Abstract.
Drug-resistant Plasmodium falciparum is a major threat to global malaria control and elimination efforts. In Botswana, a southern African country approaching malaria elimination, P. falciparum molecular data are not available. Parasites were assessed through pollymerase chain reaction (PCR) for confirmation of positive rapid diagnostic tests, multiplicity of infection (MOI), and drug resistance markers among isolates from clinical uncomplicated malaria cases collected at health facilities. Of 211 dried blood spot samples selected for the study, 186 (88.2%) were PCR positive for P. falciparum. The mean MOI based on MSP1 genotyping was 2.3 and was not associated with age. A high prevalence of wild-type parasites for pfcrt and pfmdr1 was found, with a haplotype frequency (K76/N86) of 88.8% and 17.7% of the isolates having two copies of the pfmdr1 gene. For pfATPase6, all the parasites carried the wild-type S769 allele. Sequencing showed no evidence of non-synonymous mutations associated with reduced artemisinin derivative sensitivity in the P. falciparum k13 gene. In conclusion, we found that P. falciparum parasites in Botswana were mostly wild type for the drug resistance markers evaluated. Yet, there was a high rate of a molecular marker associated to reduced sensitivity to lumefantrine. Our results indicate the need for systematic drug efficacy surveillance to complement malaria elimination efforts.
Introduction
Malaria is a major public health problem in sub-Saharan Africa, although there is evidence of declining morbidity and mortality. In Botswana, where a considerable decline in the parasite prevalence has occurred over the last 50 years,1 there is impetus for not only reducing the burden of malaria across the country, but also optimizing efforts toward elimination. This drive has emphasized the need for molecular data on current parasite population and the risk assessments of human populations in areas that are prone to endemic or epidemic Plasmodium falciparum circulation.
Botswana is a semiarid to arid country with few permanent bodies of water and two seasons, summer and winter, with rainfall in the summer season occurring between October and April. The mean annual temperatures are between 18 and 23°C; however, in summer and winter, temperatures can be above 35°C or below 0°C, respectively. The population is sparse, with current figures standing at 2.250 million over a land area of 581,730 km2.
Malaria due to P. falciparum is highest in the northern part of the country, which has an annual rainfall of 650 mm between the months of November and April. In the southwestern part, which consists of the mostly arid Kalahari desert, an average of 250 mm of rainfall is estimated. The rainfall pattern varies from year to year and occasionally there is drought, which affects malaria disease patterns. Recently, Plasmodium vivax has been detected in Botswana in parts of the Eastern, Southern, and Central districts,2,3 with a national prevalence of 4.7% among asymptomatic children. However, most clinical malaria is due to P. falciparum infection, the focus of this report.
Chloroquine was the first-line treatment for uncomplicated P. falciparum malaria in Botswana until 1997, when it was replaced with sulfadoxine/pyrimethamine.1 In 2007, sulfadoxine/pyrimethamine was discontinued and currently combination therapy with artemether/lumefantrine is recommended as the first-line treatment.1 Molecular data on P. falciparum drug resistance from prior time points are not available for Botswana, apart from anecdotal reports of an apparent increase in chloroquine treatment failures in the nineties;1 however, neither data on sensitivity nor molecular and review studies have been published in peer-reviewed journals.
In general, antimalarial drugs act principally to eliminate the erythrocytic stages of malaria parasites that are responsible for human illness. Unfortunately, P. falciparum has developed resistance to most classes of antimalarials. The genetic basis of drug resistance is because of well-known polymorphisms in P. falciparum genes. For example, mutations in the pfcrt and pfmdr1 genes are known to confer resistance against chloroquine.4 However, the pfcrt gene has also been associated with artemisinin resistance,5 whereas parasite pfmdr1 polymorphisms also confer reduced susceptibility to other antimalarial drugs, including mefloquine, lumefantrine, and quinine.6 Several P. falciparum candidate genes have been associated with artemisinin resistance. The endoplasmic and sarcoplasmic reticulum Ca2+-ATPase ortholog of P. falciparum (pfATPase6) has been postulated to be the target of artemisinin, although evidence varies.7 Finally, genome-wide analysis of artemisinin resistance in P. falciparum has demonstrated that mutations in the propeller domain of the gene encoding the Kelch 13 protein (pfK13) are associated with delayed parasite clearance in vitro and in vivo in SE Asia.5
With this in mind, we analyzed P. falciparum isolates from uncomplicated clinically identified malaria cases diagnosed at health facilities in Botswana. In particular, we assessed the following: 1) parasite PCR confirmation of rapid diagnostic test (RDT)–positive detection, 2) multiplicity of infection (MOI), and 3) drug-resistance marker polymorphisms.
