Abstract
To identify novel disease genes for type 2 diabetes (T2D) we generated two backcross populations of obese and diabetes-susceptible New Zealand Obese (NZO/HI) mice with the two lean mouse strains 129P2/OlaHsd and C3HeB/FeJ. Subsequent whole-genome linkage scans revealed 30 novel quantitative trait loci (QTL) for T2D-associated traits. The strongest association with blood glucose [12 cM, logarithm of the odds (LOD) 13.3] and plasma insulin (17 cM, LOD 4.8) was detected on proximal chromosome 7 (designated Nbg7p, NZO blood glucose on proximal chromosome 7) exclusively in the NZOxC3H crossbreeding, suggesting that the causal gene is contributed by the C3H genome. Introgression of the critical C3H fragment into the genetic NZO background by generating recombinant congenic strains and metabolic phenotyping validated the phenotype. For the detection of candidate genes in the critical region (30–46 Mb), we used a combined approach of haplotype and gene expression analysis to search for C3H-specific gene variants in the pancreatic islets, which appeared to be the most likely target tissue for the QTL. Two genes, Atp4a and Pop4, fulfilled the criteria from our candidate gene approaches. The knockdown of both genes in MIN6 cells led to decreased glucose-stimulated insulin secretion, indicating a regulatory role of both genes in insulin secretion, thereby possibly contributing to the phenotype linked to Nbg7p. In conclusion, our combined- and comparative-cross analysis approach has successfully led to the identification of two novel diabetes susceptibility candidate genes, and thus has been proven to be a valuable tool for the discovery of novel disease genes.
Keywords: diabetes, quantitative trait loci, positional cloning, candidate disease genes, haplotypes
TYPE 2 diabetes (T2D) is a complex metabolic disease affecting nearly half a billion people worldwide (Cho et al. 2018). It is well established that both genetic factors and lifestyle contribute to the pathophysiology of the disease (Permutt et al. 2005; Das and Elbein 2006). Genome-wide association studies and familial linkage analyses in humans have led to the identification of > 100 gene variants potentially associated with the pathogenesis of T2D (Dorajoo et al. 2015; Fuchsberger et al. 2016). However, the known genetic variants that have been identified so far only account for a relatively small fraction of the interindividual variability in diabetes-related traits in humans, indicating that a considerable proportion of the underlying genetic variants still remains to be identified (Morris et al. 2012; Schwenk et al. 2013; Tsaih et al. 2014).
Mouse models have proven to be an essential experimental tool for the identification of novel disease genes and signaling pathways for human diseases (Attie et al. 2017; Kleinert et al. 2018). Inbred strains are available that differ in their prevalence toward obesity and T2D, thereby providing genetic diversity with respect to diabetes risk. In contrast to human studies, mice can be used for the introduction of targeted mutations or naturally occurring risk alleles with the help of well-established genetic tools, and tissues can be collected for the analysis of gene expression or functional assays (Attie et al. 2017; Kleinert et al. 2018). Moreover, the ability to control the environment and short generation times in combination with large litter sizes represent further advantages that emphasize the utility of mouse models over human studies.
The New Zealand Obese (NZO) mouse strain presents features of the metabolic syndrome, including early onset hyperglycemia, hyperinsulinemia, hypercholesterinemia, hyperlipidemia, and hypertension, in response to high-fat diet (HFD) consumption (Jürgens et al. 2006, 2007; Kluth et al. 2011; Joost and Schürmann 2014). In the course of the disease, ∼70% of the mice progress into islet cell failure and develop overt diabetes (Jürgens et al. 2006). Consequently, NZO mice have been utilized in several studies as a polygenic model for obesity-driven human T2D (Joost and Schürmann 2014).
Previous linkage analyses with outbred populations derived from obese, diabetes-prone NZO and lean, diabetes-resistant mice, such as C57BL/6J, NON, and SJL, have contributed substantially to the understanding of the genetic architecture of T2D, and the underlying gene–diet interactions that determine the onset and progression of the disease. Several genomic regions, designated quantitative trait loci (QTL), that show linkage to disease-related phenotypes have been identified and, in some cases, subsequent positional cloning has led to the identification of causal, strain-specific gene variants including Tbc1d1, Zfp69, and Ifi202b (Chadt et al. 2008; Scherneck et al. 2009; Vogel et al. 2012). Analysis of the risk alleles, and their interaction with genetic and environmental factors in experimental mouse breeding studies, provides a relevant picture of the genetic architecture of T2D and related traits. Importantly, clinical and experimental studies have provided evidence that genes identified from mouse studies are also linked to the onset and progression of obesity, insulin resistance, and diabetes in humans (Dash et al. 2009; Scherneck et al. 2009; Vogel et al. 2012).
Despite successful positional cloning of diabetes risk genes in rodents, many causal gene variants for the majority of diabetes-related QTL are unknown and the complexity of T2D genetics is not well understood. However, current genome and phenome databases, as well as modern bioinformatics resources, may be used for data mining and subsequent reduction of the number of candidate genes identified by linkage scans (Keane et al. 2011; Yalcin et al. 2011).
To identify novel risk loci for obesity and T2D, we conducted crossbreeding experiments using the obese and diabetes-susceptible NZO, and two different lean mouse strains. Genome-wide linkage analysis and subsequent QTL mapping revealed two novel diabetes-related gene variants that might be involved in the regulation of insulin secretion in pancreatic β-cells.
Materials and Methods
Animals and breeding strategy
All experiments were in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals, and were approved by the Ethics Committee (references: 84-02.04.2013.A118 and 84-02.04.2015.A354) of the State Ministry of Agriculture, Nutrition and Forestry (State of North Rhine-Westphalia, Germany). Diabetes-prone NZO/Hl [NZO; German Diabetes Centre Duesseldorf (Herberg and Coleman 1977)], and diabetes-resistant 129P2/OlaHsd (129P2; German Institute of Human Nutrition, Nuthetal, Germany) and C3HeB/FeJ [C3H; Helmholtz Center Munich, Germany (Gailus-Durner et al. 2005)] mice were housed at three to six mice per cage (Macrolon type III) at a constant temperature of 22° and a 12-hr light–dark cycle (lights on at 6 am). Animals had free access to food and water ad libitum. Female NZO, and male 129P2 or C3H mice, respectively, were used to generate a F1 generation (NZOx129P2; NZOxC3H), and male F1 offspring were subsequently backcrossed with NZO females (N2: NZOxF1). For each backcross generation, N2(NZOx F1(NZOxC3H)) and N2(NZOx F1(NZOx129P2)), ∼300 males and 300 females were generated, designated N2(NZOxC3H) and N2(NZOx129P2). For the introgression of heterozygous C3H alleles on chromosome 7 on the NZO background, repeated backcrosses were performed until the generation of an N5/N6 generation. Subsequently, intercrosses (brother–sister breeding) were conducted to produce homozygous C3H alleles on chromosome 7. Homozygous N5F1 and N6F1 mice with the same genotype were intercrossed to obtain 100% homozygous mice in the N5F2/N6F2 generation, which were metabolically phenotyped in this study. After weaning at the age of 21 days, all experimental animals received a HFD containing 45 kcal% fat, 20 kcal% protein, and 35 kcal% carbohydrates with 4.73 kcal/gm energy (D12451; Research Diets, New Brunswick, NJ). All backcross (week 21 of age) and recombinant congenic strains (RCSs) (week 17 of age) mice were fasted for 6 hr before they were killed by cardiac puncture under isoflurane anesthesia. For the collection of the pancreatic islets, parental and RCS mice were killed at the age of 6 weeks by cervical dislocation. Due to severe β-cell loss with the progression of the disease in parental NZO and NZO allele carriers for chromosome 7, we could not collect sufficient islets from older animals.
Genotyping
Genomic DNA was isolated from mouse tail tips using the InViSorb Genomic DNA Kit II (Invitek, Berlin, Germany). The genotyping was performed by Kompetitive Allele-Specific PCR using appropriate SNP assays (LGC Genomics, Teddington, UK). Informative SNP markers [118 for N2(NZOx129P2) and 115 for N2(NZOxC3H)] polymorphic between NZO and 129P2 or C3H, respectively, were selected at a distance of 20 Mbp for each chromosome.
Body weight and body composition
Body weight was determined weekly with an electronic scale, and body composition was measured at weeks 3, 6, 10, and 15 by noninvasive nuclear magnetic resonance spectroscopy (EchoMRI-100 System; Echo Medical Systems, Houston, TX).
Blood glucose levels, determination of T2D prevalence, and survival rate
Blood glucose was measured weekly in the morning between 8 and 10 am using a CONTOUR XT glucometer (Bayer Consumer Care AG, Leverkusen, Germany). T2D prevalence was calculated by determining the cumulative number of diabetic animals (N2: blood glucose > 300 mg/dl for 3 consecutive weeks) and expressing the percentage of affected mice in relation to the total number of animals. The survival rate (living animals/total animals) was calculated accordingly.
Analysis of plasma insulin and C-peptide
Insulin and C-peptide levels were measured in plasma samples (N2: week 21 of age and RCS: week 17 of age) by ELISA (Insulin: Mouse Ultrasensitive ELISA Kit; DRG, Marburg, Germany; C-Peptide: Mouse C-Peptide ELISA Kit; CrystalChem, Chicago, IL) according to the manufacturer’s recommendations.
Pancreatic islet isolation
At the age of 6 weeks, experimental mice were killed by cervical dislocation and the pancreatic islets were isolated by ductal collagenase perfusion of the pancreas, as previously described (Yesil et al. 2009). Subsequently, the islets were cultivated overnight in CMRL (Connaught Medical Research Laboratories) islet medium (see cell culture) at 37° with 5% CO2.
RNA extraction and microarray analysis
Total RNA from pancreatic islets (collected from 6-week-old animals) and MIN6 cells was isolated using the RNeasy mini kit (QIAGEN, Valencia, CA) including DNase digestion, according to the manufacturer’s instructions. For microarray analysis of the pancreatic islets, the quality of the isolated RNA was tested using an RNA 6000 nano kit (Agilent Technologies, Taufkirchen, Germany). Only samples with RNA integrity number (RIN) values > 8 were selected for the subsequent microarray analysis. Genome-wide expression analyses (n = 5 per genotype) were performed with 150 ng RNA according to the Ambion WT Expression Kit and the WT Terminal Labeling Kit (Thermo Fisher Scientific, Darmstadt, Germany). All protocol steps were monitored using an RNA 6000 nano kit (Agilent). Mouse Gene 1.0 ST arrays were hybridized with labeled complementary RNA samples. Data were collected with the GeneChip scanner 3000 7G and analyses of primary data were performed with the GDAS 1.4 package, [Affymetrix, (Thermo Fisher Scientific)]. Data were analyzed with Expression ConsoleTM v1.1 and Transcriptome Analysis ConsoleTMv2.0 (Affymetrix) as previously described (Knebel et al. 2015).
cDNA synthesis and quantitative real-time PCR
cDNA was synthesized using the GoScript Reverse Transcriptase Kit (Promega, Madison, WI) using 500 ng RNA. For quantitative real-time (qRT)-PCR, the GoTaq qPCR Master Mix (Promega) on a QuantStudio 7 Flex PCR System (Applied Biosystems, Foster City, CA) was used. For the three genes Ffar2, Ffar3, and Zfp719, TaqMan probes (Thermo Fisher Scientific) were used. TATA box binding protein (Tbp) for pancreatic islets and β-actin (Actb) for MIN6 cells was used as an endogenous control, and gene expression was quantified using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Cell culture
Isolated pancreatic islets were cultivated in CMRL medium (Thermo Fisher Scientific) containing 15% fetal calf serum, 0.05 mM β-mercaptoethanol (Thermo Fisher Scientific), 1% penicillin/streptomycin, 10 mM glucose, and 39.2 mM NaHCO3. MIN6 cells (a gift from S. Baltrusch from the University of Rostock, Germany) were cultured in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific) with 25 mM glucose, supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. Both cell types were constantly cultivated at 37° with 5% CO2.
