Abstract
Objective
To identify the genetic cause of hypomyelinating leukodystrophy in 2 consanguineous families.
Methods
Homozygosity mapping combined with whole-exome sequencing of consanguineous families was performed. Mutation consequences were determined by studying the structural change of the protein and by the RNA analysis of patients' fibroblasts.
Results
We identified a biallelic mutation in a gene coding for a Pol III–specific subunit, POLR3K (c.121C>T/p.Arg41Trp), that cosegregates with the disease in 2 unrelated patients. Patients expressed neurologic and extraneurologic signs found in POLR3A- and POLR3B-related leukodystrophies with a peculiar severe digestive dysfunction. The mutation impaired the POLR3K-POLR3B interactions resulting in zebrafish in abnormal gut development. Functional studies in the 2 patients' fibroblasts revealed a severe decrease (60%–80%) in the expression of 5S and 7S ribosomal RNAs in comparison with control.
Conclusions
These analyses underlined the key role of ribosomal RNA regulation in the development and maintenance of the white matter and the cerebellum as already reported for diseases related to genes involved in transfer RNA or translation initiation factors.
RNA polymerase III (Pol III) mutations have been implicated in autosomal recessive hypomyelinating leukodystrophies (HLD7 [MIM 607694] and HLD8 [MIM 614381]). HLDs are characterized by a normal T1 abnormal hyper T2/fluid-attenuated inversion recovery (FLAIR) signal of the white matter on magnetic resonance imaging (MRI).1,2 The clinical presentation is variable from infantile to juvenile/adult-onset forms with motor decline manifest as progressive cerebellar dysfunction and mild cognitive regression. Other features may include hypo/oligodontia, myopia, dysmorphia, and hypogonadotropic hypogonadism.
Pol III, composed of 17 subunits, is the largest eukaryotic RNA polymerase. It transcribes small untranslated RNAs involved in cellular processes including the regulation of transcription (7SK RNA; Alu RNA), RNA processing (U6 RNA; H1 RNA), and translation (tRNA; 5S RNA).3 Promoters driving transcription of these genes have been identified, cloned, and characterized.4,5 Mutations causing HLD have been reported first in the POLR3A1 (MIM 614258) and POLR3B2,6 (MIM 614366) genes. More recently, mutations in the POLR1C gene (MIM 616494) encoding a subunit shared by Pol I and Pol III complexes have been reported as HLD11.7 In addition, a homozygous mutation of POLR1A (NM_616404) encoding the largest subunit of Pol I, RPA194, has been described in a family affected by a demyelinating form of leukodystrophy.8
Here, we report a homozygous mutation of POLR3K (NM_606007) in 2 unrelated HLD-affected patients. POLR3K encodes the RPC11 subunit of Pol III, which has been implicated in the processes of transcription termination and reinitiation.9,10 We demonstrate that the mutation affected the POLR3K-POLR3B interactions and decreased the 5S and 7SL RNA levels.
Methods
Standard protocol approvals, registrations, and patient consents
Consent was obtained from patients and their parents according to the LEUKOFRANCE research program for undetermined leukodystrophies (authorization CPP AU788; CNIL 1406552; AFSSAPS B90298-60).
Patients
Patients were referred to the French reference center for leukodystrophies, LEUKOFRANCE, for diagnosis and follow-up. DNA was extracted from white blood cells of the affected patients and unaffected family members. Fibroblasts were obtained from skin biopsy according to our previously reported protocol.11
DNA analysis
We performed homozygosity mapping in all family members using GeneChip Human Mapping 250K Nsp Array, and whole-exome sequencing (IntegraGen, Evry, France) using the SureSelect V4 capture kit (Agilent, Massy, France) and the HighSeq2000 sequencer (Illumina, San Diego, CA).12
Structural model
To get insight on the mutation effect on POLR3K protein (UniProtKB: Q9Y2Y1) structure, we performed a molecular modeling analysis. The Protein Data Bank (PDB) files and 2D structures were predicted using the PSIPRED server (bioinf.cs.ucl.ac.uk/psipred/).13
To predict the interaction between POLR3K and POLR3B, we used Phyre 2 (sbg.bio.ic.ac.uk/phyre2) to have the PDB file of POLR3B (UniProtKB: Q9NW08), and we performed the protein-protein docking with ClusPro server (cluspro.org).14–16
Fibroblasts analysis
Cell culture
Cell lines were grown in Dulbecco Modified Eagle Medium supplemented with 15% fetal bovine serum (Invitrogen, Illkirch, France), 1% minimum essential medium nonessential amino acid solution (Sigma, Saint-Quentin-Fallavier, France), 100 U/ml penicillin, and 100 µg/mL streptomycin (Invitrogen). Cell lines were maintained at 37°C in a humidified 5% CO2 atmosphere. All extracts were made from subconfluent cells in the exponential phase of growth.
