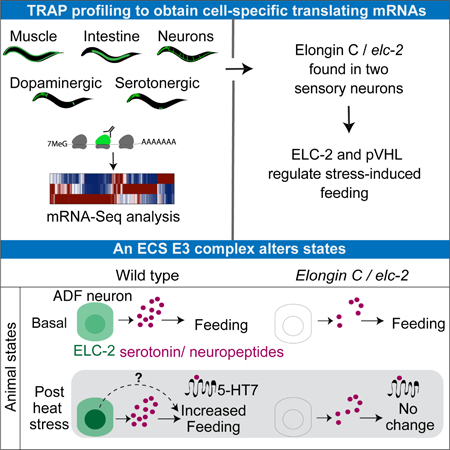
Summary
Neuromodulatory cells transduce environmental information into long lasting behavioral responses. However, the mechanisms governing how neuronal cells influence behavioral plasticity are difficult to characterize. Here, we adapted the Translating Ribosome Affinity Purification (TRAP) approach in C. elegans to profile ribosome-associated mRNAs from three major tissues and the neuromodulatory dopaminergic and serotonergic cells. We identified elc-2, an Elongin C ortholog, specifically expressed in stress-sensing ADF serotonergic sensory neurons, and found that it plays a role in mediating a long-lasting change in serotonin-dependent feeding behavior induced by heat stress. We demonstrate that ELC-2 and the von Hippel-Lindau protein VHL-1, components of an Elongin-CullinSOCS-box (ECS) E3 ubiquitin ligase, modulate this behavior after experiencing stress. Also, heat stress induces a transient redistribution of ELC-2, becoming more nuclearly enriched. Together, our results demonstrate dynamic regulation of an E3 ligase, and a role for an ECS complex in neuromodulation and control of lasting behavioral states.
Graphical Abstract
Introduction
Animals alter their behavior to cope with the environment. One common type of environment-induced behavioral change is the response to stressors — stimuli that pose or signal a threat to the fitness of an individual. In vertebrates upon exposure to stress, endocrine glands and the brain release chemicals that often serve as “molecular representations” of the external environment (Joels and Baram, 2009, McEwen et al., 2015). These molecules include a broad class of neuromodulators. Fundamental work in the stomatogastric ganglion in the crab Cancer borealis and other animals showed that neuromodulators widely impact function of neurons and neural networks, which often result in short-term or long-term changes in behavior (Katz and Harris-Warrick, 1990, Marder, 2012, Bargmann and Marder, 2013).
In mammals an increased release in the neuromodulator serotonin is thought to be instrumental in regulating fear, anxiety, and lasting states such as social behaviors and food intake in response to stress (Chaouloff, 2000, Maier and Watkins, 2005). In addition, serotonin neurons can also encode and signal positive rewards (Cohen et al., 2015, Li et al., 2016). Despite the known role of serotonin in poststress responses, the mechanisms underlying the response of serotonergic neuromodulatory cells to environmental stressors are not well understood.
Regulated protein turnover by ubiquitin and the proteasome is a rapid mechanism for activation or inhibition of signaling pathways when cells respond to the environment (Ciechanover, 2006). In the nervous system of Aplysia, ubiquitin-dependent protein degradation regulates a classic example of neuronal plasticity called long-term facilitation (Yamamoto et al., 1999, Hegde and DiAntonio, 2002). In the neurological disorder Angelman Syndrome, mutations in the E3 ubiquitin ligase UBE3A affect synapse development and they are implicated in mental retardation, highlighting the importance of E3 complexes for neuronal functions (Greer et al., 2010). Elongin C, Cullins and SOCS-box proteins form an E3 ubiquitin ligase termed the ECS complex (Pause et al., 1997, Maxwell et al., 1999, Stebbins et al., 1999). Elongin C contains an Skp1-domain, and it was initially biochemically characterized as a regulatory protein of the Elongin transcription elongation complex (Bradsher et al., 1993, Aso et al., 1995). In the ECS complex Elongin C and the small ubiquitin-like protein Elongin B connect the cullins CUL2 or CUL5 to the substrate recognition module that contains a SOCS-box domain (Kamura et al., 2004). The von Hippel-Lindau tumor suppressor protein pVHL is one of the best-characterized substrate recognition modules of the ECS complex (Okumura et al., 2012). The interaction between Elongin C and pVHL is critical for ubiquitin ligase activity (Stebbins et al., 1999). Mutations in either Elongin C or pVHL genes that disrupt their interaction and function are a leading cause of a type of renal cell cancer and blood vessel tumors of the central nervous system (Kaelin and Maher, 1998, Sato et al., 2013, Hakimi et al., 2015). Despite the critical functions of Elongin C, its essential role in early development has prevented mechanistic characterization of ECS E3 ligases in vivo in specific cell types in mature animals (Sasagawa et al., 2005, Wang et al., 2015).
The nematode C. elegans serves as an excellent model system to study stress, ubiquitin-mediated pathways, neuromodulation and serotonin (Kipreos et al., 1996, Liao et al., 2004, Avery and You, 2012, Taghert and Nitabach, 2012, Bargmann and Marder, 2013, Lemieux and Ashrafi, 2015). Several stressors induce behavioral changes in C. elegans through well-described serotonergic neurons, including a pair of sensory neurons called ADF. Serotonin production in ADF modulates different downstream networks to generate behavioral responses to various environmental stimuli, such as pathogenic bacteria, presence of novel food, starvation, or heat (Sze et al., 2000, Zhang et al., 2005, Liang et al., 2006, Anderson et al., 2013, Song et al., 2013, Tatum et al., 2015, Lemieux et al., 2015, Jin et al., 2016). Collectively, these studies suggest that similar to mammals, the C. elegans serotonergic neurons respond to a range of stressors to modulate neural circuits underlying multiple behavioral outcomes. However, how the serotonergic neurons respond to different stimuli to mount appropriate behavioral responses is not well understood.
Here, we characterize a function of an ECS E3 ubiquitin ligase complex that confers a specific response to heat stress, resulting in a lasting change in feeding, an essential behavior. We adapted the Translating Ribosome Affinity Purification (TRAP) approach in C. elegans to profile the repertoire of ribosome-associated mRNAs in specific tissues and neuronal subtypes. Using this method, we identified the Elongin C ortholog elc-2 to be expressed specifically in the ADF neurons. We demonstrated that ELC-2 and its interacting ECS complex partner VHL-1 act in this pair of neurons to modulate a change in feeding behavior after experiencing noxious heat stress. Genetic ablation of serotonin production in ADF, loss of a metabotropic serotonin receptor ser-7/5HT7, or loss of unc-31/CAPS dense core vesiclemediated neuromodulation recapitulated behavioral defects observed in ECS component mutants. In addition, we found that ELC-2 distribution is regulated by heat stress, transiently becoming enriched in the nucleus. These results together demonstrate dynamic regulation of Elongin C, and an unexpected role for ECS complex components in stress-induced modulation of behavior.
Results
Cell type-specific translational profiles in C. elegans with TRAP
To identify gene expression patterns in specific tissues and cell types in C. elegans, we adapted the Translating Ribosome Affinity Purification method (TRAP) (Heiman et al., 2008) (Figures 1A and S1). Current strategies for cell type-specific transcriptome profiling in C. elegans, such as mRNA tagging (Von Stetina et al., 2007), and sorting of dissociated cells by fluorescence (Zhang et al., 2002, Spencer et al., 2014, Kaletsky et al., 2016), require crosslinking, micro-dissection or cell sorting protocols that can disrupt physiological conditions. In TRAP, cell type-specific promoters drive expression of GFP-tagged ribosomal large subunit protein L10a, which is encoded by rpl-1 in C. elegans, in genetically defined cell types in vivo (Figure S1A–B). Populations of animals are rapidly collected in the presence of cycloheximide to stall ribosomes on their target mRNA, providing a snapshot of the subset of transcripts that are actively being translated to proteins in particular cell types and states (Heiman et al., 2008).
Figure 1. The Translating-Ribosome Affinity Purification (TRAP) method in C. elegans.
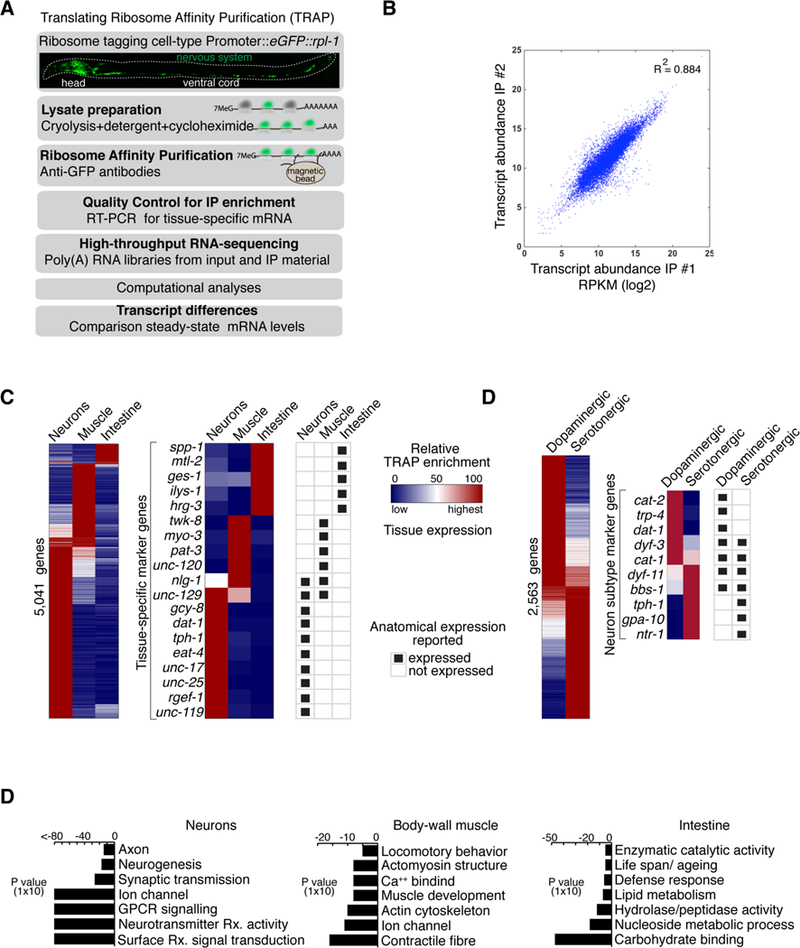
(A) TRAP method.
