Abstract
The promiscuity of G-protein-coupled receptors (GPCRs) has broad implications in disease, pharmacology and biosensing. Promiscuity is a particularly crucial consideration for protein engineering, where the ability to modulate and model promiscuity is essential for developing desirable proteins. Here, we present methodologies for (i) modifying GPCR promiscuity using directed evolution and (ii) predicting receptor response and identifying important peptide features using quantitative structure-activity relationship models and grouping-exhaustive feature selection. We apply these methodologies to the yeast pheromone receptor Ste2 and its native ligand α-factor. Using directed evolution, we created Ste2 mutants with altered specificity toward a library of α-factor variants. We then used the Vectors of Hydrophobic, Steric, and Electronic properties and partial least squares regression to characterize receptor-ligand interactions, identify important ligand positions and properties, and predict receptor response to novel ligands. Together, directed evolution and computational analysis enable the control and evaluation of GPCR promiscuity. These approaches should be broadly useful for the study and engineering of GPCRs and other protein-small molecule interactions.
Keywords: directed evolution, partial least squares regression (PLSR), receptor promiscuity
Introduction
G-protein-coupled receptors (GPCRs) are a ubiquitous class of molecular sensors able to detect a broad range of ligands including protons, small molecules, peptides, nucleotides, hormones and neurotransmitters (Lagerström and Schiöth, 2008). In addition to the diverse ligands detectable by the GPCR class, an individual GPCR may also exhibit ligand promiscuity (Civelli, 2005; Li et al., 2012). Specific residues in the GPCR alter ligand preference and modify receptor promiscuity; for example, particular regions of transmembrane domains TM3, TM6 and TM7 have been identified to modulate specificity across the entire family of rhodopsin-like GPCRs (Venkatakrishnan et al., 2013).
Mutations in GPCRs can have a wide spectrum of consequences. On one hand, spurious mutations can result in altered receptor properties that promote various cancers (Dorsam and Gutkind, 2007; O'Hayre et al., 2013) and autoimmune diseases (Schöneberg et al., 2004; Vassart and Costagliola, 2011). Even ‘neutral’ mutations can affect an individual's tolerance for, or response to, certain drugs (Zalewska et al., 2014). On the other hand, mutations altering specificity are crucial for re-engineering receptors into biosensors to recognize non-native ligands (Armbruster et al., 2007; Conklin et al., 2008). The applications for such sensors range from diagnostics to bioterrorism security to environmental monitoring (Adeniran et al., 2015; Slomovic et al., 2015). The ability to predict and control the effects of mutations on specificity is therefore essential for understanding disease, facilitating drug design and guiding biosensor development.
Directed evolution is a powerful tool for introducing mutations that alter protein function. The target protein sequence is subjected to mutagenesis to produce a diverse library of variants. The library is then screened for mutants that have acquired the desired trait and the mutations can be subsequently identified by sequencing (Cobb et al., 2013; Packer and Liu, 2015). In directed evolution, it has been observed that promiscuity predisposes enzymes toward greater success due to increased plasticity (Gould and Tawfik, 2005). In fact, a common approach for directed evolution is to improve upon the activity of a promiscuous parent, rather than evolve for novel activity (Aharoni et al., 2004; Khersonsky et al., 2006). Receptors successfully evolved to respond to a target ligand are often too promiscuous for applications such as biosensing and must be further evolved to decrease promiscuity. Thus, specifically understanding the relationship between receptor sequence and specificity would be broadly useful.
While directed evolution may alter receptor specificity, characterizing the resulting mutant receptors for activity against every ligand (e.g. peptide sequence) is infeasible. Computational tools, such as quantitative structure-activity relationship (QSAR) modeling, can identify physiochemical trends of ligands to reduce experimental effort and offer insight into the system. QSAR aims to construct predictive models of activity as a function of structure and is traditionally used for characterizing chemical compounds for drug discovery (Winkler, 2002). More recently, QSAR has been applied to characterize peptides (Cherkasov et al., 2014), such as dipeptide inhibitor interactions with GPCRs (Mei et al., 2005) and nonamer peptide binding to a Class I MHC (Pissurlenkar et al., 2007).
In this study, we demonstrate selection schemes using directed evolution capable of producing receptors with higher or lower promiscuity than the parent receptor. Using a QSAR-inspired approach, we developed a computational pipeline to elucidate the physiochemical properties and residue positions of a ligand most predictive of response by the wild-type Ste2 and representative high- and low-promiscuity receptors. We then utilized the top-performing models to predict receptor response to novel peptides that vary from the native ligand in both length and composition.
Materials and Methods
Yeast receptors and ligands
Yeast strains
Yeast strain MPY578t5 (Dong et al., 2010) was a gift from Brian Roth. yJB013 [MATa, far1::LYS2 fus1::yeGFP sst2::PSST2-G418R ste2::LEU2 fus2::PFUS2-CAN1 TRP1::mKate ura3 lys2 ade2 last five amino acids of GPA1 (KIGII) replaced with the homologous mammalian Gαi (DCGLF)] was created from MPY578t5 as previously described (manuscript in revision). In brief, fluorescent reporters were integrated at the Trp1 locus (constitutive mKate) and the Fus1 locus (yeGFP induced by the pheromone-sensitive Fus1 promoter). Strain yBA006 was created from yJB013 by knocking out the Bar1 protease with a HIS3 marker.
Receptors (DNA shuffling, epPCR)
To generate mutant receptors for directed evolution, diversity was introduced into the sequence of STE2 through error-prone PCR (epPCR) or DNA shuffling. For the low-promiscuity sort, Ste2 was subjected to epPCR using the GeneMorphII random mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. For the promiscuity sort, DNA shuffling was used to create all possible combinations of 10 mutations that we had previously discovered as potentially influencing promiscuity: M54I, F55V, S145L, S219P, A229V, L255S, S259T, S288P, K304X and A336V. Primers containing these mutations were created and listed in Supplementary data, Table S6. Wild-type Ste2 was digested as described (Stemmer, 1994) to produce ~50 bp fragments. Fragments were purified by ethanol precipitation and mixed with 1 μM equimolar ratio of the mutagenic primers in a PCR reaction with no external primers and cycled under the following conditions: [95°C/3min, (95°C/30s, 53°C/30s, 72°C/90s) × 45, 72°C/10min, 12°C hold]. 1 μl of this reaction was used as template using external primers to amplify only full-length Ste2 variants. The Mut1 receptor was created using megaprimer extension PCR with a primer containing the g126c mutation (Tyagi et al., 2004).
Libraries and individual receptors were amplified with oligonucleotides that provided 40 bp of homology on each side to the single-copy centromeric plasmid p416 (Mumberg et al., 1995) and transformed by electroporation (Benatuil et al., 2010) into yBA006 with linear p416, which was cut with XhoI and BamHI (New England Biolabs). Ste2 assembled into the cut backbone such that Ste2 was constitutively expressed from the GPD promoter. For shuffled DNA, 10 colonies were sequenced and the distribution of mutations confirms that the library is not statistically different from a library containing all possible combinations of mutations (P = 0.999, Student's t-test). For epPCR, 10 colonies were sequenced and receptors were found to have an average of 6 mutations per receptor, including silent mutations.
