Abstract
In the budding yeast Saccharomyces cerevisiae, ribosomal RNA genes are encoded in a highly repetitive tandem array referred to as the ribosomal DNA (rDNA) locus. The yeast rDNA is the site of a diverse set of DNA-dependent processes, including transcription of ribosomal RNAs by RNA polymerases I and III, transcription of noncoding RNAs by RNA polymerase II, DNA replication initiation, replication fork blocking, and recombination-mediated regulation of rDNA repeat copy number. All of this takes place in the context of chromatin, but little is known about the roles played by ATP-dependent chromatin remodeling factors at the yeast rDNA. In this work, we report that the Isw2 and Ino80 chromatin remodeling factors are targeted to this highly repetitive locus. We characterize for the first time their function in modifying local chromatin structure, finding that loss of these factors decreases the fraction of actively transcribed 35S ribosomal RNA genes and the positioning of nucleosomes flanking the ribosomal origin of replication. In addition, we report that Isw2 and Ino80 promote efficient firing of the ribosomal origin of replication and facilitate the regulated increase of rDNA repeat copy number. This work significantly expands our understanding of the importance of ATP-dependent chromatin remodeling for rDNA biology.
Keywords: Isw2, Ino80, ribosomal DNA locus, chromatin remodeling, Saccharomyces cerevisiae
IN exponentially growing cells, the enormous cellular demand for ribosomes is reflected in the proportion of resources dedicated to their production. For example, the production of ribosomal RNAs (rRNAs) accounts for an estimated 60% of all transcriptional activity in cycling yeast cells (Warner 1999). Because single genomic copies of rRNA genes would not support such large volumes of transcriptional output, eukaryotic genomes have evolved to include highly repetitive clusters of rRNA genes, termed the ribosomal DNA (rDNA) locus. In a typical cell of the budding yeast Saccharomyces cerevisiae, the rDNA locus comprises ∼150–200 tandem repeats on Chromosome XII (Figure 1A). Each repeat contains a 35S ribosomal RNA (rRNA) gene, transcribed by RNA polymerase I (Pol I), and an intergenic spacer (IGS), split into IGS1 and IGS2 regions by the 5S rRNA gene, which is transcribed by RNA polymerase III (Pol III). Due to its large size and repetitive nature, the rDNA locus has unique regulatory needs, and the IGS1 and IGS2 regions contain genetic elements that are critical to addressing these needs.
Figure 1.
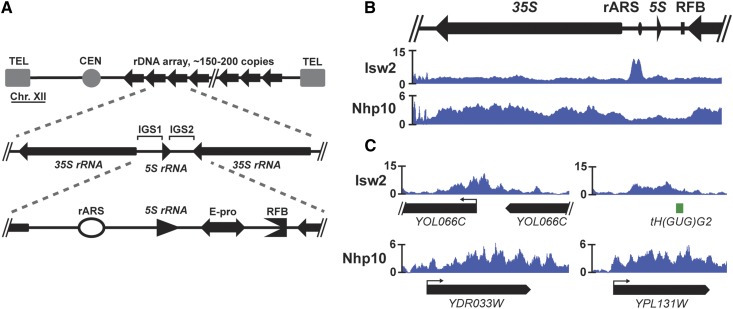
The Isw2 and Ino80 chromatin remodeling complexes are targeted to the rDNA locus. (A) A schematic drawing of the rDNA locus in S. cerevisiae. In a typical yeast cell, the rDNA locus, comprised of a tandem array of ∼150 copies of the rDNA repeat, accounts for ∼1.5 Mb of chromosome XII. Each repeat contains a 35S rRNA gene and an IGS region in between adjacent 35S genes, itself split into IGS1 and IGS2 regions by the 5S rRNA gene. IGS1 contains the ribosomal origin of replication, or autonomously replicating sequence (rARS), and IGS2 contains the bidirectional RNA Pol II promoter, E-pro, and replication fork block (RFB). (B) The Isw2 subunit of the Isw2 complex and the Nhp10 subunit of the Ino80 complex were each FLAG-tagged, chromatin immunoprecipitated, and deep-sequenced (ChIP-seq). (C) Representative ChIP-seq signals of Isw2 and Nhp10 at single copy targets outside of the rDNA.
If the rDNA locus lacked an origin of replication (autonomously replicating sequence, or ARS), replicating the rDNA array would require replication forks to traverse multiple megabases of DNA from either end of the array. To avoid this, the IGS1 contains a ribosomal ARS (rARS). As a consequence, the ∼150 ARSs in a typical rDNA array account for nearly one-third of all genomic origins of replication. Because replication factors are limiting during each S-phase (Mantiero et al. 2011), firing of too many rARSs would take vital replicative resources away from other parts of the genome, raising the risk of delayed or incomplete replication. As a result, only ∼20% of rARSs will fire in any given round of cell division (Walmsley et al. 1984; Brewer and Fangman 1988). If too few rARSs fire, replication of the rDNA array may be delayed or incomplete (Yoshida et al. 2014). Thus, properly striking this balance by regulating origin efficiency at the rDNA has critical consequences for global genome stability. Another balance must be carefully achieved in maintaining the proper size of the rDNA array. The array must be large enough to support sufficient transcription of rRNAs, but small enough to be efficiently replicated. Thus, a mechanism exists to change the size of the array by adding or removing copies of the rDNA repeat as needed, and the IGS2 region contains two genetic elements that are critical for this process: a bidirectional RNA Pol II promoter, E-pro, and a replication fork block (RFB).
All DNA-dependent processes occurring at the rDNA happen in the context of chromatin structure. The Sir2 and Rpd3 histone deacetylases (HDACs) have well-established roles in regulating rDNA chromatin structure, origin activity, and copy number maintenance (Fritze et al. 1997; Sandmeier et al. 2002; Kobayashi and Ganley 2005; Yoshida et al. 2014). In addition, the rDNA locus is regulated by ATP-dependent chromatin remodeling factors, which use the energy of ATP hydrolysis to modify the position and histone composition of nucleosomes. In humans, the nucleolar remodeling complex (NoRC) positions nucleosomes and recruits histone methyltransferase and histone deacetylase activity to promote rDNA silencing (Santoro et al. 2002; Li et al. 2006). In yeast, the SWI/SNF (Zhang et al. 2013), Isw1, Isw2, and Chd1 (Jones et al. 2007) complexes have been implicated in regulating transcription of rRNAs. However, it has not been shown how remodeling factors modify chromatin structure at the yeast rDNA or affect any DNA-dependent processes at this locus beyond rRNA transcription.
In this work, we show that the Isw2 and Ino80 ATP-dependent chromatin remodeling factors regulate chromatin structure at the rDNA. The Isw2 complex is known to slide nucleosomes over gene promoters (Fazzio and Tsukiyama 2003)—an activity that generally represses transcription, both for coding genes (Goldmark et al. 2000; Fazzio et al. 2001) and antisense transcripts (Whitehouse et al. 2007). The Ino80 complex slides and evicts nucleosomes and removes the histone variant, H2A.Z (Tsukuda et al. 2005; Papamichos-Chronakis et al. 2011; Udugama et al. 2011; Zhou et al. 2018). Ino80 is also involved in regulating the checkpoint response following DNA damage, DNA damage repair, and DNA replication (Morrison et al. 2004, 2007; Shimada et al. 2008). Isw2 and Ino80 function together to promote replication of late-replicating regions of the genome in the presence of replication stress and to attenuate the S-phase checkpoint response (Vincent et al. 2008; Au et al. 2011; Lee et al. 2015). Here, we show that both Isw2 and Ino80 are targeted to the ribosomal DNA locus. Further, we report for the first time that these remodeling factors affect local chromatin structure, as loss of the factors increases nucleosome occupancy in the 35S and alters the positioning of nucleosomes flanking the rARS. We find that loss of Isw2 and Ino80 reduces the proportion of active rDNA repeats without affecting overall transcription of rRNAs, but that Isw2 and Ino80 positively contribute both to the efficiency of the rARS and to the rate of rDNA repeat copy number increase. In sum, this study expands our understanding of how ATP-dependent chromatin remodeling factors affect both chromatin structure and essential biological processes at the ribosomal DNA locus.
Materials and Methods
Yeast strains and media
Strains used are listed in Supplemental Material, Table S1. Strains generated using standard gene replacement protocols. Unless otherwise indicated, yeast cells were grown in YPD medium (2% Bacto Peptone, 1% yeast extract, 2% glucose). All strains are congenic to MATa W303-1a. With the exception of the rDNA copy number mutants used in the copy number change experiments, all strains used have nearly identical rDNA copy number of ∼150 rDNA repeats (Figure S3).
