Abstract
Matrix metalloproteinase-14 (MMP-14) plays important roles in cancer metastasis, and the failures of broad-spectrum MMP compound inhibitors in clinical trials suggested selectivity is critical. By grafting an MMP-14 specific inhibition motif into complementarity determining region (CDR)-H3 of antibody scaffolds and optimizing other CDRs and the sequences that flank CDR-H3, we isolated a Fab 1F8 showing a binding affinity of 8.3 nM with >1000-fold enhancement on inhibition potency compared to the peptide inhibitor. Yeast surface display and fluorescence-activated cell sorting results indicated that 1F8 was highly selective to MMP-14 and competed with TIMP-2 on binding to the catalytic domain of MMP-14. Converting a low-affinity peptide inhibitor into a high potency antibody, the described methods can be used to develop other inhibitory antibodies of therapeutic significance.
Keywords: CDR grafting, inhibitory antibody, matrix metalloproteinase, phage display, synthetic library
Matrix metalloproteinases (MMPs) are a group of structurally related zinc-dependent endopeptidases capable of cleaving almost all extracellular and basement membrane proteins (Visse and Nagase, 2003). Among them, the membrane Type I matrix metalloproteinase (MT1-MMP, or MMP-14) has been recognized as one of the most crucial MMPs in cancer development and metastasis (Genís et al., 2006; Morrison et al., 2009). Several broad-spectrum MMP inhibitors have been developed in the last 20 years for evaluation as cancer treatments. However, all these small compound MMP inhibitors failed in clinical trials due to low efficacy and adverse side effects caused by their poor selectivity among the MMP family members (Turk, 2006; Zucker and Cao, 2009).
Recently, a cyclic peptide GACFSIAHECGA (Peptide G) able to selectively inhibit MMP-14 without cross-reactions to other MMPs has been reported (Suojanen et al., 2009). This peptide inhibitor effectively prevented cancer cell migration and invasion in vitro, and dramatically reduced the growth of tongue carcinoma in xenografts with prolonged survival periods. Unfortunately, Peptide G exhibited a considerably low affinity of 150 µM with a relatively short half-life, diminishing its therapeutic potential as a potent inhibitor for cancer treatments.
Emerging as promising therapeutic agents, monoclonal antibodies had notable successes in targeting cancer cell surface antigens. mAbs usually have high affinity and high specificity, given the large antigen–antibody contact surface provided by multiple complementarity determining regions (CDRs). Encouraged by numerous studies of CDR transplantation (Moroncini et al., 2004; Frederickson et al., 2006; Qin et al., 2007; Kogelberg et al., 2008; Zhang et al., 2015), we hypothesize that grafting an inhibitory motif into a CDR, especially CDR-H3, will confer binding specificity and thus the inhibition function on the antibody. In this study, we designed and synthesized human antibody Fab libraries in which Peptide G was incorporated into CDR-H3 (Fig. 1A).
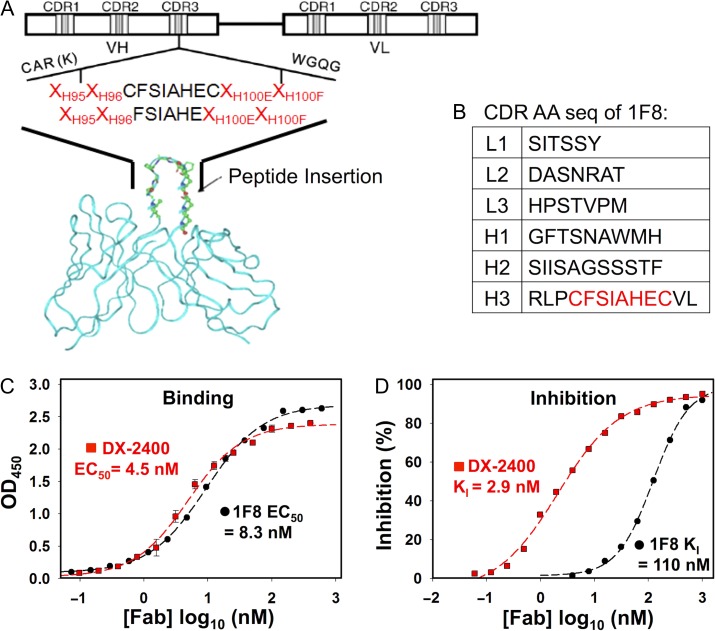
Fig. 1.
