucleotide diversity between wild and domesticated samples were estimated for 500-kb bins across the carrot genome. The average difference between groups was 1.080 with 37 potential selective sweep regions detected using the top 5% of calculated values (1.578) (Figure 4A and Table S9).
Figure 4.
Regions of the carrot genome that likely underwent a selective sweep during domestication. (A) Venn diagram represents the overlapping of 500-kb regions tested for selection signatures: top 5% of Fst and nucleotide diversity difference between wild and domesticated carrot accessions, and top 1% of cross-population composite likelihood ratio (XP-CLR) values. (B) Genomic location of potential selective sweeps identified by Fst, nucleotide diversity, and XP-CLR. The asterisk signifies the genome region carrying the candidate orange pigmentation gene Or. (C) Genome-wide linkage disequilibrium averaged across sliding windows of 100 SNPs in domesticated carrots. Regions identified as significant in (A) and (B) are highlighted in orange. The region containing the Or candidate gene for orange pigmentation in carrot is marked “Or.”
The genome-wide average Fst between domesticated and wild carrot was 0.14. We detected 38 genomic regions with Fst values above the 95% percentile differentiating wild and domesticated accessions (Figure 4A and Table S9). These regions with high levels of differentiation likely experienced selective sweeps during domestication or improvement (Wright 1951). The recently identified Y gene (Iorizzo et al. 2016), a candidate for carotenoid presence in carrot taproot, is located within one of these regions of high differentiation between wild and domesticated carrots (24.5–25.0 Mb on chromosome 5). The carotene hydroxylase DcCYP97A3 gene associated with increased α-carotene (Arango et al. 2014) is in a 500-kb bin (6.0–6.5 Mb) that has high Fst and is adjacent to a 500-kb bin (6.5–7 Mb) that is one of the 12 regions identified in all tests (Table S9).
Lastly we used the XP-CLR method to compare the wild and domesticated accessions in 10-kb bins across the genome (Chen et al. 2010). The top 1% of XP-CLR values identified 78 potential sweep bins (Figure 4A and Table S9). A candidate domestication gene associated with root thickening, DcAHLc1 (Macko-Podgorni et al. 2017), is located at 41.8 Mb on chromosome 2, near one of the regions with the highest XP-CLR scores (42.0–42.5 Mb). Another region, 33.5–34.0 Mb on chromosome 7, overlaps with the recently fine-mapped QTL, Y2, a gene associated with carotenoid accumulation (Ellison et al. 2017). Two carrot DArT markers, crPT9892544 and crPT894175, which were identified in Grzebelus et al. (2014) to show signatures of selection mapped between positions 21,070,220 and 30,397,440 on chromosome 2, and position 18,823,084 and 28,211,752 on chromosome 6, respectively. Both of these regions align with selective sweep bins identified in this study (Figure 4B).
To identify the most supported potential selective sweeps during domestication, we considered regions that were significant for all three methods of detection. Using this approach, 12 regions were identified (Figure 4, A and B and Figures S11–S13). The candidate carotenoid presence gene Or, which was identified in our GWAS, falls in 1 of these 12 regions. A genome-wide sliding window analysis of LD also identified the same region on chromosome 3 to have the slowest LD decay in domesticated carrots (Figure 4C) but not wild carrots (Figure S14). These results strongly suggest that selection pressures acted on the Or locus during carrot domestication. It is possible that high-carotene alleles at the Or locus have been fixed in most Western domesticated carrots, which may explain why it was not identified until a globally distributed data set of wild and domesticated carrots was used.