Electroporation of MIN6 cells
For the knockdown of Atp4a and Pop4, MIN6 cells were electroporated with small interfering RNA (siRNA) oligonucleotides using the SF Cell Line 4D-Nucleofector kit (Lonza, Cologne, Germany) in combination with the 4D-Nucleofector system (Lonza). Cells were electroporated according the Amaxa 4D-Nucleofector protocol for SH-SY5Y cells provided from Lonza (bio.lonza.com/go/op/290). Two million cells from each cuvette were split into six wells of a 12-well plate; three of the wells were used for glucose-stimulated insulin secretion (GSIS) (technical triplicates) and the other three for RNA isolation to confirm the knockdown efficiency by qRT-PCR. The medium was changed 1 day after the electroporation.
GSIS assay in MIN6 cells
The GSIS assay was executed 2 days after the electroporation of siRNA. For the assay, MIN6 cells (cultivated in 12-well plates) were washed three times with 500 µl of Krebs-Ringer HEPES (KRH) buffer (containing 15 mM HEPES, 5 mM KCl, 120 mM NaCl, 24 mM NaHCO3, 1 mM MgCl2, 2 mM CaCl2, and 1 mg/ml BSA). After starvation for 1 hr in glucose-free KRH buffer, the medium was removed and cells were incubated for 2 hr in 500 µl KRH buffer either containing no or 25 mM glucose, respectively. Subsequently, the supernatant was collected for the determination of the insulin concentrations. All incubation steps were conducted at 37° with 5% CO2. Finally, cells were detached in lysis buffer [containing 20 mM Tris, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, and 1% (v/v) Triton-X-100] and protein content was determined by bicinchoninic acid (BCA) assay (Pierce Chemical, Rockford, IL) for normalization of insulin levels.
Linkage analysis
Data sets for males and females from both N2 (NZOx129P2 and NZOxC3H) populations were treated separately in the linkage analysis. Distributions of phenotypic data were tested for normality by the use of the D’Agostino-Pearson omnibus test (GraphPad Software, La Jolla, CA). All data sets that were not normally distributed were log2 transformed. QTL analyses including the genetic map, genotyping errors, and linkage between individual traits and genotypes were performed on N2(NZOx129P2) (290 males and 307 females) and N2(NZOxC3H) (329 males and 310 females) mice using the R/qtl 1.40-8 package (Broman and Sen 2009) of R (version i386 3.3.2). Single-QTL genome scans were performed by interval mapping with the Expectation-maximization algorithm (Lander and Botstein 1989). The significance thresholds (P < 0.05) for linkage were estimated by 1000 permutations (Lander and Kruglyak 1995).
Sequence and haplogroup analysis
Data for mouse SNPs and SNP–gene assignments were from the Sanger Welcome Trust Institute Database (https://www.sanger.ac.uk/sanger/Mouse_SnpViewer). Coding nonsynonymous SNPs were analyzed for their potential impact on protein function using the “Sorting Tolerant From Intolerant” algorithm (http://sift.jcvi.org; (Kumar et al. 2009). As described before (Schmidt et al. 2008), intervals of 250 kbp were selected for the determination of the frequency of polymorphic SNPs between the different mouse strains, and a threshold of 100 SNPs /window was chosen to distinguish the QTL into regions that are identical-by-descent (IBD, genomic regions that are identical between individuals due to descent from a common ancestor without recombination) and polymorphic (non-IBDs) between the different mouse strains. For the determination of the total number, all SNPs annotated for the C57BL/6J reference genome with calls for C3H/HeJ, 129P2/OlaHsd, and NZO/HlLtJ were counted.
Statistical analysis
Data are presented as mean values ± SEM. Statistical significance was reported by two-tailed Student’s t-test or one/two-way ANOVA followed by post hoc Bonferroni test as appropriate. Differences were considered significant when P < 0.05. Statistical analysis was conducted by GraphPad Prism 7 (GraphPad Software).
Data availability
Supplemental Material, File S1 contains R/qtl formatted mapping information, including mouse identifiers, phenotype information, and SNP marker identifiers, locations, and genotypes (A = NZO/NZO, and H = NZO/C3H or NZO/129P2) from both N2 (NZOxC3H and NZOx129P2) populations. File S2 contains the sequences of all qRT-PCR primers and siRNAs, as well as information about the SNP markers used for genotyping of RCS.NZO.C3H-Nbg7 mice. File S3 contains further data supporting material. Microarray data are available under accession number GSE117553. Supplemental material available at Figshare: https://figshare.com/s/3666e6e39dced467c48f.
Results
Diverging prevalence for obesity and T2D in the parental strains
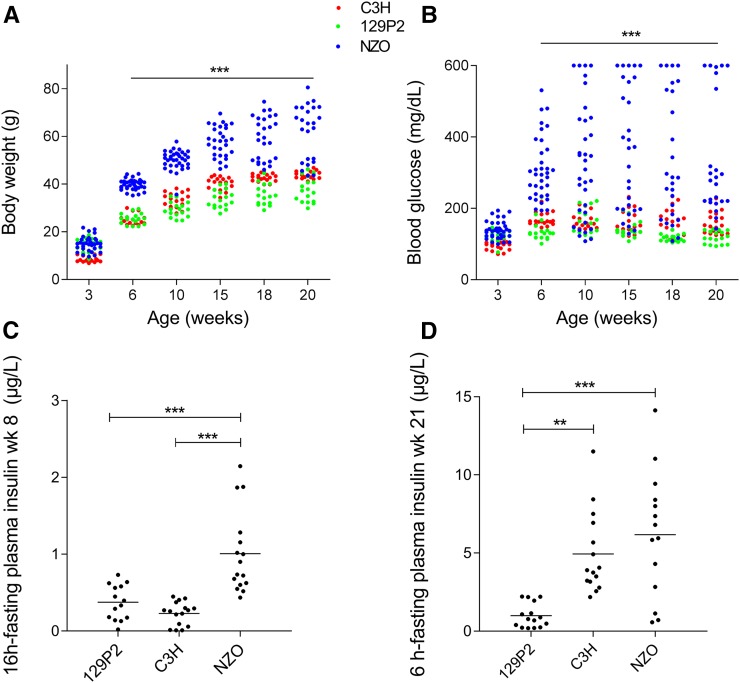
We first phenotyped the experimental animals by measuring basic metabolic features of male NZO, 129P2, and C3H mice on a HFD with 45% calories from fat. One week after weaning at the age of 4 weeks, NZO mice started to exhibit significantly higher body weight compared to C3H and 129P2 mice (NZO 25.4 ± 0.6 g, 129P2 19.9 ± 0.7 g, and C3H 15.9 ± 0.4 g; Figure 1A). At week 20, NZO mice had gained substantially more weight than C3H and 129P2 mice (NZO 62.1 ± 2.3 g, 129P2 37.6 ± 1.2 g, and C3H 44.0 ± 0.4 g), most of it due to increased body fat (week 15 of age: NZO 22.5 ± 1.2 g, 129P2 6.7 ± 0.7 g, and C3H 10.7 ± 0.3 g). Moreover, in contrast to the lean strains, NZO mice developed early-onset hyperglycemia (> 300 mg/dl) and hyperinsulinemia. On average, at the age of 6 weeks, NZO mice started to exceed the T2D threshold (> 300 mg/dl), whereas C3H and 129P2 mice maintained blood glucose levels constantly below 190 mg/dl (Figure 1B). The most significant differences in glycemia between NZO mice and the lean strains were observed at week 15, when NZO mice exhibited on average 361 ± 32 mg/dl blood glucose, compared to 160 ± 8 mg/dl observed in C3H and 142 ± 6 mg/dl in 129P2 mice. In week 8, 16-hr fasting plasma insulin levels were about threefold higher (P < 0.001) in NZO (1.0 µg/liter ± 0.13 µg/liter) compared to C3H (0.23 ± 0.04 µg/liter) and 129P2 (0.37 ± 0.06 µg/liter) mice. At 21 weeks of age, 6-hr fasting plasma insulin levels were significantly lower in the 129P2 strain (1.0 ± 0.2 µg/liter) compared to levels in the NZO (6.2 ± 1.1 µg/liter) and C3H (4.9 ± 0.7 µg/liter) strains.
Figure 1.
Metabolic characterization of the parental mouse strains 129P2, C3H, and NZO. Development of body weight (A) and blood glucose (B) measured at weeks 3, 6, 10, 15, 18, and 20. Fasting plasma insulin levels were measured at weeks 8 [16-hr fasting (C)] and 21 [6-hr fasting, (D)] by ELISA. Dots represent single male animals (129P2: n = 14–17; C3H: n = 15–18; and NZO: n = 14–35). Statistical differences between strains were calculated by one- (C and D) or two-way ANOVA (A and B) followed by post hoc Bonferroni test; ** P < 0.01 and *** P < 0.001 by comparison to NZO unless otherwise stated. NZO, New Zealand Obese; wk, week.
Genome-wide linkage analysis on both N2 populations revealed 30 novel T2D-associated QTL
Two F1 populations were generated by mating NZO females with either 129P2 or C3H males. All males from both F1 generations were metabolically phenotyped. As expected, the phenotypic data from both F1 generations, including body weight and blood glucose levels, were intermediate to those from the parental mice (Figure S1 and File S3). In general, F1(NZOxC3H) males exhibited higher mean blood glucose levels compared to F1(NZOx129P2) males [week 20 of age: F1(NZOxC3H) 377 ± 23 mg/dl and F1(NZOx129P2) 198 ± 23 mg/dl, P < 0.001], whereas body weight development was similar between the two F1 generations. Subsequently, two N2 populations were generated by breeding female NZO mice with males from the F1(NZOx129P2) cross or from the F1(NZOxC3H) cross.