Generation of MMP-14 inhibitory Fab 1F8 by CDR-H3 grafting. (A) Scheme of library design. MMP-14 inhibitory motif is inserted into CDR-H3 for library construction. Motif flanking residuals and other five CDRs are diversified. (B) CDR amino acid sequences of isolated Fab 1F8. (C) Dose–response binding affinity curves (EC50) of purified Fab 1F8 and Fab DX-2400, a potent MMP-14 inhibitor (Devy et al., 2009). (D) Inhibition function of purified Fabs 1F8 and DX-2400. An 1 μM quenched-fluorescent substrate peptide and 1 nM cdMMP-14 were used in förster resonance energy transfer (FRET) inhibition assays. KI values were calculated based on the models described in Cer et al. (2009).
The Peptide G sequences with or without terminal cysteines (CFSIAHEC or FSIAHE) were utilized as the inhibition warhead, which was flanked by two random amino acid residuals (encoded by NNS) at both ends for selection of variants able to properly present the motif within the antibody scaffold (Table S1, Supplementary data are available at PEDS online). The CDR-H3 fragments encoding Peptide G were assembled from synthetic oligonucleotides and cloned into an existing Fab library (Ge et al., 2010), which was built on a single framework of germline VH segment DP47 and Vκ segment DPK22 due to their high prevalence in human and decent expression levels in Escherichia coli (Knappik et al., 2000; Ewert et al., 2003). In addition to CDR-H3, the Fab library had randomizations on other five CDRs as previously described (Ge et al., 2010), with total theoretical diversities of 6.9 × 108 for VL and 1.6 × 105 for VH.
Electroporation of 200 optical density highly competent E. coli XL1-blue cells with 3 µg DNA ligation samples generated 5 × 108 transformants. Ninety-five colonies from the constructed library were randomly picked for VL and VH sequencing. Results indicated that 93% of sequenced VL genes and 87% of sequenced VH genes were functional with the inhibition motifs correctly incorporated at CDR-H3s. Among sequenced VH genes, 3% had a stop codon at their NNS positions, and the remaining 10% had either reading-frame shifts or non-designed mutations, likely introduced by primer mismatches during polymerase chain reaction (PCR). Analysis of the amino acid usages at the four NNS positions flanking CDR-H3s, showed distributions of all 20 amino acids as designed. Interestingly, proline and glycine were highly represented at these NNS positions compared with designs. Particularly, proline accounted for 27% and 24% at H95 and H96; and glycine accounted for 38% and 28% at H100E and H100F. Probably, proline and glycine codons have high GC contents; therefore, their fragments were assembled more efficiently than other amino acids during PCR.
The constructed library was subjected to four rounds of phage panning against immobilized MMP-14 catalytic domain (cdMMP-14), which was refolded from denatured inclusion bodies produced in E. coli. Monoclonal Fab phage ELISA of 288 randomly picked colonies from the third and the fourth rounds of panning identified 19 unique clones with significantly high signals over bovine serum albumin background. Sequencing results of these clones indicated that all these isolated 19 Fabs carried a correct Peptide G motif. Among them, 7 clones had the two cysteines flanking the inhibition warhead, while the remaining 12 clones did not have these cysteines. Genes of these 19 isolated Fabs were sub-cloned for expression in E. coli periplasm under the control of a PhoA promoter and a STII leader peptide. Except clones 1B5 and 1D11, which produced 200 µg purified Fabs per litter of culture, the majority of identified clones yielded 10 μg/L or less.
Binding affinity characterizations by ELISA with purified Fabs indicated that most Fabs showed a weak binding to cdMMP-14 with affinities at µM range. However, Fab 1F8 (its six CDR sequences shown in Fig. 1B), exhibited a high affinity of 8.3 nM (EC50 value) to cdMMP-14 (Fig. 1C). At the same conditions, Fab DX-2400 (a high potent MMP-14 inhibitory antibody; Devy et al., 2009) showed EC50 of 4.5 nM, suggesting the affinities of Fabs 1F8 and DX-2400 were in the same order of magnitude. More importantly, the inhibitory functions of purified Fabs against cdMMP-14 were then tested using an förster resonance energy transfer (FRET) peptide substrate. Among isolated MMP-14 binding Fabs, 1F8 showed a significant inhibition with KI of 110 nM (Fig. 1D). Compared with the potency of Peptide G at 150 µM, Fab 1F8 exhibited an improvement of potency by three orders of magnitude. It demonstrates that grafting an inhibition warhead to antibody scaffolds is a practical strategy able to convert a low potency peptide inhibitor into a high potent inhibitory antibody.
