Abstract
Purpose
Emerging evidence suggests that many differentially expressed long non-coding RNAs (lncRNAs) are involved in tumorigenesis. However, the functional roles of these transcripts and the mechanisms responsible for their deregulation in non-small-cell lung cancer (NSCLC) remain elusive. Here, we identified a novel lncRNA (lncRNA 1308), which was significantly upregulated in NSCLC tissues and investigated its biological function and potential molecular mechanism.
Methods
Differences in the lncRNA expression profiles between NSCLC and tumor-adjacent normal tissues were assessed by lncRNA expression microarray analysis. The microRNA in vivo precipitation (miRIP) method was used to identify the targeting microRNAs (miRNAs) on lncRNA 1308, and luciferase reporter assays were performed. Loss-of-function studies were used to explore the effect of lncRNA 1308 on lung carcinogenesis in NSCLC cells.
Results
The novel lncRNA 1308 was upregulated in NSCLC tissues and cell lines. By using biotin-labeled lncRNA 1308 for miRIP in NSCLC cells and dual-luciferase reporter assays, the results suggested that miRNA-124 was associated with lncRNA 1308. Furthermore, the expression of a disintegrin and a metalloproteinase 15 (ADAM 15) was downregulated in NSCLC cells when silencing of lncRNA 1308, the target of oncogenic miR-124, inhibits NSCLC cell proliferation and invasion. Conversely, the expression of ADAM 15 was significantly increased, when inhibiting the expression of miR-124, and alleviated cell invasion inhibition.
Conclusion
The results suggested that lncRNA 1308 may function as a competing endogenous RNA (ceRNA) for miR-124 to regulate cell invasion through the miR-124/ADAM 15 signaling pathway, indicating that lncRNA 1308 plays an important role in the disease progression of NSCLC.
Keywords: lung cancer, lncRNA 1308, miR-124, ADAM 15, competing endogenous RNA
Introduction
Lung cancer is the most common leading cause of death worldwide. In China, more than 700,000 new cases of lung cancer are diagnosed, and approximately 60,000 patients die of lung cancer each year.1 In some regions, such as Xuanwei, Qujing, or Gejiu, in the Honghe autonomous prefecture of Yunnan Province, the incidence of lung cancer is one among the highest in China and in the world.2,3 Recently, a retrospective sampling survey reported mortality rates of lung cancer in Xuanwei of 98.10/105 in men and 83.21/105 in women.4 Approximately 85% of all lung cancer cases have been diagnosed as non-small-cell lung cancer (NSCLC), and tumor metastasis occurs in more than half of all patients.5 Despite several therapies used in the clinical treatment of NSCLC, such as radiation, chemotherapy, and surgical resection, the outcomes remain unsatisfactory, with a 5-year survival rate of only 17%.6 Therefore, exploring the underlying mechanism of NSCLC and identifying potential therapeutic target become imperative requirements.
Long non-coding RNAs (lncRNAs) are a class of transcripts longer than 200 nt in size, which lack significant protein-coding capacity. Emerging data have suggested that lncRNAs are involved in a wide range of biological processes, such as cellular proliferation, apoptosis, and differentiation.7,8 In the past few years, substantial evidence indicates that some lncRNAs, such as MALAT1, MEG3, and TP73-AS1, could regulate cancer progression.9–11 Moreover, several lncRNAs have been reported to participate in NSCLC. For instance, overexpression of lncRNA HOXA11-AS could repress miR-200b and promote cell epithelial–mesenchymal transition in NSCLC,5 whereas upregulation of lncRNA 00673 promotes tumor proliferation through LSD1 interaction and repression of NCALD in NSCLC.12
Although only a small quantity of functional lncRNAs have been well studied nowadays, they have been shown to regulate gene expression at various levels, including chromatin modification, transcriptional regulation, and post-transcriptional regulation.7,13 Currently, the cross talk between lncRNAs and mRNAs as competing for shared microRNAs’ (miRNAs) response elements has been identified as a new regulatory mechanism. In other words, lncRNAs may function as competing endogenous RNAs (ceRNAs) to sponge miRNAs, thereby modulating the depression of miRNA targets and imposing an additional level of post-transcriptional regulation.14 In a previous study, lncRNA HOTAIR has been reported to be a ceRNA that regulates HER2 expression by sponging miR-331-3p in gastric cancer.15 Peng et al16 reported that lncRNA MEG3, a ceRNA of miR-181s, could regulate gastric carcinogenesis and may serve as a potential target for antineoplastic therapies. Therefore, we hypothesize that some lncRNAs may also have functions like ceRNAs, which could link miRNAs and the post-transcriptional network in the disease development of NSCLC. However, the underlying mechanisms of lncRNAs involved in the disease progression of NSCLC remain to be elucidated and characterized. Also, previous studies of occurrences of NSCLC in Xuanwei (Xuanwei lung cancer [XWLC]) are focused mainly on its epidemiology, and little is known about the underlying mechanisms.
In this study, differences in the lncRNA expression profiles between NSCLC and tumor-adjacent normal tissues were assessed by lncRNA expression microarray analysis. We first identified that lncRNA 1308, a novel lncRNA, was overexpressed in NSCLC tissues compared with adjacent normal tissues. Then, we studied the underlying mechanisms involved in this phenomenon and found that knockdown of lncRNA 1308 decreased the expression of a disintegrin and a metalloproteinase 15 (ADAM 15) by negatively regulating miR-124. Overexpression of ADAM 15 has been observed in several types of cancers and is involved in the progression of metastatic tumor,17,18 and previous literature also demonstrated that ADAM 15 can upregulate the expression of matrix metallopeptidase 9 (MM9, a key regulator of cancer invasion and metastasis), thus promoting lung cancer cell invasion.19 Taken together, this study provides the first evidence for a positive lncRNA 1308/ADAM 15 correlation and cross talk between miR-124, lncRNA 1308, and ADAM 15, shedding new light on NSCLC treatment.
