Abstract
Background
Rainfall patterns are one of the main drivers of dengue transmission as mosquitoes require standing water to reproduce. However, excess rainfall can be disruptive to the Aedes reproductive cycle by “flushing out” aquatic stages from breeding sites. We developed models to predict the occurrence of such “flushing” events from rainfall data and to evaluate the effect of flushing on dengue outbreak risk in Singapore between 2000 and 2016.
Methods
We used machine learning and regression models to predict days with “flushing” in the dataset based on entomological and corresponding rainfall observations collected in Singapore. We used a distributed lag nonlinear logistic regression model to estimate the association between the number of flushing events per week and the risk of a dengue outbreak.
Results
Days with flushing were identified through the developed logistic regression model based on entomological data (test set accuracy = 92%). Predictions were based upon the aggregate number of thresholds indicating unusually rainy conditions over multiple weeks. We observed a statistically significant reduction in dengue outbreak risk one to six weeks after flushing events occurred. For weeks with five or more flushing events, compared with weeks with no flushing events, the risk of a dengue outbreak in the subsequent weeks was reduced by 16% to 70%.
Conclusions
We have developed a high accuracy predictive model associating temporal rainfall patterns with flushing conditions. Using predicted flushing events, we have demonstrated a statistically significant reduction in dengue outbreak risk following flushing, with the time lag well aligned with time of mosquito development from larvae and infection transmission. Vector control programs should consider the effects of hydrological conditions in endemic areas on dengue transmission.
Author summary
Dengue transmission is sensitive to fluctuations in rainfall and other weather conditions because it is transmitted by the Aedes mosquito. Recent studies have identified that extreme rainfall can result in mosquito breeding site flushing. However, these rainfall conditions have neither been described nor evaluated for their potential effect on dengue transmission. In this study, we applied machine learning and regression approaches to identify rainfall thresholds associated with mosquito breeding site flushing in Singapore. We then estimated the association between the number of flushing events per week and dengue outbreaks in the following weeks. Here we demonstrate that flushing events are accurately predicted by historical rainfall patterns. We also show that flushing events are associated with a statistically significant reduction in dengue outbreak risk up to six weeks after the flushing events occurred. This research suggests that dengue predictive and early warning systems must consider hydrological conditions and other contributing factors to accurately predict near-term dengue risk.
Introduction
Dengue, a disease transmitted by Aedes mosquitoes, is a global public health problem. Dengue is endemic in more than 100 tropical and subtropical countries where 30–50% of the global population is at risk for infection [1–4]. Annually, there are an estimated 50–100 million cases of dengue, with 500,000 of these cases developing into life-threatening Dengue Hemorrhagic Fever and Dengue Shock Syndrome [5].
Various weather factors influence dengue incidence. Temperature and humidity impact dengue incidence by affecting adult feeding behavior, larvae development, and mosquito survival [2,5–16]. Small increases in average monthly temperature (e.g., 1°C) have been associated with a considerable increase in dengue incidence, leading to a 45% increase in the number of cases in subsequent months in Brazil and China [4,5,17–30].
Although mosquitoes require sufficient rainfall for breeding and larval development [2,7–9,31,32], too much rainfall can be detrimental [33–35]. Excessive rainfall can cause breeding sites to overflow, disrupting mosquito breeding and destroying developing larvae. Mosquito breeding site “flushing”, where water levels exceed a breeding site’s drainage threshold and wash away mosquito larvae, has been observed in both experimental and field settings [33–36]. In experimental studies, simulated heavy rainfall washed away the majority of mosquito larvae and resulted in significant larvae mortality [33,35,36]. The extent of the effect of flushing depended on rainfall intensity, container size, and larvae age [33,35]. A field study in Singapore demonstrated that dengue incidence is lowest following months where flushing events are most frequent suggesting that flushing events may influence how and when dengue transmission occurs [34].
Multiple studies have described the weather drivers of dengue incidence using regression and time series models [4,5,10,17–30,37–39]. While relationships between temperature and humidity and dengue incidence have been consistent, the relationship between rainfall and dengue incidence has remained unclear. Associations between rainfall and dengue incidence have ranged from weak or no connection [19,25,27–29,39], to as much as a 21% increase in dengue incidence in response to increased rainfall [10,19,26,37]. Machine learning tools have been used to predict the occurrence of dengue based on a combination of weather parameters, including rainfall [40–48]. Though successful in predicting weekly and monthly dengue incidence with over 90% accuracy, these models did not explain the relationship between rainfall and dengue.
One interesting and still unanswered question relates to the influence of flushing on dengue incidence. Observations by Seidahmed and Eltahir [34] of larvae survival in storm drains in Singapore following rainfall events have confirmed that flushing mostly occurs during the Northeast monsoon season. The authors noted that the season with the highest rainfall levels and most flushing events preceded the season with traditionally low dengue incidence. However, the study did not quantify the rainfall patterns leading to flushing nor its effect on dengue spread.
This paper proposes a quantitative approach associating flushing, expressed through rainfall patterns, with the subsequent fluctuations in dengue outbreak risk in Singapore between 2000 and 2016. The methodology uses entomological data from Singapore [34] combined with historical rainfall data to quantify the rainfall conditions associated with mosquito larvae wash-out, or “flushing”. A regression model is then used to estimate the effect of flushing events on dengue outbreaks. We show a statistically significant 16–70% reduction in dengue outbreak risk in one to six weeks following flushing events.
Materials and methods
Study area
Singapore is located on the southern-most tip of the Malay Peninsula with a population of 5.6-million people [49]. Dengue is hyper-endemic, where serotypes I-IV co-circulate, and is transmitted year-round, with peak incidence occurring between July and September [50–53]. Singapore has a tropical rainforest climate (Köppen: Af) with two monsoon seasons, the Northeast and Southwest monsoons. The former is associated with heavy rainfall between November and March while the latter occurs between June and October and is relatively drier [54]. Average annual precipitation is nearly 2.3 meters. Average daily temperature is stable throughout the year, where average daily temperatures of the hottest and coolest months differ by 1.9°C.
Surveillance data
Weekly dengue case counts in Singapore from 2000–2016 (N = 887) were obtained from the Weekly Infectious Disease Bulletin of the Singapore Ministry of Health [55]. Confirmed cases were reported by all public and private hospitals and laboratories who are mandated to report all clinically and lab diagnosed cases of dengue within 24 hours [56,57].
Entomological data
Longitudinal entomological surveys were obtained from a previously published work of Seidahmed and Eltahir [34]. These surveys were conducted in the Geylang neighborhood of Singapore, a highly urbanized neighborhood located east of the Singapore River [34]. Geylang is classified as being hyperendemic for dengue by the National Environmental Agency since dengue transmission occurs year-round [34]. Geylang is estimated to have a resident population of 32,000 and an even larger non-resident population which is a result of the large amount of cheap housing options that are primarily used by foreign laborers [34].
During entomological data collection, random aquatic surveys were completed twice a week between August 2014 and August 2015 (except between February 21st and March 10th), resulting in 107 days of entomological observations. For each survey, trained inspectors examined all potential outdoor natural and artificial mosquito breeding sites (e.g., open and closed roadside storm drains and non-drain sites such as canvas sheets, pails and flowerpots), looking for mosquito aquatic stages in randomly selected neighborhood blocks. Samples of aquatic stages were taken and evaluated for taxonomic classification. A subsample of aquatic specimens was retained until adult emergence to confirm taxonomic identification. Taxonomic keys [58–60] were used to classify sampled aquatic stages and emerged adults [34].
A total of 6,824 samples were taken from potential breeding sites (5,818 samples from open and closed storm drains) [34]. Sixty-seven breeding sites (53 occurring in storm drains) were positive for Ae. aegypti breeding [34]. Particular attention was then given to the 53 positive Ae. aegypti breeding sites that were found in open and closed storm drains [34]. Breeding sites of Ae. Aegypti were mainly found in the southern part of Geylang where denser urban drainage network and low-rise housing predominate [61].