In total, 290 males and 307 females from the N2(NZOx129P2) population, and 329 males and 310 females from the N2(NZOxC3H) population, were characterized for obesity- and T2D-associated traits, including body weight, body composition, blood glucose, and plasma insulin. As expected from their genetic diversity, huge variation of blood glucose levels and body weight was observed in both N2 populations (Figure S2 and File S3). Compared to the female N2(NZOx129P2) population, females from the N2(NZOxC3H) backcross exhibited higher blood glucose levels [week 20 of age, N2(NZOxC3H) 194 ± 4 mg/dl and N2(NZOx129P2) 141 ± 2 mg/dl, P < 0.001, Figure S2C] and body weight [week 20 of age, N2(NZOxC3H) 61 ± 1 and N2(NZOx129P2) 51 ± 1 g, P < 0.001, Figure S2D]. Also, for the N2 males of the N2(NZOxC3H) population, we measured higher blood glucose levels [week 20 of age: N2(NZOxC3H) 350 ± 9 mg/dl and N2(NZOx129P2) 252 ± 7 mg/dl, P < 0.001, Figure S2A] but lower body weight values compared to males from the other backcross [week 20 of age: N2(NZOxC3H) 68 ± 1 g and N2(NZOx129P2) 73 ± 1 g, P < 0.001, Figure S2B].
For genetic linkage analysis, as described in the Materials and Methods, a genome-wide SNP panel was used to interrogate each animal (males and females) at > 100 loci. For the N2(NZOxC3H) cross, a total number of 25 QTL (LOD > 4) were mapped, compared to only five QTL identified in the N2(NZOx129P2) cross (File S3 and Table S1). Details on the major QTL from both male N2 populations, which revealed significance for several weeks, are shown in Table 1.
Table 1. Summary of the major QTL with significance for several weeks from both male N2 (NZOxC3H and NZOx129P2) populations.
| Name | Chr | Traits | Peak Pos (cM) | 95% C.I. (cM) | Closest SNP marker (Mbp) | Max. LOD | Signif. weeks (max. effect) | Mean NZO/NZO | Mean NZO/C3H or NZO/129P2 | Cross |
|---|---|---|---|---|---|---|---|---|---|---|
| Nbg4p | 4 | BG | 28 | 23–31 | 58.1 | 5.8 | 15–20 (17) | 234 mg/dl | 307 mg/dl | Nx129 |
| Nbg4d | 4 | BG | 44 | 38–47 | 97.3 | 7.4 | 12–20 (17) | 230 mg/dl | 306 mg/dl | Nx129 |
| Nbg4d | 4 | BG | 42 | 38–52 | 119 | 6.6 | 14–20 (20) | 291 mg/dl | 389 mg/dl | NxC |
| Pancr. ins | 45 | 44–51 | 119 | 4.2 | 21 | 12.9 µg/mg | 6.2 µg/mg | NxC | ||
| Plasma ins | 40 | 44–62 | 119 | 4.3 | 21 | 10.8 µg/liter | 7.2 µg/liter | NxC | ||
| Nbw4 | 4 | BW | 32 | 16–41 | 91.0 | 8.2 | 6–20 (19) | 69.1 g | 62.7 g | NxC |
| Nbg7p | 7 | BG | 12 | 7–27 | 37.3 | 13.3 | 3, 6–20 (10) | 369 mg/dl | 247 mg/dl | NxC |
| BW | 17 | 7–22 | 37.3 | 7.4 | 12–20 (17) | 59.8 g | 65.4 g | NxC | ||
| LM | 11 | 4–21 | 37.3 | 8.6 | 15 | 31.5 g | 33.2 g | NxC | ||
| Plasma ins | 17 | 13–25 | 37.3 | 4.8 | 21 | 6.5 µg/liter | 11.3 µg/liter | NxC | ||
| Nbg7d | 7 | BG | 26 | 23–29 | 76.7 | 12.5 | 6–16 (10) | 374 mg/dl | 252 mg/dl | NxC |
| Nbg15p | 15 | BG | 23 | 18–30 | 63.3 | 6.7 | 7–20 (15) | 409 mg/dl | 310 mg/dl | NxC |
| Nbg15d | 15 | BG | 41 | 34–47 | 98 | 6.5 | 7–20 (10) | 349 mg/dl | 272 mg/dl | NxC |
LOD scores, peak positions, and 95% confidence intervals (Bayesian method) were calculated by R/qtl software. NZOxC3H: n = 269–329 males, NZOx129P2: n = 285–290 males. Chr, chromosome; Pos, Position; max., maximum; Signif., significant; NZO, New Zealand Obese; BG, blood glucose; N, NZO; C, C3H; Pancr., Pancreas; ins, insulin; BW, body weight; LM, lean mass.
Major T2D modifier QTL on chromosomes 4, 7, and 15
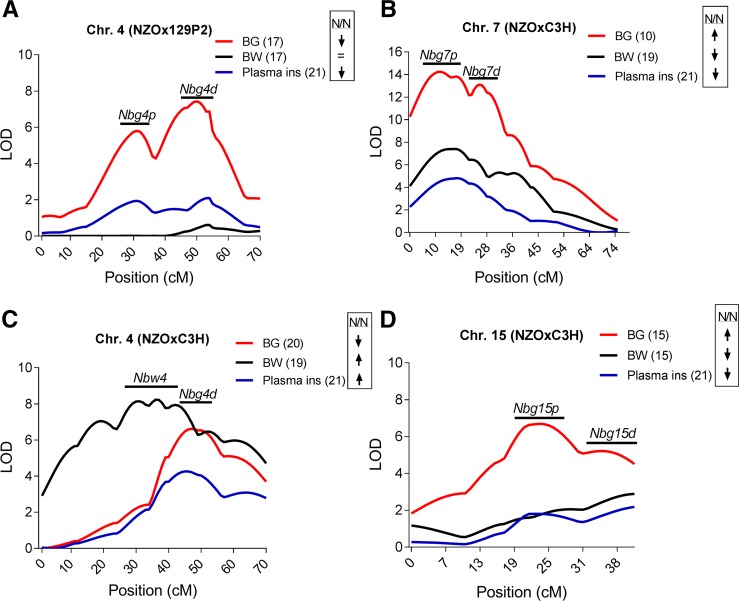
Several loci revealed significant linkage with body weight, blood glucose, or plasma insulin for several consecutive weeks in life (Table 1). In the N2(NZOx129P2) population, the strongest QTL for blood glucose levels was identified on chromosome 4 at weeks 12–20 (week 17 of age: LOD 7.4 at 44 cM, Figure 2A), this locus was designated Nbg4d (NZO blood glucose on distal chromosome 4). The NZO allele for Nbg4d was associated with lower blood glucose levels (Nbg4d is associated with decreased blood glucose levels in NZO-allele carriers). Nbg4d revealed only suggestive linkage (LOD 2.1 at 47 cM) with plasma insulin levels measured at week 21. Homozygous allele carriers exhibited significantly lower levels of plasma insulin compared to heterozygous allele carriers (Nbg4dNZO/NZO 8.3 µg/liter ± 0.6 and Nbg4dNZO/129 11.3 µg/liter ± 0.8, Δ: 3 µg/liter, P = 0.003). Starting from week 15 of age, another QTL for blood glucose (Nbg4p, NZO blood glucose on proximal chromosome 4) appeared at a more proximal position on chromosome 4 (weeks 17 of age: LOD 5.8 at 28 cM, Figure 2A) in the N2(NZOx129P2) population. Similar to what was observed for Nbg4d, the NZO allele for Nbg4p was associated with lower blood glucose levels (week 17 of age: Nbg4pNZO/NZO 234 ± 9 mg/dl and Nbg4pNZO/129 307 ± 11 mg/dl, Δ 74 mg/dl, P < 0.001).
Figure 2.
LOD score profile of the major identified QTL for blood glucose, body weight, and plasma insulin in the N2 population of the NZOx129P2 and NZOxC3H cross. In the N2(NZOx129P2) population, two QTL for blood glucose (Nbg4p and Nbg4d) were detected on chromosome 4 (A). Further QTL were detected in the N2(NZOxC3H) population. A QTL for blood glucose and plasma insulin (Nbg4d) was detected at a similar position on chromosome 4 compared to Nbg4d from the N2(NZOx129P2) population. Another locus on chromosome 4 (Nbw4) was associated with body weight (C). A QTL hotspot on proximal chromosome 7 (Nbg7p) revealed linkage with blood glucose, plasma insulin, and body weight. A second QTL for blood glucose was detected at a more distal position on chromosome 7 (Nbg7d) (B). Two further major QTL for blood glucose (Nbg15p and Nbg15d) were detected on chromosome 15 (D). Weeks of age are shown in brackets. The genotypic effects are indicated with the arrows. LOD scores were calculated using R/qtl software. Detailed information for each QTL is shown in Table 1. BG, blood glucose; BW, body weight; Chr., chromosome; ins, insulin; N/N, NZO/NZO; NZO, New Zealand Obese
Also in the N2(NZOxC3H) population, a strong QTL for blood glucose levels mapped to distal chromosome 4 at weeks 14–20 (week 20 of age: LOD 6.6 at 42 cM, Figure 2C), thus the locus was likewise designated Nbg4d. As observed in the N2(NZOx129P2) cross, homozygous NZO allele carriers exhibited lower mean blood glucose levels compared to the heterozygous NZO/C3H allele carriers for Nbg4d (week 20 of age: Nbg4dNZO/NZO 291 ± 11 mg/dl and Nbg4dNZO/C3H 389 ± 15 mg/dl, Δ: 98 mg/dl, P < 0.001). In the N2(NZOxC3H) population, Nbg4d was further significantly linked with plasma insulin levels (week 21 of age: LOD 4.2 at 45 cM). Different from the N2(129P2xNZO) population, homozygous allele carriers exhibited higher plasma insulin levels compared to heterozygous allele carriers (week 21 of age: Nbg4dNZO/NZO 10.8 ± 0.9 µg/liter and Nbg4dNZO/C3H 7.2 ± 0.7 µg/liter, Δ: 3.6 µg/liter, P = 0.005). A more proximal locus on chromosome 4 from the N2(NZOxC3H) population (Nbw4, NZO body weight on chromosome 4) revealed significant linkage with body weight at weeks 6–20 (week 19 of age: LOD 8.2 at 32 cM, Figure 2C). Homozygous allele carriers gained on average more body weight compared to heterozygous allele carriers for Nbw4 (week 19 of age: Nbw4NZO/NZO 69.1 ± 0.7 g and Nbw4NZO/C3H 62.7 ± 0.7 g, Δ: 6.4 g, P < 0.001).
