Abstract
Carboxypeptidase G2 (CPG2) is an Food and Drug Administration (FDA)-approved enzyme drug used to treat methotrexate (MTX) toxicity in cancer patients receiving MTX treatment. It has also been used in directed enzyme-prodrug chemotherapy, but this strategy has been hampered by off-site activation of the prodrug by the circulating enzyme. The development of a tumor protease activatable CPG2, which could be achieved using a circular permutation of CPG2 fused to an inactivating ‘prodomain’, would aid in these applications. We report the development of a protease accessibility-based screen to identify candidate sites for circular permutation in proximity of the CPG2 active site. The resulting six circular permutants showed similar expression, structure, thermal stability, and, in four cases, activity levels compared to the wild-type enzyme. We rationalize these results based on structural models of the permutants obtained using the Rosetta software. We developed a cell growth-based selection system, and demonstrated that when fused to periplasm-directing signal peptides, one of our circular permutants confers MTX resistance in Escherichia coli with equal efficiency as the wild-type enzyme. As the permutants have similar properties to wild-type CPG2, these enzymes are promising starting points for the development of autoinhibited, protease-activatable zymogen forms of CPG2 for use in therapeutic contexts.
Keywords: Acy1 M20 metallopeptidases family, carboxypeptidase G2; circular permutation; directed enzyme-prodrug therapy; methotrexate
Introduction
Carboxypeptidase G2 (CPG2) from Pseudomonas sp. RS-16 is a dinuclear zinc metalloenzyme that is able to cleave and inactivate methotrexate (MTX) and other structurally related compounds (Fig. 1) (Minton et al., 1983, 1984; Sherwood et al., 1985; Rowsell et al., 1997). This activity has made it an enzyme of medical interest. CPG2 is the active component of the drug glucarpidase, also known by its trade name Voraxaze, which is currently indicated for MTX toxicity rescue treatment (DeAngelis et al., 1996; Green, 2012), particularly in patients with impaired renal function, including children (Christensen et al., 2012). In addition, CPG2 is a model directed enzyme-prodrug therapy (DEPT) enzyme, owing to its ability to activate nitrogen mustard prodrugs, including N-{4-[(2-chloroethyl)(2-mesyloxyethyl)amino]benzoyl}-l-glutamic acid (CMDA) (Marais et al., 1996; Melton and Sherwood, 1996; Niculescu-Duvaz et al., 2003; Davies et al., 2005; Hedley et al., 2007). The latter potential clinical application of CPG2, which has been the subject of clinical trials (Napier et al., 2000; Francis et al., 2002), would particularly benefit from an autoinhibited version of the enzyme that is regulatable by tumor-specific environmental stimuli, such as proteolysis by a tumor-specific protease. Circular permutation has been exploited to generate autoinhibited enzymes (Plainkum et al., 2003), but, to the best of our knowledge, circular permutants of CPG2 have not been reported.
Fig. 1.
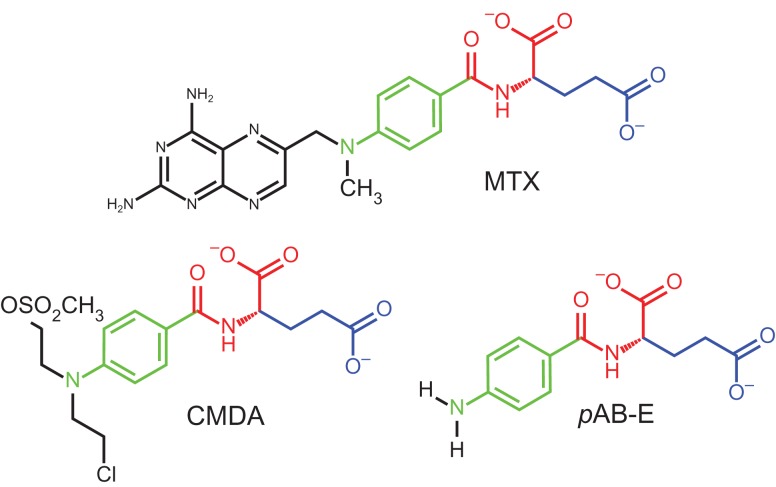
The chemical structures of three CPG2 substrates: MTX, CMDA and pAB-E. The peptide-like linkage that is hydrolyzed by CPG2, the glutamate sidechain and the para-aminobenzene group are shown with consistent colouring in all three structures.
It has been shown that some proteins with termini that are physically close in space can be modified by a ‘circular permutation’, in which the termini of the protein are linked and new termini are introduced at a different site in the polypeptide chain (Yu and Lutz, 2011). This procedure can result in a protein whose structure and function has not been substantially altered, a property that has been useful in studying protein folding (Viguera et al., 1996; Capraro et al., 2008) and evolution (Peisajovich et al., 2006). In addition, circular permutations have been shown to stabilize proteins (Whitehead et al., 2009) or alter their activity (Cheltsov et al., 2001; Qian and Lutz, 2005; Reitinger et al., 2010; Yu and Lutz, 2010; Guntas et al., 2012; Daugherty et al., 2013), suggesting that the most stable position for a protein's termini is not always the naturally observed one. The ability to modulate stability and activity has led to the use of circularly permuted proteins in a variety of biotechnology applications, including biosensors (Baird et al., 1999; Guntas et al., 2005; Pandey et al., 2016), biocatalysts (Qian and Lutz, 2005; Greenfield, 2007; Yu and Lutz, 2010; Daugherty et al., 2013; To et al., 2015) and artificial zymogens (Plainkum et al., 2003). In spite of these successes, the design of circular permutants can be unpredictable. There are few general rules for the design of the linker between the natural termini or for identifying the sites for introduction of new termini, both of which have a profound effect on the success of circular permutation (Yu and Lutz, 2011).
The enzyme CPG2 is a prototypical enzyme from the aminoacylase-1/metallopeptidase 20 (Acy1/M20) family, which is the largest family of metallopeptidases (Lindner et al., 2003, 2005). Several crystal structures of Acy1/M20 enzymes have been determined and nearly all feature a dinuclear metal-binding site and a lid domain (Rowsell et al., 1997; Håkansson and Miller, 2002; Jozic et al., 2002; Lindner et al., 2003; Lundgren et al., 2003; Unno et al., 2008; Girish and Gopal, 2010; Nocek et al., 2010, 2014; Brunger et al., 2012). The CPG2 crystal structure (Rowsell et al., 1997) features a dinuclear Zn2+-binding site, with the core catalytic activity contained in the larger, discontinuous, ~281 residue catalytic domain (residues 23–213 and 326–415), which contains all five of the zinc-coordinating residues. The smaller dimerization domain is inserted in the middle of the catalytic domain (residues 214–325) and self-associates to form the complete homodimer. Mutational, kinetic and structural studies of the closely related homologs, including mouse carnosinase 2 (mCN2) (Unno et al., 2008), Staphylococcus aureus mettalopeptidase (Sapep) (Girish and Gopal, 2010) and human aminoacylase 1 enzymes (Lindner et al., 2003, 2005), suggest that a lid-closing event mediated by the dimerization domain is necessary for catalytic activity. A ‘closed lid’ structure of inhibitor-bound mCN2 places a conserved histidine residue located on the ‘trans’ subunit (His229 in CPG2) in position to interact with the substrate (Unno et al., 2008). Mutagenesis studies in which enzyme kinetics (Unno et al., 2008) and dimer exchange (Okumura et al., 2016) were measured showed that the cross-subunit interaction of the key histidine residue is essential for mCN2 catalytic activity. This requirement for lid movement during catalysis appears to be conserved in the enzyme family, as in the crystal structure of PepV, a monomeric homolog of CPG2, the lid domain (from the same protomer) mimics elements of the dimer and contributes both the key histidine residue as well as substrate recognition elements to the active site in the closed state (Jozic et al., 2002). Conversely, a recent study of CPG2 deletion constructs suggests that while loss of the dimerization domain is deleterious to activity, the catalytic domain alone does retain some of the wild-type enzyme's activity (Jeyaharan et al., 2016).
