Abstract
Upon lowered blood glucose occurring during fasting, glucagon is secreted from pancreatic islets, exerting various metabolic effects to normalize glucose levels. A considerable portion of these effects is mediated by glucagon-activated transcription factors (TFs) in liver. Glucagon directly activates several TFs via immediate cyclic adenosine monophosphate (cAMP)– and calcium-dependent signaling events. Among these TFs, cAMP response element-binding protein (CREB) is a major factor. CREB recruits histone-modifying enzymes and cooperates with other TFs on the chromatin template to increase the rate of gene transcription. In addition to direct signal transduction, the transcriptional effects of glucagon are also influenced by dynamic TF cross talk. Specifically, assisted loading of one TF by a companion TF leads to increased binding and activity. Lastly, transcriptional regulation by glucagon is also exerted by TF cascades by which a primary TF induces the gene expression of secondary TFs that bring about their activity a few hours after the initial glucagon signal. This mechanism of a delayed response may be instrumental in establishing the temporal organization of the fasting response by which distinct metabolic events separate early from prolonged fasting. In this mini-review, we summarize recent advances and critical discoveries in glucagon-dependent gene regulation with a focus on direct TF activation, dynamic TF cross talk, and TF cascades.
This review summarizes recent discoveries in glucagon-dependent gene regulation with a focus on transcription factor cross talk.
The discovery of glucagon and its role in elevating blood glucose paved the way for decades of research into the production, secretion, and downstream effects of this hormone (1). Glucagon exerts various metabolic effects in target organs and counterregulates the effects of insulin. A considerable fraction of glucagon’s effects is brought about by regulating the rate of gene transcription through its effect on transcription factors (TFs). This mini-review will focus on transcriptional regulation by glucagon and how it supports the hormone’s metabolic effects.
Glucagon Secretion and Signaling
The proglucagon gene is expressed in α cells within pancreatic islets. Following posttranslational processing, the resulting 29-amino acid–long peptide is packaged into secretory granules. When blood glucose levels are lowered due to fasting or exercise, the rapid increase in Ca2+ influx into α cells promotes exocytotic glucagon secretion. Although low glucose is the principal signal stimulating glucagon secretion, other signals such as circulating amino acids, fatty acids, neural inputs, and glucagon-like peptides also participate in regulating glucagon secretion (2, 3).
The downstream effects of glucagon are mediated by the hormone’s binding to the glucagon receptor (GcgR), a G protein–coupled receptor highly expressed in the liver and kidney. The liver is a primary target for glucagon due to both high GcgR expression and high local hormone availability. The pancreas unloads glucagon to the portal vein shortly before it reaches the liver, leading to high portal glucagon concentration. Indeed, many of the metabolic effects of glucagon are attributable to its action on hepatocytes. GcgR is also expressed, albeit to lower extents, in brain, adipose tissue, pancreas, smooth muscle, spleen, heart, and adrenal glands. Accordingly, glucagon possesses various extrahepatic effects mediated through binding to GcgR in several tissues (4).
Upon binding to its receptor, glucagon signals through two main pathways. A Gαs-dependent pathway activates adenylate cyclase, leading to generation of cyclic adenosine monophosphate (cAMP) (5). A second pathway is activated by Gαq, by which phospholipase C (PLC) increases Ca2+ concentrations via activating inositol 1,4,5-trisphosphate (IP3) receptor on the endoplasmic reticulum (6, 7). Downstream, cAMP activates protein kinase A (PKA), which then phosphorylates several downstream kinases from the mitogen-activated protein kinase family (p38, c-Jun N-terminal kinase, and extracellular signal–regulated kinase-1/2) (8, 9). Concomitantly with PLC, PKA also increases Ca2+ levels by activating IP3 receptor (10) (Fig. 1). These signaling events have profound effects on transcriptional regulation as detailed later.
Figure 1.
Glucagon signal transduction and primary glucagon-activated TFs. Upon binding to its G protein–coupled receptor, glucagon signals through two pathways. The Gαs subunit activates adenylate cyclase (AC) that produces cAMP. Protein kinase A (PKA) is activated by cAMP and phosphorylates CREB, leading to its activation. PKA also phosphorylates p300, thereby activating FoxA2. Additionally, PKA indirectly leads to dephosphorylation of class II HDAC and activation of FoxO. Lastly, PKA activates PPARA via mitogen-activated protein kinase (MAPK) phosphorylation. The second arm of glucagon signal transduction is mediated by the Gαq subunit that activates PLC, leading to IP3 production. Binding of IP3 to its receptor on the endoplasmic reticulum (ER) increases Ca2+ concentrations and activates Ca2+/calmodulin-dependent protein kinase II (CaMKII), resulting in FoxO activation. The TFs activated by glucagon are responsible for different gene programs; CREB and FoxO promote a gluconeogenic gene program, whereas FoxA2 and PPARA support FAO/ketogenesis.
Glucagon Metabolic Effects and the Fasting Response
The major and by far most studied metabolic effect of glucagon is increasing blood glucose during fasting. Glucagon increases the breakdown of glycogen (glycogenolysis) and the de novo synthesis of glucose from noncarbohydrate precursors (gluconeogenesis). Concomitantly, glucagon inhibits glycolysis and glycogenesis (11). In this response, glucagon antagonizes the effects of insulin and was therefore termed a counterregulatory hormone. In addition to the prominent effect on glucose metabolism, glucagon also affects lipid balance. Glucagon lowers plasma cholesterol and triglycerides, reduces hepatic lipoprotein and triglyceride production, and increases lipolysis in adipose tissue. Additionally, glucagon increases fatty acid oxidation (FAO) and ketone body production (ketogenesis); both are important components of the response to fasting (2).
The rate of the three major channels for fuel production during fasting (glycogen breakdown, gluconeogenesis, and ketogenesis) is temporally regulated to optimize energy consumption and prevent organ damage. During early fasting, glycogen breakdown and gluconeogenesis produce glucose to supply the body’s energetic needs. As fasting continues, glycogen stores are exhausted, and gluconeogenesis rate is diminished to prevent muscle wasting due to depletion of amino acids (the major gluconeogenic precursors). In this protein-sparing phase, FAO followed by ketogenesis becomes the predominant fuel-producing pathway (12).