The most striking linkages with blood glucose (week 10 of age: LOD 13.3 at 12 cM) and plasma insulin levels (week 21 of age: LOD 4.8 at 17 cM) in the N2(NZOxC3H) population were detected on proximal chromosome 7 (Nbg7p, NZO blood glucose on proximal chromosome 7). The linkage of Nbg7p with blood glucose levels was already significant at 3 weeks of age and persisted until the end of the study. In addition, this locus was further linked to body weight at weeks 12–20 (week 17 of age: LOD 7.4 at 17 cM, Figure 2B) and lean mass at week 15 (LOD 8.6 at 11 cM; File S3 and Table S1). Homozygous NZO allele carriers for Nbg7p exhibited on average higher blood glucose levels (week 10 of age: Nbg7pNZO/NZO 369 ± 12 mg/dl and Nbg7pNZO/C3H 247 ± 8 mg/dl, Δ: 122 mg/dl, P < 0.001), in combination with lower plasma insulin levels (week 21 of age: Nbg7pNZO/NZO 6.5 ± 0.6 µg/liter and Nbg7pNZO/C3H 11.3 ± 1.1 µg/liter, Δ: 4.8 µg/liter, P < 0.001) and lower body weight (week 17 of age: Nbg7pNZO/NZO 59.8 ± 0.6 g and Nbg7pNZO/C3H 64.4 ± 0.6 g, Δ: 5.6 g, P < 0.001) compared to heterozygous mice. Moreover, in the same backcross, a second peak for the blood glucose QTL (week 10 of age: LOD 12.5 at 26 cM, Figure 2B) was identified at a more distal region on chromosome 7 (Nbg7d, NZO blood glucose on distal chromosome 7). Similar to Nbg7p, the NZO allele for Nbg7d was associated with increased blood glucose levels (week 10 of age: Nbg7dNZO/NZO 374 ± 12 mg/dl and Nbg7dNZO/C3H 252 ± 8 mg/dl, Δ: 122 mg/dl, P < 0.001).
Furthermore, in the N2(NZOxC3H) population, we detected two loci on chromosome 15 that were strongly associated with blood glucose levels at weeks 7–20 (week 15 of age: LOD 6.7 at 23 cM and 5.2 at 35 cM). Whereas the proximal QTL (Nbg15p, NZO blood glucose on proximal chromosome 15) was exclusively linked to blood glucose levels, the distal QTL (Nbg15d, NZO blood glucose on distal chromosome 15) showed additional suggestive linkage with body weight (week 15 of age: LOD 2.9 at 42 cM) and plasma insulin levels (week 21 of age: LOD 2.2 at 44 cM) (Figure 2D). Homozygous animals for both QTL had higher blood glucose levels compared to the heterozygous allele carriers (week 15 of age: Nbg15pNZO/NZO 409 ± 12 mg/dl and Nbg15pNZO/C3H 310 ± 12 mg/dl, Δ: 99 mg/dl, P < 0.001; week 10 of age: Nbg15dNZO/NZO 349 ± 11 mg/dl and Nbg15dNZO/C3H 272 ± 10 mg/dl, Δ: 77 mg/dl, P < 0.001) (Table 1).
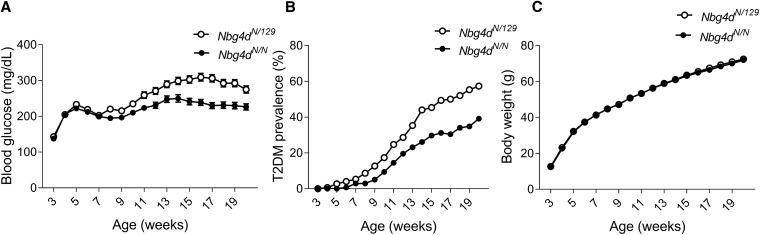
Nbg4d is associated with decreased blood glucose levels in NZO allele carriers
Figure 3 shows the development of blood glucose levels, T2D prevalence, and body weight for homozygous NZO and heterozygous allele carriers for Nbg4d from the N2(NZOx129P2) population. Starting at week 8 of age, heterozygous allele carriers exhibited significantly higher mean blood glucose levels (week 17 of age: 307 ± 10 mg/dl in Nbg4dNZO/129P2 compared to 230 ± 7 mg/dl in Nbg4dNZO/NZO, P < 0.001, Figure 3A) in line with a higher T2D prevalence (week 20 of age: 55% in Nbg4dNZO/129P2 compared to 35% in Nbg4dNZO/NZO, Figure 3B). In contrast, the development of body weight was similar between the two genotypes (Figure 3C).
Figure 3.
Quantitative effect of Nbg4d. Mean values for blood glucose levels (± SEM) (A), cumulative T2D prevalence (B), and body weight (C) for homozygous NZO and heterozygous allele carriers for Nbg4d from the N2(NZOx129P2) population. Nbg4dNZO/129P2: n = 150; Nbg4dNZO/NZO: n = 138. N, NZO; NZO, New Zealand Obese; T2D, type 2 diabetes.
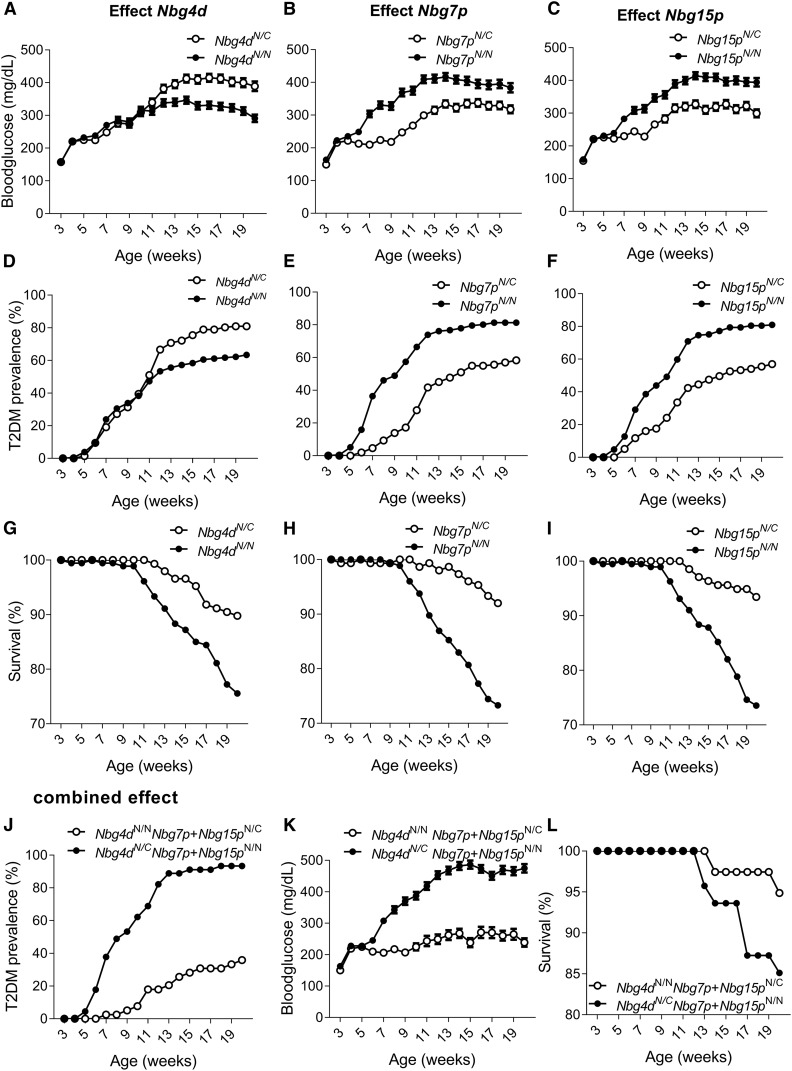
Three loci on chromosomes 4, 7, and 15 contribute to distinct steps in the onset and development of T2D in the N2(NZOxC3H) population
Next, we tested whether the loci on chromosomes 4, 7, and 15 may contribute distinctly at different stages in the development of diabetes in the N2(NZOxC3H) population. We therefore calculated mean values for blood glucose levels, cumulative T2D prevalence, and survival rates for each risk/protective allele on chromosomes 4, 7, and 15, and for the combined effect of all risk/protective alleles. The results of the analysis are illustrated in Figure 4. Starting in week 11, higher blood glucose levels were observed in mice carrying the C3H allele of Nbg4d (week 20 of age: 389 ± 15 mg/dl in Nbg4dNZO/C3H compared to 291 ± 11 mg/dl in Nbg4dNZO/NZO, P < 0.001, Figure 4A). This was accompanied by a higher T2D prevalence of C3H allele carriers of Nbg4d (week 18 of age: 79% in Nbg4dNZO/C3H compared to 61% in Nbg4dNZO/NZO, Figure 4D). However, risk allele carriers of Nbg4d exhibited a higher survival rate (week 20 of age: 90% in Nbg4dNZO/C3H compared to 76% in Nbg4dNZO/NZO, Figure 4G).
Figure 4.
Single and combined effects of major QTL in the N2(NZOxC3H) population. Mean values for blood glucose levels (± SEM) (A), cumulative T2D prevalence (D), and survival (G) for NZO/C3H and NZO/NZO allele carriers for Nbg4d. Mean values for blood glucose levels (± SEM) (B), cumulative T2D prevalence (E), and survival (H) for NZO/C3H and NZO/NZO allele carriers for Nbg7p. Mean values for blood glucose levels (± SEM) (C), cumulative T2D prevalence (F), and survival (I) for NZO/C3H and NZO/NZO allele carriers for Nbg15p. Combined effect of all three risk alleles on the development of blood glucose levels (± SEM) (J), cumulative T2D prevalence (K), and survival (L). n: Nbg4dNZO/NZO = 125–180, Nbg4dNZO/C3H = 110–147; Nbg7pNZO/NZO = 129–176, Nbg7pNZO/C3H = 139–151; Nbg15pNZO/NZO = 139–189, Nbg15pNZO/C3H = 128–137; Nbg4dNZO/NZO Nbg7pNZO/C3H Nbg15pNZO/C3H = 37–39; and Nbg4dNZO/C3H Nbg7pNZO/NZO Nbg15pNZO/NZO = 39–46. C, C3H; N, NZO; NZO, New Zealand Obese; T2D, type 2 diabetes.
In contrast, already starting from 6 weeks of age, the NZO allele for Nbg7p was associated with increased blood glucose levels (week 10 of age: 369 ± 12 mg/dl in Nbg7pNZO/NZO compared to 247 ± 8 mg/dl in Nbg7pNZO/C3H, P < 0.001, Figure 4B), an increased T2D prevalence (week 10 of age: 57% in Nbg7pNZO/NZO compared to 17% in Nbg7pNZO/C3H, Figure 4E), and a lower survival rate (week 20 of age: 73% in Nbg7pNZO/NZO compared to 92% in Nbg7pNZO/C3H, Figure 4H).
Similarly, Nbg15pNZO/NZO mice exhibited higher blood glucose levels (week 15 of age: 409 ± 12 mg/dl in Nbg15pNZO/NZO compared to 310 ± 12 mg/dl in Nbg15pNZO/C3H, P < 0.001, Figure 4C), an increased T2D prevalence (week 13 of age: 75% in Nbg15pNZO/NZO compared to 45% in Nbg15pNZO/C3H, Figure 4F), and a lower survival rate (week 20 of age: 74% in Nbg15pNZO/NZO compared to 93% in Nbg15pNZO/C3H, Figure 4I) compared to Nbg15pNZO/C3H mice.