Discussion
In this study, we genotyped a large and diverse collection of carrot accessions to determine the global structure of LD in the genome. Genome-wide coverage was ∼1 SNP per 10 kb, dense enough to give an initial assessment of the pattern of LD in carrot. The pattern of LD in a genome is a powerful signal of the population genetic processes that are structuring it, and similar LD decay rates have been found in other highly heterozygous outcrossing species, such as maize and grape (Yan et al. 2009; Myles et al. 2011). We find LD decays very rapidly in both wild and domesticated accessions, with a half-life of 67 and 6544 bp, respectively (Figure S6), and we also demonstrate that LD decline is variable across the nine chromosomes as well as between wild and domesticated accessions (Figure 4C and Figure S14). The observed rapid decay suggests that GWAS should be very useful for identifying candidate genes in carrot as long as SNP density and coverage is comprehensive. To date, only two GWAS studies have been conducted in carrot. Prior to the availability of the carrot reference genome, Jourdan et al. (2015) conducted a candidate gene association study in 380 carrots using 109 SNPs spread across 17 carotenoid biosynthesis genes. The strongest associations with pigmentation were the carotenoid genes ZEP and CRISTO. More recently, Keilwagen et al. (2017) used 85 carrot cultivars and 168,000 SNPs to identify 30 QTL for 15 terpenoid volatile organic compounds, which were further localized to four genomic regions on three different carrot chromosomes containing candidate terpene synthase genes. Future GWAS and LD projects in carrot will benefit from improved genotyping techniques, such as resequencing or two-enzyme GBS (Poland et al. 2012), to increase SNP density. There were likely some regions of the genome that did not have adequate SNP coverage in this study due to rapid linkage decay.
The primary divisions of population structure across our diverse carrot accessions are geographic distribution, west to east, and intensity of breeding effort, wild to domesticated. Previous studies have also demonstrated that wild and domesticated carrots are genetically distinct (Shim and Jorgensen 2000; Clotault et al. 2010; Baranski et al. 2012; Iorizzo et al. 2013; Rong et al. 2014), and that they separate into geographically discrete Eastern and Western groups (Baranski et al. 2012; Iorizzo et al. 2013, 2016; Grzebelus et al. 2014). However, there is evidence of continued gene flow where populations overlap geographically, such as in Western-W accessions, which are present in areas where domesticated carrot is grown. It also appears that there is significant overlap in wild and domesticated samples from the Eastern group. This may be attributed to either recent admixture or to domesticated carrots sharing many of the same alleles with wild carrots from the region. While STRUCTURE failed to identify a distinction between Eastern wild and Eastern domesticated carrots, these do appear as sister clades in the phylogeny with wild Western carrots at the root of both clades (Figure 2D), supporting recent findings that domesticated carrots are genetically closer to Eastern wild carrots than to Western wild carrots (Vavilov and Dorofeev 1992; Iorizzo et al. 2013).
Carrots from Northern Africa, Tunisian-W, form a distinct group but show the least differentiation from all other groups (Table S6). Previously, North African samples clustered closer to wild samples from the West and Middle East (Iorizzo et al. 2013) but here, using a much larger data set and number of SNPs, Tunisian-W samples appear at the base of all domesticated western carrots (Figure 2D), suggesting that carrots from this region of the world may have been important for the improvement of domesticated carrots. Future field sampling efforts and population dynamics analysis should include more representation from North Africa, to better understand carrot domestication and diversity. Finally, we observe that Portuguese-W samples are highly diverged from other accessions. Gene flow in and out of the Iberian peninsula region is likely limited because of the Pyrenees mountain range. However, crosses with Western domesticated carrot have been successful and therefore Portuguese-W samples may provide a novel source of alleles for abiotic stresses.
The analysis of an extensive and representative sample of modern domesticated, historic domesticated, and wild accessions allowed us to identify genomic regions putatively under selection. False positives can be exacerbated by large genomic data sets, so we used a conservative approach to only consider regions identified by all three detection tests (decreased nucleotide diversity, high Fst, and elevated XP-CLR scores) and identified 12 putative genomic regions under selection during domestication (Figure 4, A and B). One selective sweep located on chromosome 3 overlapped with the most significant SNPs in our GWAS analysis for carotenoid presence and contained the candidate gene Or. Analysis of the Or sequence between samples with varying carotenoid content found a nonsynonymous mutation in exon 5 that associates with the presence of α- and β-carotene, and to a lesser extent lutein. Single amino acid substitutions in the Or homologs in melon and Arabidopsis have led to increased carotenoid presence (Tzuri et al. 2015; Yuan et al. 2015). Two markers that had been previously identified to show domestication signatures (Grzebelus et al. 2014) overlapped with 2 of the 12 bins identified in this study. Interestingly, chromosome 2 was previously shown to carry Vrn1, implicated in flowering habit (Alessandro et al. 2013), which was likely a target to favor biennial growth habit during the course of carrot domestication.