RNA extraction
After several passages (<8) under tissue culture conditions, total RNA was extracted using TRIzol reagent (Invitrogen, Illkirch, France), according to the manufacturer's protocol. Genomic DNA was removed using the turboDNA free kit (Ambion). RNA concentrations were determined using a NanoDrop spectrophotometer (Nanodrop Technologies, Wilmington, DE). RNAs integrity was determined with an Agilent 2100 bioanalyzer (Palo Alto, CA). RNA measurements are automatically submitted to an algorithm that allows standardized control of RNA quality and the calculation of an RNA integrity number.17
Quantitative RT-PCR
For each sample, 2 µg of total RNA was reverse transcribed using Scientific Maxima Reverse transcriptase (Fisher Scientific, Illkirch, France) and random hexamer primers (Fermentas, Fisher Scientific, Illkirch, France). Expression of POLR3D RNA, POLR3K RNA, and U2 RNA, and that of RNA polymerase III–transcribed genes were quantified by reverse transcription and real-time PCR using the SsoAdvancedTM Universal SYBR® Green Supermix (Bio-Rad, Marnes-la-coquette, France) with gene-specific primers (table e-1, links.lww.com/NXG/A122). Real-time PCRs were run on CFX96 Real-Time PCR Detection System (Bio-Rad). Cycle conditions were 95°C for 30 seconds, followed by 40 cycles with 95°C, 5 seconds, and 60°C for 10 seconds. RNA levels were normalized with β-actin and peptidylprolyl isomerase A (PPIA) RNA, using a comparative 2ΔΔCt method, and controls were arbitrarily set at 1. RNA extractions and RT-qPCR analyses were performed with at least 3 biological replicates. Each of these biological replicates was analyzed by at least 3 technical replicates.
Northern blot
One microgram of total RNA was resolved on 8% denaturing polyacrylamide gels in 1 × tris-borate-EDTA and blotted onto a Hybond-XL nylon membrane (GE Health care, Buc, France) according to standard procedures. Oligonucleotides (Sigma-Aldrich, Saint-Quentin, Fallavier) were labeled at the 5′ end by phosphorylation with [γ-32P] adenosine triphosphate (Perkin Elmer, Villebon-sur-Yvette, France) and then purified with a microspin G-25 column (GE Health care). Membranes were incubated with, respectively, 5.8S, 5S, and U2 RNA 5′-32P-radiolabeled probes (table e-1, links.lww.com/NXG/A122) in 1X Church buffer (0.25 M NaH2PO4; 1 mM EDTA; 7% SDS; 20 mg/mL salmon sperm DNA; 0.5% bovine serum albumin) at 28°C overnight in hybridization oven. After 2 washes, one in 2× saline sodium citrate (SSC), 0.1% SDS, and the other in 1× SSC, 1% SDS, blots were analyzed by the phosphoimager and quantified using ImageQuant software (GE Health care).
Statistical analyses
Quantitative data were described and presented graphically as mean values and SDs. Group comparisons were performed using SPSS software with one-way analysis of variance and the Turkey test. A difference was considered to be statistically significant when the p value was less than 0.05 (graphically: * for p < 0.05, ** for p < 0.01, and *** for p < 0.001).
Data availability statement
The data sets analyzed during the current study are available from the corresponding author on reasonable request.
Results
Clinical characteristics
The affected patients were born from 2 distinct consanguineous families of Berber origin from Algeria (figure 1, A). Table 1 summarizes the clinical characteristics of patients.