Ligand library
A custom peptide library of α-factor and 54 single residue variants was designed to interrogate several physiochemical traits of each amino acid by scanning the α-factor peptide with alanine (small and hydrophobic), tyrosine (large, polar and aromatic), glutamate (large and negatively charged), and lysine (large and positively charged). For directed evolution experiments, we focused our efforts on residues 10–13 of the α-factor peptide, the domain responsible for binding to Ste2 (Abel et al., 1998). A complete list of all peptides and sequences may be found in Supplementary data, Table S1. Peptides 43–88 and A–F were ordered from Abbiotec (San Diego, CA); peptides 89–96 were ordered from GenScript (Piscataway, NJ). Peptides were resuspended to 1 mM in 50% V/V acetonitrile, as per the manufacturer's recommendation, sterile filtered, and stored at −20°C. All subsequent dilutions of peptides were done in RNase-free water.
Directed evolution
Directed evolution selection schemes for modifying promiscuity
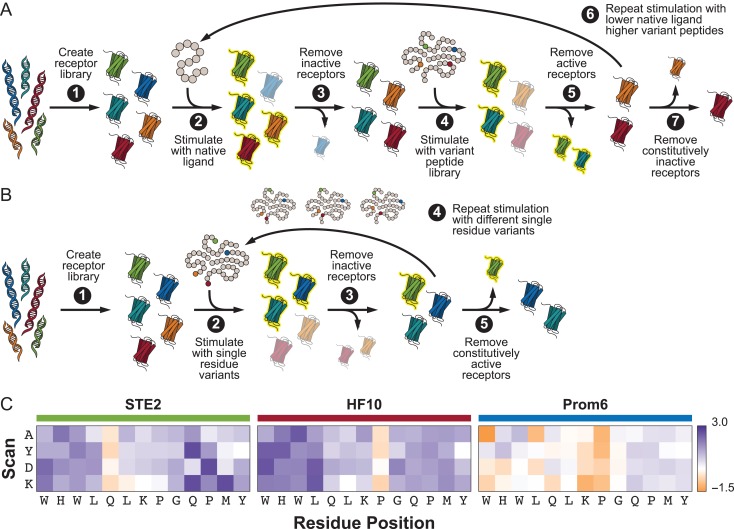
To generate a low-promiscuity receptor, the library was first selected for receptors that responded at 200nM α-factor, near the wild-type EC50 (Fig. 1A, Steps 2–3). Cells were then exposed to all peptide variants that vary from α-factor in residues 10–13, each present at a concentration that would produce a half-maximal response in the Ste2 parent (EC50). Only receptors that did not respond to any peptide were retained (Fig. 1A, Steps 4–5). Cells were next selected for a response to 25 nM α-factor, followed by a countersort against the EC80 of the C-terminal peptide variants (Fig. 1A, Step 6). A final round of sorting was performed to retain only receptors responsive to 10 nM α-factor to remove any constitutively inactive mutants (Fig. 1A, Step 7).
Fig. 1.
Directed evolution schemes for modulating receptor promiscuity. (A) Process for evolving receptors with decreased promiscuity. (B) Process for evolving receptors with increased promiscuity. (C) Heat maps demonstrating the decreased/increased promiscuity of HF10/PROM6, respectively, compared to the native Ste2 receptor against a subset of the peptide library. Values given are the log fold change in EC50 for each receptor against a novel peptide (single residue changed from the wild-type to the scan residue at the given position) from the EC50 of the receptor against α-factor. Hatched square denote the variants resulting in the wild-type α-factor sequence.
To generate a high promiscuity receptor, the library was exposed to all peptide variants at position 13 at a concentration that produced a 10% maximal response (EC10) in the Ste2 parent (Fig. 1B, Step 2). Only receptors that responded to at least one peptide were retained by FACS (Fig. 1B, Step 3). Receptors were subsequently subjected to sequential rounds of sorting with the EC10 concentration all variants at positions 12, 11 or 10 (Fig. 1B, Step 4). A final round of sorting on unstimulated cells removed any constitutively active receptors (Fig. 1B, Step 5).
For each round of cell sorting, cells were grown overnight in complete synthetic medium lacking uracil (CSM-Ura) for plasmid maintenance and then diluted to OD600 of 0.1 in CSM-Ura. The cells were stimulated as described above. After a 2.5 h stimulation, cells were collected by centrifugation at 3000 g for 3 min and resuspended in sterile 1× PBS. Cells were kept on ice until sorting. A total of 107 cells were sorted, with gates set based on wild-type Ste2 stimulated with 1 μM α-factor (positive control) or unstimulated (negative control). Cells were collected into SDCAA medium (Chao et al., 2006) lacking uracil and grown for 48–72 h. In subsequent rounds of sorting, a number of cells equivalent to a 10× oversampling of the previously retained library size were sorted.
The final library from each sorting scheme was amplified by colony PCR and retransformed into a fresh yBA006 strain with digested p416 as described above to eliminate the effects of any background mutations that may have accumulated in the genome or plasmid backbone during extended passaging. After retransformation, receptors HF10, from the low-promiscuity sort, and Prom3, Prom6 and Prom7, from the high promiscuity sort, were chosen for further characterization.
The receptors TBBI2, TBBI6 (identical to Prom6) and TBBI7 (identical to Prom3) were similarly isolated from the same shuffled library through four rounds of FACS sorting with peptide B. Cells were treated with 1 μM peptide for three rounds of sorting and with 500 nM peptide for the final round.
Dose response curves
Cells were grown and diluted as for cell sorting. 190 μl of cells were added per well of a 96-well plate. 10 μl of peptide was then added to each well to final concentrations of 50, 100, 500, 1, 5 and 10 μM. To the negative control wells, 10 μl of sterile water was added. Plates were covered with SealMate breathable barrier and incubated at 30°C with shaking for 2.5 h. Plates were spun for 5 min at 2000 g and 4°C. The culture medium was aspirated and cells were resuspended in 100 μl of 1× PBS and maintained at 4°C until they were read using the high-throughput attachment of a BD LSRII (BD Biosciences) analytic flow cytometer. mKate was excited at 561 nm and read at 620 nm; yeGFP was excited at 488 nm and read at 530 nm. For each sample, 10 000 events were collected.
The flow files (extension .fcs) were read using the FlowCytometryTools Python package (available at doi: 10.5281/zenodo.32991). For each sample, the normalized mean signal M was calculated as:
where N is the number of events where signals were not negative and the mKate signal was not below 300. The resulting M for each concentration was normalized by the 0 μM ligand control. Average across replicates for each concentration c were calculated as:
The curve_fit function from the Python package SciPy, which uses non-linear least squares, was then used to fit the resulting average normalized mean signals to the dose response equation:
where c is the EC50 and d is the hill coefficient.