(B) Scatterplot of transcript expression showing values for two biological replicates of samples collected from pan-neuronal TRAP experiments. Plot shows log2 value of RPKM (Reads Per Kilobase of transcript per Million mapped reads).
(C) Heatmaps display relative tissue enrichment (Materials and Methods) across TRAP data from indicated tissues. Left panel displays all enriched genes. Middle panel highlights a subset of marker genes. Right panel shows previously reported and validated expression patterns from the literature (Wormbase).
(D) Same as in (C) but data is from dopaminergic and serotonergic neurons.
(E) Gene ontology (GO) analysis of genes detected as enriched by TRAP in neuronal cells (left panel), muscle cells (middle panel), and intestinal cells (right panel). Selected over-represented GO terms are displayed, with False-Discovery Rate (FDR) corrected P-values plotted showing significance of enrichment.
See also Figure S1.
We purified ribosome-associated mRNAs and sequenced cDNA libraries to obtain translational profiles from fourth larval stage animals across three major cell types—neurons, intestinal cells, and body wall muscle cells—and validated the reproducibility and specificity of our procedure (Figures 1B and S1C-D; for detailed protocol see (Gracida and Calarco, 2017). We identified hundreds of tissueenriched mRNAs in these cell types (Figure 1C; Table S1). Among the genes corresponding to mRNAs enriched in each tissue, we found significant enrichment for Gene Ontology (GO) terms that reflected the functional differences between these three cell types (Figure 1E). Additionally, we surveyed the literature for reported expression patterns among genes displaying tissue-biased enrichment in our datasets, and found a strong agreement between our TRAP data and previously validated patterns (Figures 1C–D and S1C). Taken together, these observations indicate that our TRAP profiling approach can enrich for transcripts with tissue-specific and biased expression.
We also obtained translational profiles of a smaller subset of cells corresponding to two major neuromodulatory cell types, the eight dopaminergic and six serotonergic neurons (Figure S1B). Importantly, in addition to recovering known marker genes for these neuromodulatory cells (Figure 1D; Table S1), our data also finds dopaminergic or serotonergic neuron-specific enrichment patterns for hundreds of transcripts with no previously reported expression patterns (Table S1). These results indicate that TRAP in C. elegans is a powerful and complementary tool for profiling and discovering genes with tissue- and/or neuron subtype-specific expression patterns in vivo. Our datasets thus provide a systematic view of the translational profiles of two major neuronal cell types in C. elegans, serving a valuable resource for further characterization of these cells.
An ortholog of Elongin C is specifically expressed in a pair of serotonergic neurons
We searched our TRAP datasets for conserved genes that were specifically expressed in serotonergic neurons but were poorly characterized. We identified the mRNA corresponding to the Elongin C gene elc-2 as highly enriched in both the nervous system and in serotonergic neurons (Figures 2A and S2A). Other components of the Elongin and ECS complexes were also identified by TRAP in the serotonergic neurons and in other tissues (Figures 2A and S2A). We confirmed the expression pattern of ELC-2 by creating animals expressing an ELC-2::GFP fusion protein under the control of its native regulatory elements from a recombineered fosmid (Sarov et al., 2012). Intriguingly, we found that ELC-2 was detected in only two neurons in the head, overlapping with the ADF-specific marker gene srh-142 (Figure 2B).
Figure 2. ELC-2, an Elongin C ortholog, is expressed in two serotonergic sensory neurons.
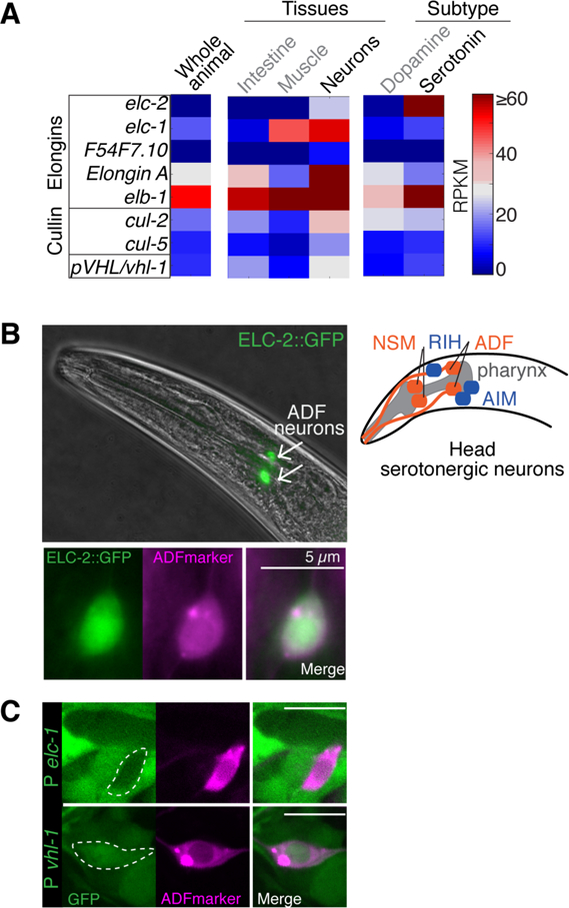
(A) Heatmap depicts relative abundance of Elongin complex and ECS components across tissues, dopaminergic neurons, and serotonergic neurons as detected by TRAP. Abundance values are RPKMs. See also Figure S2A.
(B) Expression of an elc-2 GFP-tagged fosmid in adult animals (top) and the overlapping expression (bottom) with an ADF neuron marker (Psrh-142::rfp). Bright field and fluorescence microscopy. Right: Schematic of the serotonergic neurons (red) and the serotonin-reuptaking neurons (blue) in the head.
(C) GFP transcriptional reporters driven by elc-1 or vhl-1 promoter (P).
elc-1 and elc-2 are Elongin C paralogs (Figure S2B). At the protein level, ELC-1 and ELC-2 are different only at the N-terminal region, where ELC-2 has an additional ~40 residues (Figure S2C). The remaining C terminal region containing the Skp1 domain is highly similar in both proteins, and they both contain conserved amino acids mediating physical interaction with pVHL (Sato et al., 2013) (Figure S2C). Based on transcriptional GFP reporters vhl-1 but not elc-1 is detected in ADF neurons (Figure 2C), suggesting that elc-2 is the principal Elongin C ortholog expressed in ADF. Given the essential role of the more broadly expressed elc-1 in development (Sasagawa et al., 2005), we hypothesized that characterization of elc-2 would reveal a previously unappreciated role for Elongin C in serotonergic neurons.
To characterize the function of elc-2, we generated a null mutant with CRISPR-based genome editing (Figure S3A). The ADF neurons are sensory neurons with ciliated dendritic endings directly exposed to the environment (White et al., 1986, Doroquez et al., 2014). We found that elc-2 mutants have superficially normal ADF neurons with dual ciliated endings (Figure S6A), suggesting that elc-2 does not affect gross ADF development. We also found that elc-2 mutants have a small but significant reduction in the number of head bends (WT: 17±0.5/min, elc-2: 16±0.3/min), while brood sizes are similar to wild type (Figure S3D). Since ADF is a hub for responding to different environmental stressors (Zhang et al., 2005, Hendricks et al., 2012, Anderson et al., 2013, Song et al., 2013, Lemieux et al., 2015, Tatum et al., 2015), the ELC-2 expression pattern suggested that elc-2 could play a role in the function of these neurons in response to external stimuli.