Given the multi-domain structure of the enzyme, the requirement for multiple Zn2+ cofactors, and the conformational changes required for catalytic activity, one might expect that CPG2 would not easily tolerate circular permutations. Other features of CPG2, however, suggest that it is an attractive target for circular permutation. The termini, as seen in the crystal structure (residues 26 and 414), are 15 Å apart, a distance that can be spanned by a short linker. Its catalytic domain is discontinuous, meaning that its sequence is interrupted by the dimerization domain. This suggests that the enzyme cannot completely fold until it has been fully synthesized by the ribosome, and that it may, therefore, be amenable to having its sequence re-organized. In addition, the signal peptide at the N-terminus of CPG2 suggests that the enzyme could be able to tolerate an addition to the crystallographically observed N-terminus without structural disruption.
We have employed an experimentally driven conceptual framework to predict tolerated sites for circular permutations, and then successfully produced circular permutants at two sites in CPG2, for a total of six circular permutant variants. To the best of our knowledge, these are the only reported circular permutants of CPG2, a prototypical enzyme in the large, evolutionarily ubiquitous and functionally diverse Acy1/M20 family, and the first circular permutants for an Food and Drug Administration (FDA)-approved protein-based drug. As a target for circular permutation, CPG2 is structurally complex: it is a dimer with two dinuclear metal-ion centers that relies on global conformational changes for activity. In spite of these complexities, the circular permutants show robust expression and activity, and analysis of their structure and stability show similar properties to the wild-type enzyme. Expressing these permutants in a MTX-sensitive strain of Escherichia coli also showed reduced sensitivity to MTX than in controls, indicating that they function in vivo. As the new termini for both of these circular permutation sites are close to the active site, we believe that this work has set the foundation for designing an auto-regulated, ‘proenzyme’ form of CPG2 for use in clinical DEPT applications for cancer chemotherapy.
Experimental Procedures
Creation of the CPG2 circular permutation constructs
The DNA sequence of the full-length cpg2 gene, including its signal peptide, was codon-optimized using the method of Boël et al. (2016), and ordered as a gBlock from Integrated DNA Technologies. Gibson assembly (Gibson et al., 2009) was used to insert the gene into the pET29b(+) vector (Novagen), linearized with NdeI and XhoI (New England Biolabs). This plasmid was used to transform E. coli DH5α cells. Plasmid DNA from positive clones was prepared using the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific), and the sequence was confirmed using DNA sequencing (GenScript).
In order to produce CPG2 circular permutants, a ‘tandem dimer’ construct was produced with a linker between two copies of the CPG2 gene. The CPG2 gene was amplified twice using Q5 polymerase (New England Biolabs), with different Gibson homology regions on each: one matching the linker sequence, and the other matching one end of the pET29b(+) vector linearized with NdeI and XhoI (New England Biolabs) (Table S1). The fragments were purified by gel extraction using the E.Z.N.A. MicroElute Gel Extraction Kit (Omega Bio-tek), and Gibson assembly was used to combine the two fragments into the pET29b(+) vector as described above.
Circular permutants were generated by amplifying the desired region from the tandem dimer construct (Table S1), purifying the desired fragment by gel extraction, and inserting it into either pET29b(+) or pET15b (Novagen) linearized with NdeI and XhoI (New England Biolabs) using Gibson assembly as described above.
For the MTX sensitivity assay, a cis-repressed vector under control of the T5 promoter was necessary, as the MTX-sensitive CAG12184 cell line does not have the (DE3) lysogen required for pET system expression. We obtained the iLID C530M gene in the pQE-80 L expression vector from Addgene (plasmid #60408), which we use as a negative control for our MTX sensitivity experiments. To make the CPG2 constructs, the plasmid was linearized and the gene removed using BamHI and HindIII (New England Biolabs), followed by gel extraction. The CPG2 gene and its circular permutants were amplified from the pET vector constructs, purified by gel extraction, and inserted into the resulting pQE-80 L vector backbone using Gibson assembly as described above (Table S1). Subsequently, the N-terminal His-tag was removed and replaced with the pelB or natural CPG2 signal peptide using primer extension polymerase chain reaction to generate a double-stranded megaprimer, which was incorporated into the template using the QuikChange mutagenesis protocol.
Protein expression and purification
Wild-type and circular permutant CPG2 plasmids were transformed into BL21(DE3) cells. Individual colonies were used to inoculate a 1 ml culture of Luria-Bertani (LB) containing either 100 μg/ml ampicillin or 50 μg/ml kanamycin. Cultures were grown overnight at 37°C, and 50 μl of the cultures were subcultured into 5 ml of autoinduction media (Studier, 2005) and grown at 37°C for another 3 h. The cultures were then transferred to 18°C and grown for ~20 h.
All subsequent steps were performed at 4°C unless otherwise specified. The cells were harvested by centrifugation at 3220×g for 20 min. The cell pellets were resuspended in 600 μl of 50 mM Tris 300 mM NaCl 30 mM imidazole pH 7.4 containing 1.5× BugBuster (EMD Millipore), lysozyme, DNase I and phenylmethylsulfonyl fluoride. This was incubated for 15 min at room temperature, followed by 45 min at 4°C. The cell lysates were centrifuged at 20 000×g for 15 min, the supernatants were transferred to fresh tubes, and centrifuged again for 30 min. The second supernatant was applied to 150 μl of NiNTA agarose resin (Qiagen) equilibrated with 50 mM Tris 300 mM NaCl 30 mM imidazole pH 7.4 and shaken for 1 h. The resin was washed twice with 500 μl of the same buffer, and the protein was then eluted in 300 μl of 50 mM Tris 300 mM NaCl 300 mM imidazole pH 7.4. The purified samples were dialyzed against 50 mM Tris 100 mM NaCl pH 7.4. The purity of the samples was assessed using sodium dodecyl sulfate polyacrylamide gel electrophoresis, and the protein was quantified using a Bradford assay.
Activity assay
The activity of CPG2 and its circular permutants was measured using both MTX and para-aminobenzoyl-glutamate (pAB-E) as substrates. All experiments were performed in triplicate. The substrates were prepared at a concentration of 100 μM in 50 mM Tris 50 mM NaCl 0.1 mM ZnSO4 pH 7.4, and 90 μl of substrate solution was pre-incubated at 37°C in a Greiner Bio-One UV-Star µCLEAR 96 half area well microplate. After performing initial spectral scans on the substrates using a SpectraMax M3 (Molecular Devices), the reaction was started by adding 10 μl of 10 μg/ml enzyme to 90 μl of substrate. The absorbance at 320 nm (for MTX) or 280 nm (for pAB-E) was measured continuously in the SpectraMax M3 (Molecular Devices) for 1 h at 37°C, and the initial rate was determined using linear regression. These values were converted to specific activities in units/mg, where one unit is the amount of enzyme required to convert 1 μmol of substrate in 1 min.
Circular dichroism
Samples of CPG2 and its circular permutants were analyzed using circular dichroism (CD). After dialysis against 50 mM Tris 100 mM NaCl pH 7.4, the protein concentration was verified and the samples were diluted to 100 μg/ml for CD. CD data were recorded in an Aviv model 420SF CD spectrometer with a five cuvette rotor set to 4°C. Cuvettes with a 1.0 mm pathlength were used. First, five buffer CD spectra from 197 to 260 nm were measured in 1 nm increments using the five cuvettes to be used for each of the five protein samples. A measurement averaging time of 10 s was used. Following this, identical measurements were made for the five CPG2 samples being analyzed. The CD spectra of both the samples and buffer were smoothed using the Savitzky–Golay filter (Savitzky and Golay, 1964), and the buffer spectra were subtracted from the sample spectra measured in the same cuvette. For ease of comparison, the baseline signal recorded from 250 to 260 nm, where no CD signal was obtained, was subtracted from the entire data set to ensure all samples had a matching baseline.
As all of the samples had a minimum in the CD spectrum at or around 217 nm, this wavelength was selected for analysis in thermal melt experiments. Over temperatures ranging from 4°C to 95°C, the temperature of the samples was incremented by 0.5°C. Once the target temperature was reached, after 1 min of equilibration, the CD signal was measured at 217 nm with a 10 s measurement averaging time.