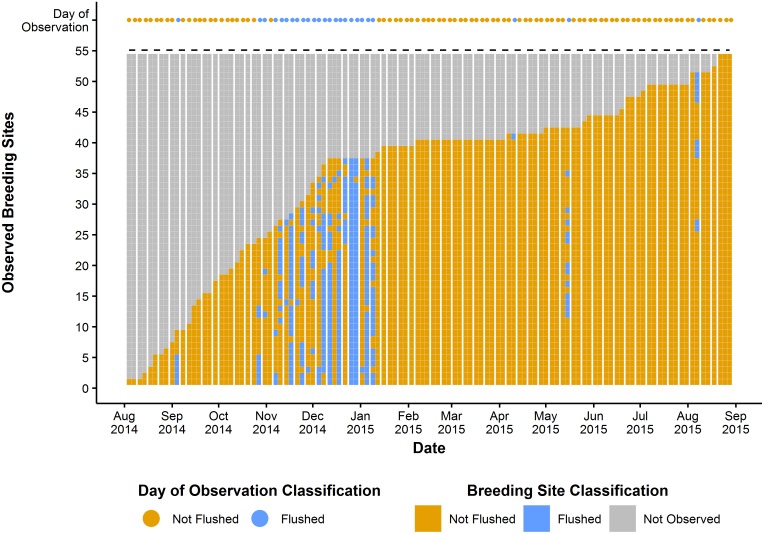
The 53 positive breeding sites occurring in storm drains were continuously monitored for hydrological conditions and changes in the presence of mosquito larvae. For each visit during the monitoring phase, the following four conditions were observed: 1) stagnant water and positive for aquatic stages, 2) stagnant water and negative for aquatic stages, 3) dry and negative for aquatic stages, and 4) flushed and negative for aquatic stages [34]. No sites were identified as flushed and positive for aquatic stages or dry and positive for aquatic stages. We then classified each day of observation as “Flushed” if at least one breeding site was classified as “flushed and negative”, meaning that water had exceeded the drainage threshold (indicated by the storm drain overflowing, or evidence of an overflow) for the breeding site and mosquito larvae were not present. Of the 107 days of entomological observations, 25 were classified as flushed and 82 as non-flushed (Fig 1). The majority of flushing events (84%) occurred during the Northeast monsoon, while only 23% of non-flushing events happened during this time. This data was used to develop a model to predict flushing occurrence for the entire study period.
Fig 1. Timeline of observed Ae. aegypti breeding sites in Geylang, Singapore, August 2014 –August 2015.
There were 107 days of entomological observations where 53 breeding sites, once identified as positive for Ae. aegypti breeding, were monitored for changes in hydrological conditions. Breeding sites where water exceeded the drainage threshold were classified as “Flushed”. For each day of observation, if at least one breeding site was observed as flushed, the day of observation was defined as flushed. If no breeding sites were observed as flushed then the day of observation was classified as not flushed. This figure, “Timeline of observed Ae. aegypti breeding sites in Geylang, Singapore, August 2014 –August 2015 is a derivative of “Timeline of the breeding drains of Aedes aegypti in Geylang, Singapore: August 2014 –August 2015” by Seidahmed and Eltahir [34], used under CC BY.
Weather data
Daily weather data, including rainfall and temperature, were obtained from Tanjong Katong weather station (selected for its proximity to Geylang) [62]. Humidity data were extracted from remote sensed reanalysis data [63] and El Niño Southern Oscillation (ENSO) data were obtained from the Climate Prediction Center [64], both of which are operated by the National Oceanic and Atmospheric Administration. The ENSO index used was the normalized Oceanic Niño Index for Niño region 3.4 [65–68] and used to define the ENSO phase (i.e., El Niño, La Niña, Neutral). Weather data were obtained from 1/1/1999–12/31/2016. Missing weather data were imputed using multiple imputations through chained equations using the MICE R package [69].
Exposure assessment: Modeling and predicting flushing event occurrence
Entomological data detailing the hydrological conditions of breeding sites in Singapore were only available for a single year (August 2014 –August 2015) during the study period (2000–2016). We developed the Predictive fLUshing-Mosquito model (PLUM) which predicts the occurrence of flushing events based on the temporal variation in daily rainfall over several weeks preceding the day of interest and allows the extension of the prediction to dates when no entomological observations are available. The objective was not only to make a daily prediction but to identify more general ‘flushing’ conditions leading to drains’ overflow. Early flooding warning systems use rainfall thresholds to predict flooding occurrence [70]. The PLUM model operates in a similar fashion by identifying a set of variables and their thresholds associated with flushing occurrence.
The PLUM model was developed using the entomological observations and the corresponding rainfall data between August 2014 to August 2015. The general model framework can be found in S1 Fig. The proposed approach identifies rainfall thresholds associated with a higher likelihood of flushing occurrence. We used supervised machine learning models to associate the identified thresholds with flushing occurrence. Each model was trained using a balanced training set where non-flushed observations were randomly under-sampled to generate a 1:1 ratio of flushed to non-flushed observations in the training set to prevent the model from classifying all observations as the majority class [71]. Each model was evaluated on unseen data using leave-one-out cross validation. The PLUM model was then extended to the entire study period, to predict the occurrence of flushing events in Singapore between 2000 and 2016.
Data preprocessing
The PLUM model operates on a daily basis, with an outcome, “flushing”, defined as a day with at least one flushed breeding site recorded in the entomological data. Rainfall variables were created to summarize trends in rainfall. We created 38 time-varying rainfall variables from the weeks preceding the day of interest reflecting frequency, intensity, and total rainfall. Rainfall was characterized using several different variables to capture the different mechanisms by which flushing may occur (i.e., very intense rainfall directly preceding the flushing event or elevated rainfall over several weeks followed by moderate rainfall directly preceding the flushing event). These variables were selected based upon their association with mosquito abundance and because they are conventionally used in studies of mosquito breeding ecology (Table 1) [36,72–76].
Table 1. Rainfall variables created for PLUM model development.
These variables were created to characterize rainfall several different ways in order to capture the different mechanisms by which storm drains may be flushed.
| Variables | Total number of variables created |
|---|---|
| Daily rainfall variables | |
| The number of rainy days in the last 7-day period | 1 variable |
| Average daily rainfall per rainy day in the last 7, 14, 21, and 28-day period | 4 variables |
| The ranked order (1st highest, 2nd highest,…, 7th highest) daily total rainfall in the previous 7-day period | 7 variables |
| Peak daily total rainfall in the previous 1-, 2-,…, 6-day period | 6 variables |
| Cumulative rainfall variables | |
| Cumulative total rainfall covering a period of 1, 2, 3,…, 20 weeks prior | 20 variables |
Identifying thresholds associated with flushing occurrence
One of the objectives of the PLUM model was to identify flushing conditions expressed through the ‘thresholds’ of various combinations of rainfall parameters. We used the Univariate Flagging Algorithm (UFA) [64] to identify conditions leading to flushing.
UFA is a threshold detection algorithm that identifies an optimal cutpoint for a single continuous variable (e.g., 1-week cumulative rainfall) that is associated with a statistically significant higher (“high risk”) likelihood of the outcome (flushing). The algorithm evaluates candidate thresholds along the variable space, selecting the threshold that optimizes the difference in the outcome rate for observations that fall outside of the threshold and a baseline rate. The baseline rate is defined as the outcome rate within the interquartile range of the evaluated variable. Two such thresholds can be identified, one above and one below the median. As recommended by the algorithm developers, statistically significant thresholds were identified using a p-value of 0.01. A total of 36 high risk thresholds were identified. The identified flushing thresholds are specific to storm drains that are similar to the sampled storm drains (see section 2.3 Entomological Data) and are invariant to time because the hydrological characteristics and drainage thresholds will remain constant unless the storm drains are rebuilt using new dimensions, are damaged, or are obstructed by accumulated rubbish or sediment.
PLUM model development and evaluation
The PLUM model predicts daily flushing occurrence based upon two variables, the aggregate number of high risk thresholds that were met per day for both cumulative and daily rainfall variables (all models that were investigated are described in section 1.1 of S1 Text). The general formula for the PLUM model is given as follows:
| (1) |
Where FlushedD is the flushing status for day D; Thresholds MetD, Daily-High Risk is the number of high risk daily rainfall thresholds met for day D; Thresholds MetD, Cumulative-High Risk is the number of high risk cumulative rainfall thresholds met for day D; β1 and β2 are the model coefficients; and β0 is the model intercept. This model used a classification threshold of 0.52, meaning that when predicted values where greater than or equal to 0.52, the model predicted “flushed”, otherwise the model predicted “not flushed”.
We performed a sensitivity analysis to evaluate how different definitions of the outcome variable “flushing” affected PLUM model performance. The sensitivity analysis is described in detail in section 1.2 of S1 Text.