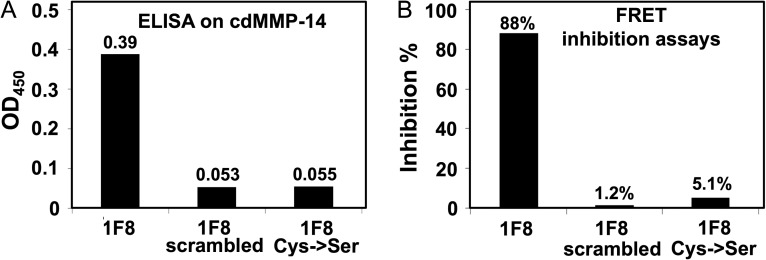
To test whether the grafted motif is crucial for MMP-14 binding and inhibition, two 1F8 mutants at its CDR-H3 were constructed. It has been suggested that a randomly scrambled sequence (CGAAPEACGIHS) and the cysteine to serine mutation (SFSIAHES) of Peptide G dramatically lost their binding and inhibition abilities (Suojanen et al., 2009). Therefore, 1F8 CDR-H3 was replaced with these two designs, and mutated Fabs were produced in E. coli for characterizations by ELISA and FRET inhibition assays. Results showed that both constructed 1F8 mutants exhibited background ELISA signals to cdMMP-14 (Fig. 2A) without significant inhibition activities (Fig. 2B). These results clearly indicated that Peptide G motif was required for the binding and inhibition capabilities of 1F8, and the cysteine residues flanking the motif were also crucial, likely due to the formation of a disulfide bridge to stabilize the inhibitory loop.
Fig. 2.
Binding and inhibition tests of 1F8 mutants. (A) In ELISA, 10 nM Fab 1F8 or its mutants was incubated with cdMMP-14 immobilized on the plates and detected by anti-Fab-HRP. Signal was recorded upon reaction with TMB and stopped by addition of 1 M H2SO4. (B) In FRET inhibition tests, activities of 1 nM cdMMP-14 were measured with 1 µM of quenched-fluorescent peptide substrate at the presence of 500 nM Fab 1F8 or its mutants.
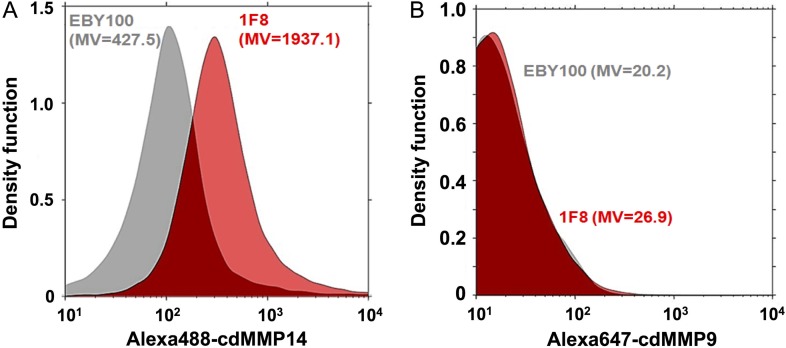
To further characterize 1F8, i.e. selectivity among MMPs and epitope determination, milligrams of purified Fab 1F8 are required. However, these efforts were hampered by limited expression level of Fab 1F8 in E. coli. Expressions under a strong pLac promoter, and with facilitation of periplasmic molecular chaperones DsbA/C co-expression were attempted, but results showed marginal improvement of yields. Given the advanced protein synthesis machineries of eukaryotic systems, 1F8 was cloned for display on yeast cell surface and characterizations by flow cytometry. To achieve effective display, scFvs of 1F8 and control clones were constructed with N-terminal Aga2 fusion and C-terminal c-Myc tag (Boder and Wittrup, 1997; Kondo and Ueda, 2004; Chao et al., 2006). The expression of scFvs on yeast surface was confirmed by labeling with primary chicken anti-c-Myc IgY and secondary Alexa647-goat anti-chicken IgG. For selectivity tests, cdMMP-14 and cdMMP-9 were chemically cross-linked with Alexa488 and Alexa647, respectively, and the activities of resulted conjugates were verified using their FRET peptide substrates. After incubation with 200 nM dye conjugated cdMMP-14/-9, yeast cells displaying scFvs were analyzed by fluorescence-activated cell sorting (FACS). Results showed that when labeled with Alexa488-cdMMP-14, cells displaying 1F8 showed 4-fold higher signals than host cell line (EBY100) without scFv expression (Fig. 3A), while the signals on Alexa647-cdMMP-9 were approximately same for 1F8 cells and EBY100 (Fig. 3B), suggesting high selectivity of 1F8 toward MMP-14 over MMP-9.