Quantification of receptor promiscuity
The EC50 (concentration at which the half-maximal response is achieved) was calculated for each receptor/peptide pair. The promiscuity of each receptor was calculated using an equation adapted from (Nath and Atkins, 2008):
where 1/EC50 to the ith peptide in the test library. The Nath and Atkins equation examines the catalytic efficiency of an enzyme toward a library of substrates by comparing values of kcat and KM between substrates. We replaced the e = kcat/KM term with e = 1/EC50, a measurement more suited to characterizing receptor signaling. The value calculated from this equation depends on the ligand library against which the receptor is assayed; different libraries will produce different values. Because all receptors were assayed against the same peptide library, promiscuity values can be compared between receptors. The value of P ranges from 0 for a perfectly faithful receptor (does not respond to any non-native ligands) to 1 for a perfectly promiscuous receptor (responding to all peptides in the library equally well).
Grouping-exhaustive PLSR
Data pre-processing
For the response data, three values were extracted from the dose response curves for each receptor/peptide combination: EC50, EC10 and response at 10 μΜ peptide concentration. For each of these values, the natural logarithm and inverse were also calculated. For the predictor data, the 13 amino acid sequence of each peptide was converted to a 104-element vector using the eight-component VHSE system (Mei et al., 2005) with each component normalized between 0 and 1.
Grouping schemes
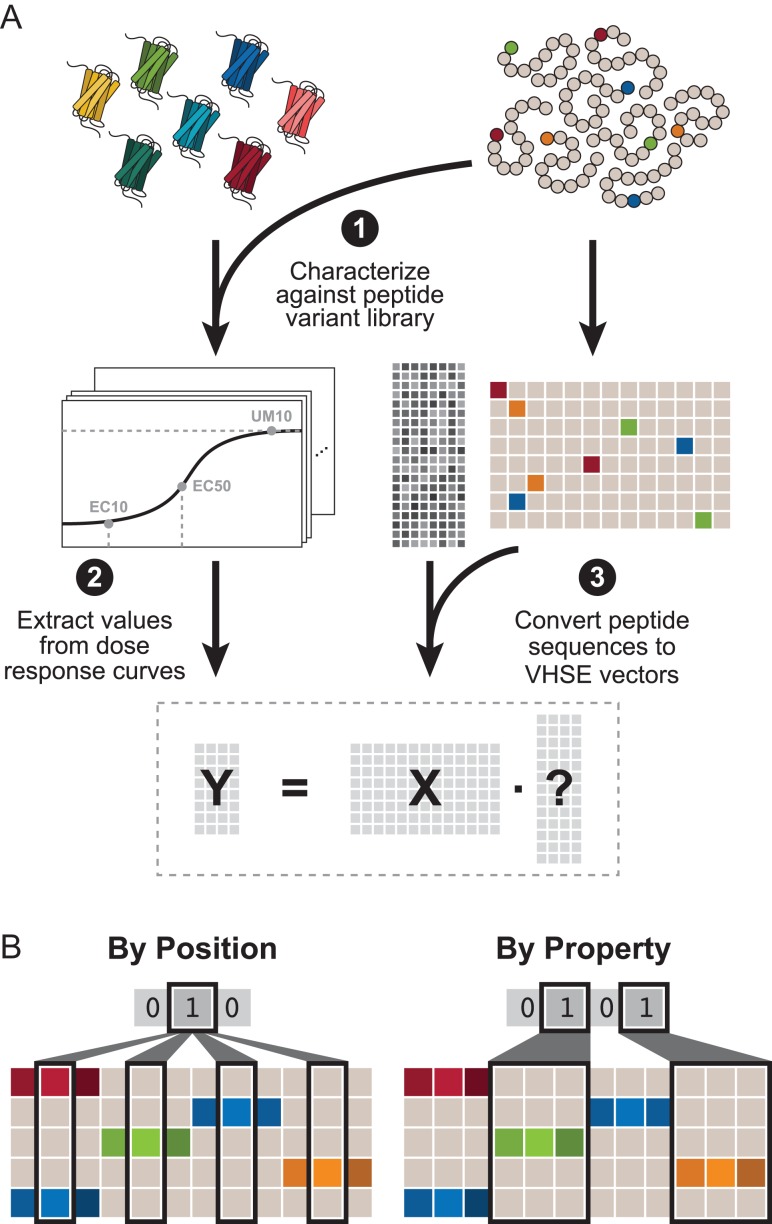
Two grouping schemes were used to reduce the search space required for an exhaustive search: By Position and By Property. (Fig. 2B). For the By Position grouping scheme, the 104-element vector was broken down into 13 groups in which each group includes the eight VHSE properties for that position. The exhaustive search for By Residue contains combinations where each residue is or is not included and can be represented by a 13-element binary vector. For example, the combination [1 1 0 0 0 0 0 0 0 0 0 0 1] contains the eight VHSE properties for residues 1, 2 and 13 for a total of 24 features. For simplicity, combinations are stored as the corresponding decimal number; the previous example would be, for example, stored as 6145. Analogously, the By Property grouping scheme, the 104-element vector was broken down into eight groups in which each group includes the property value for all 13 positions. The exhaustive search contains combinations represented by an eight-element binary vector.
Fig. 2.
Computational pipeline for data collection and processing. (A) Seven receptors (wild-type Ste2, lower promiscuity HF10, and higher promiscuity Prom6, Prom7, Prom3, Mut1 and TBBI2) were selected. Each of the 55 peptides was converted to vectors using the eight-property VHSE system for a predictor (X) matrix of 104 features. Each receptor was characterized against the peptide library to obtain a dose response curve. Three metrics (EC50, EC10 and response at 10 μM peptide) were obtained from each curve, and further expanded using inverse/logarithm manipulations to obtain the response (Y) matrix. (B) Example grouping schemes for a representative system in which peptides are four residues long and each peptide has three properties (note that our system has 13-residue long peptides with eight properties).
Partial least squares regression
For each receptor and each grouping of both grouping schemes, PLSR was performed against each of the 9 response variables. Both the Matlab plsregress function, which uses the SIMPLS algorithm to perform the regression (de Jong, 1993) and an implementation of the NIPALS algorithm (Wold et al., 2001) in Matlab plsnipals by (Li et al., 2014) were used. The built-in plsregress was used to calculate the regression coefficient and the mean-squared error MSE from a leave-one-out cross-validation. The predictive squared correlation coefficient (Consonni et al., 2010) can be calculated using:
where n is the number of observations. The plsnipals script was used to recover component weights in order to determine VIP score (Wold, 1995). VIP scores for the i-th feature is calculated by:
where n is the number of features, M is the number of components, is the weight of the i-th feature in the m-th component and is the percentage of variance in y explained by the m-th component.
Shuffled data analysis
The predictor data were randomly permuted column-wise to generate 50 different shuffled data sets. The grouping-exhaustive PLSR described above was performed using each shuffled predictor data against the EC50 response of Ste2. Resulting Q2 values for a given grouping were averaged across the 50 different shuffled data sets.