Much of these metabolic effects are mediated by glucagon-dependent gene regulation. In the following text, we will review the TFs regulated by glucagon and their cross talk with other TFs. We will start with cAMP response element-binding protein (CREB), which is the most heavily studied in that respect, and continue to other glucagon-regulated TFs. The liver is the major target for glucagon and also the main organ providing alternative fuels during fasting. Therefore, we will focus on glucagon transcriptional regulation in the liver. Additionally, we will examine the possibility that glucagon governs the temporal organization of the fasting response by activating several TFs via different mechanisms. First, glucagon triggers signal transduction pathways, leading to direct TF activation by posttranslational modifications. Second, the signal from glucagon and other fasting-related cues is integrated by dynamic cross talk between TFs, bringing about discrete transcriptional programs. Lastly, a delayed response to glucagon is achieved by TF cascades in which a direct glucagon-activated TF promotes a second wave of transcriptional regulation by inducing genes encoding secondary TFs.
Glucagon Regulation of Transcription via CREB
CREB belongs to the basic leucine zipper (bZIP) class of TFs. It regulates a variety of genes by binding to cAMP response elements (CREs; TGACGTCA) or to a CRE half-site (TGACG or CGTCA). The same motifs are bound by the two other members of the CREB-related TF family, cAMP responsive element modulator and activating transcription factor 1 (13). The glucagon-PKA-cAMP axis provides the primary CREB-activating signal (Fig. 1). PKA phosphorylates CREB at serine 133 (abbreviated to pCREB), leading to increased activity and association with cofactors and histone-modifying enzymes (14). In addition to glucagon, CREB is activated by various other signals including epinephrine, another hormone secreted during fasting (14–16). Although most of the actions of hepatic CREB during fasting are attributed to glucagon-dependent activation, it is possible that epinephrine also plays a role in CREB activation during fasting.
Rodent models of CREB perturbations
Fitting with its critical role in metabolism, glucagon was implicated in the development of diabetes, a disease in which high blood glucose levels and dyslipidemia are major driving forces of pathogenesis. Glucagon is abnormally secreted in type 1 diabetes, contributing to ketoacidosis, one of the diseases’ principal life-threatening conditions. Glucagon levels are also abnormal in type 2 diabetes, possibly mediating hyperglycemia (17). These findings and other evidence led to the hypothesis that alterations in glucagon secretion and signaling are the principal events driving diabetes (18). These pathological effects of glucagon are partially mediated by CREB, as shown in several studies manipulating its expression or activity in rodent models.
An effective dominant-negative peptide is commonly used to inhibit CREB. The leucine zipper in this peptide was designed to have an extension of acidic amino acids replacing the basic amino acids needed for DNA binding. Thus, this acidic CREB (A-CREB) dimerizes with wild-type CREB through the intact leucine zipper but prevents the dimer from binding DNA (19). Fasted transgenic mice expressing A-CREB only in hepatocytes were severely hypoglycemic and showed reduced expression of gluconeogenic genes, demonstrating that CREB is a critical TF controlling glucagon’s response in liver (20). Diabetic mice expressing adenovirally infected A-CREB normalize their glucose levels and their gluconeogenic gene expression and show a reduction in hyperinsulinemia, suggesting a CREB-dependent increase in glucose levels contributes to a diabetic phenotype (20). Because these mice expressed A-CREB systemically, the contribution of extrahepatic CREB to this phenotype is also possible.
In addition to its effect on carbohydrate metabolism, CREB was shown to regulate lipid metabolism during fasting. Mice fed a high-fat diet and systemically infected with adenoviral A-CREB have increased triglycerides and lipid accumulation in the liver. These mice also show an increase in hepatic lipogenic genes (21). In diabetic rat models, CREB antisense oligonucleotides lowered insulin and glucose levels as well as the expression of gluconeogenic genes. In contrast to the observations described in Herzig et al. (21), the antisense-based CREB knockdown lowered plasma triglycerides (22).
Motivated by these partially contradicting observations, an elegant study reported findings with a hepatocyte-specific CREB knockout (23). Although whole-body CREB knockout mice die immediately after birth (24), hepatocyte-specific CREB knockout mice were viable. Surprisingly, mice lacking hepatocyte CREB show completely normal glucose homeostasis, insulin sensitivity, and gluconeogenic gene expression. Also, CREB knockout had a minimal effect on global gene expression, with only 21 genes showing differential expression compared with wild-type mice. These perplexing observations can be reconciled with previous data showing the importance of hepatic CREB in the fasting response by two explanations. First, extrahepatic CREB definitely plays a role in the fasting response. For example, the study employing antisense nucleotides reported that adipocyte CREB is also perturbed, suggesting some of the effects reported might be attributable to CREB’s function in adipose tissue (22). Therefore, extrahepatic CREB (e.g., in adipose and kidney) might compensate for the absence of hepatic CREB. Second, redundancy between CRE-binding TFs and possibly other related bZIP proteins may occur. This explanation is supported by the fact that A-CREB is not CREB specific and targets similar proteins (19). Therefore, in addition to blocking CREB, the studies using A-CREB have potentially blocked all CRE-binding TFs and possibly more bZIP TFs. The unintended targets of A-CREB may also be responsible for its different effect on triglycerides as compared with the more specific CREB inhibition by antisense nucleotides.
CREB coactivators and changes in histone modifications
CREB activity is strictly regulated by several coactivators. The family of coactivators termed cAMP-regulated transcriptional coactivators (CRTCs) modulates CREB activity and mediates its transcriptional program in response to signals. There are three members in the CRTC family (CRTC1, CRTC2, and CRTC3), and they all bind the bZIP domain of CREB. CRTCs are sensitive to the glucagon-dependent increases in both intracellular cAMP and Ca2+ levels. With rising glucagon levels, CRTCs are dephosphorylated, enter the nucleus, and bind CREB, leading to its increased DNA binding and activity. Conversely, in low-glucagon, high-insulin states, CRTCs are phosphorylated and localized to the cytoplasm by 14-3-3 proteins. CRTCs also serve to bridge between CREB and histone-modifying enzymes as detailed later. The interplay between CREB and CRTCs, as well as their fine-tuned response to endocrine and nutritional cues, is a rich field of research. This area has been comprehensively reviewed elsewhere (14) and will not be a focus of this review.