Or is important for chromoplast development, a necessary precursor to the accumulation of carotenoids (Lu et al. 2006). Or differentiates noncolored plastids into chromoplasts, which provide the deposition sink for carotenoid accumulation (Lu et al. 2006). Or also post-transcriptionally regulates PSY, the most important regulatory enzyme in the carotenoid pathway (Li et al. 2012; Zhou et al. 2015; Park et al. 2016). This post-transcriptional effect may be why Or has not been identified in previous carrot studies that have looked at carotenoid accumulation mechanisms at the transcription level (Simpson et al. 2016). Mutations in the Or gene are associated with increased chromoplast formation, thereby providing more storage capability for carotenoids (Yuan et al. 2015). In the closest significant GBS SNP to the Or gene, we find that the wild-type allele (A) appears nearly fixed in all wild STRUCTURE groups as well as the eastern domesticated nonorange samples. The domestication allele (T) was found at high frequency in the eastern domesticated orange samples as well as western domesticated nonorange samples, and is fixed in all western domesticated orange samples. The allele frequency distribution of Or across STRUCTURE groups aligns well with the historical record that the first domesticated samples were yellow and only later did orange carrot roots become common (Banga 1963; Stolarczyk and Janick 2011). We hypothesize that a mutation in Or enhanced carotenoid sequestration by optimizing chromoplast formation. It was likely selected after the initial domestication of carrot in conjunction with y2 to increase carotenoid formation and storage in the taproot. The resulting orange phenotype was then rapidly selected in Western domesticated carrots.
This study brings us one step closer to understanding the presence of carotenoids in carrots. Future work should analyze Or expression at the transcript and protein levels, and verify the effect of disrupting its functionality on carotenoid presence. Additionally, the 11 other genomic regions showing consistent signatures of selection (Figure 4, A and B) should be explored for candidate domestication genes, and be considered in tandem with GWAS and mapping studies. Understanding the genetic consequences of domestication and selection on carrot can inform future plant breeding efforts, and allow us to achieve greater gains from selection.
Acknowledgments
The authors thank Rob Kane (deceased), Paul Miller, Brian Emerson, Thomas Hickey, Steve Pincus, Beth Kazmar, Eric Elderbrock, and Holly Ruess for field assistance, Terri Theisen for phenotyping historic accessions, David Spooner for technical comments on the manuscript, the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing genotyping-by-sequencing library preparation and sequencing services, and the many seed companies and plant breeders that provided seed for this project. S.L.E. was supported by the National Science Foundation under grant 1202666 and the National Institute of Food and Agriculture, U. S. Department of Agriculture (NIFA-USDA), under award number 2016-51181-25400. M.I. was supported by NIFA-USDA Hatch project 1008691, and K.E.C. was supported by NIFA-USDA Hatch project 1002863 and NIFA-USDA award number 2017-67013-26187. C.H.L. was supported by The Clif Bar Family Foundation Seed Matters Fellowship and NIFA-USDA North Central Region-SARE graduate student grant (GNC13-175). K.M.C. was supported by a University of Wisconsin–Madison Science and Medicine Graduate Research Scholars Fellowship.