elc-2 in the ADF serotonergic neurons is required for a post-stress change in feeding behavior
The pharyngeal pumping rate in C. elegans is a readout of its feeding behavior. The extent to which an animal eats and the rate at which an animal feeds depend on internal states, external context and previous experiences (You et al., 2008, Lemieux and Ashrafi, 2015). For example, a mild increase in the ambient temperature, from 20°C to 26.7°C, immediately increases the pumping rate (Tatum et al., 2015). We tested if elc-2 is required in two paradigms: increased pumping after starvation (Figure 3A) or in response to mild temperature increase (Figure 3B). Before and after stimulation, wild type and elc2 mutant animals had similar feeding rates, suggesting that elc-2 is dispensable in both of these paradigms (Figure 3A–B). To test whether elc-2 is required in stress-induced behavior, we developed a behavioral paradigm that measured the effect of noxious heat on feeding behavior (Figure 3C). We found that exposure to a higher temperature, a noxious heat at 34°C for 30 minutes first decreases the pumping rate (Figure 3C). Then, after a one-hour recovery at room temperature, the wild-type pumping rate rebounds to a level significantly higher than that in pre-treated animals. elc-2 mutants however, do not change their pumping rate (Figure 3D–E). To test whether loss of elc-2 abolishes or delays the poststress response, we measured the pumping rates of elc-2 mutant animals at different time intervals after recovery and did not find any detectable change even after a three-hour recovery (Figure 3E). These results reveal that elc-2 mutant animals are defective in generating the heat-induced increase in pumping. This change is observed even two hours after the exposure to heat stress, suggesting this poststress effect is long lasting (Figure 3D). Henceforth, we measured the post-heat stress pumping rate after one-hour recovery unless otherwise specified. Introduction of an elc-2::gfp single-copy transgene expressed specifically in ADF was sufficient to restore the heat stress-induced pumping response, whereas a similarly expressed control gfp transgene had no effect (Figures 3E and S3C). Importantly, expressing elc-1 or a human Elongin C transgene specifically in ADF also efficiently rescued the defects in the elc-2 mutant (Figure 3E). These results demonstrate a cell autonomous role in ADF, and an evolutionary conserved function of the Elongin C orthologs in the plasticity of an essential behavior following heat stress. Additionally, our data suggest that elc-2 has a specific role in the response to noxious heat, rather than general role in response to stress.
Figure 3. elc-2 in ADF regulates sustained change in stress-induced feeding behavior.
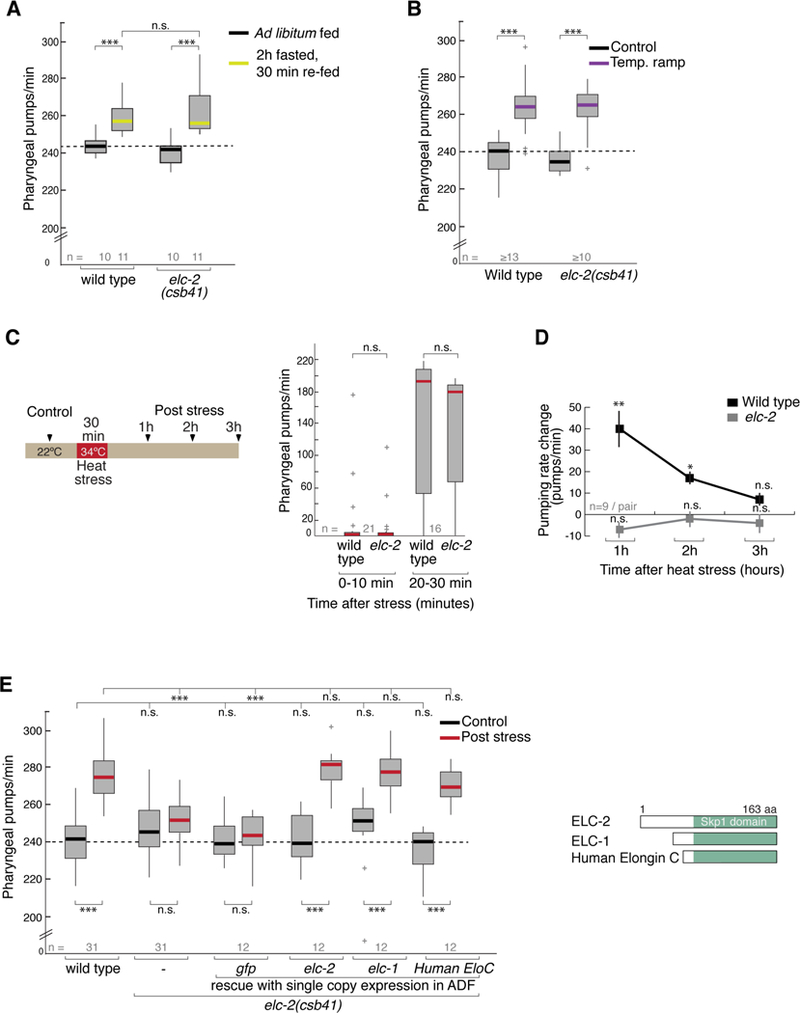
(A) Starvation-induced change in feeding behavior. Pharyngeal pumping rates in control (ad libitum fed) and 30 min after 2 hour fasting (re-fed). Box plot shows 25th and 75th percentiles, bar shows median number of pumps per minute. Genotype is indicated on x-axis. Student’s t test is used *** p<0.0005, ** p<0.005, *p<0.05, n.s. not significant.
(B) Temperature ramp-induced change in feeding behavior. Pharyngeal pumping rates in control (22ºC) and in animals subjected to a temperature ramp from 22ºC to ~26.7 in 5 minutes (Temp. ramp).
(C) Schematic of heat stress paradigm. Control animals are grown at 22ºC. On the right: Pharyngeal pumping rate in the first hour post heat exposure. Box plot outliers +.
(D) Pharyngeal pumping rates before (control) and after recovery from heat shock in wild type and elc2(csb41) knockout mutants. Graph represents the increase in pumps per minute at three time points after heat stress recovery. Student’s t test for each genotype, ** p<0.005, *p<0.05, n.s., not significant, Mean ± SE of the difference (n= 9 animals each condition).
(E) Pharyngeal pumping rates before (control) and after one hour recovery from heat shock (post stress). Genotype is indicated on x-axis. Transgene single-copy expression under ADF-specific promoter (srh-142) in elc-2 mutants is indicated. 1-way ANOVA with Dunnet’s correction is used for comparing across genotypes; Student’s t test is used for comparison within genotype. *** p<0.0005, ** p<0.005, *p<0.05, n.s. not significant. Inset panel depicts schematic of Elongin C protein orthologs used to rescue.
ELC-2 transiently redistributes to the nucleus upon heat stress in a reversible manner
After observing the defect in behavioral plasticity in elc-2 mutants, we analyzed the subcellular distribution of the elc-2::gfp reporter transgene, which rescues the heat-stress response in mutant animals (Figure 4A), before and after exposure to noxious heat. At temperatures between 15°C and 25°C, ELC-2 is distributed throughout the ADF soma in both the nucleus and the cytoplasm (Figures 4A and S4B). Unexpectedly however, we found that upon heat stress ELC-2 becomes enriched in the nucleus (Figure 4A). This nuclear enrichment is transient, as the nuclear-to-cytoplasmic distribution goes back to its basal state ~50 minutes after the experienced stress (Figure 4B). To test if this redistribution was dependent on signaling from other neurons, we assessed ELC-2 localization in unc13/Munc-13 and unc-31/CAPS mutants, which disrupted small vesicle release of neurotransmitters and dense-core vesicle release of neuropeptides, respectively (Richmond et al., 1999, Speese et al., 2007). In each mutant background, ELC-2 nuclear enrichment induced by heat stress still occurs (Figure S4C), indicating that the redistribution of ELC-2 is largely cell-autonomous and independent of intercellular synaptic or peptidergic signals. (Figure S4C). Consistently, the heat-induced redistribution of ELC-2 occurs in mutants that are defective in thermosensory response of the major thermosensory neurons AFD (Mori and Ohshima, 1995, Takeishi et al., 2016) (Figure S4D), indicating that ELC-2 responds to the noxious heat independent of the canonical thermosensory circuit. While it is still possible that other non-autonomous signals, such as electrical communication, may trigger nuclear enrichment, these results raise the interesting possibility that the ADF sensory neurons are capable of directly sensing noxious heat. These observations suggest that a dynamic change in ELC-2 in ADF correlates with a change in behavior.
Figure 4. ELC-2 subcellular localization is sensitive to heat stress and correlates with behavior.
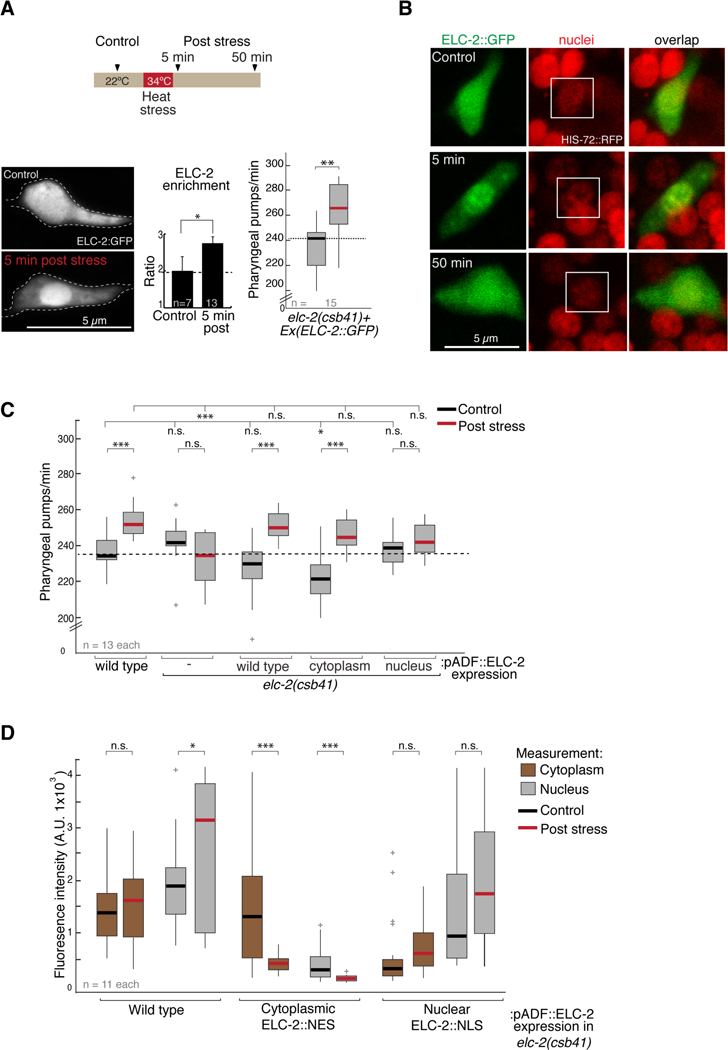
(A) Top: schematic of heat stress paradigm. Images were captured ~5 or 50 minutes after heat shock. Bottom left: confocal microscopy images show ELC-2::GFP distribution in the ADF soma before (control) and after heat stress. Middle plot shows quantification of data collected from images on left. Right: Pumping in elc-2 mutant animals rescued with extrachromosomal array containing ELC2::GFP fosmid. Black and red bar on box: control and 1h post stress, respectively. Student t test, ** p < 0.005, * p < 0.05.