Modeling the association between flushing events and dengue outbreaks
We developed a distributed lag nonlinear logistic regression model [77] to estimate the effect of the number of flushing events per week on the risk of dengue outbreak occurrence, controlling for potential confounders. The model is given as follows:
| (2) |
Where Outbreakw is a binary variable reflecting outbreak status during week W; Weekly flushingw-l is the number of flushing events per week predicted by the PLUM model for week W-l (l [1-20]); Tempw-i is the average weekly temperature for week W-i (i [1-20]); AHw-j is the average weekly absolute humidity for week W-j (j [1-20]) and was estimated using the approach described by Xu et al. [5]; Outbreakw-1 is a binary variable reflecting the outbreak status during week W-1; Monsoonw is the season (Northeast monsoon, Southwest monsoon, Non-monsoon) for week W; and ENSOw is the current ENSO period (El Niño, La Niña, Neutral) for week W.
We define the outcome, Outbreakw, as follows:
| (3) |
where the outbreak threshold for each year is defined as:
| (4) |
The exposure of interest, “Weekly flushingw” was created by predicting whether or not a flushing event occurred for each day in the study period with the PLUM model and aggregating the number of predicted flushing events per week. In Eq (2), a natural cubic spline function was used to model the nonlinear weekly flushing association with 3 degrees of freedom and the lagged association using 3 degrees of freedom.
Potential confounders and modifiers, average weekly temperature, average weekly absolute humidity, monsoon season, ENSO phase, and the previous week’s outbreak status, were selected based upon their known relationship with rainfall or dengue [2,5–16,31,32,78–80]. Data for these variables were obtained from the weather and surveillance data sets. We utilized a simple confounder model which assumed a linear relationship between each confounder and dengue outbreak risk. A simple confounder model was selected because we are interested in the relationship between flushing and dengue outbreak risk and model estimates are relatively insensitive to the choice of confounder model [81]. In this analysis, we included a lag period of one to 20 weeks for the weekly flushing, temperature and humidity variables. This period was selected based upon a literature review on the effects of weather factors on vectors and dengue transmission [51,82–88].
Results
Rainfall conditions associated with flushing events
We applied UFA to identify thresholds associated flushing. Running UFA for all 38 rainfall variables, we identified 36 thresholds (20 for cumulative rainfall variables, 16 for daily rainfall variables) associated with an increased likelihood of flushing occurrence. The likelihood of flushing occurrence was found to increase when variable values were greater than or equal to the associated high risk thresholds. All UFA identified thresholds can be found in S1 Table.
Predicting flushing events from rainfall pattern
In total, there were 107 days of entomological observations, 25 were defined as flushed and 82 were defined as not flushed. We used the PLUM model (described in section 2.5) to classify each entomological observation using leave-one-out cross validation. The PLUM model made predictions based upon two variables, the aggregate number of high risk thresholds that were met per day for both cumulative and daily rainfall variables. The PLUM model achieved 92% accuracy and demonstrated a strong ability to discriminate between flushed and non-flushed observations (Table 2).
Table 2. Performance of PLUM model classification on the unseen test data.
The model used the aggregate number of high risk thresholds that were met per day for both cumulative and daily rainfall variables. Evaluation measures include F1-score (F1), accuracy (Acc), positive predictive value (PPV), Sensitivity (Se), area under the receiver operating characteristics curve (AUC), specificity (Sp), negative predictive value (NPV).
| Model | Variables | Acc | F1 | PPV | Se | AUC | Sp | NPV |
|---|---|---|---|---|---|---|---|---|
| Logistic | # cumulative rainfall high risk thresholds # daily rainfall high risk thresholds |
92% | 0.84 | 0.77 | 0.92 | 0.94 | 0.91 | 0.97 |
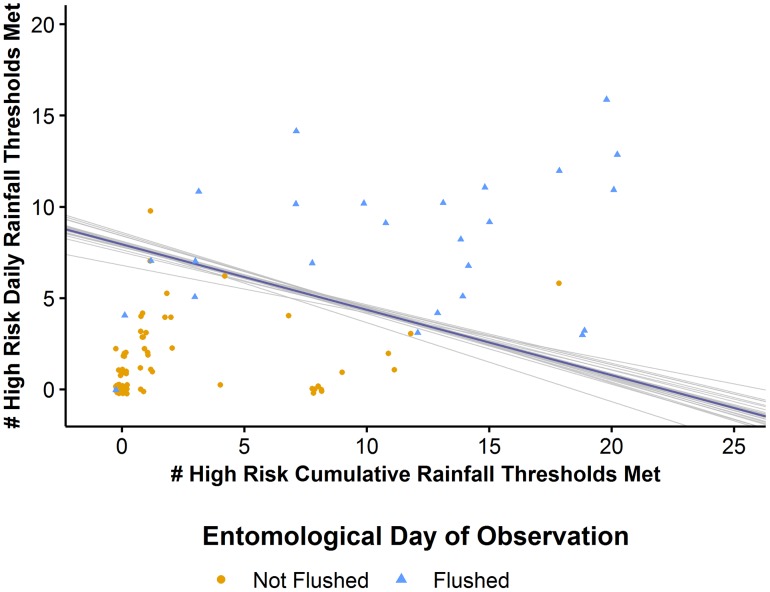
There is a well-defined threshold resulting in nearly perfect separation between flushed and non-flushed observations (Fig 2). We fit Eq (1) to each cross validation sample and identified the mean value for each model coefficient resulting in the following model:
| (5) |
Fig 2. Results of PLUM model associating flushing with the number of high risk thresholds met for cumulative and daily rainfall variables.
There is a clearly defined threshold that almost perfectly separates flushed (blue) and non-flushed (orange) observations based upon the aggregate number of high risk thresholds that were met per day for both cumulative and daily rainfall variables. Each gray line represents Eq (1) fit to each leave-one-out cross validation sample while the blue line represents the mean fit from all of the leave-one-out cross validation samples.
We observed that the fit of Eq (1) for each cross validation sample was stable indicating good generalizability of the PLUM model.
We extended the PLUM model to the entire study period. Using Eq (5) we assigned a value indicating whether or not flushing occurred on each day within the study period. For the study period, 1,242 (21.2%) days were classified as flushing, a similar proportion to the number of flushed days (25, 23.4%) in the observed entomological data time period. These results were used in creating the “weekly flushing” variable which is an aggregator of daily flushing events per week.
Measuring the association between flushing and dengue outbreaks
We used a distributed lag nonlinear logistic regression model to evaluate the association between flushing occurrence and the risk of a dengue outbreak in the weeks following the flushing events.
Dengue incidence was reported in 887 weeks between the years 2000 and 2016. Summary statistics for outbreak occurrence, flushing occurrence, and other weather variables during the study period are presented in Table 3. During the study period, 138 (15.6%) weeks were defined as an Outbreak week based upon the selected criteria. There is also evidence of seasonal variation in outbreak occurrence and rainfall. Outbreak weeks were at least three times as likely to occur during the Southwest monsoon compared with the Northeast and Non-monsoonal periods. Total weekly rainfall shows a seasonal pattern where weeks during the Northeast monsoon have on average 15-20mm more rainfall than other weeks. Due to increased rainfall, flushing events were most likely to occur during the Northeast monsoon as compared with any other season.
Table 3. Descriptive statistics for weekly data on weather variables and dengue outbreak occurrence in Singapore, 2000–2016.
| Variable | Northeast monsoon | Non-monsoon | Southwest monsoon | All Weeks |
|---|---|---|---|---|
| Number of total weeks | 368 | 148 | 371 | 887 |
| Number of outbreak weeks | 32 (8.7%) | 3 (2.0%) | 103 (27.8%) | 138 (15.6%) |
| Total weekly rainfall (Mean ± SD) | 52.6 (60.4) | 38.2 (33.7) | 33.6 (30.4) | 42 (46.5) |
| Daily temperature (Mean ± SD) | 27.2 (0.7) | 28.4 (0.7) | 28.1 (0.7) | 27.8 (0.9) |
| Daily absolute humidity (Mean ± SD) | 25.3 (1.5) | 26.7 (1.0) | 25.8 (1.2) | 25.8 (1.4) |
| Number of flushing events per week (Mean ± SD) | 2.3 (2.5) | 1.2 (1.9) | 0.6 (1.3) | 1.4 (2.1) |
Abbreviations: SD, standard deviation
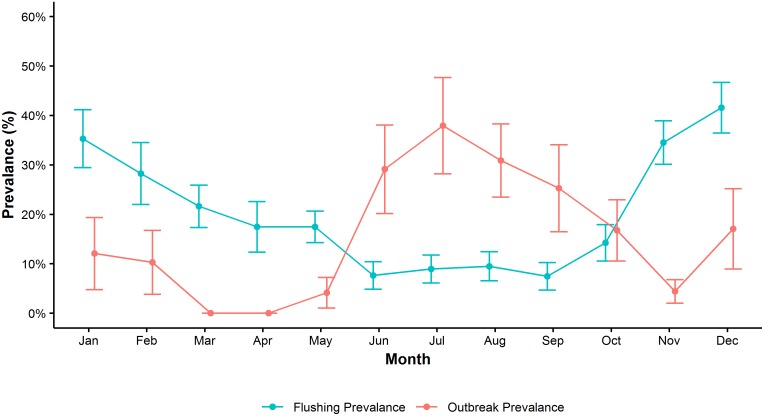
Fig 3 shows the prevalence of flushing events and the prevalence of outbreak weeks by month. Here we observe a negative association in which months where flushing event prevalence is highest (November to February) the outbreak week prevalence is low. Moreover, in months where flushing event prevalence is low (June to September), outbreak week prevalence is highest.
