Abstract
Whole-exome sequencing (WES) has become a standard method for detecting genetic variants in human diseases. Although the primary use of WES data has been the identification of single nucleotide variations and indels, these data also offer a possibility of detecting copy number variations (CNVs) at high resolution. However, WES data have uneven read coverage along the genome owing to the target capture step, and the development of a robust WES-based CNV tool is challenging. Here, we evaluate six WES somatic CNV detection tools: ADTEx, CONTRA, Control-FREEC, EXCAVATOR, ExomeCNV and Varscan2. Using WES data from 50 kidney chromophobe, 50 bladder urothelial carcinoma, and 50 stomach adenocarcinoma patients from The Cancer Genome Atlas, we compared the CNV calls from the six tools with a reference CNV set that was identified by both single nucleotide polymorphism array 6.0 and whole-genome sequencing data. We found that these algorithms gave highly variable results: visual inspection reveals significant differences between the WES-based segmentation profiles and the reference profile, as well as among the WES-based profiles. Using a 50% overlap criterion, 13–77% of WES CNV calls were covered by CNVs from the reference set, up to 21% of the copy gains were called as losses or vice versa, and dramatic differences in CNV sizes and CNV numbers were observed. Overall, ADTEx and EXCAVATOR had the best performance with relatively high precision and sensitivity. We suggest that the current algorithms for somatic CNV detection from WES data are limited in their performance and that more robust algorithms are needed.
Keywords: CNV prediction, somatic alterations, the cancer genome atlas, CNV algorithms
Introduction
Copy number variations (CNVs) in the human genome can affect gene expression by altering gene dosage, disrupting regulatory or coding sequences or causing structural changes [1–3]. Many CNVs have been shown to be associated, directly or indirectly, with various diseases, such as cancer, neuropsychiatric disorders, and Down syndrome [4–6]. In particular, cancer genomes are often characterized by somatic CNVs, with amplification of oncogenes or deletion of tumor suppressor genes [7]. CNVs can be detected using techniques such as fluorescent in situ hybridization and comparative genomic hybridization (CGH). With the development of array technology, genome-wide approaches using array comparative genomic hybridization (aCGH) and single nucleotide polymorphism (SNP) arrays have become popular. These array-based technologies sample copy number along the genome (a median resolution of ∼10–100 K between probes for high-density platforms) and ‘segmentation’ approaches are used to partition the genome into segments of different copy numbers.
More recently, the development of sequencing technology has led to a widespread use of whole-exome sequencing (WES). Compared with whole-genome sequencing (WGS), WES allows for high coverage (greater sequencing depth) at a relatively low cost by targeting only the protein-coding regions in the genome [8]. The primary use of WES data has been to identify disease-associated single nucleotide variants and indels. Using these data also for identification of CNVs is an enticing proposition, as it offers additional information at no additional cost, but CNV estimation based on WES data has been more difficult. The main difficulty comes from the noise that arises in the hybridization-capture step, in which probes (either in solution or on array) are used to ‘pull-down’ the fragments that correspond to the exonic regions. Despite significant efforts in designing better target-capture probes and better hybridization protocols, the differential efficiencies of the probes result in highly variable read depth along the genome.
A number of CNV detection tools for WES data have been developed [9–24]. Some of these methods are designed for detection of germ line CNVs (single samples without control), while others are for detection of somatic CNVs (with matched controls). These two types of approaches are related in that a germ line CNV calling indirectly uses the rest of the samples as a control. The existing methods vary in their complexity, from simple comparison (e.g. Poisson model-based) of read counts found in equal-size bins with more sophisticated hidden Markov models (HMMs). Nearly all are straightforward adaptations of the approaches already applied to aCGH/SNP array data sets (we had tested 11 such methods previously and had found a wide range of sensitivity and specificity [25, 26]). Normalization for GC bias can and should be performed for most type of high-throughput sequencing data, but these are not sufficient to correct for probe-to-probe difference in hybridization efficiency. As a result, estimation of copy number profiles for WES has been challenging.
