Abstract
Over the years, structural characterizations of α(2–8)-polysialic acid (polySia) in solution have produced inconclusive results. Efforts for obtaining detailed information in this important antigen have focused primarily on the α-linked residues and not on the distinctive characteristics of the terminal ones. The thermodynamically preferred anomeric configuration for the reducing end of sialic acids is β, which has the [I]CO2– group equatorial and the OH ([I]OH2) axial, while for all other residues the CO2– group is axial. We show that this purportedly minor difference has distinct consequences for the structure of α(2–8)-polySia near the reducing end, as the β configuration places the [I]OH2 in a favorable position for the formation of a hydrogen bond with the carboxylate group of the following residue ([II]CO2–). Molecular dynamics (MD) simulations predicted the hydrogen bond, which we subsequently directly detected by NMR. The combination of MD and residual dipolar couplings shows that the net result for the structure of Sia2-βOH is a stable conformation with well-defined hydration and charge patterns, and consistent with experimental NOE-based hydroxyl and aliphatic inter-proton distances. Moreover, we provide evidence that this distinct conformation is preserved on Sia oligosaccharides, thus constituting a motif that determines the structure and dynamics of α(2–8)-polySia for at least the first two residues of the polymer. We suggest the hypothesis that this structural motif sheds light on a longtime puzzling observation for the requirement of 10 residues of α(2–8)-polySia in order to bind effectively to specific antibodies, about four units more than for analogous cases.
Keywords: hydrogen bond, molecular dynamics, residual dipolar couplings, sialic acid structure, stereospecific assignments
Introduction
Sialic acids (Sias) are glycans constituted by nine carbons and a suite of chemical moieties playing diverse and important roles in cell and molecular interactions (Cohen and Varki 2010). Sias are expressed abundantly in vertebrates and higher invertebrates (Angata and Varki 2002); outside these groups, Sias are found only in a small number of species (Vimr et al. 2004). Among the latter, the majority is constituted by disease-causing gram-negative bacteria to human and animal hosts (Varki 2008). Moreover, microbial Sia metabolism is a virulence determinant in a range of infectious diseases (Vimr et al. 2004). Such is the case for the meningitis-causing bacteria N. meningitidis serogroup B and E. coli K1, whose capsular polysaccharides (CPs) are important virulence factors (Schiffer et al. 1976). The N. meningitidis group B (NMB) CP is a linear homopolymer of (2–8)-α-Neu5Ac (N-acetylneuraminic acid) units, also known as α(2–8)-polysialic acid (polySia), and in bacteria can reach a degree of polymerization of up to 230 Sia units (Pelkonen et al. 1988; Schnaar et al. 2014). Because CPs are less prone to variation among bacterial strains as compared to proteins, they are excellent candidates for vaccine antigens, as has been the case with the development of CP conjugate vaccines against N. meningitidis serogroups A, C, W135, and Y (Cohn et al. 2010). The exception has been the NMB CP that elicits a poor immune response in humans (Wyle et al. 1972). The reason for poor immunogenicity is not known; however, it has been attributed to the fact that humans express α(2–8)-polySia in neural cell-adhesion molecules. Therefore, apprehension that a NMB CP-based vaccine has the potential to induce autoimmune reactions has hampered research and development of a NMB CP-based vaccine (Finne et al. 1983; Saukkonen et al. 1986). However, failure for this concern to materialize among persons carrying anti-CP NMB antibodies has prompted some researchers to call for a NMB CP vaccine development (Robbins et al. 2011). In order to make well-supported decisions in this regard, a detailed understanding of NMB structure and dynamics on varied conditions is among the highest priorities.
Structural characterizations of polySia in solution with varying number of units have been advanced mainly using a combination of NMR experimental techniques and molecular modeling tools; however, no clear consensus has been reached, with proposals ranging from a variety of helical conformations to random coils (Michon et al. 1987; Yamasaki and Bacon 1991; Brisson et al. 1992; Henderson et al. 2003; Battistel et al. 2012). With the goal of obtaining greater detail, studies on short oligomers (two and three residues) with the reducing end capped with a methyl group have been reported as well (Yongye et al. 2008). Similar to the internal residues of α(2–8)-polySia, these oligomers have the anomeric carbon of the end residue locked in the α-configuration, with the axial (Figure 1) making it a good model system to study α(2–8)-polySia. However, potential effects arising from the end-cap as compared with the βOH group have not been explored in previous studies. Differences are to be expected, at least in the neighborhood of the reducing end, since the α-configuration represents only about 10% of the population for monomeric Sia in solution. However free α(2–8)-polySia is found mostly (over 99%) with the reducing end in the β configuration (Supplementary data, Figure S1), that is, with the equatorial and the OH axial (Figure 1). The biological implication or relevance of the β configuration is unknown since the vast majority of Sias are linked in the α configuration. Nevertheless, most in vitro studies utilize free polySia, hence the findings reported herein are relevant primarily to in vitro studies but may have potential in vivo applications. Hints of the potential importance of the different configuration at the anomeric end were noted as early as 1987 by Jennings and co-workers (Michon et al. 1987). These workers hypothesized that the reducing end structure was one of the reasons why NMB PS (α(2–8)-polySia) needed more Sia units (~10 residues) than the PS from NMC (α(2–9)-polySia, 6 or more residues) to bind optimally to specific antibodies, but provided scarce experimental and theoretical support for their speculative structural model of α(2–8)-polySia at the reducing residue.
Fig. 1.
Chemical structures of SiaN, corresponding to α(2–8)-linked (Neu5Ac)N oligomers (with N = 2, 3, 4 or 6), constructed for MD studies. Residue numbers throughout are indicated by roman numerals between square brackets. The adopted torsion angle definitions used for analysis are indicated: ϕ = C1’−C2’−O8−C8; ψ = C2’−O8−C8−C7; ω8 = O8−C8−C7−O7; ω7 = O7−C7−C6−O6. The terminal residue was capped with either hydrogen (R = H) or methyl (R = CH3). Two configurations for the anomeric carbon C2 at residue [I] were considered, resulting in: (A) SiaN-αOR; (B) SiaN-βOR.
In the present work we confirm that the reducing end configuration has distinct consequences on the structure of α(2–8)-polySia-βOH in the neighborhood of the reducing end by favoring hydrogen bond (Hbond) formation between the reducing OH and the carboxylate group of the next residue. The existence of this Hbond in a Sia α(2–8)-dimer, indicated as Sia2-βOH, was first predicted by molecular dynamics (MD) studies and subsequently confirmed by NMR spectroscopy. Direct and indirect support for this Hbond is provided by long range HSQMBC (Williamson et al. 2014) and HSQC-ROESY (Davis 1989) experiments, respectively, and a detailed comparison of NMR residual dipolar couplings (RDCs) with predicted RDCs for Sia2 conformers from MD trajectories. The image arising from these studies is that of a stable conformation with well-defined hydration and charge patterns. We then explore the persistence of this structural feature by MD on SiaN oligosaccharides (OS), with N = (2, 3, 4 and 6), and conclude that it remains mostly unaltered for all studied oligomers, forming a motif that is comprised of at least the first two units of the OS. This general conclusion is supported by previously reported OH exchange rates on Sia oligosaccharides (Shinar et al. 2014a).
Results
MD and conformational analysis of Sia2
To characterize the influence of the reducing end on the structure and dynamics of Sia OS, four Sia2-OR (R = H or methyl) analogs were generated in-silico with varying configurations at the anomeric end as follows: (1) Sia2-αOH, (2) Sia2-βOH, (3) Sia2-αOMe and (4) Sia2-βOMe (Figure 1). The βOH form constitutes the major anomeric population in aqueous solutions of PS and OS resulting from chemical or enzymatic hydrolysis of polySia, hence, the isomer is readily observable by NMR experiments. Sia2-αOH, the minor anomer in aqueous solution, accounts for less than 1% of the population (Supplementary data, Figure S1). Sia2-αOMe is the molecule previously studied to model the core structure of polySia (Yongye et al. 2008). Sia2-βOMe is a chimeric form (at least not detected in solution) that allows us to directly gauge the importance of the terminal OH in defining Sia OS shapes by eliminating the partial positive charge associated with the hydroxyl hydrogen while keeping the equatorial orientation of the negatively charged carboxylate group. For analysis, 100,000 snapshots were taken over each of the 1.0 μs MD trajectories.