Fig 3. The average prevalence of flushing events and dengue outbreak weeks by month, Singapore, 2000–2016.
In months where flushing event prevalence is highest (blue) dengue outbreak week prevalence (red) is lowest, and vice versa.
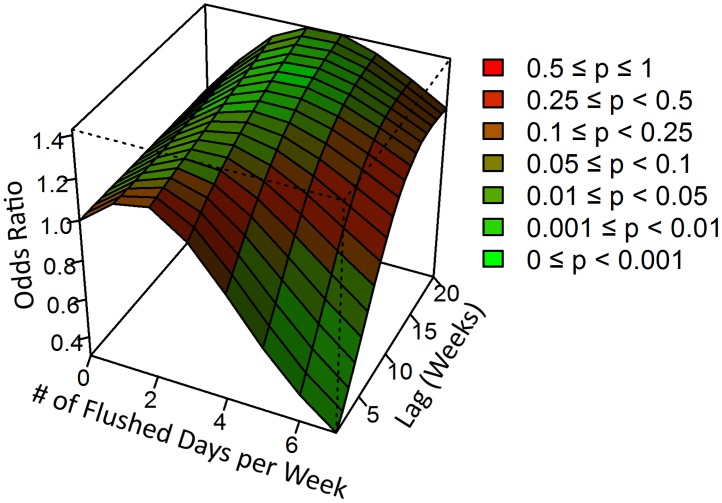
Regression analysis, Eq (2), demonstrates a negative association between flushing events and dengue outbreak risk (Fig 4). We identified a nonlinear association between the number of flushing events per week and dengue outbreak risk that varied over the lag dimension. The risk of an outbreak occurring was significantly lower for weeks where five or more flushing events occurred compared with weeks with zero flushing events; this relationship remained significant up to six weeks after the flushing events occurred (Table 4). Weeks where seven flushing events occurred, there was between a 30–70% reduction in the risk of an outbreak up to six weeks after the flushing events occurred. Smaller reductions in dengue outbreak risk were also observed when five (16–38% reduction in risk up to four weeks after the flushing events) and six (24–56% reduction in risk up to five weeks after the flushing events) flushing events occurred in a week.
Fig 4. Association between number of flushing events per week and dengue outbreak occurrence over 20 lag weeks.
The risk of an outbreak occurring within one to six weeks after the week of consideration was significantly lower if five or more flushing events occurred during the considered week.
Table 4. Association between number of flushing events per week and dengue outbreak occurrence over 20 lag weeks.
When five or more flushing events occurred in a week, compared with 0 flushing events, the risk of a dengue outbreak occurring in the subsequent weeks was significantly reduced up to six weeks after the flushing events occurred.
| Weeks after flushing events | 1 Day Flushed OR (95%CI) |
2 Day Flushed OR (95%CI) |
3 Day Flushed OR (95%CI) |
4 Day Flushed OR (95%CI) |
5 Day Flushed OR (95%CI) |
6 Day Flushed OR (95%CI) |
7 Day Flushed OR (95%CI) |
|---|---|---|---|---|---|---|---|
| 1 Week | 1.13 (0.88–1.44) | 1.16 (0.77–1.74) | 1.04 (0.67–1.62) | 0.84 (0.54–1.30) | 0.62 (0.38–1.02) | 0.44 (0.22–0.87) | 0.30 (0.11–0.79) |
| 2 Weeks | 1.13 (0.93–1.37) | 1.17 (0.85–1.60) | 1.07 (0.76–1.51) | 0.89 (0.64–1.25) | 0.69 (0.47–1.02) | 0.51 (0.29–0.88) | 0.37 (0.17–0.80) |
| 3 Weeks | 1.13 (0.97–1.30) | 1.18 (0.93–1.49) | 1.11 (0.86–1.43) | 0.95 (0.74–1.22) | 0.76 (0.57–1.03) | 0.59 (0.38–0.90) | 0.44 (0.24–0.82) |
| 4 Weeks | 1.13 (1.00–1.27) | 1.19 (0.98–1.44) | 1.14 (0.93–1.39) | 1.01 (0.83–1.22) | 0.84 (0.67–1.05) | 0.67 (0.48–0.94) | 0.53 (0.32–0.86) |
| 5 Weeks | 1.13 (1.01–1.26) | 1.20 (1.01–1.42) | 1.17 (0.97–1.40) | 1.06 (0.89–1.25) | 0.91 (0.75–1.10) | 0.76 (0.57–1.01) | 0.62 (0.40–0.94) |
| 6 Weeks | 1.13 (1.01–1.26) | 1.21 (1.01–1.44) | 1.19 (0.99–1.44) | 1.11 (0.93–1.31) | 0.98 (0.81–1.18) | 0.84 (0.64–1.10) | 0.70 (0.47–1.05) |
| 7 Weeks | 1.13 (1.00–1.27) | 1.22 (1.01–1.47) | 1.22 (1.00–1.49) | 1.15 (0.96–1.39) | 1.04 (0.86–1.27) | 0.91 (0.69–1.20) | 0.79 (0.53–1.19) |
| 8 Weeks | 1.13 (1.00–1.28) | 1.23 (1.00–1.50) | 1.24 (1.00–1.54) | 1.19 (0.98–1.46) | 1.10 (0.89–1.35) | 0.99 (0.74–1.31) | 0.87 (0.57–1.32) |
| 9 Weeks | 1.14 (1.00–1.29) | 1.24 (1.00–1.52) | 1.26 (1.01–1.58) | 1.23 (1.00–1.51) | 1.15 (0.93–1.43) | 1.05 (0.78–1.41) | 0.95 (0.62–1.45) |
| 10 Weeks | 1.14 (1.00–1.29) | 1.25 (1.01–1.53) | 1.29 (1.03–1.61) | 1.26 (1.02–1.56) | 1.20 (0.96–1.49) | 1.11 (0.82–1.49) | 1.01 (0.66–1.54) |
| 11 Weeks | 1.14 (1.01–1.29) | 1.26 (1.03–1.54) | 1.30 (1.05–1.63) | 1.29 (1.05–1.59) | 1.24 (1.00–1.53) | 1.16 (0.87–1.54) | 1.07 (0.71–1.61) |
| 12 Weeks | 1.15 (1.02–1.29) | 1.26 (1.04–1.54) | 1.32 (1.07–1.63) | 1.32 (1.08–1.61) | 1.27 (1.03–1.57) | 1.20 (0.91–1.58) | 1.11 (0.75–1.65) |
| 13 Weeks | 1.15 (1.03–1.29) | 1.27 (1.06–1.53) | 1.34 (1.09–1.64) | 1.34 (1.10–1.63) | 1.30 (1.06–1.59) | 1.23 (0.94–1.60) | 1.15 (0.80–1.66) |
| 14 Weeks | 1.15 (1.03–1.29) | 1.28 (1.07–1.54) | 1.35 (1.11–1.65) | 1.36 (1.12–1.65) | 1.32 (1.08–1.60) | 1.25 (0.98–1.61) | 1.17 (0.83–1.66) |
| 15 Weeks | 1.16 (1.04–1.30) | 1.29 (1.07–1.56) | 1.36 (1.11–1.68) | 1.37 (1.13–1.67) | 1.34 (1.10–1.63) | 1.27 (0.99–1.62) | 1.19 (0.85–1.66) |
| 16 Weeks | 1.16 (1.03–1.31) | 1.30 (1.07–1.59) | 1.38 (1.10–1.73) | 1.39 (1.12–1.72) | 1.35 (1.09–1.66) | 1.28 (1.00–1.64) | 1.20 (0.85–1.69) |
| 17 Weeks | 1.17 (1.02–1.34) | 1.31 (1.04–1.65) | 1.39 (1.07–1.80) | 1.40 (1.10–1.79) | 1.36 (1.08–1.71) | 1.29 (0.98–1.69) | 1.20 (0.83–1.74) |
| 18 Weeks | 1.17 (1.00–1.38) | 1.32 (1.01–1.73) | 1.40 (1.04–1.89) | 1.41 (1.06–1.87) | 1.36 (1.05–1.78) | 1.29 (0.94–1.75) | 1.20 (0.79–1.83) |
| 19 Weeks | 1.18 (0.98–1.43) | 1.33 (0.97–1.83) | 1.41 (0.99–2.01) | 1.42 (1.02–1.97) | 1.37 (1.01–1.86) | 1.29 (0.90–1.84) | 1.19 (0.73–1.96) |
| 20 Weeks | 1.19 (0.95–1.48) | 1.34 (0.93–1.94) | 1.42 (0.95–2.14) | 1.43 (0.97–2.09) | 1.37 (0.96–1.96) | 1.29 (0.85–1.95) | 1.19 (0.67–2.12) |
Bold values highlight statistically significant (p < 0.05) results
Abbreviations: OR, odds ratio; CI, confidence interval
Discussion
Rainfall is an important factor contributing to dengue incidence, in part due to mosquitoes’ reliance upon stagnant water pools to reproduce [7,31,32]. Excessive rainfall can flush out these breeding sites resulting in larvae death [33–35], potentially causing a reduction in dengue incidence in the following weeks.