Fig. 3.
Binding selectivity tests. Yeast cells displaying scFv 1F8 and host cell line (EBY100) were labeled with (A) Alexa488-MMP-14 or (B) Alexa647-MMP-9, then analyzed by FACS. The event count distributions are represented by density function curves. MV, mean value.
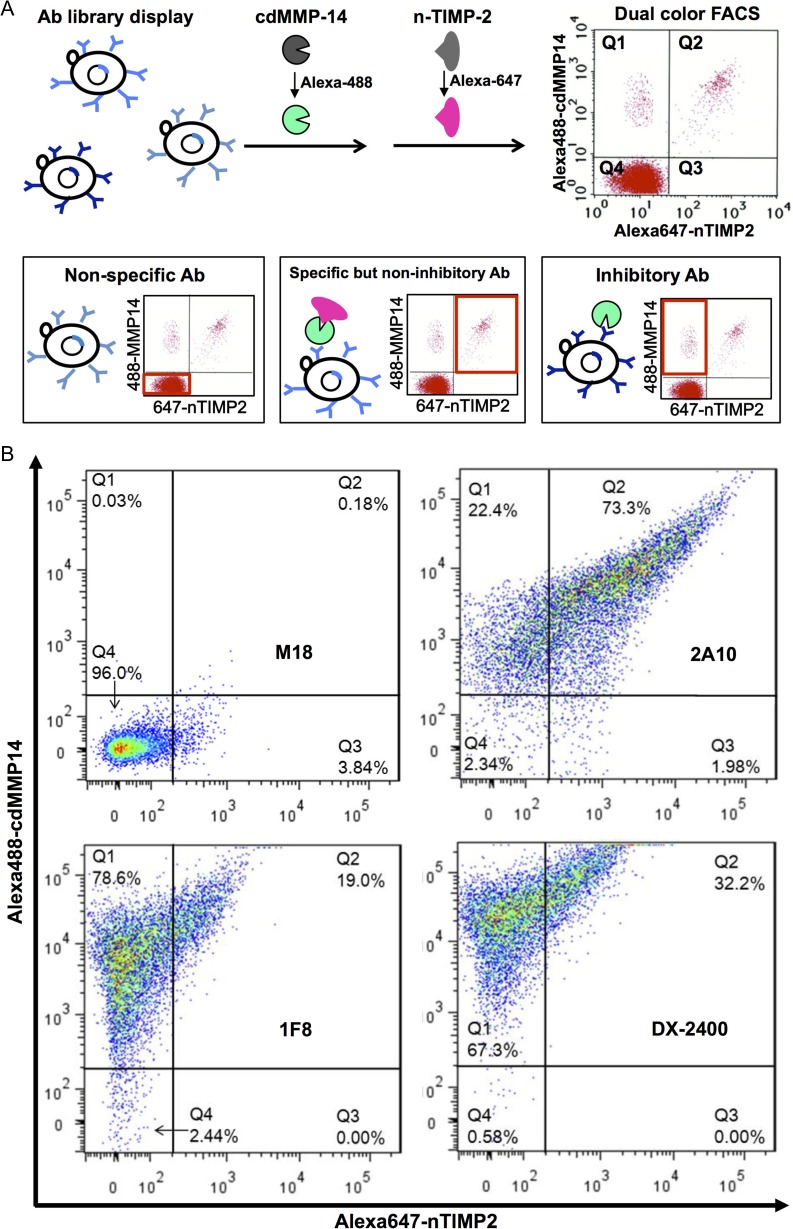
We next profiled the epitope of 1F8, by dual color FACS using the N-terminal domain of tissue inhibitor of metalloproteinase-2 (n-TMIP-2), a native inhibitor of MMP-14 competitively targeting at the reaction cleft of MMP-14 with a KI of 1.2 nM (Fernandez-Catalan et al., 1998; Butler et al., 1999). n-TIMP-2 was produced without refolding via soluble expression in the periplasma of E. coli (Nam and Ge, 2016), and fluorescently conjugated with Alexa647. MMP-14 inhibitory scFv DX-2400 (Devy et al., 2009; Ager et al., 2015) was displayed on yeast surface as a positive control. Irrelevant scFv M18 (anti-PA; Hayhurst et al., 2003) and MMP-14 specific but non-inhibiting scFv 2A10 (isolated in this study) were also cloned to serve as negative controls. The yeast cells displaying these antibody fragments were incubated with Alexa488-conjugated cdMMP-14 and Alexa647-conjugated n-TIMP-2 sequentially (Fig. 4A). Three scenarios are expected: (Scenario 1) observation of non-specific antibody clones (Fig. 4A left) that do not bind to target antigen (cdMMP-14) or native inhibitor (n-TIMP-2), therefore producing low signals on both fluorophores (region Q4 on the scatterplot); (Scenario 2) observation of specific but non-inhibitory antibodies (Fig. 4A middle) that bind to cdMMP-14 at epitopes far from the catalytic pocket, hence not interfering with binding of n-TIMP-2 and generating high signals on both fluorophores (Q2) and (Scenario 3) observation of inhibitory antibodies (Fig. 4A right) that bind to desired epitopes and block n-TIMP-2, resulting in a high signal on cdMMP-14 but a low signal on n-TIMP-2 (Q1).
Fig. 4.