The first task performed was a statistical analysis of the torsional angles over the MD trajectories of the glycerol linker (ϕ, ψ, ω8, ω7) on Sia2 OS, as defined in Figure 1A. The population analysis, expressed in kcal/mol using a Boltzmann expression, is presented in Figure 2, with two panels per dimer showing the energy maps associated with two dihedral angles each, (ϕ, ψ) on the left and (ω8, ω7) on the right. Each horizontal pair of panels corresponds to each of the probed Sia2 molecules, identified by labels. The torsion energy maps of Sia2-βOH are placed at the top because it is the predominant form found in solution. They reveal a single deep energy minimum with two higher energy regions, populated at much lower ratios. This is not a typical result for glycans, as numerous reports show that sugar dimers have great freedom of movement about the glycosidic linkages to explore the conformational space, and consequently, are highly flexible (Bush et al. 1999; DeMarco and Woods 2008; Sattelle and Almond 2012; Widmalm 2013). It could be expected that because the presence of four torsional angles, rather than the standard two (ϕ, ψ) glycosidic torsion angles in carbohydrates, the compounded degrees of freedom would further favor flexibility. Indeed, this expectation is fulfilled for the other three Sia2 models, rows 2–4 in Figure 2, showing the existence of multiple regions with equivalent energy minimums, typical of flexible OS. Furthermore, the overall patterns of the energy maps of Sia2-αOH and Sia2-(α/β)OMe are alike, indicative of similar conformers and dynamics being produced by the MD runs.
Fig. 2.
Energy (ϕ, ψ) and (ω8, ω7) maps for Sia2 linkages from MD population ratios (1 μs, 300 K). The color scale in kcal/mol is the same for the four Sia2 analogs. The plots imply that Sia2-βOH is largely found in one conformation while the other three analogs are found in multiple conformations. This figure is available in black and white in print and in color at Glycobiology online.
A more detailed examination of the maps reveals that the (ω8, ω7) torsions are not affected by the terminal configuration, since for both Sia2-αOMe and Sia2-βOMe the energy landscapes are mostly identical. The (ϕ, ψ) energy landscapes show small differences around two local minima [(ϕ, ψ) ~(305, 275) and ~(270, 270)]; however, in these cases, the maps for Sia2-βOMe are very similar to those of Sia2-αOH, indicating that the effect is not related to the end group orientation. For Sia2-αOMe, the energy maps can also be compared with results from Yongye et al. (2008). We observe that our torsional values nearly overlap those reported by Yongye et al. However, regions around ω8 ~290° and ψ ~280°, shown here as having relatively low energy (bottom panel in Figure 2), were reported as completely unpopulated in research from Yongye et al. We speculate that this difference is due to under-sampling as in the cited work the MD duration was 100 ns, 10 times shorter than in the current report. In conclusion, the results shown in Figure 2 indicate that the orientation of the anomeric OH in Sia2 is critical for defining the conformation and stabilize the reducing ring's configuration in the β-anomer, while showing little influence over the same characteristics in the α-configuration.
Analysis of Hbond interactions over the MD trajectories of the Sia2 models revealed geometries consistent with an Hbond (distances and angles) between the anomeric βOH on Sia[I] and the negatively charged carboxylate group on Sia[II] (Figure 3A) for 92.4% of the time, an extraordinarily high fraction of the trajectory time. The average distance between Sia[I](O)H2 and the closest O in Sia for Hbonded conformers was 1.8 ± 0.2 Å. On the other hand, the Sia2-αOH isomer only formed this Hbond intermittently ~20% of the time. As a donor, αOH also formed Hbonds with Sia[I]O7 (~10%). Despite these interactions, no additional differences were noticed when Sia2-αOH was compared with the Sia[I]HO2-lacking Sia2-(α/β)OMe, except for minor differences in depth for the different local minimum on the dihedral maps. These results support the notion of the anomeric αOH having a minor role in determining the molecular conformation.
Fig. 3.
Sia2-βOH conformation from MD. (A) Sia2-βOH MD snapshot indicating the predicted Hbond between Sia[I]HO2 and Sia (yellow dashed lines). (B) Average structure of Sia2-βOH over the entire MD trajectory, highlighting the average solvent density (mesh, level 4.0) surrounding the dimer. This figure is available in black and white in print and in color at Glycobiology online.
An important consequence of the steady structure of Sia2-βOH is that it provides a meaningful average model derived from the whole MD trajectory (as described in Materials and methods). In general, this procedure leads to virtual structures with seriously distorted geometries due to the variety of conformations adopted by molecules over long MD simulations. Indeed, the average structures obtained from the other three Sia2 trajectories yielded nonphysical virtual structures. However, the average model of Sia2-βOH has an almost canonical geometry for the rings and linkage, nearly identical to the single model represented in Figure 3A. Another important consequence of the steady structure of Sia2-βOH, is the solvation layer established contains a network of Hbonds between the solvent and Sia2-βOH in the neighborhood of the carboxylate groups, which can be revealed by calculating the density grid for the solvent over the trajectory. This calculation is null for flexible molecules because the water distribution is homogeneous. However, the stabilized Sia2-βOH structure as result of the Sia[I]OH2–Sia Hbond, provides a matrix of localized partially charged atoms to interact with water and thus leading to a specific solvation pattern. Figure 3B shows the differential density obtained for O (red grid) and H (blue grid) atoms from water molecules in interaction with Sia2-βOH.
To quantify similarities and differences on overall three-dimensional structure, we performed a total intra/inter-trajectory comparison of the positions for the 12 ring atoms for the four Sia2 analogs. Since the force field potentials are slightly different for residue I in each analog, they are not adequate as structural reference. Hence, root mean-squared distance (RMSD) fitting minimization of each conformer was performed to the six ring atoms of the Sia[II] residue of a reference conformer. Then the RMSD for the 12 ring atoms in both residues and for all possible pair of conformers was calculated, this time without fitting, to produce results independent of the model selected as reference. The result of this calculation is plotted in Figure 4, where the diagonal and off diagonal quadrants depict intra- and inter-trajectory RMSD comparisons, respectively. The plot clearly shows that, for the whole trajectory, the core structure of Sia2-βOH (bottom-left quadrant) remains well defined through almost the complete duration of the MD run. As evidenced by the small intra-trajectory RMSD values (lower than 2.0 Å), conformational changes during the MD simulation are minor or fleeting. In contrast, the remaining Sia2 analogs (second to fourth diagonal quadrants) lack a preferred conformation evidenced by the high RMSD values. Furthermore, the off-diagonal quadrants show that the Sia2-βOH structure is well differentiated from the other three Sia2 models, again for the whole trajectories.
Fig. 4.
Intra- and inter-MD trajectories’ RMSD of ring atoms (diagonal and off-diagonal quadrants, respectively) of Sia2 analogs. To obtain maps independent of the selected reference structure, models were distance minimized to the six ring atoms of the parametrically identical residue Sia[II] in the first frame of the Sia2-βOH trajectory before calculating the RMSD. Low intra-RMSD values for Sia2-βOH (<2 Å) indicate a rigid-like molecular frame. The other dimers, in contrast, show high flexibility. The plot is diagonally symmetrical, hence data under the scale in the Sia2-βOH column can be observed along the Sia2-βOH row. This figure is available in black and white in print and in color at Glycobiology online.