To evaluate the relationship between excessive rainfall and flushing of larvae from their breeding sites, we developed the PLUM model to identify rainfall conditions that are associated with flushing events and to predict flushing occurrence. The PLUM model predicts daily flushing occurrence based upon the temporal rainfall pattern expressed through aggregation of excessively high cumulative and peak daily rainfall conditions. Our analysis has shown that a variety of temporal rainfall patterns is associated with flushing events. These patterns, highlighted in Fig 2, ranged from heavy rainfall directly preceding the flushing event to the accumulation of excess rainfall over several weeks followed by moderate rainfall directly preceding the flushing event. For example, one flushed observation exceeded 11 daily rainfall thresholds and three cumulative rainfall thresholds (indicating heavy rainfall preceding the event) while another flushed observation exceeded 3 daily rainfall thresholds and 19 cumulative rainfall thresholds (indicating the accumulation of excess rainfall over time followed by a smaller rainfall event that finally triggered flushing). Sensitivity analysis further emphasized the importance of the temporal rainfall pattern rather than single rainfall events. When using only one daily rainfall variable (e.g., peak daily total rainfall in the preceding two days) we observed a marked decrease in model accuracy and its ability to discriminate between flushed and non-flushed observations (Accuracy: 88%; AUC: 0.75) as compared to the PLUM model (Accuracy: 92%; AUC: 0.94; Table 2).
Flushing events were negatively associated with dengue outbreak risk in Singapore between 2000 and 2016 (Figs 3 and 4). This association was observed when five or more flushing events occurred in a week, and persisted up to six weeks after the flushing events occurred. The strongest association was found for weeks where flushing events occurred each day, reducing the risk of a dengue outbreak by as much as 70%. When there were five or six flushing events per week, the observed association was marginally attenuated (a maximum reduction of 48% and 56% respectively). These results provide support for the hypothesis put forth by Seidahmed and Eltahir that in addition to typical climatic, human, and vector drivers, monsoon-driven flushing events affect seasonal abundance of Ae. aegypti via a process of flushing and drying phases. Through this mechanism, excess rainfall flushes out Aedes larvae from the breeding site and subsequent dry periods impede Aedes breeding by preventing the development of adequate breeding sites [34]. Together, these flushing and drying periods can have a harmful effect on dengue transmission.
Our findings, that excess rainfall negatively affects dengue spread, are supported by multiple studies which have observed a similar inverse association between rainfall and mosquito borne disease incidence or dengue virus isolation [29,39,89,90]. For example, a study on the relationship between rainfall and malaria incidence in China found an inverse association (high rainfall was associated with a 15–50% reduction in malaria incidence) when incidence was modeled as a function of total rainfall at the 4th and 6th, 9th, or 12th lagged weeks, hinting at a similar flushing mechanism [90]. In Curaçao, monthly rainfall above 200mm, as compared with mean monthly rainfall at 54mm was associated with a decrease in dengue incidence [29], while in Colombo Sri Lanka, where annual rainfall (2.4m) is similar to Singapore (2.3m), weekly total rainfall was observed to have a weak negative association with dengue incidence [39]. In Senegal, annual dengue virus isolates from mosquitoes decreased 9.7% for each additional 1 in of rainfall from baseline [89].
In contrast, several studies have identified an increase in the risk of dengue associated with increasing rainfall. These reports used statistical models that assumed a linear relationship between rainfall and dengue incidence [19,30,37,39,48]. This approach may obscure a high non-linearity in the studied association, where a positive relationship is observed for low to moderate levels of rainfall and a negative relationship for high rainfall patterns. As excessively high rainfall patterns are rare as compared with more moderate and close to average rainfall patterns, linear models tend to reflect a strong positive relationship between rainfall and dengue incidence observed at low and moderate levels of rainfall [4,30]. Further, several of these and other studies characterized rainfall using a single variable (monthly or weekly average rainfall) [4,10,19,25–30,37,39]. Our analysis has shown that no single variable was sufficient to describe flushing and the associated reduction in dengue outbreak risk. The association was made visible through using a combination of variables reflecting the temporal rainfall pattern over several weeks.
Interestingly, we observed that when fewer than five flushing events occurred per week there was an increase in the risk of a dengue outbreak six to 19 weeks after the flushing events occurred. We believe this association is due to the increased rainfall associated with flushing, but not flushing itself. For example, with increased rainfall new breeding sites may be created, increasing Ae. aegypti abundance and subsequent dengue outbreak risk. This finding highlights the role of moderate rainfall in promoting dengue transmission which has been previously observed [19,30,37,39,48].
There are several limitations to the study. In developing the PLUM model, we could not account for the potential effect of El Niño Southern Oscillation because entomological data were obtained from a single year and the majority of flushed observations (88%) occurred during an El Niño period. Future studies should account for this possibility by analyzing entomological data from El Niño, La Niña, and neutral periods.
The PLUM model was developed using data obtained from 53 breeding sites from a single neighborhood and during a 12-month period. If the drainage thresholds for the roadside storm drains significantly varied across the time or location, PLUM model accuracy and generalizability would be negatively affected resulting in a biased model. Despite these potential limitations, we validated the generalization of the PLUM model on unseen data and found its performance to be robust (Table 2). Furthermore, the drainage conditions should be relatively invariant over time unless the storm drain is damaged or physically changed.
In the current study, we were only able to estimate the effect of the flushing out of breeding sites occurring in road side storm drains. There are other key-breeding sites (e.g., roof gutters, flower pots, and domestic containers) defined by the National Environmental Agency of Singapore that were not evaluated due to ethical and legal considerations [34]. Additional work is needed to evaluate how the flushing of other breeding habitats influence dengue spread.
We did not have information on vector control campaigns during the study period, an important confounding factor that is missing in our study. To compensate for its effect, we statistically adjusted the models for season, as vector control measures vary by season in Singapore.
The developed model associating flushing and dengue outbreak incidence relies on weekly flushing occurrence variable values, which have been inferred though the PLUM model. Even though the PLUM model achieved very high accuracy on the unseen test data (92% correct; Table 2), it still introduced misclassification error in defining days with flushing in the data, though we estimate the influence of such misclassification on the final results to be small.
As with any epidemiological data, the dengue incidence might have been significantly underreported. We assumed this bias be non-differential; at worst we would expect that the effect of underreporting would make results biased towards no association—indicating that the magnitude of the unbiased protective association would be stronger [91].
Conclusions
We proposed a non-linear approach to understanding the relationship between excessive rainfall, flushing, and dengue outbreak occurrence in Singapore. According to the PLUM model, flushing conditions are characterized by rainfall patterns indicating excess rainfall. We demonstrated that rainfall-induced flushing is associated with a statistically significant decreased risk of dengue outbreak, with association being significant up to six weeks after the week when flushing occurred.
Supporting information
The model identifies flushing events of the dengue mosquito Ae. aegypti using variables describing patterns of rainfall in the study areas and entomological data.
(TIF)
(DOCX)
(DOCX)
Data Availability
Relevant data are publically available online. The dataset of entomological observations is available online from Seidahmed and Eltahir (Seidahmed, O. M., & Eltahir, E. A. (2016). A sequence of flushing and drying of breeding habitats of Aedes aegypti (L.) prior to the low dengue season in Singapore. PLoS neglected tropical diseases, 10(7), e0004842). The dataset of weather observations are available from the National Environmental Agency (http://www.weather.gov.sg/climate-historical-daily/) and National Oceanic and Atmospheric Administration (https://rda.ucar.edu/#!lfd?nb=y&b=proj&v=NCEP%20Climate%20Forecast%20System%20Reanalysis and http://origin.cpc.ncep.noaa.gov/products/analysis_monitoring/ensostuff/ONI_v5.php). The dataset of dengue cases is available from (https://www.moh.gov.sg/content/moh_web/home/statistics/infectiousDiseasesStatistics/weekly_infectiousdiseasesbulletin.html).