MTX sensitivity assay
CPG2 and its circular permutants in the pQE-80 L vector were transformed into MTX-sensitive CAG12184 cells obtained from the Yale Coli Genetic Stock Center (CGSC #7437). An unrelated engineered light-induced dimer (iLID C530M) and mCN2 in the same vector (Guntas et al., 2015) were used as negative controls. For each construct, two individual colonies were used to inoculate duplicate 2 ml LB cultures supplemented with 100 μg/ml ampicillin (omitted from the untransformed CAG12184 control) and 15 μg/ml tetracycline. These cultures were allowed to grow shaking overnight at 37°C. The OD600nm of the starter cultures was measured. These cultures were then subcultured into 100 μl of M9 media containing 0.2% casamino acids and 0.4% glucose and supplemented with 100 μg/ml ampicillin, 15 μg/ml tetracycline, 10 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) and in half the cultures, 100 μM MTX in a sterile Greiner Bio-one flat-bottom polystyrene 96-well microplate. A sufficient quantity of starter culture was used to be equivalent to an OD600nm of 0.05 at a 1.0 cm pathlength (generally ~1 μl). Each of the duplicate starter cultures was subcultured four times, creating a total of eight replicates for each construct, four of which contained MTX. The plate was sealed with Breathe-Easy sealing membrane (Diversified Biotech) and transferred to a Tecan Infinite M200 Pro plate reader preset to 37°C. Regular OD600nm measurements were made, with the plate being shaken at 37°C in ‘orbital’ mode with a 3 mm magnitude for 15 min in between measurements. For every well, four readings in a small grid were made in order to reduce the likelihood of outliers caused by unevenness in the cell suspension. This measurement/shaking cycle was repeated continuously for 24 h.
Computational modeling of the CPG2 circular permutant termini
Relaxed structures of CPG2 were altered to include a linker between the natural N- and C-termini, and with a chainbreak between the termini introduced in the circular permutations. To sample the energy landscape available to the termini, a set of Monte Carlo simulations were performed using Rosetta in which random perturbations were applied to the termini. In one set of simulations, the phi and psi torsion angles of the terminal residue segments were all set to zero, allowing global sampling of the termini conformations (forward-folding simulations). In a second set of simulations, this step was omitted and simulations were begun from the crystallographic conformation of the termini, allowing finer sampling of near-native conformations. In either case, a random residue between either terminus and the first element of secondary structure (up to 10 residues away from the terminus) was selected to have its phi and psi angles set to Ramachandran plot-biased values. The geometry of the termini and residues within 8 Å of the termini was then optimized by repacking their sidechains. This process was repeated at least 30 times, with each cycle being accepted or rejected according to the Metropolis criterion. A temperature of 0.6 Rosetta temperature units was used for simulations with torsions preset to 0, and a higher temperature (T = 10) was used in simulations where the native structure was used in order to enhance sampling efficiency. This process was repeated to generate at least 1500 decoys for each simulation. The command lines and RosettaScripts (Fleishman et al., 2011) files used for modeling are provided in Supplementary Information.
Results
Rationale for the CPG2 circular permutation linker sequences and termini site
For successful creation of circular permutations of proteins, careful consideration must be given to two parameters (Yu and Lutz, 2011): the nature of the linker to connect the natural termini (both linker length and sequence) and the location in the structure of the ‘new’ termini. CPG2 has a natural signal peptide that is disordered in the crystal structure. As this sequence is not believed to have a functional or structural role outside of cellular trafficking, we reasoned that this sequence could be safely eliminated from the CPG2 circular permutation linker. In order to maximize the compatibility of the sequence with CPG2’s structure, however, we used the signal peptide sequence as a base for designing the linker, while making it more flexible and less hydrophobic. Based on the distance between the terminal residues with clear density in the crystal structure, we reasoned that the minimum linker length would be four residues. To add some flexibility to the newly generated loop, we extended the linker length to six residues. A C-terminal Lys is disordered in the crystal structure, and was therefore selected as the most N-terminal residue of the interdomain linker. The five residues immediately N-terminal to the most N-terminal ordered residue of CPG2 are GTALA, with the signal peptide cleavage occurring between the Thr and Ala. To improve the hydrophilicity and flexibility of this sequence, the Leu was changed to a Ser. When this sequence is added to the previously mentioned Lys residue, it results in a linker with the sequence KGTASA. Stated another way, the circular permutation linker would be obtained by fusing the natural N- and C-termini, deleting residues 1–20, and mutating Leu24 to Ser (Fig. 2).
Fig. 2.
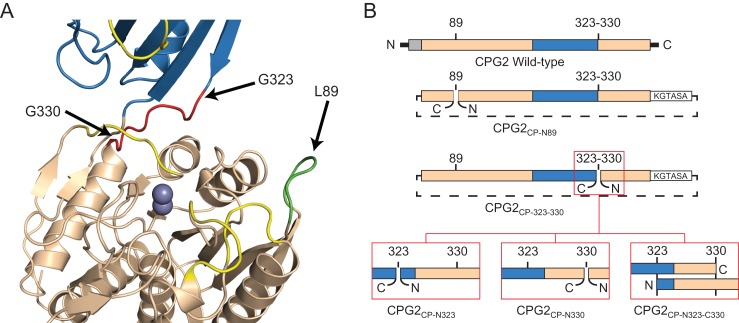
Summary of the CPG2 circular permutations. (A) Crystal structure of CPG2 (PDB ID 1CG2), with the positions of three circular permutation sites indicated. The dinuclear zinc active site is shown as two spheres. The catalytic domain is shown in wheat, the dimerization domain in blue, the CPG2CP-N89 loop in green and the CPG2CP-323-330 loop in red. The four loops that, when modified, resulted in an inactive enzyme are shown in yellow. (B) Schematic of the circular permutations. The wild-type sequence is shown with the signal peptide in gray, the catalytic domain in wheat and the dimerization domain in blue. For the circular permutants, the linker sequence, KGTASA, is indicated as a white box, with a dotted line indicating that the original N- and C-termini have been linked. The new termini are indicated with a gap in the original sequence. For the CPG2CP-323-330 series of permutants, ‘zoomed in’ images showing the difference between the permutants are shown in red boxes.
The second parameter that we considered was the position in the sequence and structure of the new termini. Our ultimate goal is to be able to use an insertion or fusion with CPG2 in order to obtain an autoinhibited enzyme that can be re-activated by proteolysis. Several loop segments that are close to the active site, but do not seem to directly affect the enzyme's catalytic machinery, appear to be attractive sites for either domain insertion or circular permutation; however, the catalytic cycle of CPG2 involves considerable conformational dynamics, and rewiring the protein simply based on the crystal structure of one state of the protein could disrupt enzyme activity, especially when the change is introduced close to the active site. To improve the success rate in selecting insertion or circular permutation sites, we developed a new approach to identify sites for modification. We chose sites on the protein close to the active site that could be extensively modified by a protease cleavage site substitution without impacting CPG2 enzymatic activity. As proteolysis is necessary for relieving the envisioned auto-inhibition, we identified five loop segments (residues 55–59, 86–91, 116–121, 271–277 and 356–361) in close proximity to the active site and mutated them to the canonical tobacco etch virus (TEV) protease sequence (Fig. 2). Our hypothesis was that if the mutant enzyme is active, it is a good candidate for either insertion of the prodomain or circular permutation followed by prodomain fusion to a terminus. If, in addition, the enzyme proved to be susceptible to proteolysis by TEV protease and retained activity, an insertion would be promising. Alternatively, if the enzyme could not be proteolysed or loses activity following proteolysis, the circular permutation followed by prodomain fusion strategy would be more likely to be successful.
Of the five sites we initially identified, only one of the sites proved to be active, though the enzyme was not cleavable by TEV protease at this position (not shown). We decided to proceed with a circular permutation at this position, referred to hereafter as CPG2CP-N89 (circular permutation, N-terminus residue 89) (Fig. 2).
Another position of interest for an insertion or circular permutation for the purposes of creating an autoinhibited CPG2 is the interdomain linker running from residues 323 to 330. In our initial screen for target positions, however, we avoided that location, as loss of Arg324 has been shown to lead to a much less active enzyme, likely due to a role in substrate recognition (Rowsell et al., 1997). A recent paper that showed that this region of the enzyme could be substantially disrupted while retaining some activity (Jeyaharan et al., 2016), however, encouraged us to proceed with this location as another circular permutation site, which we refer to as the CPG2CP-323-330 series of circular permutants. In order to maximize the possibility of an active enzyme variant, we decided to make three variations for this particular permutant. In one, the termini were introduced at the N-terminal side of the linker, residue 323, which we refer to as CPG2CP-N323 (circular permutation, N-terminus residue 323) (Fig. 2). In the second, the termini were introduced at the C-terminal side, residue 330, which we refer to as CPG2CP-N330 (circular permutation, N-terminus residue 330) (Fig. 2). In the third, we duplicated the interdomain linker on both the N-terminus and C-terminus of the circular permutant, allowing either position to contribute to substrate binding. We refer to this permutant as CPG2CP-N323-C330 (circular permutation, N-terminus residue 323, C-terminal residue 330) (Fig. 2).
