Abstract.
Microsporidia are obligate intracellular parasitic fungi causing chronic diarrhea, particularly among immunocompromised patients. The current method used for diagnosis is based on conventional microscopy, which does not differentiate parasites at the species level. The present study was carried out to identify microsporidian species in immunocompromised patients. From March 2016 to March 2017, a total of 289 archived stool samples were examined microscopically for microsporidian spores using Gram-chromotrope Kinyoun (GCK) stain. Positive stool samples by microscopy were subjected to polymerase chain reaction and sequencing for species identification. Based on microscopy examination, the overall prevalence of microsporidian infections was 32.2% (93/289; 95% CI = 27.1–37.8). Of these stool samples, 45 samples were successfully amplified and confirmed as Enterocytozoon bieneusi. No Encephalitozoon intestinalis was detected. Accurate identification of species might help clinicians to decide appropriate management strategies as dissemination risks and treatment response vary for different species, hence improving the management of microsporidian infections.
INTRODUCTION
Microsporidia are obligate intracellular parasitic fungi known to infect both vertebrate and invertebrate hosts worldwide.1,2 Microsporidia are recognized as one of the most common intestinal opportunistic parasites responsible for significant gastrointestinal illness, particularly among immunocompromised hosts, with prevalence ranging from 0.1% to 50%.3,4 To date, there are up to 150 genera with more than 1,300 species recorded. Of these, eight genera have been reported to infect humans. These include Enterocytozoon, Encephalitozoon, Pleistophora, Trachipleistophora, Vittaforma, Brachiola, Nosema, and Microsporidium.4,5 Enterocytozoon bieneusi is the most common species associated with chronic diarrhea, weight loss, malabsorption, and nausea among immunocompromised hosts.4,6 In immunocompetent hosts, it may cause self-limiting diarrhea.7
Currently, most studies conducted on the epidemiology of microsporidian infections were traditionally dependent on direct visualization of the parasites in a stool sample by using the conventional microscopy technique. The benefits of this method are mainly technical simplicity and low cost. However, utilization of microscopy alone is limited by the fact that it cannot differentiate the various species. In addition, microscopy technique is also laborious and time-consuming and requires relatively skilled personnel. The sensitivity and specificity of the microscopy technique alone are not known as it is highly dependent on the expertise and skill of the microscopist. In Malaysia, several studies have been conducted to determine the occurrence of microsporidian infections among immunocompromised patients.8–14 With the exception of only three studies,8–10 most of these reports were based on microscopy techniques. Identification of specific species, especially in clinical samples, necessitates the importance of treatment assessment, prognosis, and reducing dissemination risks among patients. Within this context, the present study was carried out to determine the microsporidian species using molecular techniques among a varied group of immunocompromised patients.
METHODS
Study population.
The stool samples were obtained from the Parasite Southeast Asian Diagnostic (Para:SEAD) Laboratory, Department of Parasitology, Faculty of Medicine, University of Malaya, Kuala Lumpur, over a period of 12 months (March 2016–March 2017). Briefly, the stool samples were sent to the Para:SEAD laboratory from various wards and units in the University Malaya Medical Center (UMMC) for routine stool examination. Only stool samples from immunodeficiency patients as stated in the request form without discrimination to age groups and genders were included in the study. Stool samples were excluded if the selection criterion was not fulfilled. The samples in this study were not randomized. Other information such as demographic and clinical symptoms were obtained from the same request form.
Ethical clearance and sample size calculation.
The study protocol was approved by the Medical Ethics Committee, University Malaya Medical Center (UMMC) (IRB Ref. No. 655.17), considering there is no risk against the physical well-being, integrity, or right to anonymity of the participants. All data generated in this study were kept strictly confidential, and to the extent permitted by law, and the information was accessible only to the researchers. The sample size was calculated according to the anticipated prevalence of microsporidia among immunocompromised patients in Malaysia. According to a previous local study among HIV patients, the prevalence of microsporidian infections was 8.5%.8 By using a significance level of 5% and confidence level of 95%, a minimum sample size of 120 was required in this study.
Sample collection and parasitology examination.