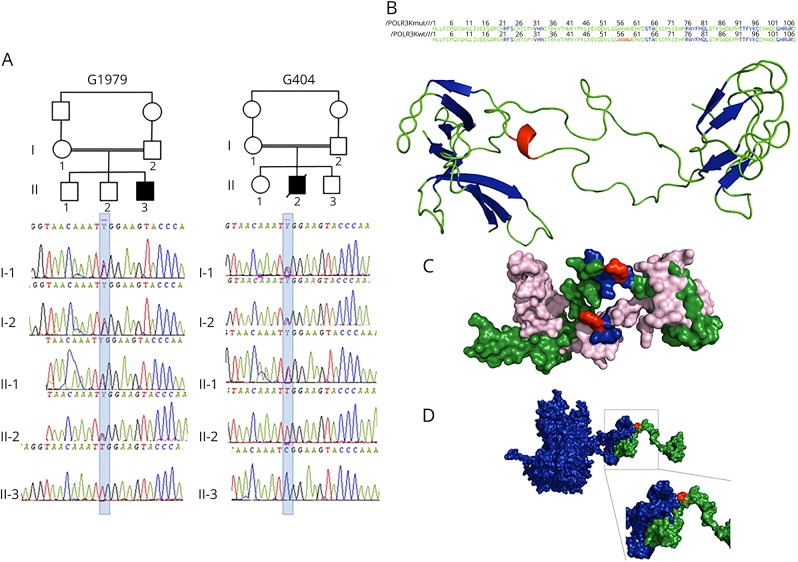
Figure 1. POLR3K mutated families, structural model of POLR3K and of POLR3K-POLR3B interactions.
(A) Pedigree and electropherograms of family G1979 (patient 1) and family G404 (patient 2). (B) Amino acid (AA) sequence and 2-dimensional structure of the mutated and wild-type POLR3K. The sheets, loops, and α-helix motifs are colored in blue, green, and red, respectively. The loop (AA 34–55), α-helix (AA 56–59), and loop (AA60–63) motifs of the wild-type protein are replaced by a unique loop (34–63) in the mutated protein. (C) Three-dimensional structure of the wild-type (in green) and mutated (in pink) POLR3K. The AAs at position 41 (arginine in the wild type and tryptophan in the mutant) are in red. The residues located within 4 Å around the Arg41, responsible for the stability of the protein (Asn40, Lys42, and Tyr43), are colored in blue. The Trp41 change induces a modification in the interactions of Tyr43 with Asn40 and Lys42 decreasing protein stability. (D) Three-dimensional structure of POLR3K (in green) and POLR3B (in blue) interactions. The mutated residue 41 colored in red is important in POLR3K-POLR3B interactions: the N-terminal part (1–41) of POLR3K interacting with the C-terminal part (1079–1133) of POLR3B.
Table 1.
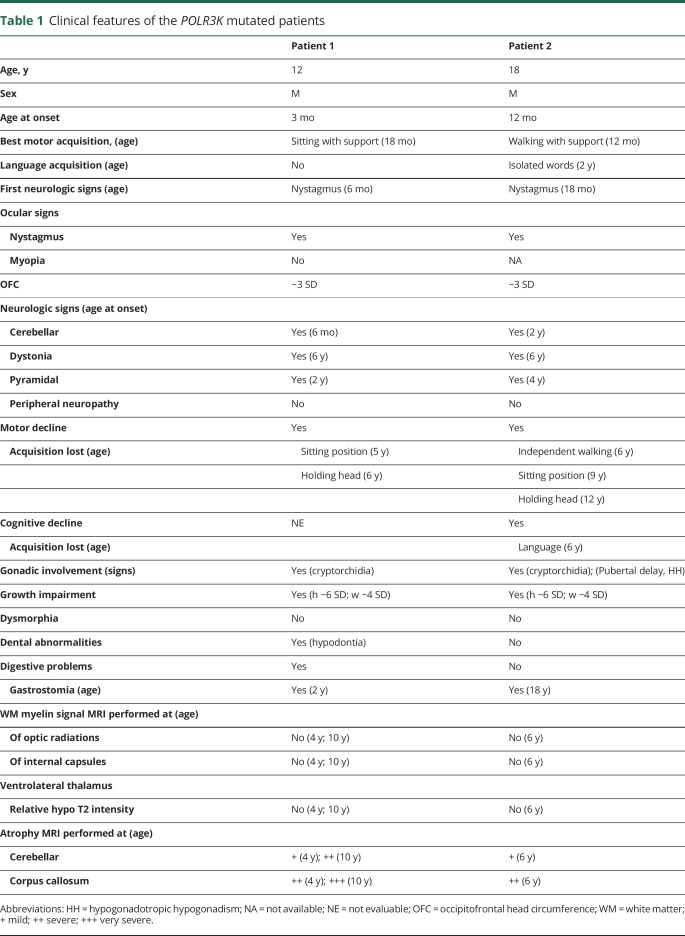
Clinical features of the POLR3K mutated patients
Patient 1 presents severe feeding difficulties with recurrent vomiting associated with constipation starting during the first months of life leading to failure to thrive (height and weight <−3SD) despite nutrition through gastrostomia at age 2 years and absence of hormones deficiency. Abnormal motor acquisitions with hypotonia leading only to sitting position with support at age 18 months were associated with the progression of neurologic signs. Acute episodes of vomiting with hypoglycemia and ketosis were observed during banal infections up to age 2–3 years, whereas transitory comas with hypothermia induced by emotions were frequent after age 10 years. At 10 years, episodic seizures, dystonia, quadriparesis, optic atrophy, and pseudobulbar signs were observed with persistence of communication with the mother.