Methods
Study area.
The study was primarily conducted in Ngamiland (North-West) and Central districts where P. falciparum malaria prevalence is the highest. However, samples were also collected in other parts of Botswana where malaria is present. Samples were collected from patients with febrile illness, seeking health care in the public sector between November 2012 and April 2016. Permission was granted by the National Malaria Programme at the Botswana Ministry of Health and Wellness to analyze dry blood spots (DBS) from those testing positive for P. falciparum using RDT. Two hundred and eleven subjects with uncomplicated malaria symptoms who tested positive on the RDT at day 0 were selected for the study. Sample sizes differ slightly between analyses because of different efficiencies of molecular tests.
DNA extraction and MSP1 genotyping.
Total genomic DNA was extracted from DBS using the EasyMag (Biomerieux, Lyons, France) automated platform. Allelic families (K1, MAD20, and RO33) of parasite MSP1 gene were analyzed using nested-PCR.8 Appropriate controls for all three allelic families (3D7, HB3, and RO33) were run with the field samples. The detection of a single PCR fragment of an allelic family was considered as an infection with one genotype. Length variations of PCR fragments were detected by gel electrophoresis and analyzed through Image Lab software (version 4.1; Bio-Rad, Hercules, CA). Alleles with closely similar sizes were grouped around an average size. Multiple infections of P. falciparum were defined as the presence of more than one genotype of MSP1 in a single isolate. The MOI or number of genotypes per infection was calculated by dividing the total number of fragments detected in one antigenic marker by the number of samples positive for the same marker.
Assessment of P. falciparum drug-resistant polymorphisms.
DNA samples were amplified by PCR-restriction fragment length polymorphism technique to identify K76T polymorphism in pfcrt and N86Y in pfmdr1 gene, according to existing protocols.9–12 Appropriate P. falciparum controls (3D7 and Dd2, wild-type and mutant strains, respectively) were used for each analysis. Copy number of pfmdr1 gene was determined by comparing the ratio of pfmdr1/β-tubulin on the LightCycler 480 V1.5.0 software (Roche Diagnostic, Basel, Switzerland), taking into account the efficiency of each PCR. The primers were obtained from Price et al.13 The reaction was carried out in a final volume of 20 µL (10 µL of LightCycler 480 Probe master mix [Roche Diagnostics], 300 nM each forward and reverse primer, 100 nM each probe, and 2–3 µL of template DNA). The amplification program was as follows: 10 minutes at 95°C; and 45 cycles, with one cycle consisting of 10 seconds at 95°C, 20 seconds at 55°C, and 5 seconds at 72°C. In each experiment, DNA from the laboratory strains 3D7 (one copy of pfmdr1) and Dd2 (three copies of pfmdr1) were used as controls. The efficiency of each PCR (pfmdr1 and β-tubulin) was determined using a scale dilution of the 3D7 DNA. Each sample was analyzed three times (three replicate assays). The PCR results of all the samples considered in this study could be localized to the linear portion of the efficiency curve in terms of Cp (crossing point = CT, threshold cycle). The copy number was calculated with the comparative ΔΔCT method.13 Assays were repeated if any of the following results were obtained: Cp value was > 35; ΔΔCt (cycle threshold) spread was > 1.5; and the 95% confidence interval around the estimation was > 0.4.13 The cutoff value of a multicopy was considered to be > 1.6.
For pfATPase6 polymorphism S769N, we designed a nested-PCR using different combinations of primers from Ferreira et al.14 and Menegon et al.15 Briefly, parasite DNA was amplified using the following primers: first, outer forward 5′-TCACCAAGGGGTATCAACAA-3′15 and outer reverse 5′-AATTATCCTTTTCATCATCTCC-3′14 to amplify an 808–base pair (bp) fragment; and then by applying nested-PCR using primers: nested forward 5′-ACTTAGCTTTGCTTATAAAAAAcTTAA-3′14 and nested reverse 5′-ACGTGGTGGATCAATAATACCT-3′15 to amplify a 125-bp fragment. An artificial mismatch (nucleotide c) was introduced in the nested forward sequence to generate a restriction site (AflII) for the wild-type form.14 Substitutions at position S769N were subsequently distinguished through enzyme digestion with AflII. Undigested PCR product was used to illustrate the behavior of a mutant genotype, as control samples of mutant genotype were unavailable.