Chromatin immunoprecipitation and micrococcal nuclease digestion followed by deep sequencing
Chromatin immunoprecipitation (ChIP) and micrococcal nuclease (MNase) digestion were performed as described previously (Rodriguez et al. 2014). For H3-ChIP experiments, anti-H3 C-term antibody (catalog # ab1791; Abcam) was used; for all other ChIPs, the targeted protein was epitope-tagged with FLAG and immuno-precipitated using an anti-FLAG monoclonal antibody (catalog # F3165; Sigma). All Isw2 ChIP-seq was performed on a FLAG-tagged, catalytically inactive allele of ISW2 as previously described (Gelbart et al. 2005). All libraries were constructed using the Nugen Ovation Ultralow System V2 (catalog # 0344-32) and then single-end (ChIP-seq) or paired-end (MNase-seq) sequenced, with 50 bp read length, on Illumina Hi-Seq 2500. Ribbon plots, bar graphs, and line graphs were generated with the ggplot2 R package (http://ggplot2.org/). For MACS2 peaks-calling, the following parameters were used: macs2 callpeak -t <filename>.bam -c <filename_sorted>.bam -f BAM -g 1.21e7 -B–nomodel–extsize 147.
For all depictions of deep-sequencing data at the rDNA, a single copy of the rDNA locus is shown. Our reference genome contains two copies of the rDNA, and any read mapping to the rDNA is randomly assigned to one of these two copies. Thus, sequencing data reflects the average signal across all rDNA repeats in all cells sampled. For ChIP-seq analyses, IP and input DNA from the same chromatin prep were both sequenced. Normalization of IP to input was done using these matched samples, thus controlling for any minor variation in rDNA copy number that might otherwise affect direct comparison between different samples.
Reverse transcription- and ChIP-quantitative PCR
RNA was isolated using hot acid phenol, then cleaned up with the Qiagen RNeasy Cleanup Kit (catalog # 74204) plus on-column treatment with DNase I (catalog # 79254; Qiagen). cDNA was generated from the RNA using Superscript III Reverse Transcriptase (catalog # 18080093; ThermoFisher). Quantitative PCR was performed on both cDNA and ChIP DNA using 2× Power SYBR Master Mix (catalog # 4367659; Fisher Scientific) run on the ABI QuantStudio5 Real Time PCR System machine.
Psoralen crosslinking
The psoralen crosslinking assay was performed as previously described (Dammann et al. 1993; Smith and Boeke 1997; Sandmeier et al. 2002). Cells were grown to midlog phase (OD660 = 0.5–0.7), ∼3 × 108 cells were collected, washed twice with ice-cold water, and then resuspended in 1.4 ml cold TE buffer. Cells were transferred to six-well plates, and 70 μl of psoralen (200 μg/ml in 100% ethanol) was added to the cells. On ice, the plates were irradiated with 365 nm UV for 5 min. Psoralen addition followed by UV irradiation was repeated four additional times, for a total of five rounds. Cells were collected, washed in water, spheroplasted with zymoylase 100T, and washed in spheroplast buffer. The pellet was lysed by resuspension in TE buffer with 0.5% SDS, and then treated with Proteinase K overnight at 50°. DNA was extracted with phenol:chloroform:IAA, ethanol precipitated, and then digested for 3 hr with EcoRI-HF. Samples were treated with RNase A at 37° for 30 min, ethanol precipitated, quantified, and then run in 1.3% LE agarose gels in 0.5× TBE for 24 hr at 60 V. Gels were irradiated for 2 min per side with 254 nm UV, transferred to a GeneScreen Plus membrane in 10× SSC, and then hybridized with a probe contained within a EcoRI restriction fragment in the rDNA ETS1. Membranes were visualized using a Typhoon Phosphor Imager, and images were visualized using ImageJ software.
Two-dimensional gel electrophoresis
DNA sample preparation based on the Brewer/Raghuraman laboratory protocol (http://fangman-brewer.genetics.washington.edu/plug.html). Cells were grown to midlog phase (OD660 = 0.5–0.7), sodium azide added to 0.1% final concentration, and then cultures were washed in water. Cell pellets were resuspended in 50 mM EDTA, mixed with an equal volume of 1.0% Low-Melt Agarose (catalog # 161−3111; Bio-Rad), and pipetted into plug molds. Cells in plugs were spheroplasted with 0.5 mg/ml Zymolyase 20-T, thoroughly washed, and stored in TE at 4°. Plugs were digested with NheI for 5 hr at 37°, then run in 0.4% agarose gels in TBE at 1 V/cm for 22 hr at room temperature. Gels were stained with ethidium bromide (EtBr), visualized with UV, and the desired size range for each sample was identified in the gel and physically cut out. This piece of gel was then rotated 90° and placed in a new gel tray, and warm 1.1% agarose in TBE was poured around it. This gel was then run at 5 V/cm for 6 hr at 4°. After running, the gel was visualized, transferred onto a GeneScreen Plus membrane (catalog # NEF986001PK; Perkin Elmer), and hybridized with a probe encompassing the RFB.
rDNA copy number change assay
Strains were made from YSI102 (Ide et al. 2010), in which the endogenous FOB1 gene had been deleted, and the number of rDNA repeats reduced to 20 copies. From the 20-rDNA-copy fob1 parent, isw2∆, nhp10∆, and isw2∆ nhp10∆ strains were generated. Separately, the FOB1 gene was Gibson cloned into the pRS426 plasmid. Either this FOB1-pRS426 plasmid or a pRS426 plasmid with no FOB1 gene was then transformed into each 20-copy strain and plated on yeast complete (Bywater et al.) –URA medium with 2% glucose. Individual transformants were restreaked on selective medium, the presence of the desired plasmid was confirmed by PCR, and then transformants were inoculated into liquid YC –URA + 2% glucose. Cultures were allowed to reach saturation, and then aliquots were collected, washed in cold 50 mM EDTA, and cell pellets frozen in liquid nitrogen and stored at −80°. From the remaining saturated cultures, all strains were diluted by the same factor, then allowed to grow back to saturation, at which point the next time point would be collected, up to ∼200 generations. Generations were calculated from the base 2 log of the dilution factor applied at each passage (for example, a saturated culture diluted by a factor of 1024 into the same volume of medium would require 10 generations to return to saturation).
Clamped homogenous electric field gel electrophoresis and southern blotting
Samples for clamped homogenous electric field (CHEF) gels were prepared in agarose based on a previously described method (Kwan et al. 2016). Frozen cell pellets were thawed, resuspended in 100 mM EDTA, then mixed with 0.8% low-melt agarose and 25 mg/ml zymolyase 20T. This mixture was pipetted into plug molds, allowed to solidify at 4°, then washed with a series of buffers (Solution V: 500 mM EDTA pH 7.5, 10 mM Tris pH 7.5; Solution VI: 5% sarcosyl, 5 mg/ml proteinase K, 500 mM EDTA pH 7.5; Solution VII: 2 mM Tris pH 7.5, 1 mM EDTA, pH 8.0). Before being run, plugs were incubated for ∼30 min in 0.5× TBE running buffer at 4° before being placed on gel comb teeth, positioned in gel mold, and then warm 0.8% 0.5× TBE was poured. The CHEF gel was run on a CHEF-DR II device with a program adapted from Ide et al. 2007: Block 1 = 2.0 V/cm, pulse time of 1200–1400 sec, total run time 72 hr; Block 2 = 6.0 V/cm, pulse time of 25–146 sec, total run time 7.5 hr. After electrophoresis, gels were incubated with 0.5 μg/ml EtBr in running buffer for 30–45 min, UV-irradiated with a Stratagene Stratalinker to nick DNA, transferred onto HyBond N+ positively charged membrane (catalog # RPN303B; GE), and hybridized with a probe targeting the RFB.
URA3 recombination assay
Strains were generated by integration of linearized plasmid pRS406 containing the URA3 gene. Cells were streaked onto YC-URA plates, single colonies were inoculated into liquid YC-URA medium, and cultures were grown to saturation. Culture concentrations were measured, and a known number of cells were inoculated into YPD. Cultures were grown for 24 hr, and the concentration was measured and used to calculate the number of generations elapsed without selection. Cells were diluted to yield a countable number of colonies, and then plated on both YPD and 5-fluoroorotic acid (5-FOA) plates. 5-FOA-resistant colonies were interpreted as the product of recombination events that led to the loss of URA3, and the number of recombination events per cell per generation was calculated.