In patient 2, walking capacities with support were acquired at 12 months when a nystagmus started. Progressive ataxia impaired further motor acquisitions, whereas isolated dysarthric words were obtained between ages 2 and 3 years. Cognitive capacities remained poor. Progressive spasticity, athetosis, and dyskinesia were noticed after age 6 years when walking capacities were lost and subsequently sitting position (9 years) and head control (12 years). Swallowing difficulties with recurrent respiratory infections leading to gastrostomia occurred at age 12 years. Failure to thrive was noticed at 18 months, leading to growth hormone treatment between ages 4 and 6 years without efficacy (height −6 SD, weight −4 SD). Bilateral cryptorchidia was treated by surgery at age 6 years. Delay in puberty with hypogonadotropic hypogonadism was reported. He suddenly died at age 18 years, a few days after he returned from Algeria, because of an acute gastrointestinal infection.
In both cases, a diffuse hypomyelinating aspect of the white matter was observed on MRI excepted in the early first myelinated area of the brainstem, associated with atrophy of the corpus callosum and cerebellum (figure 2, and table 1). In patient 1, subsequent MRI performed between ages 4 and 10 years demonstrated a progressive atrophy with a loss of 20% of the cerebellum volume. In the corpus callosum, atrophy was more pronounced in the posterior (35%) than in the anterior part (20%). The decrease in the N-acetylaspartate (NAA)/creatine associated with decreased in choline/creatine and increased myoInositol/creatine content was observed on magnetic resonance spectroscopy, confirming the severity of the brain dysfunction. Sanger sequencing and gene panel next generation sequencing analysis of POLR3A, POLR3B exons did not reveal abnormalities.
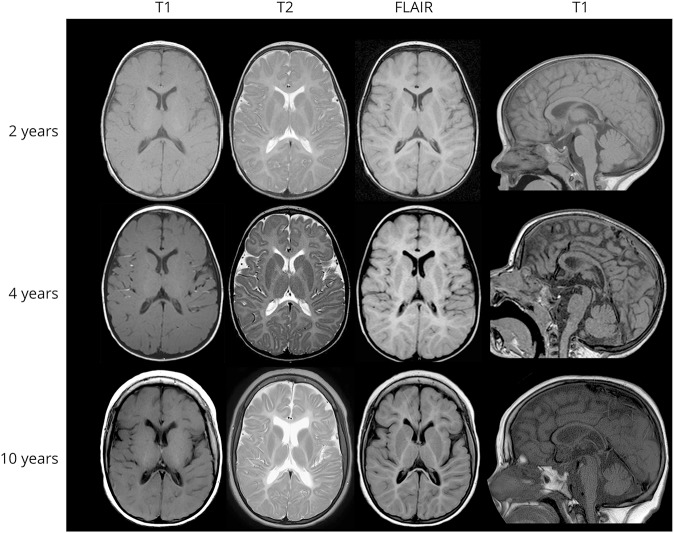
Figure 2. MRI progression in patient 1.