NMR experiments on Sia2-βOH
The unexpected prediction for the structure of Sia2-βOH with the presence of an Hbond over 90% of the time requires experimental support in order to be confirmed. Isomers of monomeric Sia in solution are present in an approximate proportion of 90:10, Sia-βOH to Sia-αOH, as easily determined by integrating the isolated C3 signals of the α/β isomers from 1D 13C-NMR experiments (Supplementary data, Figure S1). For dimer and longer OS, this ratio increases up to ~99:1 proportion as obtained by 1D 13C-NMR (Supplementary data, Figure S1). Thus, for the purpose of discussing experimental results we consider all Sia OSs to be present in solution solely in the β-configuration, as the other anomer is mostly unobserved.
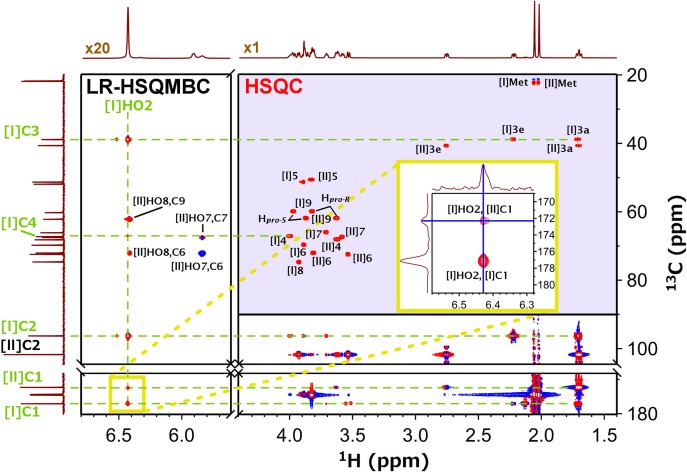
The most direct evidence for Hbonds in solution would be the detection of through-Hbond NMR correlation signals, in this particular case a Sia[I]HO2–Sia[II]C1 correlation. We used optimum conditions for observation of OH signals of anomeric carbons (T = 263 K, pH = 7.0) (Battistel et al. 2017) and ran a modified long-range HSQMBC experiment (Williamson et al. 2014) at 700 MHz to detect the Hbond (Materials and methods; SI). Figure 5 depicts a portion of the spectrum showing the correlation signals of Sia[I]HO2 with its immediate carbon, Sia[I]C2, and both Sia[I]C1 and Sia[II]C1 (assignments obtained from an HSQC-TOCSY experiment, Supplementary data, Figure S2). The inset highlights the region of interest and includes the slices (positions indicated in blue) through Sia[I]HO2 and Sia[II]C1 for clarity. The slice in the 1H dimension shows an asymmetric peak that indicates an additional overlapping correlation for Sia[II]HO8–Sia[II]C1 (see Supplementary data, Figure S3 for a better comparison). Indeed, this additional Hbond is present in the MD trajectory in a ~2% ratio so it is surprising to observe it at low temperatures. The Sia[I]HO2–Sia[II]C1 correlation unequivocally confirms the existence of an Hbond between Sia[I]HO2 and Sia , albeit the Hbonded population and its persistence at higher temperatures have yet to be established. Moreover, although the conformational space is greatly restricted due to this Hbond, there might be additional conformations compatible with this restraint, which may still differ from the one predicted by MD.
Fig. 5.
LR-HSQMBC spectrum with overlaid HSQC for CH crosspeak identification. The external 1D 1H (top, with water suppression) and 13C spectra (left) were obtained separately. The Sia[I]HO2 chemical shift is 6.429 ppm, which shows Sia[I] intra-residue correlations to C2 (96.3 ppm), C3 (38.9 ppm), C4 (67.2 ppm), and C1 (177.1 ppm). Importantly, Sia[I]HO2 also shows a through-Hbond inter-residue correlation to Sia[II]C1 at 172.4 ppm (inset). The blue lines in the inset locate the traces through the Sia[I]HO2–Sia[II]C1 signal. Sia[II]C1 shows an additional correlation with Sia[II]HO8 at 6.416 ppm, albeit weaker than with Sia[I]HO2. (*: minor conformation, <5%). This figure is available in black and white in print and in color at Glycobiology online.
A highly prevalent conformation as predicted from MD for Sia2-βOH should result in distinct RDCs (Battistel et al. 2014), readily differentiated from alternative conformations, due to its restrained conformation. We predicted that RDCs would fit well for Sia2-βOH and yield poor fits to the other structural analogs, due to their flexibility. We used a 13C,15N-enriched sample of Sia2 weakly oriented in a stretched polyacrylamide gel at 300 K to measure 13C-1H, 15N-1H, and 13C-13C RDC (denoted DCH, DNH, and DCC, respectively), totaling 38 experimental values (Figure 6A, Table I). Then we fit each individual MD model to the measured RDCs using a singular value decomposition (SVD) approach (Losonczi et al. 1999), and assessed the goodness of fits with the quality factor Q (Eq. (1)) (Clore and Garrett 1999). Figure 6B shows the result obtained for the four trajectories of Sia2 over the length of the simulations. Because of the large variations in Q values from frame to frame (~ ±0.2) due to normal molecular vibrations and internal motion, plus the large number of frames taken into consideration, the curves were smoothed to reveal the individual trajectory trends at predicting the experimental results. The plot clearly shows that the RDC fits to Sia2-βOH models are significantly and consistently closer to the experimental values over the whole trajectory relative to the other three simulations.
Fig. 6.
Comparison of RDC measurements on Sia2-βOH with MD results. (A) Measured D on Sia2-βOH are represented by (nongray) colored bonds in the 3D scheme (values given in Table I). (B) Calculated Q factors for the four Sia2 MD trajectories (smoothed curves). (C) Experimental vs. calculated RDCs for the average structure of Sia2-βOH (left, included in Table I) and a snapshot resulting in high Q value (right). The MD major conformation predicts well the experimental RDCs. This figure is available in black and white in print and in color at Glycobiology online.
Table I.