Footnotes
Communicating editor: A. Charcosset
Literature Cited
- Alessandro M. S., Galmarini C. R., Iorizzo M., Simon P., 2013. Molecular mapping of vernalization requirement and fertility restoration genes in carrot. Theor. Appl. Genet. 126: 415–423. 10.1007/s00122-012-1989-1 [DOI] [PubMed] [Google Scholar]
- Arango J., Jourdan M., Geoffriau E., Beyer P., Welsch R., 2014. Carotene hydroxylase activity determines the levels of both alpha-carotene and total carotenoids in orange carrots. Plant Cell 26: 2223–2233. 10.1105/tpc.113.122127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbizu C., Ruess H., Senalik D., Simon P., Spooner D., 2014. Phylogenomics of the carrot genus (Daucus, Apiaceae). Am. J. Bot. 101: 1666–1685. 10.3732/ajb.1400106 [DOI] [PubMed] [Google Scholar]
- Arbizu C., Ellison S., Senalik D., Simon P., Spooner D., 2016. Genotyping-by-sequencing provides the discriminating power to investigate the subspecies of Daucus carota (Apiaceae). BMC Evol. Biol. 16: 234 10.1186/s12862-016-0806-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arscott S., Tanumihardjo S., 2010. Carrots of many colors provide basic nutrition and bioavailable phytochemicals acting as a functional food. Compr. Rev. Food Sci. Food Saf. 9: 223–239. 10.1111/j.1541-4337.2009.00103.x [DOI] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M., 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296. 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- Banga O., 1957a. The development of the original European carrot material. Euphytica 6: 64–76. [Google Scholar]
- Banga O., 1957b. Origin of the European cultivated carrot. Euphytica 6: 54–63. [Google Scholar]
- Banga O., 1963. Main Types of the Western Carotene Carrot and Their Origin. W. E. J. Tjeenk Willink, Zwolle, The Netherlands. [Google Scholar]
- Baranski R., Maksylewicz-Kaul A., Nothnagel T., Cavagnaro P. F., Simon P. W., et al. , 2012. Genetic diversity of carrot (Daucus carota L.) cultivars revealed by analysis of SSR loci. Genet. Resour. Crop Evol. 59: 163–170. 10.1007/s10722-011-9777-3 [DOI] [Google Scholar]
- Boiteux L., Fonseca M., Simon P., 1999. Effects of plant tissue and DNA purification method on randomly amplified polymorphic DNA-based genetic fingerprinting analysis in carrot. J. Am. Soc. Hortic. Sci. 124: 32–38. [Google Scholar]
- Bradbury P., Zhang Z., Kroon D., Casstevens T., Ramdoss Y., 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Branca A., Paape T., Zhou P., Briskine R., Farmer A., et al. , 2011. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. USA 108: E864–E870. 10.1073/pnas.1104032108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothwell D., Brothwell P., 1969. Food in Antiquity: A Survey of the Diet of Early Peoples. Thames & Hudson, London, UK. [Google Scholar]
- Browning B., Browning S., 2016. Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98: 116–126. 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buishand J., Gabelman W., 1979. Investigations on the inheritance of color and carotenoid content in phloem and xylem of carrot roots (Daucus carota L.). Euphytica 28: 611–632. 10.1007/BF00038928 [DOI] [Google Scholar]
- Chen H., Patterson N., Reich D., 2010. Population differentiation as a test for selective sweeps. Genome Res. 20: 393–402. 10.1101/gr.100545.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotault J., Geoffriau E., Lionneton E., Briard M., Peltier D., 2010. Carotenoid biosynthesis genes provide evidence of geographical subdivision and extensive linkage disequilibrium in the carrot. Theor. Appl. Genet. 121: 659–672. 10.1007/s00122-010-1338-1 [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D. A., vonHoldt B. M., 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4: 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Ellison S., Senalik D., Bostan H., Iorizzo M., Simon P., 2017. Fine mapping, transcriptome analysis, and marker development for Y2, the gene that conditions β-carotene accumulation in carrot (Daucus carota L). G3 (Bethesda) 7: 2665–2675. 10.1534/g3.117.043067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire R., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Gouder J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Fernandes Santos C. A., Simon P. W., 2006. Heritabilities and minimum gene number estimates of carrot carotenoids. Euphytica 151: 79–86. 10.1007/s10681-006-9130-7 [DOI] [Google Scholar]