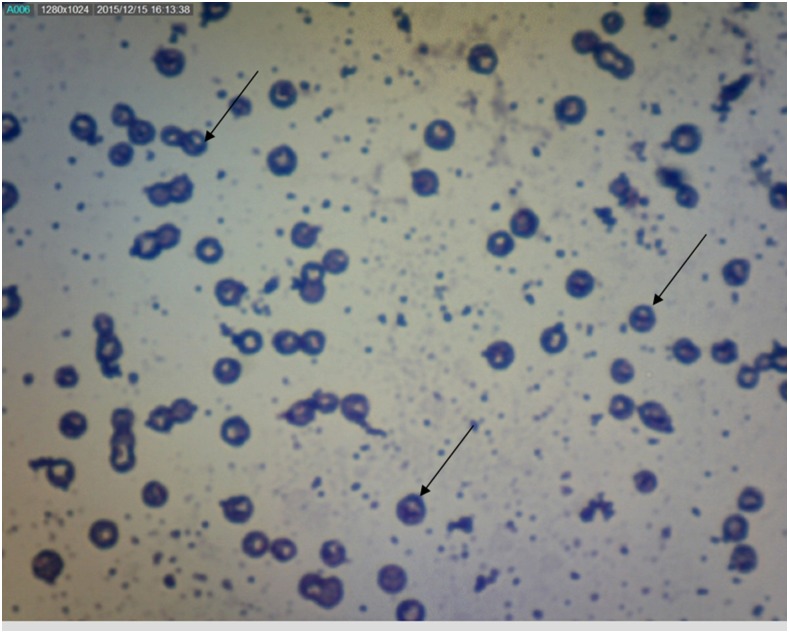
All stool samples were subjected to microscopy examination for the presence of microsporidian spores using Gram-chromotrope Kinyoun (GCK) stain.15 Microsporidia were diagnosed based on the specific morphological characteristics, which include pinkish-blue ovoid spores with a belt-like stripe across the spores against a relatively clean or pale pink background. The slide was examined under a light microscope at high magnification (×100) with oil immersion (Figure 1). The spores were graded based on the number of spores per high power field as follows: 1+ (number of spores 1–10), 2+ (number of spores 11–20), and 3+ (number of spores more than 21).11
Figure 1.
Microsporidian spores detected in a stool sample using Gram-chromotrope Kinyoun staining under light microscopy (magnification ×1,000). This figure appears in color at www.ajtmh.org.
Genomic DNA extraction.
For DNA extraction, all microscopy-positive samples were concentrated by the water–ether concentration technique. Approximately, 500 μL of the concentrated stool samples was subjected to DNA extraction using the NucleoSpin® soil DNA isolation kit (MACHEREY-NAGEL GmbH & Co. KG, Duren, Germany) according to the manufacturer’s protocol. Briefly, approximately 0.2–0.3 g of the stool sample was added to a PowerBead Tube® and incubated at 70°C for 10 minutes in the presence of cell lysis and disruption agent. The stool sample was subjected to homogenization and lysis procedure for complete cell lysis by mechanical shaking (vortexing). Final elution of DNA was performed in 50 μL with elution buffer. The extracted DNA was stored at –20°C until required for polymerase chain reaction (PCR) amplification.
DNA amplification and sequencing.
The extracted DNA was subjected to amplification targeting the conserved region of a small subunit rRNA (SSU rRNA) gene of E. bieneusi and E. intestinalis according to the previously published protocols with minor modification.16,17 The forward primer EBIEF1 (5′-GAA ACT TGT CCA CTC CTT ACG-3′) and reverse primer EBIER1 (5′-CCA TGC ACC ACT CCT GCC ATT-3′) were used to amplify a 607-bp region of the E. bieneusi. The primer pair of SINTF F (5′-TTT CGA GTG TAA AGG AGT CGA-3′) and SINTR R (5′-CCG TCC TCG TTC TCC TGC CCG-3′) was used to amplify 560 bp of the E. intestinalis. The PCR was performed in a 20-μL reaction mixture containing 10 μL of the master mix (Genet Bio, Daejeon, South Korea), 2 μL of each forward and reverse primer, 4 μL of distilled water, and 2 μL of DNA template. Both negative control (reagent mixture without template DNA) and positive control were included in each set of the PCR. The amplification steps for E. bieneusi included 5 minutes of initial denaturation at 94°C, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 minute 30 seconds. A 10-minute final extension at 72°C was used after 35 cycles. The amplification step for E. intestinalis was similar to that of E. bieneusi except for the annealing temperature (45°C instead of 55°C). DNA amplification was performed on a MyCycler thermal cycler (Bio-Rad, Hercules, CA). Amplified DNA was analyzed by gel electrophoresis in 1.5% (w/v) agarose gel and stained with SYBR® Safe DNA (Invitrogen, Auckland, New Zealand) to visualize the amplified PCR product under UV illumination.