Funding Statement
This research was supported by internal funding provided by the Charles Stark Draper Laboratory Inc. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Beatty ME, Letson W, Edgil DM, Margolis HS. Estimating the total world population at risk for locally acquired dengue infection. American Journal of Tropical Medicine and Hygiene. AMER SOC TROP MED & HYGIENE 8000 WESTPARK DR, STE 130, MCLEAN, VA 22101 USA; 2007. pp. 221–221. [Google Scholar]
- 2.Hales S, De Wet N, Maindonald J, Woodward A. Potential effect of population and climate changes on global distribution of dengue fever: an empirical model. The Lancet. 2002;360: 830–834. [DOI] [PubMed] [Google Scholar]
- 3.Rezza G. Aedes albopictus and the reemergence of Dengue. BMC Public Health. 2012;12: 72 10.1186/1471-2458-12-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hii YL, Zhu H, Ng N, Ng LC, Rocklöv J. Forecast of dengue incidence using temperature and rainfall. PLoS Negl Trop Dis. 2012;6: e1908 10.1371/journal.pntd.0001908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H-Y, Fu X, Lee LKH, Ma S, Goh KT, Wong J, et al. Statistical modeling reveals the effect of absolute humidity on dengue in Singapore. PLoS Negl Trop Dis. 2014;8: e2805 10.1371/journal.pntd.0002805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan M, Johansson MA. The incubation periods of dengue viruses. PloS One. 2012;7: e50972 10.1371/journal.pone.0050972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christophers S, others. Aedes aegypti (L.) the yellow fever mosquito: its life history, bionomics and structure. Aëdes Aegypti Yellow Fever Mosq Its Life Hist Bionomics Struct. 1960; https://www-cabdirect-org.ezproxy.bu.edu/cabdirect/abstract/19602901825
- 8.Focks DA, Haile DG, Daniels E, Mount GA. Dynamic life table model for Aedes aegypti (Diptera: Culicidae): analysis of the literature and model development. J Med Entomol. 1993;30: 1003–1017. [DOI] [PubMed] [Google Scholar]
- 9.Hopp MJ, Foley JA. Global-scale relationships between climate and the dengue fever vector, Aedes aegypti. Clim Change. 2001;48: 441–463. [Google Scholar]
- 10.Johansson MA, Cummings DA, Glass GE. Multiyear climate variability and dengue—El Nino southern oscillation, weather, and dengue incidence in Puerto Rico, Mexico, and Thailand: a longitudinal data analysis. PLoS Med. 2009;6: e1000168 10.1371/journal.pmed.1000168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rueda LM, Patel KJ, Axtell RC, Stinner RE. Temperature-dependent development and survival rates of Culex quinquefasciatus and Aedes aegypti (Diptera: Culicidae). J Med Entomol. 1990;27: 892–898. [DOI] [PubMed] [Google Scholar]
- 12.Chien L-C, Yu H-L. Impact of meteorological factors on the spatiotemporal patterns of dengue fever incidence. Environ Int. 2014;73: 46–56. 10.1016/j.envint.2014.06.018 [DOI] [PubMed] [Google Scholar]
- 13.Brady OJ, Johansson MA, Guerra CA, Bhatt S, Golding N, Pigott DM, et al. Modelling adult Aedes aegypti and Aedes albopictus survival at different temperatures in laboratory and field settings. Parasit Vectors. 2013;6: 351 10.1186/1756-3305-6-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surtees G. Effects of irrigation on mosquito populations and mosquito‐borne diseases in man, with particular reference to ricefield extension. Int J Environ Stud. 1970;1: 35–42. 10.1080/00207237008709393 [Google Scholar]
- 15.Russell RC. Mosquito-borne arboviruses in Australia: the current scene and implications of climate change for human health. Int J Parasitol. 1998;28: 955–969. 10.1016/S0020-7519(98)00053-8 [DOI] [PubMed] [Google Scholar]
- 16.Faull KJ, Williams CR. Intraspecific variation in desiccation survival time of Aedes aegypti (L.) mosquito eggs of Australian origin. J Vector Ecol. 2015;40: 292–300. 10.1111/jvec.12167 [DOI] [PubMed] [Google Scholar]
- 17.Karim MN, Munshi SU, Anwar N, Alam MS. Climatic factors influencing dengue cases in Dhaka city: A model for dengue prediction. Indian J Med Res. 2012;136: 32–39. [PMC free article] [PubMed] [Google Scholar]
- 18.Depradine CA, Lovell EH. Climatological variables and the incidence of Dengue fever in Barbados. Int J Environ Health Res. 2004;14: 429–441. 10.1080/09603120400012868 [DOI] [PubMed] [Google Scholar]
- 19.Gomes AF, Nobre AA, Cruz OG. Temporal analysis of the relationship between dengue and meteorological variables in the city of Rio de Janeiro, Brazil, 2001–2009. Cad Saude Publica. 2012;28: 2189–2197. [DOI] [PubMed] [Google Scholar]
- 20.Fuller DO, Troyo A, Beier JC. El Niño Southern Oscillation and vegetation dynamics as predictors of dengue fever cases in Costa Rica. Environ Res Lett ERL Web Site. 2009;4: 140111–140118. 10.1088/1748-9326/4/1/014011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart-Ibarra AM, Lowe R. Climate and non-climate drivers of dengue epidemics in southern coastal ecuador. Am J Trop Med Hyg. 2013;88: 971–981. 10.4269/ajtmh.12-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gharbi M, Quenel P, Gustave J, Cassadou S, La Ruche G, Girdary L, et al. Time series analysis of dengue incidence in Guadeloupe, French West Indies: forecasting models using climate variables as predictors. BMC Infect Dis. 2011;11: 166 10.1186/1471-2334-11-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurtado-Díaz M, Riojas-Rodríguez H, Rothenberg SJ, Gomez-Dantés H, Cifuentes E. Short communication: impact of climate variability on the incidence of dengue in Mexico. Trop Med Int Health TM IH. 2007;12: 1327–1337. 10.1111/j.1365-3156.2007.01930.x [DOI] [PubMed] [Google Scholar]
- 24.Wu P-C, Guo H-R, Lung S-C, Lin C-Y, Su H-J. Weather as an effective predictor for occurrence of dengue fever in Taiwan. Acta Trop. 2007;103: 50–57. 10.1016/j.actatropica.2007.05.014 [DOI] [PubMed] [Google Scholar]
- 25.Pinto E, Coelho M, Oliver L, Massad E. The influence of climate variables on dengue in Singapore. Int J Environ Health Res. 2011;21: 415–426. 10.1080/09603123.2011.572279 [DOI] [PubMed] [Google Scholar]
- 26.Cheong YL, Burkart K, Leitão PJ, Lakes T. Assessing weather effects on dengue disease in Malaysia. Int J Environ Res Public Health. 2013;10: 6319–6334. 10.3390/ijerph10126319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan J, Lin H, Wang C, Bai L, Yang S, Chu C, et al. Identifying the high-risk areas and associated meteorological factors of dengue transmission in Guangdong Province, China from 2005 to 2011. Epidemiol Infect. 2014;142: 634–643. 10.1017/S0950268813001519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu L, Lin H, Tian L, Yang W, Sun J, Liu Q. Time series analysis of dengue fever and weather in Guangzhou, China. BMC Public Health. 2009;9: 395 10.1186/1471-2458-9-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limper M, Thai KTD, Gerstenbluth I, Osterhaus A, Duits AJ, Gorp ECM. Climate factors as important determinants of dengue incidence in Curaçao. Zoonoses Public Health. 2016;63: 129–137. 10.1111/zph.12213 [DOI] [PubMed] [Google Scholar]
- 30.Promprou S, Jaroensutasinee M, Jaroensutasinee K. Climatic Factors Affecting Dengue Haemorrhagic Fever Incidence in Southern Thailand. 2005; http://apps.who.int/iris/bitstream/10665/164135/1/dbv29p41.pdf