In addition to CRTCs, CREB was reported to associate with coactivators with histone-modifying capabilities. Histone modifications (acetylation in particular) are associated with open chromatin and a chromatin environment that facilitates gene induction (25). Several studies detailed later have described the CREB-mediated recruitment of histone-modifying enzymes to fasting-related DNA regulatory regions (enhancers), leading to increased gene expression.
Following CREB phosphorylation by PKA, the association of CREB with the histone acetyl transferases (HATs) p300 and CREB-binding protein (CBP) is increased (26, 27). CRTCs are also instrumental in facilitating the interaction between CREB and p300/CBP (28). CBP and p300 are highly similar proteins; both acetylate histone tails, primarily the H3K27 lysine residue at enhancers. This leads to a chromatin environment more accessible to TF binding (29) and to increased rate of transcript elongation by RNA polymerase II (30).
In addition, p300/CBP directly acetylate regulatory lysine residues in TFs, thereby increasing their activity. For example, the activities of p53 (31) and signal transducer and activator of transcription 3 (32) are increased by p300/CBP-dependent acetylation. Thus, in addition to the direct effect p300/CBP has on chromatin accessibility, recruitment of p300/CBP to enhancers by CREB may also lead to direct acetylation of neighboring TFs, thereby locally increasing their activity. This option was not explored but may be a supporting mechanism for TF cooperativity during fasting. For example, p53 was shown to promote gluconeogenic gene expression and hepatic glucose production (33). The possibility exists that CREB recruits p300/CBP to gluconeogenic enhancers where p53 is bound, leading to local p53 acetylation and increased activity.
The early paradigm of CREB-associated HATs was recently complemented by two studies highlighting additional CREB-dependent changes in histone modifications. Work by Ravnskjaer et al. (34) revealed a third HAT recruited by CREB, lysine acetyltransferase 2B (KAT2B; also known as PCAF). Levels of H3K9 acetylation, catalyzed by KAT2B, were increased around gluconeogenic genes upon fasting. KAT2B was recruited to these regulatory regions by CRTC2 following nuclear localization of CRTC2 and physical interaction between the two proteins. Upon recruitment, KAT2B created a chromatin environment more conducive to TF binding and gene induction by increasing H3K9 acetylation. Reciprocally, reducing KAT2B levels led to a decrease in CRTC2 and CREB recruitment to gluconeogenic genes, presumably due to a chromatin environment less favorable to binding. This was associated with perturbed glucagon-dependent gene induction and reduced circulating fasting glucose (34).
In addition to histone acetylation, histone methylation by protein arginine methyltransferase 5 (PRMT5) plays a role in gluconeogenic gene expression. As for KAT2B, CRTC2 interacts with PRMT5 and recruits it to gluconeogenic genes. Glucagon stimulated increases in the PRMT5 product (H3R2me2) around gluconeogenic genes. Conversely, reducing PRMT5 levels decreased circulating glucose concentrations and gluconeogenic gene expression. The mechanism facilitating gene expression was proposed to be reduced nucleosome occupancy at PRMT5-modified histones (35).
In sum, CREB actively engages with a range of coactivators that mediate and facilitate its gene regulatory effects. These coactivators exert their effect by physical interaction with other proteins and regulation of the chromatin environment. Collectively, these studies reveal an extra layer of complexity in glucagon regulation of gene transcription and highlight the importance of histone modifications and chromatin structure in CREB activity (Fig. 2).
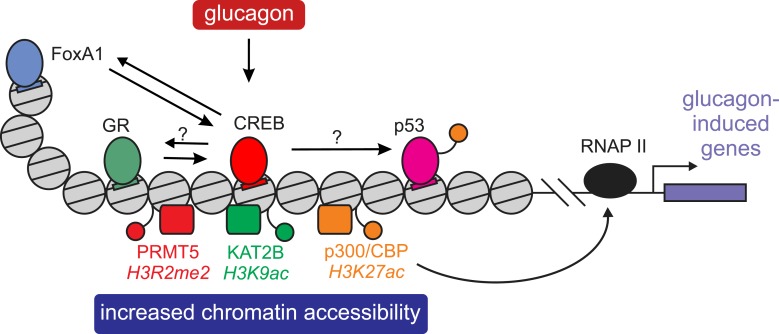
Figure 2.
CREB-mediated effects on chromatin and activation of companion TFs. Binding of the CREB/CRTC complex to chromatin initiates several events eventually promoting glucagon-dependent gene expression. CREB recruits histone-modifying enzymes (p300/CBP, KAT2B, and PRMT5) that increase the accessibility of chromatin to TF binding. Additionally, CREB-recruited p300/CBP promotes RNA polymerase II (RNAP II) activity and might activate other TFs by acetylation (e.g., p53). CREB also cooperates with other TFs via dynamic assisted loading, by which the binding of one TF facilitates binding of more TFs in the same enhancer via increased chromatin accessibility and enhancer activity. GR assists the loading of CREB onto chromatin, and so does FoxA1 (of note, CREB-FoxA1 cross talk was only described in prostate cells and not in the presence of glucagon). An assisted loading mechanism is often not unidirectional, and two TFs might enhance the binding of each other, as was shown in CREB-FoxA1 cross talk (and is also probable in the GR-CREB case).
Genome-wide occupancy of CREB
Early attempts at defining the genomic pattern of CREB occupancy in rat PC12 cells, HEK293T, and primary mouse or human hepatocytes provided some insight regarding CREB activity (36–38). Importantly, CREB was found to preferentially bind promoter-proximal regulatory regions, as compared with distal ones. Remarkably, CREB occupied as much as 20% of total promoter-proximal regions in unstimulated conditions. Although informative, these early studies used promoter-centric methods [e.g., chromatin immunoprecipitation (ChIP)-on-chip] to map CREB occupancy, and most distal regions were not included in the analyses. Another critical finding was that although CREB was phosphorylated at virtually all binding sites following treatment, only a small fraction of the genes driven by CREB-bound promoters was responsive to CREB activation. This genomic perspective, together with other studies (14), led to the conclusion that the combination of CREB phosphorylation with recruitment of coactivators is critical to CREB-dependent gene induction, whereas CREB binding per se is not indicative of gene regulation (36).