Data availability
All strains (Table S1) and plasmids (Table S3) used in this study are available upon request, and oligonucleotide sequences are included in Table S2. All genomics data are available at GEO under the accession number GSE112465. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, supplemental material, and genomics data. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7243292.
Results
The Isw2 and Ino80 chromatin remodeling complexes are targeted to the ribosomal DNA locus
All of the DNA-dependent processes that take place at the rDNA locus occur in the context of chromatin. Although HDACs such as Rpd3 and Sir2 have well-characterized functions in regulating chromatin structure, transcription, and copy number maintenance at the S. cerevisiae rDNA (Fritze et al. 1997; Smith and Boeke 1997; Sandmeier et al. 2002; Kobayashi and Ganley 2005), comparatively little is known about the roles played by ATP-dependent chromatin remodeling factors at this vital genomic locus. To address this, we performed ChIP followed by deep sequencing (ChIP-seq) to map where the Isw2 and Ino80 chromatin remodeling factors are targeted at the rDNA. We found that the catalytic subunit of Isw2 and Nhp10—a subunit unique to the Ino80 complex (Morrison et al. 2004)—were both targeted to the rDNA (Figure 1B). The ChIP-seq signal for Isw2 was slightly above the genome average throughout the 35S gene body. The pattern of targeting in the IGS included small peaks flanking the 5S gene and the region containing E-pro and the RFB, but the most prominent signal was a striking, bimodal peak on top, and to one side, of the rARS. Nhp10 was also present throughout the 35S gene body and showed a small peak around the 5S. The ChIP-seq patterns of both factors at the rDNA were consistent with peaks elsewhere in the genome with regard to both shape and magnitude: Isw2 tended to have fairly defined peaks that rise well above the genome average, located in intergenic regions, and Nhp10 peaks were generally less prominent relative to the genome average and more diffusely spread throughout a transcription unit (Figure 1C). Out of 830 peaks identified genome-wide by the MACS peaks-calling algorithm in this Nhp10 ChIP-seq data set, this rDNA peak had the third-lowest P-value (P = 4.81E−73). Given these distinct targeting patterns, we hypothesized that these ATP-dependent chromatin remodeling factors might have previously unknown functions at this highly repetitive, unique genomic locus.
Isw2 and Ino80 affect nucleosome occupancy over the 35S rRNA gene
In light of the established functions of the Isw2 and Ino80 complexes, we first asked whether these chromatin remodeling factors affect nucleosome occupancy within the rDNA locus. Individual rDNA repeats exist in one of two discrete states, being either highly occupied with nucleosomes and transcriptionally inactive, or heavily depleted of nucleosomes and highly transcriptionally active (Conconi et al. 1989; French et al. 2003; Merz et al. 2008). We assessed how nucleosome occupancy at the rDNA is affected by these two chromatin remodeling factors with ChIP-seq of histone H3 in wild-type, isw2∆, nhp10∆, and isw2∆ nhp10∆ strains. This analysis revealed that nucleosome occupancy throughout the 35S gene body is appreciably increased in the isw2∆ nhp10∆ double mutant compared to wild-type and single deletion strains (Figure 2A). Notably, this is the part of the rDNA in which the ChIP-seq signals of both chromatin remodeling factors most significantly overlap, suggesting the possibility that these factors may work together in this region.
Figure 2.
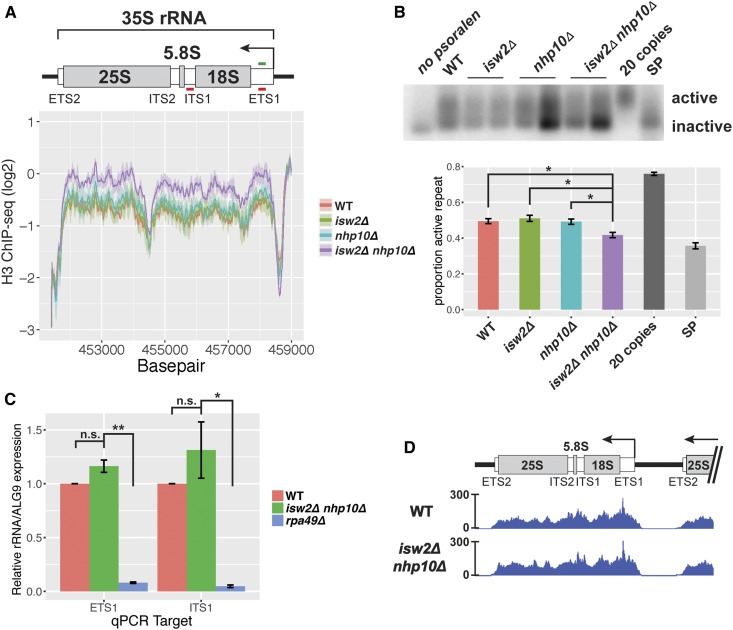
Nucleosome occupancy, but not transcription, is affected at the 35S rDNA in isw2∆ and nhp10∆ mutants. (A) Histone H3 ChIP-seq through the 35S rRNA gene. Line represents average log2 ChIP-seq signal at each base pair for two independent experiments, and the ribbon represents the SEM at each base pair. Schematic drawing of the 35S indicates transcribed spacers that are removed during processing, as well as the mature 18S, 5.8S, and 25S rRNAs that are parts of complete ribosomes. ETS1 and ITS1 qPCR primer sets are indicated with red lines, and ETS1 hybridization probe, used in the Southern blot shown in (B), indicated in green. (B) Psoralen cross-linked DNA, digested with EcoRI and hybridized with a probe to the ETS1 region. Two independent isolates of each remodeling factor mutant are shown. For quantification, signal strengths of the active and inactive bands were measured with ImageJ software, and the proportion of the total signal present in the “active” band was calculated. Values for each genotype reflect between three and five biological replicates, and error bars represent SEM. The “20 copies” sample comes from a strain with only 20 copies of the rDNA repeat, in which all 20 copies should be actively transcribed, and “SP” is the wild-type (WT) strain at stationary phase, when a large proportion of rDNA repeats should be inactive. (C) RT-qPCR measuring the ETS1 and ITS1 of the 35S pre-rRNA. For each qPCR target, expression for all strains normalized to WT. For (B) and (C), statistical significance determined by pairwise t-tests followed by Bonferroni correction for multiple testing. * P < 0.05, ** P < 0.005. (D) RNA Pol I ChIP-seq (FLAG-tagged RNA Pol I subunit RPA190).
Because individual rDNA repeats exist in one of two discrete chromatin states, we hypothesized that the increased nucleosome occupancy in isw2∆ nhp10∆ cells reflected a reduced ratio of active to inactive rDNA repeats. To test this, we used psoralen cross-linking, a well-established method to determine the ratio of active to inactive rDNA repeats (Conconi et al. 1989; Dammann et al. 1993). Occupancy of chromatin by nucleosomes blocks incorporation of psoralen. Therefore, actively transcribed, nucleosome-depleted rDNA repeats become more heavily cross-linked with psoralen, and thus run more slowly in an agarose gel, than inactive, nucleosome-occupied repeats. After digestion with appropriate restriction enzymes, Southern blotting, and hybridization with a probe targeting a region of the 35S gene unit, two discrete bands representing active and inactive repeats can be resolved (Conconi et al. 1989; Dammann et al. 1993). This method showed that, as expected, a very large proportion of repeats are active in an asynchronously growing strain with only 20 copies of the rDNA (French et al. 2003), and a very small proportion of repeats are active in a wild-type strain at stationary phase. We found that isw2∆ nhp10∆ cells have a significantly reduced proportion of active repeats compared to wild-type, isw2∆, or nhp10∆ cells (Figure 2B, P < 0.05), consistent with the observed increase in H3 occupancy in double mutant cells. Based on these results, we concluded that the Isw2 and Ino80 chromatin remodeling factors increase the fraction of active rDNA repeats.