Expression of CPG2 circular permutants
Wild-type CPG2 and all of the circular permutants used identical codon-optimized DNA sequences (excepting the alteration of the signal peptide to the linker sequence) and expression protocols. All of the circular permutants, both with N- and C-terminal His-tags, were found to have expression comparable to that of the wild-type enzyme after purification using nickel affinity chromatography (Table I), in the range of 50–90 mg/l of cell culture.
Table I.
Expression and activity data for CPG2 circular permutations
| Enzyme | Expression (mg/l cell culture)a |
MTX Activity (unitsb/mg) | pAB-E Activity (unitsb/mg) |
|---|---|---|---|
| CPG2 (wild-type, C-His-tag) | ~60 | 6.6 ± 0.3 | 4.6 ± 0.3 |
| CPG2CP-N89 (N-His-tag) | ~50 | 5.4 ± 0.9 | 4.4 ± 0.2 |
| CPG2CP-N89 (C-His-tag) | ~90 | 10.6 ± 1.5 | 4.9 ± 0.7 |
| CPG2CP-N323 (C-His-tag) | ~70 | 1.00 ± 0.06 | 0.3 ± 0.1 |
| CPG2CP-N330 (N-His-tag) | ~90 | 3.9 ± 1.0 | 1.7 ± 0.1 |
| CPG2CP-N323-C330 (N-His) | ~70 | 3.2 ± 0.6 | 0.98 ± 0.15 |
| CPG2CP-N323-C330 (C-His) | ~70 | 1.9 ± 0.2 | 0.89 ± 0.03 |
aExpression levels are extrapolated to 1 l culture size based on yields from 5 ml cultures.
bOne unit is defined as the amount of enzyme required to convert 1 μmol of substrate in 1 min.
Effects of CPG2 circular permutations on global structure
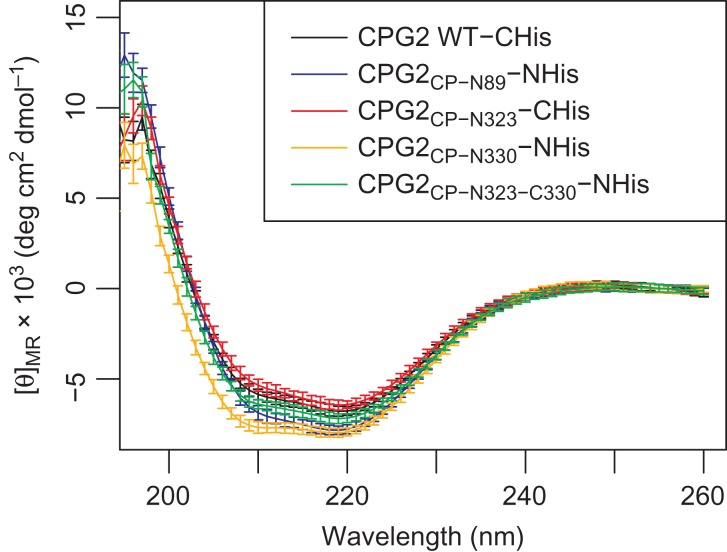
To assess whether the circular permutations had an impact on the ability of CPG2 to fold, the structures of the permutants were assessed using CD. The five permutants all showed very similar CD spectra, indicating that the global structure is likely largely unaffected (Fig. 3). The spectra all show the double negative bands at ~208 and ~222 nm that is characteristic of proteins with α-helical content (Greenfield, 2007). Given that wild-type CPG2 has an α-helical content of 38%, as determined by the program Define Secondary Structure of Proteins (Kabsch and Sander, 1983), these spectra are consistent with properly folded CPG2. We did not collect short wavelength data due to the difficulties in obtaining sufficiently low buffer and salt concentrations for these measurements while maintaining protein stability, making reliable secondary structure deconvolution impractical. Nonetheless, the similarity between the CD spectra are sufficient to suggest that the circular permutants have very similar structures to the wild-type enzyme.
Fig. 3.
CD spectra of the CPG2 and its circular permutants. The buffer signal has been subtracted from each spectrum, and the signal at 250–260 nm was used to baseline-correct the resulting spectra. The vertical axis shows the mean residue ellipticity ([θ]MR) in units of 103 deg cm2 dmol−1.
Activity of CPG2 circular permutants
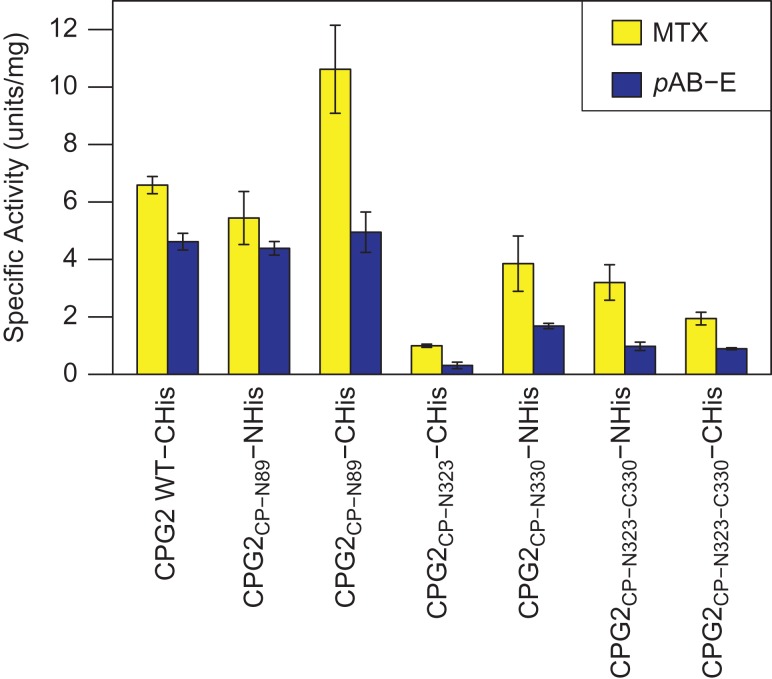
The activity of CPG2 and its circular permutants was assessed with two substrates: MTX and pAB-E (Fig. 1). In all cases, CPG2 and its circular permutants were found to hydrolyze MTX faster than pAB-E, though the degree of this difference varied depending on the specific construct (Fig. 4, see also Table I and Fig. S2). Of the two circular permutation sites considered, the CPG2CP-N89 showed no loss of activity compared to the wild-type. In fact, the C-terminal His-tagged variant of this permutant appeared to have improved activity toward MTX compared to the wild-type and its N-terminal His-tagged counterpart. In general, the series of permutants built around residues 323–330 showed somewhat reduced activity compared to the wild-type. It is, however, notable that for these permutants, the pAB-E activity was more strongly diminished in all cases than the corresponding MTX activity. Interestingly, the N-terminally His-tagged permutant with the duplicated region, CPG2CP-N323-C330, as well as the CPG2CP-N330 permutant, have similar activities. In contrast, the CPG2CP-N323 permutant and the C-terminally His-tagged permutant with the duplicated region show more greatly reduced activity, with the former showing substantial reduction for both substrates and the latter showing significantly reduced activity toward MTX.
Fig. 4.
Specific activity of CPG2 and its circular permutants toward MTX and pAB-E. One unit is defined as the amount of enzyme required to convert 1 μmol of substrate in 1 min. Error bars indicate the standard deviation of the three replicates. See also Table I.