- 31.Morrison AC, Gray K, Getis A, Astete H, Sihuincha M, Focks D, et al. Temporal and geographic patterns of Aedes aegypti (Diptera: Culicidae) production in Iquitos, Peru. J Med Entomol. 2004;41: 1123–1142. [DOI] [PubMed] [Google Scholar]
- 32.Hammond SN, Gordon AL, Lugo Edel C, Moreno G, Kuan GM, López MM, et al. Characterization of Aedes aegypti (Diptera: Culcidae) production sites in urban Nicaragua. J Med Entomol. 2007;44: 851–860. [DOI] [PubMed] [Google Scholar]
- 33.Koenraadt C. J. M., Harrington LC. Flushing effect of rain on container-inhabiting mosquitoes Aedes aegypti and Culex pipiens (Diptera: Culicidae). J Med Entomol. 2008;45: 28–35. [DOI] [PubMed] [Google Scholar]
- 34.Seidahmed OM, Eltahir EA. A Sequence of Flushing and Drying of Breeding Habitats of Aedes aegypti (L.) Prior to the Low Dengue Season in Singapore. PLoS Negl Trop Dis. 2016;10: e0004842 10.1371/journal.pntd.0004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dieng H, Rahman GS, Hassan AA, Salmah MC, Satho T, Miake F, et al. The effects of simulated rainfall on immature population dynamics of Aedes albopictus and female oviposition. Int J Biometeorol. 2012;56: 113–120. 10.1007/s00484-011-0402-0 [DOI] [PubMed] [Google Scholar]
- 36.Paaijmans KP, Wandago MO, Githeko AK, Takken W. Unexpected High Losses of Anopheles gambiae Larvae Due to Rainfall. PLOS ONE. 2007;2: e1146 10.1371/journal.pone.0001146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su GLS. Correlation of Climatic Factors and Dengue Incidence in Metro Manila, Philippines. Ambio. 2008;37: 292–294. [DOI] [PubMed] [Google Scholar]
- 38.Sari Kovats R. El Niño and human health. 2000; http://apps.who.int/iris/bitstream/10665/57525/1/bu0699.pdf [PMC free article] [PubMed]
- 39.Goto K, Kumarendran B, Mettananda S, Gunasekara D, Fujii Y, Kaneko S. Analysis of Effects of Meteorological Factors on Dengue Incidence in Sri Lanka Using Time Series Data. PLOS ONE. 2013;8: e63717 10.1371/journal.pone.0063717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang Z-H, Tzeng J-S, Chen CC, Chou T-C. A Study of Machine Learning Models in Epidemic Surveillance: Using the Query Logs of Search Engines. PACIS. 2010. p. 137. https://pdfs.semanticscholar.org/59b5/cc10cd7c471a146bde9420bf1607b296d0c6.pdf
- 41.Zhu G, Hunter J, Jiang Y. Improved Prediction of Dengue Outbreak Using the Delay Permutation Entropy. 2016 IEEE International Conference on Internet of Things (iThings) and IEEE Green Computing and Communications (GreenCom) and IEEE Cyber, Physical and Social Computing (CPSCom) and IEEE Smart Data (SmartData). 2016. pp. 828–832. 10.1109/iThings-GreenCom-CPSCom-SmartData.2016.172
- 42.Yusof Y, Mustaffa Z. Dengue outbreak prediction: A least squares support vector machines approach. Int J Comput Theory Eng. 2011;3: 489. [Google Scholar]
- 43.Wickramaarachchi W, Perera SSN, Jayasinghe S. Investigating the impact of climate on dengue disease transmission in urban Colombo: A Fuzzy logic model. International Conference on Computational Mathematics, Computational Geometry & Statistics (CMCGS) Proceedings. Global Science and Technology Forum; 2015. p. 20. http://search.proquest.com/openview/aaa45699bc210d7facc17887ca68e9bd/1?pq-origsite=gscholar&cbl=1536342
- 44.Torres C, Barguil S, Melgarejo M, Olarte A. Fuzzy model identification of dengue epidemic in Colombia based on multiresolution analysis. Artif Intell Med. 2014;60: 41–51. 10.1016/j.artmed.2013.11.008 [DOI] [PubMed] [Google Scholar]
- 45.Buczak AL, Koshute PT, Babin SM, Feighner BH, Lewis SH. A data-driven epidemiological prediction method for dengue outbreaks using local and remote sensing data. BMC Med Inform Decis Mak. 2012;12: 124 10.1186/1472-6947-12-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buczak AL, Baugher B, Babin SM, Ramac-Thomas LC, Guven E, Elbert Y, et al. Prediction of high incidence of dengue in the Philippines. PLoS Negl Trop Dis. 2014;8: e2771 10.1371/journal.pntd.0002771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu K, Wang T, Yang Z, Huang X, Milinovich GJ, Lu Y, et al. Using Baidu Search Index to Predict Dengue Outbreak in China. Sci Rep. 2016;6 10.1038/srep38040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang X, Clements AC, Williams G, Milinovich G, Hu W. A threshold analysis of dengue transmission in terms of weather variables and imported dengue cases in Australia. Emerg Microbes Infect. 2013;2: e87 10.1038/emi.2013.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singapore Department of Statistics. Population Trends 2016 [Internet]. 2016 Sep. Report No.: 1793–2424. http://www.singstat.gov.sg/docs/default-source/default-document-library/publications/publications_and_papers/population_and_population_structure/population2016.pdf
- 50.Koh BK, Ng LC, Kita Y, Tang CS, Ang LW, Wong KY, et al. The 2005 dengue epidemic in Singapore: epidemiology, prevention and control. Ann Acad Med Singap. 2008;37: 538 [PubMed] [Google Scholar]
- 51.Hii YL, Rocklöv J, Ng N, Tang CS, Pang FY, Sauerborn R. Climate variability and increase in incidence and magnitude of dengue incidence in Singapore. Glob Health Action. 2009;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng L-C, Koo C, Mudin RNB, Amin FM, Lee K-S, Kheong CC. 2013 dengue outbreaks in Singapore and Malaysia caused by different viral strains. Am J Trop Med Hyg. 2015;92: 1150–1155. 10.4269/ajtmh.14-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee K-S, Lo S, Tan SS-Y, Chua R, Tan L-K, Xu H, et al. Dengue virus surveillance in Singapore reveals high viral diversity through multiple introductions and in situ evolution. Infect Genet Evol. 2012;12: 77–85. 10.1016/j.meegid.2011.10.012 [DOI] [PubMed] [Google Scholar]
- 54.Loo YY, Billa L, Singh A. Effect of climate change on seasonal monsoon in Asia and its impact on the variability of monsoon rainfall in Southeast Asia. Geosci Front. 2015;6: 817–823. 10.1016/j.gsf.2014.02.009 [Google Scholar]
- 55.Ministry of Health Singapore. Weekly Infectious Diseases Bulletin [Internet]. [cited 21 Apr 2017]. https://www.moh.gov.sg/content/moh_web/home/statistics/infectiousDiseasesStatistics/weekly_infectiousdiseasesbulletin.html
- 56.Ong B. Communicable Diseases Surveillance in Singapore 2015. In: Communicable Diseases Surveillance in Singapore 2015 [Internet]. 11 Nov 2016 [cited 31 May 2017]. https://www.moh.gov.sg/content/moh_web/home/Publications/Reports/2016/communicable-diseases-surveillance-in-singapore-2015.html
- 57.Althouse BM, Ng YY, Cummings Derek A. T. Prediction of Dengue Incidence Using Search Query Surveillance. PLoS Negl Trop Dis. 2011;5: e1258 10.1371/journal.pntd.0001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattingly PF. Contributions to the mosquito fauna of Southeast Asia. XII. Illustrated keys to the genera of mosquitoes (Diptera, Culicidae). SMITHSONIAN INSTITUTION WASHINGTON DC; 1971.
- 59.Huang Y-M. A pictorial key to the mosquito genera of the world, including subgenera of Aedes and Ochlerotatus (Diptera: Culicidae). CISInsecta Koreana. 2002;19.
- 60.Rueda LM. Pictorial keys for the identification of mosquitoes (Diptera: Culicidae) associated with dengue virus transmission. Walter Reed Army Inst Of Research Washington Dc Department Of Entomology; 2004.