Dual color FACS for identification of inhibitory antibodies and epitope profiling. (A) Antibody-displaying cells are incubated with Alexa488-cdMMP14 and Alexa647-nTIMP2 sequentially, and subjected to FACS scanning. Three scenarios are expected: non-specific, specific but non-inhibitory and inhibitory antibodies. (B) Distinguishment of binding and inhibitory clones by dual color FACS. Non-specific antibody M18 (anti-PA) and specific but non-inhibitory antibody 2A10 are used as negative controls. DX-2400, inhibitory Ab directly competing with n-TIMP-2, is used as a positive control. 1F8 competes with n-TIMP-2 on MMP-14 inhibition.
As experimental results shown in Fig. 4B, yeast cells displaying scFv M18 had 96% of its population located in Q4, therefore it is a non-specific antibody to MMP-14 as expected (Scenario 1). Dual color FACS scanning of yeast cells displaying scFv 2A10 showed 73% of its population located in Q2 (high signals in both Alexa488 and Alexa647 channels), confirming that 2A10 was a specific but non-inhibitory antibody (Scenario 2). The FACS results of 1F8, which exhibits a high affinity in ELISA (Fig. 1C) and inhibitory function in FRET assays (Fig. 1D), showed that 78% of its population located in region Q1 (Scenario 3) with only 19% in region Q2, indicting 1F8 was significantly different from the specific but non-inhibitory clones such as 2A10. A known MMP-14 inhibitory antibody DX-2400 exhibited similar scatterplot as that of 1F8, e.g. 67% in Q1 and 32% in Q2. Notable, DX-2400 showed a higher mean of Alex488-cdMMP-14 signals than that of 1F8, presumably due to its decent expression level and high potency (Devy et al., 2009; Ager et al., 2015). Collectively these FACS scanning results suggested that 1F8 was an inhibitory antibody, and it competed with n-TIMP-2 on binding to cdMMP-14, likely either directly interacting with the vicinity of cdMMP-14 reaction cleft, or allosterically acting as an exosite inhibitor (as examples demonstrated in Wu et al., 2007; Farady et al., 2008).