Transcription of 35S ribosomal RNA is not affected by loss of Isw2 or Nhp10
Based on the reduced proportion of nucleosome-depleted rDNA repeats in the isw2∆ nhp10∆ mutant, we hypothesized that these cells would also show reduced levels of 35S rRNA transcription. The 35S is transcribed as a single long transcript before being cleaved and folded in a series of processing steps to yield mature 18S, 5.8S, and 25S RNAs (Woolford and Baserga 2013). Because mature rRNAs are components of ribosomes and thus highly stable and abundant, nascent RNA needs to be measured to assess the transcription rate of rRNAs. The external transcribed spacer 1 (ETS1) and internal transcribed spacer 1 (ITS1) sections of the 35S gene are transcribed but removed at early stages of rRNA processing. Levels of these RNA sequences thus reflect levels of nascent rRNA and are used to measure the rate of 35S transcription (Bywater et al. 2012; Laribee et al. 2015). Adopting this approach, we performed reverse-transcription quantitative PCR (RT-qPCR) targeting parts of the ETS1 and ITS1 regions of the 35S pre-rRNA (Figure 2A, 2C). As expected, we found significantly reduced levels of both ETS1 and ITS1 in an rpa49 deletion mutant (P = 0.0021 and P = 0.037, respectively), a strain known to have a reduced rate of RNA Pol I transcription (Beckouet et al. 2008; Laribee et al. 2015). In contrast, we did not see evidence of a significant difference in rates of 35S transcription in isw2∆ nhp10∆ compared to wild type (P = 0.19 and P = 1.00). To confirm this result by an independent method, we performed ChIP-seq analysis of the Pol I subunit RPA190, and observed virtually identical profiles in isw2∆ nhp10∆ and wild-type strains, with regard to both shape and overall levels (Figure 2D). Based on these results, we concluded that isw2∆ nhp10∆ cells exhibit no significant defects in the rate of 35S transcription despite the differences in nucleosome occupancy and the proportion of nucleosome-occupied rDNA repeats in these mutants.
Isw2 and Ino80 affect nucleosome positioning in the rDNA IGS
In addition to nucleosome occupancy, nucleosome positioning is known to be affected by both of these chromatin remodeling factors (Fazzio and Tsukiyama 2003; Udugama et al. 2011). ChIP-seq of histone H3 is suitable for measuring nucleosome occupancy, but because it relies on sonication-based fragmentation of chromatin, it lacks sufficient resolution to accurately reveal nucleosome positioning. Therefore, we assessed nucleosome positioning at the rDNA by micrococcal nuclease (MNase) digestion followed by deep sequencing (MNase-seq). During the preparation of MNase-digested DNA, we gel purified mononucleosome-sized fragments of ∼150 bp and then interpreted each size-selected, paired-end read as coming from a nucleosome-protected fragment of DNA. From each paired-end read, the nucleosomal dyad center was inferred and plotted. By this method, nucleosome positions appeared strongly shifted at known Isw2 targets in isw2∆ and isw2∆ nhp10∆ mutants (Figure S1). In contrast, no gross differences in nucleosome positions were observed throughout the 35S gene body (Figure S2A) or in the rDNA IGS region (Figure 3A). However, sequencing data must be interpreted carefully within the highly repetitive rDNA, as such data represent an average of the signal at all ∼150 rDNA repeats in all cells sampled, and nucleosomes in only a fraction of those repeats may change positions in any given cell.
Figure 3.
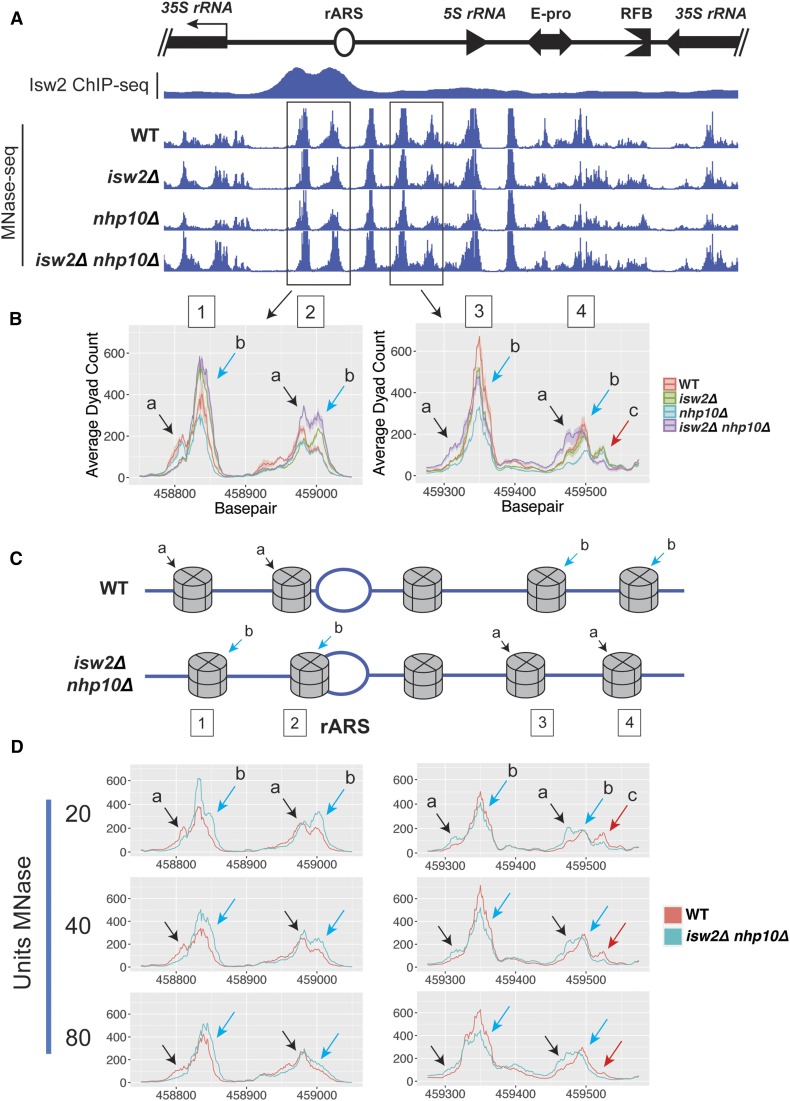
Isw2 and Ino80 affect nucleosome positioning in the rDNA IGS. (A) Micrococcal nuclease digestion followed by deep-sequencing (MNase-seq) profiles in the IGS, with Isw2 ChIP-seq data overlaid. From each paired end sequencing read, the nucleosome dyad was inferred and plotted. (B) Ribbon plots, generated as described in Figure 2A, focused on two pairs of nucleosomes, indicated with boxes in (A). Different subspecies of nucleosome positions are indicated with colored arrows and letters. (C) Cartoon diagram depicting five nucleosomes flanking the rARS, illustrating possible arrangements of nucleosomes reflecting different predominant subspecies of positions for each nucleosome in WT vs. isw2∆ nhp10∆ cells. (D) MNase-seq comparing WT and isw2∆ nhp10∆ cells across three different strengths of MNase digestion.
To refine our analysis, we compared MNase-seq profiles for the tested strains using ribbon plots in which the primary line shows the average signal at each base pair across multiple biological replicates, and the ribbon represents the SEM for those replicates (Figure 3B). This method revealed highly reproducible, strain-specific differences in nucleosome positioning at the rDNA for two pairs of nucleosomes. One pair is in between the 35S promoter and the rARS, with each nucleosome substantially overlapping one of the two subpeaks of the highly prominent Isw2 peak (Figure 3B, left panel, identified as nucleosomes 1 and 2). The other pair of affected nucleosomes is in the region between the rARS and the 5S gene, overlapping half of the short, broad Isw2 peak encompassing the 5S (Figure 3B, right panel, nucleosomes 3 and 4). Each of these four MNase-seq dyad peaks has two or three subspecies of nucleosome positions. We interpret each of these distinct subspecies as representing one of two or three distinct positions occupied by that nucleosome in different individual rDNA repeats in the array. Each of the four genotypes tested had a characteristic pattern of the relative heights of these two subspecies, which we propose reflects different proportions of rDNA arrays containing nucleosomes at each possible position. The overall trend among these mutants is that in isw2∆ nhp10∆ cells, any given rDNA repeat is more likely to have nucleosomes positioned such that they are encroaching on the rARS (Figure 3, B and C). In contrast, in both nhp10∆ and wild-type cells, these same nucleosomes are more likely to be positioned farther away from the rARS, and in isw2∆ cells these nucleosomes have profiles somewhere in between wild type and the double mutant.