Blood glucose levels increased with the number of risk alleles. Mice carrying all three risk alleles (Nbg4dNZO/C3H, Nbg7pNZO/NZO, and Nbg15pNZO/NZO) exceeded the T2D threshold of 300 mg/dl already at 7 weeks of age and reached a maximum average blood glucose level of 486 mg/dl in life-week 15. Conversely, mice carrying the protective alleles for all three QTL (Nbg4dNZO/NZO, Nbg7pNZO/C3H, and Nbg15pNZO/C3H) exhibited substantially lower mean blood glucose levels (Δ: 238 mg/dl) in the same week of life and did not exceed the T2D threshold until the end of the study (Figure 4J). The T2D prevalence reached 93% in the risk allele carriers but only 36% in the protective allele carriers for the three QTL (Figure 4K). However, we did not observe that a combination of the three protective alleles improved the survival rate of the animals (week 20 of age: 85% in risk allele combination compared to 95% in protective allele combination, Figure 4L).
Validation for the linkage of Nbg7p with blood glucose and plasma insulin in recombinant congenic mice
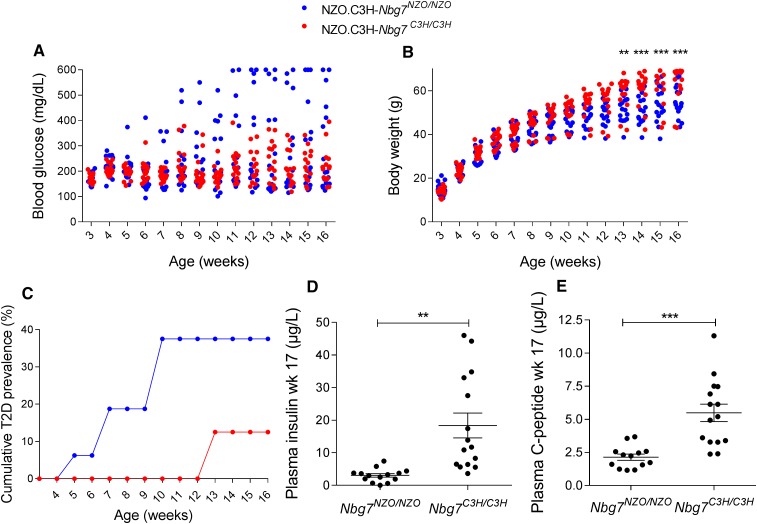
To validate linkage of the Nbg7p locus with blood glucose and plasma insulin, we generated recombinant congenic mouse lines (RCS.NZOxC3H.Nbg7C3H/C3H and RCS.NZOxC3H.Ngb7NZO/NZO) that harbor the Nbg7p locus on an NZO background (see Materials and Methods). Both genotypes were fed a 45% HFD and were metabolically characterized at weeks 3–16. Blood glucose levels were lower in RCS.NZOxC3H.Nbg7C3H/C3H mice compared to RCS.NZOxC3H.Nbg7NZO/NZO mice (week 12 of age: 219 ± 17 mg/dl vs. 299 ± 44 mg/dl, not significant). At 10 weeks of age, 38% of the RCS.NZOxC3H.Nbg7NZO/NZO mice were already diabetic (blood glucose > 300 mg/dl), whereas all RCS.NZOxC3H.Nbg7C3H/C3H mice were normoglycemic (blood glucose < 300 mg/dl) (Figure 5B). Moreover, starting at 13 weeks of age, RCS.NZOxC3H.Nbg7C3H/C3H animals gained more body weight compared to RCS.NZOxC3H.Nbg7NZO/NZO mice (week 16 of age: 63.7 ± 2.5 g vs. 55.0 ± 1.8 g, P < 0.001, Figure 5C). In addition, RCS.NZOxC3H.Nbg7C3H/C3H mice exhibited significantly higher plasma levels of insulin (18.3 ± 4 µg/liter vs. 3.0 ± 0.5 µg/liter, P = 0.0012; Figure 5D) and C-peptide (5.5 ± 0.7 µg/liter vs. 2.1 ± 0.2 µg/liter, P < 0.001; Figure 5E) than RCS.NZOxC3H.Nbg7NZO//NZO mice.
Figure 5.
Characterization of the recombinant congenic line NZO.C3H.Nbg7. Introgression of chromosome 7 (17–111 Mb) from C3H into the NZO genome led to lower blood glucose levels (A), higher body weight (B), a lower T2D prevalence (C), increased plasma insulin (D), and increased plasma C-peptide levels (E). Insulin and C-peptide levels were measured by ELISA. Data represent single animals or mean values (n = 16–17). The lines in (D) and (E) represent mean values ± SEM. Statistical differences between the genotypes were calculated by two-way ANOVA followed by post hoc Bonferroni test (B) or Student’s t-test, two-tailed (D) or unpaired (E); ** P < 0.01 and *** P < 0.001. NZO, New Zealand Obese; T2D, type 2 diabetes.
Selection of candidate genes for Nbg7p by combined haplotype and gene expression analysis
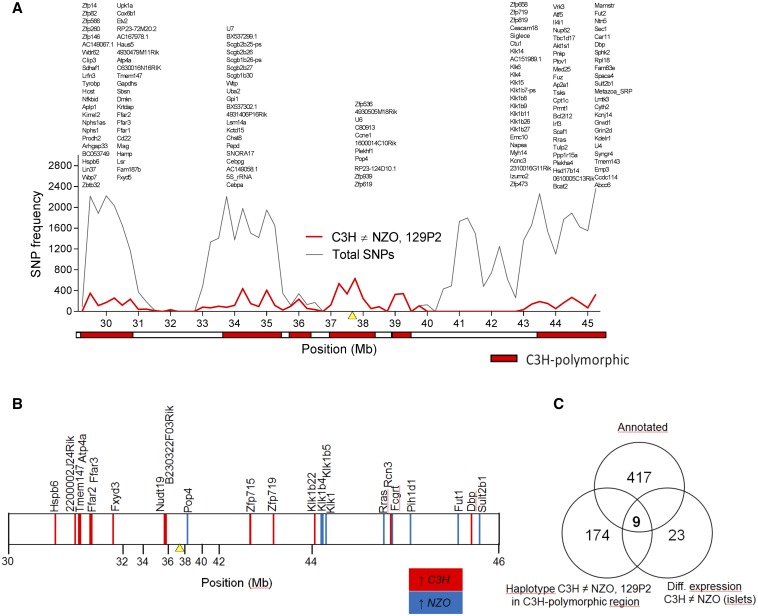
For the selection of candidate genes for Nbg7p, we analyzed SNPs of the parental strains by using the database from the Wellcome Trust Sanger Institute (Keane et al. 2011; Yalcin et al. 2011). Since the genomic sequence for our C3H substrain is not available in the database, the sequence from the closely related C3H/HeJ strain was used instead. As Nbg7p was exclusively detected in N2(NZOxC3H) progeny and thus likely underlies a C3H-specific gene variant, we searched for C3H polymorphisms in the critical region (30–46 Mb) that differed from NZO and 129P2. We identified 26 coding nonsynonymous SNPs (File S3 and Table S2) and 6 indel (insertion/deletion) polymorphisms (File S3 and Table S3) where C3H differs from both NZO and 129P2, but none of these variants are likely to affect protein function. Further haplotype analyses were conducted to narrow down regions of interest. For the dissection of the QTL peak into regions that are IBD between the strains and C3H polymorphic regions, we determined the number of C3H polymorphic SNPs each 250 kbp. As described before (Schmidt et al. 2008), regions exceeding a threshold of 100 SNPs/window were considered as C3H polymorphic (Figure 6A). Only genes with C3H-specific SNPs located in C3H polymorphic regions were considered as candidates for the QTL. Hence, out of 417 genes annotated in the database for the region, the number of candidates for Nbg7p could be narrowed down to 174 genes.
Figure 6.
Combined approach of haplotype and gene expression analysis in the islets of parental strains for the identification of C3H-specific gene variants within Nbg7p. (A) Haplotype analysis using Genome Reference Consortium Mouse Build 38 provided by the Wellcome Trust Sanger Institute. The red line represents the number of C3H-specific SNPs, whereas the gray line shows the total number of SNPs (all SNPs annotated for C57B6/J reference genome with calls for C3H, 129, and NZO). Both lines overlap in the region between 37.25 and 39.5 Mb. Genomic regions containing < 100 C3H polymorphic SNPs per window (250 kb) are considered as identical to 129P2 or/and NZO (identical-by-descent) and are represented by the white boxes. In contrast, regions with > 100 C3H polymorphic SNPs per window are designated as C3H polymorphic regions and are highlighted by red boxes. Genes carrying C3H-specific SNPs located in a C3H polymorphic region are listed above. For a better overview, gene models are not included. (B) Significantly (P < 0.05) differential gene expression at the age of 6 weeks in the pancreatic islets between C3H and NZO revealed from the microarray analysis. Higher expression in C3H (ratio C3H/NZO > 1) is shown by red vertical lines and higher expression in NZO (ratio C3H/NZO < 1) is shown by blue vertical lines. Differences between the strains were calculated by one-sided Wilcoxon signed rank test. (C) Venn diagram showing the total number of annotated genes (top), the number of genes revealed from the haplotype analysis (left), the number of genes revealed from the microarray analysis (right), and the overlap of genes (center). The yellow triangles in (A) and (B) mark the position of the QTL peak marker (rs3724525). NZO, New Zealand Obese.
Next, to further reduce the number of candidates, we integrated genome-wide transcriptome data from a microarray analysis of the islets of the parental strains C3H and NZO. This analysis revealed 23 genes with differential expression and thus potential candidates for Nbg7p (Figure 6B).