The diffuse hypomyelinating aspect of the white matter characterized by an isosignal T1, hypersignal T2, and flair of the white matter in comparison with the gray matter did not change during the 8 years of evolution (MRI performed at age 2, 4, and 10 years) despite the clinical progression of the disease observed after age 4 years. In contrast, a progressive atrophy is observed in the supratentorial and subtentorial structures between ages 4 and 10 years: deeper cortical sulci, increased subarachnoid spaces, frontal ventricular dilatation and white matter atrophy, corpus callosum atrophy (18% loss in the anterior part and 35% in the posterior part), and cerebellar atrophy (20% loss of the vermis and cerebellar hemispheres).
Whole-exome sequencing identified a new POLR3K mutation
Whole-exome sequencing combined with homozygosity mapping revealed only 1 homozygous variant in POLR3K (NM_016310.4): c.121C>T/p.Arg41Trp common to the 2 patients. Sanger sequencing confirmed that the variant segregated in the 2 families in consistence with autosomal recessive inheritance (figure 1, A). This mutation was (1) predicted to be deleterious by SIFT (score 0.00) and align Grantham variation and Grantham deviation (C35), disease causing by MutationTaster (p = 1.00), benign by PolyPhen-2 (score 0.143); (2) affecting a nucleotide highly conserved among species; (3) not found in the dbSNP, 1000 Genomes Project, or Exome Aggregation Consortium databases; and (4) not detected in 500 ethnically (North African Berbers) matched control chromosomes in neither a homozygous nor a heterozygous state.
Structural model of POLR3K-POLR3B interactions
In silico protein analysis shown structural differences between wild-type and mutated POLR3K. The positively charged and hydrophilic arginine was replaced by a neutral and hydrophobic tryptophan. The 2D and 3D structure analyses (figure 1, B and C) demonstrate that the loop (34–55), α-helix (56–59), and loop (60–63) motifs of the wild-type protein are replaced by a unique loop (34–63) in the mutated protein. The residues located within 4 Å around the Arg41, responsible for the stability of the protein, are Asn40, Lys42, and Tyr43. The Trp41 change induces a modification in the interactions of Tyr43 with Asn40 and Lys42 decreasing protein stability (figure 1, C).
In addition, the protein-protein docking analysis (figure 1, D) showed that the residue 41 is important in POLR3K-POLR3B interactions: the N-terminal part (1–41) of POLR3K interacting with the C-terminal part (1079–1133) of POLR3B.
Certain Pol III RNAs involved in translational control are decreased in the POLR3K mutated fibroblasts
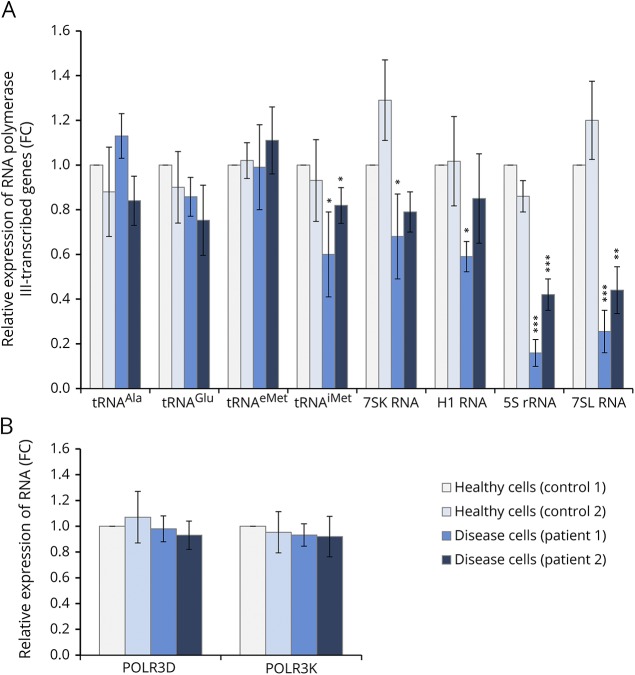
To determine whether the expression of RNA polymerase III–transcribed genes is altered in individuals carrying the POLR3K mutation, we compared the relative RNA level of the skin fibroblasts from the 2 affected patients in comparison to those derived from control individuals using PPIA and β-actin expression levels as standard. We found that expression levels of 3 of the 4 distinct tRNAs analyzed are not significantly affected by the POLR3K mutation (figure 3A). In