More recent studies profiled CREB binding by ChIP-sequencing (ChIP-seq) that covers both distal and proximal regulatory regions. A study employing ChIP-seq to examine CREB binding in mouse liver following fasting confirmed the strong promoter bias of CREB and the finding that CREB binding does not markedly increase upon an activating signal (39). Moreover, most CREB binding sites did not reside near fasting-regulated genes. Overall, this study strengthened the notion that CREB binding is not a regulatory step in CREB-dependent gene regulation.
A recent study from our laboratory also examined CREB occupancy following fasting and revealed a more complex picture. We used a computational method for detecting TF motifs that are accompanied by two telltale changes in chromatin. (1) The motif bound by the TF is sometimes protected from DNase cleavage by virtue of TF occluding DNase access, leaving a detectable footprint (40). (2) Concomitantly, TFs can lead to a broad increase in DNase accessibility in the regions flanking the motifs due to recruitment of chromatin remodeling complexes and histone-modifying enzymes (41). Measuring chromatin accessibility following fasting in mouse liver genome-wide, we examined all known motifs for changes in both footprint capacity and flanking accessibility. The CRE motif showed a deeper footprint and a profound increase in flanking accessibility following fasting, suggesting increased CREB binding and activity. Indeed, in ChIP-seq profiling of both CREB and pCREB in mouse liver, they were found to increase in binding in a predefined set of fasting enhancers (1481 enhancers showing a marked increase in enhancer accessibility following fasting) (42). Supporting the role of CREB in mediating these changes (rather than other CRE-binding TFs), CRE motifs unbound by CREB showed no increase in either footprint depth or flanking accessibility (42).
These findings stand in contrast to those of Everett et al. (39), which did not detect substantial fasting-inducible binding. Although differential binding in the study by Everett et al. (39) was weak when measured by changes in read counts (i.e., the number of sequenced reads per site), the authors did report an increase in the total number of CREB binding sites following fasting (from 5357 to 6835) and a 1.3-fold increase in the median intensity of CREB binding. Although fasting-induced changes in CREB binding may be mild and often difficult to detect, they do occur and can be indicative of gene regulation. We conclude that auxiliary methods such as footprinting and an a priori focus on regions where enhancer activity and accessibility are increased following fasting can improve the chances of detecting these changes in CREB binding.
Cross talk between CREB and other TFs: integrating multiple signals by TF dynamics
Another level of CREB activity is the cross talk with other TFs at the chromatin template. This cross talk can be antagonistic or synergistic to CREB activity, depending on the stimulus and interacting TF. We will focus on CREB cross talk with other factors in the context of a glucagon signal. Early studies of regulatory regions near the gluconeogenic gene Pck1 showed a synergistic pattern of gene expression between the cAMP-CREB axis and the glucocorticoid receptor (GR) pathway. The mechanism for synergy was assumed to be a physical interaction between the two TFs (43). In contrast, CREB and GR were found to have antagonistic relationship in other genomic loci (44).
A recent paper sheds light on these contradictory findings (42). The changes in hepatic transcriptome relating to gluconeogenic gene expression were characterized by a synergistic relationship between glucagon and corticosterone (the glucocorticoid hormone activating GR in rodents). The synergistic effect was due to GR facilitating CREB binding and activity. Upon short treatment with glucagon in primary hepatocytes, CREB binding increased next to its target genes. Cotreatment with corticosterone led to augmented CREB binding in many enhancers across the genome and to a synergistic pattern of gene expression (42). These sites were cobound by GR and CREB. Furthermore, GR treatment made the sites more accessible. Importantly, the increase in CREB binding was enhancer specific, as some CREB binding sites next to CREB-induced genes were unaffected by corticosterone, as was the gene expression pattern. This enhancer-specific effect would argue against a mode of action requiring physical interaction between the two TFs because such an interaction would indiscriminately affect all CREB binding sites cobound by GR. Indeed, CREB binding sites unaffected by corticosterone were also bound by GR (albeit to a lower extent). This enhancer specificity also excludes a stimulatory effect of corticosterone upstream the cAMP-PKA axis that would increase CREB activity at all bound enhancers. This kind of TF cross talk was termed dynamic assisted loading (45). Several such cases were reported and shown to be enhancer specific (42, 46–52) (Fig. 2). The assisted loading of CREB by GR observation was supported by tracking single CREB molecules following activation by cAMP. Compared with only a cAMP signal, coactivation of GR led to more DNA-bound CREB molecules, and their residence time on DNA increased (42). Thus, the CREB-GR interaction is complex and may lead to different outcomes in a cell type– and enhancer-specific manner. A recent study adds to this intricacy; in addition to promoting CREB activity, CRTC2 acts as a GR coactivator as well, facilitating GR’s effect on gluconeogenic genes (53).
In light of the finding that CREB binding is associated with increases in chromatin accessibility, the possibility exists that CREB assists the loading of companion TFs. This hypothesis might also explain the high number of CREB sites that seem not to directly regulate gene expression. CREB binding to these sites might function to recruit histone-modifying enzymes (CBP, p300, KAT2B, PRMT5, etc.), increase chromatin accessibility, and prime the chromatin environment to binding of other TFs. Indeed, CREB binding was recently reported to facilitate binding of forkhead box (Fox) A1 to a subset of genomic loci in prostate cancer cells (54) (Fig. 2). Of note, although CREB was activated in these cells as shown by serine 133 phosphorylation, the effect of glucagon was not tested in this study, and the CREB-FoxA1 cross talk in the context of fasting is still unexplored.
An antagonistic relationship between CREB and TFs in the context of glucagon signaling was also described. For example, pregnane X receptor was reported to bind glucagon-activated CREB and repress its function in both primary hepatocytes and fasted mice (55). Also, transcription factor 7 binds to regulatory elements next to gluconeogenic genes and perturbs CREB activity (56). A recent report describes an antagonistic cross talk between CREB and farnesoid X receptor (FXR) (57). FXR binds next to autophagy-related CREB-binding sites and represses CREB activity by dissociation of the CREB-CRTC2 complex. As observed for the CREB-GR case, cross talk between FXR and CREB is not straightforward. Downregulation of CREB led to an increase of Cyp7a1, a tightly regulated gene related to bile acid synthesis that is also repressed by FXR (22, 58). Therefore, the antagonism between CREB and FXR might be context specific, and although it is evident in autophagy gene regulation, bile acid metabolism might be mutually inhibited by both TFs.