Feature selection
The first step in selecting the top features was identifying which of the response types (EC50, EC10, response at 10 μΜ, etc.) produced the highest percentage of models with greater than 0.25 (see Supplementary data, Fig. S5, Step 1). Using this top response type, top-performing models were selected based on a greater than 0.75 where is the highest obtained for that response type (see Supplementary data, Fig. S5, Steps 2–3). For a receptor, the weighted frequency f of the i-th residue or property was calculated using:
where G is the grouping type (residue or property), is the number of top models for the grouping type (see Supplementary data, Fig. S5, Step 4).
For a given receptor and grouping scheme, the weighted feature frequencies for all 104 features were calculated. For each of the top N models, the corresponding list of features was sorted according to VIP score. The top features in this list were selected based on the location of the elbow, determined objectively as the feature with the first minimum distance d. For feature k in a list of n features sorted by decreasing VIP:
where is the max VIP score, i.e. VIP score of the first element in the list (see Supplementary data, Fig. S5, Steps 5–7). For a receptor, the weighted feature frequency F for the i-th feature was calculated using:
where G is the grouping type (residue or property), is the number of top models for the grouping type (see Supplementary data, Fig. S5, Step 8). Finally, features were selected based on average weighted feature frequency (see Supplementary data, Fig. S5, Steps 9–11):
Receptor response predictions
Dose response curves for peptides A–F against all seven receptors were collected as described above. For each receptor, responses (EC50, EC10 and response at 10 μM) were normalized against response of the receptor to α-factor. For model predictions, the amino acid sequences of peptides A–F were first vectorized as described above. For peptides shorter than 13 residues, the sequence was aligned at the C-terminal and the properties for the missing N-terminal positions were all assigned zeros. For peptides longer than 13 residues, the sequence was truncated at the N-terminal to be 13 residues before vectorizing. Predictions were obtained from models built using only the selected top features. The top response type for each receptor was use: EC50 for Ste2, response at 10 μM for HF10, EC10 for Prom6, EC50 for Prom7, log(EC50) for Mut1, EC50 for Prom3 and log(EC50) for TBBI2. As with the experimental data, predictions were then normalized to the model prediction for α-factor. Both experimental and predicted normalized responses were then binned into four groups: high sensitivity (receptor is more sensitive to the peptide than to α-factor), medium sensitivity (receptor is equally sensitive to the peptide as it is to α-factor), low sensitivity (receptor is less sensitive to the peptide than to α-factor) and no response (receptor does not respond to the peptide). Supplementary data, Table S7 summarizes the binning thresholds.
Results
Directed evolution modulates promiscuity of yeast G-protein-coupled receptor Ste2
Our work focuses on using directed evolution to generate variants of Ste2 responsive to peptide biomarkers for a variety of disease states. Because α-factor, the native peptide ligand of Ste2, differed significantly from these biomarkers, generating a responsive receptor variant from Ste2 in a single evolutionary step is infeasible. We therefore use a stepwise approach in which we design a series of intermediate ‘chimera’ peptides with sequences progressively transitioning from that of α-factor to that of the biomarker. Receptors evolved for each of these peptides are treated as evolutionary intermediates and undergo further rounds of directed evolution for the next peptide in the series, until a final receptor is obtained. We found that the evolutionary intermediate receptors that further evolved to successful receptors tended to have increased promiscuity compared to that of the wild-type parent, Ste2. To quantitatively compare ligand promiscuity, we introduced a promiscuity index, P, by adapting an equation for enzyme substrate promiscuity (Nath and Atkins, 2008),where P = 0 indicates no promiscuity (i.e. no response to non-native ligands) and P = 1 indicates full promiscuity (i.e. equal response to all ligands in the library). Using this metric, we quantified the response of evolutionary intermediate receptors from our directed evolution pathway against a library of peptides differing from the native α-factor ligand at each single position. These intermediate receptors (TBBI2, TBBI6 and TBBI7) showed elevated promiscuity (P = 0.83 ± 0.06) compared to that of wild-type Ste2 (P = 0.728). Based on this observation, we sought to determine whether promiscuity could be controlled through selection.
To investigate if directed evolution could be used to produce receptors of decreased promiscuity (higher fidelity), we employed selection criteria to identify receptors that respond only to the target ligand (α-factor) and not similar peptides. Mutant libraries of Ste2 were generated using error-prone PCR and subjected to sequential rounds of fluorescence-activated cell sorting (FACS) in which receptors that did not respond to single-amino-acid variants of α-factor were retained (Fig. 1A). These selection conditions could allow the retention of constitutively inactive mutants, so a final round of sorting was used to ensure that retained receptors could be activated by a moderate concentration of α-factor. An estimated three receptors were retained from an initial library of 106 variants.
Similarly, we sought to determine if directed evolution could produce receptors with increased promiscuity. Here, we employed selection criteria to identify receptors that respond to many different single-amino-acid variants of α-factor. Mutant libraries of Ste2 were generated through DNA shuffling of selected mutations and iterative rounds of FACS selection were used to isolate receptors that responded to stimulation by at least one of the four single-amino-acid variants at each residue tested (Fig. 1B). The sorting conditions could also allow for the retention of constitutively active mutants. Therefore, a final round of sorting was used to discard receptors that were active in the absence of any peptide. An estimated 55 receptors were retained from an initial library of 106 variants. For more detailed information on sorting conditions for both the decreased and increased promiscuity cases, see Materials and Methods section.
After sorting, receptors from each experiment were isolated at random for further study (HF10 from the low-promiscuity sort; Prom3, Prom6 and Prom7 from the high promiscuity sort). High-fidelity receptor HF10 was found to have a truncation at residue 310 (S310X) and promiscuous receptor Prom6 harbored mutations M54I, S145L and a truncation at residue 304 (K304X), all of which have been previously reported. Receptors Prom3 and Prom7 had combinations of these mutations: M54I/K304X and M54I/S145L, respectively. Truncations such as the S310X and K304X mutations are known to increase sensitivity to α-factor (Reneke et al., 1988) due to removal of regulatory domains required for pheromone desensitization. We report here that the S310X mutation also increases the discriminatory power of the receptor toward the first four residues in α-factor. The M54I and S145L mutations found in Prom6 have previously been reported to make the receptor sensitive to the S. kluyveri α-factor (Marsh, 1992), so it is unsurprising that we found that they increase promiscuity. M54 lies in a region of TM1 previously known to directly interact with peptide residues Q10–Y13 (Umanah et al., 2009; Mathew et al., 2011). The exact mechanism by which S145L increases promiscuity remains unclear; however, S145 is located near a region known to be involved in activating the conformational change of the receptor to bring it close to residue Y266 (Sommers and Dumont, 1997). It is possible that the S145L mutation predisposes the receptor toward activation even when the peptide is not an exact match to α-factor.