(B) Left: ELC-2::GFP and nuclei at three time points: prior to heat stress, 5 and 50 minutes post-heat stress. Nuclei visualized using a strain expressing Histone H2B/HIS-72 tagged with RFP.
(C) Pumping rates measured in wild type or elc-2 mutants expressing different extrachromosomal array versions of ELC-2::GFP tagged with a NES (cytoplasm) or a NLS (nucleus) using ADF-specific promoter. 1-way ANOVA with Dunnet’s correction is used for comparisons across genotypes; Student’s t test is used for comparisons within genotype. *** p<0.0005, ** p<0.005, *p<0.05, n.s. not significant. Box plot outliers +.
(D) Measurement of confocal Z-stack GFP intensity of the nucleus and cytoplasm before and after heat stress in the strains used for assessing behavior. Expression pattern in S4E. Student’s t test: *** p<0.0005, ** p<0.005, *p<0.05, n.s. not significant. Box plot outliers +.
See also Figure S4.
To test if ELC-2 redistribution is required for the modulation of behavior after stress, we prevented ELC-2::GFP bulk redistribution by expressing it mostly in either the nucleus by fusing a nuclear localization signal (ELC-2::NLS) or in the cytoplasm by fusing a nuclear export signal (ELC2::NES) (Figure S4E). We tested feeding behavior before and after heat stress in elc-2 mutants expressing a wild type or one of these two ELC-2 variant transgenes. Both the wild type protein and cytoplasm-localized ELC-2, but not the nuclear variant, were sufficient to allow increased pumping after stress (Figure 4C), suggesting that presence of ELC-2 in the cytoplasm is critical for modulation of behavior. We also measured fluorescence intensity of ELC-2::GFP in these strains in the nucleus and cytoplasm (Figure 4D). In wild type, ELC-2 levels remain similar in the cytoplasm, but significantly increase in the nucleus upon heat shock (Figure 4D and S4F). Interestingly, in the ELC-2::NES variant, in which nuclear accumulation is prevented, we observe a reduction of fluorescence in the cytoplasm following heat stress (Figure 4D). This reduction could indicate that a failure to accumulate ELC-2 in the nucleus post-stress leads to its degradation in the cytoplasm. Together these results suggest that presence of ELC-2 in the cytoplasm and a dynamic change upon heat stress, possibly involving both relocalization to the nucleus and cytoplasmic degradation, is required to modulate pharyngeal pumping behavior. Other stressors, such as starvation, do not induce ELC-2 redistribution (Figure S4G). Taken together, these data indicate that a dynamic change of elc-2 is specifically sensitive to heat stress.
ECS ubiquitin ligase complex components modulate stress-induced behavioral change
Elongin C has a dual role as a regulatory subunit of the SIII/Elongin transcription elongation complex (Bradsher et al., 1993, Aso et al., 1995), and as a component of the ECS E3 ubiquitin ligase complex (Okumura et al., 2012). To characterize the function of ELC-2, we investigated whether additional members of these protein complexes were required for the behavioral change. Elongin A is an essential subunit of the SIII/Elongin complex, while the tumor-suppressor von Hippel-Lindau protein pVHL is the substrate recognition subunit of an ECS complex (Figure 5A). We generated a deletion in the C. elegans Elongin A ortholog R03D7.4 (Figure S3B), and found that these mutants had wild-type pharyngeal pumping behavior before and after the heat stress (Figure 5A), showing that the Elongin transcription elongation complex is dispensable for this behavior. Intriguingly however, the null mutants of vhl-1 displayed defective stress-induced feeding (Figure 5A). These defects are rescued by singlecopy expression of VHL-1 in the ADF neurons, suggesting that the function of vhl-1 in ADF is both necessary and sufficient for the heat-induced behavioral change (Figure 5A). These results suggest that ELC-2 acts together with the substrate recognition subunit VHL-1 in an E3-ligase complex in ADF, and predict that the loss of other components of the ECS complex would lead to defects in stress-enhanced feeding.
Figure 5. Components of an ECS E3-ligase are required for stress-induced feeding.
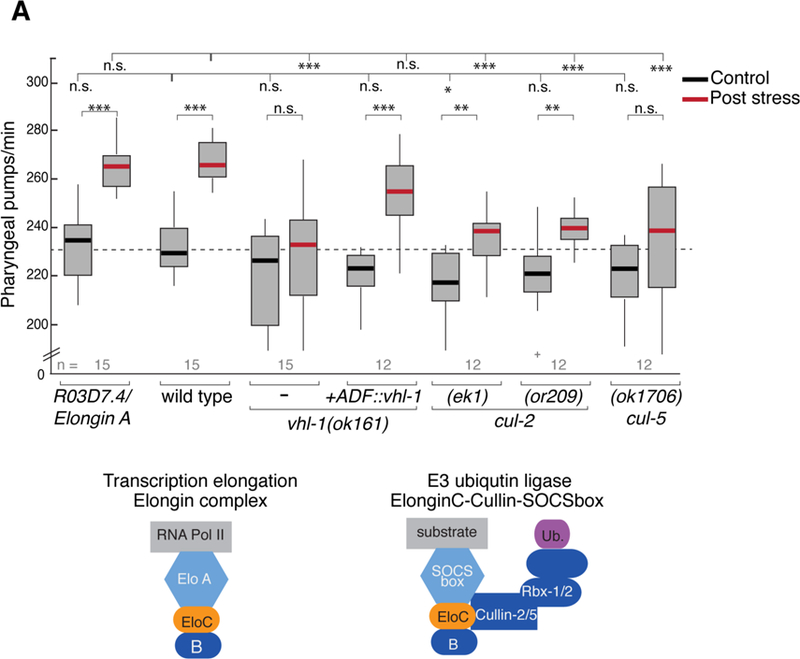
(A) Pharyngeal pumping rates in Elongin A R03D7.4(csb42) mutants and ECS mutants before and after 1 hour recovery from heat shock. Genotypes are listed on the x-axis. Transgene single-copy expression of vhl-1 under ADF-specific promoter (Psrh-142) is indicated. 1-way ANOVA with Dunnet’s correction is used for comparisons across genotypes; Student’s t test is used for comparisons within genotype. *** p<0.0005, ** p<0.005, *p<0.05, n.s. not significant. Bottom: schematic of Elongin complex and ECS complex. Elongin A and pVHL-1 represent distinct protein subunits in each of these complexes. See also Figures S2C, S3 and S5A.
We analyzed members of the Cullin family, which serve as the scaffolding proteins in E3 complexes. In ECS complexes, either CUL2 or CUL5 have been reported to serve as the scaffolding protein (Pause et al., 1997, Stebbins et al., 1999, Kamura et al., 2001). As such, we tested available cul-2 and cul-5 mutants for stress-induced pumping phenotypes. In both cul-2(ek1) null and cul-2(or209) hypomorphic mutants, an increase in pumping was observed post-stress, but to a lesser degree than in wild-type animals (Figure 5A). Mutation of cul-5 leads to a more pronounced defect in the ability of animals to increase pumping after heat stress (Figure 5A). In mammalian cells VHL interacts specifically with CUL-2 and RBX-1 to form a functional complex; SOCSbox-CUL-5-based complexes require RBX-2 (Kamura et al., 2004). We tested rbx-2 mutants in our pumping paradigm and found that these animals can increase pumping after stress similar to wild type (Figure S5A), suggesting that a complex formed by CUL-5/RBX-2 is likely dispensable in the stress-induced feeding, and that defects observed in cul-5 mutants could be through an as of yet uncharacterized complex. Taken together, our genetic data suggest that components of an ECS E3-ligase complex formed in ADF neurons regulates plasticity of a behavioral response.