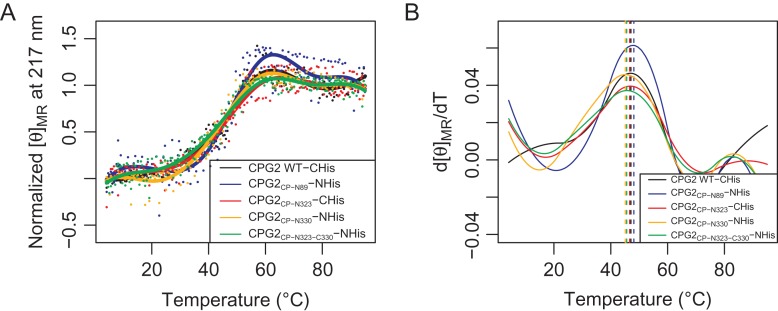
Thermal stability of CPG2 circular permutants
Having demonstrated that the circular permutants were not negatively affected in terms of structure, and that the effects on activity were minor, we next assessed whether any loss of thermal stability had occurred as a result of circular permutation. The permutants were gradually heated in the CD instrument, while measuring the signal at 217 nm to detect when the loss of secondary structure occurred, indicating the denaturation of the protein (Fig. 5). For all of the circular permutants, as with the wild-type, the proteins appeared quite thermally stable up to ~30°C. Some slight indications of loss of structure could be seen between 30°C and 40°C, with significant unfolding occurring between 40°C and 60°C. By 60°C, the CD signal had re-stabilized, suggesting that the proteins were completely unfolded at that temperature. The first derivative plot of the thermal melting data (Fig. 5B) shows a single major peak centered at a temperature of 45–48°C, corresponding to the melting temperature. That the melting point for the circular permutants is not significantly changed from the wild-type demonstrates that circular permutation has not destabilized CPG2.
Fig. 5.
Thermal melts of CPG2 and its circular permutants. (A) Normalized thermal melt data, where the initial mean residue ellipticity ([θ]MR) at 217 nm is set to 0, and the final [θ]MR is set to 1. Prior to normalization, [θ]MR is in units of deg cm2 dmol−1. Data points show the individual readings, and a fitting curve is drawn as a line for each data set. (B) First derivative of the fitting curve shown in (A). The maximum, indicating the melting temperature, is indicated using a dashed, vertical line.
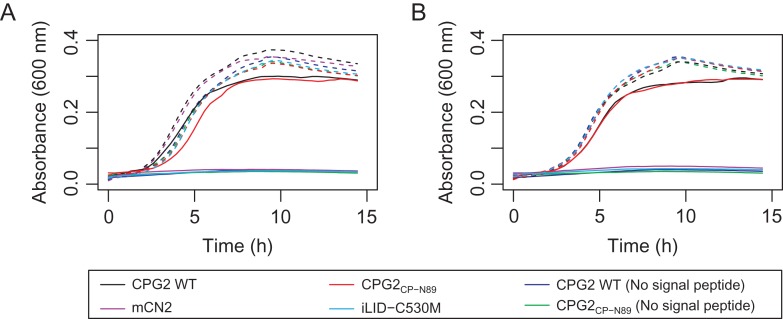
Reversing MTX sensitivity in whole cell assays
To further characterize the function of our circular permutants, we developed an in vivo assay to evaluate the ability of CPG2 and its circular permutants to protect MTX-sensitive cells from MTX toxicity in E. coli. The design of the assay was based on a similar assay used to evolve MTX-resistant human dihydrofolate reductase mutants in E. coli (Volpato et al., 2007, 2009). The tolC knockout strain CAG12184 (Yale Coli Genetic Stock Center), which lacks a multi-drug efflux pump that confers MTX resistance (Kopytek et al., 2000), was used for these experiments. Our initial studies, which were performed in LB, demonstrated that the wild-type enzyme conferred complete resistance to MTX; however, the circular permutants appeared to perform inconsistently, and with less efficacy than the wild-type (described in the Supplementary Information, see Fig. S1). To investigate this observed difference further, we altered our assay to use the more stringent M9 media, and added a signal peptide to the N-terminus of the constructs. Under these experimental conditions, we observed that both the wild-type enzyme and the CPG2CP-N89 circular permutant robustly confer MTX resistance, but only if they are fused with an N-terminal pelB or native CPG2 signal peptide (Fig. 6). In contrast, the CPG2CP-N323, CPG2CP-N330 and CPG2CP-N323-C330 circular permutants (not shown), and all of the proteins without a signal peptide did not confer MTX resistance.
Fig. 6.
MTX resistance assay. The growth of the MTX-containing (solid lines) and MTX-free (dashed lines) cultures is shown for CPG2 and its circular permutants with (A) the pelB signal peptide or (B) the natural CPG2 signal peptide attached to the N-terminus.
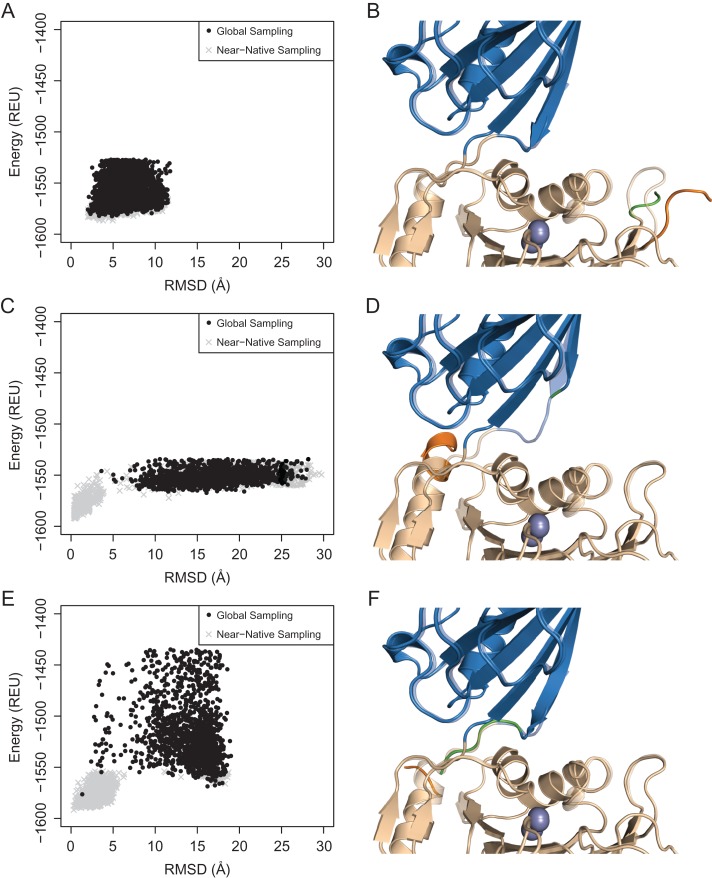
Computational modeling of the circular permutation termini
The sampling of the circular permutant conformational landscapes provides insights into the relative energetic and structural impact of the new termini on the various permutants. We generated models of the circularly permuted constructs starting from the crystal structure of CPG2, connecting the N- and C- terminal residues by linker residues and introducing new chainbreaks in the appropriate sequence locations. Using Rosetta, we investigated the energy landscapes of the resulting models of the permutants by performing (i) random conformational perturbations starting from a wild-type-like conformation at the newly generated termini and (ii) forward-folding simulations of these termini structure starting from a fully extended conformation (Fig. 7) (Hu et al., 2007). The former simulations were aimed at investigating the local stability of the wild-type conformation upon introduction of chainbreak in the region, while the latter simulations were aimed at identifying alternative stable structures that could be accessed in the context of the rest of the wild-type-like structure. In each case, between zero and 10 residues at both termini were allowed to sample alternative conformations (see Experimental Procedures for details of sampling).
Fig. 7.
Computational modeling of the (A, B) CPG2CP-N89, (C, D) CPG2CP-N323 and (E, F) CPG2CP-N330 circular permutant termini. The energy landscapes are shown on the left, with the energy calculated using the talaris2014 scorefunction. The lowest energy decoy from the global sampling simulations are shown on the right. The transparent structure is the native, relaxed structure of CPG2.
All three simple circular permutants showed global minima at lower root-mean-square deviations (RMSDs), suggesting that the most stable conformation of the circular permutants is the native conformation; however, the local energy landscape differed significantly between the most active permutant, CPG2CP-N89 (Fig. 7A), and the CPG2CP-N323 (Fig. 7C) and CPG2CP-N330 (Fig. 7E) permutants. The latter two permutants showed funneling toward the low RMSD structures, whereas CPG2CP-N89 showed a relatively flat energy landscape with only a minor energetic penalty for higher-RMSD structures. These observations suggest that the CPG2CP-N89 termini are likely flexible, with little impact on the local structure, as would be expected of the termini of many natural proteins. In contrast, the data suggest that the CPG2CP-N323 and CPG2CP-N330 termini are likely more structured in their near-native conformations, explaining the clearer energetic preference for those conformations. The introduction of chainbreaks at the latter two positions may, therefore, have the potential to entropically disrupt these stable, native conformations in a manner that negatively impacts the enzyme.