Taken together, the studies described previously portray a multifaceted pattern of cross talk between CREB and other TFs. This pattern is expected to drive different gene expression programs under various conditions as well as fine-tune and integrate multisignal responses such as fasting/feeding. However, this assumption is supported by data collected from different experimental approaches. A concentrated and systematic effort to study CREB cross talk with other TFs within a single experimental system is therefore warranted.
The Expanding Realm of Glucagon-Activated TFs and the Temporal Organization of the Fasting Response
Directly activated TFs beyond CREB
As described previously, the response to fasting is temporally organized, and gluconeogenesis is prominent in the early stages of fasting, whereas ketogenesis assumes a more major role at prolonged fasting. Many of the genes involved in gluconeogenesis, FAO, and ketogenesis are readily induced during fasting and regulated by various TFs. Nonetheless, the possibility that the temporal organization of these fuel-producing pathways is governed by TFs was given little attention in the literature. Although glucagon is undoubtedly involved in the early gluconeogenic stage, an accumulation of data suggests a role for glucagon in transcriptional regulation of metabolic events also during prolonged fasting.
In addition to the direct activation of CREB by the glucagon-cAMP axis, glucagon also activates TFs from the Fox family. The activity of FoxO and FoxA proteins is negatively regulated by insulin in the fed state through phosphorylation and cytoplasmic retention (59). Reciprocally, several studies revealed a stimulatory effect of glucagon on these TFs during fasting. In drosophila, the FoxO protein was found to be deacetylated by the class II histone deacetylase (HDAC) 4, leading to its activation and subsequent induction of fasting-related genes. In the fed state, insulin-dependent salt-inducible kinase 3 phosphorylates HDAC4 and sequesters it in the cytoplasm (60). A similar phenomenon is seen in mice in which class II HDACs 4, 5, and 7 are dephosphorylated during fasting, enter the nucleus, and recruit HDAC3 to gluconeogenic promoters. HDAC3 then deacetylates and activates FoxO at these regions (61). Evidence from a third study suggests that FoxO deacetylation (in this case by the Sirtuin 1 deacetylase) plays a role in temporal organization of fasting by which FoxO is only activated by deacetylation in later stages of fasting (62). Most evidence suggests a role for FoxO proteins in gluconeogenesis regulation, an event beginning early upon fasting. Therefore, the later activation of FoxO (62) might serve to maintain a certain level of gluconeogenesis during prolonged fasting. Indeed, gluconeogenic gene expression does not stop and even increases in prolonged fasting (42). This increase in gene expression might be aimed at increasing gluconeogenic efficiency in light of dwindling substrates. In addition to regulation of acetylation, FoxO nuclear localization is also promoted by glucagon-activated Ca2+/calmodulin-dependent protein kinase II (63) (Fig. 1).
In contrast to the gluconeogenic TF FoxO, FoxA2 is an important regulator of FAO and ketogenesis that is predominant in later stages of fasting (59). Similarly to FoxO, FoxA2 is also regulated by acetylation and activated by glucagon. However, in this case, acetylation by p300 activates FoxA2, leading to induction of ketogenic genes in a glucagon-dependent manner (Fig. 1). Reciprocally, FoxA2 activity is reduced by the Sirtuin 1 deacetylase (64). It is tempting to speculate that the reciprocal pattern of regulation by acetylation of the different Fox proteins might reflect temporal organization of the fasting response (i.e., FoxO regulates early induction and late maintenance of gluconeogenesis, whereas FoxA regulates later FAO/ketogenesis).
In addition to activating FoxA2, glucagon was also shown to regulate FAO/ketogenesis through peroxisome proliferator–activated receptor α (PPARA), another ketogenic TF (59). In contrast to wild-type mice, mice lacking GcgR did not increase FAO or FAO-related genes following fasting. The contribution of glucagon to FAO gene expression was dependent on PPARA activation by p38 and AMP-activated protein kinase (Fig. 1). These effects were only seen in prolonged fasting, in line with the role of FAO/ketogenesis in the protein-sparing phase (65).
Taken together, these studies reveal that in addition to CREB, glucagon activates FoxO, FoxA2, and PPARA via signal transduction. Glucagon-activated TFs play a role both in early activation of gluconeogenesis (CREB and FoxO) and in FAO/ketogenesis (FoxA and PPARA) during prolonged fasting (Fig. 1).
TF cascades: the delayed response
As discussed previously, the direct activation of TFs by glucagon-initiated signaling pathways and the dynamic cross talk between TFs are prominent ways to regulate gene expression. Additionally, glucagon’s reach on gene regulation extends to secondary events. In those TF cascades, a primary glucagon-activated TF induces the expression of secondary TFs that, in turn, mediate a second wave of glucagon-initiated transcription. Evidence from multiple studies suggests this is not a negligible aspect of glucagon-dependent transcriptional regulation. Indeed, TFs are the biggest functional group among CREB target genes (36). cAMP/CREB-induced TFs (and coactivators) include the orphan nuclear receptor family NR4A (NUR77, NURR1, and NOR1), EGR-1, estrogen-related receptor γ, Krüppel-like factor 15, Yin Yang 1, TF EB (TFEB), and PPARγ coactivator 1α (PGC1A). Later and in Fig. 3, we describe these TF cascades and their effect on metabolic regulation. Many of these TF cascades modulate metabolic events characterizing prolonged fasting [FAO, ketogenesis, and fibroblast growth factor 21 (FGF21) expression], suggesting this mechanism mediates, at least in part, the temporal organization of the fasting response.