ELC-2 regulates post-stress pumping behavior by modulating ADF serotonergic signaling
Pumping rates are positively regulated by serotonin: the more serotonin, the faster the pumping (Niacaris and Avery, 2003, Lemieux et al., 2015, Lemieux and Ashrafi, 2015, Tatum et al., 2015). The stress-induced modulation of pharyngeal pumping suggests that ELC-2 and VHL-1 could modulate serotonin levels in the ADF neurons or alter the release properties of these cells. Immunostaining of wild type, elc-2, and vhl-1 mutant animals with anti-serotonin antibodies shows no obvious differences in serotonin in the ADF soma in these strains (Figure S5B), suggesting that these mutants are capable of producing serotonin. This observation is consistent with our analysis of another known ADF-mediated behavior: elc-2 mutant animals were normal in the aversive olfactory learning of pathogenic bacteria (Figure S6), which is regulated by serotonin production in ADF (Zhang et al., 2005, Jin et al., 2016).
Next, we asked whether secretion of serotonin was altered in the elc-2 mutants. To address this question, we employed an assay used as a proxy for measuring dense core vesicle secretion from ADF, because like neuropeptides, serotonin can be secreted from dense core vesicles (De-Miguel and Trueta, 2005). In this assay, a fluorescently-tagged neuropeptide of the insulin family (DAF-28::mCherry) is expressed under an ADF-specific promoter. Released DAF-28 through dense-core vesicles accumulates in scavenger cells called coelomocytes, and the intensity of the fluorescence in these cells is measured as a readout for neuropeptide secretion of ADF (Lemieux et al., 2015). We found that specifically in the pre-stress state, elc-2 mutants accumulate significantly less DAF-28 neuropeptide than wild-type animals (Figure 6A and S5D), suggesting that the elc-2 mutants may have constitutive defects in dense core vesicle release from ADF even prior to exposure to heat stress.
Figure 6. Serotonin produced in ADF and components of a serotonergic circuit are required for stress-induced pumping behavior and act downstream of ECS ubiquitin ligase complex.
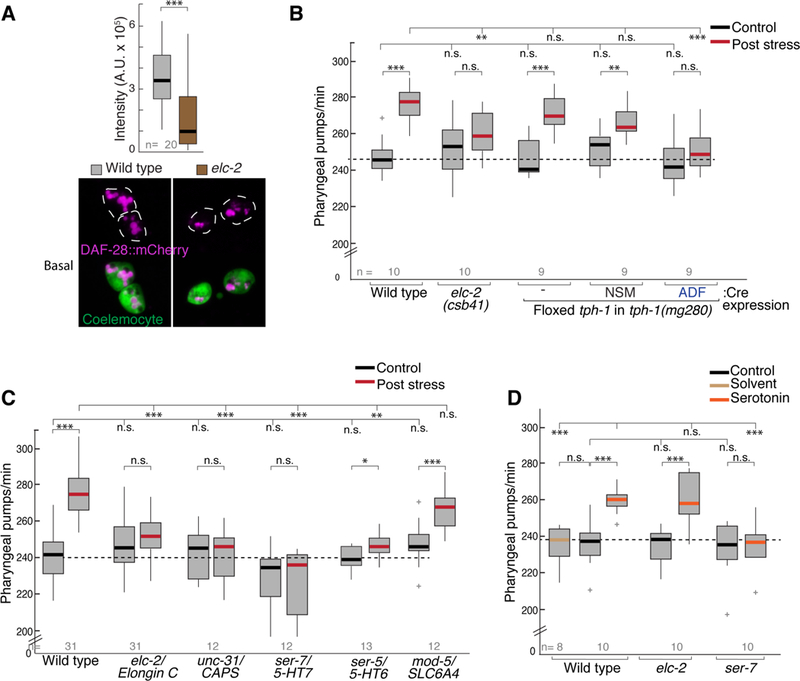
(A) Micrograph of the Insulin-like peptide (ILP) DAF-28 reporter gene fused to mCherry (magenta) is ectopically produced and secreted by ADF. Plot shows fluorescence intensity for DAF-28::mCherry accumulation in coelomocyte scavenger cells (green) at control temperature. ** p<0.005, Student’s t test.
(B) Pumping rates measured pre- and post 1 hour heat stress in tph-1(mg280) mutants rescued with a single-copy of tph-1 flanked by LoxP sites. Cre expression excises tph-1 and ablates gene function in the indicated cells.
(C) Pumping rates measured pre- and post-stress testing components of a serotonergic circuit in different mutants; mammalian orthologs are indicated.
(D) Effect of exogenous serotonin on pharyngeal pumping of different mutants.
For B-D, 1-way ANOVA with Dunnet’s correction is used for comparisons across genotypes; Student’s t test is used for comparisons within genotype, *** p<0.0005, ** p<0.005, *p<0.05, n.s. not significant.
We then tested whether reduced serotonin levels from ADF could affect the heat-induced change in pharyngeal pumping. We measured pumping rates in animals with impaired serotonin production in specific neurons after genetic ablation of the rate limiting serotonin biosynthetic enzyme TPH-1. In C. elegans hermaphrodites, serotonin is produced in the ADF, NSM and HSN neurons (Figure 2B) (Sze et al., 2000, Jafari et al., 2011). Of the serotonin-producing cells, the NSM and ADF pairs are located in the head of the animal. Using engineered strains that ablate tph-1 either in NSM or ADF (Flavell et al., 2013), we found that tph-1 disruption in ADF but not in NSM, significantly reduced the ability of the animals to increase pharyngeal pumping rate following heat stress (Figure 6B). However, both transgenic animals displayed normal pumping rate under the control condition, indicating that the serotonin production in ADF neurons is required specifically for the increased pumping evoked by heat stress.
If serotonin were released from ADF in response to stress, one prediction would be that components involved in serotonergic neuromodulation, from its neuronal source of release to the target cells where it acts, would be required for heat stress-induced pumping. ADF is not presynaptic to the MC motor neurons that innervate pharyngeal muscles to drive pumping behavior (Albertson and Thomson, 1976). Thus, serotonin reaching these motor neurons would be released extra-synaptically from dense-core vesicles (De-Miguel and Trueta, 2005). We tested this prediction genetically by measuring pumping rates in unc-31/CAPS mutants defective in the release of dense-core vesicles (Speese et al., 2007), and ser-7/5-HT7 mutants which lack the G-protein coupled serotonin receptor expressed on MC neurons (Hobson, 2005). Consistent with our prediction, both unc-31 and ser-7 mutants fail to increase pumping after stress (Figure 6C). We tested the role of ser-5/5-HT6 serotonin receptor in our paradigm, which is required in AVJ interneurons for increased feeding after starvation (Cunningham et al., 2012, Lemieux et al., 2015). In contrast to ser-7 mutants, ser-5 mutants show a small but significant increase in pumping after stress, yet the magnitude of the response differs from wild type (Figure 6C), suggesting that ser-5 may play a partial role in the full behavioral response after heat stress. We asked whether the serotonin-recapturing neurons (NSM, AIM, RIH) play a role in the induced behavior by testing mutants lacking the serotonin reuptake transporter SLC64A/mod-5 (Jafari et al., 2011). In this case, these animals displayed normal pumping rates after stress (Figure 5C), indicating these neurons are dispensable for mediating this behavior.
Finally, we tested the effect of exogenous serotonin on the pumping rate in elc-2 mutants. We found that addition of exogenous serotonin was sufficient to significantly increase the pharyngeal pumping rate in a similar way in non-stressed wild type and elc-2 mutant animals. The stimulatory effect of exogenous serotonin is blocked by the mutation of ser-7 (Figure 5D). Taken together, our results show that increasing extracellular serotonin can bypass the requirement of the ECS complex, and that the serotonin/SER-7 pathway acts downstream of the ECS complex in ADF to generate the heat-induced increase in the pumping rate.
Discussion
Components of a conserved E3 ubiquitin ligase complex modulate an essential behavior
Feeding is an essential behavior for all animals. Both internal and external cues, as well as experience, can modulate feeding to generate adaptive changes (Gruninger et al., 2007). Here, we uncovered that components of a highly conserved ECS ubiquitin ligase complex play a role in modulating heat-stress induced changes in feeding by acting in a pair of serotonergic neurons. In our paradigm, noxious heat causes the animal to stop feeding for ~15 minutes, and likely also induces cellular damage and protein misfolding that would impose an energetic cost to repair the damage. We speculate that this increased feeding may serve as a homeostatic mechanism to recover energy.
Since human Elongin C can substitute for ELC-2 function in regulating the post-stress pumping behavior, and Elongin C is widely expressed, including the brain (Consortium, 2013), together, these findings could suggest a deeply conserved role of human Elongin C in modulating stress-induced sustained changes in the brain. In a broader context, our observations suggest a novel molecular link between an ubiquitin-mediated proteolysis pathway and neuromodulation in the serotonergic system. It will be interesting to determine if a similar mechanism might be playing a permissive role for the increased serotonin efflux observed after acute stress seen in mammals (Chaouloff, 2000).
A neuron-specific Elongin C ortholog generates a stressor-dependent response
While the heterogeneity in the response of serotonergic neurons to different types of stressors has been long documented in the mammalian brain, the underlying mechanisms are not well understood (Lee et al., 1987, Petty, 1996). Upon investigating a role for elc-2 in other known ADF mediated behaviors, we found that loss of ELC-2 was dispensable for a model of aversive olfactory learning (Figure S6) and that starvation, another stressor that mediates pumping in an-ADF-dependent manner (Lemieux et al., 2015), does not require elc-2, nor does it induce ELC-2 nuclear-enrichment (Figures 3A and S4G). We propose that the heterogeneous and stressor-dependent response observed allows for fine-tuning of serotonergic signals to modulate behavioral and physiological events in ways that are appropriate for the internal and external context.