In this study, we carry out a comparative study of the algorithms for somatic CNVs using paired tumor/normal samples. There are two major issues for comparison of these algorithms. First, the performance of existing methods varies greatly depending on the scale of the CNVs. Many algorithms, for instance, will perform reliably when the size of the CNVs is large (e.g. hundreds of kilobases) but give erratic result for small CNVs (e.g. exon-level). Thus, it is often too simplistic to draw a general conclusion unless the ranges of CNV sizes are specified. Second and related issue is the lack of true CNV profiles with which the results of the algorithm are to be compared. CGH and SNP array profiles have been used in most cases, but the accuracy of these profiles themselves depend on the algorithms used and the scale of CNVs examined. In particular, although it is possible to estimate tumor fraction using SNP array data and use that information to determine the sample-specific thresholds for amplification and deletion calls, most analytical pipelines ignore this information, which results in incorrect classification of some regions. Simulated data are sometimes used to circumvent this problem, but they can give biased results, especially if the generative model bears resemblance to the model on which an algorithm is based.
We evaluated six somatic CNV detection algorithms using 50 Kidney Chromophobe (KICH), 50 Bladder Urothelial Carcinoma (BLCA), and 50 Stomach adenocarcinoma (STAD) samples from The Cancer Genome Atlas (TCGA) project [27–29]. We chose these data sets because they are among the most recent data sets from the consortium (hence high quality) and both WGS and SNP6.0 profiles were generated on the same set of DNA samples. A brief description of each algorithm is given in ‘Materials and Methods’ section. Compared with previous comparative studies [30–34], a novel aspect of this article is the use of WGS data in addition to SNP 6.0 array data to derive the truth set. By using the overlapping CNVs between those two platforms, the reference set we derived should be more accurate (mostly fewer false positives) than those used in previous comparisons.
Materials and methods
Six algorithms tested
We selected some commonly used CNV tools for paired WES data. They are briefly described below and also summarized in Table 1. These tools essentially contain two steps: normalization for GC content and other biases and segmentation of the log-ratios into discrete regions, each with the same copy number. They differ on their specifics—which and how biases are accounted for, how initial bins are defined, what approaches and criteria are used to separate and merge adjacent regions for segmentation, and how to use other information such as genotype data. In addition, we measured the running time for each algorithm (Supplementary Table S4). With a single processor, these algorithms took between 1.5 and 8 h per sample on average, with EXCAVATOR being the fastest, followed by ADTEx and Control-FREEC.
Table 1.
A summary of the WES CNV detection tools examined in this study
| Tool | Year | Language | Paired or pooled data | Input file type | Segmentation | Feature |
|---|---|---|---|---|---|---|
| ADTEx | 2014 | Python, R | Both | BAM, BEDTools DepthOfCoverage | HMM | Noise reduction |
| Ploidy estimation | ||||||
| CONTRA | 2012 | Python, R | Both | SAM, BAM | CBS | GC correction |
| Control-FREEC | 2011 | C++, R | Paired | SAM, BAM, Pileup, Eland, BED, SOAP, Arachne, BLAT, Bowtie | LASSO | GC correction, mappability |
| EXCAVATOR | 2013 | Perl, R | Both | BAM | HSLM | GC correction, mappability, exon-size correction |
| ExomeCNV | 2011 | R | Paired | BAM, Pileup, GATK DepthOfCoverage | CBS | GC correction, mappability |
| Varscan2 | 2012 | Java, Perl, R | Paired | Pileup | CBS | GC correction |
ADTEx (v.2.0) [24]: ADTEx uses two HMMs to predict copy numbers and genotypes. Depth of coverage ratios are used to predict CNVs, and B allele frequency (BAF) signals are used to estimate the ploidy of tumor and to predict the absolute copy number.
CONTRA (v.2.0.4) [18]: CONTRA uses the basepair-level log ratio to maximally remove the GC-content bias and to correct for an imbalanced library size when read lengths of case and control samples are different. Region-level log ratios are calculated by taking the mean of the basepair-level log ratio in the target region. Large CNVs are predicted using Circular Binary Segmentation (CBS) with the region-level log ratio.
Control-FREEC (v.6.7) [10]: Control-FREEC first calculates the raw copy number profile by counting reads and normalizes the profile based on GC content, ploidy and mappability. A LASSO-base algorithm is used to perform segmentation of the normalized profile.
EXCAVATOR (v.2.2) [20]: EXCAVATOR accounts for the nonuniform read depths of the capture regions. A three-step normalization is performed to reduce the GC-content, mappability and exon size effects. A novel algorithm for segmentation, which takes into account the distance between consecutive exons, was developed to improve the detection of small and large CNV regions.