Materials and methods
Specimens and cell lines
This research was in accordance with the requirements of the Declaration of Helsinki. Forty lung cancer tissues and the corresponding non-tumor tissues were obtained in accordance with the protocol approved by the ethics committee of the Third Affiliated Hospital of Kunming Medical University, Kunming, China. The patients received the necessary information concerning the study, and the written informed consent was obtained from all included individuals. The human lung cancer cell lines A549, H1975, and H1299, and the human lung normal cell line BESA-2B, were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in the supplemented media according to the ATCC’s recommendation. XWLC and Gejiu lung cancer (GLC), the local cell lines, were isolated from the cancer tissues of the patients who lived in Yunnan Province and were cultured in regular DMEM containing 10% FBS (vol/vol) as HEK-293T cells (ATCC). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR) analysis
Total RNA was extracted from lung cancer tissues, adjacent non-tumor tissues, and lung cancer cell lines by using TRIzol according to the manufacturer’s instructions. qRT-PCR assay was performed using SYBRgreen PCR Master Mix in a Fast Real-time PCR 7500 System (Thermo Fisher Scientific, Waltham, MA, USA). The sequences of gene-specific primers are listed in Table S1. β-Actin was used as an internal control for normalization and quantification of lncRNA 1308 expression. The relative expression level was calculated using the 2−ΔΔCt method.
Human lncRNA microarray assay
Total RNA from 15 pairs of NSCLC and adjacent noncancerous lung tissues was isolated using the TRIzol agent (Thermo Fisher Scientific) following the manufacturers’ instruction. Total RNA was quantified by the NanoDrop ND-2000 (Thermo Fisher Scientific), and the RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Analysis of the expression profile of NSCLC-related lncRNAs was performed by a commercially available oligonucleotide lncRNA microarray (Agilent Human lncRNA microarray, 4*180K, Design ID: 062918; Agilent Technologies) by following the manufacturer’s recommendations. Briefly, total RNA was transcribed to double strand cDNA, then synthesized into cRNA and labeled with Cyanine-3-CTP. The labeled cRNAs were hybridized onto the microarray. After washing, the arrays were scanned by the Agilent Scanner G2505C (Agilent Technologies). The array images were further analyzed by Agilent Feature Extraction software (version_10.7.1.1; Agilent Technologies). The raw data were normalized using the quantile algorithm. The threshold set for up- and downregulated genes was a fold change of ≥2.0 and a P-value of <0.05.
Luciferase assays
Luciferase reporter plasmid was constructed by cloning human ADAM 15 and lncRNA 1308 mRNA sequence into pMIR-Report construct (Ambion, Austin, TX, USA). All the primers are listed in Table S1. Luciferase reporter assays were performed according to the protocol. HEK 293 T cells plated in a 96-well plate were co-transfected with 50 nmol/L miR-124 mimics or negative control oligonucleotides, 20 ng of firefly luciferase reporter, and 10 ng of pRL-TK (Promega Corporation, Fitchburg, WI, USA) using the INTERFERin reagent (Polyplus-transfection, Illkirch France). Cells were collected 36 hours after transfection and analyzed using the Dual-Luciferase Reporter Assay System (Promega Corporation).
Oligonucleotide and plasmid transfection
RNA oligos were chemically synthesized and purified by GenePharma Co., Ltd. (Shanghai, China). The sequences of lncRNA 1308 siRNA were as follows: 5′-CGG AGA GGU CAG AGG UAG ATT-3′ (sense) and 5′- UCU ACC UCU GAC CUC UCC GTT-3′ (antisense). The sequences of control siRNA were as follows: 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (sense) and 5′-ACG UGA CAC GUU CGU AGA ATT-3′ (antisense). The sequence of human miR-124 mimics was 5′- UAA GGC ACG CGG UGA AUG CC-3′. The sequence of negative control oligonucleotides for miRNA was 5′-CAG UAC UUU UGU GUA GUA CAA-3′. The sequence of human miR-124 inhibitors was 5′- GGC AUU CAC CGC GUG CCU UA-3′. The sequence of negative control oligonucleotides for miRNA was 5′-CAG UAC UUU UGU GUA GUA CAA-3′. The transfections were performed with INTERFERin reagent (Polyplus-transfection). The final concentration of miRNA mimics was 50 nmol/L, the final concentration of miRNA inhibitors was 100 nmol/L, and the final concentration of siRNAs was 20 nmol/L.
To generate pGL3-lncRNA 1308 constructs, the sequence of lncRNA 1308 mRNA was amplified by the primers listed in Table S1. The fragments were inserted into pGL3 with the designed cutting sites KpnI and XhoI. The transfections were performed with INTERFERin reagent (Polyplus-transfection). The final concentration of plasmids was 100 ng.
microRNA in vivo precipitation (miRIP) assay
All the following experiments were performed as previously described.20 The probes that were used are listed in Table S2. Cells were transfected by the biotin-tagged specific probe or control probe and harvested 24 hours after transfection. Cells were cross-linked by formaldehyde, equilibrated in glycine buffer, and scraped with lysis buffer. Cell samples were sonicated and then centrifuged. The supernatant was transferred to a 2 mL tube and 50 µL was separately saved for analysis. The supernatant lysate was incubated with M-280 beads for 1 hour while rotating. The beads–sample mixture was washed and incubated to reverse the formaldehyde cross-links. Subsequently, RNAs were repurified by TRIzol. A multiplex miRNA array was used to analyze the sequences of 184 miRNAs simultaneously from a single RT-PCR reaction to discover the associated miRNAs that would target lncRNA 1308 in XWLC cells (the primers used for multiplex miRNA array are provided in Table S3).
In vitro invasion assays
The 24-well transwell chambers coated with Matrigel (BD Biosciences, San Jose, CA, USA) were used to determine the invasive ability of lung cancer cells.21 Transfected cells in serum-free medium were seeded at the concentration of 5×104 in the top chamber, and the RPMI-1640 containing 10% FBS as a chemoattractant was filled in the bottom chamber. The chamber was sustained at 37°C in a humidified incubator containing 5% CO2. Cells that migrated to the underside of the membrane were fixed with 4% paraformaldehyde (Sigma-Aldrich Co., St Louis, MO, USA), stained with crystal violet (Beyotime, Shanghai, China), imaged, and counted using a microscope (Leica Microsystems, Wetzlar, Germany) after 24 hours. All experiments were performed in triplicate.