Receptors Prom6 and Prom3 were identical to previously isolated evolutionary intermediates TBBI6 and TBBI7, respectively, suggesting that promiscuity is favored in evolutionary pathways even when not under direct selective pressure. Because all of the characterized promiscuous receptors contained the M54I mutation, but the single mutant had not been observed in our screen, we also created the M54I single mutant receptor (Mut1) for further study. These promiscuous receptors (Prom3, Prom6, Prom7, TBBI2 and Mut1) along with the high-fidelity HF10 and wild-type Ste2 were characterized in technical triplicate by a dose response curve assay against the entire single-amino-acid variant peptide library (see Supplementary data, Figs. S1 andS2, Supplementary data, Table S1) and the promiscuity index P was calculated for each receptor (see Supplementary data, Table S2). The values of P were as expected: the promiscuous receptors (P = 0.84 ± 0.05) higher than Ste2 (P = 0.728) and high-fidelity HF10 (P = 0.609) lower than Ste2. HF10 shows increased EC50 (i.e. more difficult to stimulate) in response to stimulation with peptide variants compared to its response to α-factor, particularly in the first four residue positions (Fig. 1C). Prom6, on the other hand, shows greatly reduced EC50 (i.e. easier to stimulate) when stimulated with peptide variants compared to its response to α-factor (Fig. 1C). Prom7, Prom3, Mut1 and TBBI2 all show similarly reduced EC50 (see Supplementary data, Fig. S3).
PLSR identifies predictive groupings of residue properties and positions
While a single value like P gives a sense of receptor promiscuity, it is insufficient to predict a receptor's response to a novel peptide. We hypothesize that there exists (i) a quantitative relationship between peptide structure (positions and physiochemical properties of the residues) and activity (receptor response when stimulated by the peptide) and (ii) a qualitative relationship between receptor promiscuity and predictive peptide features. We sought to capture the former using multi-dimensional QSAR models and the latter using a novel feature selection approach.
Figure 2A briefly outlines the computational pipeline; Supplementary data, Figs. S4 and S5 describe the pipeline in greater detail. The 13-residue long peptide ligands were quantified using the Vectors of Hydrophobic, Steric, and Electronic properties (VHSE) system (Mei et al., 2005) in which each amino acid is assigned eight properties. These properties are orthogonal components derived by Principal Component Analysis of 50 physiochemical properties. VHSE components 1–2, 3–4 and 5–8 reflect hydrophobic, steric and electronic characteristics of the each amino acid, respectively. Representing each residue of our peptide using VHSE properties forms a vector of (8 properties) (13 residues) = 104 features. Because we did not know the nature of the relationship between peptide sequence and receptor response, we considered nine possible response variables: EC50 (concentration at 50% activation), EC10 (concentration at 10% activation), UM10 (fluorescence-average of the population when exposed to 10 μM of a specific peptide), as well as the natural logarithm and inverse values of each, in hopes of identifying the most linear relationship. Together, this resulted in a predictor matrix (X) of 104 features for each of the 55 peptide variants and a response matrix for each receptor () of the nine response variables for each of the 55 peptide variants.
We selected partial least squares regression (PLSR) to build predictive models for receptor response against a novel ligand and identify predictive features because it can account for noisy and correlated data (Wold et al., 2001). The number of possible combinations of features was large ( so exhaustive feature selection was computationally infeasible. Instead, we used two orthogonal grouping schemes (Fig. 2B): (i) By Position, where a group consists of all of the VHSE properties corresponding to a given position in the peptide, and (ii) By Property, where a group consists of the values for a single VHSE property at every position. This allowed us to perform a computationally tractable ‘grouping exhaustive’ search in which each group of features is either fully included or excluded from the predictor matrix.
The performance of the resulting models was quantified by the predictive squared correlation coefficient (Consonni et al., 2010). measures the predictive power of a model using leave-one-out validation, in which the model attempts to predict the response of a sample on which it was not trained. This is in contrast to the coefficient of determination , which simply measures how well the model fits the entire data set (i.e. no external validation). To further validate our approach, we also considered 50 different shuffled data sets of wild-type Ste2 with its EC50 response. The average for the shuffled data sets is narrowly distributed in the range of while the distribution of for the true, unshuffled data is broader and covers a range of (see Supplementary data, Fig. S6). Our vectorized peptides have significantly higher predictive capability than a random data set of the same distribution.
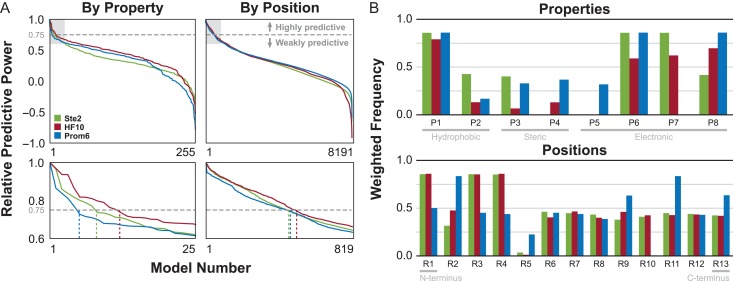
Distributions of the resulting values for all possible groupings ( using the By Position scheme, using the By Property scheme) against all nine response variables are given in Supplementary data, Fig. S7. For a given receptor/grouping scheme pair, the most predictive response type was selected based on the fraction of models with a higher than 0.25. Each receptor was best modeled by a different response type−EC50 for Ste2, EC10 for Prom6, and response at 10 μM for HF10−and best response type was consistent between schemes for a given receptor. Analysis of additional promiscuous receptors Prom3, Prom7, Mut1 and TBBI2 show a preference for EC50-derived response types, with consistency between schemes for all but TBBI2. We then analyzed each pair by sorting the models for the best response type by . From the sorted models, the top performers were selected using a threshold of 0.75 (i.e. models with a no more than 25% lower than the best for that pair). All but the two most promiscuous receptors (Prom3 and Prom7) showed similar trends (Fig. 3A, see Supplementary data, Fig. S8A). For these receptors, we selected 2.7 ± 1.2% of 255 total models using the By Property scheme and 4.3 ± 2.1% of 8191 total models using the By Position scheme (see Supplementary data, Table S3). In contrast, Prom3 and Prom7 showed a much steeper drop in model performance. Using the same threshold for these two receptors, we selected 23.2 ± 0.8% and 37.8 ± 17.2% of models using the By Property and By Position schemes, respectively (see Supplementary data, Table S3).
Fig. 3.
Characteristics of top-performing models. (A) Relative predictive power (sorted by Q2/Q2max) for each receptor/scheme pair for the top-performing response type. Dashed gray line indicates the selection threshold of Q2/Q2max = 0.75. Plots in the bottom row expand the light gray area of the corresponding top row, with dotted lines indicating the number of selected top models. (B) Weighted frequencies of each property (for the By Property grouping schemes) or position (for the By Position grouping schemes) in the selected top models for each receptor.