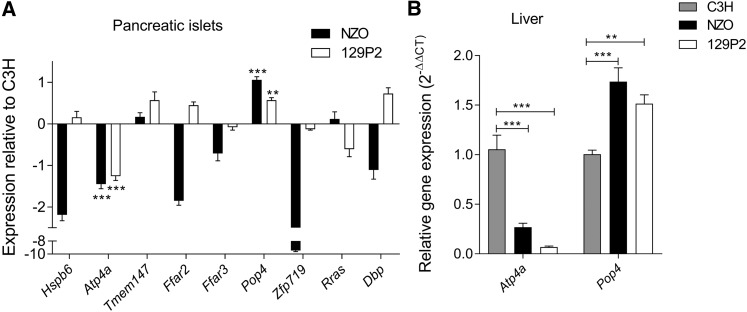
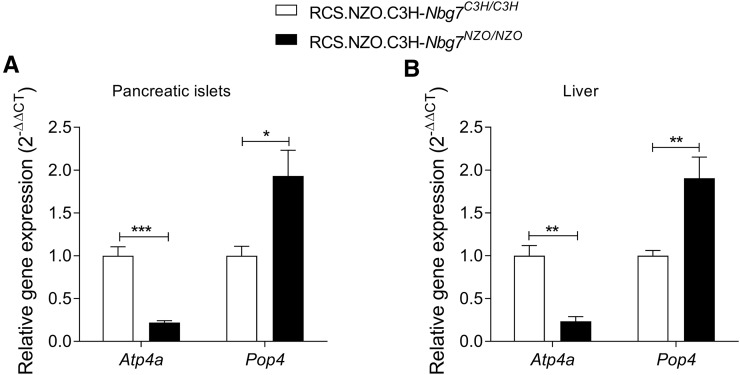
In total, we found an overlap of nine genes [Heat shock protein, α-crystallin-related, B6 (Hspb6); Gastric hydrogen-potassium exchanging ATPase α (Atp4a); Transmembrane protein 147 (Tmem147); Free fatty acid receptor 2 (Ffar2); Free fatty acid receptor 3 (Ffar3); Zinc finger protein 719 (Zfp719); D site albumin promoter binding protein (Dbp); and Related RAS viral (r-ras) oncogene (Rras)] from our candidate gene approaches (Figure 6C). We further analyzed the expression of these genes by qRT-PCR and could validate the expression differences of seven genes, whereas differential expression for Tmem147 and Rras could not be confirmed. In addition, we integrated pancreatic islets from 129P2 in the qRT-PCR analysis to search for C3H-specific expression. Only the two candidates Atp4a and Pop4 were distinctively regulated in islets from C3H, thus representing the most likely candidates for Nbg7p. mRNA levels of Atp4a were significantly higher in islets from C3H compared to NZO and 129P2. In contrast, expression of Pop4 was downregulated in C3H compared to the other two strains (Figure 7A). For both genes, a similar expression pattern as in the pancreatic islets was further detected in liver tissue of the parental strains (Figure 7B). We further analyzed the expression of Atp4a and Pop4 in the pancreatic islets (Figure 8A) and liver (Figure 8B) of our RCS.NZO.C3H.Nbg7 mice, and found the same expression differences between NZO and C3H allele carriers for chromosome 7 as observed in the parental strains.
Figure 7.
Quantitative real-time PCR of candidate genes in the parental strains. (A) Expression of nine candidate genes in isolated pancreatic islets is shown relative to C3H. Genes were selected based on differential expression between C3H and NZO in the microarray analysis, and location in C3H polymorphic regions. Significance is indicated for genes with similar expression patterns between NZO and 129P2, but differing from C3H. (B) Expression of Pop4 and Atp4a in liver tissue. Both tissues were collected at the age of 6 weeks and Tbp was used as endogenous control. Data represent mean values ± SEM from five to eight mice. Statistical differences between strains were calculated by one-way ANOVA followed by post hoc Bonferroni test; ** P < 0.01 and *** P < 0.001 (in (A): by comparison to C3H). NZO, New Zealand Obese.
Figure 8.
Quantitative real-time PCR of Pop4 and Atp4a in the recombinant congenic strains. Relative expression in C3H/C3H and NZO/NZO allele carriers for chromosome 7 in isolated pancreatic collected at 6 weeks of age (A) and liver tissue collected at 17 weeks of age (B). Tbp was used as endogenous control. Data represent mean values ± SEM from five to eight mice. Statistical differences between the genotypes were calculated by Student’s t-test, two-tailed, unpaired; * P < 0.05, ** P < 0.01, and *** P < 0.001. NZO, New Zealand Obese; RCS, recombinant congenic strain.
Atp4a and Pop4 alter GSIS in MIN6 cells
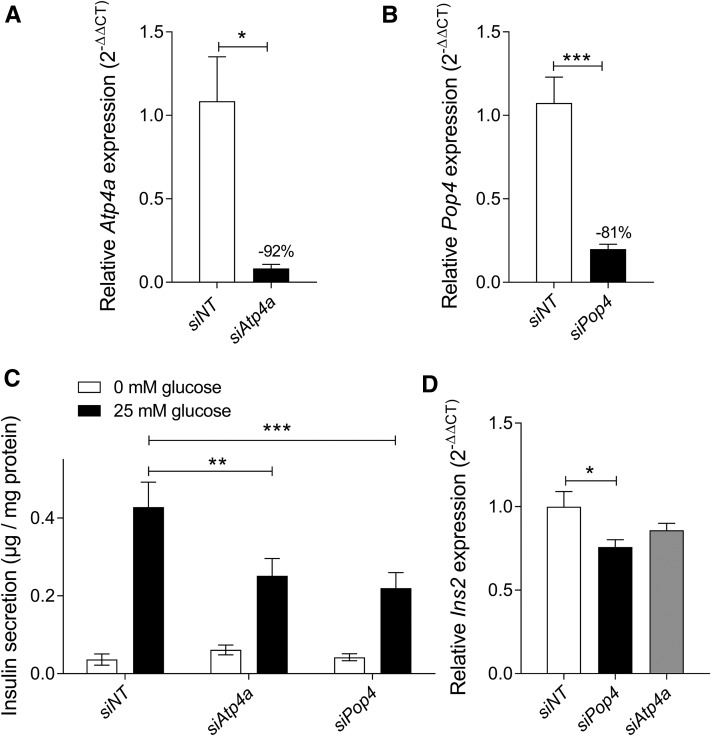
We investigated the roles of Atp4a and Pop4 in insulin secretion using MIN6 cells. By applying specific siRNA oligonucleotides, mRNA levels could be reduced by 92% for Atp4a (Figure 9A) and by 81% for Pop4 (Figure 9B). Insulin secretion without glucose was not different between control (nontarget siRNA-transfected) and knockdown (target siRNA-transfected) cells. In contrast, upon glucose stimulation (25 mM glucose), significantly lower insulin secretion was observed for both Atp4a- (∼40% reduction) and Pop4- (∼50% reduction) knockdown cells (Figure 9C). Moreover, we measured insulin mRNA levels in our knockdown cells and observed a significant reduction in Pop4-deficient cells compared to controls (Figure 9D).
Figure 9.
GSIS in MIN6 cells after knockdown of Pop4 and Atp4a. MIN6 cells were electroporated with siRNA oligonucleotides 2 days prior the GSIS assay. The knockdown efficiency of Atp4a (A) and Pop4 (B) in MIN6 cells was determined by the quantification of mRNA levels via qRT-PCR. For the assay (C), cells were supplemented with Krebs-Ringer HEPES buffer containing no or high glucose (25 mM), and secreted insulin in the supernatants was measured by ELISA. Insulin mRNA levels were quantified by qRT-PCR with nontarget siRNA-transfected cells as controls (D). Data represent mean values ± SEM from four to eight independent experiments. Statistical differences were calculated by Student’s t-test, two-tailed, unpaired (A and B), and two- (C) or one-way (D) ANOVA followed by post hoc Bonferroni test; * P < 0.05, ** P < 0.01, and *** P < 0.001. GSIS, glucose-stimulated insulin secretion; NT, nontarget; qRT-PCR, quantitative real-time-PCR; siRNA, small interfering RNA.
Discussion
Numerous inbred mouse strains are available that exhibit substantial genetic diversity and may reflect susceptibility to various metabolic diseases in humans, including obesity and diabetes (Clee and Attie 2007). Previous studies have investigated the genetic architecture of diabetes and related diseases by taking advantage of different physiological properties that segregate in outbred populations, advanced sequencing resources, and publicly available data sets (Bogue et al. 2018).
Most linkage studies used a single outcross population to search for the genetic determinants for phenotypic distinctions between two strains. However, the genetic comparison of only two strains usually results in a high nomination of candidate genes. The use of more than one inbred cross may substantially improve the mapping resolution of candidates and thus can facilitate gene discovery (Li et al. 2005; Vogel et al. 2018). In our study, we combined linkage data from two different backcross populations generated from the breeding of obese and T2D-prone NZO mice with two widely used lean strains, C3H and 129P2, thereby allowing the identification of strain-specific linkage signals. The strongest QTL for blood glucose and plasma insulin on proximal chromosome 7 (designated Nbg7p), presumably directly influencing pancreatic function, appeared exclusively in the NZOxC3H crossbreeding. Through a combination of sequence and expression data from the three different parental inbred strains, we identified Pop4 and Atp4a as candidates in C3H polymorphic regions with C3H-specific expression patterns in the pancreatic islets, and thus as likely candidates for the QTL Nbg7p.
In response to a HFD, NZO mice develop obesity and insulin resistance, with rapid progression of β-cell failure and severe hyperglycemia (Jürgens et al. 2006, 2007; Joost 2010; Kluth et al. 2011; Joost and Schürmann 2014). In contrast, 129P2 mice are known to be protected from the development of T2D (Clee and Attie 2007). C3H mice show an intermediate phenotype, as they exhibit mild features of the metabolic syndrome, including relatively high body fat content in combination with high plasma glucose, cholesterol, and triglyceride levels (Champy et al. 2008). Notably, embryonic stem cells from 129P2 have been widely used for the generation of targeted mutations in mice, thus rendering many existing lines partially congenic with this strain. Similarly, C3H-derived stem cells were used to generate multiple mutant lines carrying ENU-induced mutations (Coghill et al. 2002; Sabrautzki et al. 2012; van Buerck et al. 2012). We took advantage of the metabolic diversity of the three different inbred strains, and generated two backcross populations by breeding NZO females with 129P2 and C3H males, respectively.
As expected from the genetic diversity of the used parental strains, the subsequent linkage analysis revealed numerous QTL associated with metabolic traits. A locus on distal chromosome 4 (Nbg4d) revealed linkage with blood glucose in both backcross populations, indicating that both QTL underlie the same gene variant derived from the NZO strain, used as a breeding partner in both crosses (NZOx129P2 and NZOxC3H). Interestingly, the NZO allele for Nbg4d was associated with decreased blood glucose levels, suggesting that the obese NZO strain carries a T2D-protective gene variant at this locus. On the other hand, it may also be possible that the QTL might underlie different diabetogenic gene variants from the two lean strains. Interestingly, the Nbg4d locus overlaps partially with a previously reported diabetes QTL, Nidd1/SJL, where Zfp69 (Scherneck et al. 2009) and likely additional variants (Chung et al. 2015) in the region were identified to contribute to the hyperglycemia phenotype. Further studies are required to identify shared candidates in the Nbg4d region. Surprisingly, despite a higher diabetes risk, C3H allele carriers for Nbg4d from the N2(NZOxC3H) backcross had better survival compared to NZO allele carriers. This contradictory phenotype could be explained by additional gene variants within the locus itself or elsewhere in the genome that might regulate survival through glucose-independent mechanisms.