All positive amplicons were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). The purified PCR products were then subjected to DNA sequencing in both directions. Sequence chromatograms were viewed using Sequence Scanner version 1.0 program (Applied Biosystems, Waltham, MA). Both forward and reverse nucleotide sequences were manually aligned using the BioEdit Sequence Alignment Editor version 7.0.9 program.18 Homology searches were carried out using reference sequences on the Basic Local Alignment Search Tool (BLAST), hosted by the National Center for Biotechnology Information (NCBI). Enterocytozoon bieneusi and Encephalitozoon intestinalis sequences were determined, and multiple alignments were performed using the ClustalW program.19 All sequences generated in this study were submitted and deposited to the GenBank (GenBank accession number: MH027421–MH027470).
Statistical analysis.
All the data entry was cross-checked for consistency before analysis. The Statistical Package for the Social Sciences software program version 21 was used to determine the relationship between the parasitism, immunocompromised condition, age, gender, and clinical manifestations. The degree of association was determined using the χ2 test. A significance level of P < 0.05 was used for all tests.
RESULTS
General characteristics.
A total of 289 stool samples from immunocompromised patients were examined for the presence of microsporidian spores by the microscopy technique. These included 119 (41.2%) stool samples from HIV/AIDS, 69 cancer (23.9%), 51 end-stage renal failure (ESRF) (17.6%), 27 organ transplant recipient (9.3%), 15 leukemia (5.2%), and eight autoimmune disease (2.8%) patients. Of these, more than half of the patients (174/289; 60.2%) presented with diarrhea. Other symptoms included nausea (7/289; 2.4%), fever (6/289; 2.1%), weight loss (1/289; 0.3%), and having more than one symptom (47/289; 16.3%). Meanwhile, 18.7% (54/289) of the patients were asymptomatic. Most of the patients were males (203; 70.2%), whereas females constituted 29.8% (86). The age of the patients ranged between 1 and 93 years (median age, 42 years). Based on ethnic group, Malay was the highest (48.8%), followed by Chinese (34.9%), Indian (13.5%), and other races (2.8%) (Table 1).
Table 1.
Prevalence of microsporidiosis among immunocompromised patients as determined by microscopy based on a demographic characteristic (N = 289)
| Variables | Number examined | Positive microscopy | Prevalence (%) | P value |
|---|---|---|---|---|
| Gender | ||||
| Male | 203 | 72 | 35.5 | |
| Female | 86 | 21 | 24.4 | 0.047 |
| Age groups (years) | ||||
| ≥ 18 | 245 | 81 | 33.1 | |
| < 18 | 44 | 12 | 27.3 | 0.449 |
| Race | ||||
| Malay | 141 | 46 | 32.6 | |
| Chinese | 101 | 32 | 31.7 | |
| Indian | 39 | 14 | 35.9 | |
| Others | 8 | 1 | 12.5 | 0.639 |
| Total | 289 | 93 | 32.2 | |
Microscopy examination.
Of the 289 stool samples, 93 (32.2%; 95% CI = 27.1–37.8) were positive for microsporidia by microscopic examination. The overall prevalence of microsporidian infections was significantly higher among males compared with females (35.5% versus 24.4%). The prevalence was also higher among patients aged 18 years and older compared with those aged less than 18 years (33.1% versus 27.3%) (Table 1). Based on the type of immunocompromised groups, the highest prevalence was reported among autoimmune disease patients (62.5%; 95% CI = 30.6–86.3), followed by ESRF (52.9%; 95% CI = 39.5–66.0), cancer (31.9%; 95% CI = 22.1–43.6), HIV/AIDS (27.7%; 95% CI = 20.5–36.4), and organ transplant recipient (22.2%; 95% CI = 10.6–40.8) patients. No microsporidia infection was found among leukemia patients (Table 2).
Table 2.