contrast, tRNAimet, 7SK, and H1 RNA expression was significantly reduced in patient 1 and tRNAimet expression only in patient 2 fibroblasts. Most strikingly, the expression of 5S rRNA and 7SL RNA genes was strongly reduced (60%–80%) in fibroblasts of the 2 patients with leukodystrophy (figure 3A). We verified that the reduced 5S and 7SL RNA expression was not due to an impaired expression of Pol III subunits by quantifying in parallel POLR3K and POLR3D mRNA expression levels (figure 3B). To verify the decrease in Pol III RNA expression levels by an independent method, we performed Northern blot analyses. By comparing 5.8S rRNA with U2 snRNA levels, we observed a relative increase in U2 snRNA levels, indicating that the overall levels of 5.8S rRNA were slightly lower in patients with leukodystrophy than in controls (figure e-1A, links.lww.com/NXG/A121). Comparing 5S rRNA levels with U2 snRNA levels confirmed significantly lower expression of 5S rRNA in patient fibroblasts compared with control fibroblasts (figure e-1B). These results clearly indicate that the (p.Arg41Trp) POLR3K mutation reduces 5S and 7SL RNA levels, potentially contributing to the development of the disease.
Figure 3. RNA levels determined by RT-qPCR analysis.
Total RNAs were isolated from healthy (control cells) or POLR3K mutated (disease cells) patients' fibroblasts. Relative gene expression levels were normalized with β-actin and PPIA genes as an internal control and compared with control cells. Each bar represents the mean ± SD of at least 3 independent experiments. *Indicated a significant difference of disease cells compared with healthy cells. *p < 0.05, **p < 0.01, ***p < 0.001. (A) POLR3K and POLR3D mRNA expression levels. (B) Expression of RNA polymerase III–transcribed genes.
Discussion
In this article, we reported a novel homozygous missense variant in the POLR3K gene in 2 HLD-affected patients from 2 consanguineous families using whole exome sequencing and functional analysis. Following the latest the American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines,18 this variant has 1 strong (functional analysis), at least 2 moderate (highly conserved and very low allele frequency), and 2 supporting (bioinformatics and segregation analysis) criteria of pathogenicity. The variant is not found in polymorphisms databases and in controls of the same Berber ethnic background. The p.Arg41Trp substitution found is located within domain II of the POLR3K protein, a highly conserved region from yeast to human.9 Docking analysis of the POLR3K missense substitution p.Arg41Trp suggested less stability in the interactions of the RPC128 subunits encoded by POLR3B and RPC11 encoded by POLR3K. Of interest, a mutation of the POLR3B gene has been reported in zebrafish with impaired proliferation of digestive organs. The zebrafish mutation reduced RPC11 association with Pol III and the transcription of tRNA and 7SL genes. Overexpression of RPC11 in this model system could rescue some of these defects.19
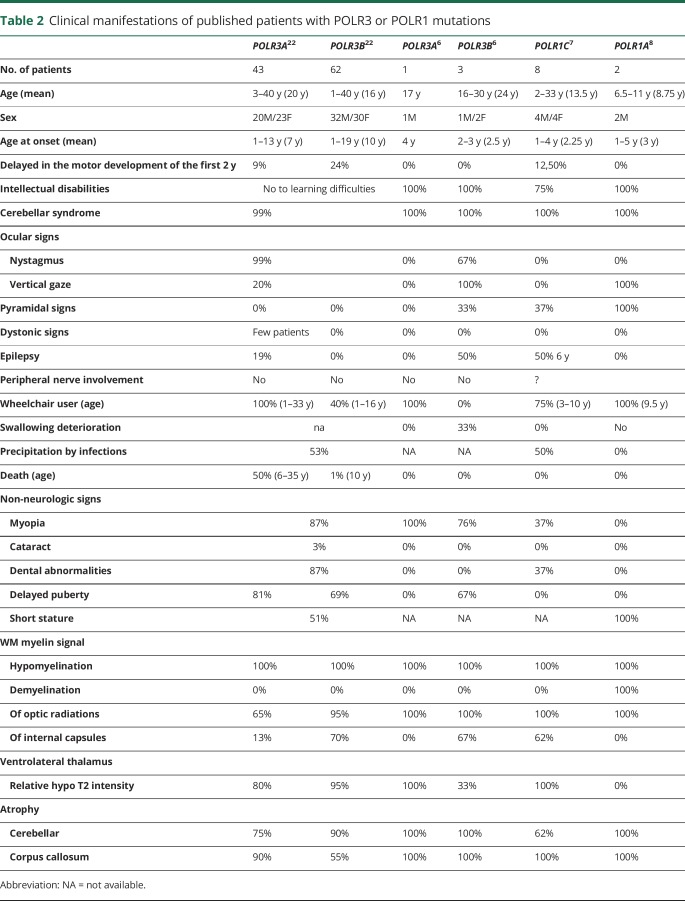