It has been shown that the strength of MNase digestion can affect nucleosome mapping results, especially for nucleosomes that are highly MNase sensitive (Weiner et al. 2010). Because the differences in MNase-seq signal at the rDNA locus were more subtle than what is typically observed at single-copy loci, we sought to ensure that these differences are not due to differential MNase sensitivity of these nucleosomes. To this end, we compared the MNase-seq profiles for these nucleosomes in wild-type and isw2∆ nhp10∆ strains using three different concentrations of MNase (Figure 3D and Figure S2B). The overall shapes of the MNase-seq profiles varied depending on the MNase concentrations used. However, at any specific degree of digestion, the relative heights of nucleosomal subspecies for wild-type vs. isw2∆ nhp10∆ cells matched the patterns described above. These results confirmed that the observed differences in nucleosome positions in mutants were not due to differential MNase sensitivity of these nucleosomes.
Isw2 and Ino80 facilitate efficient firing of rDNA origin of replication
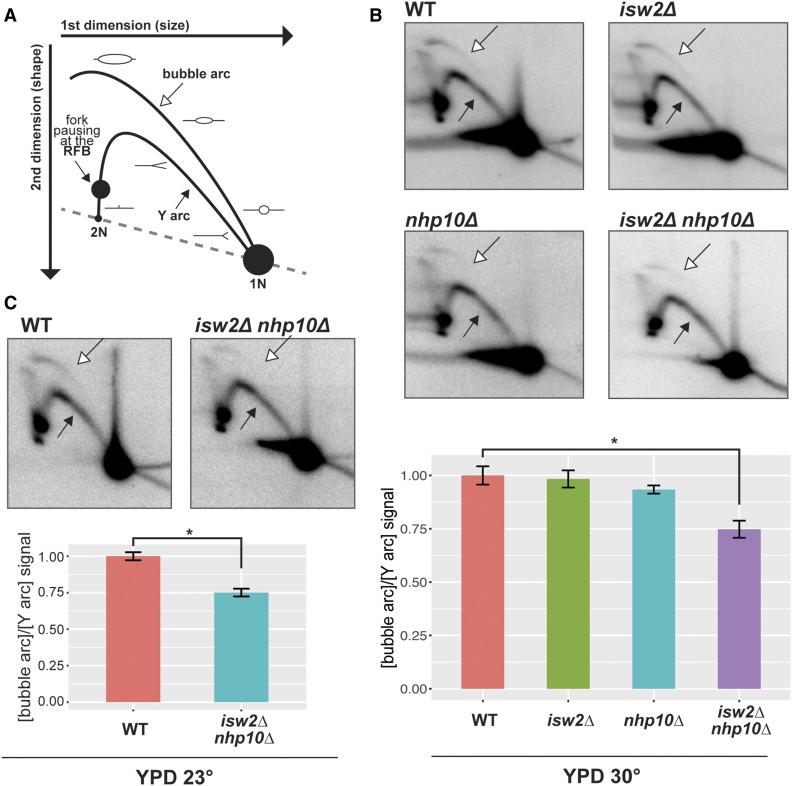
The prominent Isw2 peak around the rARS coupled with the shrinkage of the rARS-containing nucleosome-depleted region (NDR) in chromatin remodeling factor mutants led us to ask whether origin activity is affected by these factors. To address this question, we performed two-dimensional (2D) gel electrophoresis probing activity of the rARS (Brewer and Fangman 1988). The Y arc of the 2D gel is comprised of restriction fragments in the process of being passively replicated, and the bubble arc of restriction fragments in which an origin of replication has actively fired. Therefore, the ratio of bubble to Y arc signals from asynchronously growing cells reflects the ratio of actively to passively replicated restriction fragments, and thus of origin efficiency (Figure 4A). By this method, the ratio of bubble arc to Y arc signal, and thus rARS origin efficiency, was greatest in the wild-type and isw2∆ cells. In contrast, origin efficiency was moderately reduced in nhp10∆ cells, by ∼10%, and even more reduced, by nearly 30%, in isw2∆ nhp10∆ double mutants (Figure 4B, P = 0.04). This reduction in origin efficiency in the double mutant is approximately half the magnitude of the reported increase in rARS efficiency in cells lacking SIR2 (Pasero et al. 2002), the best-characterized chromatin regulator of rARS activity. To test whether Isw2 and Nhp10 also affect rARS firing under a suboptimal growth condition, we performed 2D gels of wild type and double mutant cells at 23°. Consistent with our model, robust reduction in origin efficiency was also detectable in the double mutant under this condition (Figure 4C). Collectively, these results indicate that the Isw2 and Ino80 chromatin remodeling factors promote the efficient firing of the ribosomal origin of replication.
Figure 4.
Isw2 and Ino80 facilitate efficient firing of rDNA origin of replication. (A) Schematic drawing of 2D gel with features annotated. The 1N spot is comprised of restriction fragments that are not in the process of replicating; the Y arc of restriction fragments that are being passively replicated; and the bubble arc of restriction fragments in which an origin of replication has actively fired. Replication fork pausing at the RFB causes an accumulation of restriction fragments with a specific size and shape, visible as a dark spot on the left arm of the Y-arc. The ratio of bubble arc to Y arc signal is indicative of the ratio of actively to passively replicated restriction fragments, and thus of origin efficiency. (B) Representative 2D gels of NheI-digested DNA over rARS and RFB, from cells grown in YPD at 30°. Exposures of the blots have been adjusted so that the Y arc is of comparable intensity for each blot, such that direct comparison of bubble arc intensity across images is equivalent to a comparison of bubble-to-Y ratio. Bubble arc indicated by empty arrow, Y arc indicated by filled arrow. Quantification based on measurement of average intensity of arcs using ImageQuantTL software, and reflects at least two independent experiments for each genotype. All values normalized to the bubble:Y ratio for wild-type. Error bars show SEM. Statistical significance determined by pairwise t-tests followed by Bonferroni correction for multiple testing. * P < 0.05. (C) As in (B), but from cells grown in YPD at 23°. Statistical significance determined by Welch’s t-test. * P < 0.05.
Isw2 and Ino80 affect the rate of rDNA copy number increase
Each yeast strain maintains a particular number of rDNA repeats, typically ∼150–200 repeats. The lower limit on this number is imposed both by the demand for transcription of rRNAs (Warner 1999) and by the benefit to genome stability of having enough rDNA repeats such that not all are transcribed simultaneously (Ide et al. 2010). An upper limit on the number of repeats comes from balancing the above needs against the burden of replicating a lengthy rDNA array in a context of limiting replication factors (Salim et al. 2017). All of the strains used in the previously described experiments maintain approximately the same steady-state rDNA copy number of ∼150 copies (Figure S3), indicating that loss of Isw2 and Nhp10 does not affect the steady-state size of the rDNA array. This, however, does not necessarily mean these remodeling factors play no roles in rDNA copy number change, because a population of cells with optimally sized rDNA arrays are unlikely to visibly alter their size during exponential growth without any perturbation. To determine whether Isw2 and Ino80 affect the process of regulated rDNA copy number change, we employed an established experimental system for this purpose. This approach uses a strain background in which endogenous FOB1 has been deleted and the rDNA array reduced to 20 repeats. In the absence of Fob1, there is no pausing at the RFB, stabilizing rDNA copy number. These cells can survive with 20 copies of the rDNA, but because this is a suboptimal number of repeats, introduction of Fob1 via a plasmid causes a rapid increase in the number of rDNA repeats via homologous recombination until the rDNA array reaches a more optimal size of ∼150 copies (Kobayashi and Ganley 2005).
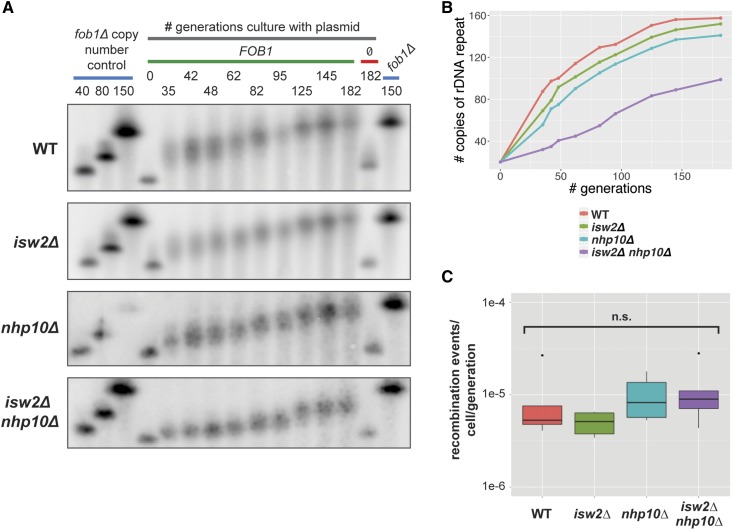
Starting with a fob1∆ strain with 20 copies of the rDNA, ISW2, NHP10, or both genes were deleted. FOB1 was then reintroduced on a plasmid, and the cells were cultured continuously under selection for almost 200 generations. Samples were taken at multiple time points, synchronized such that time points were taken for all tested strains at the same number of generations after reintroduction of Fob1, thus controlling for any small differences in growth rate between the strains. The copy number of rDNA repeats was monitored by CHEF gel electrophoresis followed by Southern blot analysis using a probe against Chromosome XII. This experiment thus reveals the regulated process of rDNA copy number increase by the cell, distinct from the small, likely stochastic fluctuations around the optimal copy number for any given strain.