Experimental RDCs (Dmeas) and SVD calculated RDCs (DSVD) for Sia2-βOH models from the MD trajectory
| [residue]bond | Dmeas [Hz] | DSVD Sia2-βOH MD Model Best Q, Q = 0.105 | <DSVD> Sia2-βOH MD traj.a <Q> = 0.23 ± 0.06 | Back-calculated <DCC/XH> Sia2-βOH MD traj.bQ = 0.34 | DSVD for <Sia2-βOH>cQ = 0.19 |
|---|---|---|---|---|---|
| [I]C3–H3a | 6.93 ± 1.0 | 8.32 | 4.7 ± 1.46 | 2.73 | 5.40 |
| [I]C3–H3e | 2.54 ± 0.5 | 1.37 | 3.95 ± 2.13 | 5.92 | 4.53 |
| [I]C4–H4 | 8.46 ± 1.2 | 6.4 | 4.97 ± 1.43 | 2.81 | 7.52 |
| [I]C5–H5 | 6.47 ± 1.2 | 4.83 | 4.49 ± 1.55 | 2.57 | 6.10 |
| [I]C6–H6 | 7.64 ± 0.9 | 6.5 | 4.74 ± 1.34 | 2.89 | 5.26 |
| [I]C7–H7 | 3.74 ± 1.4 | 2.17 | 5.2 ± 2.1 | 7.23 | 5.95 |
| [I]C8–H8 | 9.41 ± 1.0 | 5.99 | 5.3 ± 2.25 | 7.27 | 6.87 |
| [I]C9–H9R | 3.94 ± 1.1 | 4.66 | 4.7 ± 3.28 | 3.57 | NA |
| [I]C9–H9S | −9.41 ± 1.0 | −13.04 | −8.87 ± 6.75 | −9.88 | NA |
| [II]C3–H3a | 8.71 ± 1.2 | 14.03 | 9.08 ± 1.33 | 12.95 | 11.98 |
| [II]C3–H3e | −3.29 ± 1.0 | −9.39 | −7.17 ± 2.58 | −9.11 | −9.80 |
| [II]C4–H4 | 12.25 ± 2.1 | 10.85 | 9.03 ± 1.38 | 12.73 | 12.49 |
| [II]C5–H5 | 13.44 ± 1.1 | 13.04 | 9.18 ± 1.6 | 13.29 | 11.27 |
| [II]C6–H6 | 11.35 ± 1.1 | 9.95 | 9.06 ± 1.49 | 12.95 | 11.77 |
| [II]C7–H7 | −13.06 ± 1.3 | −9.88 | −8.6 ± 2.33 | −9.88 | −12.45 |
| [II]C8–H8 | −4.52 ± 3.6 | −4.58 | −6.09 ± 5.44 | −6.81 | NA |
| [II]C9–H9R | 3.45 ± 1.5 | 3.08 | −1.79 ± 7.23 | −1.92 | NA |
| [II]C9–H9S | 5.25 ± 1.5 | 5.39 | 4.93 ± 5.32 | 6.48 | NA |
| [I]N5–H5N | −3.9 ± 0.4 | −3.99 | −2.22 ± 1.08 | −1.07 | −3.48 |
| [II]N5–H5N | −0.4 ± 0.4 | −2.54 | −3.94 ± 1.19 | −5.82 | NA |
| [I]C1–C2 | 0.38 ± 0.2 | 0.37 | 0.42 ± 0.16 | 0.4 | 0.41 |
| [I]C2–C3 | −1.47 ± 0.1 | −1.2 | −1.2 ± 0.16 | −1.13 | −1.38 |
| [I]C3–C4 | 0.31 ± 0.3 | 0.39 | 0.36 ± 0.18 | 0.37 | 0.44 |
| [I]C4–C5 | −0.03 ± 0.2 | 0.23 | 0.34 ± 0.18 | 0.26 | 0.28 |
| [I]C5–C6 | −1.35 ± 0.3 | −1.22 | −1.21 ± 0.16 | −1.14 | −1.35 |
| [I]C6–C7 | 0.12 ± 0.3 | 0.05 | 0.2 ± 0.19 | 0.16 | 0.25 |
| [I]C7–C8 | −1.46 ± 0.4 | −1.16 | −1.16 ± 0.16 | −1.1 | −1.42 |
| [I]C8–C9 | 0.27 ± 0.2 | 0.16 | 0.24 ± 0.21 | 0.22 | 0.35 |
| [I]C5N–CMe | 0.16 ± 0.5 | 0.66 | 0.48 ± 0.24 | 0.45 | NA |
| [II]C1–C2 | 1.26 ± 0.1 | 1.18 | 0.66 ± 0.14 | 0.59 | 0.70 |
| [II]C2–C3 | 0.3 ± 0.2 | 0.41 | 0.46 ± 0.17 | 0.35 | 0.68 |
| [II]C3–C4 | −1.13 ± 0.3 | −0.89 | −0.68 ± 0.2 | −0.63 | −0.87 |
| [II]C4–C5 | −0.77 ± 0.2 | −0.64 | −0.68 ± 0.21 | −0.6 | −0.88 |
| [II]C5–C6 | 0.19 ± 0.4 | 0.39 | 0.42 ± 0.15 | 0.31 | 0.68 |
| [II]C6–C7 | −0.79 ± 0.3 | −0.56 | −0.4 ± 0.2 | −0.32 | −0.41 |
| [II]C7–C8 | 0.12 ± 0.3 | 0.42 | 0.44 ± 0.17 | 0.33 | NA |
| [II]C8–C9 | −0.82 ± 0.2 | −0.14 | −0.4 ± 0.3 | −0.35 | NA |
| [II]C5N–CMe | −0.76 ± 0.1 | −0.8 | −0.42 ± 0.29 | −0.32 | NA |
RDCs are grouped by residue ([I], [II]) and bond type (CH, NH, CC).
aMean values ± standard deviation over the whole MD trajectory. <Q> is the mean of all Q values. Q calculated using Eq. (1) with <DaNH> = −21585.2 * <Aa> and <R > . Q = 0.27 using <DSVD> .
bColumn contains two complementary sets of data: (i) DCH and DNH (indicated as DXH) back-calculated using measured DCC for SVD; and (ii) DCC (indicated as DXH) back-calculated using measured DCH and DNH for SVD.
cCalculated RDCs for the resulting RMSD minimized average structure from the whole Sia2-βOH MD trajectory. Shortened bonds in the resulting virtual structure due to motion were not considered (NA).
The above described RDC predictions are derived from single structure models; thus the overall agreement reflected by the sustained low Q values over the trajectory strongly supports that the MD conformations closely represent the state of Sia2-βOH in solution. An important consequence of the good fittings is that it allowed stereospecific resonance assignments for H9 pro-R’s and H9 pro-S’s, which cannot be performed by traditional NMR methods. Thus, the assignment was achieved by predicting the RDCs for the CH9pro-R/pro-S bonds and comparing with the measured values which was particularly clear in the case of Sia[I]9 (Table I), indicating that H9 1H resonances (in ppm) are: δ(Sia[I]H9 pro-R) = 3.813, δ(Sia[I]H9 pro-S) = 3.968, δ(Sia[II]H9 pro-R) = 3.651, and δ(Sia[II]H9 pro-S) = 3.876 (HSQC, Figure 5).
As mentioned earlier, the Sia2-βOH MD trajectory yielded a well-defined structure for the rings and linker associated with the minimum potential energy represented in Figure 2. However, the glycerol tail of residue Sia[II] was predicted to be much more flexible, which would result in significantly reduced RDC magnitudes. Furthermore, in small molecules like Sia2, lateral groups’ motions can significantly affect the overall alignment tensor, and thus all RDC values, preventing the separate analysis of molecular regions (Almond and Axelsen 2002; Azurmendi and Bush 2002; Azurmendi et al. 2002; Zweckstetter 2008). Our approach to deal with this problem was to include all measured RDCs and to perform a statistical analysis on the calculated RDCs over the trajectory. Subsequently, mean and standard deviation of the predicted values are compared to the experimental ones to draw inferences from it. Thus, along with (i) experimental and (ii) calculated RDCs for the model with lowest Q from the MD trajectory, Table I lists (iii) the mean and standard deviation for the calculated RDCs over the whole Sia2-βOH trajectory, (iv) mean back-calculated RDCs (obtained by using either just DCC or DCH and DNH), and (v) calculated for the RMSD minimized averaged structure over the whole trajectory of Sia2-βOH (Materials and methods).
An initial assessment reveals good overall agreement in all cases, despite the inclusion of RDC values with relatively large experimental error (Sia[II]C8–H8 or Sia[I]C7–C8) or affected by internal motion (Sia[II]C8–H8,C9–H9), that a priori could be problematic. The good fits are further verified by both back-calculating DCH and DNH from the best fit alignment tensor parameters derived with just 18 DCC, as well as predicting the measured DCC from parameters derived using only the 18 measured DCH. Given the small number of RDCs used in each case and the relatively high error for some of them, the value of 0.34 for Q is remarkably good. Finally, we back-calculated RDCs for the undistorted bonds of the averaged model shown in Figure 3B, giving an exceptionally good Q value of 0.19 (Figure 6C, left panel; Table I). Clearly, there is a single outlier value (−3.3, −9.8 Hz), corresponding to Sia[II]C3–H3e, most likely due to an underestimate of the RDC averaging from MD. Otherwise, the predicted RDCs lend support to the MD averaged structure. To illustrate the power of Q in discriminating between structural models, we included the right panel in Figure 6C, showing the calculated vs. experimental RDCs for a trajectory model with relatively high Q value. Models with Q ≥ 0.5 are infrequent for the Sia2-βOH MD trajectory (~1%), but are present on the order of 10% or more for the other three cases. The plot insets are expansions of the DCC range of values for both models. They show W scaling in Eq. (1) at work (Clore and Garrett 1999), by making Q similarly sensitive to RDC values from bonds containing both low and high γ (magnetogyric ratio) nuclei. In this way all measured RDCs can be analyzed simultaneously. This is particularly relevant in glycans where specific types of RDCs may be insufficient for effective analysis.