Although we did not investigate why ELC-2 expression is largely restricted to a single neuronal pair, it remains possible that elc-2 has evolved to perform a new function in the ADF neurons that cannot be compensated for by other Elongin C paralogs. Importantly, our identification of an Elongin C ortholog with restricted expression allowed us to uncover a novel role for components of an E3 ubiquitin ligase in neurons in vivo by circumventing early developmental requirements of Elongin C.
Elongin C transiently enriches in the neuronal nucleus in response to stress
The finding that Elongin C transiently changes its subcellular distribution in an experiencedependent manner is intriguing. Our data suggest that the primary site of activity for ELC-2 in the context of this behavior is in the cytoplasm, although at present we cannot exclude that the addition of the NLS to ELC-2 renders it non-functional. Our microscopy data for ELC-2 variants suggest two potential mechanisms controlling the dynamic redistribution of ELC-2 in response to heat: through its accumulation in the nucleus, or, if artificially prevented from localizing to the nucleus, through its degradation in the cytoplasm. Both mechanisms may have evolved to ensure robustness in controlling the level of ELC-2 in the cytoplasm. For example, it is possible that ELC-2 expression may increase after heat stress, and the nucleus may act as a temporary sink for this extra source of ELC-2, maintaining tightly controlled and unchanged levels in the cytoplasm. However, there could also be an additional role for nuclear ELC-2 during this redistribution, such as in the regulation of transcription, where its presence may lead to other long lasting changes beyond the timescales observed in this study.
In human cells pVHL also displays active nuclear shuttling, and its nuclear to cytoplasmic distribution ratio is affected by the cellular context (Lee et al., 1996, Lewis and Roberts, 2003, Zheng et al., 2006). pVHL can target substrates for degradation in either the nucleus or cytoplasm (Berra et al., 2001), and CUL2 also undergoes nuclear shuttling (Pause et al., 1997). Moreover, proteasomes can undergo dynamic changes in subcellular localization and composition in response to neuronal activity. These results suggest that ECS components can be dynamically regulated by subcellular localization, suggesting a potential molecular mechanism for transducing signals from the environment.
A model for the role of Elongin C in neuromodulation
We propose a model where ELC-2 regulates aspects of neuromodulation that manifest into effects on feeding behavior after exposure to heat stress (Figure 7). Taking all of our results together, our model considers two non-mutually exclusive possibilities for the role of Elongin C in modulating behavior. First, loss of elc-2 may lead to steady-state reduced release of serotonin and/or neuropeptides, altering the ability of animals to properly respond to noxious heat when they are presented with the stress. Second, it is possible that heat stress induces the alteration of activity and/or distribution of ELC2 in the cytoplasm, leading to additional effects on neuromodulation through its influence on production or release of serotonin or neuropeptides. At present, it is unclear whether the redistribution of ELC-2 plays a direct role in modulating behavior by acting in the same or a parallel pathway as the dense core vesicle release pathway. It may also be true that the dynamic localization of ELC-2 is not related to modulating feeding behavior, but rather represents a response to alter other aspects of animal physiology acting at distinct timescales as described above. Future experiments will be directed to better resolve some of these possibilities.
Figure 7. Possible mechanism of ELC-2 regulating behavioral plasticity in response to heat stress. Basal state:
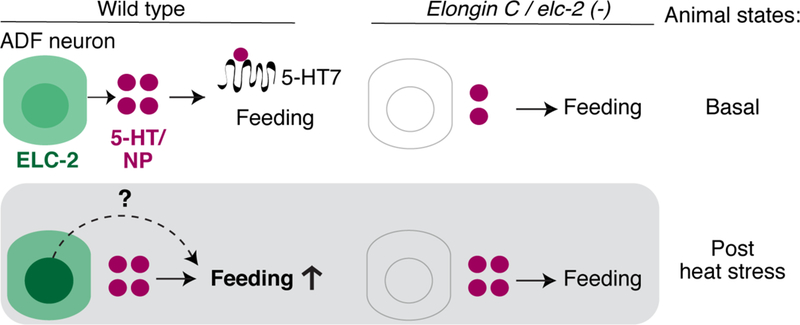
ELC-2 is expressed in ADF sensory neurons, which release serotonin and neuropeptides through dense-core vesicles. Serotonin from ADF and from other neurons (top circles) regulate pharyngeal pumping feeding behavior. The basal state in elc-2 mutants is different from wild type, with reduced dense core vesicle release, but its basal feeding behavior is normal. Post stress state: Heat stress experience induces both a dynamic localization of ELC-2 in ADF nucleus, and increased feeding in the animal. Serotonin binds metabotropic receptor SER-7/5-HT7 (scribble; omitted in top panels) to regulate stimulus-dependent pharyngeal pumping feeding behavior. elc-2 and other ECS mutants are unable to alter feeding behavior in response to stress. The role of ELC-2 dynamic redistribution is unclear, and may serve to alter the activity of ELC-2 in the cytoplasm to increase feeding (dotted line) and/or contribute to a parallel function in the nucleus that is initiated by noxious heat. See also Figures 4 and S4–5.
A versatile and powerful cell-type specific profiling method in C. elegans
Our adaptation of the translating-ribosome affinity purification method provides additional advantages to a growing list of profiling methods available in in C. elegans (Spencer et al., 2014). One strength of TRAP is that the rapid freezing of translating mRNAs in their native context contrasts with emerging methods using tissue dissociation and cell sorting of populations of fluorescent cells followed by transcriptome sequencing. These latter methods, while proving highly useful in identifying stably expressed transcripts specific to cell types (Spencer et al., 2014, Kaletsky et al., 2016), often require longer manipulations to isolate sorted cells for further processing. These extended processing times can complicate the analysis of transcriptomes isolated from animals undergoing plasticity in behavior on timescales ranging from minutes to hours. However, these approaches do have the advantage in that total RNA from a sorted population of cells can be collected, enabling the detection and analysis of nonpolyadenylated transcripts and small RNA populations. Since TRAP is an immuno-precipitation-based technique there will be mRNAs from other tissues that can non-specifically bind to resins used, giving rise to false positive hits. As in other genome-wide techniques, performing this technique with biological replicates and independent validation of potential candidates is necessary. Overall, TRAP will serve as a complementary strategy in the growing toolbox of available methods for profiling transcript populations in specific cell types in C. elegans.
Experimental Procedures
Maintenance of C. elegans and strains used in present study
Animals were grown using standard conditions on NGM plates (Brenner, 1974), seeded with OP50–1, and grown at 22ºC, unless noted otherwise.
Translating Ribosome Affinity Purification and RNA-seq libraries
Bleach-synchronized animals grown at 22Cº in standard NGM 15-cm plates seeded with OP50–1. L4 larvae collected 46–48 h past L1 release. Worm lysis and immunoprecipitations were performed as previously described (Heiman et al., 2014). Detailed protocol in (Gracida and Calarco, 2017). RNA was extracted using TRI reagent (Sigma T9424). Semi-quantitative RT-PCR (Qiagen 210210) was performed to evaluate quality and enrichment of IP. cDNA libraries were made using TruSeq RNA Library Kit (Illumina RS-122–2001). Two libraries corresponding to two independent biological replicate experiments per genotype were sequenced at >150 million 100–150 base paired-end reads each using an Illumina HiSeq2000 machine.
Post-processing and transcriptome analyses
RNA-seq reads were mapped to C. elegans genome WS235 using STAR alignment software (Dobin et al., 2012). Gene-centered counts for all genes were obtained using the HTSeq package (Anders et al., 2015). Gene counts were re-scaled using upper quartile normalization, and fold changes (TRAP immuno-precipitation counts / input counts) were calculated for genes with conservatively detectable expression (RPKM value >= 1). Relative tissue enrichment was calculated by comparing fold change values across relevant tissue; the cell type with the highest fold change value was normalized to 100%. For GO enrichment analysis, statistically overrepresented Gene Ontology (GO) terms were calculated with GOstat using datasets with the top-enriched mRNAs (neuronal data >= 10 fold enrichment; muscle data >=10 fold enrichment; intestinal data >= 2 fold enrichment).
Pharyngeal pumping assays
Assays were performed 50–52 hours post L1 food release at 22ºC (bleach synchronized). For heat stress 5–10 animals were transferred to pre-warmed (35ºC) standard NGM plate with food. Plate was sealed with parafilm and submerged in a water bath at 34ºC for 30 min. After treatment parafilm was removed and lids briefly opened and closed. Plates were incubated at room temperature for 60–70 min before counting pumping rates. Pumping rates were measured using a Zeiss Axiozoom macroscope. Pumps were counted for 20 seconds three times. Each genotype was tested in at least three independent replicates. Each replicate included a wild-type positive control.
Genome editing and single-copy transgene genome integration
CRISPR-mediated deletions of elc-2 and elongin A and tagging of endogenous his-72 were conducted as reported in (Norris et al., 2015). Single-copy transgene genome integrations were done using “miniMos” approach as described in (Frokjaer-Jensen et al., 2014).