Prevalence of microsporidiosis as determined by microscopy and polymerase chain reaction (PCR) among various groups of immunocompromised patients
| Diseases | Number examined | Microscopy | PCR | ||
|---|---|---|---|---|---|
| n | %* | n | %† | ||
| Autoimmune diseases | 8 | 5 | 62.5 | 5 | 100.0 |
| ESRF | 51 | 27 | 52.9 | 9 | 33.3 |
| Cancer | 69 | 22 | 31.9 | 9 | 41.0 |
| HIV/AIDS | 119 | 33 | 27.7 | 16 | 48.5 |
| Organ transplant recipients | 27 | 6 | 22.2 | 6 | 100.0 |
| Leukemia | 15 | – | – | – | – |
| Total | 289 | 93 | 32.2 | 45 | 48.4 |
n = number positive.
Based on the number examined.
Based on the number positive by microscopy (N = 93).
With regard to symptoms, 58 of 174 (33.3%; 95% CI = 26.8–40.6) patients with diarrhea were positive with microsporidian spores. In patients with fever, two of six (33.3%; 95% CI = 9.7–70) had microsporidian infections. In patients with more than one symptom, 38.3% (18/47; 95% CI = 25.8–52.6) were infected with microsporidia. In addition, 15 of 54 (27.8%; 95% CI = 17.6–40.9) asymptomatic patients were infected with microsporidia. Meanwhile, no microsporidian infections were detected among patients who suffered from weight loss and nausea.
Molecular diagnosis.
Of the 93 positive stool samples by microscopy, 45 were successfully amplified by PCR. Enterocytozoon bieneusi DNA was amplified in all 45 samples by PCR. No E. intestinalis was detected by PCR among immunocompromised patients. Of these, a high prevalence of E. bieneusi was reported among autoimmune disease patients and organ transplant recipients, followed by HIV/AIDS and cancer patients. Meanwhile, the lowest prevalence of E. bieneusi was detected among ESRF patients (Table 2). Polymerase chain reaction analysis was repeated twice in all the 48 microscopically positive stool samples that failed to amplify. Nevertheless, no DNA amplification was observed in these samples. In addition, pooled microscopy-negative samples were also randomly screened by PCR for microsporidian species identification, with no DNA amplification seen. This finding further confirmed that these samples were negative for microsporidian infections. All 45 amplicons of the E. bieneusi were successfully sequenced and analyzed using BLAST analysis. The results demonstrated that all sequences from this study were found to be 99–100% homologous to previously published sequences of E. bieneusi in the NCBI database.
DISCUSSION
In Malaysia, microsporidian infections in immunocompromised hosts are not well documented, especially at the molecular level. To date, there were only a few reports that highlighted the prevalence of infections among these patients. To the best of our knowledge, this is the first study to determine the microsporidian species among various types of immunocompromised patients. The overall prevalence of microsporidian infections among immunocompromised patients was slightly higher compared with previous local studies.8–14,20–22 Comparing the findings of the present study with studies from other countries showed that the prevalence was higher than for those reported in Spain (17%),23 Czech Republic (15%),24 and India (1.8%).25 By contrast, studies in Iran reported a high prevalence of microsporidiosis (more than 30%) among immunocompromised hosts.2,26,27
In the present study, the overall prevalence of microsporidiosis was significantly higher among males compared with the females. The findings are consistent with a study conducted among HIV-positive hospital patients in Nigeria.28 By contrast, no significant differences between other demographic characteristics such as age groups and races were reported by previous local studies.11,20,21 The clinical manifestations of microsporidiosis are very diverse, varying according to the causal species, with diarrhea being the most common mainly in individuals with a weakened immune system such as immunocompromised patients.7,29 In the present study, more than half of the patients infected with microsporidia presented with diarrhea. In microsporidiosis-related diarrhea cases, chronic diarrhea is more commonly reported compared with acute diarrhea. This is likely due, at least in part, to the unreported cases because the symptoms are usually self-limiting, particularly in immunocompetent hosts.8 Several studies have also suggested that individuals may be asymptomatic carriers or that infections may reactivate under the immunocompromised condition.5,30
It is also essential to determine if asymptomatic and persistent microsporidian infections occur in humans, and if so, improved and reliable diagnostic methods are needed to prevent the transmission to others at risk. In addition, the results of the present study highlight the importance of evaluation of immunocompromised individuals with diarrhea for microsporidian infections, which may help in better management of these patients. However, the etiology of diarrhea could not be precisely determined in the present study, suggesting a need for comprehensive etiological studies covering bacterial, viral, and other parasitic causes of diarrhea among immunocompromised patients. Thus, the association between microsporidiosis and diarrhea requires a more in-depth study in the future.