Considering the more global conformational landscape of the 323–330 permutants, it is notable that the CPG2CP-N323 permutant has a flat, featureless landscape at RMSDs >5 Å (Fig. 7C), whereas CPG2CP-N330 shows a bistable landscape with a second energy minimum at RMSD ~17 Å (Fig. 7E). Compared to the CPG2CP-N89 landscape, which effectively samples the near-native, low RMSD conformations, the 323–330 permutant global simulations rarely (CPG2CP-N330) or never (CPG2CP-N323) reach the low energy/low RMSD states. This observation may imply that the 323–330 permutants are more likely to get kinetically trapped in alternative conformations. In accord with this expectation, several of the 323–330 circular permutants are less active than the wild-type enzyme or CPG2CP-N89, are more sensitive to the positioning of the His-tag, and may be incompatible with a functional signal peptide. While these simulation data provide indirect insights into possible reasons for the observed differences in the relative activity levels of the permutants, experimental determination of the structure and dynamics of the permutants is necessary to draw definitive conclusions.
Discussion
As an FDA-approved drug, CPG2 is a useful enzyme for treatment of patients suffering from MTX toxicity (DeAngelis et al., 1996; Christensen et al., 2012; Green, 2012). Its use in DEPT as a cancer therapeutic further widens the scope of its utility (Napier et al., 2000; Francis et al., 2002), but our inability to regulate CPG2’s activity leads to off-target prodrug activation and toxicity during DEPT treatment. An attractive method of generating a regulatable form of CPG2 is by adding an inhibitory ‘prodomain’ that can be proteolytically cleaved by tumor-overexpressed proteases, such as matrix metalloproteases (Gerspach et al., 2006; Mason and Joyce, 2011; Puskas et al., 2011). As the natural termini of CPG2 are distant from the active site, this prodomain needs to be introduced in the middle of the sequence, either as an insertion within the canonical polypeptide chain or by engineering new termini through circular permutation. The latter method allows greater flexibility in the selection of a prodomain, and also avoids the requirement for the enzyme to maintain its structural integrity with a break mid-sequence in the polypeptide chain following protease-based activation. A broken-chain ‘nicked’ enzyme would be entropically predisposed to self-dissociation and unfolding to a greater extent than a circular permutant. We developed an experimental approach to assess various insertion sites for feasibility for either insertion, circular permutation, or both, and we have used this approach to successfully identify a circular permutant of CPG2 that retains wild-type activity, structure, stability and the ability to confer MTX resistance to MTX-sensitive E. coli. Another circular permutant that retains wild-type structure and stability while exhibiting only slight loss of activity provides insight into the role of the dimerization domain in substrate recognition by CPG2, and possibly other Acy1/M20 family members.
We utilized our protease-sensitivity approach to assess the feasibility of insertion and/or circular permutation at five sites that are in proximity to the active site, allowing for the introduction of an auto-inhibitory domain. Of the five sites that we assessed, only one, spanning residues 86–92 (Fig. 2A), retained its activity, indicating that CPG2 is able to withstand substantial sequence modification at that position without loss of activity. As such, this site was selected as ideal for introduction of a future prodomain; however, this site was not susceptible to proteolytic cleavage by TEV protease. We believe that the inability to cleave the enzyme at this loop may be caused by its restrictive β-hairpin geometry. While it is possible that lengthening of that loop will allow it to adopt an extended conformation consistent with protease cleavage, allowing us to reassess its utility for mid-sequence insertions, a circular permutation would also allow us to use a fusion protein with a flexible linker that can adopt an extended, protease cleavable conformation.
A recent paper (Jeyaharan et al., 2016) showed that CPG2 is able to retain its activity even with the loss of the dimerization domain, unlike what has been observed in homologs of CPG2 (Unno et al., 2008). Their observations led us to try a second circular permutation at the site of the interdomain linker, residues 323–330, which is positioned ideally for access to the substrate binding site. Because of the suspected role of Arg324 is substrate recognition, we decided to make three variations of this circular permutant.
All of the circular permutant variants proved to express at a similar level to the wild-type enzyme, and solubility did not prove to be a significant challenge during protein expression. Our success in producing soluble, full-length (including signal peptide) CPG2 in E. coli is in contrast to the experience of Jeyaharan et al. (2016); we believe this to be attributable to our use of autoinduction media (Studier, 2005), as we have observed falling cell densities and very small cell pellets when using IPTG-based induction. In addition, our assessment of overall structure and thermal stability by CD demonstrated that all of the permutants exhibited similar overall structure and stability to the wild-type. This demonstrates that CPG2 is structurally robust, with no appreciable deterioration in its structure despite substantial rearrangements in its sequence.
The activity data provide insight into the local structural effects of these sequence rearrangements on the active site of CPG2. The circular permutant identified by our selection strategy, CPG2CP-N89, proved to have an essentially identical activity profile to the wild-type. In fact, the only notable deviation for either MTX or pAB-E was the apparent 1.6× increase in activity for the C-terminally His-tagged variant of CPG2CP-N89. These data suggest that CPG2CP-N89 is ideally suited for addition of a prodomain for the purposes of creating an autoinhibited form of CPG2.
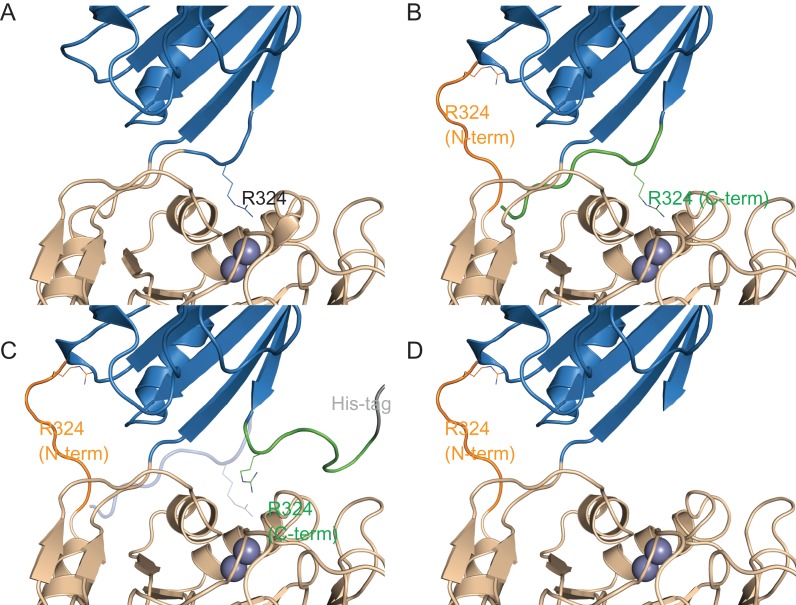
The activity data for CPG2CP-N323, CPG2CP-N330 and CPG2CP-N323-C330 show that there is a reduction in all cases toward both substrates, although they retain sufficient activity to still be usable as starting points for prodomain addition. The activity profiles, however, do highlight the role of the sequence from 323 to 330 in substrate recognition. First, the CPG2CP-N323 permutant and the C-terminally His-tagged variant of CPG2CP-N323-C330 have substantially lower activity than CPG2CP-N330 and the N-terminally His-tagged variant of CPG2CP-N323-C330. Based on these data, we propose that for the 323–330 segment to occupy the ‘canonical’ position of the wild-type structure (Fig. 8A), it must be appended C-terminally to residue 322, while attachment to the N-terminal residue 331 does not allow it fulfill its functional role (Fig. 8B). In the case of CPG2CP-N323-C330, the C-terminal repeat of residues 323–330 occupies the wild-type position (Fig. 8B), resulting in an active permutant. Likewise, the CPG2CP-N330 permutant loses only the non-functional, N-terminal repeat of 323–330 in comparison to CPG2CP-N323-C330, leading to a similarly active enzyme. Addition of a C-terminal His-tag to CPG2CP-N323-C330 disrupts the positioning of the C-terminal repeat of 323–330 (Fig. 8C), explaining the reduced activity of that variant. This effect is even more pronounced in the CPG2CP-N323 permutant, where the residues 323–330 are only present on the N-terminus, where it is not effective (Fig. 8D). We believe that for this reason, the C-terminally His-tagged CPG2CP-N323-C330 permutant and, especially, the CPG2CP-N323 permutant have significantly reduced activity. In contrast, the CPG2CP-N330 permutant has the sequence in the ‘correct’ position, and the N-terminally His-tagged form of CPG2CP-N323-C330 has the His-tag attached to the segment that is non-functional anyway. These data highlight the role of the 323–330 linker, which contributes the interaction between Arg324 and the glutamate sidechain of the substrates. It is noteworthy that while this segment has an extended conformation, one of the two anchor points appears to be far more important for the correct structure, and therefore function, of this loop. That the required anchor point is two residues away from Arg324 provides additional support for the role of that residue in substrate recognition.