ExomeCNV (v.1.4) [22]: ExomeCNV firstly calculates the log adjusted ratio and the optimized cutoff based on read coverage, exon length and estimated admixture rate. CNV is called on each exon, and CBS is used to merge individual segments for the final CNV detection.
Varscan2 (v.2.3.6) [16]: By only accepting at least one of a tumor sample and a matched normal reached at the minimum coverage requirement, Varscan2 calculates the depth for the samples individually. Fisher’s exact test is used to determine if the ratio of tumor and normal depth changes significantly. CBS is applied to each target region to merge adjacent small segments into large segments.
Data sets analyzed
We downloaded 50 KICH, 50 BLCA and 50 STAD samples (tumor/normal pairs for WES and WGS) from the Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/), which contains controlled-access sequencing data from TCGA. The list of samples used is in Supplementary Table S1.
Generation of reference CNVs
We downloaded the Affymetrix SNP Array 6.0 Level 3 data via the TCGA data portal (https://tcga-data.nci.nih.gov) for the samples. ‘Level 3’ refers to the copy number profiles obtained using a standard TCGA SNP array processing protocols, which include segmentation by CBS [35]. The data for the three tumor types were processed in the same way; details are described, for example, in the Supplement of the kidney chromophobe paper [27]. We used ‘nocnv’ segmentation, which excluded germ line CNV events (http://www.broadinstitute.org/cancer/software/genepattern/affymetrix-snp6-copy-number-inference-pipeline). CNVs were detected from the WGS data using BIC-seq2 (bin size = 100, lambda = 3), which contains an additional GC and mappability normalization step compared with the original BIC-seq [36]. The ‘true’ CNVs were assumed to be the regions that overlap between the CNVs found in the SNP array and WGS data. We evaluated the agreement of CNVs obtained from WGS and SNP arrays and found that they overlapped by 80.2% (SD: 10.9%). Non-overlapping CNVs between SNP array and WGS were in the regions with low probe density in the SNP array. Gain and loss events were analyzed separately. We obtained a total of 2592 CNVs (1155 gains and 1437 losses) in the 50 KICH samples, 6233 CNVs (3073 gains and 3160 losses) in the 50 BLCA samples, and 3599 CNVs (2101 gains and 1498 losses) in the STAD samples.
Parameters for the algorithms
We used the default parameter settings for each CNV tool (see ‘Discussion’ section). Additional information was provided for the CNV tools when necessary. For example, read length was required for Control-FREEC and ExomeCNV. CONTRA was set to use the largeDeletion option to detect large segmentations. ADTEx, CONTRA, Control-FREEC and EXCAVATOR used the original BAM files as input, whereas ExomeCNV and Varscan2 required a file conversion process. DepthofCoverage from the Genome Analysis Toolkit (GATK v.2.4-7) [37] was used for file conversion for ExomeCNV, and the mpileup format from SAMtools (v.0.1.19) [38] was used for file conversion for Varscan2. ADTEx and Control-FREEC also computed BAF data, which we do not evaluate in this study. We assigned a target region as a gain (log2 ratio ≥ 0.25) or loss (log2 ratio ≤ −0.25).
Results
Variation in CNV counts and sizes among algorithms
We first calculated the CNV counts and the sizes of the gain and loss events for each algorithm. Compared with the 2592 reference CNVs in KICH, the number of CNVs identified by WES CNV algorithms spanned a wide range, from 1163 (EXCAVATOR) to 22 129 (Varscan2) across the 50 KICH samples as shown in Figure 1 and Supplementary Figure S1 (BLCA: 6233 reference CNVs, from 2357 using EXCAVATOR to 52 434 using Varscan2; STAD: 3599 reference CNVs, from 1104 using EXCAVATOR to 19 051 using ExomeCNV). The fractions of gain and loss events were also variable, with the number of loss events ranging from 13.1% (ADTEx) to 57.0% (Varscan2) in KICH (BLCA: from 11.1% (Control-FREEC) to 53.5% (Varscan2); STAD: from 8.0% (Control-FREEC) to 56.9% (Varscan2). In the KICH reference set, 55.4% of the detected CNVs were losses (BLCA: 50.7%; STAD: 41.6%). ADTEx, CONTRA, Control-FREEC and EXCAVATOR identified more gains, while ExomeCNV and Varscan2 identified more losses across the three tumor types (see Figure 1 and Supplementary Figure S1). There could be many reasons for this variation, such as how the data were normalized for the different library sizes between the tumor and normal samples. For example, one approach is to use a multiplicative scaling factor to equalize the library sizes. A more sophisticated version is to do an initial CNV calling to identify the non-CNV regions and use only those regions to compute the multiplicative factor. Another approach is to compute the distribution of log-ratios between tumor and normal using bins and then shift the distribution so that its mode is at zero.