Enterocytozoon bieneusi is the most common species reported among immunocompromised individuals,2,9,31,32 as in the present study. Enterocytozoon bieneusi was reported in one of the most recent studies conducted among HIV/AIDS patients in Malaysia.9 Likewise, E. bieneusi also was previously reported among the Malaysian aborigine community.10 A study in Portugal also indicated a high prevalence of E. bieneusi among HIV/AIDS groups.31 A study conducted in Nigeria also showed that E. bieneusi was the most common species infecting HIV/AIDS patients, followed by E. intestinalis and E. cuniculi.32 Meanwhile, a study in Iran reported that E. bieneusi was detected in bone marrow transplantation patients.2,6
The inability to amplify some microscopy-positive stool samples was most likely due, at least in part, to the presence of a stool inhibitor that was not completely removed during DNA extraction. It is highly recommended that extra steps to optimize the elimination of these inhibitors during the extraction process be carried out in a future study. Other than that, the failure to amplify the microscopically positive samples could also be because of the presence of other microsporidian species.4 Thus, the utilization of another set of primers targeting different genes as genetic markers for species differentiation should be taken into consideration in the future study.
We acknowledge several limitations in the present study. First, a single stool sample from each patient was examined. This may underestimate the actual prevalence of microsporidia as it is recommended to examine multiple stool samples or at least three stool samples per patient.33 Studies have suggested that examination of three stool samples resulted in the detection of up to 90% more infections.33,34 Thus, it is highly recommended to collect at least three stool samples on different days in the future study. Second, because this study evaluated archived samples, information on CD4+ cell count and antiretroviral drug history was not available due to privacy ruling. We acknowledge that the information on antiretroviral drug history is crucial to monitor the presence of microsporidian spores before and after treatment with antiretroviral drugs. A study on microsporidian infections among HIV/AIDS patients in Iran demonstrated that the CD4+ cell count improved after the patients received antiretroviral drugs.35 Another study also described that effective antiretroviral drugs might help in the restoration of the immune response to wipe out opportunistic parasitic infections including microsporidia.36 In addition, the resolution of diarrhea seemed to be related to an increase in CD4+ cell count.36 Thus, a comprehensive study on antiretroviral drug history, CD4+ count, and their association with microsporidian infections among immunocompromised patients is highly needed for future investigations.
In conclusion, this study reported the presence of microsporidian species among various groups of immunocompromised patients. Only E. bieneusi was detected among the immunocompromised patients. As the dissemination risks and treatment response vary for different species, accurate identification of species might help clinicians to improve the management strategies. Such information will provide useful insights into the epidemiology of microsporidiosis, particularly among high-risk groups.
Acknowledgments:
We thank the staff of the Para:SEAD Laboratory for the technical assistance rendered in this study.