Gel electrophoresis and immunoblotting
Cells were lysed in RIPA lysis buffer (Beyotime) containing 1% protease inhibitor Cocktail (Sigma-Aldrich Co.). Protein extract (20 µg) from the cell lysis was separated in a 4%–20% Tris-glycine gel and then transferred to a polyvinylidene fluoride (PVDF) membrane (Immobilon-P transfer membranes; EMD Millipore, Billerica, MA, USA). After blocking with 5% non-fat dry milk in PBS with 0.1% Tween-20 for 2 hours at room temperature, the blots were probed with 1:200 dilution of primary antibody against ADAM 15 (ab12698; Abcam, Cambridge, UK) and β-actin (c-11; Santa Cruz Biotechnology Inc., Dallas, TX, USA) overnight at 4°C. The blots were then probed with a 1:750 dilution of secondary antibodies for 1 hour at room temperature, followed by washing in PBS with 0.1% Tween-20 and detected using an Odyssey Scanning system.
Statistical analyses
SPSS 16.0 statistical software was used to perform statistical analyses. Data are shown as mean ± SD. Student’s t-test or non-parametric Mann–Whitney U test was used to analyze the differences between the 2 groups, and multiple comparisons were assessed by 1-way ANOVA followed by Dunnett’s test. Statistical significance was defined as the P-value less than 0.05.
Results
The novel lncRNA 1308 is upregulated in NSCLC tissues and cell lines
We performed a human lncRNA expression microarray on 15 pairs of NSCLC and adjacent non-cancerous lung tissues. Since the threshold had been set for upregulated and downregulated genes at a fold change of ≥2.0 and a P-value of <0.05, a total of 913 upregulated and 2,253 downregulated lncRNAs were identified in NSCLC samples compared with adjacent normal lung tissues, as depicted in the heat map (Figure 1). The top 10 upregulated lncRNAs in NSCLC tissues are listed in Table 1. We selected lncRNA 1308 for the subsequent analysis. For further validation, the expression levels of lncRNA 1308 were also investigated by using qRT-PCR assay in 40 pairs of lung cancerous, paracancerous tissues, and 5 cell lines. As shown in Figure 2A, the expression level of lncRNA 1308 was significantly increased in NSCLC tissues compared with that in adjacent normal tumor tissues. In addition, the expression of lncRNA 1308 was also upregulated in NSCLC cell lines, including A549, H1299, H1975, XWLC, and GLC, compared with that in the human lung normal cell line BESA-2B (Figure 2B).
Figure 1.
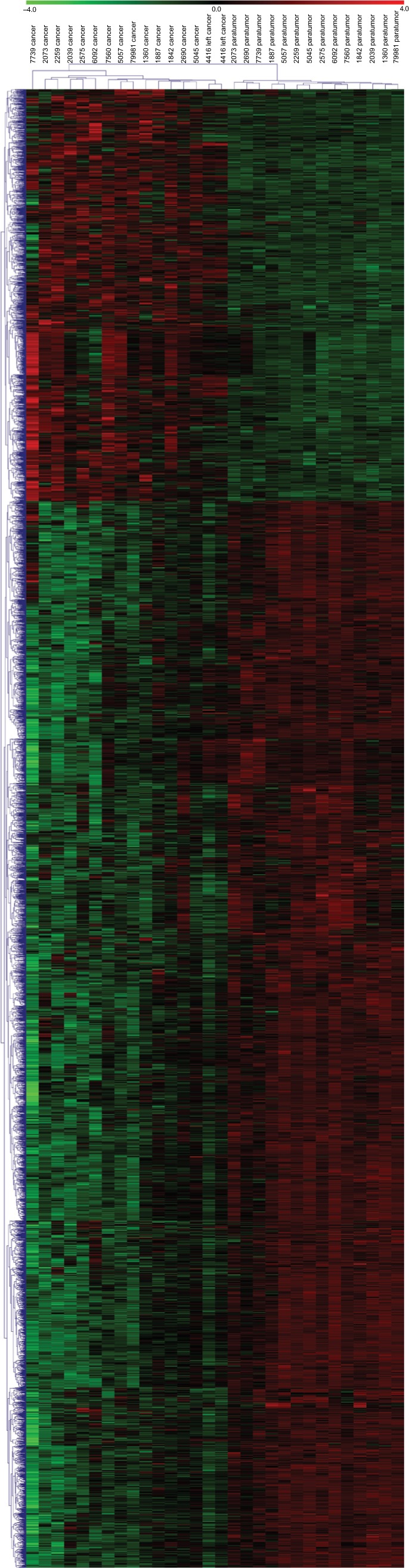
lncRNAs are systemically dysregulated in NSCLC tissues.
Notes: Heatmap of expression profiles for the 3,166 lncRNAs showed significant expression changes (913 upregulated and 2,253 downregulated).
Abbreviations: lncRNA, long non-coding RNA; NSCLC, non-small-cell lung cancer.
Table 1.
Top 10 upregulated lncRNAs in NSCLC tissues
| Probe name | Gene symbol | P-value | FC | Regulation |
|---|---|---|---|---|
|
| ||||
| C_15547 | lncRNA NONHSAG038390 | 2.03E–06 | 28.39 | Up |
| C_79834 | lncRNA 1308 | 7.35E–09 | 23.60 | Up |
| C_1346 | lncRNA SLC32A1 | 8.15E–05 | 11.10 | Up |
| C_64647 | lncRNA ELTD1-2 | 2.55E–05 | 8.69 | Up |
| C_16443 | lncRNA PVT1-108 | 2.10E–08 | 6.37 | Up |
| C_44036 | lncRNA PIK3C2G-2 | 1.59E–06 | 5.90 | Up |
| C_87304 | lncRNA NONHSAG047350 | 3.60E–06 | 5.68 | Up |
| C_2609 | lncRNA TSSK2-2 | 1.92E–05 | 5.62 | Up |
| C_89598 | lncRNA OR4Q3-3 | 0.001056 | 5.05 | Up |
| C_33744 | lncRNA EFR3A-5 | 1.12E–07 | 4.98 | Up |
Abbreviations: FC, fold change; lncRNA, long non-coding RNA; NSCLC, non-small-cell lung cancer.