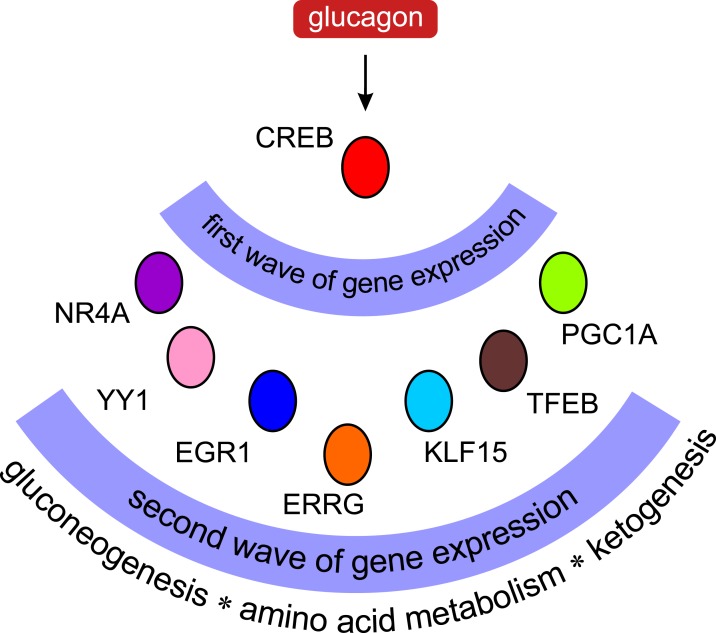
Figure 3.
TF cascades under glucagon control. Upon activation by glucagon, the cAMP/CREB pathway induces a group of genes that themselves encode TFs. These secondary TFs lead to a second wave of gene transcription, thus expanding the scope of glucagon-related gene regulation. Evidence suggests that these waves of transcription facilitate the temporal organization of the fasting response in liver. ERRG, estrogen-related receptor γ; KLF15, Krüppel-like factor 15; YY1, Yin Yang 1.
The gene levels of all three members of the NR4A family are induced in liver during fasting through the cAMP-PKA-CREB pathway. In turn, NR4As induce a set of gluconeogenic genes, thereby enhancing gluconeogenesis and increasing blood glucose levels (66). In addition to regulating gluconeogenesis, NUR77 induces the expression of FGF21, a fasting-related hepatokine with far-reaching roles in fasting (67). In accordance, glucagon was shown to increase FGF21 levels in rodents and humans (68), although the effect of glucagon on FGF21 could also be mediated by PPARA and FoxA2 (59).
Another possible axis involves CREB induction of EGR-1 (69). EGR-1 induces the Cebpa gene, encoding a critical fasting-related TF (59, 70). Estrogen-related receptor γ is another fasting-related TF induced by the glucagon-cAMP-CREB axis (71). Moreover, cAMP induces Krüppel-like factor 15 (72), which plays a role in regulation of gluconeogenic genes but also in regulating genes that facilitate amino acid uptake and catabolism in hepatocytes to serve as gluconeogenic precursors (73).
In addition to direct CREB-GR cross talk described previously, CREB also facilitates the activity of GR and PPARA by several TF cascades. CREB induces YY1 that in turn leads to increased levels of GR (74). Additionally, CREB induces the gene levels of CCAAT/enhancer binding protein β (75), a TF known to augment GR binding by dynamic assisted loading (47). Finally, CREB induces PGC1A, a cofactor that facilitates the activity of GR (20) as well as the activity of PPARA and other TFs (76). PPARA activity might be further augmented via CREB-dependent induction of TFEB (57). TFEB induces PGC1A gene levels and promotes the FAO/ketogenic effect of PPARA (77). These PPARA-promoting TF cascades are complementary to the direct activation of PPARA by glucagon (65).
Considering these various reports, it appears that by exerting a second wave of gene expression, TF cascades are a major arm in glucagon gene regulation. This delayed mechanism may be instrumental in bringing about the temporal organization of the fasting response. This is in agreement with the various fasting-related metabolic pathways regulated by TF cascades (amino acid metabolism, FAO, ketogenesis, etc.) (Fig. 3).
Conclusions
Glucagon is a dominant hormone that regulates metabolism. Much of the hormone’s downstream effects are mediated by glucagon-activated TFs affecting gene expression. Glucagon can directly activate TFs through signaling events, foster dynamic cooperation between TFs through assisted loading, and activate TFs in a delayed mechanism via TF cascades. These actions are critical components of the glucagon pathway, enabling a fine-tuned, temporally organized, and integrated cellular response to glucagon. Understanding how glucagon regulates TFs, coactivators, and chromatin is critical to characterizing its metabolic effects and restoring them when dysregulated in disease.
Acknowledgments
Disclosure Summary: The authors have nothing to disclose.
Abbreviations:
- A-CREB
acidic cyclic adenosine monophosphate response element-binding protein
- bZIP
basic leucine zipper
- cAMP
cyclic adenosine monophosphate
- CBP
cyclic adenosine monophosphate response element-binding protein–binding protein
- ChIP
chromatin immunoprecipitation
- ChIP-seq
chromatin immunoprecipitation sequencing
- CRE
cyclic adenosine monophosphate response element
- CREB
cyclic adenosine monophosphate response element-binding protein
- CRTC
cyclic adenosine monophosphate–regulated transcriptional coactivator
- FAO
fatty acid oxidation
- FGF21
fibroblast growth factor 21
- Fox
forkhead box
- FXR
farnesoid X receptor
- GcgR
glucagon receptor
- GR
glucocorticoid receptor
- HAT
histone acetyl transferase
- HDAC
histone deacetylase
- IP3
inositol 1,4,5-trisphosphate
- KAT2B
lysine acetyltransferase 2B
- PGC1A
peroxisome proliferator–activated receptor γ coactivator 1α
- PKA
protein kinase A
- PLC
phospholipase C
- PPARA
peroxisome proliferator–activated receptor α
- PRMT5
protein arginine methyltransferase 5
- TF
transcription factor.