Figure 1.
Boxplots of the total numbers of copy number gains (red) and losses (blue) from the reference set and the six WES CNV detection tools. Empty circles represent the number of CNVs in each KICH sample. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
The distribution of the CNV sizes detected from the three tumor types varied across the six algorithms. In Figure 2, we classified the CNVs into bins of different sizes on a logarithmic scale, with gains in Figure 2A and losses in Figure 2B (BLCA and STAD data in Supplementary Figure S2). Whereas the most frequent size range in the reference set is 10–100 M (KICH), it is <1 K for CONTRA, Control-FREEC, ExomeCNV, Varscan2. For example, the total CNV counts from CONTRA and the reference set were similar, but 38.5% of the CNVs detected by CONTRA were smaller than 1 K despite applying the ‘largeDeletion’ option. Although these algorithms appeared to be limited in detecting arm-level CNVs, it is likely that portions of these arm-level CNVs were detected and classified into other bins. This ‘hypersegmentation’ is a common feature, and a heuristic re-merging step is often used with varying effectiveness in different algorithms. In contrast, EXCAVATOR identified larger CNVs between 1 Mb and 100 Mb often but failed to detect CNVs below 10 Kb. Control-FREEC, ExomeCNV and Varscan2 tended to detect smaller CNVs, while ADTEx most frequently detected medium-size CNVs.
Figure 2.
The number of CNVs stratified by CNV lengths from the reference set and the six WES CNV detection tools for CNV gain (A) and loss events (B). A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
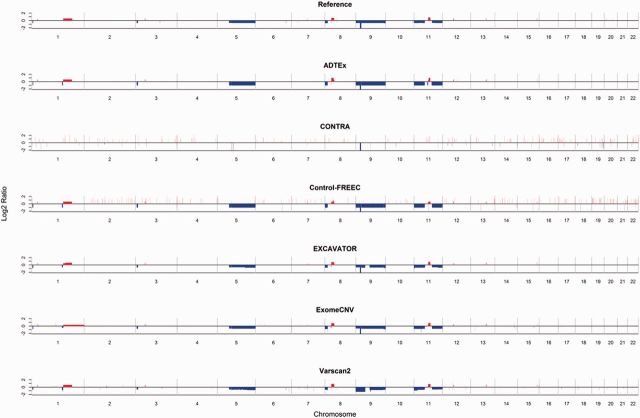
As an illustrative example, Figure 3 shows the results of applying the six tools to one of the BLCA samples (TCGA-4Z-AA7O). All tools were able to detect prominent CNVs in the reference except for CONTRA. The known recurrent homozygous deletion region (9p21; CDKN2A) [39] and several focal or large amplifications/deletions were detected by most of the tools. However, CONTRA and Control-FREEC also called many more focal amplifications and deletions (see other samples in Supplementary Figure S3). For a higher resolution view, Chromosomes 8 and 9 of Figure 3 (TCGA-4Z-AA7O) are shown in Supplementary Figure S4. Our results suggest that most WES CNV tools can reliably detect homozygous deletions or high-level amplifications but not heterozygous deletions or low-level amplifications.
Figure 3.