In our previous study, high concentrations of n-TIMP-2 were used as an eluent to release the antigen-binding phages from the cdMMP-14 bait, resulted in the discovery of 14 inhibitory antibodies (Nam et al., 2016). In the current study, we investigated the binding profiles of isolated antibodies by applying the similar competition between n-TIMP-2 and antibodies, not in ELISA plates but on yeast cell surface. We found that when antibodies like 1F8 and DX-2400 occupy the active site, n-TIMP-2 at low concentrations loses its ability to bind cdMMP-14; when high concentrations (i.e. 2 µM) of n-TIMP-2 were applied, cdMMP-14 will be released from the surface of 1F8/DX-2400 displaying cells. These results consistently suggested that 1F8 and DX-2400 directly competed with n-TIMP-2. In addition, this epitope-specific dual color FACS has potentials to be applied as a novel function-based high-throughput screening method to directly mine synthetic or naïve antibody libraries for inhibitory antibodies.
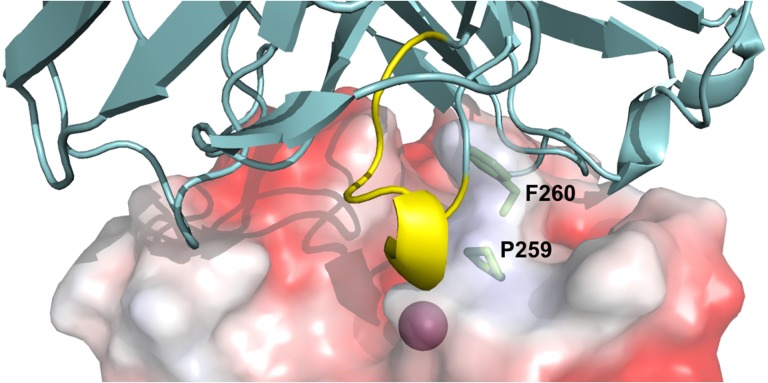
To further elucidate the inhibition model of 1F8, computational simulations were performed to predict Fab 1F8/cdMMP-14 complex structure. Using the multiple functions provided at Structural Antibody Prediction Server (SAbPred; Dunbar et al., 2016), Fab 1F8 structure was generated, and applied for paratope prediction and epitope mapping with the aid of reported structure of cdMMP-14 (PDB ID 1bqq; Fernandez-Catalan et al., 1998). Fab 1F8 and cdMMP-14 were then docked using ZDOCK (Pierce et al., 2014) with the generated paratope/epitope prediction results as restraints. As shown in Fig. 5, structure simulation indicates that the grafted Peptide G motif (yellow in Fig. 5) penetrates into the reaction cleft of cdMMP-14, suggesting the inhibition function of Fab 1F8 is likely given by direct interaction with MMP-14 active site. In addition, Pro259 and Phe260 of MMP-14 (green in Fig. 5), as the key residues forming MMP-specific S1′ substrate binding site (Nagase, 2001; Gupta and Patil, 2012), were identified as the possible epitopes, and thus may partially explain the high selectivity of 1F8.
Fig. 5.
Structural prediction of Fab 1F8/cdMMP-14 complex. The active site of cdMMP-14 (electrostatic potential surface model, PDB ID 1bqq, Fernandez-Catalan et al., 1998) was shown with the catalytic Zn2+ (magenta) at the bottom of the reaction pocket. Fv 1F8 (cartoon, cyan) binds to vicinity of cdMMP-14 reaction cleft through direct interaction between the grafted Peptide G motif (yellow) and the active site. MMP-14 residues Pro259 and Phe260 (sticks, green) form the S1′ MMP-specific substrate binding pocket. Model of Fab 1F8 was generated using SAbPred (Dunbar et al., 2016). Fab 1F8 and cdMMP-14 were docked using ZDOCK (Pierce et al., 2014). Images were generated using PyMOL.