Figure 2.
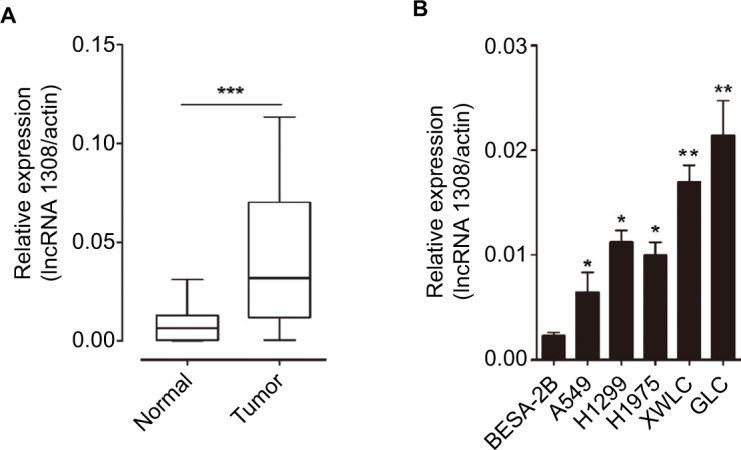
lncRNA 1308 is upregulated in lung cancer samples and cell lines.
Notes: (A) Expression of lncRNA 1308 was measured in 40 lung cancer samples and matched to adjacent normal tissues by qRT-PCR, and the expression levels of lncRNA 1308 were normalized to actin RNA expression for subsequent analyses. (B) The expression levels of lncRNA 1308 were measured in human BESA-2B cell lines and 5 lung cancer cell lines (A549, H1299, H1975, XWLC, and GLC) by qRT-PCR, and the expression levels of lncRNA 1308 were normalized to actin RNA expression for subsequent analyses. Results are presented as mean ± SD. *P<0.05, **P<0.01, and ***P<0.001.
Abbreviations: GLC, Gejiu lung cancer; lncRNA 1308, long non-coding RNA 1308; qRT-PCR, quantitative reverse transcription PCR; XWLC, Xuanwei lung cancer.
Downregulation of lncRNA 1308 inhibits cell invasion in NSCLC cells
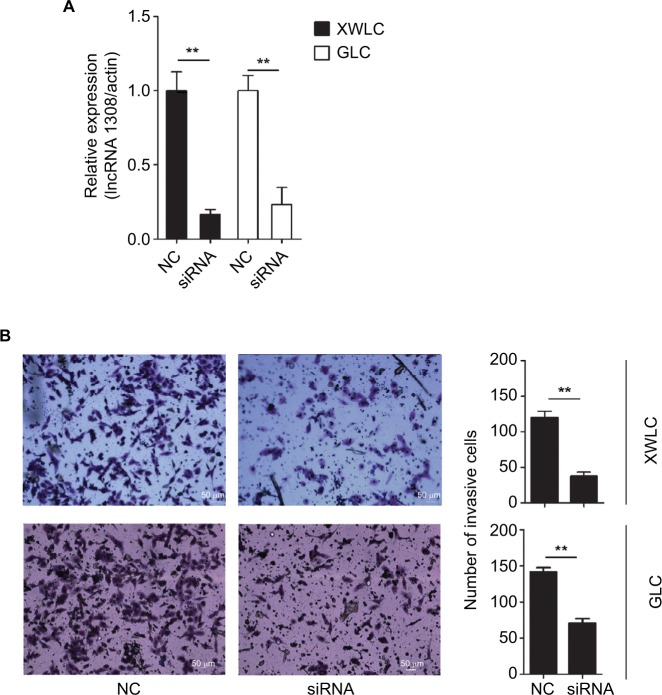
Loss-of-function studies were performed to investigate the effect of lncRNA 1308 on lung carcinogenesis in NSCLC cells, XWLC and GLC cell lines, which were isolated from the NSCLC tissues of the patients from Xuanwei and Gejiu, respectively. lncRNA 1308 siRNA and negative control oligonucleotides were transiently transfected into 2 specific lung cancer cell lines: XWLC and GLC cell lines. Results from qRT-PCR revealed a significant decrease in the expression level of lncRNA 1308 (Figure 3A). Furthermore, the functional role of lncRNA 1308 in the invasion of NSCLC cells was also investigated by performing invasion assays. Silencing of lncRNA 1308 in XWLC and GLC cells resulted in significant suppression of cell invasion capacity (Figure 3B). These data suggest that lncRNA 1308 modulated NSCLC cell viability and that the downregulation of lncRNA 1308 inhibited NSCLC cell invasion capacity.
Figure 3.
Downregulation of lncRNA 1308 suppresses cell invasion in lung cancer cells.
Notes: (A) lncRNA 1308 expression was quantified by qRT-PCR analysis. lncRNA 1308 expression was interfered in both XWLC and GLC cells. (B) The invasive ability of XWLC and GLC cells was evaluated by in vitro invasion assays after transfection of NC and siRNA of lncRNA 1308. Results are presented as mean ± SD. **P<0.01. Magnification 100×.