Our patients expressed neurologic signs classically found in patients mutated for POLR3A, POLR3B, or POLR1C (table 2) as early hypotonia, nystagmus, ataxia associated with hypomyelinated white matter, and cerebellar/corpus callosum atrophy. Extraneurologic signs such as dental abnormalities, short stature, and hypogonadism are also frequently reported. Disease progression for both patients appeared in the most severe range of Pol III–related leukodystrophies in terms of age at onset (<18 months) and death (<20 years) and motor and cognitive development (no independent walking) and degradation (age 4–6 years). However, the progressivity of the microcephaly and the severe spasticity and dystonia observed before age 10 years, particularly in patient 1 with low NAA, reflect the severity of the neurodegenerative process. In addition, the severity of the upper and lower digestive dysfunctions leading to early gastrostomia or cachexia has not yet been reported in POLR3A, POLR3B, and POLR1C mutated patients but could be related to the impaired proliferation of digestive organs reported in the zebrafish POLR3B mutant affecting the RPC128-RPC11 interaction.
Table 2.
Clinical manifestations of published patients with POLR3 or POLR1 mutations
To determine the effects of the variant on Pol III transcriptional activity, we compared the expression levels of some Pol III–transcribed RNAs in patient and control fibroblasts: transcription of both 5S rRNA and 7SL RNA was most severely reduced. 5S rRNA is a component of the large subunit of the ribosome and therefore important for ribosomal functioning. 7SL RNA is part of the signal recognition particle required for associating the ribosome nascent peptide chain with the endoplasmic reticulum. Of interest, a reduction in 7SL RNA was also reported in zebrafish with a POLR3B mutation affecting the RPB128-RPC11 interaction.19 Disruption in ribosomal regulation of mRNA translation may contribute to white matter developmental abnormalities observed in our patients. 7SL RNA seems to play a role in the expression of myelin basic protein, which is tightly needed for myelin development and stability.20 Abnormal RNA regulation has also been reported in leukodystrophies related to mutations in the mitochondrial or cytoplasmic tRNA synthetases21 and in the 5 subunits of the eukaryotic initiation factor EIF2B (childhood ataxia with central nervous system hypomyelination/vanishing white matter).11 Stress-induced acute neurologic distress has particularly been reported in this latter form of leukodystrophy, whereas neurologic degradation has been also reported after infections in 50% of patients with POLR3A, POLR3B, and POLR1C mutations (table 2), suggesting that altered tRNA and rRNA synthesis associated common dysfunctional pathways.
Here, we demonstrated the involvement of a hitherto unknown RNA polymerase III mutation of the POLR3K gene in the development of HLD, supporting the evidence that RNA polymerase III plays a crucial role in white matter and cerebellar integrity.
Acknowledgment
The authors thank the cytogenetic department of the CHU de Clermont-Ferrand responsible for the LEUKOFRANCE Biobank. They also thank Pr Judith Melki (Inserm UMR-1169, Le Kremlin Bicêtre) for her help in homozygosity mapping.