In summary, with the hypothesis that grafting an inhibitory peptide into CDR-H3 confers binding specificity and inhibition effect to the antibody, in this study, a synthetic antibody library was generated to optimize the sequences flanking CDR-H3 and other five CDRs of both the heavy and light variable domains. After phage panning, among dozens of affinity binders, one inhibitory antibody, Fab 1F8, with binding affinity (EC50) of 8.3 nM and inhibition potency (KI) of 110 nM was isolated, demonstrating the successful conversion of a low-affinity peptide inhibitor to an inhibitory antibody with high selectivity and >1000-fold enhancement of potency. Tests with 1F8 mutants confirmed that the grafted Peptide G motif played an important role for MMP-14 inhibition. Yeast cell surface display and followed FACS analysis indicated 1F8 direct competed with n-TIMP-2 on binding with MMP-14. And computational simulation suggested the interaction is likely through direct binding to the vicinity of MMP-14 reaction cleft.
The MMP family members are promising drug targets in many states of pathologies (Cook et al., 2000; Elkington et al., 2005; Overall and Kleifeld, 2006; Dev et al., 2010; Castro and Tanus-Santos, 2013). Besides MMP-14, peptide inhibitors toward other MMP family members have also been identified (Koivunen et al, 1999; Heikkilä et al., 2006). It is highly likely that the methodology of this study could be readily applied for the generation of highly selective inhibitory antibodies targeting additional individual MMPs. In addition to therapeutic potentials, these inhibitors with high selectivity can also be exploited as research tools to shed more light on the MMP functionality in normal and patho-physiological conditions. Other than MMP family members, we also expect that the techniques described here, i.e. motif grafting (Moroncini et al., 2004; Frederickson et al., 2006; Qin et al., 2007; Kogelberg et al., 2008; Zhang et al., 2015), CDR optimization and epitope-specific FACS, are valuable on development of high potency inhibitory antibodies based on peptide inhibitors.
Supplementary Material
Supplementary data
Supplementary data are available at Protein Engineering, Design & Selection online.
Funding
This research was supported by National Science Foundation [1453645] University of California Cancer Research Coordinating Committee, and Dissertation Year Program Fellowship from UC Riverside. We appreciate Holly Eckelhoefer at UCR Genomic Center for her helps on FACS experiments.
References
- Ager E.I., Kozin S.V., Kirkpatrick N.D. et al. (2015) J. Natl. Cancer Inst., 107: djv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder E.T. and Wittrup K.D. (1997) Nat. Biotechnol., 15, 553–557. [DOI] [PubMed] [Google Scholar]
- Butler G.S., Hutton M., Wattam B.A., Williamson R.A., Knäuper V., Willenbrock F. and Murphy G. (1999) J. Biol. Chem., 274, 20391–20396. [DOI] [PubMed] [Google Scholar]
- Castro M.M. and Tanus-Santos J.E. (2013) Curr. Drug Targets, 14, 335–343. [DOI] [PubMed] [Google Scholar]
- Cer R.Z., Mudunuri U., Stephens R. and Lebeda F.J. (2009) Nucleic Acids Res., 37, W441–W445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M. and Wittrup K.D. (2006) Nat. Protoc., 1, 755–768. [DOI] [PubMed] [Google Scholar]
- Cook H., Stephens P., Davies K.J., Harding K.G. and Thomas D.W. (2000) J. Invest. Dermatol., 115, 225–233. [DOI] [PubMed] [Google Scholar]
- Dev R., Srivastava P.K., Iyer J.P., Dastidar S.G. and Ray A. (2010) Expert Opin. Investig. Drugs, 19, 455–468. [DOI] [PubMed] [Google Scholar]
- Devy L., Huang L., Naa L. et al. (2009) Cancer Res., 69, 1517–1526. [DOI] [PubMed] [Google Scholar]