Although all four strains began to increase their rDNA copy number immediately following introduction of plasmid-borne Fob1, each of the strains behaved differently (Figure 5, A and B). In wild-type and isw2∆ cells, and to a slightly lesser degree in nhp10∆ cells, there was a strong jump in copy number at ∼35 generations after Fob1 reintroduction, the earliest time point we were able to sample. In contrast, isw2∆ nhp10∆ cells exhibited only a very small increase in copy number at 35 generations. After nearly 200 generations in the presence of Fob1, both the wild-type and isw2∆ strains had recovered essentially wild-type rDNA copy number of ∼150 copies, and nhp10∆ was close to this number. In contrast, isw2∆ nhp10∆ had barely reached 100 copies by this time point. Based on this data, we conclude that Isw2 and Ino80 facilitate the regulated increase of rDNA copy number in the rDNA array, and that their loss reduces the rate at which rDNA copy number can be increased in a population of cells.
Figure 5.
Isw2 and Ino80 affect the rate of rDNA copy number change. (A) rDNA copy number change assay. Blue bars indicate fob1∆ copy number control strains that maintain the indicated number of rDNA repeats (identical 150-copy control samples run on both ends of the gel to facilitate comparison of band migration). The gray bar indicates samples grown in a time course for the indicated number of generations, in selective medium to ensure retention of either a plasmid containing FOB1 (green bar) or the plasmid backbone pRS426 without FOB1 (red bar). (B) Quantification of the copy number change assay. Average copy number at each time point was calculated based on migration of bands relative to controls. (C) URA3 recombination assay. Cells were grown under selection, then without selection for ∼10 generations, then plated on 5-FOA plates. Number of recombination events per cell per generation determined based on number of 5-FOA-resistant colonies. Statistical significance tested by pairwise t-tests followed by Bonferroni correction for multiple testing. n.s., not significant.
Given the established role for Ino80 in the DNA damage response, we wondered whether the reduced rate of rDNA copy number increase in the isw2∆ nhp10∆ cells was unique to the rDNA locus or the result of a more general defect in recombination. To address this question, we measured the recombination efficiency of remodeling factor mutants at a locus outside of the rDNA. We first integrated the URA3 gene into WT, isw2∆, nhp10∆, and isw2∆ nhp10∆ strains with the ura3-1 allele. These strains were cultured first under selection to ensure a pure population of URA3+ cells, then switched to growth without selection, during which time the URA3 gene would recombine with the mutant ura3-1 allele at some low frequency, rendering the cell able to survive in the presence of 5-FOA. The cells were then plated on 5-FOA medium, and the number of 5-FOA-resistant cells, the product of these recombination events, was calculated. Based on this assay, none of the tested strains significantly differed in their baseline rates of recombination (Figure 5C). We therefore concluded that the reduced rate of rDNA copy number increase observed in isw2∆ nhp10∆ cells resulted not from a general defect in recombination, but rather from the loss of a unique function of these chromatin remodeling factors in promoting rDNA copy number change.
Discussion
The ribosomal DNA locus is the evolutionarily conserved site of many different DNA-dependent processes, all of which must be carefully balanced. Sufficient rRNA must be transcribed to support ribosome biogenesis, but without interfering with faithful replication of the rDNA (Warner 1999). The rDNA array must be fully replicated, while still allowing for the replication of other parts of the genome (Yoshida et al. 2014). The size of the rDNA array must be carefully maintained through recombination, yet the array must be protected from unintended recombination despite its highly repetitive nature. Many studies have detailed these complex processes, but relatively little is known about how ATP-dependent chromatin remodeling factors dynamically regulate chromatin structure at the S. cerevisiae rDNA locus to allow for these processes to occur. It has been shown that the SWI/SNF complex localizes to the rDNA and that deletion of its Snf6 subunit reduces 35S rRNA transcription (Zhang et al. 2013). In addition, it was shown that Isw2, Isw1, and Chd1 are present at the rDNA, and that their simultaneous deletion reduces 35S rRNA transcriptional termination (Jones et al. 2007). However, the nature of chromatin regulation by these remodeling factors at the rDNA locus has remained unknown, as has their involvement in processes beyond transcription of rRNA. In this study, we show that in addition to Isw2, the Ino80 ATP-dependent chromatin remodeling factor is targeted to the yeast rDNA. We show for the first time that these factors modify local chromatin structure at the levels of nucleosome occupancy, the ratio of nucleosome-occupied to nucleosome-depleted rDNA repeats, and nucleosome positioning. In addition, we find that these chromatin remodeling factors affect two critical activities that take place at the rDNA, replication initiation from the ribosomal ARS, and rDNA array amplification.
Our data indicate that Isw2 and Ino80 do not affect overall levels of 35S rRNA transcription, a result that initially surprised us. According to one part of the accepted model, nucleosome occupancy through the 35S gene body dictates 35S transcription, as rDNA repeats that are heavily occupied with nucleosomes are transcriptionally silent, while repeats that are depleted of nucleosomes are transcriptionally active. Thus, based on the increased nucleosome occupancy and reduced proportion of psoralen-accessible rDNA repeats observed in isw2∆ nhp10∆ cells, we expected that 35S rRNA transcription would be correspondingly decreased. The lack of an effect on transcription may be explained by the robustness of 35S transcriptional regulation: when one element of this system is disrupted, another element is adjusted to maintain the desired level of transcription. For example, in a S. cerevisiae strain in which the rDNA array has been reduced from a normal size of ∼150 copies down to ∼40 copies, loading of RNA Pol I on any given active repeat is increased, such that there is no net decrease in 35S transcriptional output (French et al. 2003). Similarly, in mammalian cells, inducing silencing of some rDNA repeats by depletion of upstream binding factor (UBF) leads to a compensatory increase in transcription per active repeat (Sanij et al. 2008). We therefore speculate that the robust homeostatic regulation of rRNA transcription overcomes changes in nucleosome occupancy in isw2∆ nhp10∆ cells, reacting to a reduced proportion of active repeats by increasing RNA Pol I transcription in each active unit. This would produce no net alteration in rRNA production compared to wild-type cells.
A critical transcriptional regulator at the mammalian rDNA is the nucleolar remodeling complex (NoRC), which contains SNF2h, the mammalian ortholog of yeast Isw2. Among other activities that influence rRNA transcription, this complex shifts the nucleosome at the promoter of the 45S rRNA gene, the mammalian ortholog of the yeast 35S, into a transcriptionally repressive position (Li et al. 2006). Notably, we see nearly identical nucleosome positioning profiles at the comparable nucleosome in isw2∆ and isw2∆ nhp10∆ cells compared to wild-type cells (Figure S2C). This finding, in conjunction with our observing no differences in rRNA transcription in these deletion strains, distinguishes the Isw2-mediated regulation of the yeast rDNA from the NoRC-mediated regulation of the mammalian rDNA.
While we find that loss of Isw2 and Ino80 does not affect net rRNA transcription, we do find that their loss reduces the activity of the rARS. There are multiple reports that chromatin structure around replication origins significantly affects DNA replication. Blocking an ARS with a nucleosome reduces the efficiency of that ARS (Simpson 1990), and proper positioning of nucleosomes adjacent to an ARS is important for replication initiation (Lipford and Bell 2001). Compared to naked DNA, chromatinized DNA facilitates much greater origin selectivity at the stage of origin licensing, suggesting that chromatin structure regulates which origins fire during S-phase (Kurat et al. 2017). Consistent with these findings, ATP-dependent chromatin remodeling factors contribute to regulating replication initiation. For example, the SWI/SNF complex is targeted to a subset of origins in HeLa cells (Euskirchen et al. 2011) and facilitates replication initiation at one out of four natural ARSs tested in a mini-chromosome maintenance assay in S. cerevisiae (Flanagan and Peterson 1999). By applying an in vitro replication assay to nucleosomal templates remodeled by different chromatin remodeling factors, a recent study found that most factors permitted origin licensing, but that Isw2 and Chd1 prevented it (Azmi et al. 2017). As far as we know, however, there have been no reports of how chromatin remodeling factors change chromatin structure to affect activity of replication origins at their native loci. Our work therefore established the first example in which chromatin remodeling factors affect both chromatin structure and replication initiation at a specific origin of replication at its natural genomic locus in vivo.