- 61.Seidahmed OM, Lu D, Chong CS, Ng LC, Eltahir EA. Patterns of urban housing shape dengue distribution in Singapore at neighborhood and country scales. GeoHealth. 2018;2: 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.National Environment Agency, Singapore. Historical Daily Records [Internet]. [cited 21 Apr 2017]. http://www.nea.gov.sg/weather-climate/climate/historical-daily-records
- 63.Saha S, Moorthi S, Pan H-L, Wu X, Wang J, Nadiga S, et al. The NCEP climate forecast system reanalysis. Bull Am Meteorol Soc. 2010;91: 1015–1057. [Google Scholar]
- 64.Climate Prediction Center (CPC)Oceanic Nino Index—Data.gov [Internet]. [cited 23 Jan 2018]. https://catalog.data.gov/dataset/climate-prediction-center-cpcoceanic-nino-index
- 65.Trenberth KE. The definition of el nino. Bull Am Meteorol Soc. 1997;78: 2771–2777. [Google Scholar]
- 66.Huang B, Banzon VF, Freeman E, Lawrimore J, Liu W, Peterson TC, et al. Extended Reconstructed Sea Surface Temperature Version 4 (ERSST.v4). Part I: Upgrades and Intercomparisons. J Clim. 2014;28: 911–930. 10.1175/JCLI-D-14-00006.1 [Google Scholar]
- 67.Huang B, Thorne PW, Smith TM, Liu W, Lawrimore J, Banzon VF, et al. Further Exploring and Quantifying Uncertainties for Extended Reconstructed Sea Surface Temperature (ERSST) Version 4 (v4). J Clim. 2015;29: 3119–3142. 10.1175/JCLI-D-15-0430.1 [Google Scholar]
- 68.Liu W, Huang B, Thorne PW, Banzon VF, Zhang H-M, Freeman E, et al. Extended Reconstructed Sea Surface Temperature Version 4 (ERSST.v4): Part II. Parametric and Structural Uncertainty Estimations. J Clim. 2014;28: 931–951. 10.1175/JCLI-D-14-00007.1 [Google Scholar]
- 69.Buuren S, Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J Stat Softw. 2011;45 http://doc.utwente.nl/78938/ [Google Scholar]
- 70.Martina MLV, Todini E, Libralon A. A Bayesian decision approach to rainfall thresholds based flood warning. Hydrol Earth Syst Sci Discuss. 2005;2: 2663–2706. [Google Scholar]
- 71.Olson DL. Data Set Balancing. Data Mining and Knowledge Management. Springer, Berlin, Heidelberg; 2005. pp. 71–80. 10.1007/978-3-540-30537-8_8
- 72.DeGaetano AT. Meteorological effects on adult mosquito (Culex) populations in metropolitan New Jersey. Int J Biometeorol. 2005;49: 345–353. 10.1007/s00484-004-0242-2 [DOI] [PubMed] [Google Scholar]
- 73.Galardo AKR, Zimmerman RH, Lounibos LP, Young LJ, Galardo CD, Arruda M, et al. Seasonal abundance of anopheline mosquitoes and their association with rainfall and malaria along the Matapi River, Amapi, Brazil. Med Vet Entomol. 2009;23: 335–349. 10.1111/j.1365-2915.2009.00839.x [DOI] [PubMed] [Google Scholar]
- 74.Lebl K, Brugger K, Rubel F. Predicting Culex pipiens/restuans population dynamics by interval lagged weather data. Parasit Vectors. 2013;6: 129 10.1186/1756-3305-6-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shone SM, Curriero FC, Lesser CR, Glass GE. Characterizing population dynamics of Aedes sollicitans (Diptera: Culicidae) using meteorological data. J Med Entomol. 2014;43: 393–402. [DOI] [PubMed] [Google Scholar]
- 76.Azil AH, Long SA, Ritchie SA, Williams CR. The development of predictive tools for pre-emptive dengue vector control: a study of Aedes aegypti abundance and meteorological variables in North Queensland, Australia. Trop Med Int Health. 2010;15: 1190–1197. 10.1111/j.1365-3156.2010.02592.x [DOI] [PubMed] [Google Scholar]
- 77.Gasparrini A, Armstrong B, Kenward MG. Distributed lag non-linear models. Stat Med. 2010;29: 2224–2234. 10.1002/sim.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hales S, Weinstein P, Woodward A. Dengue fever epidemics in the South Pacific: driven by El Nino Southern Oscillation? The Lancet. 1996;348: 1664–1665. 10.1016/S0140-6736(05)65737-6 [DOI] [PubMed] [Google Scholar]
- 79.Hales S, Weinstein P, Souares Y, Woodward A. El Niño and the dynamics of vectorborne disease transmission. Environ Health Perspect. 1999;107: 99 10.1289/ehp.9910799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gagnon AS, Bush AB, Smoyer-Tomic KE. Dengue epidemics and the El Niño southern oscillation. Clim Res. 2001;19: 35–43. [Google Scholar]
- 81.Welty LJ, Zeger SL. Are the acute effects of particulate matter on mortality in the National Morbidity, Mortality, and Air Pollution Study the result of inadequate control for weather and season? A sensitivity analysis using flexible distributed lag models. Am J Epidemiol. 2005;162: 80–88. 10.1093/aje/kwi157 [DOI] [PubMed] [Google Scholar]
- 82.Yang HM, Macoris MLG, Galvani KC, Andrighetti MTM, Wanderley DMV. Assessing the effects of temperature on dengue transmission. Epidemiol Infect. 2009;137: 1179–1187. 10.1017/S0950268809002052 [DOI] [PubMed] [Google Scholar]
- 83.Yang HM, Macoris MLG, Galvani KC, Andrighetti MTM, Wanderley DMV. Assessing the effects of temperature on the population of Aedes aegypti, the vector of dengue. Epidemiol Infect. 2009;137: 1188–1202. 10.1017/S0950268809002040 [DOI] [PubMed] [Google Scholar]
- 84.Watts DM, Burke DS, Harrison BA, Whitmire RE, Nisalak A. Effect of temperature on the vector efficiency of Aedes aegypti for dengue 2 virus. Am J Trop Med Hyg. 1987;36: 143–152. [DOI] [PubMed] [Google Scholar]
- 85.Focks DA, Brenner RJ, Hayes J, Daniels E. Transmission thresholds for dengue in terms of Aedes aegypti pupae per person with discussion of their utility in source reduction efforts. Am J Trop Med Hyg. 2000;62: 11–18. [PubMed] [Google Scholar]
- 86.Chaves LF, Morrison AC, Kitron UD, Scott TW. Nonlinear impacts of climatic variability on the density-dependent regulation of an insect vector of disease. Glob Change Biol. 2012;18: 457–468. [Google Scholar]
- 87.Fouque F, Carinci R, Gaborit P, Issaly J, Bicout DJ, Sabatier P. Aedes aegypti survival and dengue transmission patterns in French Guiana. J Vector Ecol. 2006;31: 390–399. [DOI] [PubMed] [Google Scholar]
- 88.Earnest A, Tan SB, Wilder-Smith A. Meteorological factors and El Nino Southern Oscillation are independently associated with dengue infections. Epidemiol Infect. 2012;140: 1244–1251. 10.1017/S095026881100183X [DOI] [PubMed] [Google Scholar]
- 89.Althouse BM, Hanley KA, Diallo M, Sall AA, Ba Y, Faye O, et al. Impact of climate and mosquito vector abundance on sylvatic arbovirus circulation dynamics in Senegal. Am J Trop Med Hyg. 2015;92: 88–97. 10.4269/ajtmh.13-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu Y, Qiao Z, Wang N, Yu H, Feng Z, Li X, et al. Describing interaction effect between lagged rainfalls on malaria: an epidemiological study in south–west China. Malar J. 2017;16: 53 10.1186/s12936-017-1706-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brenner H, Savitz DA. The effects of sensitivity and specificity of case selection on validity, sample size, precision, and power in hospital-based case-control studies. Am J Epidemiol. 1990;132: 181–192. 10.1093/oxfordjournals.aje.a115630 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The model identifies flushing events of the dengue mosquito Ae. aegypti using variables describing patterns of rainfall in the study areas and entomological data.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
Relevant data are publically available online. The dataset of entomological observations is available online from Seidahmed and Eltahir (Seidahmed, O. M., & Eltahir, E. A. (2016). A sequence of flushing and drying of breeding habitats of Aedes aegypti (L.) prior to the low dengue season in Singapore. PLoS neglected tropical diseases, 10(7), e0004842). The dataset of weather observations are available from the National Environmental Agency (http://www.weather.gov.sg/climate-historical-daily/) and National Oceanic and Atmospheric Administration (https://rda.ucar.edu/#!lfd?nb=y&b=proj&v=NCEP%20Climate%20Forecast%20System%20Reanalysis and http://origin.cpc.ncep.noaa.gov/products/analysis_monitoring/ensostuff/ONI_v5.php). The dataset of dengue cases is available from (https://www.moh.gov.sg/content/moh_web/home/statistics/infectiousDiseasesStatistics/weekly_infectiousdiseasesbulletin.html).