- Dunbar J., Krawczyk K., Leem J., Marks C., Nowak J., Regep C., Georges G., Kelm S., Popovic B. and Deane C.M. (2016) Nucleic Acids Res., 44, W474–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkington P.T.G., O'Kane C.M. and Friedland J.S. (2005) Clin. Exp. Immunol., 142, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewert S., Huber T., Honegger A. and Pluckthun A. (2003) J. Mol. Biol., 325, 531–553. [DOI] [PubMed] [Google Scholar]
- Farady C.J., Egea P.F., Schneider E.L., Darragh M.R. and Craik C.S. (2008) J. Mol. Biol., 380, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Catalan C., Bode W., Huber R., Turk D., Calvete J.J., Lichte A., Tschesche H. and Maskos K. (1998) EMBO J, 17, 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson S., Renshaw M.W., Lin B. et al. (2006) Proc. Natl Acad. Sci. USA, 103, 14307–14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Mazor Y., Hunicke-Smith S.P., Ellington A.D. and Georgiou G. (2010) Biotechnol. Bioeng., 106, 347–357. [DOI] [PubMed] [Google Scholar]
- Genís L., Gálvez B.G., Gonzalo P. and Arroyo A.G. (2006) Cancer Metastasis Rev., 25, 77–86. [DOI] [PubMed] [Google Scholar]
- Gupta S.P. and Patil V.M. (2012) Gupta S.P.(ed),. Matrix Metalloproteinase Inhibitors. Basel, Springer, pp. 35–56. [Google Scholar]
- Hayhurst A., Happe S., Mabry R., Koch Z., Iverson B.L. and Georgiou G. (2003) J. Immunol. Methods, 276, 185–196. [DOI] [PubMed] [Google Scholar]
- Heikkilä P., Suojanen J., Pirilä E., Väänänen A., Koivunen E., Sorsa T. and Salo T. (2006) Int. J. Cancer, 118, 2202–2209. [DOI] [PubMed] [Google Scholar]
- Knappik A., Ge L.M., Honegger A., Pack P., Fischer M., Wellnhofer G., Hoess A., Wolle J., Pluckthun A. and Virnekas B. (2000) J. Mol. Biol., 296, 57–86. [DOI] [PubMed] [Google Scholar]
- Kogelberg H., Tolner B., Thomas G.J. et al. (2008) J. Mol. Biol., 382, 385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen E., Arap W., Valtanen H. et al. (1999) Nat. Biotechnol., 17, 768–774. [DOI] [PubMed] [Google Scholar]
- Kondo A. and Ueda M. (2004) Appl. Microbiol. Biotechnol., 64, 28–40. [DOI] [PubMed] [Google Scholar]
- Moroncini G., Kanu N., Solforosi L., Abalos G., Telling G.C., Head M., Ironside J., Brockes J.P., Burton D.R. and Williamson R.A. (2004) Proc. Natl Acad. Sci. USA, 101, 10404–10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C.J., Butler G.S., Rodríguez D. and Overall C.M. (2009) Curr. Opin. Cell. Biol., 21, 645–653. [DOI] [PubMed] [Google Scholar]
- Nagase H. (2001) Clendeninn N.J. and Appelt K. (eds),. Matrix Metalloproteinase Inhibitors in Cancer Therapy. Humana Press, pp. 403–420. [Google Scholar]
- Nam D.H. and Ge X. (2016) Biotechnol. Bioeng., 113, 717–723. [DOI] [PubMed] [Google Scholar]
- Nam D.H., Rodriguez C., Remacle A.G., Strongin A.Y. and Ge X. (2016) Proc. Natl Acad. Sci. USA, doi:10.1073/pnas.1609375114.
- Overall C.M. and Kleifeld O. (2006) Nat. Rev. Cancer, 6, 227–239. [DOI] [PubMed] [Google Scholar]
- Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T. and Weng Z. (2014) Bioinformatics, 30, 1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Feng J., Li Y., Lin Z. and Shen B. (2007) Mol. Immunol., 44, 2355–2361. [DOI] [PubMed] [Google Scholar]
- Suojanen J., Salo T., Koivunen E., Sorsa T. and Pirilä E. (2009) Cancer Biol. Ther., 8, 2362–2370. [DOI] [PubMed] [Google Scholar]
- Turk B. (2006) Nat. Rev. Drug Discov., 5, 785–799. [DOI] [PubMed] [Google Scholar]
- Visse R. and Nagsase H. (2003) Circ. Res., 92, 827–839. [DOI] [PubMed] [Google Scholar]
- Wu Y., Eigenbrot C., Liang W.C., Stawicki S., Shia S., Fan B., Ganesan R, Lipari M.T. and Kirchhofer D. (2007) Proc. Natl Acad. Sci. USA, 104, 19784–19789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zou H., Wang Y.. et al. (2015) Angew. Chem. Int. Ed. Engl., 54, 2126–2130. [DOI] [PubMed] [Google Scholar]
- Zucker S. and Cao J. (2009) Cancer Biol. Ther., 8, 2371–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.