Immunofluorescence and image acquisition
Microscopy images were processed using confocal microscopy (FV1000MPE Olympus). Anti-serotonin staining as described in (Loer and Kenyon, 1993) using rabbit serum S5545 (Sigma-Aldrich), and AntiGFP chicken IgG: GFP-1020 (Aves Labs). Animals expressing ELC-2::GFP were mounted live on agar pads and immobilized with 1% sodium azide. For cytoplasmic and nuclear ELC-2::GFP measurements a single z-stack was quantified by taking two cytoplasmic measurements and two nuclear per neuron. DAF-28::mCherry accumulation was calculated by encircling the mCherry aggregate in the coelemocyte and then multiplying the mean fluorescence intensity by the area of the aggregate. Two z-Stacks were projected and summed for these quantifications.
Temperature ramp
Temperature ramp conditions from 22ºC to 26.7ºC were set up for 60 mm NGM plates using a FLIR infrared camera (FLIR-E64501). Water bath temperature was set to 29ºC; plates reached ~26.7ºC in 5 minutes as reported in Tatum et al. 2015. Plates containing five worms were kept at 22ºC in incubator, then they were removed, parafilmed and submerged in water bath. Pumping rates were counted on Zeiss Axiozoom as described above.
Exogenous serotonin supplementation
50 µl of 20 mM serotonin hydrochloride (Sigma H9523) in water were added on top of animals on NGM plate with food. Pharyngeal pumping was counted 15 minutes after.
Statistical analyses
Pharyngeal pumping data is reported as box plots. Box is 25th and 75th percentile, whisker are data extremes not considered outliers (Matlab). Statistical significance was calculated across genotypes using 1-way ANOVA with Dunnet’s correction (GraphPad Prism7). Two-tailed Student’s t test was used when comparing conditions of the same genotype in behavioral assays, and microscopy quantifications.
Supplementary Material
Acknowledgments
We thank Andrew Murray, Arneet Saltzman, Adam Norris, and Luisa Cochella for critical reading of the manuscript. We thank He Liu for help in experiments not shown, and for the code to analyze movement. Bicheng Han and Wenxing Yang for advice on experiments. We would also like to thank Peter Rodriguez, Gustavo Gonzalez, JC. Martínez, Claire Reardon, Christian Daly, and Jeff Offermann, Ed Soucy and Steve Turney for assistance and technical support. We thank Cori Bargmann, Kaveh Ashrafi, and Piali Sengupta for sharing strains with us. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by an NIH Director’s Early Independence Award (DP5OD009153), by the Bauer Fellows Program at Harvard University, and by startup funds from the University of Toronto (to J.A.C), and NIH Grant 1P01GM103770 (to Y.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number for RNA-Seq datasets
Our raw RNA-Seq datasets (.fastq files) can be obtained from GEO under the accession number GSE106374.
References
- ALBERTSON DG & THOMSON JN 1976. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci, 275, 299–325. [DOI] [PubMed] [Google Scholar]
- ANDERS S, PYL PT & HUBER W 2015. HTSeq--a Python framework to work with highthroughput sequencing data. Bioinformatics, 31, 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANDERSON A, LAURENSON-SCHAFER H, PARTRIDGE FA, HODGKIN J & MCMULLAN R 2013. Serotonergic chemosensory neurons modify the C. elegans immune response by regulating G-protein signaling in epithelial cells. PLoS Pathog, 9, e1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASO T, CONAWAY JW & CONAWAY RC 1995. The RNA polymerase II elongation complex. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 9, 1419–1428. [DOI] [PubMed] [Google Scholar]
- AVERY L & YOU YJ 2012. C. elegans feeding. WormBook, 1–23. [DOI] [PMC free article] [PubMed]
- BARGMANN CI & MARDER E 2013. From the connectome to brain function. Nat Methods, 10, 483–90. [DOI] [PubMed] [Google Scholar]
- BERRA E, ROUX D, RICHARD DE & POUYSSEGUR J 2001. Hypoxia-inducible factor-1 alpha (HIF-1 alpha) escapes O(2)-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep, 2, 615–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRADSHER JN, TAN S, MCLAURY HJ, CONAWAY JW & CONAWAY RC 1993. RNA polymerase II transcription factor SIII. II. Functional properties and role in RNA chain elongation. J Biol Chem, 268, 25594–603. [PubMed] [Google Scholar]
- BRENNER S 1974. The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAOULOFF F 2000. Serotonin, stress and corticoids. J Psychopharmacol, 14, 139–51. [DOI] [PubMed] [Google Scholar]
- CIECHANOVER A 2006. The ubiquitin proteolytic system: from a vague idea, through basic mechanisms, and onto human diseases and drug targeting. Neurology, 66, S7–19. [DOI] [PubMed] [Google Scholar]
- COHEN JY, AMOROSO MW & UCHIDA N 2015. Serotonergic neurons signal reward and punishment on multiple timescales. Elife, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONSORTIUM GT 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet, 45, 580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUNNINGHAM KA, HUA Z, SRINIVASAN S, LIU J, LEE BH, EDWARDS RH & ASHRAFI K 2012. AMP-activated kinase links serotonergic signaling to glutamate release for regulation of feeding behavior in C. elegans. Cell metabolism, 16, 113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE-MIGUEL FF & TRUETA C 2005. Synaptic and extrasynaptic secretion of serotonin. Cell Mol Neurobiol, 25, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOBIN A, DAVIS CA, SCHLESINGER F, DRENKOW J, ZALESKI C, BATUT P, CHAISSON M & GINGERAS TR 2012. STAR : ultrafast universal RNA-seq aligner 1–7. [DOI] [PMC free article] [PubMed]
- DOROQUEZ DB, BERCIU C, ANDERSON JR, SENGUPTA P & NICASTRO D 2014. A highresolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife, 3, e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLAVELL SW, POKALA N, MACOSKO EZ, ALBRECHT DR, LARSCH J & BARGMANN CI 2013. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell, 154, 1023–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FROKJAER-JENSEN C, DAVIS MW, SAROV M, TAYLOR J, FLIBOTTE S, LABELLA M, POZNIAKOVSKY A, MOERMAN DG & JORGENSEN EM 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods, 11, 529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRACIDA X & CALARCO JA 2017. Cell type-specific transcriptome profiling in C. elegans using the Translating Ribosome Affinity Purification technique. Methods [DOI] [PubMed]
- GREER PL, HANAYAMA R, BLOODGOOD BL, MARDINLY AR, LIPTON DM, FLAVELL SW, KIM TK, GRIFFITH EC, WALDON Z, MAEHR R, PLOEGH HL, CHOWDHURY S, WORLEY PF, STEEN J & GREENBERG ME 2010. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell, 140, 704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRUNINGER TR, LEBOEUF B, LIU Y & GARCIA LR 2007. Molecular signaling involved in regulating feeding and other motivated behaviors. Mol Neurobiol, 35, 1–20. [DOI] [PubMed] [Google Scholar]
- HAKIMI AA, TICKOO SK, JACOBSEN A, SARUNGBAM J, SFAKIANOS JP, SATO Y, MORIKAWA T, KUME H, FUKAYAMA M, HOMMA Y, CHEN YB, SANKIN AI, MANO R, COLEMAN JA, RUSSO P, OGAWA S, SANDER C, HSIEH JJ & REUTER VE 2015. TCEB1-mutated renal cell carcinoma: a distinct genomic and morphological subtype. Mod Pathol, 28, 845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEGDE AN & DIANTONIO A 2002. Ubiquitin and the synapse. Nat Rev Neurosci, 3, 854–61. [DOI] [PubMed] [Google Scholar]
- HEIMAN M, KULICKE R, FENSTER RJ, GREENGARD P & HEINTZ N 2014. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nature protocols, 9, 1282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEIMAN M, SCHAEFER A, GONG S, PETERSON JD, DAY M, RAMSEY KE, SUÁREZ-FARIÑAS M, SCHWARZ C, STEPHAN DA, SURMEIER DJ, GREENGARD P & HEINTZ N 2008. A translational profiling approach for the molecular characterization of CNS cell types. Cell, 135, 738–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENDRICKS M, HA H, MAFFEY N & ZHANG Y 2012. Compartmentalized calcium dynamics in a C. elegans interneuron encode head movement. Nature, 487, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOBSON RJ 2005. SER-7, a Caenorhabditis elegans 5-HT7-like Receptor, Is Essential for the 5-HT Stimulation of Pharyngeal Pumping and Egg Laying. Genetics, 172, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAFARI G, XIE Y, KULLYEV A, LIANG B & SZE JY 2011. Regulation of extrasynaptic 5-HT by serotonin reuptake transporter function in 5-HT-absorbing neurons underscores adaptation behavior in Caenorhabditis elegans. J Neurosci, 31, 8948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIN X, POKALA N & BARGMANN CI 2016. Distinct Circuits for the Formation and Retrieval of an Imprinted Olfactory Memory. Cell, 164, 632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOELS M & BARAM TZ 2009. The neuro-symphony of stress. Nat Rev Neurosci, 10, 459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAELIN WG JR. & MAHER ER 1998. The VHL tumour-suppressor gene paradigm. Trends Genet, 14, 423–6. [DOI] [PubMed] [Google Scholar]