Smoothing procedures, as implemented in Figure 6B, yield general trends for trajectories; however, they may obscure important details happening at shorter times than the smoothing range in the system analyzed (100 ns here). This is particularly true for RDCs, because of the ambiguous nature of the technique in relation to the symmetric properties of the alignment tensor solutions may not be unique when utilizing a single orienting medium (Canales et al. 2012). We found that this is the case when trying to correlate the short periods during which the Sia[I]HO2–Sia[II]O1 Hbond breaks with increase in Q values, revealing that there is a second narrow set of open structures compatible with the set of measured RDCs, i.e., with Q values as low as the Hbonded model using the 38 measured RDCs. Figure 7A depicts the Sia[I]HO2–Sia[II]O1 distance over the MD trajectory, which shows the transitions from “closed” Hbonded structures to “open” conformations and back. We expected such transitions to be reflected in Q values obtained by SVD with all the experimental data (panel 7E), however this was not evident for some cases. Interestingly, a correlation between Hbond “breakage” and increase in Q values is obtained when using RDCs from just one of the rings (five DCH and six DCC) to determine the overall alignment tensor parameters and then, back-calculating the 38 RDCs and the corresponding Q values (Figure 7, plots F and G). The effect on Q is observed in both cases, but is more pronounced when determining the alignment tensor from Sia[I]-ring RDCs (Figure 7F) over Sia[II]-ring RDCs (Figure 7G), perhaps thanks to the equatorial C1–C2 bond, which is parallel to four out of the five CH RDCs per ring. In summary, RDCs provide essential experimental support at ambient temperature for the existence of the predicted Hbonded structure of Sia2, but further evidence was desirable.
Fig. 7.
Hbond breakage/formation in Sia2-βOH correlated to predicted ROEs and RDCs over the MD trajectory. (A) Sia[I]HO2−Sia[II]O1 distance; (B–D) H−H interatomic distances, between structurally diagnostic atoms; (E–G) Q values obtained considering three sets of RDCs (all, nine ring-[I], and nine ring[II], respectively). Both instantaneous (gray) and “smoothed” (colored) values are shown in all panels. Hbonded structures correlate with diagnostic H–H distances <5 Å (predicting specific ROE observations), and yield good RDC predictions. This figure is available in black and white in print and in color at Glycobiology online.
Another strong indication for the presence of an HO2 Hbond in Sia2-βOH is the recently published OH exchange rate constants in Sia OSs (Shinar et al. 2014a). The reported OH/OD exchange rate constant for HO2 in the dimer was 3.4 s−1, which is two-fold smaller than the next lowest value, and at least an order of magnitude smaller than the rapidly exchanging OHs at pH 7–7.5, 268 K. Based on this rate constant, we measured ROE data (a better alternative than NOE for molecules this size) under varied conditions in order to detect short internuclear distances between Sia[I]HO2 and nearby CHs to compare to structural data from MD simulations. Figure 8A shows the average MD structure of Sia2-βOH with HO2–H distances that are less than 5 Å, indicated with dashed lines (Supplementary data, Table S1). Predicted inter-residue distances between Sia[I]H6 and Sia[II]H3a,e are distinctive for the MD predicted conformation. For instance, evaluating averaged distances for Sia2-αOH trajectories, yields 5.07 ± 0.95 Å and 5.21 ± 0.93 Å for Sia[I]H6 and Sia[II]H3a,e, respectively. Furthermore, distance analysis over the trajectory of Sia2-βOH shows excellent correlation between breakage of the [I]HO2–[II]O1 Hbond (H–O distance > 3 Å; Figure 7A) and departure from the interatomic distance necessary to observe Sia[I]HO2/H6–Sia[II]H3a,e ROE cross peaks (H–H distance limit ~5 Å, Figure 7B–D). Thus, simultaneous observation of 1H–1H ROEs between all indicated atoms in Figure 8A is diagnostic support of the averaged MD structure. The HSQC-ROESY spectrum (Figure 8B) shows 1H correlations to other nearby 1H's, separated by the chemical shifts of the 13C atoms to which the latter are attached. Strips of the HO2 and H3a,e chemical shift regions show (dashed lines) five main 13C correlations of HO2 with: Sia[I]C6, Sia[I]C4, Sia[I]C5, Sia[I]C3, and Sia[II]C3. In addition, Sia[I]C6 correlates with both Sia[I]H3 and Sia[II]H3, completing the set of predicted correlations from the structure in Figure 8A. On the other hand Sia[II]C6 correlates only with [II]H3, which serves as control that hydrogen atoms farther than 5 Å do not produce spurious signals. Whole spectra and peak assignments are included in SI. In summary, the discussed HSQC-ROESY results require both, a slow Sia[I]HO2 exchange rate value (i.e., Hbonded) and a restrained conformation with concomitant short distances between Sia[II]H3 and both Sia[I]OH2 and Sia[I]H6, providing strong support for the predicted structure of Sia2-βOH.
Fig. 8.
Verification of predicted ROEs for Sia2-βOH. (A) Sia2-βOH average structure from the MD trajectory. Indicated H–H distances (in parentheses) are given for diagnostic atoms expected to produce NMR through-space correlation signals. (B) HSQC-ROESY spectrum of 270 mM Sia2 in 10% D2O/H2O at 263 K. Dashed lines are used as aids in identifying diagnostic cross peaks from the MD predicted structure of Sia2-βOH. This figure is available in black and white in print and in color at Glycobiology online.
Observing OH signals requires running experiments at low temperature to slow down OH exchange. In the present case, the highest temperature still compatible with good quality ROE OH signals was 273 K, but evidence of slow exchange at higher temperatures for Sia[I]HO2 comes from temperature coefficients derived from HSQC-TOCSY experiments where the Sia[I]HO2–C2 correlation is observed up to 283 K (Supplementary data, Figure S4). At higher temperatures, slow exchange is still evident from broadening of the 13C signal in 13C 1D experiments performed in 1:1 solutions of H2O:D2O up to 310 K (Supplementary data, Figure S5). All these observations, together with the room temperature RDC experiments, indicate that the averaged MD conformation is a good representation of the actual structure of free Sia2-βOH in solution, or at least the great majority of the population, directly implying the presence of the stabilizing Sia[I]OH2–Sia Hbond. It is noteworthy that as Sia[I]HO2 exchange rate increases with higher temperatures diagnostic ROE's disappear, consistent with an increased molecular flexibility, yet, none of the experimental data contradicts the MD structure.
MD and conformational analysis of SiaN
An important question arising from the discussed results is whether the conformation adopted by Sia2-βOH is conserved in longer OS, namely SiaN-βOH, with N > 2. To answer this question we performed MD simulations under the same conditions used for Sia2 on SiaN oligosaccharides with N = 3, 4 and 6. We then performed intra- and inter-trajectory RMSD analysis similar to the one shown for Sia2 in Figure 4. As expected, the longer the OS the less the structure is preserved for all residues (higher RMSD, not shown). However, when restricting the analysis to the common 12 ring-atoms in the first two residues of each OS, the intra-trajectory RMSD results for SiaN-βOH stay below 1.0 Å for almost the whole trajectory in each of the four cases (diagonal quadrants in Figure 9A), indicating that the first two residues are equally restrained in all cases. The inter-trajectory comparisons (off-diagonal quadrants) show no significant difference in the structures adopted by residues [I–II] for SiaN>2 OS, but there is a detectable difference between those and the conformation adopted by Sia2, with inter-trajectory RMSD values increasing from ~1.0 to ~2.0 Å. This increase corresponds to a minor change in the average relative orientations between residues Sia[I] and Sia[II], with the Sia[I]OH2–Sia Hbond still effectively stabilizing the anomeric end in the OS. Figure 9B depicts an overlay of the average structures of Sia2-βOH (in green) and the first two residues of the tetramer ([I–II]Sia4-βOH, in blue) showing the small difference obtained between the average structures on the common two residues, yielding an RMSD value of 0.006 for the 12 ring atoms. The last two residues in the tetramer model (colored gray) belong to a single model and are included as visual aid, because the spatial average of these residues results in distorted Sia geometries. The nearly perfect overlay of residues [I–II] as well as the presence of Sia[I]OH2 Hbonded to Sia indicates that these residues are a stable structural motif of free Sia OS in solution. Experimental support for this prediction derived from MD comes again from the work by Shinar et al. (2014a), that contains OH exchange values for SiaN OS with N = [1–6]. The Sia[I]OH2 rates for all OS (i.e., excepting monomer) ranges between ∼3 s−1 and 7 s−1, with the minimum corresponding to the hexamer, suggesting that the Hbond is present on Sia PS as well. By comparison, OH4 rates for all hexamer residues range between 11 s−1 and 17 s−1 (all data obtained at pH 7–7.5, 268 K).