Glossary
- HLD
hypomyelinating leukodystrophy
Author contributions
I. Dorboz: study concept and design, data analysis, and manuscript writing. H. Dumay-Odelot: acquisition and interpretation of data and critical revision of the manuscript. K. Boussaid: collection and analysis of clinical data. Y. Bouyacoub, P. Barreau, S. Samaan, and H. Jmel: acquisition and interpretation of molecular data. E. Eymard-Pierre: fibroblast cultures and DNA biobank. C. Cances, C. Bar, A.-L. Poulat, C. Rousselle, and F. Renaldo: acquisition of clinical data. M. Elmaleh-Bergès: analysis and interpretation of radiologic data. M. Teichmann: acquisition and interpretation of data and critical revision of the manuscript. O. Boespflug-Tanguy: study concept and design, analysis and interpretation of data, and manuscript writing.
Study funding
This study was supported by the European Leukodystrophy Association (ELA), grant number ELA 2009-007I4, and by the European Union FP7 RD Connect project.
Disclosure
I. Dorboz has received research support from the European Leukodystrophy Association (ELA). H. Dumay-Odelot has received research support from INSERM, CNRS UMR 5320, and Ligue contre le cancer. K. Boussaid, Y. Bouyacoub, P. Barreau, S. Samaan, H. Jmel, E. Eymard-Pierre, C. Cances, C. Bar, A. Poulat, C. Rousselle, F. Renaldo, and M. Elmaleh-Bergès report no disclosures. M. Teichmann has received research support from INSERM, CNRS UMR 5320, and Ligue contre le cancer. O. Boespflug-Tanguy reports no disclosures. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Bernard G, Chouery E, Putorti ML, et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet 2011;89:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tétreault M, Choquet K, Orcesi S, et al. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am J Hum Genet 2011;89:652–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet 2007;23:614–622. [DOI] [PubMed] [Google Scholar]
- 4.Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle 2010;9:3687–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumay-Odelot H, Durrieu-Gaillard S, El Ayoubi L, Parrot C, Teichmann M. Contributions of in vitro transcription to the understanding of human RNA polymerase III transcription. Transcription 2014;5:e27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitsu H, Osaka H, Sasaki M, et al. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet 2011;89:644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiffault I, Wolf NI, Forget D, et al. Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III. Nat Commun 2015;6:7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kara B, Köroğlu Ç, Peltonen K, et al. Severe neurodegenerative disease in brothers with homozygous mutation in POLR1A. Eur J Hum Genet 2017;25:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev 1998;12:3857–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J 2006;25:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huyghe A, Horzinski L, Hénaut A, et al. Developmental splicing deregulation in leukodystrophies related to EIF2B mutations. PLoS One 2012;7:e38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbier M, Gross MS, Aubart M, et al. MFAP5 loss-of-function mutations underscore the involvement of matrix alteration in the pathogenesis of familial thoracic aortic aneurysms and dissections. Am J Hum Genet 2014;95:736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res 2013;41:W349–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015;10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozakov D, Hall DR, Xia B, et al. The ClusPro web server for protein-protein docking. Nat Protoc 2017;12:255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res 2004;32:W96–W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder A, Mueller O, Stocker S et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 2006;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical genetics and genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yee NS, Gong W, Huang Y, et al. Mutation of RNA Pol III subunit rpc2/polr3b leads to deficiency of subunit Rpc11 and disrupts zebrafish digestive development. PLoS Biol 2007;5:e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tretiakova A, Gallia GL, Shcherbik N, et al. Association of Puralpha with RNAs homologous to 7 SL determines its binding ability to the myelin basic protein promoter DNA sequence. J Biol Chem 1998;273:22241–22247. [DOI] [PubMed] [Google Scholar]
- 21.Scheper GC, van der Klok T, van Andel RJ, et al. Mitochondrial aspartyltRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet 2007;39:534–539. [DOI] [PubMed] [Google Scholar]
- 22.Wolf NI, Vanderver A, van Spaendonk RM et al. Clinical spectrum of 4H leukodystrophy caused by POLR3A and POLR3B mutations. Neurology 2014;83:1898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets analyzed during the current study are available from the corresponding author on reasonable request.