An example of CNVs detected by the reference set and six WES CNV tools (sample ID: TCGA-4Z-AA7O). The red and blue bars indicate gain and loss events, respectively. All six tools were able to detect the recurrent homozygous deletion in 9p21. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
Overlap between WES CNVs and reference CNVs
To examine the accuracy of the WES CNV tools at the segment level, we first conducted an overlap analysis, measuring the fraction of WES CNVs covered by reference CNVs. We divided the CNVs into gain and loss events, and examined 50% and 90% overlaps (by base pair) of WES CNVs with reference CNVs (Figure 4 and Supplementary Figure S5). We only considered the length of WES CNVs and marked each CNV as a ‘match’ when a WES CNV region overlapped the reference CNVs at greater than or equal to the specified percentage, ‘mismatch’ when the specified minimum overlap was not detected, or ‘opposite direction’ when regions overlapped but a loss region were identified as a gain region or vice versa. For example, in the 50% overlap analysis of KICH samples, 73% and 53% of the gain events in EXCAVATOR and ADTEx had at least 50% covered by the reference CNVs, respectively (77% and 70% for loss events, respectively). The overlaps were less for the other four tools; as we described above, those tools predicted too many CNVs. The results across the different overlap percentages are similar for each algorithm except for ADTEx and EXCAVATOR, because the CNV calls tend to be wholly contained within the reference CNVs (see Figure 2). Overall, ADTEx and EXCAVATOR have higher true positive rates compared with the other algorithms. However, in the 90% overlap analysis, the overlap of the ADTEx and EXCAVATOR-detected CNVs with the reference CNVs decreased to 46% and 53% for gain events and 58% and 57% for loss events, respectively. Because the majority of the CNVs detected from ADTEx and EXCAVATOR were large, some relatively small CNVs from the reference set were included under the 50% criterion but were removed under the 90% criterion. Strikingly, a large number of ‘opposite direction’ CNVs were observed with all six tools. Approximately 1% of the CNVs detected by EXCAVATOR and >10% of the CNVs detected by each of the other tools were classified as ‘opposite direction’.
Figure 4.
The percentages of WES-based CNVs overlapping with the reference CNV set. Match: a CNV region overlaps with the reference at the specified level. Mismatch: no overlapping area is found. Opposite direction: an overlapping gain region was called as a loss, and vice versa. The mean percentages across the 50 KICH samples are shown. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
We also examined the fraction of reference CNV regions covered by the WES CNV tools. For this analysis, the reference CNVs and the six WES CNV results were divided into groups of 50% and 90% reciprocal overlap [40] for comparison. Reciprocal overlap was defined as an instance in which a CNV region from the reference set additionally showed 50% and 90% overlap with the lengths of the WES CNV regions (base pair). In the 50% reciprocal overlap analysis of KICH samples, 194 of the 641 CNVs (30.3%) that EXCAVATOR detected as gains matched with 16.8% of the 1155 gain events from the reference CNVs. Of the 522 CNVs that EXCAVATOR detected as losses, 206 (39.5%) matched with 14.3% of the 1437 loss events from the reference CNVs (Supplementary Table S2A). In other tumor types, the results of the six tools were similar to KICH (Supplementary Table S2B, Supplementary Data). A lower coverage rate was obtained with the 90% overlap criteria.
Precision, recall and the F1-score
To further assess the performance of the six algorithms, we estimated the precision (positive predictive value), recall (sensitivity) and F1-scores. True positive CNVs are defined as concordant CNVs between reference CNVs and WES CNVs, false negatives are reference-only CNVs and false positives are WES-only CNVs. The precision was calculated as the ratio of the number of correctly detected CNVs (i.e. the overlap between each tool and the reference set) to the total number of CNVs detected by a specific tool. The recall was calculated as the ratio of the number of correctly detected CNVs to the total number of CNVs in the reference set. The F1-score was estimated as a weighted average of the precision and recall, with 1 as the best score and 0 as the worst score. We applied the 50% and 90% reciprocal overlap criteria. Figure 5 and Supplementary Figure S5 show the precision, recall and F1-scores under the two overlap percentages in the three tumor types. Under the 50% overlap criterion, the F1-scores across the five algorithms are highly variable, but under the 90% overlap criterion, the differences of the F1-scores become smaller.
Figure 5.
The precision-recall plots of the six WES CNV tools according to the reciprocal overlap criteria. The gray curve indicates constant F1-scores. The best F1-score is 1 and the worst F1-score is 0. The up-triangles represent gain events and the down-triangles indicate loss events. (A) 50% overlap criterion; (B) 90% overlap criterion. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
ADTEx and EXCAVATOR had good performance based on the F1-score using the 50% overlap criterion. ADTEx had slightly higher recall than EXCAVATOR, whereas EXCAVATOR had higher precision than ADTEx. Although the recall rates exhibited by Control-FREEC, ExomeCNV and Varscan2 were high owing to the large number of CNVs detected, the F1-scores were low owing to the small number of true positives identified. The features of the six tools are summarized in Table 2 and Supplementary Table S3.
Table 2.