Fig. 9.
The structural motif of the leading two residues of SiaN-βOH oligomers is conserved. (A) Intra- and inter-MD trajectory RMSD of ring atoms of the two terminal residues for four SiaN-βOH oligomers (N = 2,3,4 and 6). (B) Overlay of the average structure of Sia2-βOH and the average structure of the first two residues of Sia4-βOH (the last two residues belong to a single model and are included only for reference). The plot is diagonally symmetrical, hence data under the scale in the Sia2-βOH column can be observed along the Sia2-βOH row. This figure is available in black and white in print and in color at Glycobiology online.
Comparison with crystallographic results
Geometries similar to the ones derived from MD can be found in the X-ray crystallographic database among the few structures reported of SiaN-βOH. A survey of deposited crystallographic structures on the PDB revealed one entry with oligomers (dimer, trimer, and pentamer) of α(2–8)Neu5Ac bound to the neuroaminidase endoNF (wild-type and two mutants) (Schulz et al. 2010). It might be expected that a small OS bounded to a protein would have its conformation affected by multiple interactions with amino acids, but to our surprise, a 0.211 Å RMSD was obtained between the Sia2 averaged MD structure and the Sia2 ligand in endoNF-wt (PDB entry 3GVL), using 15 heavy atoms, six from each ring and three from the linker (Supplementary data, Figure S6). The two mutants reported in the same work (PDB entries 3GVJ and 3GVK) had additional Sia oligomers bound to different sites, but in most cases with the anomeric Sia residue interacting weakly with the protein, and hence poorly defined. The exception was the Sia3 bound at the active site, with a structure in the anomeric end clearly different from the Hbonded motif predicted in this work. This is not surprising because this Sia3 is the final product of the catalyzed reaction at the active site, interacting with various amino acids, which may distort the trimer's torsional angles.
Discussion
The availability of better force fields and computational power for implementation of MD techniques open opportunities to investigate aspects of molecular interactions at the atomic level and sometimes to predict unexpected structural features guiding researchers to new experimental challenges that either support or disprove theoretical predictions. A unique feature of MD and related techniques based in theory is that it allows the generation of chimeric compounds or sparsely populated isomers that are experimentally inaccessible to perform comparative studies. In this work we presented examples of both aspects of MD, first predicting an unexpectedly stable conformation for Sia2-βOH associated with the presence of an Hbond involving the OH on the anomeric Sia[I]C2 and Sia . Second, we generated related compounds in-silico, including an isomer with a very low population in aqueous solution (Sia2-αOH) and a structural analog, which to our knowledge has not been synthesized (Sia2-βOMe), to analyze the importance of both the reducing-residue's hydroxyl group and the anomeric configuration, concluding that the two aspects contribute to yield a well-defined conformation for the dimer.
The MD predictions of the Hbond and a stable conformation for Sia2-βOH led us to seek experimental support by NMR. The presence of the Sia[I]OH2–Sia Hbond was confirmed by a long-range scalar-coupling mediated NMR correlation between Sia[I]HO2–Sia[II]C1. Evidence from RDCs and ROEs strongly support the MD predicted three-dimensional structures. Moreover, diminished OH/OD exchange rate constants obtained by NMR indicate that Sia[I]OH2 exists in an Hbonded state at low temperatures. In addition, the observed H6–H3a,e inter-residue ROEs seem to be possible only for the Hbonded structure. This evidence along with other findings discussed in this report point to the MD predictions as an accurate representation of the dimer in solution. Looking forward, we studied Sia OS of the four forms (SiaN-[α/β]O[H/Met], N = 2, 3, 4 and 6) to explore if the Hbond and the conformation of the two terminal residues remained stable. Again the MD results indicate that the Hbond and conformation of the two-terminal residues are retained. This result is found to be consistent with slow exchange values previously reported for Sia[I]HO2 in OS from dimer up to hexamer (Shinar et al. 2014b). We conclude that by virtue of the Sia[I]OH2–Sia Hbond presence, the two terminal residues at the reducing end of α(2–8)-polySia constitute a stable structural motif in free solution, distinct from the extended polymer structure. A longtime puzzling observation is the requirement of 10 residues of α(2–8)-polySia in order to bind effectively to specific antibodies, about four units more than for analogous cases (Michon et al. 1987). The reported findings suggest that the unique structural motif of the β-reducing end on α(2–8)-polySia may be central in understanding this conundrum and has implications for future in vitro studies when purified polySia OS are used.
Materials and methods
Computational methods
MD simulations were performed using the AMBER 14 software package (Case et al. 2015) in a workstation equipped with four GeForce GTX 780 3GB Graphic Processing Units. The SiaN structures (Figure 1) and parameter files (N = 2, 3, 4 and 6) were created using the XLEaP module included in AMBER with the force-field Glycam-06 h (Kirschner et al. 2008). The initial conformation was produced by energy minimization with the AMBER tool minab using an implicit solvent option. Then, the conformer was charge-neutralized with the appropriate number of Na+ counterions (i.e., N Na+) and solvated with TIP3P (Jorgensen et al. 1983) explicit water models (or TIP5P (Mahoney and Jorgensen 2000), where indicated), again with the XLEaP module. Cubic periodic boundary conditions were used for dimer (Sia2), trimer (Sia3) and tetramer (Sia4) OS. For hexamer (Sia6) OS octahedral periodic conditions were used to reduce the number of water molecules required for solvation. The number of water molecules employed was proportional to the spatial extension of the OS and ranged between 682 for Sia2 to 3759 for Sia6. Nonbonded van der Waals and electrostatic scaling factors for 1–4 interactions were set to unity (SCEE = SCNB = 1) as required by the Glycam force field. Long range electrostatic interactions were computed with particle-mesh Ewald summation (York et al. 1993), with a nonbonded cutoff distance of 8 Å. The systems were energy minimized and then slowly equilibrated at the target temperature of 300 K finalizing the preparation stage with 800 ps MD run using NPT conditions and an integration time for the equations of motion of 2 fs, with hydrogen-containing covalent bonds constrained to their equilibrium lengths using the SHAKE algorithm (van Gunsteren and Berendsen 1977). The same conditions were used for the 1.0 μs production runs. Independence of initial conditions for the production runs were verified by exchanging the configurations for Sia2-OH from α to β and vice versa while keeping the respective dihedrals constant and then running a production run after a short energy minimization and equilibration cycle. Likewise, comparisons of trajectories were performed for Sia2-OH and Sia6-OH solvated with either TIP3P or TIP5P water models. In none of the cases were meaningful differences uncovered upon analysis.
Processing of trajectories was performed using cpptraj, included in AMBER, and python in-house scripts. For analysis, Hbonds were geometrically defined with cutoff values for donor–acceptor distances (dDA) of 3.0 Å and angles (θD-H-A) of 135°. Average structures of Sia[I–II] from MD trajectories of SiaN OS were produced by RMSD fitting of 15 atoms comprising the 2×6 ring atoms of the first two SiaN residues plus the three atoms of the glycerol linker (C7, C8 and O8), and averaging all the atoms positions through the MD trajectory. Figures were created with OriginPro 2015 (www.OriginLab.com; Northampton, MA), PyMol (www.schrodinger.com/pymol), and GIMP (www.gimp.org).