Abbreviations: GLC, Gejiu lung cancer; lncRNA 1308, long non-coding RNA 1308; qRT-PCR, quantitative reverse transcription PCR; NC, negative control oligonucleotides; XWLC, Xuanwei lung cancer.
lncRNA 1308 directly interacts with miR-124
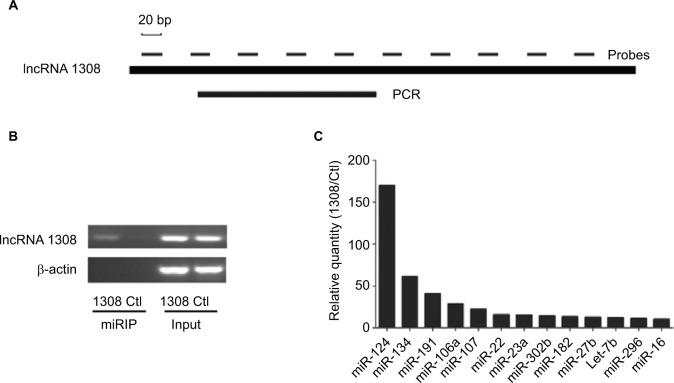
lncRNAs could serve as ceRNAs to sponge miRNAs, thereby regulating gene expression.14 To understand the mechanism by which lncRNA 1308 suppresses lung cancer cell invasion capacity, the miRIP method was used to identify the targeting miRNAs on lncRNA 1308.20 Ten probes of lncRNA 1308 were transfected into XWLC cells to hybridize with lncRNA 1308 mRNA for 24 hours (Figure 4A). The associated mRNAs of lncRNA 1308 were purified using Streptavidin Dynabeads (M-280) and isolated by TRIzol. RT-PCR detection of mRNA extracted from the transfected XWLC cells demonstrated a specific enrichment for the mRNAs of lncRNA 1308 (Figure 4B). Moreover, a multiplex miRNA array was used to analyze the sequences of 184 miRNAs simultaneously from a single RT-PCR reaction to discover the associated miRNAs that would target lncRNA 1308 in XWLC cells. miR-124, which interacted with lncRNA 1308 mRNA most robustly, was selected for the next immediate research (Figure 4C; Table 2).
Figure 4.
Affinity purification of lncRNA mRNA by miRIP in XWLC cells.
Notes: (A) lncRNA 1308 mRNA, probe binding sites, and region analyzed by qRT-PCR. (B) RT-PCR analysis of lncRNA 1308 mRNAs, from lncRNA 1308 probe affinity purification, in XWLC cells. (C) qRT-PCR of multiplex miRNA data of lncRNA 1308 probe affinity purification, in XWLC cells. Data shown are normalized values for the enriched miRNAs (>10 fold).
Abbreviations: Ctl, control probe; lncRNA 1308, long non-coding RNA 1308; miRIP, microRNA in vivo precipitation; miRNA, microRNA; qRT-PCR, quantitative reverse transcription PCR; XWLC, Xuanwei lung cancer.
Table 2.
Relative miRIP isolation of miRNAs in XWLC cells using lncRNA 1308 probes (lncRNA1308/control >10)
| miRNA | lncRNA 1308/control |
|---|---|
|
| |
| miR-124 | 170.1 |
| miR-134 | 61.4 |
| miR-191 | 40.8 |
| miR-106a | 28.4 |
| miR-107 | 22.3 |
| miR-22 | 15.5 |
| miR-23a | 15.2 |
| miR-302b | 13.8 |
| miR-182 | 13.0 |
| miR-27b | 12.6 |
| let-7b | 11.9 |
| miR-296 | 10.9 |
| miR-16 | 10.1 |
Abbreviations: lncRNA, long non-coding RNA; miRIP, microRNA in vivo precipitation; miRNA, microRNA.
lncRNA1308 is a target of miR-124 and controls the miR-124 target ADAM 15
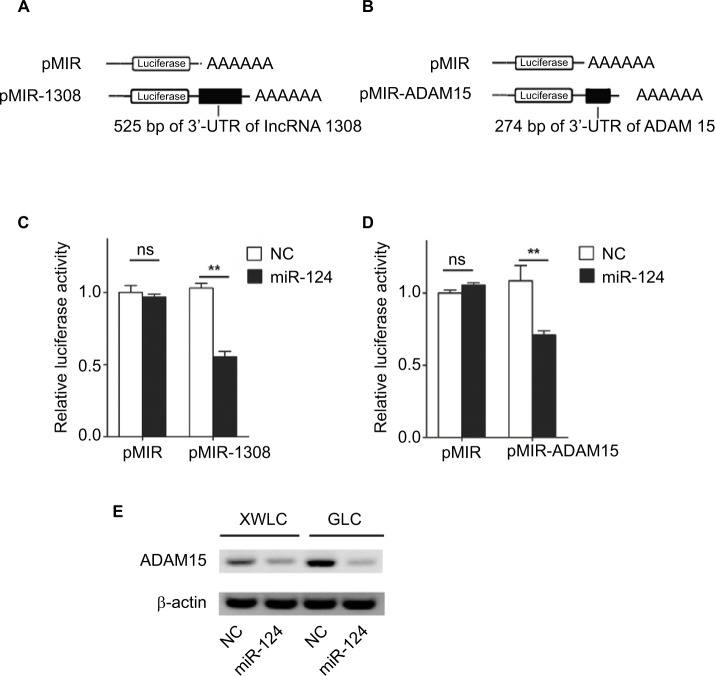
According to the data from DIANA-TarBase v8.0 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex/), miR-124 was found to target ADAM 15, which was associated with multiple cellular functions including metastatic progression.22 Therefore, we assumed that lncRNA 1308 could act as a ceRNA for ADAM 15 in NSCLC cells. To validate our hypothesis, first, luciferase reporter assays were performed in HEK 293 T cells. We cloned lncRNA 1308 and ADAM 15 3¢-UTR sequence, as indicated in Figure 5A and B, into the luciferase reporter plasmid. Second, these recombinant luciferase reporter plasmids were cotransfected with miR-124 mimics or negative control oligonucleotides into HEK 293 T cells for 36 hours, and the luciferase activity was measured in these transfected cells. As shown in Figure 5C and D, miR-124 transfection caused a significant decrease in luciferase activity in cells transfected with the reporter plasmid with lncRNA 1308 and ADAM 15 3¢-UTR sequence, but no change in the cells transfected with reporter plasmids without the targeting sequence. Third, we explored whether endogenous ADAM 15 in lung cancer cells was regulated by a similar mode. XWLC and GLC cells were transfected with miR-124 mimics or negative control oligonucleotides. The results of Western blotting showed that the ADAM 15 protein level was consistently and substantially downregulated by miR-124 (Figure 5E). These results suggest that both lncRNA 1308 and ADAM 15 are directly targeted by the shared microRNA, which is miR-124.