Polymorphism analysis of the propeller domain of the pfK13 gene was performed by PCR amplification and sequencing, with PCR conditions described by Taylor et al.16 All PCR products were subsequently sequenced. The obtained sequences were compiled and analyzed by Discovery Studio Gene software (Accelrys Inc., San Diego, CA). PlasmoDB gene identification no. PF3D7_1343700 (P. falciparum 3D7 strain) was used as reference in the numbering of nucleotide and amino acid positions.
All stated activities are prescribed by the Revised Guidelines for the Diagnosis and Treatment of Malaria in Botswana 1, and permission was granted by the Ministry of Health and Wellness research ethics board.
Results
Rapid diagnostic test accuracy and patient characteristics.
Of the 211 DBS samples selected for the study, 186 (88.2%) were PCR positive for P. falciparum by at least one molecular assay. Age of the participants ranged from 7 months to 80 years, median age was 24.2 years, and 52.9% were male. The most represented age groups were 16–50 years (38.0%), followed by 5–15 years (32.8%), with children < 5 years old representing only 15% of the sample, and adults > 50 years being 14.2%. The more represented district of Botswana was Ngamiland (50.7%) with others as follows: Central, 37.7%; South-East, 6.5%; Ghanzi, 2.2%; Kgalagadi, 1.5%; Kgatleng, 0.7%; and North-East, 0.7%.
MSP1 genotyping and MOI.
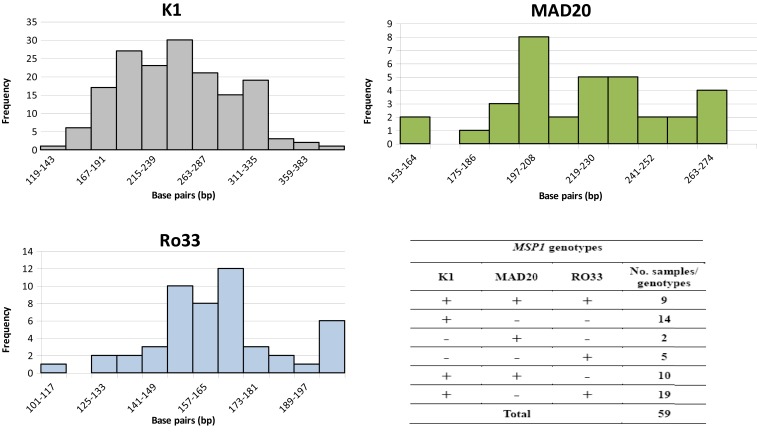
MSP1 gene amplification was performed on a subset of 59 samples in which all three MSP1 allelic families (K1, MAD20, and RO33) were detected. The K1 allelic family was found most frequently (88.1%), whereas RO33 and MAD20 were observed in 55.9% and 35.6% of the samples, respectively. Among the analyzed isolates, 49.2% (29/59) harbored mixed genotypes belonging to two different allelic families. In addition, nine samples (15.2%) harbored all three MSP1 allelic families (Figure 1). Most samples (43/59) showed multiple size variants from the same allelic family: 21 samples harbored two size variants, 10 samples harbored three variants, 10 samples harbored four variants, and five and six size variants were identified in the two different isolates, for a total of 139 size alleles detected in 59 samples (Figure 1). The overall mean MOI was 2.3. There was no significant association between age and MOI (Spearman rank coefficient = 0.02; P = 0.8).
Figure 1.
Size variant allele frequency for each allelic family observed in the analyzed samples and number of samples harboring each MSP1 allelic family. For K1, the mean size ± SE of PCR fragments was 247 base pairs (bp) ± 4 bp, K-S D statistic = 0.18, P = n.s.; for MAD20, the mean size ± SE was 222 bp ± 5 bp, K-S D statistic = 0.22, P = n.s.; and for RO33, the mean size ± SE was 165 bp ± 3 bp, K-S D statistic = 0.23, P = n.s. This figure appears in color at www.ajtmh.org.
Pfcrt and pfmdr1 polymorphisms.