NMR spectroscopy
NMR experiments were performed on natural abundance and uniformly 13C,15N-enriched samples of Sia2. The natural abundance sample was purchased from Nacalai (Tokyo, Japan) and used without further purification. The isotope enriched polySia sample was produced in-house as described previously, hydrolyzed to dimer and purified by ion-exchange chromatography (Azurmendi et al. 2007; Battistel et al. 2012). NMR samples were prepared at pH 7.0 containing 0.1 (wt %) DSS-d6 and 0.1 (wt %) NaN3. The solvent used for measuring RDCs, HSQC-ROESY and LR-HSQMBC was 10% 2H2O/1H2O. Data were collected on a 700 MHz Bruker Avance III equipped with a TCI xyz-gradient CryoProbe. For RDC measurements, 15% acrylamide stretched gels were used as orienting media (Tycko et al. 2000). To circumvent 1H-1H strong coupling effects on 1H-13C RDC, the RDCs were measured by the J-modulation method using CTi-HSQC experiments (Yu et al. 2013). 1H, 13C 1J (1JHC) modulation curves were obtained in isotropic and aligned media and RDC values were derived from the curve-fitted 1JHC values obtained with the samples in aligned or isotropic media. The spectral windows, carrier frequencies, and digital resolution were 12 and 30 ppm, 4.75 and 65 ppm and 16 and 83 Hz/point in the 1H and 13C dimensions, respectively. For each t1 point 8 transients were accumulated. 1H–15N RDC values were determined from 1JHN measured by frequency difference in the 1H and 15N dimension of 1H, 15N coupled HSQC experiments. The spectral windows, carrier frequencies, and digital resolution in the 1H and 15N dimensions were 12 and 5 ppm, 4.75 and 124 ppm and 16 and 3 Hz/point, respectively. For each t1 point 32 transients were accumulated. From these spectra, 1H and 15N 1D traces were extracted and fitted to Gaussian line-shapes in Topspin 3.2. 13C–13C RDC were obtained using 13C-constant time-COSY experiments (Azurmendi and Freedberg 2013), performed using 197 ppm spectral window centered at 100 ppm and 8 and 271 Hz/point resolution for the direct and indirect 13C dimensions, respectively.
Individual MD trajectory models were fitted to measured RDCs using a SVD approach (Losonczi et al. 1999), which we implemented as a Python script, to derive alignment tensor parameters that for the given rigid model best reproduce the experimental RDC values. The goodness of fit for each model is assessed with the quality factor Q (Clore and Garrett 1999), as follows (Eq. (1)):
| (1) |
where the sum is taken over all N measured RDCs, W is a scaling factor for different atom pairs, and the denominator results from assuming a random uniform distribution of RDC. The values of the scaling factor W in Eq. (1) were 1.0, 0.5 and 5.0 for DNH, DCH, and DCC, respectively (as per the current version of NMRPipe (Delaglio et al. 1995)). The denominator results from assuming a random uniform distribution of RDC (Clore and Garrett 1999), with Da = DaNH = −21585*Aa Hz with the adopted scaling, and R the rhombicity of the alignment tensor. Curve smoothing in Figure 6 was done with a percentile function and a window of 500 points (equivalent to 100 ns) available in OriginPro. NMR data were processed and analyzed using TopSpin 3.2 (www.bruker.com), NMRPipe (http://ibbr.umd.edu/nmrpipe), and MestReNova (www.mestrelab.com).
A 100 mM Sia2 sample in natural isotopic abundance, pH 7.0, was used for 13C-edited ROESY experiments (HSQC-ROESY) (Davis 1989). The spectral windows, carrier frequencies, and digital resolution in the 1H and 13C dimensions were 10 and 58 ppm, 5.1 and 50 ppm, and 3 and 80 Hz/point, respectively. For each t1 point 128 transients were accumulated. A 100 ms ROESY spinlock was used. For Hbond detection a 200 mM Sia2 sample, pH 7.0, was used. The spectral windows, carrier frequencies, and digital resolution in the 1H and 13C dimensions were 10 and 180 ppm, 5.1 and 100 ppm, and 3 and 124 Hz/point, respectively. For each t1 point 320 transients were accumulated. The long-range HSQMBC pulse sequence (Williamson et al. 2014) was adapted to include more efficient water suppression with coherence selection gradients at the first INEPT module and during the refocusing INEPT module at the end of the pulse sequence. Additionally, the water trim pulse was removed from the original pulse sequence to avoid exchangeable proton saturation. Finally, shaped water-selective flip-back pulses were introduced before and after the final simultaneous 1H and 13C 180° pulses in the refocusing INEPT pulse sequence element (Sklenar 1990).
Supplementary Material
Acknowledgments
We thank Gary Martin and Josep Sauri for providing the original LR-HSQMBC NMR pulse sequence and Martin Frank for helpful comments.
Supplementary data
Supplementary data is available at Glycobiology online.
Conflict of interest statement
None declared.
Abbreviations
CP, capsular polysaccharide; NMB (NMC), N.meningitidis group B (or C); Hbond, hydrogen bond; HON, label of hydrogen attached to oxygen N (natural number); HSQC, heteronuclear single quantum coherence; MD, molecular dynamics; Neu5Ac, N-acetylneuraminic acid; NMR, nuclear magnetic resonance; OH, hydroxyl chemical group; OS, oligosaccharide; PS, polysaccharide; polySia, polysialic acid; RDC, residual dipolar couplings; ROESY, rotating-frame nuclear Overhauser effect correlation spectroscopy; Sia, sialic acid; SiaN, OS formed with N Sia units; Sia[N], Nth residue of polySia.