In addition to Nbg4d, we found two further major T2D modifier QTL (Nbg7p and Nbg15p) exclusively in the NZOxC3H crossbreeding where the C3H allele was associated with lower blood glucose, indicating the contribution of two T2D-protective genes from the C3H genome. The effects mediated by the chromosomes 4, 7, and 15 showed different time courses of linkage, indicating that the loci contribute distinctly at different stages in the development of T2D in the N2(NZOxC3H) population. Linkage of blood glucose with Nbg7p was already significant at 3 weeks of age, clearly indicating that the locus targets early steps of T2D development. In contrast, linkage of Nbg4d was not observed before week 14, suggesting that the locus contributes at late stages to the progression of the disease. Interestingly, we could observe a combined effect of the three loci on the development of blood glucose levels, which almost seems to fully explain the incidence of T2D within the population. This observation is remarkable when considering that T2D is assumed to be influenced by dozens of genes scattered all across the genome, each of them contributing to a different extent to its pathophysiology (Clee and Attie 2007; Ali 2013; Prasad and Groop 2015). The onset of T2D due to three loci clearly emphasizes the important impact of the underlying genes. The QTL Nbg7p revealed the most significant linkage with blood glucose and plasma insulin levels from the N2(NZOxC3H) population. Whereas NZO.C3H-Nbg7pNZO/C3H animals were revealed to be widely protected from T2D, NZO.C3H-Nbg7pNZO/NZO animals displayed hyperglycemia already at early age, progressing to severe pancreatic β-cell loss as indicated by the lack of plasma insulin and body weight reduction at final stages of age. The onset of hyperglycemia was independent from the body weight, indicating that the locus directly targets the pancreatic islets rather than the peripheral tissues. We generated RCSs for the validation of the linkage (Darvasi 1997; Brockmann and Neuschl 2012). In accordance with the phenotype from the backcross population, C3H allele carriers exhibited lower mean blood glucose values, and markedly higher plasma insulin and C-peptide levels compared to controls (NZO allele carriers on chromosome 7). However, due to the variation in the control group, the differences in glycemia between the genotypes did not reach statistical significance.
A survey of genetic polymorphisms listed in the Sanger database did not identify sequence variants likely to affect protein function and thus failed to reveal strong candidate genes in the Nbg7p locus. By a combined approach of haplotype and gene expression analysis, we identified C3H-specific sequence and expression variants differing from NZO and 129P2, and thus were able to reduce the number of candidates from 417 annotated genes to two candidates: Potassium-transporting ATPase α chain 1 (Atp4a) and Ribonuclease P protein subunit p29 (Pop4). Differential expression for both genes could further be confirmed in the pancreatic islets from the RCSs, demonstrating that the expression differences associate with the critical fragment from C3H on the NZO background. Interestingly, two genes in the Nbg7p locus, Ffar2 and Ffar3, have already been associated with diabetes-related phenotypes in islets (Priyadarshini and Layden 2015; Priyadarshini et al. 2015) and show lower expression in islets of the diabetes-prone NZO strain. However, both lean strains showed higher expression of Ffar2 and Ffar3, indicating that both genes are unlikely candidates for the phenotype contributed by the C3H allele. Moreover, a mutation of Abcc8 located close to the Nbg7p locus that results in strongly reduced expression of the gene has been reported previously to impair insulin secretion in the related NZO/Wehi strain (Andrikopoulos et al. 2016). Abcc8 was not differentially expressed between our strains, suggesting that this gene is rather an unlikely candidate for the Nbg7p QTL.
Using the murine β-cell line MIN6 (Miyazaki et al. 1990), the knockdown of our strongest candidates, Pop4 and Atp4a, was associated with a decreased GSIS, indicating that the genes may function in the process of insulin release. For Pop4, whose mRNA levels were higher in nondiabetic NZO islets, the observed function is contradictory to the phenotype, as NZO allele carriers exhibited lower levels of plasma insulin. However, mRNA levels were measured in 6-week-old animals, whereas plasma insulin levels were determined at weeks 21 (N2) and 17 (RCS). It might be possible that Pop4-related changes in insulin secretion at early stages of life might contribute to late-onset failure of the islets. Pop4 (also known as Rpp29) encodes one of the protein subunits of the ribonuclease P complex, a protein that was originally discovered as the endoribonuclease that processes the 5′ leader of precursor transfer RNA (tRNA) (Guerrier-Takada et al. 1983). Whereas critical defects in this processing machinery are lethal, alterations in tRNA processing have been associated with impaired insulin secretion and diabetes (Wei et al. 2011; Palmer et al. 2017). In recent years, it has become increasingly clear that the different subunits of the ribonuclease P complex are engaged in other important roles. Pop4 has recently been shown to be involved in the DNA damage response machinery (Abu-Zhayia et al. 2017), and to participate in the regulation of chromatin structure and function (Newhart et al. 2016), which is known to play a fundamental role in the epigenetic modification of the genome and likely also in β-cell biology (Paul et al. 2016). In humans, copy number variants of POP4 have been associated with breast and ovarian cancer (Wrzeszczynski et al. 2011; Natrajan et al. 2012). Our finding that knockdown of Pop4 reduces mRNA for insulin supports a role of this gene in β-cells and insulin secretion. We speculate that elevated levels of Pop4 in young, nondiabetic NZO mice may be compensatory to early-onset insulin resistance, whereas gene expression could be reduced in overt diabetes. Further studies of Pop4 in β-cell function are therefore required.
Another candidate for Nbg7p is Atp4a, as its insulin-stimulating impact is in accordance with increased mRNA and plasma insulin levels in the C3H strain. The gene encodes the α subunit of the heterodimeric gastric proton pump H+/K+-ATPase, which has been reported to function in proton exchange in gastric parietal cells (Spicer et al. 2000). Recently, the gastric H+/K+-ATPase has also been shown to be expressed in the human pancreas where it contributes to pancreatic exocrine secretion (Wang et al. 2015), but its potential role in the secretion of insulin is unknown. It is well established that GSIS in pancreatic islets involves the closure of KATP channels to allow the depolarization of the cell membrane followed by calcium influx through voltage-dependent Ca2+ channels, which subsequently triggers the exocytosis of insulin vesicles. The observed impact of the H+/K+-ATPase on GSIS in MIN6 cells suggests that this proton pump may participate in this process, possibly by contributing to the regulation of the intracellular ion concentration. Interestingly, the autoimmune disease atrophic body gastritis, which is characterized by the persistent presence of ATP4A autoantibodies, has been associated with type 1 diabetes mellitus in humans (Chobot et al. 2014), indicating that the gene may also play an important role in human diabetes.
In conclusion, our study demonstrates that the comparative analysis of multiple inbred populations generated with one common breeding partner allows the identification of strain-specific linkage signals, thereby substantially improving the mapping resolution of disease genes. While T2D is polygenic, the nature of the onset and progression of the disease can be almost fully reconstituted by the interaction of only three different genetic loci, suggesting that in the NZOxC3H backcross only a limited number of variants are contributing to T2D. Nevertheless, considerable evidence exists that QTL regions may include multiple genes that contribute to the phenotype (Buchner and Nadeau 2015). Moreover, a number of genetic variants, including noncoding SNPs, indels, copy number polymorphisms, and yet unknown de novo mutations, may exert effects on regulatory circuits in the locus, thereby affecting islet cell function and glycemic control. Consequently, future studies, including characterization of congenic and tissue-specific knockout lines, will be needed to determine the contribution of Pop4 and Atp4a, and potentially other genetic variations in the locus, to the impact of Nbg7p on blood glucose and plasma insulin levels.
Acknowledgments
We thank Angela Pelligra, Janek Masuch, Angelika Horrighs, Anette Kurowski, Annette Schober, Peter Herdt, and Andrea Scheffel-Clausewitz for technical assistance. This work was supported by grants from the German Ministry of Education and Research, the State of North-Rhine-Westphalia and the State of Brandenburg (grants 82DZD00202 to H.A.-H. and 82DZD00302 to A.S.), and the German Federal Ministry of Health (Bundesministerium für Gesundheit). D.A. was supported by a Deutscher Akademischer Austauschdienst Fellowship and T. Stermann received a stipend from the research training group “vivid” of the Heinrich-Heine University Duesseldorf.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7066604.