Of 186 P. falciparum–positive isolates, wild-type pfcrt K76T and pfmdr1 N86Y polymorphisms were amplified in 167 and 166 samples, respectively. The mutant allele pfcrt 76T was identified in only 10 (6.0%) isolates, six (3.6%) of which were pure mutants and four (2.4%) mixed with both K and T alleles. In pfmdr1, mutant 86Y allele was identified in only 13 (7.8%) isolates, seven (4.2%) being pure form and six (3.6%) associated with codon N86 as mixed infections. Most examined isolates were found to carry K76 and N86 alleles in pfcrt and pfmdr1 genes, respectively (Table 1). The combination of pfcrt and pfmdr1 genotypes (total number of successfully genotyped isolates for both genes being 152) showed seven distinct haplotypes (Table 1). In addition, pfmdr1 gene copy number was determined in 96 isolates. Of these, 79 (82.3%) had one and 17 (17.7%) had two copies of the gene.
Table 1.
Frequencies of the single nucleotide polymorphisms in the pfcrt and pfmdr1 genes associated with drug resistance among samples collected in Botswana
| pfcrt K76T | pfmdr1 N86Y | No. of pfcrt/pfmdr1 haplotypes (%) |
|---|---|---|
| K | N | 135 (88.8) |
| K | Y | 3 (2.0) |
| K | N/Y | 4 (2.6) |
| T | N | 3 (2.0) |
| T | Y | 3 (2.0) |
| K/T | N | 2 (1.3) |
| K/T | N/Y | 2 (1.3) |
| Total | 152 (100.0) | |
pfATPase6 and pfK13 polymorphisms.
The pfATPase6 S769N polymorphism was investigated in 152 samples. All examined isolates were found to carry the wild-type allele S769. The pfK13 gene was amplified and sequenced from 127 samples. Compared with the pfK13 wild-type reference sequence PF3D7_1343700, the synonymous mutations R513R and V555V were detected in two and one P. falciparum isolates, respectively. All the other samples were classified as wild type when compared to the PF3D7_1343700 sequence.
Discussion
Antimalarial drugs are a key tool for the control and elimination of malaria. Resistance to most of the currently available antimalarials, including the artemisinins, which have played a central role in the global decreases in malaria burden since 2000, has already been confirmed in Southeast Asia (Greater Mekong subregion).17,18 Currently, Botswana represents an ecosystem similar to the Greater Mekong subregion, as it includes areas of low to very low malaria transmission. In such low transmission areas advancing toward elimination, the incidence of infection is too infrequent for the development of partial malaria immunity needed to compensate for failing treatments. In fact, it has been demonstrated that the capacity to clear P. falciparum drug-resistant infections is proportional to the level of acquired immunity.19 In addition, the low level of multiple infections increases inbreeding, which may accelerate the rate of spread of resistance. Drug pressure is often increased in areas of low transmission intensity because most cases are symptomatic, although asymptomatic infections occur. Thus, drug resistance is most likely to emerge, establish, and spread in these areas. The high prevalence of human immunodeficiency virus in Botswana (18.5% for the general population), leading to a high coinfection rate, may further increase the risk of antimalarial drug resistance selection and spread.20
Our results showed that in Botswana, most of the P. falciparum parasites carry wild-type genotypes for the drug resistance markers evaluated. The most important finding is that the pfK13 gene does not show evidence of non-synonymous mutations associated with artemisinin derivative sensitivity. The three samples with synonymous mutations are all from Shakawe village near the Namibian border and were collected between February and March 2016, at the peak of the transmission season. All the other parasites analyzed in this study showed the wild-type sequence of pfK13. Moreover, a high prevalence of 95.8% (pure and mixed infections) of pfmdr1 N86 was found. This polymorphism is involved in the development of reduced sensitivity to lumefantrine,4 while conferring sensitivity to 4-aminoquinolines (chloroquine and amodiaquine),2 both not actually used as antimalarials in Botswana. Moreover, we observed a consistency between wild-type pfcrt and pfmdr1 haplotypes in our samples, with N86 and K76 alleles occurring together. Several studies reported that in the absence of drug pressure, chloroquine-resistant strains have been replaced by sensitive ones,21,22 and that could be happening in Botswana, given the national policy for malaria treatment and the widespread usage. The switch to artemisinin-based antimalarial treatment has been already shown to co-select these alleles in Uganda,23,24 where genotypes with decreased sensitivity to artemether/lumefantrine components increased over time. Also, the finding of 17.7% of isolates with amplification of pfmdr1 (two copies) is remarkable for Africa, where