- Glaubitz J., Casstevens T., Lu F., Harriman J., Elshire R., et al. , 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J., 2005. Hierfstat, a package for r to compute and test hierarchical f-statistics. Mol. Ecol. Notes 5: 184–186. 10.1111/j.1471-8286.2004.00828.x [DOI] [Google Scholar]
- Grzebelus D., Iorizzo M., Senalik D., Ellison S., Cavagnaro P., et al. , 2014. Diversity, genetic mapping, and signatures of domestication in the carrot (Daucus carota L.) genome, as revealed by Diversity Arrays Technology (DArT) markers. Mol. Breed. 33: 625–637. 10.1007/s11032-013-9979-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorizzo M., Senalik D., Ellison S., Grzebelus D., Cavagnaro P., et al. , 2013. Genetic structure and domestication of carrot (Daucus carota subsp. sativus) (Apiaceae). Am. J. Bot. 100: 930–938. 10.3732/ajb.1300055 [DOI] [PubMed] [Google Scholar]
- Iorizzo M., Ellison S., Senalik D., Zeng P., Satapoomin P., et al. , 2016. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48: 657–666. 10.1038/ng.3565 [DOI] [PubMed] [Google Scholar]
- Jourdan M., Gagné S., Dubois-Laurent C., Maghraoui M., Huet S., et al. , 2015. Carotenoid content and root color of cultivated carrot: a candidate-gene association study using an original broad unstructured population. PLoS One 10: e0116674 10.1371/journal.pone.0116674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just B. J., Santos C. A., Fonseca M. E., Boiteux L. S., Oloizia B. B., et al. , 2007. Carotenoid biosynthesis structural genes in carrot (Daucus carota): isolation, sequence-characterization, single nucleotide polymorphism (SNP) markers and genome mapping. Theor. Appl. Genet. 114: 693–704. 10.1007/s00122-006-0469-x [DOI] [PubMed] [Google Scholar]
- Keilwagen J., Lehnert H., Berner T., Budahn H., Nothnagel T., et al. , 2017. The terpene synthase gene family of carrot (Daucus carota L.): identification of QTLs and candidate genes associated with terpenoid volatile compounds. Front. Plant Sci. 8: 1930 10.3389/fpls.2017.01930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Plagnol V., Hu T., Toomajian C., Clark R., et al. , 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39: 1151–1155. 10.1038/ng2115 [DOI] [PubMed] [Google Scholar]
- Lam H., Xu X., Liu X., Chen G., Yang W., et al. , 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42: 1053–1059. 10.1038/ng.715 [DOI] [PubMed] [Google Scholar]
- Li L., Paolillo D., Parthasarathy M., Dimuzio E., Garvin D., 2001. A novel gene mutation that confers abnormal patterns of beta-carotene accumulation in cauliflower (Brassica oleracea var. Botrytis). Plant J. 26: 59–67. 10.1046/j.1365-313x.2001.01008.x [DOI] [PubMed] [Google Scholar]
- Li L., Yang Y., Xu Q., Owsiany K., Welsch R., et al. , 2012. The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Mol. Plant 5: 339–352. 10.1093/mp/ssr099 [DOI] [PubMed] [Google Scholar]
- Lu S., Van Eck J., Zhou X., Lopez A., O’Halloran D., et al. , 2006. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18: 3594–3605. 10.1105/tpc.106.046417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass D., Arango J., Wüst F., Beyer P., Welsch R., 2009. Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS One 4: e6373 10.1371/journal.pone.0006373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macko-Podgórni A., Machaj G., Stelmach K., Senalik D., Grzebelus E., et al. , 2017. Characterization of a genomic region under selection in cultivated carrot (Daucus carota subsp. sativus) reveals a candidate domestication gene. Front. Plant Sci. 8: 12 10.3389/fpls.2017.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M., Thompson W., 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4326. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S., Boyko A., Owens C., Brown P., Grassi F., et al. , 2011. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 108: 3530–3535. 10.1073/pnas.1009363108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. 10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kim H., Jung Y., Kim S., Ji C., et al. , 2016. Orange protein has a role in phytoene synthase stabilization in sweet potato. Sci. Rep. 6: 33563 10.1038/srep33563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J., Brown P., Sorrells M., Jannink J., 2012. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7: e32253 10.1371/journal.pone.0032253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J., Lammers Y., Strasburg J., Schidlo N., Ariyurek Y., et al. , 2014. New insights into domestication of carrot from root transcriptome analyses. BMC Genomics 15: 895 10.1186/1471-2164-15-895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N., 2004. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4: 137–138. 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- Santos C. A., Simon P. W., 2002. QTL analyses reveal clustered loci for accumulation of major provitamin A carotenes and lycopene in carrot roots. Mol. Genet. Genomics 268: 122–129. 10.1007/s00438-002-0735-9 [DOI] [PubMed] [Google Scholar]