A summary of the features of the six WES CNV tools in KICH samples
| Tool | Reference | ADTEx | CONTRA | Control-FREEC | EXCAVATOR | ExomeCNV | Varscan2 |
|---|---|---|---|---|---|---|---|
| Gain | |||||||
| Count | 1155 | 8886 | 2075 | 12 456 | 641 | 4454 | 9515 |
| Precision | – | 0.23 | 0 | 0.03 | 0.39 | 0.13 | 0.08 |
| Recall | – | 0.38 | 0 | 0.3 | 0.32 | 0.35 | 0.38 |
| Loss | |||||||
| Count | 1437 | 1340 | 1179 | 4804 | 522 | 4915 | 12 614 |
| Precision | – | 0.42 | 0.06 | 0.05 | 0.48 | 0.11 | 0.04 |
| Recall | – | 0.3 | 0.02 | 0.23 | 0.3 | 0.32 | 0.25 |
The numbers in bold correspond to the highest precision or recall score. Precision and recall were calculated at 50% overlap criterion.
Discussion
We evaluated the capability of six WES CNV algorithms to detect somatic CNVs from 150 paired TCGA tumor samples. Overall, our results are consistent with previous analyses [30–34] that suggested variable performance of the available methods. We confirmed that the CNV counts obtained from each tool varied significantly from the reference set. We found that there may be a bias in detection of gain versus loss—for example, the predictions from ADTEx, CONTRA and Control-FREEC seemed to be biased toward detecting copy gains. We also found that some methods, notably CONTRA and Control-FREEC, tend to give hyper-segmented profile, with most likely a large fraction of false positives. Although CONTRA uses the CBS algorithm on the region-level log ratio to detect large CNVs, we found that CONTRA detected mainly small CNVs and therefore may be unsuitable for large CNV detection. Control-FREEC had a higher accuracy for losses than gains among its detected CNVs. For these tools, parameters for conservative segmentation and a filter with a high log2 ratio may be helpful in reducing false positives. The high recall of Control-FREEC, ExomeCNV and Varscan2 may be attributed to the higher number of CNVs that were predicted. In addition, the considerably larger number of small CNVs detected by these algorithms suggests that their normalization and segmentation algorithms were not applied properly.
The CNVs called by ADTEx and EXCAVATOR had a higher proportion matching with the reference set, while the other CNV tools only had <50% of CNV calls matching the reference set. ADTEx and EXCAVATOR also had a relatively low rate of opposite-direction CNVs compared with the other tools. However, ADTEx and EXCAVATOR tended to detect large CNVs, and thus, the matching rates dropped when a 90% overlap criterion was used. In terms of precision and recall, ADTEx and EXCAVATOR had the best performance based on the F1-score. Although ADTEx and EXCAVATOR appear to be the best choice for somatic CNV detection based on our analysis, we do note that results are likely to vary depending on the specific data sets and parameters.
There are three important limitations to the current study. The first is that we did not attempt to obtain optimal performance for each algorithm by tuning its parameters. For instance, in Control-FREEC, there is a parameter called ‘minCNAlength’ that specifies the minimum number of consecutive windows. We used the default value of 1 in our runs, but setting this value larger removes smaller segments (a related parameter is ‘window’, for which we used the 500 bp recommended for exome data; setting this parameter larger would also remove smaller segments). Although such tuning might improve the performance of each algorithm, it may also make the comparisons more subjective and prone to bias. In addition, these algorithms often have multiple parameters (Control-FREEC has >15 parameters), and attempting to obtain an optimal combination of these parameters is difficult for general users. The second limitation is that there are multiple ways to measure overlap between two segmentation profiles and that none is perfect. We chose to use two measures based on how much a CNV in one profile is covered by CNVs from the other profile (and vice versa). Another possible way is to measure overlap based only on exonic regions, as CNVs covering genes are often most relevant. The third limitation is that, although better than other choices, the ‘reference’ CNV profiles we generated using SNP array and WGS data are not perfect. In particular, the use of SNP array profiles reduces the resolution of CNVs, and it becomes difficult to evaluate the correctness of small CNVs identified from exome data.
We note that, regardless of the method chosen, it would be important to experiment with its parameters to check if the resulting profiles are reasonable (e.g. no hypersegmentation) and to confirm at least a subset of the final call set using additional data from wet-lab experiments or orthogonal platforms. Finally, while some methods do perform more reliably than others, it is clear that more accurate and robust approaches are needed for the growing number of exome data sets.
Key Points
Somatic copy number variants (CNVs) can be detected using paired (tumor and matched normal) whole-exome sequencing (WES) data, but current methods give highly variable results.