Communicating editor: E. Chesler
Literature Cited
- Abu-Zhayia E. R., Khoury-Haddad H., Guttmann-Raviv N., Serruya R., Jarrous N., et al. , 2017. A role of human RNase P subunits, Rpp29 and Rpp21, in homology directed-repair of double-strand breaks. Sci. Rep. 7: 1002 10.1038/s41598-017-01185-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali O., 2013. Genetics of type 2 diabetes. World J. Diabetes 4: 114–123. 10.4239/wjd.v4.i4.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrikopoulos S., Fam B. C., Holdsworth A., Visinoni S., Ruan Z., et al. , 2016. Identification of ABCC8 as a contributory gene to impaired early-phase insulin secretion in NZO mice. J. Endocrinol. 228: 61–73. 10.1530/JOE-15-0290 [DOI] [PubMed] [Google Scholar]
- Attie A. D., Churchill G. A., Nadeau J. H., 2017. How mice are indispensable for understanding obesity and diabetes genetics. Curr. Opin. Endocrinol. Diabetes Obes. 24: 83–91. 10.1097/MED.0000000000000321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue M. A., Grubb S. C., Walton D. O., Philip V. M., Kolishovski G., et al. , 2018. Mouse phenome database: an integrative database and analysis suite for curated empirical phenotype data from laboratory mice. Nucleic Acids Res. 46: D843–D850. 10.1093/nar/gkx1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann G. A., Neuschl C., 2012. Positional cloning of diabetes genes. Methods Mol. Biol. 933: 275–289. 10.1007/978-1-62703-068-7_18 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Sen S., 2009. A Guide to QTL Mapping with R/qtl. Springer, New York. [Google Scholar]
- Buchner D. A., Nadeau J. H., 2015. Contrasting genetic architectures in different mouse reference populations used for studying complex traits. Genome Res. 25: 775–791. 10.1101/gr.187450.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadt A., Leicht K., Deshmukh A., Jiang L. Q., Scherneck S., et al. , 2008. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat. Genet. 40: 1354–1359. 10.1038/ng.244 [DOI] [PubMed] [Google Scholar]
- Champy M. F., Selloum M., Zeitler V., Caradec C., Jung B., et al. , 2008. Genetic background determines metabolic phenotypes in the mouse. Mamm. Genome 19: 318–331. 10.1007/s00335-008-9107-z [DOI] [PubMed] [Google Scholar]
- Cho N. H., Shaw J. E., Karuranga S., Huang Y., da Rocha Fernandes J. D., et al. , 2018. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 138: 271–281. 10.1016/j.diabres.2018.02.023 [DOI] [PubMed] [Google Scholar]
- Chobot A., Wenzlau J., Bak-Drabik K., Kwiecien J., Polanska J., et al. , 2014. ATP4A autoimmunity and Helicobacter pylori infection in children with type 1 diabetes. Clin. Exp. Immunol. 177: 598–602. 10.1111/cei.12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B., Stadion M., Schulz N., Jain D., Scherneck S., et al. , 2015. The diabetes gene Zfp69 modulates hepatic insulin sensitivity in mice. Diabetologia 58: 2403–2413. 10.1007/s00125-015-3703-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clee S. M., Attie A. D., 2007. The genetic landscape of type 2 diabetes in mice. Endocr. Rev. 28: 48–83. 10.1210/er.2006-0035 [DOI] [PubMed] [Google Scholar]
- Coghill E. L., Hugill A., Parkinson N., Davison C., Glenister P., et al. , 2002. A gene-driven approach to the identification of ENU mutants in the mouse. Nat. Genet. 30: 255–256. 10.1038/ng847 [DOI] [PubMed] [Google Scholar]
- Darvasi A., 1997. Interval-specific congenic strains (ISCS): an experimental design for mapping a QTL into a 1-centimorgan interval. Mamm. Genome 8: 163–167. 10.1007/s003359900382 [DOI] [PubMed] [Google Scholar]
- Das S. K., Elbein S. C., 2006. The genetic basis of type 2 diabetes. Cellscience 2: 100–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S., Sano H., Rochford J. J., Semple R. K., Yeo G., et al. , 2009. A truncation mutation in TBC1D4 in a family with acanthosis nigricans and postprandial hyperinsulinemia. Proc. Natl. Acad. Sci. USA 106: 9350–9355. 10.1073/pnas.0900909106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorajoo R., Liu J., Boehm B. O., 2015. Genetics of type 2 diabetes and clinical utility. Genes (Basel) 6: 372–384. 10.3390/genes6020372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C., Flannick J., Teslovich T. M., Mahajan A., Agarwala V., et al. , 2016. The genetic architecture of type 2 diabetes. Nature 536: 41–47. 10.1038/nature18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailus-Durner V., Fuchs H., Becker L., Bolle I., Brielmeier M., et al. , 2005. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat. Methods 2: 403–404. 10.1038/nmeth0605-403 [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S., 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. 10.1016/0092-8674(83)90117-4 [DOI] [PubMed] [Google Scholar]
- Herberg L., Coleman D. L., 1977. Laboratory animals exhibiting obesity and diabetes syndromes. Metabolism 26: 59–99. 10.1016/0026-0495(77)90128-7 [DOI] [PubMed] [Google Scholar]
- Joost H. G., 2010. The genetic basis of obesity and type 2 diabetes: lessons from the new zealand obese mouse, a polygenic model of the metabolic syndrome. Results Probl. Cell Differ. 52: 1–11. 10.1007/978-3-642-14426-4_1 [DOI] [PubMed] [Google Scholar]
- Joost H. G., Schürmann A., 2014. The genetic basis of obesity-associated type 2 diabetes (diabesity) in polygenic mouse models. Mamm. Genome 25: 401–412. 10.1007/s00335-014-9514-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens H. S., Schürmann A., Kluge R., Ortmann S., Klaus S., et al. , 2006. Hyperphagia, lower body temperature, and reduced running wheel activity precede development of morbid obesity in New Zealand obese mice. Physiol. Genomics 25: 234–241. 10.1152/physiolgenomics.00252.2005 [DOI] [PubMed] [Google Scholar]
- Jürgens H. S., Neschen S., Ortmann S., Scherneck S., Schmolz K., et al. , 2007. Development of diabetes in obese, insulin-resistant mice: essential role of dietary carbohydrate in beta cell destruction. Diabetologia 50: 1481–1489. 10.1007/s00125-007-0662-8 [DOI] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinert M., Clemmensen C., Hofmann S. M., Moore M. C., Renner S., et al. , 2018. Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol. 14: 140–162. 10.1038/nrendo.2017.161 [DOI] [PubMed] [Google Scholar]
- Kluth O., Mirhashemi F., Scherneck S., Kaiser D., Kluge R., et al. , 2011. Dissociation of lipotoxicity and glucotoxicity in a mouse model of obesity associated diabetes: role of forkhead box O1 (FOXO1) in glucose-induced beta cell failure. Diabetologia 54: 605–616. 10.1007/s00125-010-1973-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebel B., Hartwig S., Haas J., Lehr S., Goeddeke S., et al. , 2015. Peroxisomes compensate hepatic lipid overflow in mice with fatty liver. Biochim. Biophys. Acta 1851: 965–976. 10.1016/j.bbalip.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P. C., 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Lander E., Kruglyak L., 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Lyons M. A., Wittenburg H., Paigen B., Churchill G. A., 2005. Combining data from multiple inbred line crosses improves the power and resolution of quantitative trait loci mapping. Genetics 169: 1699–1709. 10.1534/genetics.104.033993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., et al. , 1990. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127: 126–132. 10.1210/endo-127-1-126 [DOI] [PubMed] [Google Scholar]
- Morris A. P., Voight B. F., Teslovich T. M., Ferreira T., Segrè A. V., et al. , 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44: 981–990. 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R., Mackay A., Wilkerson P. M., Lambros M. B., Wetterskog D., et al. , 2012. Functional characterization of the 19q12 amplicon in grade III breast cancers. Breast Cancer Res. 14: R53 10.1186/bcr3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhart A., Powers S. L., Shastrula P. K., Sierra I., Joo L. M., et al. , 2016. RNase P protein subunit Rpp29 represses histone H3.3 nucleosome deposition. Mol. Biol. Cell 27: 1154–1169. 10.1091/mbc.e15-02-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C. J., Bruckner R. J., Paulo J. A., Kazak L., Long J. Z., et al. , 2017. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol. Metab. 6: 1212–1225. 10.1016/j.molmet.2017.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P. K., Rabaglia M. E., Wang C. Y., Stapleton D. S., Leng N., et al. , 2016. Histone chaperone ASF1B promotes human beta-cell proliferation via recruitment of histone H3.3. Cell Cycle 15: 3191–3202. 10.1080/15384101.2016.1241914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permutt M. A., Wasson J., Cox N., 2005. Genetic epidemiology of diabetes. J. Clin. Invest. 115: 1431–1439. 10.1172/JCI24758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R. B., Groop L., 2015. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 6: 87–123. 10.3390/genes6010087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini M., Layden B. T., 2015. FFAR3 modulates insulin secretion and global gene expression in mouse islets. Islets 7: e1045182 10.1080/19382014.2015.1045182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini M., Villa S. R., Fuller M., Wicksteed B., Mackay C. R., et al. , 2015. An acetate-specific GPCR, FFAR2, regulates insulin secretion. Mol. Endocrinol. 29: 1055–1066. 10.1210/me.2015-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabrautzki S., Rubio-Aliaga I., Hans W., Fuchs H., Rathkolb B., et al. , 2012. New mouse models for metabolic bone diseases generated by genome-wide ENU mutagenesis. Mamm. Genome 23: 416–430 [corrigenda: Mamm. Genome 25: 648 (2014)] 10.1007/s00335-012-9397-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherneck S., Nestler M., Vogel H., Bluher M., Block M. D., et al. , 2009. Positional cloning of zinc finger domain transcription factor Zfp69, a candidate gene for obesity-associated diabetes contributed by mouse locus Nidd/SJL. PLoS Genet. 5: e1000541 10.1371/journal.pgen.1000541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Gonzaludo N. P., Strunk S., Dahm S., Schuchhardt J., et al. , 2008. A meta-analysis of QTL for diabetes-related traits in rodents. Physiol. Genomics 34: 42–53. 10.1152/physiolgenomics.00267.2007 [DOI] [PubMed] [Google Scholar]
- Schwenk R. W., Vogel H., Schürmann A., 2013. Genetic and epigenetic control of metabolic health. Mol. Metab. 2: 337–347. 10.1016/j.molmet.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer Z., Miller M. L., Andringa A., Riddle T. M., Duffy J. J., et al. , 2000. Stomachs of mice lacking the gastric H,K-ATPase alpha -subunit have achlorhydria, abnormal parietal cells, and ciliated metaplasia. J. Biol. Chem. 275: 21555–21565. 10.1074/jbc.M001558200 [DOI] [PubMed] [Google Scholar]
- Tsaih S. W., Holl K., Jia S., Kaldunski M., Tschannen M., et al. , 2014. Identification of a novel gene for diabetic traits in rats, mice, and humans. Genetics 198: 17–29. 10.1534/genetics.114.162982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buerck L., Schuster M., Rathkolb B., Sabrautzki S., Hrabe de Angelis M., et al. , 2012. Enhanced oxidative stress and endocrine pancreas alterations are linked to a novel glucokinase missense mutation in ENU-derived Munich Gck(D217V) mutants. Mol. Cell. Endocrinol. 362: 139–148. 10.1016/j.mce.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Vogel H., Scherneck S., Kanzleiter T., Benz V., Kluge R., et al. , 2012. Loss of function of Ifi202b by a microdeletion on chromosome 1 of C57BL/6J mice suppresses 11beta-hydroxysteroid dehydrogenase type 1 expression and development of obesity. Hum. Mol. Genet. 21: 3845–3857. 10.1093/hmg/dds213 [DOI] [PubMed] [Google Scholar]
- Vogel H., Kamitz A., Hallahan N., Lebek S., Schallschmidt T., et al. , 2018. A collective diabetes cross in combination with a computational framework to dissect the genetics of human obesity and Type 2 diabetes. Hum. Mol. Genet. 27: 3099–3112. 10.1093/hmg/ddy217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Barbuskaite D., Tozzi M., Giannuzzo A., Sorensen C. E., et al. , 2015. Proton pump inhibitors inhibit pancreatic secretion: role of gastric and non-gastric H+/K+-ATPases. PLoS One 10: e0126432 10.1371/journal.pone.0126432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F. Y., Suzuki T., Watanabe S., Kimura S., Kaitsuka T., et al. , 2011. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 121: 3598–3608. 10.1172/JCI58056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzeszczynski K. O., Varadan V., Byrnes J., Lum E., Kamalakaran S., et al. , 2011. Identification of tumor suppressors and oncogenes from genomic and epigenetic features in ovarian cancer. PLoS One 6: e28503 10.1371/journal.pone.0028503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M., et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. 10.1038/nature10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesil P., Michel M., Chwalek K., Pedack S., Jany C., et al. , 2009. A new collagenase blend increases the number of islets isolated from mouse pancreas. Islets 1: 185–190. 10.4161/isl.1.3.9556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental Material, File S1 contains R/qtl formatted mapping information, including mouse identifiers, phenotype information, and SNP marker identifiers, locations, and genotypes (A = NZO/NZO, and H = NZO/C3H or NZO/129P2) from both N2 (NZOxC3H and NZOx129P2) populations. File S2 contains the sequences of all qRT-PCR primers and siRNAs, as well as information about the SNP markers used for genotyping of RCS.NZO.C3H-Nbg7 mice. File S3 contains further data supporting material. Microarray data are available under accession number GSE117553. Supplemental material available at Figshare: https://figshare.com/s/3666e6e39dced467c48f.