We report that loss of ISW2 and NHP10, individually and together, reduced the efficiency of the rARS during logarithmic growth conditions in rich medium. We found that isw2∆ nhp10∆ cells have the most robust differences in nucleosome positioning compared to wild-type cells, with a clear trend of an enrichment for nucleosomes in positions that encroach on the rARS. These same cells have the most reduced efficiency at this ARS compared to wild type. This effect is opposite that of Isw2 at Pol II-transcribed genes. At such genes, when ISW2 is deleted, NDRs at the end of the gene targeted by Isw2 tend to widen, and nearby coding and noncoding transcription increases, suggesting that this remodeling factor typically functions to narrow these NDRs and repress transcription (Whitehouse et al. 2007). Our data suggest that the NDR containing the rARS overall becomes narrower and origin efficiency goes down in isw2∆ nhp10∆ cells, suggesting a normal function of these factors in keeping this NDR wide and thus permissive to replication initiation. Given that both Isw2 and Nhp10 are present at the rARS and alter positions of nucleosomes around rARS, it is plausible that they play direct roles in rARS activity. In addition, reduced rARS efficiency in our mutants may also be partially due to the altered ratio of transcriptionally active to inactive rDNA repeats. It has been shown that rARSs are more likely to fire when they are adjacent to actively transcribed rDNA repeats (Muller et al. 2000). The proportion of actively transcribed repeats is reduced in isw2∆ nhp10∆ cells, and thus a reduced proportion of rARSs in the array are adjacent to actively transcribed repeats, possibly contributing to the reduced origin efficiency we observe in these mutants. We also note that, despite this significant reduction in rARS efficiency, isw2∆ nhp10∆ cells do not have an obvious growth defect in rich medium at 30°. As described earlier, sir2∆ strains have been shown to have an even greater magnitude change in rARS activity and are similarly healthy under such conditions. These results collectively suggest the robustness of replication control mechanisms at the rDNA locus under unstressed conditions.
In addition to regulating rARS activity, a cell must carefully calibrate the size of the rDNA array. This highly repetitive locus must be large enough to allow for the transcription of sufficient ribosomal RNA to satisfy a cell’s demand for ribosomes. In a typical yeast cell, ∼75 copies of the rDNA are actively transcribed to satisfy this demand (Warner 1999). However, those 75 copies of the rDNA repeat must be insufficient under some circumstances, as a typical yeast rDNA array contains ∼150 copies of the rDNA repeat. These additional copies are believed to be necessary to maximize genome stability. Active ribosomal RNA genes are transcribed at extremely high levels, with densely loaded transcriptional machinery. This presents an obstacle to the repair of damage to the underlying DNA, and persistent, unrepaired damage to the rDNA array delays complete replication of the genome and progression through S-phase (Ide et al. 2010). Thus, to maximize genome stability, the rDNA array must be large enough to support sufficient rRNA transcription without requiring all repeats to be actively transcribed. This requirement imposes a lower limit on the optimal size of the rDNA array. Similarly, the array cannot exceed a certain size. If the rDNA grows too large, its complete replication would require an excessively large proportion of the finite pool of replisome components available during each S-phase, depriving other parts of the genome of those replication factors (Yoshida et al. 2014). In addition, having a smaller rDNA array improves growth during persistent replication stress, perhaps by making more of the limiting replication factors available to other parts of the genome (Salim et al. 2017). Thus, the number of repeats in the rDNA locus must be actively managed by the cell to facilitate optimal transcriptional output and maximize genome stability. During unperturbed growth, changes in rDNA copy number are subtle and take place in only a small fraction of cells within the population, making it difficult to detect these changes or to investigate their underlying mechanisms. As a consequence, most of our knowledge about the mechanism of rDNA copy number change comes from studying the cellular response to a significant perturbation in copy number, a situation in which copy number change is readily detectable. For example, if an rDNA array is artificially truncated, it will steadily increase until it reaches a normal size (Kobayashi et al. 1998). Conversely, the rDNA array will shrink when the RPA135 subunit of RNA Pol I is deleted (Brewer et al. 1992; Kobayashi et al. 1998), when the activity of the origin recognition complex is compromised (Sanchez et al. 2017), or when a number of other replication factors are lost (Salim et al. 2017). Together, these studies demonstrate that maintenance of the size of the rDNA is a vital process that is actively regulated by the cell.
In this study, we describe a nearly twofold reduction in the rate of copy number increase in isw2∆ nhp10∆ cells relative to wild-type cells and moderate reductions in the rate of increase in isw2∆ and nhp10∆ cells. A critical step in the process of rDNA copy number change is the repair of the targeted DNA double-strand break (DSB) that takes place at RFB-paused replication forks. Although Ino80 plays roles in DSB repair (Papamichos-Chronakis et al. 2006, 2011; Lademann et al. 2017), the mutants we used did not exhibit a general recombination defect. This argues against the possibility that isw2∆ and nhp10∆ mutations affect rDNA array expansion indirectly through recombination itself or transcription of factors involved in recombination and demonstrates that Isw2 and Ino80 play special roles at the rDNA locus. It is possible that the reduced proportion of transcriptionally active rDNA repeats in isw2∆ nhp10∆ cells indirectly affects the rate of rDNA expansion. It has been shown that copy number change events require firing of the rARS adjacent to the RFB at which a replication fork is paused, a DSB is induced, and then that DSB repaired. Consistent with this notion, the efficiency of the ARS in the IGS correlates with the rate of copy number increase (Ganley et al. 2009). Accordingly, it is possible that the rARS efficiency decreases at least partly due to the reduced ratio of active to inactive repeats in the double mutant (see above), which in turn reduces the frequency of copy number change events, thus accounting for the reduced rate of copy number increase in the double mutant cells. Despite the strong reduction in the rate of rDNA copy number expansion, isw2∆ nhp10∆ cells have normal-sized rDNA arrays. We suspect this is likely due to the fact that fluctuation in rDNA copy number during exponential growth is small, making detection of defects in copy number change difficult. However, we cannot exclude the possibility that rDNA copy number maintenance and expansion have currently unknown mechanistic differences. Though the complexities of this highly repetitive locus create challenges in demonstrating direct, causal mechanisms, this work establishes a novel role for ATP-dependent chromatin remodeling factors in strongly influencing multiple aspects of rDNA biology, including the process of rDNA copy number change.
Acknowledgments
We are grateful to members of the Brewer/Raghuraman laboratory, especially Bonny Brewer, M. K. Raghuraman, Joe Sanchez, and Liz Kwan, for help with experimental design and troubleshooting, as well as conceptual discussion and feedback, and members of the Smith laboratory, especially Randy Hyppa and Andrew Taylor, for help with CHEF gels. We thank Takehiko Kobayashi for strains used in this study. Finally, we thank members of the Tsukiyama laboratory for helpful discussion and commentary on this manuscript. This work is supported by National Science Foundation Fellowship DGE-1256082 to S.C. and grant R01GM058465 to T.T.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7243292.