References
- 1. Ahrén B. Glucagon--Early breakthroughs and recent discoveries. Peptides. 2015;67:74–81. [DOI] [PubMed] [Google Scholar]
- 2. Habegger KM, Heppner KM, Geary N, Bartness TJ, DiMarchi R, Tschöp MH. The metabolic actions of glucagon revisited. Nat Rev Endocrinol. 2010;6(12):689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell JE, Drucker DJ. Islet α cells and glucagon--critical regulators of energy homeostasis. Nat Rev Endocrinol. 2015;11(6):329–338. [DOI] [PubMed] [Google Scholar]
- 4. Authier F, Desbuquois B. Glucagon receptors. Cell Mol Life Sci. 2008;65(12):1880–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ravnskjaer K, Madiraju A, Montminy M. Role of the cAMP pathway in glucose and lipid metabolism. Handb Exp Pharmacol. 2016;233:29–49. [DOI] [PubMed] [Google Scholar]
- 6. Jelinek LJ, Lok S, Rosenberg GB, Smith RA, Grant FJ, Biggs S, Bensch PA, Kuijper JL, Sheppard PO, Sprecher CA, O’Hara PJ, Foster D, Walker KM, Chen LHJ, Mckernan PA, Kindsvogel W. Expression cloning and signaling properties of the rat glucagon receptor. Science. 1993;259(5101):1614–1616. [DOI] [PubMed] [Google Scholar]
- 7. Wakelam MJ, Murphy GJ, Hruby VJ, Houslay MD. Activation of two signal-transduction systems in hepatocytes by glucagon. Nature. 1986;323(6083):68–71. [DOI] [PubMed] [Google Scholar]
- 8. Chen J, Ishac EJ, Dent P, Kunos G, Gao B. Effects of ethanol on mitogen-activated protein kinase and stress-activated protein kinase cascades in normal and regenerating liver. Biochem J. 1998;334(Pt 3):669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang Y, Cypess AM, Muse ED, Wu CR, Unson CG, Merrifield RB, Sakmar TP. Glucagon receptor activates extracellular signal-regulated protein kinase 1/2 via cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2001;98(18):10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Li G, Goode J, Paz JC, Ouyang K, Screaton R, Fischer WH, Chen J, Tabas I, Montminy M. Inositol-1,4,5-trisphosphate receptor regulates hepatic gluconeogenesis in fasting and diabetes. Nature. 2012;485(7396):128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab. 2003;284(4):E671–E678. [DOI] [PubMed] [Google Scholar]
- 12. Cahill GF., Jr Fuel metabolism in starvation. Annu Rev Nutr. 2006;26(1):1–22. [DOI] [PubMed] [Google Scholar]
- 13. Servillo G, Della Fazia MA, Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM. Exp Cell Res. 2002;275(2):143–154. [DOI] [PubMed] [Google Scholar]
- 14. Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68(1):821–861. [DOI] [PubMed] [Google Scholar]
- 16. Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. [DOI] [PubMed] [Google Scholar]
- 17. D’Alessio D. The role of dysregulated glucagon secretion in type 2 diabetes. Diabetes Obes Metab. 2011;13(Suppl 1):126–132. [DOI] [PubMed] [Google Scholar]
- 18. Unger RH, Cherrington AD. Glucagonocentric restructuring of diabetes: a pathophysiologic and therapeutic makeover. J Clin Invest. 2012;122(1):4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18(2):967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–183. [DOI] [PubMed] [Google Scholar]
- 21. Herzig S, Hedrick S, Morantte I, Koo SH, Galimi F, Montminy M. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-gamma. Nature. 2003;426(6963):190–193. [DOI] [PubMed] [Google Scholar]
- 22. Erion DM, Ignatova ID, Yonemitsu S, Nagai Y, Chatterjee P, Weismann D, Hsiao JJ, Zhang D, Iwasaki T, Stark R, Flannery C, Kahn M, Carmean CM, Yu XX, Murray SF, Bhanot S, Monia BP, Cline GW, Samuel VT, Shulman GI. Prevention of hepatic steatosis and hepatic insulin resistance by knockdown of cAMP response element-binding protein. Cell Metab. 2009;10(6):499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee D, Le Lay J, Kaestner KH. The transcription factor CREB has no non-redundant functions in hepatic glucose metabolism in mice. Diabetologia. 2014;57(6):1242–1248. [DOI] [PubMed] [Google Scholar]
- 24. Rudolph D, Tafuri A, Gass P, Hämmerling GJ, Arnold B, Schütz G. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc Natl Acad Sci USA. 1998;95(8):4481–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16(4):258–264. [DOI] [PubMed] [Google Scholar]
- 26. Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–859. [DOI] [PubMed] [Google Scholar]
- 27. Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374(6517):85–88. [DOI] [PubMed] [Google Scholar]
- 28. Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR III, Montminy M. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 2007;26(12):2880–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114(Pt 13):2363–2373. [DOI] [PubMed] [Google Scholar]
- 30. Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, Kimura H. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516(7530):272–275. [DOI] [PubMed] [Google Scholar]
- 31. Reed SM, Quelle DE. p53 Acetylation: regulation and consequences. Cancers (Basel). 2014;7(1):30–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Icardi L, De Bosscher K, Tavernier J. The HAT/HDAC interplay: multilevel control of STAT signaling. Cytokine Growth Factor Rev. 2012;23(6):283–291. [DOI] [PubMed] [Google Scholar]
- 33. Goldstein I, Yizhak K, Madar S, Goldfinger N, Ruppin E, Rotter V. p53 promotes the expression of gluconeogenesis-related genes and enhances hepatic glucose production. Cancer Metab. 2013;1(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravnskjaer K, Hogan MF, Lackey D, Tora L, Dent SY, Olefsky J, Montminy M. Glucagon regulates gluconeogenesis through KAT2B- and WDR5-mediated epigenetic effects. J Clin Invest. 2013;123(10):4318–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsai WW, Niessen S, Goebel N, Yates JR III, Guccione E, Montminy M. PRMT5 modulates the metabolic response to fasting signals. Proc Natl Acad Sci USA. 2013;110(22):8870–8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102(12):4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119(7):1041–1054. [DOI] [PubMed] [Google Scholar]
- 38. Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2:2006.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Everett LJ, Le Lay J, Lukovac S, Bernstein D, Steger DJ, Lazar MA, Kaestner KH. Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics. 2013;14(1):337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goldstein I, Hager GL. Dynamic enhancer function in the chromatin context. Wiley Interdiscip Rev Syst Biol Med. 2017;e1390. [DOI] [PMC free article] [PubMed]
- 41. Baek S, Goldstein I, Hager GL. Bivariate genomic footprinting detects changes in transcription factor activity. Cell Reports. 2017;19(8):1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldstein I, Baek S, Presman DM, Paakinaho V, Swinstead EE, Hager GL. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 2017;27(3):427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Imai E, Miner JN, Mitchell JA, Yamamoto KR, Granner DK. Glucocorticoid receptor-cAMP response element-binding protein interaction and the response of the phosphoenolpyruvate carboxykinase gene to glucocorticoids. J Biol Chem. 1993;268(8):5353–5356. [PubMed] [Google Scholar]