Among the six evaluated CNV tools, ADTEx and EXCAVATOR showed the most reliable results for the data sets tested.
Incorporation of whole-genome data is helpful in evaluating the performance of WES-based CNV methods.
More accurate and robust approaches are needed to take full advantage of the large number of exome data sets.
Funding
Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea [grant number: HI13C-2096].
Supplementary Material
References
- 1. Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet 2004;36:949–51. [DOI] [PubMed] [Google Scholar]
- 2. Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science 2004;305:525–8. [DOI] [PubMed] [Google Scholar]
- 3. Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature 2006;444:444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shlien A, Malkin D. Copy number variations and cancer. Genome Med 2009;1:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Megarbane A, Ravel A, Mircher C, et al. The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet Med 2009;11:611–16. [DOI] [PubMed] [Google Scholar]
- 7. Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010;42:30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Backenroth D, Homsy J, Murillo LR, et al. CANOES: detecting rare copy number variants from whole exome sequencing data. Nucleic Acids Res 2014;42:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012;28:423–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coin LJ, Cao D, Ren J, et al. An exome sequencing pipeline for identifying and genotyping common CNVs associated with disease with application to psoriasis. Bioinformatics 2012;28:i370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng X. SeqGene: a comprehensive software solution for mining exome- and transcriptome- sequencing data. BMC Bioinformatics 2011;12:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 2012;91:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gusnanto A, Wood HM, Pawitan Y, et al. Correcting for cancer genome size and tumour cell content enables better estimation of copy number alterations from next-generation sequence data. Bioinformatics 2012;28:40–7. [DOI] [PubMed] [Google Scholar]
- 15. Klambauer G, Schwarzbauer K, Mayr A, et al. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res 2012;40:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res 2012;22:1525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J, Lupat R, Amarasinghe KC, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics 2012;28:1307–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Mysickova A, Sun R, et al. Modeling read counts for CNV detection in exome sequencing data. Stat Appl Genet Mol Biol 2011;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magi A, Tattini L, Cifola I, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol 2013;14:R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plagnol V, Curtis J, Epstein M, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 2012;28:2747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sathirapongsasuti JF, Lee H, Horst BA, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 2011;27:2648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi Y, Majewski J. FishingCNV: a graphical software package for detecting rare copy number variations in exome-sequencing data. Bioinformatics 2013;29:1461–2. [DOI] [PubMed] [Google Scholar]
- 24. Amarasinghe KC, Li J, Hunter SM, et al. Inferring copy number and genotype in tumour exome data. BMC Genomics 2014;15:732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai WR, Johnson MD, Kucherlapati R, et al. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics 2005;21:3763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lai W, Choudhary V, Park PJ. CGHweb: a tool for comparing DNA copy number segmentations from multiple algorithms. Bioinformatics 2008;24:1014–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014;26:319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo Y, Sheng Q, Samuels DC, et al. Comparative study of exome copy number variation estimation tools using array comparative genomic hybridization as control. Biomed Res Int 2013;2013:915636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tan R, Wang Y, Kleinstein SE, et al. An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat 2014;35:899–907. [DOI] [PubMed] [Google Scholar]
- 32. Samarakoon PS, Sorte HS, Kristiansen BE, et al. Identification of copy number variants from exome sequence data. BMC Genomics 2014;15:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alkodsi A, Louhimo R, Hautaniemi S. Comparative analysis of methods for identifying somatic copy number alterations from deep sequencing data. Brief Bioinform 2015;16:242–54. [DOI] [PubMed] [Google Scholar]
- 34. Kadalayil L, Rafiq S, Rose-Zerilli MJ, et al. Exome sequence read depth methods for identifying copy number changes. Brief Bioinform 2015;16:380–92. [DOI] [PubMed] [Google Scholar]
- 35. Olshen AB, Venkatraman ES, Lucito R, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5:557–72. [DOI] [PubMed] [Google Scholar]
- 36. Xi R, Hadjipanayis AG, Luquette LJ, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci USA 2011;108:E1128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sasaki S, Kitagawa Y, Sekido Y, et al. Molecular processes of chromosome 9p21 deletions in human cancers. Oncogene 2003;22:3792–8. [DOI] [PubMed] [Google Scholar]
- 40. Pinto D, Darvishi K, Shi X, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 2011;29:512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.