Fig. 8.
Structural basis for the difference in activity of the CPG2CP-323-330 series of circular permutants. The catalytic domain is shown in wheat, and the dimerization domain is shown in blue. The residue 323–330 sequence is shown in orange when attached to the N-terminus of the circular permutant, and in green when attached to the C-terminus. The position of catalytically important R324 is shown as sticks. The dinuclear zinc active site is shown as two spheres. The His-tag is shown in gray. Differences from the CPG2 crystal structure (PDB ID 1CG2) are for illustrative purposes only, and do not represent computationally modeled or experimentally determined structures. (A) Wild-type CPG2. (B) The CPG2CP-N323-C330-N-His circular permutant (His-tag is not shown). The C-terminal sequence is believed to occupy the ‘canonical’ position, allowing only a slight reduction of activity. The N-terminal sequence is shown adopting a random conformation, or in the case of CPG2CP-N330 (not shown), is deleted. (C) The CPG2CP-N323-C330-C-His circular permutant. The C-terminal His-tag disrupts the positioning of the C-terminal 323–330 sequence, preventing it from occupying the ‘canonical’ position (transparent blue cartoon). As the C-terminal sequence is not available to occupy the ‘canonical’ position and the N-terminal sequence is unable to compensate, the activity is reduced. (D) The CPG2CP-N323 circular permutant (His-tag is not shown). As the C-terminal sequence is not available to occupy the ‘canonical’ position and the N-terminal sequence is unable to compensate, the activity is greatly reduced.
In order to further probe the likely structures of the circular permutants, Monte Carlo simulations in which the termini are first perturbed and then minimized were performed to sample energetically favorable conformations using Rosetta (Fig. 7). According to these simulations, CPG2CP-N89 produces a flat energy landscape (Fig. 7A), suggesting that perturbing these termini has little impact on the enzyme's structure. In contrast, simulations of CPG2CP-N323 and CPG2CP-N330 both show energetic funneling toward the native conformation. As the 323–330 segment forms the interdomain linker, it stands to reason that these termini have an energetically preferred conformation, and that disruption of that conformation could result in a loss of activity, as observed in our activity assays. If we assume that the conformation of the 323–330 segment is important for activity, this could explain the sensitivity of CPG2 to being modified at these positions.
For CPG2CP-N323-C330, it is difficult to interpret the simulated energy landscape (not shown), as the introduction of duplicated termini precludes the comparison to a native structure. It should be noted, though, that in the only one of the five lowest energy structures to have its terminus in the native position (the lowest energy structure), it was the C-terminal duplicate that had the native conformation. As proposed based on our activity data (Fig. 8), this may suggest that the C-terminal repeat is more likely to occupy the wild-type position than the N-terminal repeat.
The series of 323–330 permutants, which are modified close to the glutamate moiety binding site, also show consistent trends with regards to selectivity toward the two substrates under study. The loss of activity toward pAB-E is always more pronounced than the loss of activity toward MTX in these permutants. We speculate that this trend may be due to MTX's larger size. The pterin ring system of MTX can form interactions deep in the active site, at a site that is more distant from the disruption introduced in the 323–330 permutants. This second anchoring point may be ‘rescuing’ MTX activity to a greater extent than what is observed for pAB-E, leading to the greater effect of these permutants on pAB-E activity.
The therapeutic utility of CPG2 circular permutants naturally relies not only on their in vitro activity, but also their ability to degrade MTX while being either expressed by or added to an in vivo system. The strict requirement for the presence of a signal peptide at the N-terminus of either wild-type CPG2 or the CPG2CP-N89 circular permutant suggests that to confer MTX resistance, the MTX neutralizing activity must be localized to the periplasm. While the target of MTX, dihydrofolate reductase, is a cytosolic enzyme, the periplasmic requirement for CPG2 to provide MTX resistance can be explained by the extremely high affinity for MTX to its target (Summerfield et al., 2006). Once in the cytosol, it is likely that MTX binds to its target too rapidly to be hydrolyzed by CPG2, and does not dissociate from its target at a significant rate. As such, the only way to provide MTX toxicity is to prevent its arrival in the cytosol by neutralizing it in the periplasm.
It is also noteworthy that the Pseudomonad CPG2 signal peptide is functional in E. coli, in spite of conflicting statements in the literature (Minton et al., 1983; Jeyaharan et al., 2016). While we do not assess the efficiency of periplasmic export in a quantitative way, both pelB and the CPG2 signal peptide are able to confer complete resistance to 100 μM MTX under our experimental conditions, suggesting a significant concentration of CPG2 is exported to the periplasm.
While it is possible that the inability of the CPG2CP-N323, CPG2CP-N330 and CPG2CP-N323-C330 circular permutants to confer MTX resistance is caused by their lower activity, these differences are unlikely to account for their observed behavior, especially for the latter two cases that have near-wild-type activities in vitro. More likely, the position of the termini at the interdomain interface causes the signal peptide to interact with the rest of the enzyme, precluding its normal recognition and processing by the periplasmic export machinery. That the activity of these permutants is sensitive to deletions and the positioning of a His-tag also supports this explanation.
Activation of biological cascades by proteolysis is ubiquitous in regulated biological processes, such as blood clotting and apoptosis. In this type of regulation, a ‘propeptide’, typically located at either protein terminus, occludes the active site, thereby keeping the enzyme inhibited in its zymogen state. Inhibition is relieved upon proteolysis of the propeptide or prodomain. Proteolytic activation is also an attractive general approach for designing zymogens from enzymes with termini in proximity to their active sites, allowing the fusion of a designed inhibitory peptide or domain. Circular permutation leads to the generation of new termini, and has previously been utilized to build autoinhibited versions of RNase (Plainkum et al., 2003). Our approach, in which, first, a short protease recognition motif is inserted into various solvent accessible loops and, second, circular permutation is performed on those variants which tolerate the insertion without compromising activity, should provide a general strategy for productively generating active circular permutants. As these circular permutants are chosen based on their ability to tolerate the protease site, they also provide a natural starting point for fusion of a protease-cleavable, inhibitory propeptide/domain.
Conclusion
We have developed an experimentally driven, conceptual framework for identifying sites in a protein that are amenable to circular permutation for the purpose of adding an auto-inhibitory domain. Using this method, we have designed and made a series of circular permutations of the medically important enzyme, CPG2, that show similar structure and stability to the wild-type, while retaining equivalent enzymatic activity in most cases. We also demonstrated that one of the circular permutants confers MTX resistance when expressed in E. coli similarly to wild-type CPG2. This represents the first case, to the best of our knowledge, of artificial circular permutations being made to CPG2, a protein that has been approved as a drug, and a prototype enzyme from the large Acy1/M20 enzyme family. These circular permutants provide a promising starting point for generating a protease regulatable form of CPG2, which will be valuable in advancing DEPT chemotherapy technology. These circular permutants also provide mechanistic insight into substrate recognition by CPG2, emphasizing the role of the interdomain interface of the enzyme in substrate binding and catalysis.
Supplementary Material
Acknowledgements
We would like to thank the members of the Khare lab for their help and suggestions. We would like to also thank Prof. Vikas Nanda and Jose James for allowing us to use their Aviv model 420SF CD spectrometer, and Prof. Joseph Marcotrigiano for allowing us to use his SpectraMax M3 plate reader.
Conflict of interest
The authors declare that they have no competing financial interests.
Funding
This work was supported by grants from the Rutgers Cancer Institute of New Jersey [P30CA072720-18] and the National Science Foundation [MCB-1330760] awarded to S.D.K. B.J.Y. holds a Canadian Institutes of Health Research Post-Doctoral Fellowship.