- 44. Ratman D, Vanden BW, Dejager L, Libert C, Tavernier J, Beck IM, De Bosscher K. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol Cell Endocrinol. 2013;380(1-2):41–54. [DOI] [PubMed] [Google Scholar]
- 45. Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146(4):544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, Vinson C, Stamatoyannopoulos JA, Hager GL. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43(1):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grøntved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013;32(11):1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miranda TB, Voss TC, Sung MH, Baek S, John S, Hawkins M, Grøntved L, Schiltz RL, Hager GL. Reprogramming the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res. 2013;73(16):5130–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Madsen MS, Siersbæk R, Boergesen M, Nielsen R, Mandrup S. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol. 2014;34(6):939–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soccio RE, Chen ER, Rajapurkar SR, Safabakhsh P, Marinis JM, Dispirito JR, Emmett MJ, Briggs ER, Fang B, Everett LJ, Lim HW, Won KJ, Steger DJ, Wu Y, Civelek M, Voight BF, Lazar MA. Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell. 2015;162(1):33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis LD, Grimm JB, Morisaki T, Grøntved L, Presman DM, Hager GL. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell. 2016;165(3):593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O’Malley BW. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol Cell. 2015;60(5):769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hill MJ, Suzuki S, Segars JH, Kino T. CRTC2 is a coactivator of GR and couples GR and CREB in the regulation of hepatic gluconeogenesis. Mol Endocrinol. 2016;30(1):104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sunkel B, Wu D, Chen Z, Wang CM, Liu X, Ye Z, Horning AM, Liu J, Mahalingam D, Lopez-Nicora H, Lin CL, Goodfellow PJ, Clinton SK, Jin VX, Chen CL, Huang TH, Wang Q. Integrative analysis identifies targetable CREB1/FoxA1 transcriptional co-regulation as a predictor of prostate cancer recurrence. Nucleic Acids Res. 2016;44(9):4105–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kodama S, Moore R, Yamamoto Y, Negishi M. Human nuclear pregnane X receptor cross-talk with CREB to repress cAMP activation of the glucose-6-phosphatase gene. Biochem J. 2007;407(3):373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oh KJ, Park J, Kim SS, Oh H, Choi CS, Koo SH. TCF7L2 modulates glucose homeostasis by regulating CREB- and FoxO1-dependent transcriptional pathway in the liver. PLoS Genet. 2012;8(9):e1002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516(7529):108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13(4):213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goldstein I, Hager GL. Transcriptional and chromatin regulation during fasting - The genomic era. Trends Endocrinol Metab. 2015;26(12):699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR III, Fischer WH, Thomas JB, Montminy M. A hormone-dependent module regulating energy balance. Cell. 2011;145(4):596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145(4):607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J III, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ozcan L, Wong CC, Li G, Xu T, Pajvani U, Park SK, Wronska A, Chen BX, Marks AR, Fukamizu A, Backs J, Singer HA, Yates JR III, Accili D, Tabas I. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 2012;15(5):739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. von Meyenn F, Porstmann T, Gasser E, Selevsek N, Schmidt A, Aebersold R, Stoffel M. Glucagon-induced acetylation of Foxa2 regulates hepatic lipid metabolism. Cell Metab. 2013;17(3):436–447. [DOI] [PubMed] [Google Scholar]
- 65. Longuet C, Sinclair EM, Maida A, Baggio LL, Maziarz M, Charron MJ, Drucker DJ. The glucagon receptor is required for the adaptive metabolic response to fasting. Cell Metab. 2008;8(5):359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12(9):1048–1055. [DOI] [PubMed] [Google Scholar]
- 67. Min AK, Bae KH, Jung YA, Choi YK, Kim MJ, Kim JH, Jeon JH, Kim JG, Lee IK, Park KG. Orphan nuclear receptor Nur77 mediates fasting-induced hepatic fibroblast growth factor 21 expression. Endocrinology. 2014;155(8):2924–2931. [DOI] [PubMed] [Google Scholar]
- 68. Arafat AM, Kaczmarek P, Skrzypski M, Pruszyńska-Oszmalek E, Kołodziejski P, Szczepankiewicz D, Sassek M, Wojciechowicz T, Wiedenmann B, Pfeiffer AF, Nowak KW, Strowski MZ. Glucagon increases circulating fibroblast growth factor 21 independently of endogenous insulin levels: a novel mechanism of glucagon-stimulated lipolysis? Diabetologia. 2013;56(3):588–597. [DOI] [PubMed] [Google Scholar]
- 69. Lee HJ, Mignacca RC, Sakamoto KM. Transcriptional activation of egr-1 by granulocyte-macrophage colony-stimulating factor but not interleukin 3 requires phosphorylation of cAMP response element-binding protein (CREB) on serine 133. J Biol Chem. 1995;270(27):15979–15983. [DOI] [PubMed] [Google Scholar]
- 70. Shen N, Jiang S, Lu JM, Yu X, Lai SS, Zhang JZ, Zhang JL, Tao WW, Wang XX, Xu N, Xue B, Li CJ. The constitutive activation of Egr-1/C/EBPa mediates the development of type 2 diabetes mellitus by enhancing hepatic gluconeogenesis. Am J Pathol. 2015;185(2):513–523. [DOI] [PubMed] [Google Scholar]
- 71. Kim DK, Ryu D, Koh M, Lee MW, Lim D, Kim MJ, Kim YH, Cho WJ, Lee CH, Park SB, Koo SH, Choi HS. Orphan nuclear receptor estrogen-related receptor γ (ERRγ) is key regulator of hepatic gluconeogenesis. J Biol Chem. 2012;287(26):21628–21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Takashima M, Ogawa W, Hayashi K, Inoue H, Kinoshita S, Okamoto Y, Sakaue H, Wataoka Y, Emi A, Senga Y, Matsuki Y, Watanabe E, Hiramatsu R, Kasuga M. Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes. 2010;59(7):1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, Jain MK. Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 2007;5(4):305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lu Y, Xiong X, Wang X, Zhang Z, Li J, Shi G, Yang J, Zhang H, Ning G, Li X. Yin Yang 1 promotes hepatic gluconeogenesis through upregulation of glucocorticoid receptor. Diabetes. 2013;62(4):1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Niehof M, Manns MP, Trautwein C. CREB controls LAP/C/EBP beta transcription. Mol Cell Biol. 1997;17(7):3600–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116(3):615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]