increased copy number is uncommon. In general, amplification of pfmdr1 has been associated not only to decreased susceptibility to artemisinin derivatives and resistance to aryl amino alcohols, particularly mefloquine, but also to lumefantrine, in the field and in vitro.13,25,26 In addition, the detection of multicopy pfmdr1 in African parasites suggests a high potential for rapid selection for resistance.27 Nevertheless, copy number assays are challenging, such that comparison of prevalence between studies with varied methods may be problematic. For pfATPase6, we found all the parasites carrying S769 allele, considered the wild-type form. Moreover, the mean MOI (2.3 different parasite clones per isolate) demonstrates the circulation of malaria parasites of different MSP1 genotypes among endemic areas of Botswana. The relatively high diversity of the parasite population was surprising, although it was consistent with other studies from low-transmission areas.28,29 Such diversity could be caused by circulation of genotypically diverse parasites due to population movement in the region, mostly from Angola, northern Namibia, and Zambia.30,31 Another possibility that we did not assess in this study is that transmission is not homogeneous among all the sampled areas with possible hotspots of higher entomological inoculation rate.32 In this study, there was no association between MOI and age, likely because of low or absent antimalarial immunity due to low malaria transmission.33,34 Taken together, these results suggest that in Botswana, P. falciparum parasites are molecularly sensitive to artemisinin but with a high rate of a molecular marker associated to reduced susceptibility to lumefatrine, also because of the absence of selective pressure from 4-aminoquinolines. However, a limitation to the study is the lack of historical data that could directly illustrate changing allele frequencies, and thus positive selection. The results illustrate the need for systematic drug efficacy surveillance to be performed in selected areas of Botswana to sustain and complement the efforts for malaria elimination.
Acknowledgments:
We would like to thank Margaret Mokomane, chief medical laboratory scientist at the National Health Laboratory in Gaborone, Botswana, who assisted us in setting the study and screening the DBS samples, and Talitah Eyman, procurement officer at the School of Medicine, University of Botswana, for assisting the research team with acquisition of laboratory supplies. Finally, we acknowledge Robert Gross, associate professor of Medicine and Epidemiology at the Perelman School of Medicine of the University of Pennsylvania, for a critical review of this paper.
REFERENCES
- 1.Botswana Ministry of Health, Department of Public Health, National Malaria Programme , 2015. Revised Guidelines for the Diagnosis and Treatment of Malaria in Botswana. Gaborone, Botswana: Botswana Ministry of Health. [Google Scholar]
- 2.Howes RE, et al. 2015. Plasmodium vivax transmission in Africa. PLoS Negl Trop Dis 9: e0004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motshoge T, et al. 2016. Molecular evidence of high rates of asymptomatic P. vivax infection and very low P. falciparum malaria in Botswana. BMC Infect Dis 16: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR, 2002. Epidemiology of drug-resistant malaria. Lancet Infect Dis 2: 209–218. [DOI] [PubMed] [Google Scholar]
- 5.Miotto O, et al. 2015. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 47: 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wurtz N, et al. 2014. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob Agents Chemother 58: 7032–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David-Bosne S, Clausen MV, Poulsen H, Møller JV, Nissen P, le Maire M, 2016. Reappraising the effects of artemisinin on the ATPase activity of PfATP6 and SERCA1a E255L expressed in Xenopus laevis oocytes. Nat Struct Mol Biol 23: 1–2. [DOI] [PubMed] [Google Scholar]
- 8.Tanabe K, Mackay M, Goman M, Scaife JG, 1987. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol 195: 273–287. [DOI] [PubMed] [Google Scholar]
- 9.Fidock DA, et al. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 6: 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djimdé A, et al. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344: 257–263. [DOI] [PubMed] [Google Scholar]
- 11.Duah NO, Wilson MD, Ghansah A, Abuaku B, Edoh D, Quashie NB, Koram KA, 2007. Mutations in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance genes, and treatment outcomes in Ghanaian children with uncomplicated malaria. J Trop Pediatr 53: 27–31. [DOI] [PubMed] [Google Scholar]