The feature groups (i.e. properties or positions) present in the selected top-performing models demonstrate differences across the seven receptors. The frequencies, weighted by , of each property in the By Property and for each position in the By Position schemes in the top models are given in Fig. 3B and Supplementary data, Fig. S8B. Properties 1 (related to hydrophobicity) and 6 (related to electronic character) occur frequently in the top-performing models for Ste2, HF10 and Prom6. Property 8 (electronic character) is important for all receptors except Ste2, which suggests that receptor sensitivity to some aspects of a peptide's electronic character may be affected by mutations from the wild-type. Property 7 (electronic character) is important for Ste2, HF10 and TBBI2, but not Prom6, Prom3, Prom7 or Mut1, suggesting that this aspect of a peptide's electronic character may be related to the absence of mutation M45I. Properties 2, 3, 4 and 5 are consistently infrequent for all receptors and are therefore weakly predictive of receptor response. Positions 1, 3 and 4 are important for Ste2 and HF10, while a dramatically different set (2, 9, 11 and 13) is predictive for Prom6. The important positions for receptors Mut1 (2, 4, 9, 11 and 13) and TBBI2 (1, 4, 9, 11 and 13) lie between these two sets. Position 11 (electronic character) is highly important for all of the higher promiscuity receptors (Prom6, Prom3, Prom7, Mut1 and TBBI2).
Weighted VIP-based feature scores select top predictive peptide features
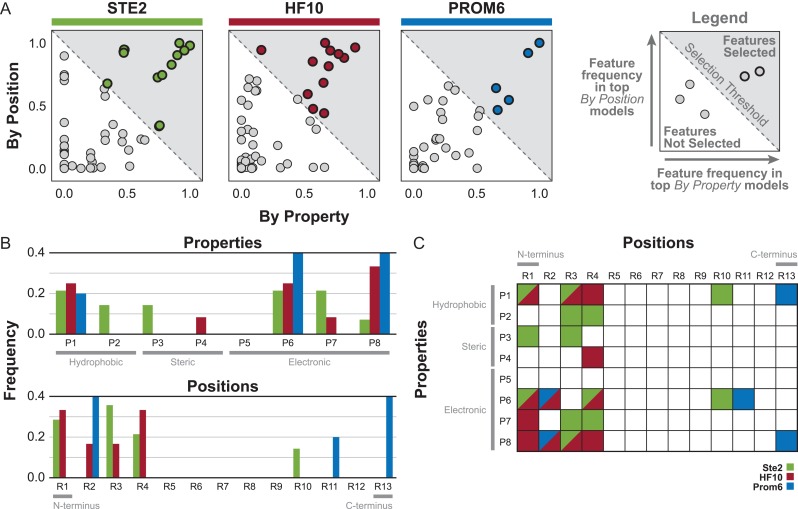
Previously, we had only considered top groupings of physical properties (across the entire peptide) or a residue position (regardless of physical property) for predicting peptide response (see Supplementary data, Fig. S5, Steps 1–4). To identify the specific features (i.e. a particular physical property at a particular position) that are most predictive, we employed the Variable Importance in Projection (VIP) score, which quantifies the importance of a feature in terms of both weighting in the regression and percentage of variance explained (Wold, 1995). For a given top model, the VIP score was used to rank the importance of each individual feature within the set of features for that model. From this list, the top features were selected based on the ‘elbow’ of the ranking versus VIP score curve, calculated as the feature with the first minimal distance from the origin. This process was repeated for all the top models for each receptor/scheme pair and the frequency of each feature in the resulting top feature lists was calculated (see Supplementary data, Fig. S5). For a given receptor, features with an average frequency between the By Property and By Position scheme greater than 0.5 were selected (Fig. 4A, see Supplementary data, Fig. S9A). This threshold captures the general trends of the feature characteristics and results in a reasonable number of final features (see Supplementary data, Fig. S10).
Fig. 4.
Characteristics of top features selected using grouping schemes. (A) Scatter of weighted feature frequencies based on VIP score for the top-performing models across both grouping schemes. Features (darker outline) with a weighted average frequency across both schemes greater than 0.5 (gray region) were selected. (B) Frequency of properties or positions in the top features. (C) Grid of selected top features for each receptor. Diagonal grid cells indicate that the feature was present in the top feature list for multiple receptors.
Building a PLSR model using only these top features further increased () or only slightly decreased () the from the highest obtained using the By Position and By Property grouping schemes for the top response type (see Supplementary data, Fig. S11) for all cases except TBBI2 against the By Property scheme (). This suggests that the majority of the predictive power of the grouped models resulted from the inclusion of these top features. Given the agreement on important features between the two grouping schemes and the minimal impact on of the resulting models, we were confident that our feature selection approach was able to identify a highly predictive subset of residue properties and positions in a computationally feasible manner (see Supplementary data, Table S4).
As expected based on our initial characterization (Fig. 1C, see Supplementary data, Fig. S3), the distributions of the top features reflect some of the trends seen at the grouping level, such as the importance of properties 6 and 8 (Fig. 4B, see Supplementary data, Fig. S9B). All features for HF10 occur in the first four residues, again emphasizing the importance of the N-terminus for determining specificity for that receptor (Fig. 4C). No predictive features for any of the seven receptors occur in residues 6, 7 or 8 (see Supplementary data, Fig. S12A), the ‘bend’ domain of the peptide which is not thought to physically contact the receptor (Abel et al., 1998). In addition, we anticipated that promiscuous receptors would have fewer highly predictive features because of their reduced ability to distinguish differences between α-factor and the variant peptides. The promiscuous receptors indeed had only 4–7 predictive features, compared to 14 and 12 for Ste2 and HF10, respectively (Fig. 4A, see Supplementary data, Fig. S9A). Promiscuous receptors also have few features in common with HF10 and Ste2, the two of which have a majority of features in common with each other (Fig. 4C, see Supplementary data, Fig. S9C). Interestingly, but unsurprisingly, the M54I mutation was directly correlated with identifying residue 11 as being highly predictive (see Supplementary data, Fig. S12), since M54 is known to be positioned near peptide residues Q10–Y13 (Umanah et al., 2009; Mathew et al., 2011). It appears that much of the effect of the M54I mutation on promiscuity is due to changing the specificity toward the binding domain of the peptide. In contrast, there was no clear pattern of predictive features correlated with the S145L and truncation mutations (see Supplementary data, Fig. S12). These mutations are far from regions of the receptor that contact the peptide and likely have more general effects on receptor structure and energetics rather than direct effects on any specific peptide residue.