- Shim S., Jorgensen R., 2000. Genetic structure in cultivated and wild carrots (Daucus carota L.) revealed by AFLP analysis. Theor. Appl. Genet. 101: 227–233. 10.1007/s001220051473 [DOI] [Google Scholar]
- Simon P., 1990. Carrots and other horticultural crops as a source of provitamin a carotenes. HortScience 25: 1495–1499. [Google Scholar]
- Simon P., 2000. Domestication, historical development, and modern breeding of carrot. Plant Breed. Rev. 19: 157– 190. [Google Scholar]
- Simon, P., and I. Goldman, 2007 Carrot, pp. 497–517 in Genetic Resources, Chromosome Engineering and Crop Improvement: Vegetable Crops, edited by R. J. Singh. CRC Press, Boca Raton, FL. [Google Scholar]
- Simon P., Wolff X., 1987. Carotenes in typical and dark orange carrots. J. Agric. Food Chem. 35: 1017–1022. 10.1021/jf00078a038 [DOI] [Google Scholar]
- Simon P., Wolff X., Peterson C., Kammerlohr D., 1989. High carotene mass carrot population. HortScience 24: 174–175. [Google Scholar]
- Simon, P., R. Freeman, J. Vieira, L. Boiteux, M. Briard et al., 2008 Carrot, pp. 327–357 in Handbook of Crop Breeding, Vol. 1, Vegetable Breeding. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- Simon P., Pollak L., Clevidence B., Holden J., Haytowitz D., 2009. Plant breeding for human nutrition. Plant Breed. Rev. 31: 325–392. [Google Scholar]
- Simpson K., Cerda A., Stange C., 2016. Carotenoid biosynthesis in Daucus carota. Subcell. Biochem. 79: 199–217. [DOI] [PubMed] [Google Scholar]
- Soufflet-Freslon V., Jourdan M., Clotault J., Huet S., Briard M., et al. , 2013. Functional gene polymorphism to reveal species history: the case of the CRTISO gene in cultivated carrots. PLoS One 8: e70801 10.1371/journal.pone.0070801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolarczyk J., Janick J., 2011. Carrot: history and iconography. Chron. Hortic. 51: 13–18. [Google Scholar]
- Turner S., 2014. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. Available at: https://www.biorxiv.org/content/early/2014/05/14/005165. 10.1101/005165 [DOI] [Google Scholar]
- Tzuri G., Zhou X., Chayut N., Yuan H., Portnoy V., et al. , 2015. A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis Melo). Plant J. 82: 267–279. 10.1111/tpj.12814 [DOI] [PubMed] [Google Scholar]
- Vavilov N., Dorofeev V., 1992. Origin and Geography of Cultivated Plants. Cambridge University Press, Cambridge [England]; New York. [Google Scholar]
- Vos P. G., Paulo M. J., Voorrips R. G., Visser R. G., van Eck H. J., et al. , 2017. Evaluation of LD decay and various LD-decay estimators in simulated and SNP-array data of tetraploid potato. Theor. Appl. Genet. 130: 123–135. 10.1007/s00122-016-2798-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B., Cockerham C., 1984. Estimating f-statistics for the analysis of population structure. Evolution 38: 1358–1370. 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- Wickham H., 2009. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Wright S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. 10.1111/j.1469-1809.1949.tb02451.x [DOI] [PubMed] [Google Scholar]
- Yan J., Shah T., Warburton M., Buckler E., McMullen M., et al. , 2009. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One 4: e8451 10.1371/journal.pone.0008451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Zhang J., Nageswaran D., Li L., 2015. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2: 01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C., Eizenga G., Wright M., Ali M., et al. , 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2: 467 10.1038/ncomms1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Welsch R., Yang Y., Álvarez D., Riediger M., et al. , 2015. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 112: 3558–3563. 10.1073/pnas.1420831112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Carrot sequences used for the Or gene alignment and D. syrticus samples used as an outgroup for phylogenetic analysis are available under the National Center for Biotechnology Information Bioproject accession PRJNA291976. All supplementary tables and data sets necessary to reproduce the analyses for this manuscript are available at Figshare: 10.6084/m9.figshare.6177110.