Communicating editor: J. Surtees
Literature Cited
- Au T. J., Rodriguez J., Vincent J. A., Tsukiyama T., 2011. ATP-dependent chromatin remodeling factors tune S phase checkpoint activity. Mol. Cell. Biol. 31: 4454–4463. 10.1128/MCB.05931-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I. F., Watanabe S., Maloney M. F., Kang S., Belsky J. A., et al. , 2017. Nucleosomes influence multiple steps during replication initiation. eLife 6: pii: e22512 10.7554/eLife.22512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckouet F., Labarre-Mariotte S., Albert B., Imazawa Y., Werner M., et al. , 2008. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol. Cell. Biol. 28: 1596–1605. 10.1128/MCB.01464-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L., 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643. 10.1016/0092-8674(88)90222-X [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Lockshon D., Fangman W. L., 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71: 267–276. 10.1016/0092-8674(92)90355-G [DOI] [PubMed] [Google Scholar]
- Bywater M. J., Poortinga G., Sanij E., Hein N., Peck A., et al. , 2012. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22: 51–65. 10.1016/j.ccr.2012.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Widmer R. M., Koller T., Sogo J. M., 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57: 753–761. 10.1016/0092-8674(89)90790-3 [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J. M., 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21: 2331–2338. 10.1093/nar/21.10.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G. M., Auerbach R. K., Davidov E., Gianoulis T. A., Zhong G., et al. , 2011. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7: e1002008 10.1371/journal.pgen.1002008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio T. G., Tsukiyama T., 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12: 1333–1340. 10.1016/S1097-2765(03)00436-2 [DOI] [PubMed] [Google Scholar]
- Fazzio T. G., Kooperberg C., Goldmark J. P., Neal C., Basom R., et al. , 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21: 6450–6460. 10.1128/MCB.21.19.6450-6460.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J. F., Peterson C. L., 1999. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res. 27: 2022–2028. 10.1093/nar/27.9.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S. L., Osheim Y. N., Cioci F., Nomura M., Beyer A. L., 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23: 1558–1568. 10.1128/MCB.23.5.1558-1568.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze C. E., Verschueren K., Strich R., Easton Esposito R., 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16: 6495–6509. 10.1093/emboj/16.21.6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley A. R., Ide S., Saka K., Kobayashi T., 2009. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol. Cell 35: 683–693. 10.1016/j.molcel.2009.07.012 [DOI] [PubMed] [Google Scholar]
- Gelbart M. E., Bachman N., Delrow J., Boeke J. D., Tsukiyama T., 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19: 942–954. 10.1101/gad.1298905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T., 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–433. 10.1016/S0092-8674(00)00134-3 [DOI] [PubMed] [Google Scholar]
- Ide S., Watanabe K., Watanabe H., Shirahige K., Kobayashi T., et al. , 2007. Abnormality in initiation program of DNA replication is monitored by the highly repetitive rRNA gene array on chromosome XII in budding yeast. Mol Cell Biol 27: 568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S., Miyazaki T., Maki H., Kobayashi T., 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696. 10.1126/science.1179044 [DOI] [PubMed] [Google Scholar]
- Jones H. S., Kawauchi J., Braglia P., Alen C. M., Kent N. A., et al. , 2007. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 14: 123–130. 10.1038/nsmb1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Ganley A. R., 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584. 10.1126/science.1116102 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck D. J., Nomura M., Horiuchi T., 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12: 3821–3830. 10.1101/gad.12.24.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C. F., Yeeles J. T. P., Patel H., Early A., Diffley J. F. X., 2017. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 65: 117–130. 10.1016/j.molcel.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan E. X., Wang X. S., Amemiya H. M., Brewer B. J., Raghuraman M. K., 2016. rDNA copy number variants are frequent passenger mutations in Saccharomyces cerevisiae deletion collections and de novo transformants. G3 (Bethesda) 6: 2829–2838. 10.1534/g3.116.030296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lademann C. A., Renkawitz J., Pfander B., Jentsch S., 2017. The INO80 complex removes H2A.Z to promote presynaptic filament formation during homologous recombination. Cell Rep. 19: 1294–1303. 10.1016/j.celrep.2017.04.051 [DOI] [PubMed] [Google Scholar]
- Laribee R. N., Hosni-Ahmed A., Workman J. J., Chen H., 2015. Ccr4-not regulates RNA polymerase I transcription and couples nutrient signaling to the control of ribosomal RNA biogenesis. PLoS Genet. 11: e1005113 10.1371/journal.pgen.1005113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Rodriguez J., Tsukiyama T., 2015. Chromatin remodeling factors Isw2 and Ino80 regulate checkpoint activity and chromatin structure in S phase. Genetics 199: 1077–1091. 10.1534/genetics.115.174730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Langst G., Grummt I., 2006. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 25: 5735–5741. 10.1038/sj.emboj.7601454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J. R., Bell S. P., 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7: 21–30. 10.1016/S1097-2765(01)00151-4 [DOI] [PubMed] [Google Scholar]
- Mantiero D., Mackenzie A., Donaldson A., Zegerman P., 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 30: 4805–4814. 10.1038/emboj.2011.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz K., Hondele M., Goetze H., Gmelch K., Stoeckl U., et al. , 2008. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 22: 1190–1204. 10.1101/gad.466908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A. J., Highland J., Krogan N. J., Arbel-Eden A., Greenblatt J. F., et al. , 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775. 10.1016/j.cell.2004.11.037 [DOI] [PubMed] [Google Scholar]
- Morrison A. J., Kim J. A., Person M. D., Highland J., Xiao J., et al. , 2007. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130: 499–511. 10.1016/j.cell.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Muller M., Lucchini R., Sogo J. M., 2000. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol. Cell 5: 767–777. 10.1016/S1097-2765(00)80317-2 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Krebs J. E., Peterson C. L., 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20: 2437–2449. 10.1101/gad.1440206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Watanabe S., Rando O. J., Peterson C. L., 2011. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144: 200–213. 10.1016/j.cell.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P., Bensimon A., Schwob E., 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 16: 2479–2484. 10.1101/gad.232902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., McKnight J. N., Tsukiyama T., 2014. Genome-wide analysis of nucleosome positions, occupancy, and accessibility in yeast: nucleosome mapping, high-resolution histone ChIP, and NCAM. Curr. Protoc. Mol. Biol. 108: 21.28.1–21.28.16. 10.1002/0471142727.mb2128s108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim D., Bradford W. D., Freeland A., Cady G., Wang J., et al. , 2017. DNA replication stress restricts ribosomal DNA copy number. PLoS Genet. 13: e1007006 10.1371/journal.pgen.1007006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J. C., Kwan E. X., Pohl T. J., Amemiya H. M., Raghuraman M. K., et al. , 2017. Defective replication initiation results in locus specific chromosome breakage and a ribosomal RNA deficiency in yeast. PLoS Genet. 13: e1007041 10.1371/journal.pgen.1007041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier J. J., French S., Osheim Y., Cheung W. L., Gallo C. M., et al. , 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21: 4959–4968. 10.1093/emboj/cdf498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T. P., et al. , 2008. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 183: 1259–1274. 10.1083/jcb.200805146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R., Li J., Grummt I., 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32: 393–396. 10.1038/ng1010 [DOI] [PubMed] [Google Scholar]
- Shimada K., Oma Y., Schleker T., Kugou K., Ohta K., et al. , 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 18: 566–575. 10.1016/j.cub.2008.03.049 [DOI] [PubMed] [Google Scholar]
- Simpson R. T., 1990. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature 343: 387–389. 10.1038/343387a0 [DOI] [PubMed] [Google Scholar]
- Smith J. S., Boeke J. D., 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11: 241–254. 10.1101/gad.11.2.241 [DOI] [PubMed] [Google Scholar]
- Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A., 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438: 379–383. 10.1038/nature04148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama M., Sabri A., Bartholomew B., 2011. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 31: 662–673. 10.1128/MCB.01035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. A., Kwong T. J., Tsukiyama T., 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 15: 477–484. 10.1038/nsmb.1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley R. M., Johnston L. H., Williamson D. H., Oliver S. G., 1984. Replicon size of yeast ribosomal DNA. Mol. Gen. Genet. 195: 260–266. 10.1007/BF00332757 [DOI] [PubMed] [Google Scholar]
- Warner J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- Weiner A., Hughes A., Yassour M., Rando O. J., Friedman N., 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20: 90–100. 10.1101/gr.098509.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I., Rando O. J., Delrow J., Tsukiyama T., 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035. 10.1038/nature06391 [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Baserga S. J., 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Bacal J., Desmarais D., Padioleau I., Tsaponina O., et al. , 2014. The histone deacetylases sir2 and rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol. Cell 54: 691–697. 10.1016/j.molcel.2014.04.032 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Anderson S. J., French S. L., Sikes M. L., Viktorovskaya O. V., et al. , 2013. The SWI/SNF chromatin remodeling complex influences transcription by RNA polymerase I in Saccharomyces cerevisiae. PLoS One 8: e56793 10.1371/journal.pone.0056793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. Y., Johnson S. L., Lee L. J., Longhurst A. D., Beckwith S. L., et al. , 2018. The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol. Cell 69: 677–688.e9. 10.1016/j.molcel.2018.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains (Table S1) and plasmids (Table S3) used in this study are available upon request, and oligonucleotide sequences are included in Table S2. All genomics data are available at GEO under the accession number GSE112465. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, supplemental material, and genomics data. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7243292.