References
- Baird G.S., Zacharias D.A. and Tsien R.Y. (1999) Proc. Natl. Acad. Sci. U.S.A., 96, 11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boël G., Letso R., Neely H. et al. (2016) Nature., 529, 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T., Das D., Deacon A.M. et al. (2012) Acta Crystallogr. Sect. D Biol. Crystallogr., 68, 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capraro D.T., Roy M., Onuchic J.N. and Jennings P.A. (2008) Proc. Natl. Acad. Sci. U.S.A., 105, 14844–14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheltsov A.V., Barber M.J. and Ferreira G.C. (2001) J. Biol. Chem., 276, 19141–19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A.M., Pauley J.L., Molinelli A.R. et al. (2012) Cancer, 118, 4321–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty A.B., Govindarajan S. and Lutz S. (2013) J. Am. Chem. Soc., 135, 14425–14432. [DOI] [PubMed] [Google Scholar]
- Davies L.C., Friedlos F., Hedley D., Martin J., Ogilvie L.M., Scanlon I.J. and Springer C.J. (2005) J. Med. Chem., 48, 5321–5328. [DOI] [PubMed] [Google Scholar]
- DeAngelis L.M., Tong W.P., Lin S., Fleisher M. and Bertino J.R. (1996) J. Clin. Oncol., 14, 2145–2149. [DOI] [PubMed] [Google Scholar]
- Fleishman S.J., Leaver-Fay A., Corn J.E. et al. (2011) PLoS. ONE., 6, e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R.J., Sharma S.K., Springer C. et al. (2002) Br. J. Cancer., 87, 600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerspach J., Müller D., Münkel S. et al. (2006) Cell. Death. Differ., 13, 273–284. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. and Smith H.O. (2009) Nat. Methods., 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Girish T.S. and Gopal B. (2010) J. Biol. Chem., 285, 29406–29415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J.M. (2012) Ther. Clin. Risk. Manag., 8, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N.J. (2007) Nat. Protoc., 1, 2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G., Hallett R.A., Zimmerman S.P., Williams T., Yumerefendi H., Bear J.E. and Kuhlman B. (2015) Proc. Natl. Acad. Sci. U.S.A., 112, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G., Kanwar M. and Ostermeier M. (2012) PLoS. ONE., 7, e35998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G., Mansell T.J., Kim J.R. and Ostermeier M. (2005) Proc. Natl. Acad. Sci. U.S.A., 102, 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson K. and Miller C.G. (2002) Eur. J. Biochem., 269, 443–450. [DOI] [PubMed] [Google Scholar]
- Hedley D., Ogilvie L. and Springer C. (2007) Nat. Rev. Cancer., 7, 870–879. [DOI] [PubMed] [Google Scholar]
- Hu X., Wang H., Ke H. and Kuhlman B. (2007) Proc. Natl. Acad. Sci. U.S.A., 104, 17668–17673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaharan D., Aston P., Garcia-Perez A., Schouten J., Davis P. and Dixon A.M. (2016) Protein. Expr. Purif., 127, 44–52. [DOI] [PubMed] [Google Scholar]
- Jozic D., Bourenkow G., Bartunik H., Scholze H., Dive V., Henrich B., Huber R., Bode W. and Maskos K. (2002) Structure., 10, 1097–1106. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander C. (1983) Biopolymers., 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kopytek S.J., Dyer J.C., Knapp G.S. and Hu J.C. (2000) Antimicrob. Agents. Chemother., 44, 3210–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Alary A., Boju L.I., Sulea T. and Ménard R. (2005) Biochemistry., 44, 15645–15651. [DOI] [PubMed] [Google Scholar]
- Lindner H.A., Lunin V.V., Alary A., Hecker R., Cygler M. and Ménard R. (2003) J. Biol. Chem., 278, 44496–44504. [DOI] [PubMed] [Google Scholar]
- Lundgren S., Gojković Z., Piskur J. and Dobritzsch D. (2003) J. Biol. Chem., 278, 51851–51862. [DOI] [PubMed] [Google Scholar]
- Marais R., Spooner R.A., Light Y., Martin J. and Springer C.J. (1996) Cancer. Res., 56, 4735–4742. [PubMed] [Google Scholar]
- Mason S.D. and Joyce J.A. (2011) Trends. Cell. Biol., 21, 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton R.G. and Sherwood R.F. (1996) J. Natl. Cancer. Inst., 88, 153–165. [DOI] [PubMed] [Google Scholar]
- Minton N.P., Atkinson T., Bruton C.J. and Sherwood R.F. (1984) Gene., 31, 31–38. [DOI] [PubMed] [Google Scholar]
- Minton N.P., Atkinson T. and Sherwood R.F. (1983) J. Bacteriol., 156, 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier M.P., Sharma S.K. and Springer C.J. (2000) Clin. Cancer. Res., 6, 765–772. [PubMed] [Google Scholar]
- Niculescu-Duvaz D., Niculescu-Duvaz I., Friedlos F., Martin J., Lehouritis P., Marais R. and Springer C.J. (2003) J. Med. Chem., 46, 1690–1705. [DOI] [PubMed] [Google Scholar]
- Nocek B., Starus A., Makowska-Grzyska M. et al. (2014) PLoS. ONE., 9, e93593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocek B.P., Gillner D.M., Fan Y., Holz R.C. and Joachimiak A. (2010) J. Mol. Biol., 397, 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura N., Tamura J. and Takao T. (2016) Protein. Sci., 25, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N., Kuypers B.E., Nassif B., Thomas E.E., Alnahhas R.N., Segatori L. and Silberg J.J. (2016) Biochemistry., 55, 3763–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisajovich S.G., Rockah L. and Tawfik D.S. (2006) Nat. Genet., 38, 168–174. [DOI] [PubMed] [Google Scholar]
- Plainkum P., Fuchs S.M., Wiyakrutta S. and Raines R.T. (2003) Nat. Struct. Biol., 10, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskas J., Skrombolas D., Sedlacek A., Lord E., Sullivan M. and Frelinger J. (2011) Immunology., 133, 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z. and Lutz S. (2005) J. Am. Chem. Soc., 127, 13466–13467. [DOI] [PubMed] [Google Scholar]
- Reitinger S., Yu Y., Wicki J. et al. (2010) Biochemistry., 49, 2464–2474. [DOI] [PubMed] [Google Scholar]
- Rowsell S., Pauptit R.A., Tucker A.D., Melton R.G., Blow D.M. and Brick P. (1997) Structure., 5, 337–347. [DOI] [PubMed] [Google Scholar]
- Savitzky A. and Golay M.J.E. (1964) Anal. Chem., 36, 1627–1639. [Google Scholar]
- Sherwood R.F., Melton R.G., Alwan S.M. and Hughes P. (1985) Eur. J. Biochem., 148, 447–453. [DOI] [PubMed] [Google Scholar]
- Studier F.W. (2005) Protein. Expr. Purif., 41, 207–234. [DOI] [PubMed] [Google Scholar]
- Summerfield R.L., Daigle D.M., Mayer S. et al. (2006) J. Med. Chem., 49, 6977–6986. [DOI] [PubMed] [Google Scholar]
- To T.L., Piggott B.J., Makhijani K., Yu D., Jan Y.N. and Shu X. (2015) Proc. Natl. Acad. Sci. U.S.A., 112, 3338–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno H., Yamashita T., Ujita S., Okumura N., Otani H., Okumura A., Nagai K. and Kusunoki M. (2008) J. Biol. Chem., 283, 27289–27299. [DOI] [PubMed] [Google Scholar]
- Viguera A.R., Serrano L. and Wilmanns M. (1996) Nat. Struct. Biol., 3, 874–880. [DOI] [PubMed] [Google Scholar]
- Volpato J.P., Fossati E. and Pelletier J.N. (2007) J. Mol. Biol., 373, 599–611. [DOI] [PubMed] [Google Scholar]
- Volpato J.P., Yachnin B.J., Blanchet J., Guerrero V., Poulin L., Fossati E., Berghuis A.M. and Pelletier J.N. (2009) J. Biol. Chem., 284, 20079–20089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead T.A., Bergeron L.M. and Clark D.S. (2009) Protein. Eng. Des. Sel., 22, 607–613. [DOI] [PubMed] [Google Scholar]
- Yu Y. and Lutz S. (2010) Biotechnol. Bioeng., 105, 44–50. [DOI] [PubMed] [Google Scholar]
- Yu Y. and Lutz S. (2011) Trends. Biotechnol., 29, 18–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.