Figure 5.
lncRNA 1308 is a target of miR-124 and miRNA that represses protein levels of ADAM 15
Notes: (A and B) Schematic representation of pMIR firefly luciferase reporter construction. (A) A 525-bp lncRNA 1308 sequence was cloned and constructed in pMIR firefly luciferase reporter. (B) 274 bp of ADAM 15 3′-UTR sequence was cloned and constructed in pMIR firefly luciferase reporter. (C and D) Analysis of luciferase activity. HEK 293T cells were co-transfected with pMIR firefly luciferase reporter plasmids and pTK-Renilla luciferase plasmids, together with negative control and miR-124 mimics. After 36 h, firefly luciferase activity was measured and normalized by Renilla luciferase activity. (E) Effects of miR-124 mimics on the endogenous ADAM 15 protein levels. XWLC and GLC cells were cotransfected with miR-124 mimics or NC. Forty-eight hours after transfection, the cells were isolated, and the expression of ADAM 15 was analyzed by Western blotting. Results were represented as mean ± SD. **P<0.01.
Abbreviations: ADAM15, a disintegrin and a metalloproteinase 15; GLC, Gejiu lung cancer; lncRNA 1308, long non-coding RNA 1308; miRNA, microRNA; NC, negative control oligonucleotides; ns, not significant; XWLC, Xuanwei lung cancer.
lncRNA 1308 acted as a ceRNA for ADAM 15 to inhibit cell invasion in lung cancer cells
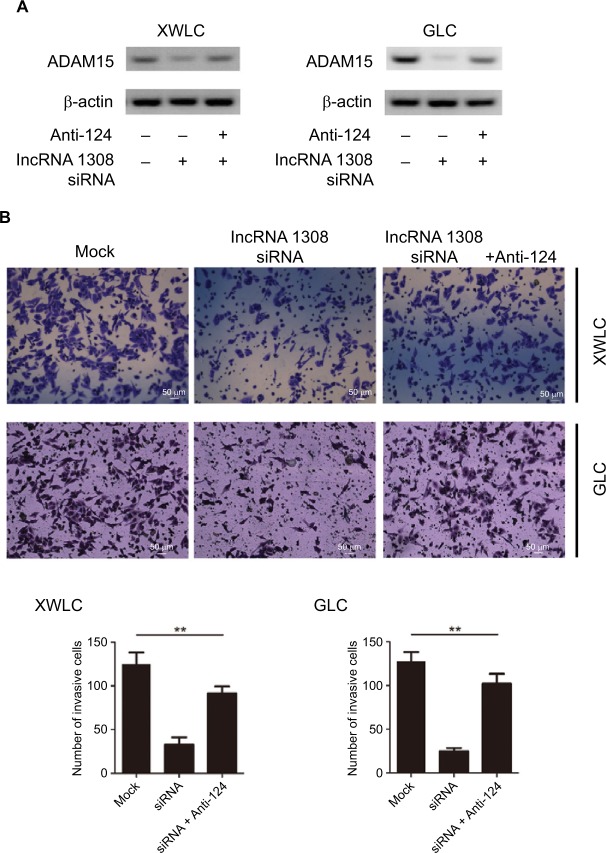
We found that lncRNA 1308 could promote cell invasion of lung cancer cells and both lncRNA 1308 and ADAM 15 are directly targeted by the shared miR-124. Therefore, we hypothesized that lncRNA 1308 could regulate cell invasion in NSCLC cells by acting as a ceRNA for ADAM 15. As expected, the interference of lncRNA 1308 in XWLC and GLC cells decreased the accumulation of ADAM 15, whereas inhibiting miR-124 expression in XWLC and GLC cells markedly increased the expression of ADAM 15 (Figure 6A). In line with these results, knockdown of lncRNA 1308 significantly inhibited cell invasion in XWLC and GLC cells, while it inhibited miR-124 expression in XWLC and GLC cells, which markedly alleviated the inhibition of lncRNA1308-induced cell invasion (Figure 6B). Taken together, these results demonstrate that lncRNA 1308 could regulate lung cancer cell invasion through acting as a ceRNA for ADAM 15.
Figure 6.
lncRNA 1308 acts as a ceRNA to regulate ADAM 15 levels and suppresses cell invasion in lung cancer cells.
Notes: (A) lncRNA 1308 acts as a ceRNA to inhibit ADAM 15 expression in lung cancer cells. XWLC and GLC cells were transfected with lncRNA 1308 siRNA and/or infected with miR-124 inhibitors, and the expression of ADAM 15 was analyzed by Western blotting. (B) lncRNA 1308 acts as a ceRNA to inhibit cell invasion. XWLC and GLC cells were transfected with lncRNA 1308 siRNA and/or infected with miR-124 inhibitors, and the invasive ability of cells was evaluated by in vitro invasion assays. Results are presented as mean ± SD. **P<0.01. Anti-124, miR-124 inhibitor. Magnification 100×.
Abbreviations: ADAM15, a disintegrin and a metalloproteinase 15; ceRNA, competing endogenous RNA; GLC, Gejiu lung cancer; lncRNA 1308, long non-coding RNA 1308; XWLC, Xuanwei lung cancer.
Discussion
Recently, the roles of lncRNAs have attracted much attention, especially carcinogenesis and cancer progression.23 As their expression and function in NSCLC development remain unclear, we screened the differentially expressed lncRNAs among NSCLCs, matched non-tumor lung tissues through chip microarray, and focused on the roles and underlying mechanisms of aberrant lncRNAs expression in NSCLC progression. We first identified lncRNA 1308, which was overexpressed in NSCLC tissue samples. Consistently, the expression level of lncRNA 1308 was obviously higher in NSCLC cell lines compared to that of the normal cell lines. Mechanistic analysis revealed that knockdown of lncRNA 1308 inhibits NSCLC cell invasion, and lncRNA 1308 affects the interaction between miR-124 and ADAM 15 3′-UTR to regulate the expression of ADAM 15 by functioning as a ceRNA, thereby contributing to NSCLC progression. These results indicate that lncRNA 1308 plays a vital role in the modulation of oncogenic properties and the disease progression of NSCLC in a direct manner; these may be new research directions and become potential therapeutic options that consider lncRNA 1308 as a novel prognostic marker and therapeutic target in NSCLC.