PLSR model predicts qualitative behavior for peptide ligands with multiple mutations and lengths
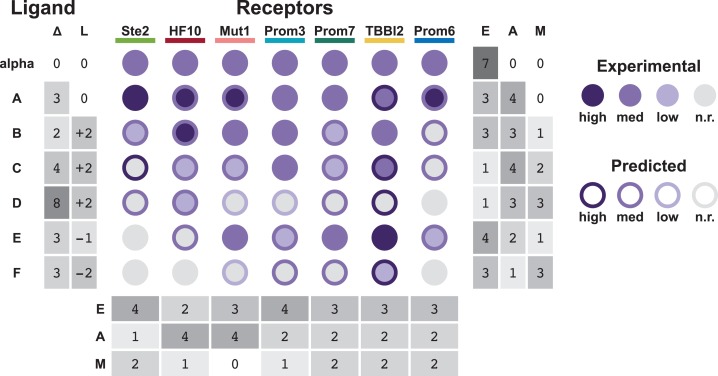
To explore the limits of our model's predictive ability, we compared the predicted and actual responses of each receptor to six novel peptides containing more than one amino acid difference from α-factor (Fig. 5). Peptides A–F vary in sequence from α-factor at 2–8 positions (denoted Δ). Several of these peptides also differ in length from α-factor (denoted L). Peptides A–D and E–F are two series of evolutionary intermediates that increasingly deviate in sequence from α-factor toward two different peptide targets (see Supplementary data, Table S5). As our model was trained using single-amino-acid variants of the same length, interactions between residues in the peptide ligand would not necessarily be captured. However, the models perform qualitatively well on novel ligands with more than one change. Predictions and experimental data were binned into four categories: high sensitivity, medium sensitivity, low sensitivity and no response. Of 42 predictions on receptor response to novel peptides, 15 predictions were exactly correct and only two receptor/peptide pairs were predicted to have high sensitivity but showed no response. Peptides C, D and F resulted in the highest number of incorrect predictions. These peptides were also the most divergent in sequence from alpha-factor, including several substitutions as well as multiple additions or deletions. Because our model was not trained on a data set that included insertions or deletions, it is not surprising that it has relatively limited predictive power in these cases.
Fig. 5.
Predictions of receptor response to novel peptides. Diagram summarizing the qualitative predicted and experimental responses of all seven receptors against six novel peptides. Variations in sequence and differences in length from α-factor are denoted Δ and L, respectively. Each circle indicates the experimental response (circle color) and the predicted response (circle border) binned into high sensitivity, medium sensitivity, low sensitivity and no response (n.r.). The number of exact matches (E), almost matches (A) and mismatches (M) are given per receptor and per peptide. Almost matches are defined as ‘off by one’ bins, i.e. high/medium, medium/low and low/n.r. combinations.
Peptide A is the most similar to α-factor, differing in only three positions and matching in length. Surprisingly, our model predicted that the wild-type receptor Ste2 would be more sensitive to Peptide A than to α-factor, a prediction that we validated experimentally (EC50 of 214 nM and 275 nM Peptide A and α-factor, respectively). These results suggest that the Ste2/α-factor pair has not been fully optimized by natural evolution for maximal sensitivity. Notably, we find that receptors evolved for promiscuity using only our single-amino-acid variant library (Prom3, Prom6 and Prom7) show a similar response profile to evolutionary intermediate peptides to those of receptors evolved directly for response to those evolutionary intermediates (TBBI2). This reinforces our previous observation that promiscuity is favored in evolutionary pathways.
While predictions were not fully quantitative, the majority of predictions produced actionable results. In our experience, peptides for which receptors have low sensitivity function well as evolutionary intermediates. Our models can be used as a tool to design the sequence of optimal evolutionary intermediate peptides. Given a library of characterized receptors, the model may also assist in selecting the best receptor to use as a starting point for directed evolution. For example, based on the predictions, receptor Prom6 (which is predicted to respond to peptide E) would be a better starting point for evolving for peptide F than Ste2 (which is not predicted to respond to peptides E or F). Similarly, for receptor Prom6, peptide C is a reasonable evolutionary step, while peptide D is unlikely to succeed. This modeling approach allows for predictive design of experiments that are more likely to succeed, shortening the design-build-test cycle for receptor engineering.
Discussion
Receptor promiscuity is a critical consideration for disease treatment, drug design and biosensor engineering. In context of directed evolution, promiscuity is a hallmark of successful pathway intermediates (Aharoni et al., 2004; Khersonsky et al., 2006). Our results demonstrate that the promiscuity of a receptor can be directly modified under appropriate selective conditions and that there exist multiple genotypes capable of conferring increased promiscuity. A single round of directed evolution comprising 1–3 non-synonymous mutations in the receptor produced changes in receptor promiscuity of up to 1.2 fold. We anticipate that additional rounds of mutagenesis or more stringent selection criteria could produce more dramatic changes. Our success in evolving both high- and low-promiscuity variants of the wild-type Ste2 receptor suggests that this method could be generally applicable for tuning the promiscuity of receptors of many sequences and specificities.
Our intuition suggested that low-promiscuity receptors would have more constraints on the physiochemical characteristics of the C-terminal peptide residues, where the wild-type receptor has evolved for high selectivity. We were surprised to find that the low-promiscuity receptor HF10 had no highly predictive features in any of the four C-terminal residues (R10–R13); instead, the primary determinant of high-fidelity by HF10 seems to be in the four N-terminal residues (R1–R4). This behavior is captured by single-amino-acid scans along the length of the peptide, where HF10 displays clear selectivity in the first four residue positions (Fig. 1C).
After we demonstrated the ability to modulate promiscuity using directed evolution, we sought to characterize the relationships between (i) peptide structure and receptor response and (ii) important peptide features and receptor promiscuity. The former was accomplished using a straightforward QSAR approach in which peptides were quantified into feature vectors and regressed against receptor response using PLSR. This approach, repeated for each receptor, allowed us to examine the peptide versus receptor relationship. The latter was accomplished using a unique grouping feature selection approach. An exhaustive search of all features combinations () was computationally infeasible; therefore, we reduced the search space by assigning features into two orthogonal grouping schemes (By Position and By Property) from which top features were selected based on importance in both schemes. Given our unique data set, which quantifies the response of seven receptors differing in promiscuity against the same library of peptides, this approach allowed us to examine receptor versus receptor relationships. The final models were found to be generally predictive of responses to the peptides assayed.
To further test our models, we compared the experimental and predicted responses of receptors to peptides with multiple residue changes and differing lengths. We were surprised to find that our relatively simple model, trained on measurements from single-amino-acid variants of the same length, retained qualitative predictive power for ligands in which many physiochemical properties are varied simultaneously. Although the sensitivity of the response predicted by our models sometimes differed from observed values, we found only two cases in which a receptor was predicted to be highly sensitive to a peptide that it experimentally did not respond to. The analysis reveals that our model's predictions are most reliable when the peptide lacks C-terminal additions. This may indicate an important mechanistic change in the activity of peptides with additions to the C-terminal-binding domain.
The models’ primary utility, therefore, lies in qualitative predictions of response rather than exact magnitude. This approach could, for example, be applied to (i) inform searches for native ligands to orphan receptors, (ii) predict off-target drug interactions with GPCRs and (iii) aid in receptor directed evolution. A peptide QSAR model trained on orphan receptor response to a portfolio of compounds can be used to scan all known biological compounds for potential hits. The grouping-exhaustive feature selection approach can identify important features to guide drug discovery. Finally, the approach can streamline receptor directed evolution identifying trivial and infeasible ligand steps to produce a more efficient directed evolution pathway.