- 12.Dahlström S, et al. 2014. Plasmodium falciparum polymorphisms associated with ex vivo drug susceptibility and clinical effectiveness of artemisinin-based combination therapies in Benin. Antimicrob Agents Chemother 58: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price RN, et al. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364: 438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira ID, Martinelli A, Rodrigues LA, do Carmo EL, do Rosário VE, Póvoa MM, Cravo P, 2008. Plasmodium falciparum from Pará state (Brazil) shows satisfactory in vitro response to artemisinin derivatives and absence of the S769N mutation in the SERCA-type PfATPase6. Trop Med Int Health 13: 199–207. [DOI] [PubMed] [Google Scholar]
- 15.Menegon M, Sannella AR, Majori G, Severini C, 2008. Detection of novel point mutations in the Plasmodium falciparum ATPase6 candidate gene for resistance to artemisinins. Parasitol Int 57: 233–235. [DOI] [PubMed] [Google Scholar]
- 16.Taylor SM, et al. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211: 680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mita T, Tanabe K, 2012. Evolution of Plasmodium falciparum drug resistance: implications for the development and containment of artemisinin resistance. Jpn J Infect Dis 65: 465–475. [DOI] [PubMed] [Google Scholar]
- 18.Bustos MD, Wongsrichanalai C, Delacollette C, Burkholder B, 2013. Monitoring antimalarial drug efficacy in the Greater Mekong Subregion: an overview of in vivo results from 2008 to 2010. Southeast Asian J Trop Med Public Health 44: 201–230. [PubMed] [Google Scholar]
- 19.Djimde AA, et al. 2003. Clearance of drug-resistant parasites as a model for protective immunity in Plasmodium falciparum malaria. Am J Trop Med Hyg 69: 558–563. [PubMed] [Google Scholar]
- 20.González R, Ataíde R, Naniche D, Menéndez C, Mayor A, 2012. HIV and malaria interactions: where do we stand? Expert Rev Anti Infect Ther 10: 153–165. [DOI] [PubMed] [Google Scholar]
- 21.Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimdé AA, Kouriba B, Taylor TE, Plowe CV, 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 187: 1870–1875. [DOI] [PubMed] [Google Scholar]
- 22.Huang B, et al. 2016. Prevalence of crt and mdr-1 mutations in Plasmodium falciparum isolates from Grande Comore island after withdrawal of chloroquine. Malar J 15: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad MD, et al. 2014. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis 210: 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tumwebaze P, et al. 2015. Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother 59: 3018–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA, 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis 194: 528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlemann AC, Ramharter M, Lell B, Kremsner PG, Krishna S, 2005. Amplification of Plasmodium falciparum multidrug resistance gene 1 in isolates from Gabon. J Infect Dis 192: 1830–1835. [DOI] [PubMed] [Google Scholar]
- 27.Witkowski B, Nicolau ML, Soh PN, Iriart X, Menard S, Alvarez M, Marchou B, Magnaval JF, Benoit-Vical F, Berry A, 2010. Plasmodium falciparum isolates with increased pfmdr1 copy number circulate in west Africa. Antimicrob Agents Chemother 54: 3049–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agyeman-Budu A, Brown C, Adjei G, Adams M, Dosoo D, Dery D, Wilson M, Asante KP, Greenwood B, Owusu-Agyei S, 2013. Trends in multiplicity of Plasmodium falciparum infections among asymptomatic residents in the middle belt of Ghana. Malar J 12: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Congpuong K, Sukaram R, Prompan Y, Dornae A, 2014. Genetic diversity of the msp-1, msp-2, and glurp genes of Plasmodium falciparum isolates along the Thai-Myanmar borders. Asian Pac J Trop Biomed 4: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon C, Moakofhi K, Mosweunyane T, Jibril HB, Nkomo B, Motlaleng M, Ntebela DS, Chanda E, Haque U, 2013. Malaria control in Botswana, 2008–2012: the path towards elimination. Malar J 12: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chihanga S, Haque U, Chanda E, Mosweunyane T, Moakofhi K, Jibril HB, Motlaleng M, Zhang W, Glass GE, 2016. Malaria elimination in Botswana, 2012–2014: achievements and challenges. Parasit Vectors 9: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karl S, White MT, Milne GJ, Gurarie D, Hay SI, Barry AE, Felger I, Mueller I, 2016. Spatial effects on the multiplicity of Plasmodium falciparum infections. PLoS One 11: e0164054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ntoumi F, Contamin H, Rogier C, Bonnefoy S, Trape JF, Mercereau-Puijalon O, 1995. Age-dependent carriage of multiple Plasmodium falciparum merozoite surface antigen-2 alleles in asymptomatic malaria infections. Am J Trop Med Hyg 52: 81–88. [DOI] [PubMed] [Google Scholar]
- 34.Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F, 2008. Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malar J 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]