Although thousands of lncRNAs have recently been identified, the functional characterization of lncRNAs has just begun. Currently, lncRNAs have been reported to regulate various cellular functions through different molecular mechanisms such as chromatin modification, transcriptional regulation, and post-transcriptional regulation.8,24 Recently, lncRNAs have been shown to serve as ceRNAs to sponge miRNAs, consequently modulating the derepression of miRNA targets.14,23 For example, Liu et al25 found that lncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells by upregulating EZH2. lncRNA BC032469 has been identified as a tumor-related molecule in gastric cancer by interfering miR-1207-5p.26 Together with miRNAs and their target gene, lncRNA–miRNA–mRNA axis may be an extensive regulatory network in gene expression, and their dysregulation may lead to disease progression including the development of cancer. This presumption may lead to interesting future studies that elucidate the regulatory network of non-coding RNAs and coding genes in cancer biology. Currently, candidate lncRNAs, and miRNAs targeted by lncRNAs, are mostly predicted and selected by literature, which seems to lack foresight. Precisely, determining the candidate lncRNAs that are involved in NSCLC progression, and which specific miRNA binds to a certain lncRNA, is important for investigating the mechanisms of lncRNA functions. Recently, Su et al20 reported a new method, called miRIP, to identify which type of miRNAs can bind to the specific mRNA in a cell. By using the miRIP approach, the potential interaction between miR-92a and p21, and between miR-197 and STAT3, was elucidated in recent studies.20,27 In this study, by completing lncRNA expression microarray analysis, followed by performing RIP using a lncRNA 1308-specific probe, we found that miR-124 was the lncRNA 1308-binding miRNA, and we then proved that lncRNA 1308 functioned as the sponge of the miR-124. Thus, the miRIP approach would be a practical and useful method to identify the true and definite interaction between miRNAs and lncRNAs, especially considering that the lncRNA–miRNA interaction network is relatively complicated. Previous studies have shown that miR-124 is deregulated in several cancers.28,29 Sun et al28 found that downregulation of miR-124 was significantly associated with poor prognosis in patients with pancreatic ductal adenocarcinoma. Interestingly, the recent evidence further showed that miR-124 can suppress NSCLC cell migration and invasion activity by targeting zinc finger e-box binding homeobox 1 (ZEB1).29 Thus, these results suggest that miR-124 may function as an important regulatory gene in the development of NSCLC.
By using bioinformatics analysis, ADAM 15 was predicted to be a downstream target of miR-124. In this study, a luciferase reporter assay and rescue experiment confirmed that miR-124 targets 3′-UTR of ADAM 15 to suppress the expression of ADAM 15 and inhibit NSCLC invasion. ADAM 15 is a membrane-associated proteinase belonging to the ADAM family.19 The ADAMs are members of the metzincin superfamily of matrix metalloproteinases. To date, 21 functional ADAMs have been described in humans and 40 have been described in different organisms.18 Recently, specific ADAMs were implicated in many diseases, including rheumatoid arthritis, atherosclerosis, and cancer.18,30,31 As a catalytic active member of the ADAM family, ADAM 15 is the only ADAM containing an arginine–glycine–aspartic acid (RGD) motif in the disintegrin domain. The RGD motif of ADAM 15 interacts with integrins αvβ3 and α5β1 to regulate cell adhesion and motility.19 ADAM 15 is widely expressed in normal tissues and cancer cell lines. Overexpression of ADAM 15 has been reported in many solid tumors including those of breast, lung, colorectal, ovarian, and prostate cancers.17,19,32,33 It is believed that the tumor-promoting action of ADAM 15 results from disruption of cell–cell and cell–matrix adhesion and from release of membrane-bound growth factors.22,34,35 Accordingly, ADAM 15 deregulation has been linked to poor patient outcome in lung and colon cancers.19,32 Thus, there is accumulating evidence that ADAM 15 may play a key role in cancer biology. Furthermore, functional studies have indicated that ADAM 15 could support prostate cancer metastasis by modulating tumor cell–endothelial cell interaction in PC-3 prostate cancer cells,22 and Dong et al19 demonstrated that ADAM 15 directly activates MMP9 to promote cell migration and invasion in lung cancer cells. Consistently, our results showed that interference of lncRNA 1308 in XWLC and GLC cells decreased the accumulation of ADAM 15 and inhibited cell invasion, whereas inhibited miR-124 expression in XWLC and GLC cells markedly increased the expression of ADAM 15 and alleviated cell invasion inhibition. Therefore, our data not only supported a pro-metastatic role of ADAM 15 in NSCLC progression but also revealed a novel mechanism of lncRNA in promoting cancer cell invasion through indirectly targeting ADAM 15 activation.
Conclusion
We first identified that lncRNA 1308, a novel lncRNA that was significantly overexpressed in NSCLC tissues and cells, competitively binds to miR-124 and subsequently upregulated the expression of its target gene ADAM 15 to promote the invasion of NSCLC cells. This lncRNA 1308/miR-124/ADAM 15 regulatory network may shed light on tumorigenesis in NSCLCs, and it may be valuable for the development of novel diagnostic and treatment approaches for NSCLC.
Supplementary Materials
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Nos. 81860494, 81860499, 81460435, and 81860638), the Yunnan Applied Basic Research project (No. 2017FB138), and the Science Research Foundation of the Yunnan Province Education Department (No. 2016ZZX014).