Conclusions
In summary, we report two advances that enable more sophisticated understanding of peptide GPCR promiscuity. First, we demonstrate a method for using selection criteria to increase or decrease the promiscuity of a receptor. This advance allowed us to produce higher and lower promiscuity variants of Ste2. More promiscuous receptors serve as better ‘parents’ for directed evolution, and, in the context of biosensors, less promiscuous receptors improve the specificity of the sensor. Second, we establish a peptide QSAR model and grouping-exhaustive feature selection pipeline to produce informative models of receptor response. This advance allowed us to quantify the interactions between peptide features and receptor response as well as qualitatively investigate the peptide features underlying receptor promiscuity. The models have strong predictive power for single residue variants of α-factor and provide informative predictions for peptides that vary dramatically from α-factor in both length and sequence. These advances enable refined control and evaluation of receptor promiscuity.
Supplementary Material
Acknowledgements
We thank Bryan Roth at the University of North Carolina, Chapel Hill for the gift of the MPY578t5 yeast strain, the Northwestern University Sequencing Core, the Northwestern University Robert H. Lurie Flow Cytometry Core Facility, and the Northwestern University Center for Global Health Technologies.
Supplementary data
Supplementary data are available at Protein Engineering, Design and Selection online.
Funding
Northwestern University Flow Cytometry Core Facility, supported by Cancer Center Support Grant (NCI CA060553). Flow cytometry cell sorting was performed on a BD FACSAria SORP system, purchased through the support of NIH 1S10OD011996-01. National Science Foundation grant DGE-1324585, the Bill and Melinda Gates Foundation grant OPP1061177, and the Robert R. McCormick School of Engineering and Applied Science.
Conflict of interest
None declared.
References
- Abel M.G., Zhang Y.L., Lu H.-F., Naider F. and Becker J.M. (1998) J. Pept. Res., 52, 95–106. [DOI] [PubMed] [Google Scholar]
- Adeniran A., Sherer M. and Tyo K.E.J. (2015) FEMS Yeast Res., 15, 1–15. [DOI] [PubMed] [Google Scholar]
- Aharoni A., Gaidukov L., Khersonsky O., Gould S.M., Roodveldt C. and Tawfik D.S. (2004) Nat. Genet., 37, 73–76. [DOI] [PubMed] [Google Scholar]
- Armbruster B.N., Li X., Pausch M.H., Herlitze S. and Roth B.L. (2007) Proc. Natl. Acad. Sci., 104, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatuil L., Perez J.M., Belk J. and Hsieh C.-M. (2010) Protein Eng. Des. Sel., 23, 155–159. [DOI] [PubMed] [Google Scholar]
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M. and Wittrup K.D. (2006) Nat. Protoc., 1, 755–768. [DOI] [PubMed] [Google Scholar]
- Cherkasov A., Muratov E.N., Fourches D. et al. (2014) J. Med. Chem., 57, 4977–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelli O. (2005) Trends Pharmacol. Sci., 26, 15–19. [DOI] [PubMed] [Google Scholar]
- Cobb R.E., Sun N. and Zhao H. (2013) Methods, 60, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin B.R., Hsiao E.C., Claeysen S. et al. (2008) Nat. Methods, 5, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni V., Ballabio D. and Todeschini R. (2010) J. Chemom., 24, 194–201. [Google Scholar]
- Dong S., Rogan S.C. and Roth B.L. (2010) Nat. Protoc., 5, 561–573. [DOI] [PubMed] [Google Scholar]
- Dorsam R.T. and Gutkind J.S. (2007) Nat. Rev. Cancer, 7, 79–94. [DOI] [PubMed] [Google Scholar]
- Gould S.M. and Tawfik D.S. (2005) Biochemistry (Mosc)., 44, 5444–5452. [DOI] [PubMed] [Google Scholar]
- de Jong S. (1993) Chemom. Intell. Lab. Syst., 18, 251–263. [Google Scholar]
- Khersonsky O., Roodveldt C. and Tawfik D. (2006) Curr. Opin. Chem. Biol., 10, 498–508. [DOI] [PubMed] [Google Scholar]
- Lagerström M.C. and Schiöth H.B. (2008) Nat. Rev. Drug Discov., 7, 339–357. [DOI] [PubMed] [Google Scholar]
- Li H., Xu Q. and Liang Y. (2014) libPLS: An Integrated Library for Partial Least Squares Regression and Discriminant Analysis.
- Li J., Haddad R., Chen S., Santos V. and Luetje C.W. (2012) J. Neurochem., 121, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L. (1992) Mol. Cell. Biol., 12, 3959–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew E., Bajaj A., Connelly S.M., Sargsyan H., Ding F.-X., Hajduczok A.G., Naider F. and Dumont M.E. (2011) J. Mol. Biol., 409, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H., Liao Z.H., Zhou Y. and Li S.Z. (2005) Biopolymers, 80, 775–786. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller R. and Funk M. (1995) Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Nath A. and Atkins W.M. (2008) Biochemistry (Mosc)., 47, 157–166. [DOI] [PubMed] [Google Scholar]
- O'Hayre M., Vázquez-Prado J., Kufareva I., Stawiski E.W., Handel T.M., Seshagiri S. and Gutkind J.S. (2013) Nat. Rev. Cancer, 13, 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer M.S. and Liu D.R. (2015) Nat. Rev. Genet., 16, 379–394. [DOI] [PubMed] [Google Scholar]
- Pissurlenkar R.R.S., Malde A.K., Khedkar S.A. and Coutinho E.C. (2007) QSAR Comb. Sci., 26, 189–203. [Google Scholar]
- Reneke J.E., Blumer K.J., Courchesne W.E. and Thorner J. (1988) Cell, 55, 221–234. [DOI] [PubMed] [Google Scholar]
- Schöneberg T., Schulz A., Biebermann H., Hermsdorf T., Römpler H. and Sangkuhl K. (2004) Pharmacol. Ther., 104, 173–206. [DOI] [PubMed] [Google Scholar]
- Slomovic S., Pardee K. and Collins J.J. (2015) Proc. Natl. Acad. Sci., 112, 14429–14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers C.M. and Dumont M.E. (1997) J. Mol. Biol., 266, 559–575. [DOI] [PubMed] [Google Scholar]
- Stemmer W.P. (1994) Proc. Natl. Acad. Sci. U. S. A., 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi R., Lai R. and Duggleby R.G. (2004) BMC. Biotechnol., 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umanah G.K.E., Son C., Ding F., Naider F. and Becker J.M. (2009) Biochemistry (Mosc)., 48, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassart G. and Costagliola S. (2011) Nat. Rev. Endocrinol., 7, 362–372. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan A.J., Deupi X., Lebon G., Tate C.G., Schertler G.F. and Babu M.M. (2013) Nature., 494, 185–194. [DOI] [PubMed] [Google Scholar]
- Winkler D.A. (2002) Brief. Bioinform., 3, 73–86. [DOI] [PubMed] [Google Scholar]
- Wold S. (1995) van de Waterbeemd H. (ed),. Chemometric Methods in Molecular Design. VCH, Weinheim; New York, pp. 195–218. [Google Scholar]
- Wold S., Sjöström M. and Eriksson L. (2001) Chemom. Intell. Lab. Syst., 58, 109–130. [Google Scholar]
- Zalewska M., Siara M. and Sajewicz W. (2014) Acta Pol. Pharm., 71, 229–243. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.