References
- Almond A, Axelsen JB. 2002. Physical interpretation of residual dipolar couplings in neutral aligned media. J Am Chem Soc. 124:9986–9987. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 102:439–469. [DOI] [PubMed] [Google Scholar]
- Azurmendi HF, Bush CA. 2002. Tracking alignment from the moment of inertia tensor (TRAMITE) of biomolecules in neutral dilute liquid crystal solutions. J Am Chem Soc. 124:2426–2427. [DOI] [PubMed] [Google Scholar]
- Azurmendi HF, Freedberg DI. 2013. Accurate determinations of one-bond 13 C-13 C couplings in 13 C-labeled carbohydrates. J Magn Reson. 228:130–135. [DOI] [PubMed] [Google Scholar]
- Azurmendi HF, Martin-Pastor M, Bush CA. 2002. Conformational studies of Lewis X and Lewis A trisaccharides using NMR residual dipolar couplings. Biopolymers. 63:89–98. [DOI] [PubMed] [Google Scholar]
- Azurmendi HF, Vionnet J, Wrightson L, Trinh LB, Shiloach J, Freedberg DI. 2007. Extracellular structure of polysialic acid explored by on cell solution NMR. Proc Natl Acad Sci USA. 104:11557–11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistel MD, Azurmendi HF, Freedberg DI. 2017. Glycan OH exchange rate determination in aqueous solution: seeking evidence for transient hydrogen bonds. J Phys Chem B. 121:683–695. [DOI] [PubMed] [Google Scholar]
- Battistel MD, Azurmendi HF, Yu B, Freedberg DI. 2014. NMR of glycans: shedding new light on old problems. Prog Nucl Magn Reson Spectrosc. 79:48–68. [DOI] [PubMed] [Google Scholar]
- Battistel MD, Shangold M, Trinh L, Shiloach J, Freedberg DI. 2012. Evidence for helical structure in a tetramer of alpha2-8 sialic acid: unveiling a structural antigen. J Am Chem Soc. 134:10717–10720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JR, Baumann H, Imberty A, Perez S, Jennings HJ. 1992. Helical epitope of the group B meningococcal alpha(2–8)-linked sialic acid polysaccharide. Biochemistry. 31:4996–5004. [DOI] [PubMed] [Google Scholar]
- Bush CA, Martin-Pastor M, Imberty A. 1999. Structure and conformation of complex carbohydrates of glycoproteins, glycolipids, and bacterial polysaccharides. Annu Rev Biophys Biomol Struct. 28:269–293. [DOI] [PubMed] [Google Scholar]
- Canales A, Jimenez-Barbero J, Martin-Pastor M. 2012. Review: use of residual dipolar couplings to determine the structure of carbohydrates. Magn Reson Chem. 50(Suppl 1)):S80–S85. [DOI] [PubMed] [Google Scholar]
- Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE I, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW et al. 2015. AMBER 2015. San Francisco: University of California. [Google Scholar]
- Clore GM, Garrett DS. 1999. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J Am Chem Soc. 121:9008–9012. [Google Scholar]
- Cohen M, Varki A. 2010. The sialome—far more than the sum of its parts. OMICS: J Integrative Biol. 14:455–464. [DOI] [PubMed] [Google Scholar]
- Cohn AC, MacNeil JR, Harrison LH, Hatcher C, Theodore J, Schmidt M, Pondo T, Arnold KE, Baumbach J, Bennett N et al. 2010. Changes in Neisseria meningitidis disease epidemiology in the United States, 1998–2007: implications for prevention of meningococcal disease. Clin Infect Dis. 50:184–191. [DOI] [PubMed] [Google Scholar]
- Davis DG. 1989. Enhanced correlations in 1 H-detected heteronuclear two-dimensional spectroscopy via spin-locked 1 H magnetization transfer. J Magn Reson. 84:417–424. [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- DeMarco ML, Woods RJ. 2008. Structural glycobiology: a game of snakes and ladders. Glycobiology. 18:426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finne J, Leinonen M, Makela PH. 1983. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 2:355–357. [DOI] [PubMed] [Google Scholar]
- Henderson TJ, Venable RM, Egan W. 2003. Conformational flexibility of the group B meningococcal polysaccharide in solution. J Am Chem Soc. 125:2930–2939. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 79:926–935. [Google Scholar]
- Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. 2008. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J Comput Chem. 29:622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losonczi JA, Andrec M, Fischer MW, Prestegard JH. 1999. Order matrix analysis of residual dipolar couplings using singular value decomposition. J Magn Reson. 138:334–342. [DOI] [PubMed] [Google Scholar]
- Mahoney MW, Jorgensen WL. 2000. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J Chem Phys. 112:8910–8922. [Google Scholar]
- Michon F, Brisson JR, Jennings HJ. 1987. Conformational differences between linear alpha (2----8)-linked homosialooligosaccharides and the epitope of the group B meningococcal polysaccharide. Biochemistry. 26:8399–8405. [DOI] [PubMed] [Google Scholar]
- Pelkonen S, Hayrinen J, Finne J. 1988. Polyacrylamide gel electrophoresis of the capsular polysaccharides of Escherichia coli K1 and other bacteria. J Bacteriol. 170:2646–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JB, Schneerson R, Xie G, Hanson LA, Miller MA. 2011. Capsular polysaccharide vaccine for Group B Neisseria meningitidis, Escherichia coli K1, and Pasteurella haemolytica A2. Proc Natl Acad Sci USA. 108:17871–17875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattelle BM, Almond A. 2012. Assigning kinetic 3D-signatures to glycocodes. PCCP. 14:5843–5848. [DOI] [PubMed] [Google Scholar]
- Saukkonen K, Haltia M, Frosch M, Bitter-Suerman D, Leinonen M. 1986. Antibodies to the capsular polysaccharide of Neisseria meningitidis group B or E. coli K1 bind to the brains of infant rats in vitro but not in vivo. Microb Pathog. 1:101–105. [DOI] [PubMed] [Google Scholar]
- Schiffer MS, Oliveira E, Glode MP, McCracken GH Jr, Sarff LM, Robbins JB. 1976. A review: relation between invasiveness and the K1 capsular polysaccharide of Escherichia coli. Pediatr Res. 10:82–87. [DOI] [PubMed] [Google Scholar]
- Schnaar RL, Gerardy-Schahn R, Hildebrandt H. 2014. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol Rev. 94:461–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EC, Schwarzer D, Frank M, Stummeyer K, Muhlenhoff M, Dickmanns A, Gerardy-Schahn R, Ficner R. 2010. Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF. J Mol Biol. 397:341–351. [DOI] [PubMed] [Google Scholar]
- Shinar H, Battistel MD, Mandler M, Lichaa F, Freedberg DI, Navon G. 2014. a. Sialo-CEST: chemical exchange saturation transfer NMR of oligo- and poly-sialic acids and the assignment of their hydroxyl groups using selective- and HSQC-TOCSY. Carbohydr Res. 389:165–173. [DOI] [PubMed] [Google Scholar]
- Shinar H, Battistel MD, Mandler M, Lichaa F, Freedberg DI, Navon G. 2014. b. Sialo-CEST: chemical exchange saturation transfer NMR of oligo- and poly-sialic acids and the assignment of their hydroxyl groups using selective- and HSQC-TOCSY. Carbohydr Res. 389:165–173. [DOI] [PubMed] [Google Scholar]
- Sklenar V. 1990. Selective excitation techniques for water suppression in one- and two-dimensional NMR spectroscopy. Basic Life Sci. 56:63–84. [DOI] [PubMed] [Google Scholar]
- Tycko R, Blanco FJ, Ishii Y. 2000. Alignment of biopolymers in strained gels: a new way to create detectable dipole–dipole couplings in high-resolution biomolecular NMR. J Am Chem Soc. 122:9340–9341. [Google Scholar]
- van Gunsteren WF, Berendsen HJC. 1977. Algorithms for macromolecular dynamics and constraint dynamics. Mol Phys. 34:1311–1327. [Google Scholar]
- Varki A. 2008. Sialic acids in human health and disease. Trends Mol Med. 14:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 68:132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmalm G. 2013. A perspective on the primary and three-dimensional structures of carbohydrates. Carbohydr Res. 378:123–132. [DOI] [PubMed] [Google Scholar]
- Williamson RT, Buevich AV, Martin GE, Parella T. 2014. LR-HSQMBC: a sensitive NMR technique to probe very long-range heteronuclear coupling pathways. J Org Chem. 79:3887–3894. [DOI] [PubMed] [Google Scholar]
- Wyle FA, Artenstein MS, Brandt BL, Tramont EC, Kasper DL, Altieri PL, Berman SL, Lowenthal JP. 1972. Immunologic response of man to group B meningococcal polysaccharide vaccines. J Infect Dis Antimicrobial Agents. 126:514–521. [DOI] [PubMed] [Google Scholar]
- Yamasaki R, Bacon B. 1991. Three-dimensional structural analysis of the group B polysaccharide of Neisseria meningitidis 6275 by two-dimensional NMR: the polysaccharide is suggested to exist in helical conformations in solution. Biochemistry. 30:851–857. [DOI] [PubMed] [Google Scholar]
- Yongye AB, Gonzalez-Outeirino J, Glushka J, Schultheis V, Woods RJ. 2008. The conformational properties of methyl alpha-(2,8)-di/trisialosides and their N-acyl analogues: implications for anti-Neisseria meningitidis B vaccine design. Biochemistry. 47:12493–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York DM, Darden TA, Pedersen LG. 1993. The effect of long-range electrostatic interactions in simulations of macromolecular crystals – a comparison of the ewald and truncated list methods. J Chem Phys. 99:8345–8348. [Google Scholar]
- Yu B, van Ingen H, Freedberg DI. 2013. Constant time INEPT CT-HSQC (CTi-CT-HSQC) – a new NMR method to measure accurate one-bond J and RDCs with strong 1 H-1 H couplings in natural abundance. J Magn Reson. 228:159–165. [DOI] [PubMed] [Google Scholar]
- Zweckstetter M. 2008. NMR: prediction of molecular alignment from structure using the PALES software. Nat Protoc. 3:679–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.