REFERENCES
- 1.Cox FEG, 2007. Classification and introduction to the parasitic protozoa. Cox FEG, Wakelin D, Gillespie SH, eds. Topley, Wilson’s Parasitology, 10th edition. London, United Kingdom: Edward Arnold, 186–199. [Google Scholar]
- 2.Tavalla M, Mardani-Kateki M, Abdizadeh R, Nashibi R, Rafie A, Khademvatan S, 2017. Molecular identification of Enterocytozoon bieneusi and Encephalitozoon spp. in immunocompromised patients in Ahvaz, southwest of Iran. Acta Trop 172: 107–112. [DOI] [PubMed] [Google Scholar]
- 3.Navin TR, et al. 1999. Declining CD4+ T-lymphocyte counts are associated with increased risk of enteric parasitosis and chronic diarrhea: results of a 3-year longitudinal study. J Acquir Immune Defic Syndr Hum Retrovirol 20: 154–159. [DOI] [PubMed] [Google Scholar]
- 4.Matos O, Lobo ML, Xiao L, 2012. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anane S, Attouchi H, 2010. Microsporidiosis: epidemiology, clinical data, and therapy. Gastroenterol Clin Biol 3: 450–464. [DOI] [PubMed] [Google Scholar]
- 6.Agholi M, Hatam GR, Motazedian MH, 2013. HIV+/AIDS-associated opportunistic protozoal diarrhea. AIDS Res Hum Retroviruses 29: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobottka I, Albrecht H, Schottelius J, Schmetz C, Bentfeld M, Laufs R, Schwartz DA. 1995. Self limited traveller’s diarrhea due to a dual infection with Enterocytozoon bieneusi and Cryptosporidium parvum in an immunocompetent HIV-negative child. Eur J Clin Microbiol Infect Dis 14: 919–920. [DOI] [PubMed] [Google Scholar]
- 8.Lono A, Kumar S, Chye TT, 2011. Detection of microsporidia in local HIV positive population in Malaysia. Trans R Soc Trop Med Hyg 105: 409–413. [DOI] [PubMed] [Google Scholar]
- 9.Zainudin NS, Nasarudin SNS, Periyasamy P, Moktar N, Noordin R, Osman E, 2016. Diagnosis of disseminated microsporidiosis: detection of circulating Enterocytozoon bieneusi DNA in blood of HIV/AIDS patients. Trop Biomed 33: 761–770. [PubMed] [Google Scholar]
- 10.Ashikin A, Al-Mekhlafi HM, Norhayati M, Anuar TS, 2017. Molecular detection and species identification of Enterocytozoon bieneusi isolated from immunocompetent Orang Asli in Malaysia. Parasitol Int 66: 163–165. [DOI] [PubMed] [Google Scholar]
- 11.Norhayati M, Azlin M, Al-Mekhlafi MH, Anisah N, Aini UN, Fatmah MS, Rozlida AR, 2008. A preliminary study on the prevalence of intestinal microsporidiosis in patients with and without gastrointestinal symptoms in Malaysia. Trans R Soc Trop Med Hyg 102: 1274–1278. [DOI] [PubMed] [Google Scholar]
- 12.Rukman AH, Malina O, Noorhayati MI, Marlyn M, Wan Omar A, Roslaini AM, Ngah Z, Norhayati M, 2008. Intestinal microsporidiosis: a new entity in Malaysia? J Med Health Sci 4: 11–24. [Google Scholar]
- 13.Lono AR, Kumar S, Chye TT, 2008. Incidence of microsporidia in cancer patients. J Gastrointest Cancer 39: 124–129. [DOI] [PubMed] [Google Scholar]
- 14.Anuar TS, Bakar NH, Al-Mekhlafi HM, Moktar N, Osman E, 2016. Prevalence and risk factors for asymptomatic intestinal microsporidiosis among aboriginal school children in Pahang, Malaysia. Southeast Asian J Trop Med Public Health 47: 441–449. [PubMed] [Google Scholar]
- 15.Moura H, Da Silva JL, Sodre FC, Brasil P, Wallmo K, Wahlquist S, Wallace S, Croppo GP, Visvesvara GS, 1996. Gram chromotrope: a new technique that enhances detection of microsporidial spores in clinical specimens. J Eukaryot Microbiol 43: 94S–95S. [DOI] [PubMed] [Google Scholar]
- 16.da Silva AJ, Schwartz DA, Visvesvara GS, De Moura H, Slemenda SB, Pieniazek NJ, 1996. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J Clin Microbiol 34: 986–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franzen C, 2008. Microsporidia: a review of 150 years of research. Open Parasitol J 2: 1–34. [Google Scholar]
- 18.Hall TA, 1999. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norhayati M, et al. 2007. Intestinal microsporidial infections among Orang Asli (Aborigine) children from Malaysia. Ann Trop Med Parasitol 101: 547–550. [DOI] [PubMed] [Google Scholar]