- KALETSKY R, LAKHINA V, AREY R, WILLIAMS A, LANDIS J, ASHRAF J & MURPHY CT 2016. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature, 529, 92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAMURA T, BURIAN D, YAN Q, SCHMIDT SL, LANE WS, QUERIDO E, BRANTON PE, SHILATIFARD A, CONAWAY RC & CONAWAY JW 2001. Muf1, a novel Elongin BCinteracting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J Biol Chem, 276, 29748–53. [DOI] [PubMed] [Google Scholar]
- KAMURA T, MAENAKA K, KOTOSHIBA S, MATSUMOTO M, KOHDA D, CONAWAY RC, CONAWAY JW & NAKAYAMA KI 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev, 18, 3055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATZ PS & HARRIS-WARRICK RM 1990. Neuromodulation of the crab pyloric central pattern generator by serotonergic/cholinergic proprioceptive afferents. J Neurosci, 10, 1495–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIPREOS ET, LANDER LE, WING JP, HE WW & HEDGECOCK EM 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell, 85, 829–39. [DOI] [PubMed] [Google Scholar]
- LEE EH, LIN HH & YIN HM 1987. Differential influences of different stressors upon midbrain raphe neurons in rats. Neurosci Lett, 80, 115–9. [DOI] [PubMed] [Google Scholar]
- LEE S, CHEN DY, HUMPHREY JS, GNARRA JR, LINEHAN WM & KLAUSNER RD 1996. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci U S A, 93, 1770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEMIEUX GA & ASHRAFI K 2015. Neural Regulatory Pathways of Feeding and Fat in Caenorhabditis elegans. Annu Rev Genet, 49, 413–38. [DOI] [PubMed] [Google Scholar]
- LEMIEUX GA, CUNNINGHAM KA, LIN L, MAYER F, WERB Z & ASHRAFI K 2015. Article Kynurenic Acid Is a Nutritional Cue that Enables Behavioral Plasticity. Cell, 160, 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS MD & ROBERTS BJ 2003. Role of nuclear and cytoplasmic localization in the tumoursuppressor activity of the von Hippel-Lindau protein. Oncogene, 22, 3992–7. [DOI] [PubMed] [Google Scholar]
- LI Y, ZHONG W, WANG D, FENG Q, LIU Z, ZHOU J, JIA C, HU F, ZENG J, GUO Q, FU L & LUO M 2016. Serotonin neurons in the dorsal raphe nucleus encode reward signals. Nat Commun, 7, 10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIANG B, MOUSSAIF M, KUAN CJ, GARGUS JJ & SZE JY 2006. Serotonin targets the DAF16/FOXO signaling pathway to modulate stress responses. Cell Metab, 4, 429–40. [DOI] [PubMed] [Google Scholar]
- LIAO EH, HUNG W, ABRAMS B & ZHEN M 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature, 430, 345–50. [DOI] [PubMed] [Google Scholar]
- LOER CM & KENYON CJ 1993. Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J Neurosci, 13, 5407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAIER SF & WATKINS LR 2005. Stressor controllability and learned helplessness: the roles of the dorsal raphe nucleus, serotonin, and corticotropin-releasing factor. Neurosci Biobehav Rev, 29, 829–41. [DOI] [PubMed] [Google Scholar]
- MARDER E 2012. Neuromodulation of neuronal circuits: back to the future. Neuron, 76, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAXWELL PH, WIESENER MS, CHANG GW, CLIFFORD SC, VAUX EC, COCKMAN ME, WYKOFF CC, PUGH CW, MAHER ER & RATCLIFFE PJ 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature, 399, 271–5. [DOI] [PubMed] [Google Scholar]
- MCEWEN BS, BOWLES NP, GRAY JD, HILL MN, HUNTER RG, KARATSOREOS IN & NASCA C 2015. Mechanisms of stress in the brain. Nat Neurosci, 18, 1353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORI I & OHSHIMA Y 1995. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature, 376, 344–8. [DOI] [PubMed] [Google Scholar]
- NIACARIS T & AVERY L 2003. Serotonin regulates repolarization of the C. elegans pharyngeal muscle. J Exp Biol, 206, 223–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORRIS AD, KIM HM, COLAIACOVO MP & CALARCO JA 2015. Efficient Genome Editing in Caenorhabditis elegans with a Toolkit of Dual-Marker Selection Cassettes. Genetics, 201, 449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKUMURA F, MATSUZAKI M, NAKATSUKASA K & KAMURA T 2012. The Role of Elongin BCContaining Ubiquitin Ligases. Front Oncol, 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAUSE A, LEE S, WORRELL RA, CHEN DY, BURGESS WH, LINEHAN WM & KLAUSNER RD 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci U S A, 94, 2156–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETTY FKGLLAL 1996. Neurochemistry of stress : Regional brain levels of biogenic amines and metabolites with ten different stressors. Biogenic Amines, 12, 377–394. [Google Scholar]
- RICHMOND JE, DAVIS WS & JORGENSEN EM 1999. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci, 2, 959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAROV M, MURRAY JI, SCHANZE K, POZNIAKOVSKI A, NIU W, ANGERMANN K, HASSE S, RUPPRECHT M, VINIS E, TINNEY M, PRESTON E, ZINKE A, ENST S, TEICHGRABER T, JANETTE J, REIS K, JANOSCH S, SCHLOISSNIG S, EJSMONT RK, SLIGHTAM C, XU X, KIM SK, REINKE V, STEWART AF, SNYDER M, WATERSTON RH & HYMAN AA 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell, 150, 855–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SASAGAWA Y, KIKUCHI K, DAZAI K & HIGASHITANI A 2005. Caenorhabditis elegans Elongin BC complex is essential for cell proliferation and chromosome condensation and segregation during mitosis and meiotic division II. Chromosome Res, 13, 357–75. [DOI] [PubMed] [Google Scholar]
- SATO Y, YOSHIZATO T, SHIRAISHI Y, MAEKAWA S, OKUNO Y, KAMURA T, SHIMAMURA T, SATO-OTSUBO A, NAGAE G, SUZUKI H, NAGATA Y, YOSHIDA K, KON A, SUZUKI Y, CHIBA K, TANAKA H, NIIDA A, FUJIMOTO A, TSUNODA T, MORIKAWA T, MAEDA D, KUME H, SUGANO S, FUKAYAMA M, ABURATANI H, SANADA M, MIYANO S, HOMMA Y & OGAWA S 2013. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet, 45, 860–7. [DOI] [PubMed] [Google Scholar]
- SONG B-M, FAUMONT S, LOCKERY S & AVERY L 2013. Recognition of familiar food activates feeding via an endocrine serotonin signal in Caenorhabditis elegans. eLife, 2, e00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPEESE S, PETRIE M, SCHUSKE K, AILION M, ANN K, IWASAKI K, JORGENSEN EM & MARTIN TF 2007. UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci, 27, 6150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPENCER WC, MCWHIRTER R, MILLER T, STRASBOURGER P, THOMPSON O, HILLIER LW, WATERSTON RH & MILLER DM 3RD 2014. Isolation of specific neurons from C. elegans larvae for gene expression profiling. PLoS One, 9, e112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEBBINS CE, KAELIN WG JR. & PAVLETICH NP 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science, 284, 455–61. [DOI] [PubMed] [Google Scholar]
- SZE JY, VICTOR M, LOER C, SHI Y & RUVKUN G 2000. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature, 403, 560–4. [DOI] [PubMed] [Google Scholar]
- TAGHERT PH & NITABACH MN 2012. Peptide neuromodulation in invertebrate model systems. Neuron, 76, 82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKEISHI A, YU YV, HAPIAK VM, BELL HW, O’LEARY T & SENGUPTA P 2016. Receptortype Guanylyl Cyclases Confer Thermosensory Responses in C. elegans. Neuron, 90, 235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TATUM MC, OOI FK, CHIKKA MR, CHAUVE L, MARTINEZ-VELAZQUEZ LA, STEINBUSCH HW, MORIMOTO RI & PRAHLAD V 2015. Neuronal serotonin release triggers the heat shock response in C. elegans in the absence of temperature increase. Curr Biol, 25, 163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VON STETINA SE, WATSON JD, FOX RM, OLSZEWSKI KL, SPENCER WC, ROY PJ & MILLER DM 3RD 2007. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol, 8, R135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG T, BIRSOY K, HUGHES NW, KRUPCZAK KM, POST Y, WEI JJ, LANDER ES & SABATINI DM 2015. Identification and characterization of essential genes in the human genome. Science, 350, 1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITE JG, SOUTHGATE E, THOMSON JN & BRENNER S 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci, 314, 1340. [DOI] [PubMed] [Google Scholar]
- YAMAMOTO N, HEGDE AN, CHAIN DG & SCHWARTZ JH 1999. Activation and degradation of the transcription factor C/EBP during long-term facilitation in Aplysia. J Neurochem, 73, 2415–23. [DOI] [PubMed] [Google Scholar]
- YOU YJ, KIM J, RAIZEN DM & AVERY L 2008. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab, 7, 249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y, LU H & BARGMANN CI 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature, 438, 179–84. [DOI] [PubMed] [Google Scholar]
- ZHANG Y, MA C, DELOHERY T, NASIPAK B, FOAT BC, BOUNOUTAS A, BUSSEMAKER HJ, KIM SK & CHALFIE M 2002. Identification of genes expressed in C. elegans touch receptor neurons. Nature, 418, 331–5. [DOI] [PubMed] [Google Scholar]
- ZHENG X, RUAS JL, CAO R, SALOMONS FA, CAO Y, POELLINGER L & PEREIRA T 2006. Celltype-specific regulation of degradation of hypoxia-inducible factor 1 alpha: role of subcellular compartmentalization. Mol Cell Biol, 26, 4628–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