Footnotes
Author contributions
HL Li, CJ Xiao, and SY Zheng conceived and designed the experiments; HL Li, XP Guo, QT Li, and PZ Ran performed the experiments; XD Xiang, YC Yuan, TQ Dong, and B Zhu analyzed the data; L Wang, FF Li, CY Yang, DC Mu, and D Wang contributed reagents/materials/analysis tools; HL Li and SY Zheng wrote the paper. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.She J, Yang P, Hong Q, Bai C. Lung cancer in China: challenges and interventions. Chest. 2013;143(4):1117–1126. doi: 10.1378/chest.11-2948. [DOI] [PubMed] [Google Scholar]
- 2.Barone-Adesi F, Chapman RS, Silverman DT, et al. Risk of lung cancer associated with domestic use of coal in Xuanwei, China: retrospective cohort study. BMJ. 2012;345:e5414. doi: 10.1136/bmj.e5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Xue Q, Pan G, et al. Integrated Analysis of Genome-Wide Copy Number Alterations and Gene Expression Profiling of Lung Cancer in Xuanwei, China. PLoS One. 2017;12(1):e0169098. doi: 10.1371/journal.pone.0169098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H, Meng S, Xu Q, et al. Gene expression profiling of lung adenocarcinoma in Xuanwei, China. Eur J Cancer Prev. 2016;25(6):508–517. doi: 10.1097/CEJ.0000000000000214. [DOI] [PubMed] [Google Scholar]
- 5.Chen JH, Zhou LY, Xu S, Zheng YL, Wan YF, Hu CP. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017;17:64. doi: 10.1186/s12935-017-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sève P, Reiman T, Dumontet C. The role of betaIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer. 2010;67(2):136–143. doi: 10.1016/j.lungcan.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Wang SH, Zhang WJ, Wu XC, et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7(25):37857–37867. doi: 10.18632/oncotarget.9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Huang Y, Huang Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36(1):51. doi: 10.1186/s13046-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi X, Ma C, Zhu Q, et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. 2016;7(18):25558–25575. doi: 10.18632/oncotarget.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Zhang R, Zhao J. The Novel Long Noncoding RNA TUSC7 Inhibits Proliferation by Sponging MiR-211 in Colorectal Cancer. Cell Physiol Biochem. 2017;41(2):635–644. doi: 10.1159/000457938. [DOI] [PubMed] [Google Scholar]
- 14.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng W, Si S, Zhang Q, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burdelski C, Fitzner M, Hube-Magg C, et al. Overexpression of the A Disintegrin and Metalloproteinase ADAM15 is linked to a Small but Highly Aggressive Subset of Prostate Cancers. Neoplasia. 2017;19(4):279–287. doi: 10.1016/j.neo.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy MJ, Mckiernan E, O’Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin Cancer Res. 2009;15(4):1140–1144. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- 19.Dong DD, Zhou H, Li G. ADAM15 targets MMP9 activity to promote lung cancer cell invasion. Oncol Rep. 2015;34(5):2451–2460. doi: 10.3892/or.2015.4203. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Wang H, Ge W, et al. An In Vivo Method to Identify microRNA Targets Not Predicted by Computation Algorithms: p21 Targeting by miR-92a in Cancer. Cancer Res. 2015;75(14):2875–2885. doi: 10.1158/0008-5472.CAN-14-2218. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G, Jiang G, Wang C, et al. Decreased expression of microRNA-320a promotes proliferation and invasion of non-small cell lung cancer cells by increasing VDAC1 expression. Oncotarget. 2016;7(31):49470–49480. doi: 10.18632/oncotarget.9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Najy AJ, Day KC, Day ML. ADAM15 supports prostate cancer metastasis by modulating tumor cell-endothelial cell interaction. Cancer Res. 2008;68(4):1092–1099. doi: 10.1158/0008-5472.CAN-07-2432. [DOI] [PubMed] [Google Scholar]
- 23.Yang C, Wu D, Gao L, et al. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7(12):13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, Li Y, Luo G, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Lü MH, Tang B, Zeng S, et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35(27):3524–3534. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Su X, Yang M, et al. Reciprocal control of miR-197 and IL-6/STAT3 pathway reveals miR-197 as potential therapeutic target for hepatocellular carcinoma. Oncoimmunology. 2015;4(10):e1031440. doi: 10.1080/2162402X.2015.1031440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun B, Liu X, Gao Y, Li L, Dong Z. Downregulation of miR-124 predicts poor prognosis in pancreatic ductal adenocarcinoma patients. Br J Biomed Sci. 2016;73(4):152–157. doi: 10.1080/09674845.2016.1220706. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Wang X, Li W, Wu L, Chang L, Chen H. miRNA-124 modulates lung carcinoma cell migration and invasion. Int J Clin Pharmacol Ther. 2016;54(8):603–612. doi: 10.5414/CP202551. [DOI] [PubMed] [Google Scholar]
- 30.Kossmann CM, Annereau M, Thomas-Schoemann A, et al. ADAM9 expression promotes an aggressive lung adenocarcinoma phenotype. Tumour Biol. 2017;39(7):101042831771607. doi: 10.1177/1010428317716077. [DOI] [PubMed] [Google Scholar]
- 31.Dosch J, Ziemke E, Wan S, et al. Targeting ADAM17 inhibits human colorectal adenocarcinoma progression and tumor-initiating cell frequency. Oncotarget. 2017;8(39):65090–65099. doi: 10.18632/oncotarget.17780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toquet C, Colson A, Jarry A, et al. ADAM15 to α5β1 integrin switch in colon carcinoma cells: a late event in cancer progression associated with tumor dedifferentiation and poor prognosis. Int J Cancer. 2012;130(2):278–287. doi: 10.1002/ijc.25891. [DOI] [PubMed] [Google Scholar]
- 33.Maretzky T, Le Gall SM, Worpenberg-Pietruk S, et al. Src stimulates fibroblast growth factor receptor-2 shedding by an ADAM15 splice variant linked to breast cancer. Cancer Res. 2009;69(11):4573–4576. doi: 10.1158/0008-5472.CAN-08-4766. [DOI] [PubMed] [Google Scholar]
- 34.Lucas N, Day ML. The role of the disintegrin metalloproteinase ADAM15 in prostate cancer progression. J Cell Biochem. 2009;106(6):967–974. doi: 10.1002/jcb.22087. [DOI] [PubMed] [Google Scholar]
- 35.White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15(5):598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.