- 21.Lono A, Kumar GS, Chye TT, 2010. Prevalence of microsporidia in an indigenous Orang Asli community in Pahang, Malaysia. Trans R Soc Trop Med Hyg 104: 214–218. [DOI] [PubMed] [Google Scholar]
- 22.Shahrul Anuar T, Al-Mekhlafi HM, Salleh FM, Moktar N, 2013. New insights of microsporidial infection among asymptomatic aboriginal population in Malaysia. PLoS One 8: e71870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lores B, Lopez-Miragaya I, Arias C, Fenoy S, Torres J, Del Aguila C, 2002. Intestinal microsporidiosis due to Enterocytozoon bieneusi in elderly human immunodeficiency virus-negative patients from Vigo, Spain. Clin Infect Dis 34: 918–921. [DOI] [PubMed] [Google Scholar]
- 24.Sak B, Brady D, Pelikanova M, Kvetonova D, Rost M, Kostka M, Tolarova V, Huzova A, Kvac M, 2011. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. J Clin Microbiol 49: 1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khanduja S, Ghoshal U, Agarwak V, Priyannk P, Ghoshal UC, 2017. Identification and genotyping of Enterocytozoon bieneusi among human immunodeficiency virus infected patients. J Infect Public Health 10: 31–40. [DOI] [PubMed] [Google Scholar]
- 26.Mirjalali H, Mohebali M, Mirhendi H, Gholami R, Keshavarz H, Meamar AR, Rezaeian M, 2014. Emerging intestinal microsporidial infection in HIV+/AIDS patients in Iran: microscopical and molecular detection. Iran J Parasitol 9: 149–154. [PMC free article] [PubMed] [Google Scholar]
- 27.Mirjalali H, Mirhendi H, Meamar AR, Mohebali M, Askari Z, Mirsamadi Rezaeian ESM, 2015. Genotyping and molecular analysis of Enterocytozoon bieneusi isolated from immunocompromised patients in Iran. Infect Genet Evol 36: 244–249. [DOI] [PubMed] [Google Scholar]
- 28.Nyamngee A, Edungbola LD, Agbede OO, Salami AK, Nwabuisi C, Akanbi AA, 2nd, Ibrahim OOK, Tilahun M, Moser DB, 2013. Prevalence, intensity and complications of Microsporidium spores amongst HIV-positive hospital patients in Ilorin, Nigeria: original research. Afr J Lab Med 2: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endeshaw T, Kebede A, Verweij JJ, Zewide A, Tsige K, Abraham Y, Petros B, 2006. Intestinal microsporidiosis in diarrheal patients infected with human immunodeficiency virus-1 in Addis Ababa, Ethiopia. Jpn J Infect Dis 59: 306. [PubMed] [Google Scholar]
- 30.Sriaroon C, Mayer CA, Chen L, Accurso C, Greene JN, Vincent AL, 2008. Diffuse intra-abdominal granulomatous seeding as a manifestation of immune reconstitution inflammatory syndrome associated with microsporidiosis in a patient with HIV. AIDS Patient Care STDS 22: 611–612. [DOI] [PubMed] [Google Scholar]
- 31.Lobo ML, Xiao L, Antunes F, Matos O, 2012. Microsporidia as emerging pathogens and the implication for public health: a 10-year study on HIV-positive and -negative patients. Int J Parasitol 42: 197–205. [DOI] [PubMed] [Google Scholar]
- 32.Ojuromi OT, Izquierdo F, Fenoy S, Fagbenro-Beyioku A, Oyibo W, Akanmu A, Odunukwe N, Henriques-Gil N, del Aquila C, 2012. Identification and char-acterization of microsporidia from fecal samples of HIV-positive patients from Lagos, Nigeria. PLoS One 7: e35239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marti HP, Koella JC, 1993. Multiple stool examinations forova and parasites and rate of false-negative results. J Clin Microbiol 31: 3044–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinmann P, Zhou XN, Du ZW, Jiang JY, Wang LB, Wang XZ, Li LH, Marti H, Utzinger J, 2007. Occurrence of Strongyloides stercoralis in Yunnan province, China, and comparison of diagnostic methods. PLoS Negl Trop Dis 1: e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nooshadokht M, Sharifi I, Mohammadi MA, Pirestani M, Afgar A, Mahootchi A, Salari S, Babaei Z, 2017. Intestinal microsporidiosis in Iran: infection in immunocompromised and immunocompetent patients. Curr Med Mycol 3: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maggi P, Larocca AM, Quarto M, 2000. Effect of antiretroviral therapy on cryptosporidiosis and microsporidiosis in patients infected with human immunodeficiency virus type 1. Eur J Clin Microbiol Infect Dis 19: 213–217. [DOI] [PubMed] [Google Scholar]